Abstract
Cesium (Cs) is chemically similar to potassium (K). However, although K is an essential element, Cs is toxic to plants. Two contrasting hypotheses to explain Cs toxicity have been proposed: (1) extracellular Cs+ prevents K+ uptake and, thereby, induces K starvation; and (2) intracellular Cs+ interacts with vital K+-binding sites in proteins, either competitively or noncompetitively, impairing their activities. We tested these hypotheses with Arabidopsis (Arabidopsis thaliana). Increasing the Cs concentration in the agar ([Cs]agar) on which Arabidopsis were grown reduced shoot growth. Increasing the K concentration in the agar ([K]agar) increased the [Cs]agar at which Cs toxicity was observed. However, although increasing [Cs]agar reduced shoot K concentration ([K]shoot), the decrease in shoot growth appeared unrelated to [K]shoot per se. Furthermore, the changes in gene expression in Cs-intoxicated plants differed from those of K-starved plants, suggesting that Cs intoxication was not perceived genetically solely as K starvation. In addition to reducing [K]shoot, increasing [Cs]agar also increased shoot Cs concentration ([Cs]shoot), but shoot growth appeared unrelated to [Cs]shoot per se. The relationship between shoot growth and [Cs]shoot/[K]shoot suggested that, at a nontoxic [Cs]shoot, growth was determined by [K]shoot but that the growth of Cs-intoxicated plants was related to the [Cs]shoot/[K]shoot quotient. This is consistent with Cs intoxication resulting from competition between K+ and Cs+ for K+-binding sites on essential proteins.
Potassium (K) is an essential macronutrient. It is required (as K+) at concentrations of 100 to 150 mm in the cytoplasm of plant cells to activate enzymes and stabilize protein and nucleotide structure (Leigh and Wyn Jones, 1984; Marschner, 1995). The cation Cs+ shares similar chemical properties to K+ and competes with K+ for cation binding sites in proteins (Avery, 1995). Unfortunately, Cs+ does not behave identically to K+ and inhibits the activity of many K+-activated enzymes (Avery, 1995). Consequently, Cs+ is potentially toxic to plants. In addition, Cs+ inhibits the inward-rectifying K channels in the plasma membranes of plant cells (White, 1997; White and Broadley, 2000), including the major inward-rectifying K channels involved in plant K nutrition (e.g. AtAKT1 in Arabidopsis [Arabidopsis thaliana]). Excessive Cs+ in the rhizosphere could, therefore, induce K starvation in plants. The growth of a number of plant species, including bean, tomato, Arabidopsis, and rice, is inhibited by Cs+ concentrations in the rhizosphere exceeding 200 μm (Cline and Hungate, 1960; Kordan, 1987; Sheahan et al., 1993; Hasegawa, 1996; White and Broadley, 2000). The symptoms of Cs intoxication can be reversed, however, by supplying more K. This is consistent with the hypothesis that Cs is toxic because it interferes with K uptake and/or K biochemistry.
Natural soil Cs concentrations are generally low and nontoxic to plants. The stable isotope 133Cs occurs naturally in the aluminosilicate mineral pollucite and may reach concentrations of 25 μg g−1 dry soil (White and Broadley, 2000). Only in areas containing Cs-rich pollucite ores, such as those found in southeastern Manitoba (Teertstra et al., 1992), might Cs cause environmental toxicity. However, two radioisotopes of Cs (134Cs and 137Cs) are of environmental concern due to their emissions of harmful β and γ radiation, relatively long half-lives, and rapid incorporation into biological systems (White and Broadley, 2000). These isotopes arise from the manufacture and testing of thermonuclear weapons and from intentional and unintentional discharges from nuclear installations. They enter the terrestrial food chain through plants, and their presence in foodstuffs impacts upon both health and commerce. Plant roots take up Cs from the soil solution as the monovalent cation (Cs+), which is transported symplastically to the xylem. Theoretical models suggest that, in K-replete plants, Cs influx to root cells is predominantly through voltage-insensitive cation channels (VICCs), with “high-affinity” K+/H+ symporters transporting the remainder (White and Broadley, 2000). In Arabidopsis, members of the AtCNGC and AtGLR gene families are thought to encode VICCs (White and Broadley, 2000; Demidchik et al., 2002; White et al., 2002), and members of the AtKUP/AtHAK gene family encode the K+/H+ symporters. However, since plant K-status regulates the expression of genes encoding several cyclic nucleotide gated channels (CNGCs), glutamate receptors (GLRs), and K+ uptake permeases (KUPs; Kim et al., 1998; Maathuis et al., 2003; Ahn et al., 2004), it is possible that the relative contributions of VICCs and KUPs are reversed in K-deficient plants (White et al., 2004).
Here we investigate the mechanism(s) of Cs toxicity in Arabidopsis. We consider three hypotheses: (1) Cs inhibits plant growth because it reduces K+ uptake and causes K starvation; (2) intracellular Cs is toxic per se, perhaps due to irreversible binding to essential K-dependent proteins; and (3) Cs+ competes with K+ for essential biochemical functions and, therefore, Cs toxicity is related to the [Cs]shoot/[K]shoot quotient. We conclude that Cs toxicity is determined by the [Cs]shoot/[K]shoot quotient. However, we did observe that the expression of many (but not all) genes was altered similarly in response to both K starvation and Cs intoxication. Thus, although Cs intoxication was not perceived genetically solely as K starvation, some Cs-induced K deficiency may be evident. We also note a significant increase in the expression of the gene encoding the H+/Cs+ symporter AtHAK5 in K-starved plants. This resulted in an increased Cs+ influx to K-starved plants and characteristic changes in its pharmacology.
RESULTS
K Dependence of Shoot Growth
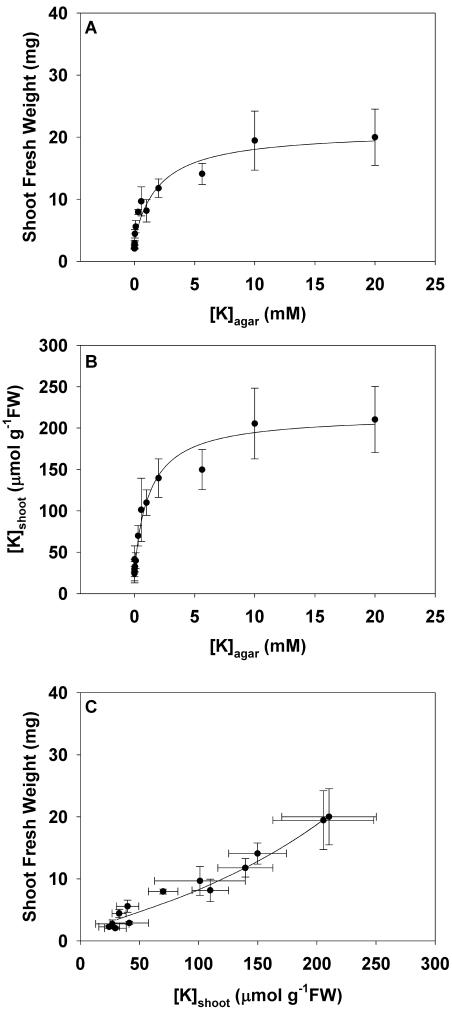
The Arabidopsis accession Wassilewskija (Ws2) was grown for 21 d on agar containing a complete mineral supplement with K concentrations ([K]agar) between 0.5 and 20,000 μm (Fig. 1). The relationships between (A) shoot fresh weight (FW) and (B) mean shoot K concentration ([K]shoot) and [K]agar fitted the equation of a rectangular hyperbola (Table I). Shoot FW increased with increasing [K]agar up to a maximum value (Fig. 1A). At the highest [K]agar assayed (20 mm), shoot FW was 20.0 ± 4.5 mg (n = 6 experiments). The critical [K]agar, at which shoot FW was 90% of its value at 20 mm [K]agar, was 9.9 mm. The [K]shoot also increased with increasing [K]agar up to a maximum value (Fig. 1B). This approximated 220 μmol g−1 FW (Table I). The [K]agar at which the [K]shoot was half-maximal was 0.93 mm. This value is similar to the Km of the low affinity mechanism for K+ uptake into plants (Epstein, 1972). The relationship between shoot FW and [K]shoot was almost linear (Fig. 1C). However, neither shoot FW nor [K]shoot can increase infinitely, and data from plants grown at the highest [K]agar cluster around the same coordinates. The increase in shoot FW as a function of [K]shoot was about 0.09 g2 mmol−1.
Figure 1.
The relationships between shoot FW versus the K concentration in the agar ([K]agar; A), shoot K concentration ([K]shoot) versus [K]agar (B), and shoot FW versus [K]shoot (C) for Arabidopsis accession Ws2 grown for 21 d on mineral-replete agar containing K concentrations between 0.0005 and 20 mm. All data represent means ± se from at least three replicate experiments. Rectangular hyperbolae were fitted to the relationships between shoot FW and [K]shoot versus [K]agar and the relationship between shoot FW and [K]shoot was derived from these. The parameters for all equations are given in Table I.
Table I.
| Relationship | Parameter (units) | Mean ± se | |
|---|---|---|---|
| Shoot FW | Fig. 1A | D (m−1) | 484 ± 238 |
| versus [K]agar | n = 72 | B (mg) | −17.6 ± 2.3 |
| A (mg) | 21.0 ± 2.3 | ||
| [K]shoot | Fig. 1B | D (m−1) | 764 ± 353 |
| versus [K]agar | n = 70 | B (μmol g−1FW) | −185 ± 21.0 |
| A (μmol g−1FW) | 216 ± 20.2 | ||
| Shoot FW | Fig. 2 | B (log μm)−1 | −8.25 ± 1.7 |
| versus [Cs]agar | n = 80 | C (mg) | 14.98 ± 0.52 |
| at 2 mm [K]agar | Ka50 (log μm) | 2.76 ± 0.028 | |
| Shoot FW | Fig. 2 | B (log μm)−1 | −9.35 ± 3.2 |
| versus [Cs]agar | n = 64 | C (mg) | 19.4 ± 0.78 |
| at 20 mm [K]agar | Ka50 (log μm) | 3.26 ± 0.038 | |
| [Cs]shoot | Fig. 3 | K (L kg−1FW) | 16.4 ± 0.49 |
| versus [Cs]agar | n = 51 | ||
| at 2 mm [K]agar | |||
| [Cs]shoot | Fig. 3 | K (L kg−1FW) | 6.3 ± 0.19 |
| versus [Cs]agar | n = 40 | ||
| at 20 mm [K]agar |
The relationships between shoot FW (g) and shoot K concentration ([K]shoot, μmol g−1 FW) versus the K concentration in agar ([K]agar, μm; Fig. 1) were fitted to a rectangular hyperbola: Y = A+B/(1+DX). The relationships between shoot FW (g) versus Cs concentration in the agar ([Cs]agar, μm; Fig. 2) were fitted to the equation of a logistic sigmoidal curve: FW = C/(1 + exp(−B(log[Cs]agar − Ka50))). The relationships between shoot Cs concentration ([Cs]shoot, nmol g−1FW) versus [Cs]agar (Fig. 3) were fitted to the equation for a straight line: [Cs]shoot = K[Cs]agar. Data are expressed as means ± se for n data.
K Alleviates the Inhibition of Shoot Growth by Cs
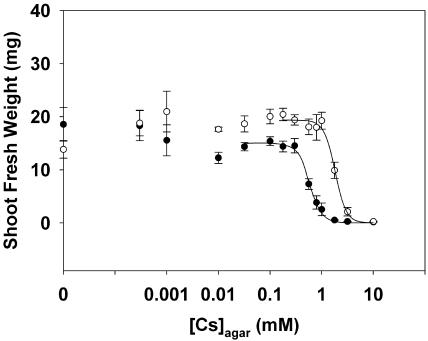
To determine the effects of Cs concentration in the agar ([Cs]agar) on growth, plants were grown for 21 d on agar containing a complete mineral supplement plus 134Cs-labeled [Cs]agar between 0 and 10 mm. Since the toxicity of Cs in the rhizosphere has been shown to depend on the rhizosphere K concentration (Kordan, 1987), this experiment was performed at [K]agar of 2 mm and 20 mm. The shoot FW, mean shoot Cs concentration ([Cs]shoot), and [K]shoot were determined on bulked shoot material. However, since it was impractical to determine [Cs]shoot from 134Cs content and [K]shoot from inductively coupled plasma optical emission spectrometry (ICP-OES) simultaneously, experiments to determine [Cs]shoot and [K]shoot were performed separately. There were no differences in shoot FW between plants grown in the presence or absence of 134Cs (data not shown). Plants were grown at each [K]agar-[Cs]agar combination at least eight times.
Arabidopsis grown at low [Cs]agar at a [K]agar of 20 mm had greater shoot FWs than those of plants grown in the absence of Cs (Fig. 2). The reason for this is unknown. There was a gradual decline in shoot FW as [Cs]agar was increased above 0.3 mm, in the presence of 2 mm [K]agar, or above 1 mm, in the presence of 20 mm [K]agar. The relationship between shoot FW and [Cs]agar fitted the equation of a logistic sigmoidal curve. The minimal shoot FW in the presence of high [Cs]agar was close to zero. The maximal shoot FW of plants grown in the presence of 20 mm K was generally greater than that of plants grown in the presence of 2 mm K (Parameter C in Table I). The [Cs]agar at which shoot FW was half-maximal (Ka50), which is a measure of the tolerance of a plant to Cs in the rhizosphere, was significantly lower for plants grown with 2 mm [K]agar than for plants grown with 20 mm [K]agar (P < 0.001). The rate of change of shoot FW as the [Cs]agar was increased (Parameter B in Table I) did not differ significantly (P > 0.05) when plants were grown in the presence of 2 mm or 20 mm [K]agar.
Figure 2.
The relationships between shoot FW versus the Cs concentration in the agar ([Cs]agar) for Arabidopsis accession Ws2 grown for 21 d on mineral-replete agar containing Cs concentrations up to 10 mm and K concentrations of either 2 mm (•) or 20 mm (○). All data represent means ± se from at least eight replicate experiments. A logistic sigmoidal curve was fitted to the relationships between shoot FW versus [Cs]agar. The parameters for these equations are given in Table I.
K Reduces Cs Accumulation in Shoots
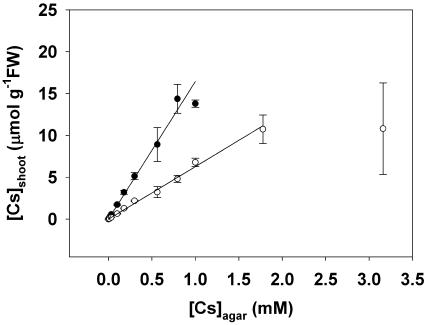
The shoot Cs concentration ([Cs]shoot) increased linearly with increasing [Cs]agar up to an apparent maximum [Cs]shoot (Fig. 3). At the maximal [Cs]shoot, shoot FW was minimal. When assayed at a specific [Cs]agar, the [Cs]shoot was higher with 2 mm [K]agar than with 20 mm [K]agar, and the rate of increase in [Cs]shoot with [Cs]agar (Parameter K in Table I) differed significantly (P < 0.001) between plants grown with 2 mm and 20 mm [K]agar. Thus increasing rhizosphere K concentration reduces the accumulation of Cs in the shoot.
Figure 3.
The relationship between the shoot Cs concentration ([Cs]shoot) versus the Cs concentration in the agar ([Cs]agar) for Arabidopsis accession Ws2 grown for 21 d on mineral-replete agar containing Cs concentrations from 0.0003 to 10 mm and K concentrations of either 2 mm (•) or 20 mm (○). All data represent means ± se from at least three replicate experiments. A linear regression was fitted to the relationship between [Cs]shoot versus [Cs]agar. The parameters for all equations are given in Table I. Data for [Cs]shoot of plants with shoot FW less than 10% of the maximal were not included in these regressions.
Cs Reduces K Accumulation in Shoots
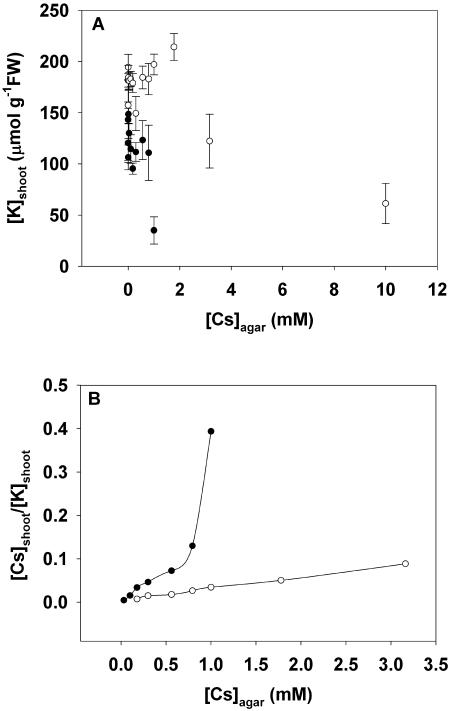
Increasing the [Cs]agar reduced [K]shoot (Fig. 4A). In general, [K]shoot was smaller in plants grown in the presence of 2 mm [K]agar than those grown in the presence of 20 mm [K]agar at the same [Cs]agar. Intriguingly, the [K]shoot of plants grown at low [Cs]agar (<0.01 mm [Cs]agar at 2 mm [K]agar, <2 mm [Cs]agar at 20 mm [K]agar) were greater than those grown in the absence of Cs. When [Cs]agar was increased beyond these concentrations, [K]shoot declined.
Figure 4.
The relationships between the shoot K concentration ([K]shoot) versus the Cs concentration in the agar ([Cs]agar; A) and the quotient of Cs to K concentrations in the shoot ([Cs]shoot/[K]shoot) versus the Cs concentration in the agar ([Cs]agar; B) for Arabidopsis accession Ws2 grown for 21 d on mineral-replete agar containing up to 10 mm Cs and either 2 mm (•) or 20 mm (○) K. All data represent means ± se from at least three replicate experiments. Data for [K]shoot below 20 μmol g−1 FW were excluded from this figure.
Since [Cs]shoot increased (Fig. 3) and [K]shoot decreased (Fig. 4A) with increasing [Cs]agar, it follows that the [Cs]shoot/[K]shoot quotient increased as [Cs]agar increased (Fig. 4B). As expected, [Cs]shoot/[K]shoot was greater in plants grown in the presence of 2 mm [K]agar than those grown in the presence of 20 mm [K]agar at the same [Cs]agar. The [Cs]shoot approximated one-fortieth that of [K]shoot when grown in the presence of 0.2 mm [Cs]agar and 2 mm [K]agar and one-twentieth that of [K]shoot when grown in the presence of 2 mm [Cs]agar and 20 mm [K]agar. The rate of increase in [Cs]shoot/[K]shoot with increasing [Cs]agar was greater in plants grown with 2 mm [K]agar than those grown with 20 mm [K]agar (Fig. 4B). The relationship between [Cs]shoot/[K]shoot versus [Cs]agar in the presence of 2 mm [K]agar also appeared to be biphasic, the rate of increase in [Cs]shoot/[K]shoot with increasing [Cs]agar increasing abruptly at [Cs]agar greater than 0.5 mm. This might indicate a change in the complement of transporters contributing to the uptake of monovalent cations as [Cs]agar was increased, but other interpretations are possible.
Three Hypotheses to Explain Cs Toxicity in Arabidopsis
From the data presented above, three hypotheses might be considered to explain Cs toxicity in Arabidopsis. The first is that [Cs]agar reduces shoot FW because it causes K starvation by lowering [K]shoot (Fig. 4A). The second is that [Cs]shoot is toxic per se (Avery, 1995). The third is that Cs+ competes with K+ for essential biochemical functions, and Cs toxicity is related to the [Cs]shoot/[K]shoot quotient. These hypotheses were tested.
Cs Toxicity Is Not Caused Solely by K Starvation
When assayed in the absence of Cs, a significant, positive, linear relationship between shoot FW and [K]shoot was observed (r2 = 0.968; Fig. 1C). However, when plants were grown in the presence of Cs, positive linear relationships between shoot FW and [K]shoot were less significant (r2 = 0.449 with 2 mm [K]agar and r2 = 0.511 with 20 mm [K]agar; Fig. 5). This implies that the reduction in shoot FW by toxic [Cs]agar is unlikely to be caused by K starvation alone.
Figure 5.
The relationship between shoot FW versus the K concentration in the shoot ([K]shoot) for Arabidopsis accession Ws2 grown for 21 d on mineral-replete agar containing between 0.0005 and 20 mm K in the absence of Cs (▴), or containing between 0.0003 and 10 mm Cs and 2 mm (•) or 20 mm (○) K. All data represent means from at least three replicate experiments. The ses for data on plants grown in the absence of Cs are shown for comparison. The line represents the relationship between shoot FW and [K]shoot in the absence of Cs derived from the rectangular hyperbolae fitted to the relationships between shoot FW versus [K]agar and [K]shoot versus [K]agar (Fig. 1). Parameters for all fitted equations are given in Table I.
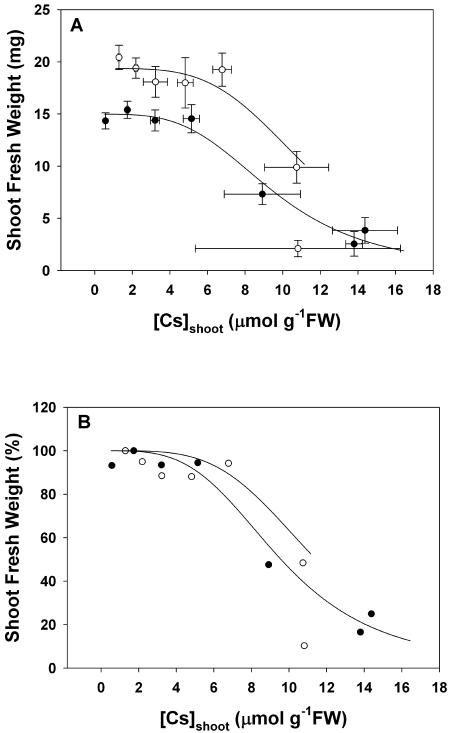
The Relationship between Shoot Cs Concentration and Shoot FW Depends on Shoot K Concentration
Shoot FW decreased as [Cs]shoot increased in the presence of either 2 mm or 20 mm K (Fig. 6A). The exact relationship between shoot FW and [Cs]shoot is derived from the logistic sigmoidal relationships between shoot FW versus [Cs]agar (Fig. 2) and the linear relationships between [Cs]shoot versus [Cs]agar (Fig. 3). It resembles a logistic sigmoidal curve. The shoot FWs of seedlings grown in the presence of 20 mm K were greater than those grown in the presence of 2 mm K at the same [Cs]shoot when plants were alive. This implies that the absolute shoot FW does not depend solely on [Cs]shoot. The [Cs]shoot at which shoot FW was half-maximal (Ks50), which is a measure of the tissue Cs tolerance of a plant, was lower for plants grown with 2 mm [K]agar than for plants grown with 20 mm [K]agar. When grown in the presence of 2 mm [K]agar, the Ks50 was 9,570 ± 285 μmol g−1 FW, and when grown in the presence of 20 mm [K]agar, the Ks50 was 11,440 ± 352 μmol g−1 FW. The observation that plants grown at 20 mm [K]agar had a greater shoot FW at the same [Cs]shoot than those grown at 2 mm [K]agar may be related to a protective effect of increased [K]shoot (Figs. 1 and 4). Indeed, when shoot FW was expressed as a percentage of the value obtained in the absence of [Cs]agar, the effect of [Cs]shoot on (maximal) shoot FW was similar irrespective of the [K]agar (Fig. 6B). This suggests that an interaction between Cs and K might determine shoot FW.
Figure 6.
The relationships between shoot FW versus the Cs concentration in the shoot ([Cs]shoot; A) and between shoot FW, expressed as a percentage of the value obtained in the absence of [Cs]agar, versus [Cs]shoot (B) for Arabidopsis accession Ws2 grown for 21 d on mineral-replete agar containing between 0.0003 and 10 mm Cs and 2 mm (•) or 20 mm (○) K. All data represent means ± se from at least three replicate experiments. The line represents the relationship between shoot FW and [Cs]shoot derived from the logistic sigmoidal curve fitted to the relationships between shoot FW versus [Cs]agar (Fig. 2) and the linear regression fitted to the relationships between [Cs]shoot versus [Cs]agar (Fig. 3). Parameters for all fitted equations are given in Table I. Data for [Cs]shoot of plants with shoot FW less than 10% of the maximal were not plotted.
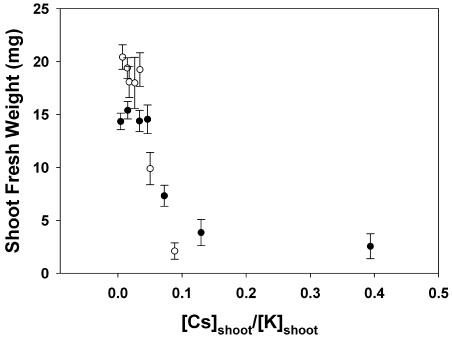
To investigate the effects of interactions between Cs and K on shoot FW, the relationship between shoot FW and the [Cs]shoot/[K]shoot quotient was determined (Fig. 7). At a low [Cs]shoot/[K]shoot, the shoot FWs of plants grown in the presence of 20 mm K were greater than those grown in the presence of 2 mm K. This implies that when [Cs]shoot does not affect growth, shoot FW is determined primarily by [K]shoot. However, the shoot FW of plants grown at 2 mm K or 20 mm K decreased with increasing [Cs]shoot/[K]shoot with the same relationship when [Cs]shoot becomes toxic. This suggests that the [Cs]shoot/[K]shoot quotient determines the reduction in shoot FW, probably because Cs+ cannot replace K+ in its biochemical functions and competes for its binding sites. Thus, Cs toxicity might be perceived by a plant cell as K deficiency.
Figure 7.
The relationship between shoot FW versus the quotient of Cs to K concentrations in the shoot ([Cs]shoot/[K]shoot) for Arabidopsis accession Ws2 grown for 21 d on mineral-replete agar containing between 0.0003 and 10 mm Cs and 2 mm (•) or 20 mm (○) K. All data represent means ± se from at least three replicate experiments. Data for [Cs]shoot of plants with shoot FW less than 10% of the maximal and for [K]shoot below 20 μmol g−1 FW were not plotted.
Transcriptional Profiles of K-Replete, K-Starved, and Cs-Intoxicated Plants
To test the hypothesis that Cs toxicity might be perceived by a plant cell as K deficiency, the transcriptional profiles of K-replete, K-starved, and Cs-intoxicated plants were compared. It was assumed that if Cs intoxication were perceived as K deficiency, then the transcriptional profiles of these two treatments would be identical. However, it is obvious that the transcriptional profiles of Arabidopsis plants subjected to K starvation or Cs toxicity will change with the magnitude and/or duration of stress. Therefore, plants were harvested 7 d after the imposition of these stresses when tissue K concentration had declined (compare with Table I; Hammond et al., 2003) or tissue Cs concentration had become toxic (C.R. Hampton, unpublished data).
When compared with K-replete plants, the expression of 1,349 genes differed significantly (P < 0.05) in roots and the expression of 3,972 genes differed significantly (P < 0.05) in shoots of K-starved plants. The 50 statistically most significant changes in gene expression (based on P values) in K-starved roots and shoots are shown in Table II. It is noteworthy that many genes involved in defense responses, and also numerous transcription factors, are among these genes. In addition, the expression of the K-transporter gene AtHAK5/AtPOT5 was increased 9-fold in Arabidopsis roots by K starvation (Table II). This transporter is likely to catalyze the uptake of both K+ and Cs+ (White et al., 2004). The expression of several genes encoding other transporters that might contribute to Cs+ fluxes was also significantly greater (P < 0.05) in roots of K-starved plants than in roots of K-replete plants. These included AtGLR1.2 and AtGLR1.3. However, no other AtKUP, AtCNGC, or AtGLR gene showed differential expression in roots of K-starved and K-replete plants. In shoots of K-starved plants, the expression of AtHAK5/AtPOT5, AtKUP9, AtCNCG1, AtCNGC13, AtGLR1.2, AtGLR1.3, and AtGLR1.4 was significantly higher (P < 0.05) and the expression of AtKUP2, AtKUP3, AtKUP5, and AtKUP8 was significantly lower (P < 0.05) than in shoots of K-replete plants.
Table II.
The 50 most significant genes expressed differentially in shoots and roots of K-starved compared to K-replete plants
| K Starved
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Shoots
|
Roots
|
||||||||
| AGIa | Nameb | P Valuec | Fold Changed | see | AGIa | Nameb | P Valuec | Fold Changed | see |
| At3g60420 | Expressed protein | 1.37E-08 | 8.0 | 0.1 | At1g12780 | UDP-Glc 4-epimerase | 1.95E-05 | 2.6 | 0.11 |
| At4g18280 | Gly-rich cell wall protein | 2.79E-07 | 23.5 | 1.1 | At3g12900 | Oxidoreductase | 5.45E-05 | 7.1 | 0.77 |
| At1g16510 | Auxin-induced protein family | 3.70E-07 | 11.0 | 0.4 | At2g46600 | Calcium-binding protein | 5.53E-05 | 2.4 | 0.11 |
| At2g23120 | Expressed protein | 4.30E-07 | 7.7 | 0.3 | At4g18950 | Protein kinase | 6.07E-05 | 2.6 | 0.14 |
| At5g10695 | Expressed protein | 7.09E-07 | 4.2 | 0.1 | At3g12820 | Myb family transcription factor | 6.11E-05 | 3.5 | 0.25 |
| At3g27210 | Expressed protein | 8.07E-07 | 4.2 | 0.1 | At5g38820 | Amino acid transporter family | 8.37E-05 | 2.5 | 0.14 |
| At4g05020 | NADH dehydrogenase | 9.05E-07 | 5.8 | 0.2 | At1g25560 | AP2 domain transcription factor | 9.64E-05 | 2.9 | 0.20 |
| At2g41010 | Expressed protein | 1.11E-06 | 4.1 | 0.1 | At1g16510 | Auxin-induced protein family | 1.33E-04 | 2.9 | 0.21 |
| At1g01560 | Mitogen-activated protein kinase (MPK11) | 1.64E-06 | 4.3 | 0.1 | At4g33666 | Expressed protein | 1.38E-04 | 3.3 | 0.27 |
| At5g26340 | Hexose transporter | 1.86E-06 | 4.0 | 0.1 | At4g19200 | Gly/Pro-rich protein | 1.40E-04 | 2.5 | 0.16 |
| At1g78410 | Expressed protein | 1.88E-06 | 10.0 | 0.5 | At1g09070 | C2 domain-containing protein | 1.56E-04 | 2.0 | 0.10 |
| At5g19875 | Expressed protein | 1.98E-06 | 5.9 | 0.2 | At4g13420 | K transporter (HAK5/POT5) | 1.95E-04 | 9.0 | 1.53 |
| At1g43910 | AAA-type ATPase family | 2.08E-06 | 6.0 | 0.3 | At5g06270 | B-type cyclin | 2.10E-04 | 2.1 | 0.12 |
| At1g13340 | Expressed protein | 2.23E-06 | 7.0 | 0.1 | At4g15610 | Membrane protein | 2.47E-04 | 2.1 | 0.12 |
| At5g52750 | Heavy-metal-associated domain-containing protein | 2.23E-06 | 5.2 | 0.2 | At4g22070 | WRKY transcription factor 31 (WRKY31) | 2.78E-04 | 2.0 | 0.12 |
| At1g65690 | Expressed protein | 2.88E-06 | 8.2 | 0.5 | At4g29190 | Zinc finger transcription factor | 2.84E-04 | 2.1 | 0.13 |
| At5g18310 | Expressed protein | 2.99E-06 | 3.8 | 0.1 | At1g23020 | Ferric-chelate reductase | 2.93E-04 | 2.2 | 0.14 |
| At5g06320 | Harpin-induced protein 1 family (NDR1/HIN1-like protein 3) | 3.01E-06 | 3.6 | 0.1 | At2g02120 | Plant defensin protein (PDF2.1 family) | 3.02E-04 | 2.6 | 0.22 |
| At4g23880 | Hypothetical protein | 3.11E-06 | 4.8 | 0.2 | At5g01720 | F-box protein family (FBL3) | 3.02E-04 | 2.7 | 0.22 |
| At2g37970 | Expressed protein | 4.11E-06 | 4.8 | 0.2 | At4g01250 | WRKY family transcription factor | 3.11E-04 | 2.7 | 0.22 |
| At1g74020 | Strictosidine synthase family | 4.14E-06 | 5.3 | 0.3 | At1g76410 | RING zinc finger protein | 3.35E-04 | 2.5 | 0.20 |
| At2g17500 | Auxin efflux carrier protein family | 4.49E-06 | 5.0 | 0.2 | At1g63720 | Expressed protein | 3.51E-04 | 2.3 | 0.17 |
| At3g01830 | Calmodulin-related protein | 5.07E-06 | 14.6 | 0.5 | At5g02780 | In2-1 protein | 3.55E-04 | 2.6 | 0.23 |
| At5g17860 | Cation exchanger (CAX7) | 5.22E-06 | 6.6 | 0.4 | At3g19580 | Zinc finger protein | 3.56E-04 | 2.0 | 0.12 |
| At2g30550 | Lipase (class 3) famil | 5.93E-06 | 3.4 | 0.1 | At2g46750 | FAD-linked oxidoreductase family | 3.65E-04 | 2.7 | 0.23 |
| At1g21520 | Expressed protein | 6.54E-06 | 7.9 | 0.5 | At2g30670 | Short-chain dehydrogenase/reductase family protein | 3.66E-04 | 2.2 | 0.16 |
| At1g19020 | Expressed protein | 7.11E-06 | 8.6 | 0.6 | At4g19810 | Glycosyl hydrolase family 18 | 3.71E-04 | 1.8 | 0.10 |
| At1g28370 | Ethylene responsive element binding factor 11 (EREBP11) | 7.90E-06 | 4.1 | 0.2 | At1g80440 | Kelch repeat containing F-box protein family | 3.73E-04 | 2.4 | 0.19 |
| At3g51860 | Cation exchanger (CAX3) | 7.96E-06 | 16.4 | 1.6 | At5g22630 | Prephenate dehydratase family | 3.84E-04 | 1.9 | 0.10 |
| At1g09070 | C2 domain-containing protein | 8.39E-06 | 5.6 | 0.3 | At1g68840 | AP2 domain protein RAP2.8 (RAV2) | 3.84E-04 | 1.8 | 0.10 |
| At1g75270 | Dehydroascorbate reductase | 9.70E-06 | 2.8 | 0.1 | At1g80590 | WRKY family transcription factor | 4.21E-04 | 2.1 | 0.15 |
| At1g18210 | Calcium-binding protein | 1.04E-05 | 5.3 | 0.3 | At1g19610 | Plant defensin protein (PDF1.4 family) | 4.26E-04 | 2.3 | 0.18 |
| At3g63380 | Calcium-transporting ATPase | 1.13E-05 | 7.8 | 0.3 | At3g13610 | Oxidoreductase | 4.33E-04 | 1.9 | 0.12 |
| At2g31980 | Cys proteinase inhibitor B (cystatin B) | 1.22E-05 | 6.4 | 0.4 | At2g30660 | 3-Hydroxyisobutyryl-Coenzyme A hydrolase (CHY1) | 4.46E-04 | 1.9 | 0.11 |
| At1g75170 | Sec14 cytosolic factor family | 1.34E-05 | 3.7 | 0.2 | At1g64660 | Methionine/cystathionine gamma lyase | 4.49E-04 | 2.4 | 0.20 |
| At2g39710 | Expressed protein | 1.37E-05 | 3.2 | 0.1 | At3g51860 | Cation exchanger (CAX3) | 4.56E-04 | 2.0 | 0.13 |
| At3g48890 | Cytochrome b5 domain-containing protein | 1.46E-05 | 2.6 | 0.1 | At5g05410 | DRE-binding protein (DREB2A) | 5.56E-04 | 2.2 | 0.17 |
| At2g28710 | C2H2-type zinc finger protein | 1.48E-05 | 7.5 | 0.3 | At5g59820 | Zinc finger protein Zat12 | 5.56E-04 | 2.1 | 0.16 |
| At3g44860 | Methyltransferase-related | 1.51E-05 | 4.1 | 0.2 | At1g56160 | Myb family transcription factor | 6.46E-04 | 5.5 | 0.24 |
| At3g47480 | Calcium-binding EF-hand family protein | 1.54E-05 | 11.8 | 1.2 | At1g21000 | Expressed protein | 6.81E-04 | 1.8 | 0.11 |
| At4g09030 | Arabinogalactan-protein (AGP10) | 1.56E-05 | 4.7 | 0.3 | At4g23420 | Oxidoreductase famly | 6.90E-04 | 2.3 | 0.21 |
| At3g46620 | Expressed protein | 1.65E-05 | 2.8 | 0.1 | At3g45300 | Isovaleryl-CoA-dehydrogenase precursor (IVD) | 7.48E-04 | 1.8 | 0.11 |
| At1g68620 | Expressed protein | 1.71E-05 | 6.7 | 0.3 | At3g61930 | Expressed protein | 7.67E-04 | 4.0 | 0.61 |
| At5g22630 | Prephenate dehydratase family | 1.76E-05 | 3.9 | 0.2 | At3g11930 | Ethylene-responsive protein | 7.73E-04 | 1.7 | 0.10 |
| At4g23610 | Expressed protein | 1.77E-05 | 4.4 | 0.3 | At5g63600 | Flavonol synthase | 8.43E-04 | 1.7 | 0.10 |
| At2g37750 | Expressed protein | 1.77E-05 | 19.2 | 0.2 | At3g10980 | Expressed protein | 8.51E-04 | 1.7 | 0.10 |
| At2g20145 | Disease resistance protein (TIR class) | 1.87E-05 | 4.6 | 0.1 | At5g27920 | F-box protein family | 8.81E-04 | 2.1 | 0.18 |
| At1g51780 | Auxin conjugate hydrolase/IAA-amino acid hydrolase (ILL5) | 1.89E-05 | 4.3 | 0.3 | At2g18050 | Histone H1 | 9.63E-04 | 2.6 | 0.29 |
| At5g27920 | F-box protein family | 1.90E-05 | 3.4 | 0.2 | At1g74760 | Hypothetical protein | 9.78E-04 | 2.2 | 0.21 |
| At4g21830 | Expressed protein | 1.92E-05 | 18.5 | 0.2 | At3g61890 | Homeobox-Leu zipper protein ATHB-12 | 9.98E-04 | 2.6 | 0.28 |
In three replicate experiments, Arabidopsis (Ws2) were grown on MS-agar for 14 d before being transferred to a hydroponics system where plants were grown in MS-solutions containing or lacking K to generate K-replete or K-starved plants. Shoots and roots were harvested after a further 7 d growth and total RNA extracted. Total RNA was then used to probe Affymetrix Arabidopsis ATH1 (22K) GeneChips.
AGI numbers.
Brief descriptions of the transcript identified by BLAST searches.
Statistical significance (P value) of differential gene expression in plants starved of K.
Fold change of transcript abundance in plants starved of K, relative to K-replete plants.
The associated se for that fold change (footnote d) based on three replicate experiments.
Several, but not all, of the genes whose expression responded to K starvation showed similar changes in expression upon Cs intoxication. When compared with K-replete plants, the expression of 964 genes differed significantly (P < 0.05) in roots and the expression of 2,551 genes differed significantly in shoots (P < 0.05) of Cs-intoxicated plants. The expression of 22% (211) of the genes whose expression was altered (P < 0.05) in roots by Cs intoxication was also altered by K starvation. Assuming that the expression of 24,000 genes was being assayed on the ATH1 array, if the genetic responses to Cs intoxication and K starvation in roots were independent, only 54 differentially expressed genes would be expected to be common to both stresses [(964/24,000) × (1,349/24,000) × 24,000 = 54]. Thus, the transcriptional responses in Arabidopsis roots to both Cs intoxication and K starvation do not appear to be independent phenomena (χ21df = 501). The expression of 43% (1,098) of the genes whose expression was altered (P < 0.05) in shoots by Cs intoxication was also altered by K starvation. Again, if the genetic responses to Cs intoxication and K starvation in shoots were independent, only 422 differentially expressed genes would be expected to be common to both [(2,551/24,000) × (3,972/24,000) × 24,000 = 422]. Thus, the transcriptional responses in Arabidopsis shoots to both Cs intoxication and K starvation do not appear to be independent phenomena (χ21df = 1,451). These observations are consistent with Cs intoxication being perceived, in part, as K deficiency. Interestingly, four (At2g021020, At1g19610, At2g46600, and At2g46750) of the 50 statistically most significant changes in gene expression in Cs-intoxicated roots and two (At5g26340 and At1g65690) of the 50 statistically most significant changes in gene expression in Cs-intoxicated shoots were also in the 50 statistically most significant changes in gene expression upon K starvation (compare Tables II and III). The expression of several genes encoding transporters that might contribute to Cs+ fluxes was increased significantly (P < 0.05) in Cs-intoxicated plants. The expression of genes encoding AtGLR1.2 and AtGLR1.3 was increased in roots of Cs-intoxicated plants. In shoots of Cs-intoxicated plants, the expression of AtKUP6, AtCNCG1, AtCNGC11, AtCNGC12, AtCNGC13, AtCNGC20, and AtGLR1.3 was increased significantly (P < 0.05).
Table III.
The 50 most significant genes expressed differentially in shoots and roots of Cs-intoxicated compared to nonintoxicated plants
| Cs Intoxicated
| |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Shoots
|
Roots
|
||||||||
| AGIa | Nameb | P Valuec | Fold Changed | see | AGIa | Nameb | P Valuec | Fold Changed | see |
| At4g11650 | Osmotin-like protein (OSM34) | 9.51E-06 | 4.6 | 0.11 | At5g38910 | Germin-like protein | 4.15E-05 | 5.1 | 0.43 |
| At2g04160 | Subtilisin-like Ser protease AIR3 | 1.58E-05 | 2.5 | 0.09 | At1g22890 | Hypothetical protein | 6.40E-05 | 2.3 | 0.11 |
| At4g11280 | 1-Aminocyclopropane-1-Carboxylate synthase 6 (ACS6) | 2.73E-05 | 2.4 | 0.09 | At2g43570 | Glycosyl hydrolase family 19 (chitinase) | 2.17E-04 | 2.0 | 0.11 |
| At2g39330 | Jacalin lectin family | 3.14E-05 | 2.6 | 0.12 | At3g51600 | Nonspecific lipid transfer protein 5 (LTP 5) | 2.26E-04 | 2.7 | 0.21 |
| At1g72830 | CCAAT-binding factor B subunit | 3.46E-05 | 2.3 | 0.09 | At2g02120 | Plant defensin protein (PDF2.1 family) | 2.34E-04 | 2.0 | 0.11 |
| At4g16660 | Heat shock protein hsp70 | 4.74E-05 | 2.1 | 0.08 | At4g17030 | Expansin-related protein 1 precursor (At-EXPR1) | 2.35E-04 | 2.3 | 0.15 |
| At4g39830 | l-Ascorbate oxidase | 5.08E-05 | 2.4 | 0.12 | At2g30000 | Expressed protein | 4.20E-04 | 1.9 | 0.11 |
| At3g14990 | Putative 4-methyl-5(b-hydroxyethyl)-thiazole monophosphate biosynthesis protein | 6.73E-05 | 2.6 | 0.15 | AtCg00750 | Ribosomal protein S11 | 4.80E-04 | 2.1 | 0.15 |
| At2g47800 | Glutathione-conjugate transporter AtMRP4 | 7.53E-05 | 2.2 | 0.10 | At2g02850 | Plantacyanin | 4.86E-04 | 1.8 | 0.11 |
| At5g26920 | Calmodulin-binding protein | 8.25E-05 | 2.9 | 0.19 | At1g19610 | Plant defensin protein (PDF1.4 family) | 5.07E-04 | 2.3 | 0.18 |
| At5g54100 | Expressed protein | 9.36E-05 | 2.1 | 0.10 | At5g44580 | Expressed protein | 5.11E-04 | 1.7 | 0.09 |
| At3g10500 | Expressed protein | 1.09E-04 | 2.3 | 0.13 | At5g09570 | Expressed protein | 5.80E-04 | 2.7 | 0.18 |
| At3g03470 | Cytochrome P450 | 1.12E-04 | 2.5 | 0.15 | At3g09440 | Heat shock protein hsc70-3 | 6.09E-04 | 1.8 | 0.11 |
| At2g40140 | CCCH-type zinc finger protein | 1.22E-04 | 2.1 | 0.10 | At1g32940 | Subtilisin-like Ser protease | 7.26E-04 | 1.7 | 0.10 |
| At5g48180 | Kelch repeats protein family | 1.29E-04 | 2.2 | 0.12 | At3g11580 | DNA-binding protein | 7.66E-04 | 1.6 | 0.09 |
| At3g05970 | AMP-binding protein | 1.31E-04 | 1.9 | 0.08 | At5g57230 | Hypothetical protein | 7.94E-04 | 1.7 | 0.09 |
| At4g11850 | Phospholipase d-gamma | 1.45E-04 | 2.2 | 0.12 | At3g47480 | Calcium-binding EF-hand family protein | 8.15E-04 | 2.8 | 0.33 |
| At2g38470 | WRKY transcription factor | 1.47E-04 | 2.3 | 0.14 | At4g32520 | Gly hydroxymethyl transferase | 8.28E-04 | 1.6 | 0.09 |
| At3g06860 | Fatty acid multifunctional protein (AtMFP2) | 1.56E-04 | 1.9 | 0.09 | At1g74590 | Glutathione transferase | 9.55E-04 | 1.9 | 0.14 |
| At4g02940 | Oxidoreductase | 1.57E-04 | 2.0 | 0.11 | At2g46680 | Homeobox-Leu zipper protein ATHB-7 | 9.69E-04 | 1.8 | 0.11 |
| At4g34135 | Glucosyltransferase | 1.77E-04 | 2.4 | 0.16 | At5g26340 | Hexose transporter | 9.77E-04 | 1.7 | 0.10 |
| At3g09350 | Expressed protein | 1.82E-04 | 2.5 | 0.17 | At1g76520 | Auxin efflux carrier protein | 9.99E-04 | 2.3 | 0.22 |
| At5g13750 | Transporter-related protein | 1.82E-04 | 2.6 | 0.18 | At2g29530 | Small zinc finger-related protein (TIM10) | 0.001196 | 1.6 | 0.09 |
| At1g77920 | bZIP transcription factor, bZIP50 | 1.84E-04 | 2.3 | 0.14 | At1g72890 | Disease resistance protein (TIR-NBS class) | 0.001209 | 2.0 | 0.16 |
| At2g14610 | Pathogenesis-related protein 1 (PR-1) | 1.88E-04 | 2.5 | 0.17 | At3g55090 | ABC transporter | 0.001297 | 2.3 | 0.25 |
| At4g36640 | Sec14 cytosolic factor family | 1.93E-04 | 2.2 | 0.13 | At4g29340 | Profilin 3 | 0.001311 | 1.6 | 0.09 |
| At2g16060 | Class 1 nonsymbiotic hemoglobin (AHB1) | 2.03E-04 | 2.3 | 0.14 | At4g30270 | Xyloglucan endotransglycosylase (meri5B) | 0.001365 | 1.8 | 0.13 |
| At4g37640 | Calcium-transporting ATPase 2 | 2.19E-04 | 1.9 | 0.09 | At2g22430 | Homeobox-Leu zipper protein ATHB-6 | 0.001383 | 1.7 | 0.11 |
| At5g54840 | SGP1 monomeric G-protein | 2.20E-04 | 2.0 | 0.11 | At3g47780 | ABC transporter | 0.001422 | 2.6 | 0.31 |
| At5g41790 | Myosin heavy chain-related protein | 2.27E-04 | 2.0 | 0.11 | At3g49120 | Peroxidase | 0.001545 | 2.8 | 0.38 |
| At2g38860 | (YLS5) proteaseI (pfpI)-like protein | 2.34E-04 | 2.5 | 0.18 | At2g46600 | Calcium-binding protein | 0.001563 | 1.6 | 0.09 |
| At2g18680 | Expressed protein | 2.39E-04 | 2.6 | 0.12 | At3g55740 | Pro transporter 2 (ProT2) | 0.001595 | 1.8 | 0.15 |
| At1g56120 | Wall-associated kinase 2 | 2.50E-04 | 2.6 | 0.21 | At3g29575 | Expressed protein | 0.001601 | 1.6 | 0.10 |
| At1g65690 | Expressed protein | 2.63E-04 | 4.3 | 0.51 | At4g34710 | Arg decarboxylase SPE2 | 0.001605 | 2.6 | 0.34 |
| At3g18830 | Mannitol transporter | 2.66E-04 | 2.2 | 0.15 | At3g28510 | Expressed protein | 0.001608 | 2.5 | 0.30 |
| At4g34200 | d-3-Phosphoglycerate dehydrogenase | 2.70E-04 | 2.2 | 0.14 | At3g09390 | Metallothionein-related protein | 0.00165 | 2.5 | 0.30 |
| At2g46680 | Homeobox-Leu zipper protein (ATHB-7) | 2.75E-04 | 3.5 | 0.37 | At1g13930 | Expressed protein | 0.001665 | 1.6 | 0.09 |
| At3g13940 | Expressed protein | 2.78E-04 | 1.9 | 0.10 | At3g47470 | Light-harvesting chlorophyll a/b-binding protein (CAB-4) | 0.001716 | 3.0 | 0.44 |
| At4g12280 | Copper amine oxidase like protein | 3.20E-04 | 2.5 | 0.20 | At2g25510 | Expressed protein | 0.001775 | 3.8 | 0.70 |
| At3g52430 | Phytoalexin-deficient 4 protein (pad4) | 3.27E-04 | 1.9 | 0.11 | At4g31470 | Pathogenesis-related protein | 0.001962 | 1.6 | 0.11 |
| At3g50910 | Expressed protein | 3.29E-04 | 2.1 | 0.14 | At1g68620 | Expressed protein | 0.002074 | 2.1 | 0.22 |
| At5g42050 | Expressed protein | 3.32E-04 | 2.1 | 0.14 | At2g18690 | Expressed protein | 0.002099 | 2.4 | 0.31 |
| At1g71330 | ABC transporter | 3.34E-04 | 3.7 | 0.42 | At5g41280 | Hypothetical protein | 0.002135 | 1.7 | 0.12 |
| At2g17040 | No apical meristem (NAM) protein family | 3.38E-04 | 2.0 | 0.12 | At2g46750 | FAD-linked oxidoreductase family | 0.002152 | 2.0 | 0.20 |
| At3g09440 | Heat shock protein hsc70-3 | 3.40E-04 | 2.0 | 0.12 | At3g17030 | Expressed protein | 0.002168 | 1.5 | 0.09 |
| At5g26340 | Hexose transporter | 3.41E-04 | 3.7 | 0.43 | At3g25830 | Myrcene/ocimene synthase | 0.002198 | 1.6 | 0.11 |
| At4g31500 | Cytochrome P450 83B1 | 3.43E-04 | 1.7 | 0.08 | At2g38210 | SOR1-related | 0.002274 | 1.8 | 0.15 |
| At5g62620 | Galactosyltransferase family | 3.61E-04 | 1.9 | 0.10 | At1g64600 | Expressed protein | 0.002305 | 1.7 | 0.12 |
| At1g12650 | Expressed protein | 3.71E-04 | 2.7 | 0.15 | At5g16010 | 3-Oxo-5-alpha-steroid 4-dehydrogenase | 0.002306 | 2.6 | 0.36 |
| At3g08720 | Ribosomal-protein S6 kinase (ATPK19) | 3.78E-04 | 2.4 | 0.18 | At5g57300 | UbiE/COQ5 methyltransferase family | 0.002317 | 1.6 | 0.11 |
In three replicate experiments, Arabidopsis (Ws2) were grown on MS-agar for 14 d before being transferred to a hydroponics system where plants were grown in MS-solutions lacking or containing 2 mm Cs to generate nonintoxicated or Cs-intoxicated plants. Shoots and roots were harvested after a further 7 d growth and total RNA extracted. Total RNA was then used to probe Affymetrix Arabidopsis ATH1 (22K) GeneChips.
AGI numbers.
Brief descriptions of the transcript identified by BLAST searches.
Statistical significance (P value) of differential gene expression in Cs-intoxicated plants.
Fold change of transcript abundance in Cs-intoxicated plants, relative to nonintoxicated plants.
The associated se for that fold change (footnote d) based on three replicate experiments.
The Pharmacology of Cs Influx Changes with Plant K Status
The transcriptional profiling of Arabidopsis roots suggests that the complement of transport proteins able to catalyze Cs+ influx to plants changes with plant K-status. In K-replete plants, Cs influx is likely to be dominated by VICCs, such as those encoded by CNGCs and GLRs (White and Broadley, 2000). However, in K-deficient plants, KUPs may contribute significantly to Cs+ influx (Table II; White et al., 2004). To test this hypothesis, advantage can be taken of the different pharmacologies of VICCs and KUPs. VICCs are inhibited by Ca2+ and lanthanides (White, 1997; White and Broadley, 2000), whereas high-affinity K+ transporters, such as KUPs, appear to be inhibited by ammonium (Spalding et al., 1999). As observed previously (Broadley et al., 2001), Cs influx to K-replete Arabidopsis was inhibited by Ca2+ and Gd3+ but not by tetraethylammonium (TEA+) or ammonium (Table IV). By contrast, Cs influx to K-starved Arabidopsis was inhibited by ammonium and Gd3+ but not by Ca2+ or TEA+. These changes in the pharmacology of Cs+ influx, as well as the larger Cs influx to K-deficient plants than into K-replete plants (Table IV, legend), are consistent with the increased expression of AtKUPs encoding H+/Cs+ symporters in plants starved of K (Table IV).
Table IV.
Effects of 1 mm K channel and transport inhibitors on Cs influx from a solution containing 50 μm CsCl
| Inhibitor
|
Relative Influx (%)
|
||
|---|---|---|---|
| Low K (0.5 μm) | Medium (100 μm) | High (2 mm) | |
| TEA | 118 ± 11 | 63 ± 14 | 89 ± 12 |
| Ammonium | 66 ± 5 | 57 ± 7 | 93 ± 35 |
| Calcium | 94 ± 21 | 104 ± 38 | 37 ± 6 |
| Gadolinium | 60 ± 7 | 61 ± 24 | 46 ± 12 |
Arabidopsis were grown previously in solutions containing high (2 mm), medium (100 μm), or low (0.5 μm) K concentrations.Cs influx is expressed as a percentage of that observed in the absence of inhibitors, which was 0.34 ± 0.069, 0.46 ± 0.040, 0.26 ± 0.026 μmol g−1 FW root for Arabidopsis grown with 0.5 μm, 100 μm, and 2 mm K, respectively. The K concentrations in the roots and shoots of Arabidopsis grown with 0.5 μm K were 5.4 ± 1.75 and 1.1 ± 0.27 μmol g−1 dry weight (DW), respectively. The K concentrations in the roots and shoots of Arabidopsis grown with 100 μm K were 4.8 ± 1.91 and 1.4 ± 0.07 μmol g−1 DW, respectively. The K concentrations in the roots and shoots of Arabidopsis grown with 2 mm K were 9.1 ± 3.37 and 3.0 ± 0.75 μmol g−1 DW, respectively. All data are expressed as mean ± se (n = 5 experiments).
DISCUSSION
Cs is toxic to Arabidopsis. The shoot FW of plants grown on agar decreased when [Cs]agar was increased above 0.3 mm in the presence of 2 mm [K]agar and when [Cs]agar was increased above 1 mm in the presence of 20 mm [K]agar. However, since the K concentration in the soil solution is commonly in the millimolar range (Marschner, 1995) and the Cs concentration in the soil solution is generally in the low micromolar range (White and Broadley, 2000), Cs is unlikely to cause environmental toxicity in most natural environments.
The relationship between shoot FW and [Cs]agar fitted the equation of a logistic sigmoidal curve. The [Cs]agar at which shoot FW was one-half its maximal value (Ka50) was greater when plants were grown at higher [K]agar (Table I). This might be explained if increasing [K]agar reduced the entry of Cs to plants and/or increased K within the plant, which protected it against cellular Cs toxicity. It was observed that increasing [K]agar lowered [Cs]shoot (Fig. 3). Thus, one mechanism whereby increasing rhizosphere K protects plants from Cs in the environment is by reducing their Cs uptake. This observation is consistent with previous studies showing that increasing rhizosphere K reduces Cs uptake (Sutcliffe, 1957; Bange and Overstreet, 1960; Handley and Overstreet, 1961) and accumulation (Menzel, 1954; Cline and Hungate, 1960; Nishita et al., 1960, 1962; Jackson et al., 1965; Jackson and Nisbet, 1990; Shaw and Bell, 1991; Shaw et al., 1992; Belli et al., 1995; Smolders et al., 1996a, 1996b; Zhu et al., 1999) by plants and provides a rationale for using K fertilization as a countermeasure in radiocesium-contaminated soils (Konoplev et al., 1993; Prister et al., 1993; Zhu et al., 2000; White et al., 2003). It was also observed that increasing [K]agar in the presence of Cs also increased [K]shoot (Fig. 4), and it is possible that this increased [K]shoot protected the plant against cellular Cs toxicity.
We considered three hypotheses to explain cellular Cs toxicity in Arabidopsis. These were: (1) increasing [Cs]agar reduced shoot FW because it inhibited K+ uptake and caused K starvation; (2) [Cs]shoot was toxic per se; and (3) Cs+ competed with K+ for essential biochemical functions and, therefore, Cs toxicity was related to the [Cs]shoot/[K]shoot quotient. Increasing [Cs]agar reduced [K]shoot (Fig. 4A). This is consistent with previous studies showing that increasing rhizosphere Cs reduces K uptake and accumulation in plants (Sutcliffe, 1957; Nishita et al., 1960; Maathuis and Sanders, 1996). However, increasing [Cs]agar also increased [Cs]shoot (Fig. 3) and the [Cs]shoot/[K]shoot quotient (Fig. 4B). To differentiate between the three hypotheses to explain cellular Cs toxicity, we examined the relationships between shoot FW and [K]shoot (Fig. 5), [Cs]shoot (Fig. 6), or [Cs]shoot/[K]shoot (Fig. 7) in detail. Since the relationships between shoot FW and [K]shoot obtained in the presence of Cs did not follow the same positive linear relationship between shoot FW and [K]shoot observed in the absence of Cs, it was concluded that Cs toxicity was unlikely to be caused by K starvation alone (Fig. 5), which is consistent with the different transcriptional profiles of K-starved and Cs-intoxicated plants (Tables II and III). The observation that the shoot FWs of seedlings grown in the presence of 20 mm K were greater than those grown in the presence of 2 mm K with the same [Cs]shoot (Fig. 6) suggested that Cs toxicity was not the result of [Cs]shoot per se but might be related to the [Cs]shoot/[K]shoot quotient. The relationship between shoot FW and [Cs]shoot/[K]shoot (Fig. 7) suggested that, at a nontoxic [Cs]shoot, plant growth was determined by [K]shoot but that the growth of Cs-intoxicated plants was related to the [Cs]shoot/[K]shoot quotient. This is consistent with Cs intoxication resulting from competition between K+ and Cs+ for binding sites on essential K+-activated proteins, as proposed by Avery (1995).
The transcriptional profiles of K-replete, K-starved, and Cs-intoxicated plants differed. In particular, the expression of several genes encoding proteins that might catalyze Cs+ transport across cell membranes was altered. In K-replete plants, Cs+ appears to be taken up largely by VICCs (White and Broadley, 2000; Broadley et al., 2001; White et al., 2003). However, when Arabidopsis are starved of K+, the expression of several AtKUP genes, such as those encoding AtKUP3 and AtHAK5 (Table II; Kim et al., 1998; Maathuis et al., 2003), is increased. These AtKUPs are able to transport Cs+ across the plasma membrane of root cells (Rubio et al., 2000; White et al., 2004), and an increase in their expression may account for the increased Cs+ influx capacity and distinct pharmacology of Cs+ influx in K-starved plants (Table IV). In Escherichia coli Cs intoxication up-regulates a high-affinity K+ transport operon (Jung et al., 2001). Although Cs intoxication does not induce a significant increase in the expression of AtKUPs in roots, it is noteworthy that the expression of two AtGLR genes (AtGLR1.2 and AtGLR1.3) was increased significantly in roots of both K-starved and Cs-intoxicated plants. It is unfortunate that the expression of genes encoding putative Cs transporters is increased in roots of Cs-intoxicated plants because this might augment the uptake of Cs leading to inexorable toxicity. An increase in the expression of genes encoding monovalent cation transporters in Cs-intoxicated plants could explain why the addition of stable Cs increased the uptake of 137Cs and K in bean plants (Nishita et al., 1962; Wallace et al., 1982) and the greater [K]shoot in plants grown at low [Cs]agar (Fig. 4A).
Since the Chernobyl accident of 1986, a variety of agricultural countermeasures have been implemented to reduce the entry of radiocesium into the food chain, and this has been the most effective means of reducing the total radiation dose to the population (Alexakhin, 1993). In a recent international review of 130 countermeasures for managing radiological contamination, selective crop breeding was one of only six countermeasures considered worthy of further exploration (http://www.strategy-ec.org.uk/). The development of plants that accumulate large amounts of Cs, for phytoremediation purposes, and Cs-excluding “safe” crops is high on the agenda (White et al., 2003; Payne et al., 2004). Knowledge of the mechanisms of Cs accumulation, the interactions between Cs and K nutrition, and the causes of Cs toxicity, will inform the development of these plant phenotypes and also assist in the management of radiocesium-contaminated land. For example, our data indicate that plants with decreased VICC activity would not exclude significant amounts of Cs in K-deficient soils, just as plants with decreased KUP activity would not exclude Cs if grown on K-replete soils. Thus, the genetic responses of plants to the mineral environment should be considered when selecting crops for a particular purpose.
MATERIALS AND METHODS
Plant Material
Seeds of Arabidopsis (Arabidopsis thaliana) L. Heynh. accession Wassilewskija (Ws2; N1601) were obtained from the Nottingham Arabidopsis Stock Centre (NASC, Nottingham, UK). Seeds were washed in 70% (v/v) ethanol/water, rinsed in distilled water, and surface sterilized using NaOCl (1% active chlorine). Seeds were rinsed again and imbibed for 3 to 6 d in sterile distilled water at 4°C to break dormancy. Following imbibation, seeds were sown into 10 cm (length) × 10 cm (width) × 9.5 cm (depth), unvented, polycarbonate culture boxes (Sigma-Aldrich, Gillingham, UK). For the analysis of plant growth and cation content, six seeds were sown directly on 75 mL of 0.8% (w/v) sterile agar medium (Murashige and Skoog [MS] agar) containing 1% (w/v) Suc and a basal salt mix at 10% of the full-strength MS formulation (Murashige and Skoog, 1962), unless the Ca, K, Cs, or Cl concentration was varied. For the transcriptional profiling and radiocesium influx experiments, seeds were sown on perforated polycarbonate discs (diameter 91 mm, thickness 5 mm) placed over 75 mL 10% MS agar. Roots grew into the agar, but shoots remained on the opposite side of the disc. Boxes were placed in a growth room set to 24°C with 16 h light/d. Illumination was provided by a bank of 100 W 84 fluorescent tubes (Philips, Eindhoven, The Netherlands) giving a photon flux density of 45 μmol photons m−2 s−1 at plant height. Shoots were harvested 21 d after sowing for the analysis of plant growth and cation content or transferred on their polycarbonate discs to a hydroponics system situated in a Saxcil growth cabinet (S.K. Saxton, ARC Works, Cheshire, UK) after 14 d for transcriptional profiling or after 7 d for radiocesium influx experiments. In the hydroponics system, plants were supported on polycarbonate discs over 450 mL of aerated nutrient solution in a light-proof 500-mL plastic beaker. The Saxcil cabinet provided a constant temperature of 22°C and constant illumination, provided by a bank of OSRAM L 58 W/23 fluorescent tubes giving a photon flux density between 400 and 700 nm of 75 μmol photons m−2 s−1 at plant height. The relative humidity was approximately 80%. Transcriptional profiling and radiocesium influx experiments were performed on plants 21 d after germination.
Analysis of Plant Growth and Cation Content
The effect of K concentration in the agar ([K]agar) on shoot FW and shoot K concentration ([K]shoot) was determined in the absence and presence of Cs. In the first experiment, plants were grown on agar containing 0.5, 1, 3, 10, 30, 100, 300, 1,000, 2,000, 10,000, and 20,000 μm K. The [K]agar in 10% MS agar was 2 mm. To raise [K]agar above 2 mm, KCl was added. To reduce [K]agar below 2 mm, KNO3 and KH2PO4 were replaced with Ca(NO3)2 and Ca(H2PO4)2. In the second experiment, the effect of Cs concentration in the agar ([Cs]agar) on shoot FW, shoot Cs concentration ([Cs]shoot), and [K]shoot was determined in the presence of 2 mm and 20 mm K. The [Cs]agar was raised to 0.3, 1, 10, 100, 178, 300, 562, 794, 1,000, 1,778, 3,162, and 10,000 μm using CsCl, and the [K]agar was raised from 2 mm to 20 mm using KCl. The shoot FW of individual plants was determined and six shoots were bulked to determine [K]shoot by inductively coupled plasma optical emission spectrometry (J Y Horiba Ultima 2 ICP-OES, Jobin Yvon, Middlesex, UK). Different plants were used to determine [K]shoot and [Cs]shoot. The radioisotope 134Cs (Radioisotope Centre Polatom, Świerk, Poland) was used to quantify [Cs]shoot. Plants on which [Cs]shoot was estimated were grown on agar spiked with 134Cs at an activity of 9.165 kBq L−1 (4 pM 134Cs). Six shoots were bulked, and their 134Cs content was determined using a Wallac 1480, Wizard gamma counter (Perkin-Elmer Life Sciences, Turku, Finland).
Transcriptional Profiling
Transcriptional profiling was performed on shoot and root tissues. Plants were transferred from 10% MS agar to hydroponics 14 d after germination. In the hydroponics system, plants were fed one of three nutrient solutions (pH 5.6): (1) a basal salt mix at 10% of the full-strength MS formulation (MS solution containing 2 mm K+) to produce K-replete plants; (2) an MS solution containing only 0.5 μm K+, in which K salts were replaced by Ca salts, to produce K-starved plants; or (3) an MS solution plus 2 mm CsCl2 to produce Cs-intoxicated plants. Plants were grown in these solutions for 7 d before plants were harvested. At each harvest, shoot and root material from 20 to 30 plants from each treatment were bulked into 1.5-mL colorless, sterile, screw-cap polypropylene tubes and snap-frozen in liquid nitrogen. Tissue samples were stored at −70°C prior to the extraction of total RNA.
Total RNA was extracted following the addition of 1 mL TRIzol reagent to tissue samples placed in liquid nitrogen according to the manufacturer's instructions (Invitrogen Life Technologies, Paisley, UK) but with the following modifications (Hammond et al., 2003): (1) After homogenization with the TRIzol reagent, the samples were centrifuged to remove any remaining plant material. The supernatant was then transferred to a clean Eppendorf tube. (2) To aid precipitation of RNA from the aqueous phase, 0.25 mL of isopropanol and 0.25 mL of a 1.2 m NaCl solution containing 0.8 m sodium citrate were added. Samples of total RNA were purified using a Qiagen RNeasy column (Qiagen, Crawley, UK). Samples of total RNA were then sent to NASC for labeling and hybridization to Affymetrix Arabidopsis ATH1 GeneChips (Affymetrix, Santa Clara, CA). The complete set of microarray data is available from NASC (http://affymetrix.arabidopsis.info/narrays/experimentbrowse.pl).
Signal and detection call values were generated by Affymetrix Microarray Analysis Suite 5.0 software (Affymetrix). Raw data were normalized and analyzed using GeneSpring version 6.1 (Silicon Genetics, Redwood City, CA). Signal values below 20 were set to 20 to limit the number of false positive results (Hammond et al., 2003). For each sample, each signal value was normalized by dividing it by the median of all signal values above 20 of genes classified as present or marginal in that sample. To determine the transcripts that were differentially expressed in K-starved and Cs-intoxicated plants, the normalized signal value for each transcript in K-starved and Cs-intoxicated plants was divided by the normalized signal value for that transcript in the corresponding sample from K-replete plants. A cross gene error model was used in the interpretation of the data, and data were filtered using a maximum confidence level of 5% for genes whose expression was significantly different from one. The annotation of filtered genes was confirmed by cross-referencing the Arabidopsis Genome Initiative (AGI) numbers given by Affymetrix with GenBank. In addition, the target nucleotide sequences used by Affymetrix to design probes on the ATH1 GeneChip (available from http://www.affymetrix.com) were used to BLAST the GenBank sequence database to confirm the fidelity of each probe (http://www.ncbi.nlm.nih.gov/BLAST/). A similar confirmation of probe fidelity was undertaken independently by Ghassemian et al. (2001).
Radiocesium Influx Experiments
Plants used for radiocesium influx experiments were transferred from 10% MS agar to hydroponics 7 d after germination. In the hydroponics system, plants were initially fed MS solution for 7 d. Plants were then transferred for a further 7 d to one of three solutions: (1) MS solution (2 mm K), to produce K-replete plants; (2) an MS solution containing only 100 μm K+; or (3) an MS solution containing only 0.5 μm K+, to produce K-starved plants. In solutions (2) and (3), K salts were replaced by Ca salts. Shoots and roots of representative plants grown in each solution were harvested and weighed and their K content determined using ICP-OES. Cs influx experiments were then performed as described by Broadley et al. (2001). Briefly, polycarbonate discs supporting Arabidopsis were placed over 455 mL of an aerated, single salt solution containing 50 μm CsCl radiolabeled with 104 kBq L−1 134CsCl with or without 1 mm TEACl, NH4Cl, CaCl2, or GdCl3. After 20 min, plants were transferred to 450 mL of a solution containing 50 μm CsCl plus 1 mm CaCl2 for 2 min to remove 134Cs from the root apoplast. Plant roots were blotted with tissue paper, and the shoots and roots of individual plants were separated and weighed and their Cs contents estimated from 134Cs activities determined using a gamma counter.
Statistics
Each experiment described was repeated at least three times. Data were fitted to a variety of equations using GenStat for Windows (sixth edition, release 6.1.0.200, VSN International, Oxford). The significance of differences in parameters for contrasting experimental treatments was tested using parallel regression analysis. The significance of commonalities in transcriptional profiles was assessed using chi-squared analysis of the two-way contingency table.
Acknowledgments
We thank Simon Elliott, Mark Powell, and Joan Yurkwich (HRI) for mineral analyses and staff at the Nottingham Arabidopsis Stock Centre for their support.
This work was supported by the Biotechnology and Biological Sciences Research Council (UK). C.R.H. was supported by an HRI/Birmingham University Studentship, J.P.H. was supported by an HRI Browning Studentship, and K.A.P. was supported by a BBSRC Committee Studentship.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.046672.
References
- Ahn SJ, Shin R, Schachtman DP (2004) Expression of KT/KUP genes in Arabidopsis and the role of root hairs in K+ uptake. Plant Physiol 134: 1135–1145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexakhin RM (1993) Countermeasures in agricultural production as an effective means of mitigating the radiological consequences of the Chernobyl accident. Sci Total Environ 137: 9–20 [Google Scholar]
- Avery SV (1995) Caesium accumulation by microorganisms: uptake mechanisms, cation compartmentalization and toxicity. J Ind Microbiol 14: 76–84 [DOI] [PubMed] [Google Scholar]
- Bange GGJ, Overstreet R (1960) Some observations on absorption of cesium by excised barley roots. Plant Physiol 35: 605–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belli M, Sansone U, Ardiani R, Feoli E, Scimone M, Menegon S, Parente G (1995) The effect of fertilizer applications on 137Cs uptake by different plant species and vegetation types. J Environ Radioact 27: 75–89 [Google Scholar]
- Broadley MR, Escobar-Gutiérrez AJ, Bowen HC, Willey NJ, White PJ (2001) Influx and accumulation of Cs+ by the akt1 mutant of Arabidopsis thaliana (L.) Heynh. lacking a dominant K+ transport system. J Exp Bot 52: 839–844 [DOI] [PubMed] [Google Scholar]
- Cline JF, Hungate FP (1960) Accumulation of potassium, cesium137 and rubidium86 in bean plants grown in nutrient solutions. Plant Physiol 35: 826–829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demidchik V, Davenport RJ, Tester M (2002) Nonselective cation channels in plants. Annu Rev Plant Biol 53: 67–107 [DOI] [PubMed] [Google Scholar]
- Epstein E (1972) Mineral Nutrition of Plants: Principles and Perspectives. John Wiley & Sons, New York
- Ghassemian M, Waner D, Tchieu J, Gribskov M, Schroeder JI (2001) An integrated Arabidopsis annotation database for Affymetrix Genechip® data analysis, and tools for regulatory motif searches. Trends Plant Sci 6: 448–449 [DOI] [PubMed] [Google Scholar]
- Hammond JP, Bennett MJ, Bowen HC, Broadley MR, Eastwood DC, May ST, Rahn C, Swarup R, Woolaway KE, White PJ (2003) Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol 132: 578–596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handley R, Overstreet R (1961) Effect of various cations upon absorption of carrier-free cesium. Plant Physiol 36: 66–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hasegawa H (1996) Selection for mutants with low nitrate uptake ability in rice (Oryza sativa). Physiol Plant 96: 199–204 [Google Scholar]
- Jackson D, Nisbet AF (1990) The effect of fertiliser treatment, soil pH and grazing on the transfer of radiocaesium to upland fell vegetation. In G Desmet, P Nassimbeni, M Belli, eds, Transfer of Radionuclides in Natural and Semi-Natural Environments. Elsevier Applied Science, London, pp 395–402
- Jackson WA, Craig D, Hector ML (1965) Effects of various cations on cesium uptake from soils and clay suspensions. Soil Sci 99: 345–353 [Google Scholar]
- Jung K, Krabusch M, Altendorf K (2001) Cs+ induces the kdp operon of Escherichia coli by lowering the intracellular K+ concentration. J Bacteriol 183: 3800–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim EJ, Kwak JM, Uozumi N, Schroeder JI (1998) AtKUP1: an Arabidopsis gene encoding high-affinity potassium transport activity. Plant Cell 10: 51–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konoplev AV, Viktorova NV, Virchenko EP, Popov VE, Bulgakov AA, Desmet GM (1993) Influence of agricultural countermeasures on the ratio of different chemical forms of radionuclides in soil solution. Sci Total Environ 137: 147–162 [Google Scholar]
- Kordan HA (1987) Reversal of caesium inhibition of growth by potassium in hypocotyls of tomato seedlings (Lycopersicon esculentum L.). New Phytol 107: 395–401 [DOI] [PubMed] [Google Scholar]
- Leigh RA, Wyn Jones RG (1984) An hypothesis relating critical potassium concentrations for growth to the distribution and functions of this ion in the plant cell. New Phytol 97: 1–13 [Google Scholar]
- Maathuis FJM, Filatov V, Herzyk P, Krijger GC, Axelsen KB, Chen S, Green BJ, Li Y, Madagan KL, Sanchez-Fernandez R, et al (2003) Transcriptome analysis of root transporters reveals participation of multiple gene families in the response to cation stress. Plant J 35: 675–692 [DOI] [PubMed] [Google Scholar]
- Maathuis FJM, Sanders D (1996) Characterization of csi52, a Cs+ resistant mutant of Arabidopsis thaliana altered in K+ transport. Plant J 10: 579–589 [DOI] [PubMed] [Google Scholar]
- Marschner H (1995) Mineral Nutrition of Higher Plants. Academic Press, London
- Menzel RG (1954) Competitive uptake by plants of potassium, rubidium, cesium, and calcium, strontium, barium from soils. Soil Sci 77: 419–425 [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15: 473–497 [Google Scholar]
- Nishita H, Dixon D, Larson KH (1962) Accumulation of Cs and K and growth of bean plants in nutrient solution and soils. Plant Soil 17: 221–242 [Google Scholar]
- Nishita H, Romney EM, Alexander GV, Larson KH (1960) Influence of K and Cs on release of Cs137 from three soils. Soil Sci 89: 167–176 [Google Scholar]
- Payne KA, Bowen HC, Hammond JP, Hampton CR, Lynn JR, Mead A, Swarup K, Bennett MJ, White PJ, Broadley MR (2004) Natural genetic variation in caesium (Cs) accumulation by Arabidopsis thaliana. New Phytol 162: 535–548 [Google Scholar]
- Prister BS, Perepelyatnikov GP, Perepelyatnikova LV (1993) Countermeasures used in the Ukraine to produce forage and animal food-products with radionuclide levels below intervention limits after the Chernobyl accident. Sci Total Environ 137: 183–198 [Google Scholar]
- Rubio F, Santa-Maria GE, Rodríguez-Navarro A (2000) Cloning of Arabidopsis and barley cDNAs encoding HAK potassium transporters in root and shoot cells. Physiol Plant 109: 34–43 [Google Scholar]
- Shaw G, Bell JNB (1991) Competitive effects of potassium and ammonium on caesium uptake kinetics in wheat. J Environ Radioact 13: 283–296 [Google Scholar]
- Shaw G, Hewamanna R, Lillywhite J, Bell JNB (1992) Radiocaesium uptake and translocation in wheat with reference to the transfer factor concept and ion competition effects. J Environ Radioact 16: 167–180 [Google Scholar]
- Sheahan JJ, Ribeiro-Neto L, Sussman MR (1993) Cesium-insensitive mutants of Arabidopsis thaliana. Plant J 3: 647–656 [Google Scholar]
- Smolders E, Kiebooms L, Buysse J, Merckx R (1996. a) 137Cs uptake in spring wheat (Triticum aestivum L. cv Tonic) at varying K supply. I. The effect in solution culture. Plant Soil 181: 205–209 [Google Scholar]
- Smolders E, Kiebooms L, Buysse J, Merckx R (1996. b) 137Cs uptake in spring wheat (Triticum aestivum L cv Tonic) at varying K supply. II. A potted soil experiment. Plant Soil 181: 211–220 [Google Scholar]
- Spalding EP, Hirsch RE, Lewis DR, Qi Z, Sussman MR, Lewis BD (1999) Potassium uptake supporting plant growth in the absence of AKT1 channel activity—inhibition by ammonium and stimulation by sodium. J Gen Physiol 113: 909–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe JF (1957) The selective uptake of alkali cations by red beet root tissue. J Exp Bot 8: 36–49 [Google Scholar]
- Teertstra DK, Cerny P, Chapman R (1992) Compositional heterogeneity of pollucite from High Grade Dyke, Maskwa Lake, southeastern Manitoba. Can Mineral 30: 687–697 [Google Scholar]
- Wallace A, Romney EM, Wood RA (1982) The role of stable cesium on plant uptake of cesium-137. Soil Sci 134: 71–75 [Google Scholar]
- White P, Bowen H, Broadley M, Hampton C, Meacham M, Payne K (2004) The mechanisms of caesium uptake by plants. In J Inaba, H Tsukada, A Takeda, eds, Proceedings of the International Symposium on Radioecology and Environmental Dosimetry, Aomori, Japan, October 2003, pp 255–262
- White PJ (1997) Cation channels in the plasma membrane of rye roots. J Exp Bot 48: 499–514 [DOI] [PubMed] [Google Scholar]
- White PJ, Bowen HC, Demidchik V, Nichols C, Davies JA (2002) Genes for calcium-permeable channels in the plasma membrane of plant root cells. Biochim Biophys Acta 1564: 299–309 [DOI] [PubMed] [Google Scholar]
- White PJ, Broadley MR (2000) Mechanisms of caesium uptake by plants. New Phytol 147: 241–256 [Google Scholar]
- White PJ, Swarup K, Escobar-Gutiérrez AJ, Bowen HC, Willey NJ, Broadley MR (2003) Selecting plants to minimise radiocaesium in the food chain. Plant Soil 249: 177–186 [Google Scholar]
- Zhu YG, Shaw G, Nisbet AF, Wilkins BT (1999) Effects of external potassium supply on compartmentation and flux characteristics of radiocaesium in intact spring wheat roots. Ann Bot 84: 639–644 [Google Scholar]
- Zhu YG, Shaw G, Nisbet AF, Wilkins BT (2000) Effect of potassium (K) supply on the uptake of 137Cs by spring wheat (Triticum aestivum cv. Tonic): a lysimeter study. Radiat Environ Biophys 39: 283–290 [DOI] [PubMed] [Google Scholar]