Abstract
In chloroplasts, the control of mRNA stability is of critical importance for proper regulation of gene expression. The Chlamydomonas reinhardtii strain Δ26pAtE is engineered such that the atpB mRNA terminates with an mRNA destabilizing polyadenylate tract, resulting in this strain being unable to conduct photosynthesis. A collection of photosynthetic revertants was obtained from Δ26pAtE, and gel blot hybridizations revealed RNA processing alterations in the majority of these suppressor of polyadenylation (spa) strains, resulting in a failure to expose the atpB mRNA 3′ poly(A) tail. Two exceptions were spa19 and spa23, which maintained unusual heteroplasmic chloroplast genomes. One genome type, termed PS+, conferred photosynthetic competence by contributing to the stability of atpB mRNA; the other, termed PS−, was required for viability but could not produce stable atpB transcripts. Based on strand-specific RT-PCR, S1 nuclease protection, and RNA gel blots, evidence was obtained that the PS+ genome stabilizes atpB mRNA by generating an atpB antisense transcript, which attenuates the degradation of the polyadenylated form. The accumulation of double-stranded RNA was confirmed by insensitivity of atpB mRNA from PS+ genome-containing cells to S1 nuclease digestion. To obtain additional evidence for antisense RNA function in chloroplasts, we used strain Δ26, in which atpB mRNA is unstable because of the lack of a 3′ stem-loop structure. In this context, when a 121-nucleotide segment of atpB antisense RNA was expressed from an ectopic site, an elevated accumulation of atpB mRNA resulted. Finally, when spa19 was placed in a genetic background in which expression of the chloroplast exoribonuclease polynucleotide phosphorylase was diminished, the PS+ genome and the antisense transcript were no longer required for photosynthesis. Taken together, our results suggest that antisense RNA in chloroplasts can protect otherwise unstable transcripts from 3′→5′ exonuclease activity, a phenomenon that may occur naturally in the symmetrically transcribed and densely packed chloroplast genome.
INTRODUCTION
Chloroplasts are photosynthetic organelles that arose from prokaryotic endosymbionts during eukaryotic evolution (reviewed in Gray, 1992). Chloroplasts contain their own DNA and gene expression machinery, which have combined features of their progenitors with evolved traits. For example, as in bacteria, chloroplast genes are often organized into operons and clusters; however, once these are transcribed into precursor transcripts, they undergo complex posttranscriptional processing events, including splicing and intercistronic cleavages (reviewed in Rochaix, 1996; Sugita and Sugiura, 1996; Monde et al., 2000). Chloroplasts also tend to use modulation of mRNA stability rather than transcriptional control as a mode of regulating mRNA accumulation, which has led since its initial discovery (Deng and Gruissem, 1987; Mullet and Klein, 1987) to intensive investigations of posttranscriptional control mechanisms.
Considerable information is available on cis-acting elements that can act as positive or negative regulators of chloroplast RNA (cpRNA) stability, some of which resemble those of their bacterial ancestors (reviewed in Hayes et al., 1999; Schuster et al., 1999). Elements that enhance transcript stability include inverted repeat (IR) sequences found at mRNA 3′ ends as well as 5′ untranslated region (UTR) elements that serve as binding sites for nucleus-encoded protein complexes that protect cpRNAs from 5′→3′ exonucleases (Higgs et al., 1999; Boudreau et al., 2000; Vaistij et al., 2000a, 2000b). The 3′ IRs form stem-loop structures and impede the progress of processive exonucleases, thereby prolonging mRNA lifetime (Stern and Gruissem, 1987; Stern et al., 1991).
Chloroplast mRNAs can also carry signals for instability, the best characterized of which are poly(A) or A-rich tails that can be added posttranscriptionally at a variety of sites, including the coding region or 3′ end (Kudla et al., 1996; Lisitsky et al., 1996; Komine et al., 2000). The polyadenylated molecules are then rapidly degraded by the 3′→5′ exonuclease polynucleotide phosphorylase (PNPase) and possibly other enzymes (Lisitsky et al., 1997a, 1997b; Walter et al., 2002). Based on these and other results (Klaff, 1995), RNA degradation in chloroplasts is currently hypothesized to begin with endonucleolytic cleavage, for example, by the IR-recognizing enzyme CSP41 (Bollenbach and Stern, 2003a, 2003b), followed by the addition of a poly(A) or poly(A)-rich tail and exonucleolytic degradation. However, genetic analysis of this system in chloroplasts is at a very early stage.
In vivo studies of chloroplast RNA stability determinants have been mainly performed using the unicellular green alga Chlamydomonas reinhardtii, a model system for molecular genetic studies of photosynthesis (Grossman et al., 2003). Our earlier work demonstrated the importance of the 3′ IR as a stabilizing element through modification of the atpB gene. Deletion of the IR in the strain atpBΔ26 caused a dramatic reduction in mRNA accumulation and a temperature-sensitive nonphotosynthetic phenotype because of insufficient accumulation of the atpB gene product, the β-subunit of ATP synthase (Stern et al., 1991). Using this conditional phenotype, the nuclear suppressor crp3 was isolated (Levy et al., 1997). crp3 displayed multiple defects in cpRNA 3′ processing, suggesting that cpRNA stability resulted from an interaction between RNA cis-elements and nucleus-encoded trans-acting factors.
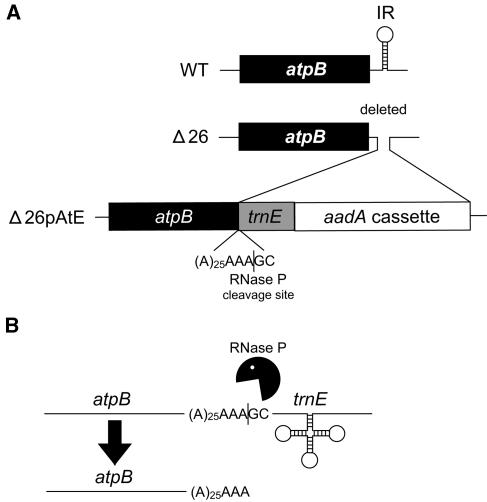
Subsequently, a derivative of atpBΔ26 was constructed to test whether poly(A) tails destabilized cpRNAs in vivo, as had already been shown in vitro. To do this, the IR-less atpB gene was followed by an A25 tract. To expose the poly(A) sequence after its transcription, an RNase P site contained within an ectopic copy of trnE1 was added downstream (Figure 1A). Exposure of the poly(A) tail further reduced mRNA stability, resulting in an obligate heterotrophic phenotype because of the absence of the ATP synthase complex. This offered an opportunity to conduct a genetic screen to identify nuclear genes or chloroplast cis-elements that were involved in polyadenylation-stimulated degradation, and initial results suggested that suppressors that accumulated atpB mRNA could be readily obtained (Komine et al., 2002).
Figure 1.
Construction and Expression of the atpB Gene in Δ26pAtE.
(A) Diagram showing the modifications made to the wild-type atpB gene. First, the 3′ UTR stem-loop was deleted to generate Δ26. Then, a poly(A) tract, the trnE1 gene including its upstream RNase P site, and an aadA selectable marker cassette were inserted, generating Δ26pAtE (Komine et al., 2002).
(B) The atpB gene, poly(A) tract, and trnE1 gene are presumed to be cotranscribed into a pre-mRNA. After processing by RNase P, an A28 tail is exposed at the 3′ end of atpB mRNA. A mature tRNAGlu molecule is also presumably generated.
Here, we report on an extensive analysis of such strains, which we have designated as suppressor of polyadenylation (spa). Genetic analysis showed that all the spa strains had chloroplast mutations and that two unusual strains had stably heteroplasmic genomes. This led to the observations that double-stranded RNA (dsRNA) formation between sense and antisense transcripts might overcome poly(A)-mediated instability and that PNPase appears to be required for rapid degradation of polyadenylated Chlamydomonas cpRNA in vivo.
RESULTS
Processing Alterations Prevent the Exposure of the atpB Poly(A) Tail in Some spa Strains
The structure of the chloroplast genome in the region surrounding atpB is shown in Figure 1A for wild-type, Δ26, and Δ26pAtE cells. In wild-type cells, atpB transcription reads through a downstream IR, followed by a two-step processing mechanism to yield the mature 3′ end, which is coincident with the stem-loop (Stern and Kindle, 1993). In Δ26pAtE, the 3′ IR, which is absent in Δ26, has been replaced by a sequence of 25 adenosines, followed by an ectopic copy of the tRNAGlu (trnE1) gene and flanking sequences and a selectable marker cassette, including the aadA coding region, which confers resistance to spectinomycin and streptomycin (Goldschmidt-Clermont, 1991).
In Δ26pAtE, atpB transcription reads through the polyadenylate tract and trnE1 moieties, yielding a presumptive precursor that may also contain aadA cassette sequences (Figure 1B). This precursor is processed intercistronically by RNase P, yielding a tRNAGlu molecule and an atpB transcript terminated by 28 adenosines. As we have shown previously (Komine et al., 2002), such atpB transcripts do not accumulate to a level detectable by filter blotting or RT-PCR because of inherent instability of 3′ polyadenylated transcripts in the chloroplast. The resultant lack of ATP synthase assembly, and consequent nonphotosynthetic phenotype of Δ26pAtE, served as a basis for the selection of photoautotrophic revertants that might be impaired in the degradation of polyadenylated atpB transcripts.
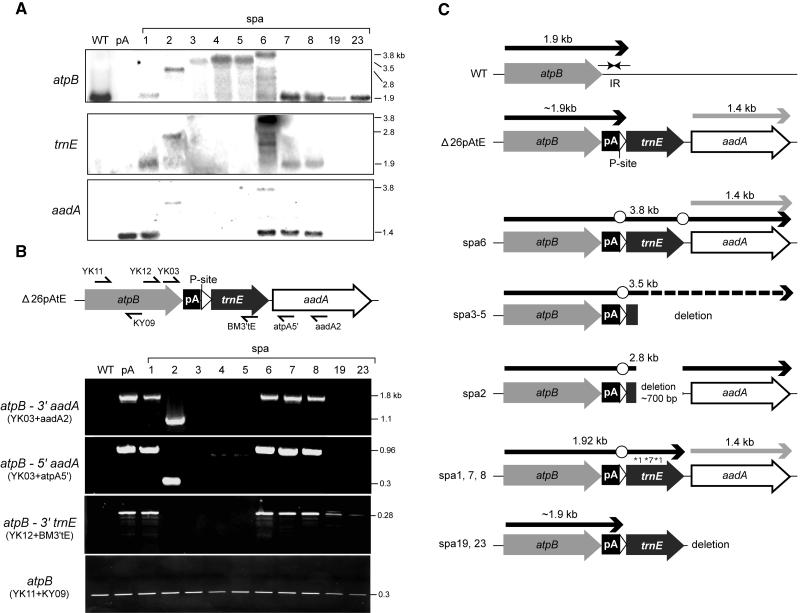
This screen identified 38 spontaneous phenotypic revertants of Δ26pAtE based on their ability to grow on minimal medium. Given the number of cells originally plated and their survival rate, a reversion rate of ∼1 in 105 cells was estimated (see Methods). This relatively high rate suggested that cpDNA-based reversion or suppression was occurring and/or that mutations in a variety of nuclear loci could confer the selected growth phenotype. Because we were interested in identifying strains defective in degrading polyadenylated mRNA, RNA filter hybridizations were performed for wild-type, Δ26pAtE, and the spa strains. As shown in Figure 2A, wild-type cells accumulated a 1.9-kb species, whereas no RNA was detected for Δ26pAtE (pA) as a result of rapid degradation after the exposure of poly(A) tail. On the other hand, all spa strains accumulated discrete atpB transcripts, but many were significantly longer than 1.9 kb (e.g., spa 2-6), suggesting a failure to expose the poly(A) tail. A second class of spa strains accumulated atpB mRNA with approximately the same size as the wild type (e.g., spa1, spa7, spa8, spa19, and spa23). All of the 38 revertants had one of the RNA patterns represented in Figure 2A.
Figure 2.
Transcription and Structure of the atpB Region in spa Strains.
(A) RNA gel blot analysis. Total RNA (5 μg) from strains shown across the top were probed, as shown at the left of each panel. The portion of the trnE1 blot showing the native tRNAGlu species (75 nucleotides) has been cropped. Sizes of transcripts, shown at right, were calibrated from RNA size standards.
(B) PCR analysis of spa DNA. The positions of the primers used are shown schematically, and their sequences are given in Table 2. To the left of each panel, the region amplified is given, and primers sets are shown in parentheses. Fragment sizes at right were based on known sequences and/or DNA size standards.
(C) Summary of RNA processing and DNA alterations in spa strains. The gray arrows represent the atpB coding regions, with the 3′ IR shown as horizontal arrows for the wild type. The poly(A) tract (pA) is represented by a closed box, the RNase P site by an open arrowhead, and the mature trnE1 sequence and aadA cassettes by labeled closed or open arrows, respectively. Note that the sequences are not fully to scale. Transcripts are shown as heavy black (atpB) or gray (aadA) arrows above the arrows/boxes representing DNA sequences. The dashed line for spa3-5 indicates that the 3′ ends of the deletion and atpB transcript have not been determined. Open circles represent the sites of processing defects and asterisks the sites of point mutations in spa1 and spa7. The sequence of spa8 has not been determined, thus its classification is tentative.
To test for the failure to expose the poly(A) tail, additional probes were used in RNA gel blot analysis. Two probes downstream of atpB were used: trnE1 and aadA (Figure 2A). The results showed that atpB transcripts of six strains (spa1, spa2, and spa6-8) hybridized with the trnE1 probe, indicating that the pre-mRNA was abnormally processed, avoiding poly(A) tail exposure. The aadA probe revealed a monocistronic mRNA emanating from the selectable marker cassette in pA, spa1, and spa6-8, corresponding to those strains in which the cassette had not been disrupted (see below). The atpB-containing transcripts in spa2 and spa6 also hybridized with aadA, confirming their cotranscription.
In spa3-5, however, the atpB transcripts hybridized with neither trnE1 nor aadA despite their lengths (∼3.5 kb), suggesting DNA deletion of most or all of trnE1 and aadA sequences. To confirm and map the apparent deletions, PCR analysis was performed (Figure 2B). Three sets of reactions were run using two upstream atpB coding region primers (YK03 and YK12) and three different downstream primers (aadA2, atpA5′, and BM3′tE), which anneal within trnE1 and the aadA cassette. As a positive control, a primer set within the atpB mRNA coding region was used (YK11 + KY09).
When PCR was performed with primer sets YK03/12 + (aadA2/atpA5′/BM3′tE), spa1 and spa6-8 yielded the same sized PCR product as Δ26pAtE. By contrast, spa3-5 DNAs failed to generate PCR products, indicating that deletion of trnE1 and at least part of the aadA cassette had occurred. A different result was obtained for spa2, which gave a shorter product than Δ26pAtE with primer sets (YK12 + aadA2/atpA5′) but not from (YK03 + BM3′E), which indicated that spa2 had a deletion within trnE1. Indeed, sequencing of the spa2 PCR product revealed a 750-bp deletion beginning within trnE1 and extending into the aadA cassette promoter, as we previously reported (Komine et al., 2002). Because the trnE1 sequence is required for efficient processing at the RNase P site and, thus, for exposure of the poly(A) tail, the deletion of trnE1 sequence is likely to be the major reason for phenotypic reversion of spa2-5 (summarized in Figure 2C).
On the other hand, the atpB mRNAs of spa1, spa7, spa8, spa19, and spa23 were close to the size of the wild-type transcript, consistent with a 3′ end near the poly(A) tract. However, only the transcripts of spa19 and spa23 failed to hybridize with trnE1 (Figure 2A). In the other cases (spa1, spa7, and spa8), an intercistronic processing defect prevents exposure of the poly(A) tail, explaining the suppression phenotype. Sequence analysis of spa1 and spa7 revealed two and one point mutations in trnE1, respectively, which might compromise RNase P site function. Taken together, these results lent support to the possibility that uniquely in spa19 and spa23, the atpB mRNA might accumulate with an exposed poly(A) tail. Therefore, we focused on how polyadenylated mRNA might be stabilized in these two suppressors. We note that because of the way in which they were recovered, spa19 and spa23 are likely to be clones of a single initial suppressor; in fact, data obtained from them were indistinguishable in every case examined.
Genetic Analysis Revealed an Exceptional Segregation Pattern for spa19/23
To determine whether the spa strains carried suppressor mutations in the chloroplast or nucleus, the mt+ suppressors were crossed to the mt− wild-type strain CC-124, and tetrad progeny were analyzed. According to classical nuclear/chloroplast genetics, if the phenotype was attributable to a single nuclear mutation, tetrad progeny would segregate 2:2 for photosynthetic to nonphotosynthetic growth. On the other hand, if the mutation were in the chloroplast, all the progeny would be photosynthetic. Indeed, genetic data for spa1-8 revealed a 4:0 segregation for photosynthesis, consistent with the suspected chloroplast location of the suppression events (Table 1).
Table 1.
Genetic Analysis of spa Strains
| Number of Tetrads Analyzeda
|
Number of Progeny Analyzedb
|
|||
|---|---|---|---|---|
| Strain | Complete | Incomplete | Photosynthetic | Nonphotosynthetic |
| spa1c | 10 | 0 | 40 | 0 |
| spa2 | 10 | 0 | 40 | 0 |
| spa3 | 10 | 0 | 40 | 0 |
| spa4 | 10 | 0 | 40 | 0 |
| spa5 | 10 | 0 | 40 | 0 |
| spa6 | 14 | 0 | 56 | 0 |
| spa7 | 10 | 0 | 40 | 0 |
| spa8 | 10 | 0 | 40 | 0 |
| spa19d | 8 | 16 | 20 (2 + 4) | 59 (6 + 12) |
| spa23d | 10 | 7 | 10 (1 + 2) | 56 (11 + 4) |
Spa strains (mt+) were crossed to CC-124 (mt−). Complete tetrads are those in which all four spores were viable on TAP medium. Incomplete tetrads had two or three surviving members.
Progeny were tested for growth on minimal medium, then classified as photosynthetic or nonphotosynthetic. The number given is the total from complete and incomplete tetrads.
The spa1 mutant was originally reported (Komine et al., 2002) to result from a nuclear mutation.
For spa19 and spa23, both photosynthetic and nonphotosynthetic progeny were obtained. However, in a given tetrad, only a single phenotype was observed. Numbers in parentheses represent the total number of (complete + incomplete) tetrads from which the progeny were analyzed.
To our surprise, the cross (spa19/23 mt+ × CC-124 mt−) yielded two classes of tetrads, one in which the four members were all photosynthetic (PS+), and the other in which all were nonphotosynthetic (PS−). Of 42 complete or partial tetrads, nine were PS+ and 33 were PS−. In all tetrads obtained, Mendelian 2:2 segregation of nuclear alleles was confirmed by PCR analysis of the nuclear mating type loci, as described in Methods (and shown in Figure 3B).
Figure 3.
Chloroplast DNA Analysis of spa19/23.
(A) Total DNAs of wild type, Δ26pAtE (pA), and spa19/23 were digested with PstI, and blotted DNAs were probed with the atpB coding region. Note that although both wild-type and spa19/23 possess atpB-containing 7.8-kb fragments, the 3′ ends of these fragments are not coincident and, thus, the comigration is coincidental.
(B) Analyses of (spa23 mt+ × WT mt−) progeny. In the top panel, parental strains and progeny of representative tetrads were plated on TAP or minimal medium to determine the photosynthetic growth phenotype. The middle panel shows cpDNA analysis as described for (A). The bottom panel shows PCR analysis of the mating type locus to confirm mating type segregation in tetrads.
Although several models could be developed to explain this exceptional segregation pattern, the most likely possibility seemed to be that spa19 and spa23 had a mixture of chloroplast genomes and that a poorly transmitted version conferred the PS+ phenotype, whereas a better transmitted one conferred the PS− phenotype. This hypothesis was confirmed by molecular analysis, as described below.
Spa19/23 Maintain Heteroplasmic cpDNA
To test whether spa19 and spa23 might bear heteroplasmic cpDNA, DNA gel blot analysis was performed, as shown in Figure 3. After digestion with PstI and probing with an atpB internal fragment, the expected single signal was seen for wild-type (7.8 kb) and Δ26pAtE (0.75 kb) DNAs (Figure 3A). Spa19/23, on the other hand, possessed both a minor 7.8-kb fragment and a second fragment ∼1.95 kb in size, but lacked the 0.75-kb fragment of their progenitor. Note that the similar ∼7.8-kb size of the wild type and spa19/23 fragments is coincidental; although the PstI site within atpB is common, the sequences downstream of atpB are entirely different because of modifications in Δ26pAtE and subsequent deletion of the region containing the aadA cassette.
When tetrad progeny were examined, two patterns were found, which correlated with the photosynthetic growth phenotype. In spa19/23 and in PS+ progeny resulting from the cross to CC-124, both 7.8- and 1.95-kb fragments were found (data for spa23 are shown in Figure 3B). Thus, the PS+ progeny resembled the original suppressor. In PS− tetrads, however, only the 7.8-kb fragment was found, leading to the conclusion that the 1.95-kb fragment cosegregated with a PS+ phenotype. We will therefore refer to cpDNA containing the 1.95-kb fragment as the PS+ genome and that containing the 7.8-kb fragment as the PS− genome. We further inferred that the PS+ genome is required for accumulation of atpB transcripts, whereas the PS− genome is insufficient for this purpose. However, because the PS− genome is invariably present, it seemed probable that it contributed essential genes absent from the PS+ genome.
Taking the genetic and molecular data together, it was immediately obvious that although the PS− genome was inherited alone more frequently than the PS+/PS− genome combination, the PS− genome is much less abundant than the PS+ genome in vegetative spa19/23 cells (Figure 3A). Indeed, a quantitative comparison of the signal intensities between 1.95-kb/7.8-kb fragments revealed an approximately fourfold molar excess of the PS+ genome (data not shown; see Methods and Figure 5). Thus, the more abundant genome is inherited poorly in genetic crosses, suggesting a defect in transmission and/or replication during zygote maturation.
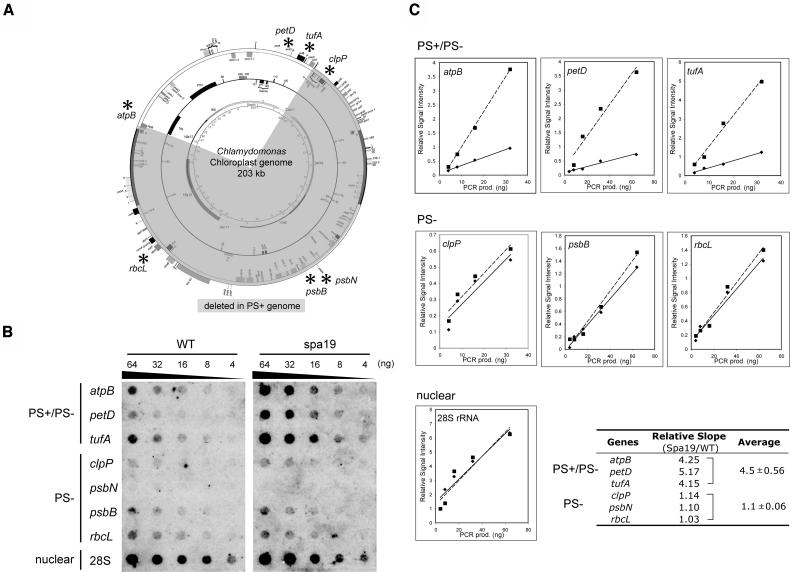
Figure 5.
Organization and Stoichiometry of the PS+ Genome in spa19/23.
(A) Seven chloroplast genes (marked by asterisks) and one nuclear gene (28S rDNA) were chosen for analysis. Representation of the PS+ genome is as in Figure 4A.
(B) The indicated amounts of PCR products corresponding to each gene were dot blotted and hybridized with radiolabeled total DNA extracted from wild-type and spa19 cells. The genome(s) on which each gene is located are indicated at left.
(C) The signal intensity was quantified for the wild type (diamond) and spa19 (square). To normalize the data, the values were divided by those obtained for 4 ng of 28S rDNA on the same blot. The corrected values were plotted against the amount of PCR products, and average relative signal intensities per nanogram of PCR product were calculated as the slopes of lines (the wild type, solid lines; spa19, dashed lines). The resultant ratios are shown in the table at the bottom right.
Inverse PCR Reveals a Large Deletion in the spa19/23 PS+ Genome
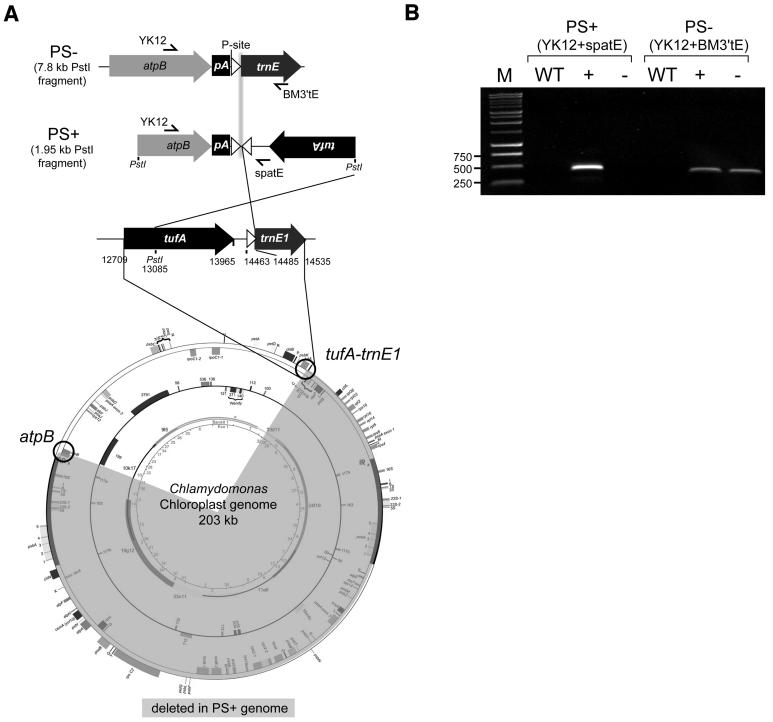
To clarify the presumed chloroplast mutation underlying the PS+ phenotype of spa19/23, an inverse PCR approach was taken to clone the sequence downstream of atpB in the PS+ genome. PCR was conducted after PstI digestion and subsequent self-ligation of spa19 DNA. Sequencing of the cloned 1.95-kb PstI fragment revealed that in addition to the aadA selectable marker cassette being deleted (Figure 2C), deletion and/or rearrangement had resulted in upstream portions of the endogenous trnE1 gene (i.e., that corresponding to the ectopic one used in Δ26pAtE) being positioned immediately downstream of atpB, but in an inverted orientation, as shown in Figure 4A.
Figure 4.
Derivation of the PS+ Genome in spa19/23.
(A) Schematic representation of the atpB regions from the PS− and PS+ genomes and the source of sequences involved in the rearrangement, with numbering according to GenBank accession number BK000554. Symbols are as described for Figure 2C. The region of the chloroplast genome believed to have been deleted during formation of the spa19/23 PS+ genome is highlighted in gray. The atpB and endogenous trnE1 genes are circled as reference points. Primer sets YK12+spatE and YK12 + BM3′tE were designed specifically for PS+ and PS− genomes, respectively.
(B) PCR was performed to test for the presence of the indicated genomes in the wild type or in photosynthetic (+) or nonphotosynthetic progeny (−) from the cross shown in Figure 3B. M, molecular weight markers, with relevant sizes in base pairs.
The presence of both PS+ and PS− genomes in phenotypically PS+, but not PS− strains, was confirmed by PCR. Primer sets specific for the PS+ (YK12+spatE) and PS− (YK12+BM3′tE) genomes were designed, as shown in Figure 4A. DNA was extracted from wild-type, photosynthetic (PS+), and nonphotosynthetic (PS−) progeny from the cross (spa19/23 mt+ × CC-124 mt−) and used as a template for PCR. The PS+ specific primer pair yielded a single PCR product with the predicted size (437 bp) only from photosynthetic progeny (Figure 4B). On the other hand, PS− specific primers yielded the 376-bp product from both types of progeny. These results confirmed that the sequence obtained by inverse PCR from spa19/23 exists in vivo and is maintained heteroplasmically with the PS− genome, consistent with results of the DNA gel blot described above. The fact that the PS+ PCR product was undetectable with DNA isolated from nonphotosynthetic cells further suggests that this configuration is not maintained at a substoichiometric level, as is often seen in plant mitochondria (reviewed in Mackenzie and McIntosh, 1999).
Because in wild-type and Δ26pAtE cpDNAs atpB is located ∼145 kb away from trnE1-tufA, yet the genes maintain their relative orientations in spa19/23, the simplest explanation is a recombination-mediated deletion of these 145 kb (shaded part of chloroplast genome at the bottom of Figure 4A). Given the fact that the PS+ genome is overrepresented (Figure 3), it is possible to verify the hypothesized deletion by measuring the stoichiometries of genes that are predicted to be present or absent within it. To do so, dot blot membranes were prepared with a dilution series of PCR products representing seven chloroplast genes and one nuclear gene (28S rRNA) and hybridized with radioactively labeled total DNA extracted either from the wild type or spa19. Among the chloroplast genes, four (clpP, psbN, psbB, and rbcL) were predicted to be PS− specific, whereas the other three (atpB, petD, and tufA) should be present in both the PS+ and PS− genomes (Figure 5A). As shown in Figure 5B, the signal intensities for PS− genome-specific genes and 28S rDNA were similar when the wild type and spa19 were compared (nuclear rDNA is naturally present in high copy number). By contrast, signals for putative PS+ specific genes were stronger in spa19 than in the wild type.
These signals were quantified, normalized to the signal for 28S rDNA on the same membrane, and plotted against the amount of PCR product present in each dot (Figure 5C). Average relative signals per nanogram of PCR product were calculated, which allowed the seven chloroplast genes tested to be categorized into two clear groups, atpB-petD-tufA and clpP-psbB-rbcL (signal from the short psbN gene was too weak to be used). The slopes for the atpB-petD-tufA group and 28S rDNA were almost identical between the wild type and spa19, with the average ratio of spa19/wild type being 1.1 ± 0.06. On the other hand, the ratio for clpP-psbB-rbcL was 4.5 ± 0.56, indicating that these genes are three to four times more abundant in spa19 than in the wild type. This stoichiometry can be most simply explained by positing that the PS+ genome arose through a deletion of 145 kb and is maintained at three to four times the copy number of the PS− genome in vegetative cells.
Transcriptional Activity of the PS+ Genome
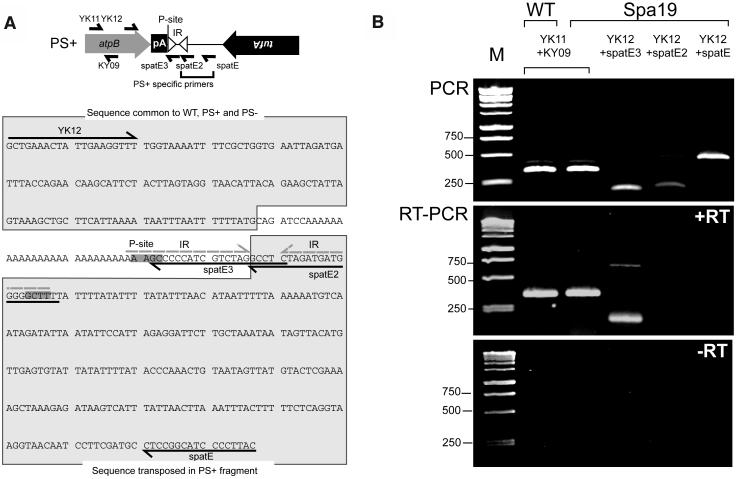
Because our data demonstrated that the PS+ genome was required for photosynthesis, it seemed likely that this genome was transcriptionally active and contributing a factor(s) necessary to confer photosynthetic competence. To address this issue, RT-PCR analysis was used, with which we could distinguish between some PS+ and PS− genome transcripts derived from the atpB region. As shown in Figure 6A, three different reverse (cDNA synthesis) primers (spatE, spatE2, and spatE3) were used with the same forward primer, YK12. Among them, spatE and spatE2 were designed to detect PS+ specific sequences, whereas spatE3 was designed to detect sequences common to atpB transcripts from PS+ and PS− genomes. As a positive control, primers corresponding to the atpB coding region (YK11+KY09) were used. All the primer sets were confirmed to work effectively by performing PCR with total DNA as a template (Figure 6B, top panel).
Figure 6.
Sequence and Expression of the PS+/PS− Genome atpB Region.
(A) Organization of the atpB region in the PS+ genome, with symbols as in Figure 2C. The tufA gene is shown inverted because it is on the opposite strand with respect to atpB. Primers are shown both on the schematic and in the sequence below. In this sequence, the 4 bp surrounding the RNase P cleavage sites (P-site) are in bold and highlighted in dark gray. Because the two sites are on opposite strands, the sequences are complementary. An IR that could be formed by sequences surrounding the RNase P cleavage site has dashed gray lines above it. Sequences common to the wild-type, PS+, and PS− genomes, as well as those downstream of atpB in the PS+ but not PS− genome, are highlighted in light gray and outlined.
(B) PCR (top panel) or RT-PCR (bottom two panels) was performed with the primer sets shown across the top. The source of the ∼740-bp product with RT-PCR of spa19 RNA and (YK12+spatE3) is unknown. The bottom panel shows the experiment without added reverse transcriptase, demonstrating the lack of DNA contamination. M, molecular weight markers, with relevant sizes in base pairs.
Close inspection of the novel PS+ genome sequence revealed a potential explanation for the accumulation of atpB mRNA. As shown in the bottom panel of Figure 6A, the apposition of two identical copies of trnE1-derived sequences surrounding the RNase P site, in an inverted orientation, could form a stem-loop if present in the atpB mRNA. This near-perfect 24-nucleotide IR is indicated with a line above it in the sequence and if formed would likely protect the 3′ end of the RNA with a classical secondary structure. Furthermore, if this stem-loop were present in vivo, both primers spatE3 and spatE2 should yield RT-PCR products with YK12 as a second primer. However, the results obtained by RT-PCR were unexpected. As shown in the middle panel of Figure 6B (+RT), we found that a product was obtained when primers common to PS+ and PS− genomes were used (spatE3-YK12) but not when spatE2, which anneals only 20 nucleotides downstream, was used. This result indicates that an atpB mRNA species accumulated in which the poly(A) tail was in fact not exposed, but was followed by a sequence of 5 to 20 nucleotides, which would be required to stably anneal the 3′ end of the spatE3 primer. Although this short sequence would internalize the poly(A) tract, it is not predicted to form a strong secondary structure, and it is known that atpB mRNA lacking an IR (e.g., strain Δ26) is unstable (Stern et al., 1991).
To map the 3′end of atpB mRNA more precisely, S1 nuclease protection analysis was performed, as shown in Figure 7. To generate probes, PCR products were amplified from the PS+ and PS− genome using primers YK14 (within atpB) and spatE and BM3′tE, respectively (Figure 7A). The 3′ end of the antisense strand of the PCR product was labeled by taking advantage of a SexAI site that is present 142 bp upstream of the atpB translation termination codon. PCR products were also prepared with three different reverse primers, spatE3, spatE4, and spatE5, and labeled in the same way. These served as precise size markers for the end of the atpB coding sequence (spatE5), the end of the poly(A) sequence (spatE4), and the end of the sequence shared between the PS+ and PS− genomes (spatE3; see Figure 7B for sequence detail).
Figure 7.
S1 Nuclease Protection Mapping of the atpB 3′ End.
(A) The positions of primers used to prepare probes and size markers and the restriction site (SexAI) used for the radiolabeling are shown in the top panel, together with schematic drawings of the atpB regions in the PS+ and PS− genomes.
(B) Sequence showing the positions of primers and elements. An arrowhead within the spatE3 primer sequence shows the location of the mRNA 3′ end as calculated from the nuclease protection data shown in (C) and (D). The dashed portion of primer spatE3 corresponds to sequences present in the PS+ but not the PS− genome.
(C) S1 protection analysis with a PS− probe. Markers (M) or the probe shown in (A) protected by the wild type or spa19 RNA were analyzed in a 7% denaturing polyacrylamide gel. Migration of each marker in nucleotides is indicated at the left. The products corresponding to the atpB 3′ end in the wild type or spa19 are indicated by arrowheads.
(D) Comparataive S1 protection analysis with PS+ and PS− probes.
(E) S1 protection analysis with a PS+ probe and synthetic RNAs prepared using PCR products T7YK14+spatE (spatE) and T7YK14+spatE3 (spatE3) as templates.
The results of S1 protection with the PS− probe are shown in Figure 7C. When wild-type RNA was used, the size of protected product was calculated to be 161 nucleotides, very close to the predicted 159 nucleotides, which corresponds to the discontinuity between the engineered sequence in Δ26pAtE and the wild-type atpB gene. Using spa19 mRNA, a single protected product was also observed. Based on its migration between the markers generated using primers spatE4 and spatE3, its size was 197 nucleotides, very close to the 200 nucleotides that would place it at the boundary of sequences shared between the PS+ and PS− genomes (arrowhead in Figure 7B). Using the PS+ probe instead of the PS− probe, the same ∼161- and ∼197-nucleotide protected products were observed for the wild type and spa19 mRNA, respectively, as shown in Figure 7D. This demonstrates that sequences derived uniquely from the PS+ genome are not present in spa19 atpB mRNA at the resolution possible with this analysis, consistent with the RT-PCR experiments shown above.
To eliminate the possibility that S1 nuclease protection had generated an artifact because of, for example, strong RNA secondary structure, we performed a control experiment with two in vitro–generated atpB transcripts, as shown in Figure 7E. A transcript having its 3′ end coincident with primer spatE3 represented the mRNA accumulating in vivo based on the experiments described above, whereas a transcript with its end coincident with primer spatE included PS+ genome-specific sequences, including the fortuitous IR created during the generation of the PS+ genome. When the transcript with its 3′end at spatE3 was used with a PS+ probe, a single protected product with the same size as the 210-nucleotide marker was observed, as expected. When the transcript with its 3′ end at spatE was used, the entire probe was protected, without any evidence of artifactual cleavage products (e.g., within the loop of the putative IR). Thus, the poly(A) sequence of atpB mRNA in spa19 is not exposed, but rather is capped with a short sequence common to the PS+ and PS− genomes. This left the open questions of how such a short and apparently unstructured sequence might stabilize the mRNA, and why atpB transcripts are detected apparently only corresponding to sequences shared between the PS+ and PS− genomes.
Antisense Transcripts for atpB Accumulate and Form Duplexes in spa19/23
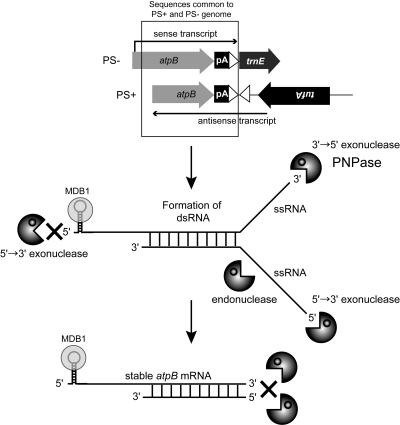
To explain the RNA phenotype of spa19/23, we elected to test the hypothesis shown in Figure 8, namely, that functional atpB transcripts are stabilized via hybridization to antisense atpB mRNA transcribed from the PS+ genome. Our logic was based on the fact that in the PS+ genome, atpB is flanked by the transposed trnE1 upstream sequence and tufA, both on the opposite strand. Because at least tufA is robustly transcribed (Hwang et al., 1996; trnE1 is a duplicated gene and the copies cannot be transcriptionally differentiated) and the PS+ genome is in elevated copy number, we considered that tufA-initiated transcription might read through the trnE1 upstream region and lead to significant accumulation of antisense atpB mRNA. If such antisense transcripts accumulated in excess over sense RNA, duplex formation with sense transcripts emanating from the PS+ or PS− genome might be favored. As illustrated in Figure 8, this dsRNA structure might stabilize the atpB mRNA by impeding 3′→5′ exoribonuclease and/or single-stranded RNA-specific endoribonucleases. The way this diagram is drawn, which is in agreement with the gel blot, RT-PCR, and S1 nuclease protection data shown above, the eventual accumulating sense transcript would have a size and 3′ end most consistent with a hybrid between sense transcripts from the PS− genome and antisense RNA from the PS+ genome (see Discussion for alternatives).
Figure 8.
Hypothesis for Mechanism to Stabilize atpB mRNA in spa19/23.
Artificially polyadenylated atpB mRNA is unstable because the 3′ end is susceptible to the attack by 3′→5′ exoribonuclease(s), such as PNPase, whereas its 5′ end would be protected from 5′→3′ exoribonuclease(s) by the stem-loop structure in its 5′ UTR (Anthonisen et al., 2001) and possibly a protein encoded by the nuclear gene Mdb1, which is mutated in the atpB mRNA stability mutant Thm24 (Drapier et al., 1992; see text). But in spa19/23, artificially polyadenylated atpB sense mRNA transcribed from the PS− genome, and atpB antisense mRNA transcribed from the PS+ genome (top), may form a dsRNA duplex (center). Unpaired regions of the duplex may be degraded by exoribonuclease(s) and/or endoribonucleases, whereas double-stranded regions would be resistant, resulting in the stabilization of atpB transcripts in spa19/23 (bottom).
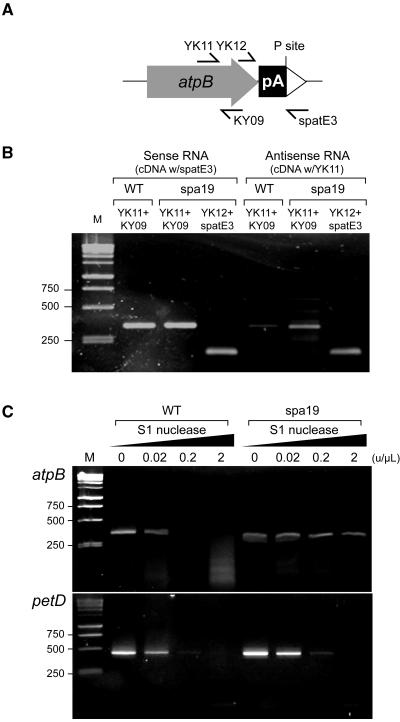
Two testable predictions arise from this hypothesis, namely, that antisense atpB mRNA should accumulate in spa19 and that duplexes should form between antisense and sense atpB transcripts. To test the first of these predictions, we performed RT-PCR so as to distinguish sense and antisense transcripts, as shown in Figure 9. Total RNA was extracted from wild-type and spa19 cells, and cDNA was synthesized using forward or reverse primers to detect antisense or sense transcripts, respectively. cDNA was then amplified by PCR with appropriate primer pairs, as shown in Figure 9A. The atpB coding region was analyzed with the primer pair (YK11+KY09). In the wild type, sense atpB transcript was clearly detected, but almost no signal was obtained representing antisense transcripts, as expected (Figure 9B). In spa19, however, PCR products corresponding to not only sense transcripts, but also to antisense transcripts, were detected. A similar result was obtained for the region between the 3′ end of atpB and tufA (YK12+spatE3), consistent with antisense transcripts spanning the PS+ genome-specific region into atpB.
Figure 9.
Analysis of Antisense and Double-Stranded RNA in Wild-Type and spa19 Cells.
(A) Locations of primers used in this figure.
(B) RT-PCR using RNA from the wild type or spa19 as shown. The primer used for cDNA synthesis is indicated above the strains, with subsequent PCR primer sets given above each lane. M, 1-kb DNA ladder.
(C) S1 nuclease protection analysis in combination with RT-PCR. Total RNA was extracted from the wild type and spa19 and treated with the concentration of S1 nuclease shown above each lane. After S1 nuclease treatment, cDNA was amplified by RT-PCR using primer sets for atpB (YK11+KY09) or petD (petF+petR).
To assess the possibility that atpB forms dsRNA in spa19, S1 nuclease protection was used, as shown in Figure 9C. S1 nuclease digests single-stranded DNA or RNA nonspecifically but will not digest fully paired duplexes. Total RNA was extracted from wild-type and spa19 cells, and equal amounts were treated with increasing concentrations of S1 nuclease. Subsequently, RT-PCR was performed using a one-step assay with primers YK11+KY09 to detect the atpB coding region or for another chloroplast gene, petD, as a control. For both genes, clear signals were obtained from the wild type and spa19 when the RNAs were not treated with S1 nuclease. But as higher concentrations of S1 nuclease were used, we found that atpB RNA from spa19 was more resistant to the action of S1 nuclease than that of the wild type. On the other hand, no such difference was observed when petD mRNA was examined using the same RNA samples. These results support the possible formation of double-stranded atpB mRNA in spa19 chloroplasts.
Ectopically Expressed Antisense RNA Suppresses the Instability of atpB mRNA Lacking a 3′ IR
The RT-PCR results described above are strong evidence for the presence of antisense atpB transcripts and the formation of double-stranded atpB mRNA in spa19 chloroplasts. To obtain more direct proof that antisense RNA can mitigate RNA degradation in chloroplasts, we created a strain expressing a specific antisense RNA. The strain atpBΔ26 was chosen as a host because atpB mRNA is unstable as a result of the lack of a 3′ IR, and any increase in stability could therefore be observed.
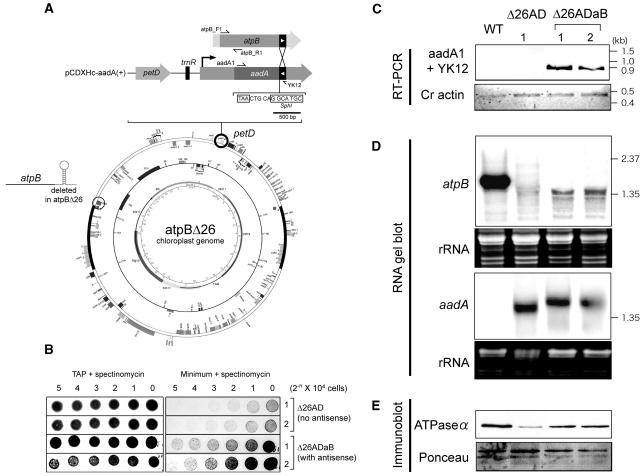
The experimental strategy is shown in Figure 10A. A 121-bp segment of the atpB coding region was amplified by PCR, with the reverse primer designed not to include any 3′ UTR/IR sequences. This 121-bp sequence was inserted at the end of the aadA coding region in an inverted orientation. The SphI site used for this insertion was 5 bp downstream of the aadA translation termination codon, allowing the expression of chimeric aadA sense–atpB antisense RNA without affecting aadA translation. This engineered aadA cassette was targeted to a site near petD, which is ∼45 kb from the native atpB gene (Figure 10A). Thus, the site of antisense RNA expression is unlinked, in contrast with the situation in spa19. This strategy renders the strain unable to reconstitute any IR sequence through any recombination events after transformation. The native atpB IR is not present in the strain, and recombination between the distant but tandemly oriented 121-bp sequences would place the atpB coding region on a different molecule than the rbcL IR, which flanks the aadA coding region downstream of the 121-bp segment.
Figure 10.
Ectopic Expression of atpB Antisense Transcripts Stabilizes mRNA Lacking a Terminal Stem-Loop.
(A) Schematic drawing of the experimental strategy. The 121-bp segment of the atpB coding region (filled box with white arrowhead) was inserted at the end of aadA coding region in pCDXHc aadA (+) in an inverted orientation. This chimeric construct was directed to a region downstream of the petD gene by chloroplast transformation. The DNA sequence shows the relationship of the SphI insertion site of the atpB moiety relative to the aadA TAA translation termination codon.
(B) Growth of control Δ26AD and antisense-expressing (Δ26ADaB) strains on TAP or minimal medium containing 100 μg/mL of spectinomycin. Numbers across the top indicate the quantity of cells originally plated in a series of twofold dilutions.
(C) RT-PCR was used to confirm expression of the chimeric aadA-atpB antisense transcripts in the strains indicated across the top. As a positive control, Chlamydomonas (Cr) actin cDNA was amplified.
(D) RNA gel blots were used to measure the accumulation of atpB and aadA mRNAs. The atpB probe was generated by PCR using the primers shown at the top of (A). The aadA probe was generated by PCR using primers aadA1 and aadA2. To show relative loading, ethidium bromide–stained images are shown below each panel. For space reasons, these images are slightly compressed in the vertical dimension.
(E) Immunoblot analysis was used to measure accumulation of the ATP synthase by probing with an antibody directed against the Chlamydomonas ATPase α-subunit. To show relative loading, a Ponceau-S stained image is shown at the bottom.
The antisense-expressing plasmid, along with a control plasmid lacking the antisense segment, were transformed into atpBΔ26 cells, and colonies were selected on Tris-acetate-phosphate (TAP) medium containing 100 μg/mL of spectinomycin. DNA of several individual transformants was analyzed to confirm correct integration of the aadA cassette and homoplasmicity of chloroplast genomes (data not shown). atpBΔ26 grows slowly on minimal medium because of the unstable atpB mRNA and a dramatic reduction of its gene product (Stern et al., 1991). We therefore compared the growth of two representative control (Δ26AD) and antisense (Δ26ADaB) strains using a dilution series of cells, as shown in Figure 10B. On TAP (acetate-containing) plates, robust and equivalent growth was observed for all strains. On minimal medium, however, the Δ26ADaB strains showed much more robust growth than the controls, as can be clearly seen by comparing the numbers of cells in the respective strains after plating of equal numbers and incubation for 9 d. This suggested that expression of the antisense RNA increased the expression of the unlinked atpB gene.
Next, the expression of the chimeric aadA-atpB mRNA was analyzed by RT-PCR, as shown in Figure 10C, with actin mRNA as a control. Products of the expected size (897 bp) were detected only from the antisense (Δ26ADaB) transformants, whereas actin mRNA was detected in all the samples. This result confirmed the expression of the chimeric transcript in the antisense strains. Further analysis was performed using RNA gel blots, as shown in Figure 10D. Accumulation of atpB mRNA was monitored using a labeled PCR product from a part of the coding region excluded from the antisense construct (primers atpB_F1 + atpB_R1; Figure 10A). Because of its lack of a stem-loop structure, strain Δ26AD accumulates much less atpB mRNA than the wild type, as has been previously reported for its close relative Δ26 (Stern et al., 1991). In the antisense-expressing strains, however, a clear band of ∼1.35 kb was observed, with a signal intensity approximately fivefold weaker than the wild type, when gel loading is taken into account. The size of this transcript is smaller than that of the wild type, presumably because of the lack of the ∼100-nucleotide 3′ UTR, and is consistent with the mRNA 3′ end being stabilized by base pairing with the antisense moiety of the aadA transcript. Finally, an aadA probe was used, which identified discrete transcripts in both Δ26AD and Δ26ADaB. The transcript size was larger in Δ26ADaB, again confirming the expression of the atpB antisense segment in these transformants.
More robust photosynthetic growth, as shown in Figure 10B, would be expected to be accompanied by an increase of the atpB gene product and would be consistent with the greater accumulation of the cognizant mRNA, as shown in Figure 10D. We therefore performed immunoblot analysis, using an antibody raised against the ATPase α-subunit, which accumulates stoichiometrically with the atpB gene product, the β-subunit (Drapier et al., 1992). Consistent with the previous report for atpBΔ26, we detected a decreased amount of ATP synthase in Δ26AD, as compared with the wild type. On the other hand, this decrease was partially restored in the Δ26ADaB strains, accounting for their faster growth on minimal medium. Taken together, these results reaffirm the possibility that antisense transcripts are able to stabilize sense transcripts in trans.
PNPase May Degrade atpB mRNA Synthesized from the PS− Genome
If the antisense transcripts can stabilize the sense mRNA, a subsequent question is how this stabilization occurs. According to the hypothesis shown in Figure 8, the formation of dsRNA might impede 3′→5′ exoribonucleases that would otherwise degrade the atpB transcript. A prime candidate for such an enzyme is PNPase, which has a high affinity for polyadenylated transcripts (Lisitsky et al., 1997a), is present in chloroplasts (Yehudai-Resheff et al., 2001; Walter et al., 2002), and is known to be involved in bulk mRNA decay in Escherichia coli (Donovan and Kushner, 1986). If PNPase were necessary for the degradation of the atpB transcript derived from the PS− genome, it could be predicted that the PS+ genome would not be required if the activity of PNPase were compromised.
In work to be published elsewhere, we have described the generation of Chlamydomonas strains depleted for PNPase (our unpublished data). This depletion was achieved via antisense technology (Figure 11A) to partially silence the nucleus-encoded PNPase gene (note that antisense RNA in the nucleus downregulates gene expression, whereas the model proposed in Figure 8 of this article evokes a stabilizing role for antisense RNA in the chloroplast). One of the antisense strains obtained was Ap6, which, based on immunoblot analysis, retains ∼30% of the wild-type level of PNPase. To attain further insight into the possible function of dsRNA in spa19, we crossed spa19 (mt+) to Ap6 (mt−) so as to introduce the spa19 chloroplast into the Ap6 nuclear background.
Figure 11.
Analysis of spa19 (mt+) × PNPase (Ap6) Antisense (mt−) Progeny.
(A) Locations of primers used in (B). The ble gene confers resistance to zeocin and is fused transcriptionally to a 365-bp segment of the Pnp gene in the reverse orientation.
(B) Spa19 was crossed to the antisense strain Ap6, and the zeocin-resistant members of each tetrad were analyzed; three representative sets are shown here. The top panel is RT-PCR to detect ble-anti-Pnp cotranscripts, and the second panel is RT-PCR to detect Pnp sense transcripts. Pnp transcripts accumulate to very low levels even in wild-type cells; hence, the PCR product is not abundant. The third panel shows amplification of actin cDNA as a control for RNA amounts. The bottom two panels show PCR analysis to assess the presence or absence of the PS+ or PS− genome in the progeny. The primer sets used are indicated at the right. M, 1-kb ladder.
Zygotes were dissected on acetate-containing medium containing 2 μg/mL of zeocin. Zeocin was included to select for expression of the antisense RNA, which is cotranscribed with the ble gene. Expression of ble confers zeocin resistance (Stevens et al., 1996), and in the absence of constant selection, Chlamydomonas nuclear transgenes are often silenced by methylation or other mechanisms (Cerutti et al., 1997). Consequently, in the 51 tetrads that were obtained, only two of the four progeny survived (i.e., those that received the ble-pnp cassette), a supposition that was confirmed by PCR analysis (data not shown). When replica-plated on minimal medium lacking acetate but containing zeocin, we found that the surviving progeny were all photosynthetic, suggesting a correlation between antisense expression and a PS+ phenotype.
RT-PCR was used to confirm expression of the dicistronic ble-pnp antisense transcript (Figure 11B, top panel) and a reduction in the level of Pnp sense transcripts, with actin as a positive control (second and third panels). Subsequently, the presence or absence of PS+ and PS− genomes was assayed by PCR (fourth and fifth panels). No positive signal was obtained for the PS+ genome in any of the 24 zeocin-resistant progeny examined, whereas the PS− genome was clearly detected in all of them. This result is consistent with the interpretation that in the Ap6 background the PS+ genome is no longer required for photosynthesis, presumably because the reduced expression of PNPase results in the stability of otherwise unstable atpB mRNA.
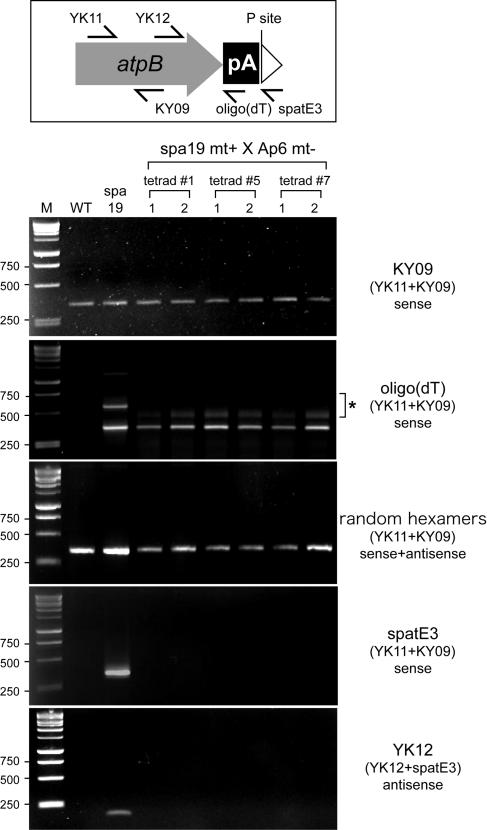
Next, the 3′ termini of atpB transcripts in the progeny were estimated by preparing cDNAs with three different reverse primers [KY09, oligo(dT), or spatE3; Figure 12, top]. KY09 was used as a positive control because it anneals to the atpB coding region and was expected to yield a product with RNA from wild-type cells. As shown in Figure 12, bottom, clear signals were obtained from the wild type, spa19, and all the progeny when the atpB mRNA coding region was amplified after cDNA synthesis directed by KY09. When an oligo(dT) primer was used for cDNA synthesis, no signal was obtained from wild-type cells, which is consistent with previous reports showing the exceedingly low abundance of polyadenylated transcripts in chloroplasts (Lisitsky et al., 1996; Komine et al., 2002). On the other hand, clear signals were obtained from spa19 and the spa19 × Ap6 progeny. Additional PCR products of unexpected sizes were also detected in this experiment (marked with an asterisk). We conclude that they are artifacts related to the oligo(dT) primer because they were not present when random hexamers were used for first-strand cDNA synthesis. Using primer spatE3, which anneals immediately downstream of the poly(A) tract, for cDNA synthesis, a product was obtained from spa19 (also see Figure 9B), but no product was obtained from the progeny. These results suggest that atpB transcripts accumulate with exposed poly(A) tails in chloroplasts containing the PS− genome but reduced levels of PNPase. Finally, the presence of atpB antisense transcripts in the progeny was assessed. cDNA synthesized by the forward primer (YK12) was amplified with the primer pair (YK12+spatE3). In this experiment, a positive signal was obtained from spa19 but not from the progeny. These data show that atpB antisense transcripts are lost in the absence of the PS+ genome. Taken together, this experiment supports the hypothesis that the PS+ genome generates antisense atpB RNA, which is required for the stabilization of sense atpB mRNA generated by the PS− genome, against degradation by a mechanism requiring PNPase.
Figure 12.
The 3′ End Mapping of atpB Transcripts in spa19/PNPase Antisense Cells.
Top, locations of primers used, with reference to the PS+ genome structure shown in Figure 2C. Bottom, atpB transcripts were analyzed in the wild type, spa19, and the zeocin-resistant progeny of (spa19 mt+ × Ap6 mt−). cDNAs were prepared using the primers shown in larger letters at the right. PCR was subsequently performed with the primer pairs shown in parentheses. When the oligo(dT) primer was used for cDNA synthesis, minor PCR products were obtained for spa19 and the progeny (asterisk). Although their molecular nature is unknown, these are probable artifacts because they were not detected when random hexamers were used for first-strand cDNA synthesis (center panel).
DISCUSSION
In this study, we have performed reversion analysis of a strain in which the chloroplast was engineered to generate an mRNA rendered unstable because of exposure of a poly(A) tail. The strain, Δ26pAtE, gave rise to spontaneous photosynthetic revertants at a remarkably high frequency, all of which turned out to have chloroplast mutations. Deliberately decreasing expression of the nuclear PNPase gene allowed photosynthesis in spite of the accumulation of polyadenylated atpB mRNA, demonstrating that nuclear factors play a role in the polyadenylation-mediated decay pathway. By intensively examining the chloroplast suppressors, we discovered unexpected features of RNA stabilization, including duplex formation between sense and antisense transcripts.
Polyadenylated mRNA Can Be Stabilized by Capping the 3′ UTR
Addition of poly(A) tails to 3′ ends has been shown to be an efficient signal for degradation of upstream mRNA in vitro and in vivo (Lisitsky et al., 1996; Komine et al., 2002). Our analysis of spa strains, however, revealed that these poly(A) signals can be masked when they are flanked by other sequences, as evidenced by the RNA processing alterations associated with renewed atpB transcript accumulation in certain spa strains (Figure 2). These capping sequences might inactivate the poly(A) signal in two ways. In most cases, the exposure of the A-tail creates a high-affinity site for binding of PNPase (Lisitsky et al., 1997a; Lisitsky and Schuster, 1999), and this exposure would be curtailed in the spa mutants. In some cases, however, RNA secondary structures in the capping sequences could contribute to protection against decay. The poly(A)-mediated degradation system is thought to be highly sensitive to RNA secondary structure because it has been shown that PNPase and RNase II, the two major 3′→5′ exoribonucleases in bacteria (Kushner, 2002), pause at stem-loop structures (Spickler and Mackie, 2000). This is also true for the chloroplast PNPase (Yehudai-Resheff et al., 2003) and is likely attributable to the narrow channel at its catalytic site (Symmons et al., 2000). At least in spa1, spa6, spa7, and spa8, the lack of processing between atpB and trnE1 yields a cotranscript that is likely to form a strong secondary structure because of the tRNA moiety. Our observations suggest that such a structure is largely impervious to exonucleases.
Polyadenylated mRNA Is Likely Stabilized by an Antisense Transcript in spa19/23
Two unusual chloroplast suppressors described in this report are spa19/23. Analysis of their RNA profiles (Figure 2A) as well as a comparison of their chloroplast genomes (Figure 3A) suggest that spa19 and spa23 are clonal mutants that arose from the division of a single mutant cell in liquid culture. Unlike the other spa strains, the atpB mRNA in spa19/23 did not hybridize with the downstream trnE1 sequence, suggesting it might terminate in a poly(A) tail. DNA gel blots (Figure 3), inverse PCR (Figure 4), and dot blot analysis (Figure 5) revealed an unusual organization of chloroplast genomes in spa19/23. In addition to possessing a chloroplast chromosome nearly identical to that of Δ26pAtE (the PS− genome), these heteroplasmic strains maintained a PS+ genome, which appeared to have been generated through recombination-mediated deletion. Because the PS+ genome always cosegregated with photosynthetic activity in genetic crosses (Figure 3), we focused on the function of the PS+ genome.
Upon close inspection of the PS+ genome, we found a fortuitous IR sequence downstream of atpB, so our initial hypothesis was that atpB mRNA transcribed from the PS+ genome would be stabilized by this IR (Figure 6). Indeed, RT-PCR showed that in at least some atpB transcripts the poly(A) tail was not exposed, but rather was flanked by a very short sequence (Figures 6 and 7). However, the 3′ end of atpB mRNA we detected was shorter than the end of this IR sequence and therefore was not predicted to form a 3′ end secondary structure. Because the primer spatE3 terminates in six G/C residues, this is probably the minimum overlap with the transcript. A maximum length would be defined by the transcript's inability to anneal to the downstream primer spatE2. Taken together, we estimated that 6 to 20 nucleotides flank the poly(A) tract in the accumulating spa19/23 atpB mRNA detected by RT-PCR (Figure 6B).
We also tried to detect this IR in atpB mRNA using S1 protection. The probes were generated from both the PS+ and PS− genomes, which, if the IR were present, would generate longer protected product(s) when the PS+ probe was used. However, such products were not observed (Figure 7C), and indeed, the sizes of the protected products were indistinguishable whether the PS+ or PS− probe was used (Figure 7D). At the resolution of this experiment, we cannot exclude that some atpB transcripts contain a few nucleotides derived from the PS+ genome; however, clearly no transcripts are long enough to contain the full IR and, thus, a 3′ stabilizing element. Thus, these observations disagreed with a classical model of IR-mediated stabilization and led us to an alternative and unexpected model presented in Figure 8. In this alternative model (Figure 8), the PS+ genome provides antisense transcripts for the atpB gene, driven by the tufA and/or trnE1 promoters, and these anneal to sense transcripts from the PS− genome. After trimming of the single-stranded 3′ regions by ribonucleases, the remaining dsRNA structure would stabilize the 3′ end and thus permit translation of the message. The presence of antisense atpB transcripts and dsRNA, shown by RT-PCR (Figure 9), both support the antisense hypothesis. However, we felt it prudent to obtain independent evidence for such an unexpected mechanism and therefore created chloroplast transformants that express artificial antisense transcripts. This experiment showed that the introduction of antisense transcripts could partially stabilize otherwise unstable atpB mRNA in the strain atpBΔ26 (Figure 10). Based on these data, we deduced that the stabilization of atpB mRNA in spa19/23 could most likely be explained by the antisense hypothesis. We emphasize that our data do not completely exclude the presence of atpB mRNA with the fortuitous stem-loop sequence, nor the stabilization of some atpB mRNA by another antisense-independent mechanism. However, it seems clear that sense–antisense RNA pairing can protect at least some cpRNA from nucleolytic attack.
Another perplexing finding is the apparent unidirectional (antisense) transcription of the PS+ genome atpB region. In principle, the sense transcripts could be generated from the PS+ as well as PS− genome because both contain the atpB-poly(A)-trnE1 (upstream) sequences present in the accumulating mRNA. However, if sense–antisense pairing occurred between transcripts both generated from the PS+ genome, we would expect the duplex—and, hence, the accumulating transcript—to extend considerably further downstream, perhaps as far as the 3′ end of tufA, where the incipient antisense transcript presumably would be processed as part of tufA 3′ end maturation. However, such a 3′ end was not observed using S1 protection analysis (Figure 7). And because we detected only one class of transcript in spa19/23 by RNA filter blot, it is likely that sense transcripts are generated only from the PS− genome and the antisense transcripts solely from the PS+ genome. The basis for this apparent exclusivity remains an open question. One possible explanation might be the predominance of tufA/trnE promoter activity over that of the atpB promoter. Transcriptional interference between convergent genes is well known (e.g., Spencer et al., 1986; Eszterhas et al., 2002; Prescott and Proudfoot, 2002), and in certain cases one gene can be dominantly expressed, as we have proposed here (Spencer et al., 1986).
On the other hand, the PS+ genome was invariably lost when spa19/23 were crossed to strains expressing nuclear antisense RNA to reduce PNPase expression (Figures 11 and 12). This indicates that the dsRNA structure imparts stability by impeding the action of PNPase and that the rapid degradation of atpB mRNA would be the selective pressure that resulted in at least some progeny from spa19/23 × CC-124 crosses retaining PS+ genomes, although the zygotes were not plated on minimal medium. It is also possible that the reduction in PNPase had indirect effects on cpDNA transmission, for example, by altering expression of some chloroplast genes. In any case, our results suggest that PNPase is the major 3′→5′ exoribonuclease that degrades polyadenylated mRNA in Chlamydomonas chloroplasts.
Natural Occurrence of Antisense RNA
Our results raise the question of whether antisense mRNA naturally occurs in chloroplasts and, if so, what its function might be. One possible example comes from the psbB operon (Westhoff et al., 1986), containing psbB, psbT, psbN, psbH, petB, and petD. Curiously, psbN is in opposite orientation to the other genes, a feature well conserved among chloroplast genomes (Westhoff et al., 1983; Ohyama et al., 1986; Shinozaki et al., 1986; Rock et al., 1987), including Chlamydomonas (Maul et al., 2002). Although the genes other than psbN are cotranscribed, the 51-kD chlorophyll a apoprotein, encoded by psbB, increases in response to light, whereas products from psbH, petB, and petD do not (Westhoff and Herrmann, 1988). Because psbB mRNA constitutes more than a dozen RNA species because of complex processing (Westhoff and Herrmann, 1988), whether this light response is regulated at a transcriptional or translational level is still controversial. Interestingly, Kohchi et al. (1988) found that psbN was also light inducible and speculated that antisense transcription from the psbN promoter might have some role in the light responsiveness of psbB. Their hypothesis was that the psbB-psbN duplex might terminate transcription downstream of psbN, thus attenuating the light response of psbH-petB/D. We would argue that this is unlikely because RNA secondary structure has been repeatedly shown not to terminate transcription in chloroplasts (Rott et al., 1996). Instead, this phenomenon could be reasonably explained if psbN mRNA, acting as an antisense transcript, stabilized psbB mRNA with respect to psbH-petB/D. Whether this occurs in vivo remains to be determined.
Apart from the special case of psbN, antisense transcripts do not appear to accumulate generally in chloroplasts. Although many such RNAs may be synthesized as a result of the compact nature of chloroplast genomes, because of the lack of efficient termination, they are likely to be unstable. For example, Chlamydomonas chloroplasts have a highly efficient mechanism for degrading atpB mRNA downstream of its eventual maturation site (Hicks et al., 2002), a phenomenon that might in fact help the chloroplast avoid the accumulation of dsRNA, whose stability might be difficult to control. Furthermore, the Chlamydomonas chloroplast genome is atypically asymmetric (i.e., although genes are encoded on both strands, they are mostly in blocks that are likely to be independently transcribed). Although this is also true for Euglena cpDNA (Hallick et al., 1993), most sequenced chloroplast genomes are not asymmetric and thus would be subject to antisense RNA synthesis.
Because dsRNA-directed gene silencing has been receiving great attention (Cerutti, 2003), it may seem counterintuitive that dsRNA would enhance stability in organelles. But considering that dsRNA-based gene silencing is likely to have evolved as an adaptive defense against viruses and transposable elements in the cytoplasm (Waterhouse et al., 2001), it is not surprising that organelles do not possess this feature. Furthermore, both chloroplasts and mitochondria have long been known to harbor apparently nonpathogenic dsRNA plasmid-like elements (Finnegan and Brown, 1986; Ishihara et al., 1992; Koga et al., 2003). Finally, antisense RNA has long been known to have regulatory functions in E. coli (Xu et al., 1993; Gubbins et al., 2003; Masse et al., 2003), although, intriguingly, it can both negatively and positively regulate the complementary species. Whether chloroplasts possess small regulatory RNAs—of either the sense or antisense variety—remains a pertinent question given the proliferation of such discoveries in both prokaryotes and eukaryotes (reviewed in Gottesman, 2002).
Dynamic Alteration of cpDNA in spa Strains
All the spa strains that we isolated were chloroplast mutants, which featured a mixture of point mutations, deletions, and rearrangements. Chloroplast-based suppression of nonphotosynthetic phenotypes is well known in Chlamydomonas. Examples include point mutations that overcome translational incompetence in the psbC 5′ UTR (Rochaix et al., 1989), a large intragenomic inversion that restored expression of a petD gene with a 5′ UTR deletion (Higgs et al., 1998) and an amber suppressor tRNA that allowed expression of an rbcL mRNA possessing a stop codon (Yu and Spreitzer, 1992). In this context, the suppression we observed in all but spa19/23 was consistent with the known mutability of cpDNA, under conditions where expression of other essential genes remains sufficient.
The spa19/23 strains, however, are novel. Stable heteroplasmicity is unknown in chloroplasts except under conditions where both genomes are being selected (Goldschmidt-Clermont, 1991; Shikanai et al., 2001), but even after more than 1 year of culture on nonselective acetate-containing medium, spa19/23 have maintained their bipartite character. In fact, the DNA filter blots in Figure 2B reveal a few additional faintly hybridizing bands, which may signify ongoing evolution of the genomes. A subtle selective pressure might be exerted by the fact that chloroplast ATPase mutants are sensitive to high light (Majeran et al., 2001), although we have maintained these strains under the same conditions that are not deleterious to the well-characterized atpB mutant CC-373 (Shepherd et al., 1979).
Our tentative scenario for the spa19/23 suppression event initiates with loss of the aadA cassette, generating the spa19/23 PS− genome, which is nearly identical to the Δ26pAtE chloroplast genome. The PS− genome would continue to generate atpB transcripts with the engineered poly(A) sequence, which would be rapidly degraded by 3′→5′ exoribonucleases such as PNPase. In a second step, the PS+ genome would be generated by a nearly 145-kb deletion apparently mediated by recombination. Although this event involves two identical copies of trnE1, the copies are on opposite strands and thus represent IRs, which in chloroplasts frequently mediate flip-flop recombination (Palmer, 1983). Because the PS+ genome was apparently created through deletion, this should involve direct repeats, which are also known to recombine actively in chloroplasts (Fischer et al., 1996). The best candidate for a role in homologous recombination is an imperfect 10-bp repeat at the deletion boundary (GTCTAGGCCT…GTCcTAGagGCCT).
Heteroplasmicity in spa19/23 has two other features of interest. The first is the imbalance between the PS− and PS+ genomes, with the latter being in threefold to fourfold excess in vegetative cells. The closest precedent for an overabundant genome type in Chlamydomonas also resulted from suppression of a photosynthetic growth phenotype. Strain Δ26, which lacks the atpB 3′ UTR stem-loop structure, grows slowly on minimal medium because of the instability of atpB mRNA. From this strain, robustly photosynthetic clones were isolated, many of which contained amplified copies of atpB DNA sequences, most probably as episomal elements (Kindle et al., 1994; Suzuki et al., 1997). Amplification of the PS+ genome could also be a consequence of its presumably reduced size relative to the PS− genome (203 kb versus 58 kb). In other words, the PS+ genome may be more rapidly replicated, contributing to its ability to suppress poly(A)-mediated mRNA instability. A possible parallel would be yeast petite strains, where large deletions in mitochondrial DNA are accompanied by amplification of the remaining sequences (Van Dyck and Clayton, 1998). Another possibility is that the PS+ genome fails to interact with nuclear gene products regulating genome dynamics. Recently, CHM, a nuclear gene that regulates substoichiometric shifting of mitochondrial genomes, was isolated from Arabidopsis thaliana (Abdelnoor et al., 2003). CHM encodes a protein related to MutS, which is involved in mismatch repair and DNA recombination. Perhaps in spa19/23 altered expression of nuclear genes encoding recombinases, DNA helicases, or DNA gyrases may have facilitated the acquisition of the suppression phenotype.
The second feature is the unusually poor transmission of the PS+ genome in genetic crosses, despite its high copy number. Presently, we do not have clear mechanistic insights into this phenomenon. However, it is known that a DNA replication step during zygote germination is required for normal inheritance of Chlamydomonas cpDNA (Umen and Goodenough, 2001). We are currently investigating whether the PS+ genome is poorly replicated at this stage and, thus, poorly transmitted.
METHODS
Selection and Genetic Analysis of Suppressors
The progenitor strain for the suppressor screen was Δ26pAtE mt+ (Komine et al., 2002). Cells were grown under constant light in TAP medium (Harris, 1989) and harvested at mid-log phase (1 to 2 × 106 cells/mL). Cells (2 × 108) were plated on minimal medium (lacking a reduced carbon source) to select for photosynthetic growth. Resulting colonies were screened by PCR with primers YK03 and B-M-3′-tE (Table 2) to confirm the presence of the atpB-trnE1 fusion in the chloroplast genome. Because the original Δ26pAtE strain was lost as a result of reversion before a rate of reversion could be calculated, the 10−5 rate reported in Results was derived from a PS− progeny of a cross of spa19 × CC-124, which differs from Δ26pAtE by its loss of the aadA cassette.
Table 2.
Oligonucleotide Primers
| Designation | Sequence (5′ to 3′) | Locationa | Directionb |
|---|---|---|---|
| atpB_F1 | GAAACAAAAAACATGGGACGTA | atpB coding | F |
| YK11 | CCTTACTGACCCTGCTCCTG | atpB coding | F |
| KY09 | GGTGAACCTGTAAATACTTC | atpB coding | R |
| YK03 | CAAGCATTCTACTTAGTAGG | atpB coding | F |
| YK14 | ATTGCTATTCTAGGTTTAGATG | atpB coding | F |
| T7-YK14 | CAGAGATGCATAATACGACTCACTATAGGGAGAATTGCTATTCTA | T7 promoter + atpB coding | F |
| sphI_YK12 | AAGCATGCGCTGAAACTATTGAA | SphI site + atpB coding | F |
| YK12 | GCTGAAACTATTGAA | atpB coding | F |
| atpB_R1 | TTCCATACCACGCATTAAACC | atpB coding | R |
| sphI_atpB3′_R | AAGCATGCATTTTAATGAAGCAG | SphI site + atpB coding | R |
| polyT18-P1 | CGACCAAGCTTGGGATCCCGTTTTTTTTTTTTTTTTTT | Poly(A) sequence | R |
| polyT3-P2 | CGACCAAGCTTGGGATCCCGTTT | 5′ of polyT18-P1 | R |
| spatE5 | ATTTAATGAAGCAG | atpB coding | R |
| spatE4 | TTTTTTTTTTTTTTTTTTTTTTTTTTTTGGATCTG | atpB 3′ end | R |
| spatE3 | GAGGCCTAGACGATGGGGGC | trnE1 5′ end | R |
| spatE2 | CCCCATCATCTAGAGGCCTA | trnE1 5′ end | F (R in spa19/23) |
| spatE | GTAAGGGGATGCCGGAG | trnE1 5′ end | F (R in spa19/23) |
| B-M3′tE | AGATCTGAATTGAGATTCTCTGACACTGTC | trnE1 3′ end | R |
| spatEGC | CTCCGGCATCCCCTTAC | trnE1 5′ end | R |
| atpX 3′ | GGACATTTTCACTTCTGGAGTG | atpA promoter | R |
| aadA 3′ | TTATTTGCCAACTACCTTAGTGAT | aadA coding | R |
| pet_F | AAACCTGATTTAAGCGATCCAGT | petD coding | F |
| pet_R | AGGGAATGTTGAACCAATACCTAAC | petD coding | R |
| MT+F | ATGCCTATCTTTCTCATT | Fus1 coding | F |
| MT+R | GCAAAATACACGTCTGGAAG | Fus1 coding | R |
| MT-F | ATGGCCTGTTTCTTAGC | Mid coding | F |
| MT-R | CTACATGTGTTTCTTGAC | Mid coding | R |
| BLE F2 | GTATCGATGGCCAAGCTGACCAGCGCCG | ble coding | F |
| BLE R2 | TTGGTCGACGTCGGTTAGTCC | ble coding | R |
| aadA1 | CGACTCAACTATCAGAGG | aadA coding | F |
| aadA2 | CGTAGTGGACAAATTCTTCC | aadA coding | R |
| PNP F | CCGGAGGCAAGGAGATCAAG | Pnp coding | F (R in antisense lines) |
| PNP R | TCGCACAGAGAGGCGATGGC | Pnp coding | R |
| CrActin_F | AATCGTGCGCGACATCAAGGAGAA | Actin coding | F |
| CrActin_R | TTGGCGATCCACATTTGCTGGAAGGT | Actin coding | R |
| TufA_F | TATCCGAATGTTAATGCTTAGT | tufA coding | F |
| TufA_R | ATTTTATACCCAAACTGTAATAGTTAT | tufA coding | R |
| ClpP_F | CTAACTATTACTTTCTTGCGTTTGT | clpP coding | F |
| ClpP_R | TAACTCATTTGGCGGTTCTG | clpP coding | R |
| PsbN_F | CAGGGCAAAATTTTGAGTAGATTA | psbN coding | F |
| PsbN_R | TGAAGGTGGACCAAAACTAACATA | psbN coding | R |
| PsbB_F | AACCACGTAAGGCGACCAC | psbB coding | F |
| PsbB_R | TATGGGTTTACCTTGGTATCGTGT | psbB coding | R |
| RbcL_F | TGAGTGAAGTAAATACCACGGCTACGG | rbcL coding | F |
| RbcL_R | GTACATGGACTACAGTATGGACTGACGGT | rbcL coding | R |
| 28S_F | CGAAAGATGGTGAACTATGCCTGAA | 28S rRNA | F |
| 28S_R | TTGCCGACTTCCCTTATCTACTACATTGTT | 28S rRNA | R |
Fus1 is a nuclear gene within the mt+ locus; Mid is a nuclear gene specific to mt− cells.
Direction specifies the orientation of the primer with respect to the gene it amplifies. F indicates the same orientation as the gene; R indicates the antisense orientation.
The wild-type strain used in backcrosses was CC-124 mt−. Mating type plus suppressor strains were crossed to CC-124 according to Harris (1989). Tetrad progeny were analyzed for photosynthetic growth on minimal medium. PCR with the mating type–specific primers MT+F, MT+R, MT-F, and MT-R (Table 2) confirmed Mendelian segregation of the nuclear mating-type genes (Werner and Dieter, 1998).
DNA/RNA/Protein Analysis
For DNA gel blot analysis, total DNA was as previously described (Rochaix, 1980). Digested DNA was resolved in 0.7% agarose gels and transferred to GeneScreen nylon filters (NEN Life Science Research Products, Boston, MA) by capillary action. All probes were labeled either by random priming or linear PCR amplification of the antisense strand in the presence of [α-32P]dCTP. DNA gel blots were hybridized with radiolabeled probes as described previously (Church and Gilbert, 1984), and visualization of radioactive bands was achieved with a Storm scanner (Molecular Dynamics, Sunnyvale, CA). For RNA gel blot analysis, total RNA was isolated using TRI reagent (Molecular Research Center, Cincinnati, OH). Five micrograms of RNA was resolved in 1% agarose/6% formaldehyde gels in Mops buffer, transferred to GeneScreen nylon filters by vacuum blotting, and hybridized and visualized in the same manner as DNA. S1 nuclease protection was conducted as previously described (Stern and Kindle, 1993). In vitro transcription was performed according to Stern and Gruissem (1987). PCR was used to prepare the templates and to add the T7 promoter sequence (5′-TAATACGACTCACTATAGGGAGA-3′).
For inverse PCR, total DNA was isolated from spa19, and a procedure adapted from Ochman et al. (1990) was used. Total DNA (2.5 μg) was digested with PstI, and 0.5 μg of this DNA was self-ligated overnight in a 40-μL reaction. Ligations were ethanol precipitated and resuspended in 20 μL of water, of which 2 μL were used in a 50-μL PCR reaction with YK12 and KY09, which are atpB gene–specific inverse primers. PCR products were cloned using the pGEM-T Easy vector (Promega, Madison, WI) and used as templates for DNA sequencing.
For DNA dot blot analysis, PCR products for atpB, petD, tufA, clpP, psbN, psbB, rbcL, and 28S rDNA were prepared using primer sets shown in Table 2. PCR products (64, 32, 16, 8, and 4 ng) were blotted on GeneScreen nylon filters using the Bio-Dot microfiltration apparatus (Bio-Rad, Hercules, CA). The DNA filter blots were hybridized with radiolabeled total DNA extracted from wild-type or spa19 cells, which were labeled by random priming. The radioactive dots were visualized using the Storm scanner and quantified using ImageQuant software (Amersham Pharmacia Biotech, Uppsala, Sweden). Microsoft Excel (version X) was used to calculate a least-squares fit line and slope for the data shown in Figure 5C and to generate an exponential curve using the migration of size markers in Figure 7; the derived equation was used to calculate the size of the protected products.
Total protein was isolated as previously described (Komine et al., 2002). Proteins were resolved in 12% SDS-PAGE gels and transferred to polyvinylidene difluoride filters with a Trans-Blot semi-dry electrophoretic transfer cell (Bio-Rad). Blots were incubated with antibodies raised against the α-subunit of the ATP synthase (generously provided by Francis-André Wollman) and detected with the ECL system (Amersham Pharmacia Biotech).
RT-PCR
For RT-PCR, 1 μg of total RNA, extracted as described above, was treated with 1 unit of RQI RNase-Free DNase I (Promega) for 40 min at 37° in a total volume of 50 μL. The DNase was inactivated by heating to 65° for 10 min, and the sample was placed on ice. Then, 10-μL aliquots were used for reverse transcription (final volume 20 μL) with Superscript III (Invitrogen, Carlsbad, CA) at 55° for 45 min. The reverse transcriptase was inactivated by boiling for 10 min, and PCR was performed with 2 μL of RT reaction products for 25 cycles (94, 55, and 72°C). PCR products were resolved in 2% agarose gels, and their sizes were estimated by comparison with a 1-kb DNA ladder (Promega). We used random hexamers or an oligo(dT) primer for reverse transcriptase reactions, but for strand-specific RT-PCR, only the forward or reverse gene-specific primer was used. In some experiments, 1 μg of total RNA was directly used for single tube RT-PCR with the Access RT-PCR system (Promega). The reaction mixture (50 μL) was constituted according to the manufacturer's instructions with 10 pmol of the forward and reverse gene-specific primers. The first step was reverse transcription at 45°C for 45 min, followed by 25 PCR cycles as above.
Sequencing was performed to ensure that PCR products were derived from the targeted genes. DNA fragments were excised from gels, purified using QiaQuick resin (Qiagen, Valencia, CA), and cloned using the TOPO-TA cloning vector (Invitrogen). DNA from positive colonies was end sequenced using vector primers.
Plasmid Construction and Chloroplast Transformation
The construct shown in Figure 10 is based on pCDXHc aadA(+) (Higgs et al., 1999), which contains petD and the aadA cassette in the same orientation. A 121-bp sequence from the 3′ part of the atpB coding region was amplified by PCR with primers sphI_YK12 and sphI_atpB3′_R and inserted into pGEM-T (Promega). After confirming the sequence, the atpB fragment was excised using the SphI sites added during PCR and inserted into the SphI site between the aadA coding region and the rbcL 3′ UTR in pCDXHc aadA(+). This construct was used for particle bombardment into the strain atpBΔ26 (Stern et al., 1991).
Acknowledgments
We gratefully acknowledge Shinya Murakami and Gadi Schuster for thoughtful insights into this work, which was supported by Japan Society for the Promotion of Science, Francis Goelet postdoctoral fellowships to Y.N., and awards from the Binational Agricultural Research and Development Fund (US-3177-99C) and the National Science Foundation (MCB 0091020) to D.B.S, including a National Science Foundation Research Experience for Undergraduates award to E.A.K.
The authors responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: Yoshiki Nishimura (yn37@cornell.edu) and David B. Stern (ds28@cornell.edu).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026203.
References
- Abdelnoor, R.V., Yule, R., Elo, A., Christensen, A.C., Meyer-Gauen, G., and Mackenzie, S.A. (2003). Substoichiometric shifting in the plant mitochondrial genome is influenced by a gene homologous to MutS. Proc. Natl. Acad. Sci. USA 100, 5968–5973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anthonisen, I.L., Salvador, M.L., and Klein, U. (2001). Specific sequence elements in the 5′ untranslated regions of rbcL and atpB gene mRNAs stabilize transcripts in the chloroplast of Chlamydomonas reinhardtii. RNA 7, 1024–1033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bollenbach, T.J., and Stern, D.B. (2003. a). Secondary structures common to chloroplast mRNA 3′-untranslated regions direct cleavage by CSP41, an endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. J. Biol. Chem. 278, 25832–25838. [DOI] [PubMed] [Google Scholar]
- Bollenbach, T.J., and Stern, D.B. (2003. b). Divalent metal-dependent catalysis and cleavage specificity of CSP41, a chloroplast endoribonuclease belonging to the short chain dehydrogenase/reductase superfamily. Nucleic Acids Res. 31, 4317–4325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudreau, E., Nickelsen, J., Lemaire, S.D., Ossenbuhl, F., and Rochaix, J.-D. (2000). The Nac2 gene of Chlamydomonas reinhardtii encodes a chloroplast TPR protein involved in psbD mRNA stability, processing and/or translation. EMBO J. 19, 3366–3376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerutti, H. (2003). RNA interference: Traveling in the cell and gaining functions? Trends Genet. 19, 39–46. [DOI] [PubMed] [Google Scholar]
- Cerutti, H., Johnson, A.M., Gillham, N.W., and Boynton, J.E. (1997). Epigenetic silencing of a foreign gene in nuclear transformants of Chlamydomonas. Plant Cell 9, 925–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church, G., and Gilbert, W. (1984). Genomic sequencing. Proc. Natl. Acad. Sci. USA 81, 1991–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng, X.W., and Gruissem, W. (1987). Control of plastid gene expression during development: The limited role of transcriptional regulation. Cell 49, 379–387. [DOI] [PubMed] [Google Scholar]
- Donovan, W.P., and Kushner, S.R. (1986). Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc. Natl. Acad. Sci. USA 83, 120–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drapier, D., Girard-Bascou, J., and Wollman, F.-A. (1992). Evidence for nuclear control of the expression of the atpA and atpB chloroplast genes in Chlamydomonas. Plant Cell 4, 283–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eszterhas, S.K., Bouhassira, E.E., Martin, D.I., and Fiering, S. (2002). Transcriptional interference by independently regulated genes occurs in any relative arrangement of the genes and is influenced by chromosomal integration position. Mol. Cell. Biol. 22, 469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finnegan, P.M., and Brown, G.G. (1986). Autonomously replicating RNA in mitochondria of maize plants with S-type cytoplasm. Proc. Natl. Acad. Sci. USA 83, 5175–5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer, N., Stampacchia, O., Redding, K., and Rochaix, J.D. (1996). Selectable marker recycling in the chloroplast. Mol. Gen. Genet. 251, 373–380. [DOI] [PubMed] [Google Scholar]
- Goldschmidt-Clermont, M. (1991). Transgenic expression of aminoglycoside adenine transferase in the chloroplast: A selectable marker for site-directed transformation of Chlamydomonas. Nucleic Acids Res. 19, 4083–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gottesman, S. (2002). Stealth regulation: Biological circuits with small RNA switches. Genes Dev. 16, 2829–2842. [DOI] [PubMed] [Google Scholar]
- Gray, M.W. (1992). The endosymbiont hypothesis revisited. Int. Rev. Cytol. 141, 233–357. [DOI] [PubMed] [Google Scholar]
- Grossman, A.R., Harris, E., Hauser, C., Lefebvre, P., Martinez, D., Rokhsar, D., Shrager, J., Silflow, C., Stern, D., Vallon, O., and Zhang, Z. (2003). Chlamydomonas reinhardtii at the crossroads of genomics. Eukaryot. Cell 2, 1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbins, M.J., Arthur, D.C., Ghetu, A.F., Glover, J.N., and Frost, L.S. (2003). Characterizing the structural features of RNA/RNA interactions of the F-plasmid FinOP fertility inhibition system. J. Biol. Chem. 278, 27663–27671. [DOI] [PubMed] [Google Scholar]
- Hallick, R.B., Hong, L., Drager, R.G., Favreau, M.R., Monfort, A., Orsat, B., Spielmann, A., and Stutz, E. (1993). Complete DNA sequence of Euglena gracilis chloroplast DNA. Nucleic Acids Res. 21, 3537–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, E.H. (1989). The Chlamydomonas Sourcebook: A Comprehensive Guide to Biology and Laboratory Use. (San Diego: Academic Press). [DOI] [PubMed]
- Hayes, R., Kudla, J., and Gruissem, W. (1999). Degrading chloroplast mRNA: The role of polyadenylation. Trends Biochem. Sci. 24, 199–202. [DOI] [PubMed] [Google Scholar]
- Hicks, A., Drager, R.G., Higgs, D.C., and Stern, D.B. (2002). An mRNA 3′ processing site targets downstream sequences for rapid degradation in Chlamydomonas chloroplasts. J. Biol. Chem. 277, 3325–3333. [DOI] [PubMed] [Google Scholar]
- Higgs, D.C., Kuras, R., Kindle, K.L., Wollman, F.A., and Stern, D.B. (1998). Inversions in the Chlamydomonas chloroplast genome suppress a petD 5′ untranslated region deletion by creating functional chimeric mRNAs. Plant J. 14, 663–671. [DOI] [PubMed] [Google Scholar]
- Higgs, D.C., Shapiro, R.S., Kindle, K.L., and Stern, D.B. (1999). Small cis-acting sequences that specify secondary structures in a chloroplast mRNA are essential for RNA stability and translation. Mol. Cell. Biol. 19, 8479–8491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang, S., Kawazoe, R., and Herrin, D.L. (1996). Transcription of tufA and other chloroplast-encoded genes is controlled by a circadian clock in Chlamydomonas. Proc. Natl. Acad. Sci. USA 93, 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara, J., Pak, J.Y., Fukuhara, T., and Nitta, T. (1992). Association of particles that contain double-stranded RNAs with algal chloroplasts and mitochondria. Planta 187, 475–482. [DOI] [PubMed] [Google Scholar]
- Kindle, K.L., Suzuki, H., and Stern, D.B. (1994). Gene amplification can correct a photosynthetic growth defect caused by mRNA instability in Chlamydomonas chloroplasts. Plant Cell 6, 187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klaff, P. (1995). mRNA decay in spinach chloroplasts: psbA mRNA degradation is initiated by endonucleolytic cleavages within the coding region. Nucleic Acids Res. 23, 4885–4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga, R., Horiuchi, H., and Fukuhara, T. (2003). Double-stranded RNA replicons associated with chloroplasts of a green alga, Bryopsis cinicola. Plant Mol. Biol. 51, 991–999. [DOI] [PubMed] [Google Scholar]
- Kohchi, T., Yoshida, T., Komano, T., and Ohyama, K. (1988). Divergent messenger RNA transcription in the chloroplast psbB operon. EMBO J. 7, 885–892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine, Y., Kikis, E., Schuster, G., and Stern, D. (2002). Evidence for in vivo modulation of chloroplast RNA stability by 3′-UTR homopolymeric tails in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 99, 4085–4090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komine, Y., Kwong, L., Anguera, M.C., Schuster, G., and Stern, D.B. (2000). Polyadenylation of three classes of chloroplast RNA in Chlamydomonas reinhardtii. RNA 6, 598–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudla, J., Hayes, R., and Gruissem, W. (1996). Polyadenylation accelerates degradation of chloroplast mRNA. EMBO J. 15, 7137–7146. [PMC free article] [PubMed] [Google Scholar]
- Kushner, S.R. (2002). mRNA decay in Escherichia coli comes of age. J. Bacteriol. 184, 4658–4665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy, H., Kindle, K.L., and Stern, D.B. (1997). A nuclear mutation that affects the 3′ processing of several mRNAs in Chlamydomonas chloroplasts. Plant Cell 9, 825–836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky, I., Klaff, P., and Schuster, G. (1996). Addition of poly(A)-rich sequences to endonucleolytic cleavage sites in the degradation of spinach chloroplast mRNA. Proc. Natl. Acad. Sci. USA 93, 13398–13403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lisitsky, I., Klaff, P., and Schuster, G. (1997. b). Blocking polyadenylation of mRNA in the chloroplast inhibits its degradation. Plant J. 12, 1173–1178. [Google Scholar]
- Lisitsky, I., Kotler, A., and Schuster, G. (1997. a). The mechanism of preferential degradation of polyadenylated RNA in the chloroplast: The exoribonuclease 100RNP-polynucleotide phosphorylase displays high binding affinity for poly(A) sequences. J. Biol. Chem. 272, 17648–17653. [DOI] [PubMed] [Google Scholar]
- Lisitsky, I., and Schuster, G. (1999). Preferential degradation of polyadenylated and polyuridinylated RNAs by the bacterial exoribonuclease polynucleotide phosphorylase. Eur. J. Biochem. 261, 468–474.10215858 [Google Scholar]
- Mackenzie, S., and McIntosh, L. (1999). Higher plant mitochondria. Plant Cell 11, 571–586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majeran, W., Olive, J., Drapier, D., Vallon, O., and Wollman, F.A. (2001). The light sensitivity of ATP synthase mutants of Chlamydomonas reinhardtii. Plant Physiol. 126, 421–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse, E., Escorcia, F., and Gottesman, S. (2003). Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17, 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maul, J.E., Lilly, J.W., Cui, L., dePamphilis, C.W., Miller, W., Harris, E.H., and Stern, D.B. (2002). The Chlamydomonas reinhardtii plastid chromosome: Islands of genes in a sea of repeats. Plant Cell 14, 2659–2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monde, R.A., Schuster, G., and Stern, D.B. (2000). Processing and degradation of chloroplast mRNA. Biochimie 82, 573–582. [DOI] [PubMed] [Google Scholar]
- Mullet, J.E., and Klein, R.R. (1987). Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 6, 1571–1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., Ajioka, J.W., Garza, D., and Hartl, D.L. (1990). Inverse polymerase chain reaction. Biotechnology 8, 759–760. [DOI] [PubMed] [Google Scholar]
- Ohyama, K., et al. (1986). Chloroplast gene organization deduced from complete sequence of liverwort (Marchantia polymorpha) chloroplast DNA. Nature 322, 572–574. [Google Scholar]
- Palmer, J.D. (1983). Chloroplast DNA exists in 2 orientations. Nature 301, 92–93. [Google Scholar]
- Prescott, E.M., and Proudfoot, N.J. (2002). Transcriptional collision between convergent genes in budding yeast. Proc. Natl. Acad. Sci. USA 99, 8796–8801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rochaix, J.-D. (1980). Restriction fragments from Chlamydomonas chloroplast DNA. Methods Enzymol. 65, 785–795. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.-D. (1996). Post-transcriptional regulation of chloroplast gene expression in Chlamydomonas. Plant Mol. Biol. 32, 327–341. [DOI] [PubMed] [Google Scholar]
- Rochaix, J.-D., Kuchka, M., Mayfield, S., Schirmer-Rahire, M., Girard-Bascou, J., and Bennoun, P. (1989). Nuclear and chloroplast mutations affect the synthesis or stability of the chloroplast psbC gene product in Chlamydomonas reinhardtii. EMBO J. 8, 1013–1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rock, C.D., Barkan, A., and Taylor, W.C. (1987). The maize plastid psbB-psbF-petB-petD gene cluster: Spliced and unspliced petB and petD RNAs encode alternative products. Curr. Genet. 12, 69–77. [DOI] [PubMed] [Google Scholar]
- Rott, R., Drager, R.G., Stern, D.B., and Schuster, G. (1996). The 3′ untranslated regions of chloroplast genes in Chlamydomonas reinhardtii do not serve as efficient transcriptional terminators. Mol. Gen. Genet. 252, 676–683. [DOI] [PubMed] [Google Scholar]
- Schuster, G., Lisitsky, I., and Klaff, P. (1999). Polyadenylation and degradation of mRNA in the chloroplast. Plant Physiol. 120, 937–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd, H.S., Boynton, J.E., and Gillham, N.W. (1979). Mutations in nine chloroplast loci of Chlamydomonas affecting photosynthetic functions. Proc. Natl. Acad. Sci. USA 76, 1353–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shikanai, T., Shimizu, K., Ueda, K., Nishimura, Y., Kuroiwa, T., and Hashimoto, T. (2001). The chloroplast clpP gene, encoding a proteolytic subunit of ATP-dependent protease, is indispensable for chloroplast development in tobacco. Plant Cell Physiol. 42, 264–273. [DOI] [PubMed] [Google Scholar]
- Shinozaki, K., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: Its gene organization and expression. EMBO J. 5, 2043–2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer, C.A., Gietz, R.D., and Hodgetts, R.B. (1986). Overlapping transcription units in the dopa decarboxylase region of Drosophila. Nature 322, 279–281. [DOI] [PubMed] [Google Scholar]
- Spickler, C., and Mackie, G.A. (2000). Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 182, 2422–2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D.B., and Gruissem, W. (1987). Control of plastid gene expression: 3′ inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell 51, 1145–1157. [DOI] [PubMed] [Google Scholar]
- Stern, D.B., and Kindle, K.L. (1993). 3′ end maturation of the Chlamydomonas reinhardtii chloroplast atpB mRNA is a two-step process. Mol. Cell. Biol. 13, 2277–2285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern, D.B., Radwanski, E.R., and Kindle, K.L. (1991). A 3′ stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell 3, 285–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevens, D.R., Rochaix, J.D., and Purton, S. (1996). The bacterial phleomycin resistance gene ble as a dominant selectable marker in Chlamydomonas. Mol. Gen. Genet. 251, 23–30. [DOI] [PubMed] [Google Scholar]
- Sugita, M., and Sugiura, M. (1996). Regulation of gene expression in chloroplasts of higher plants. Plant Mol. Biol. 32, 315–326. [DOI] [PubMed] [Google Scholar]
- Suzuki, H., Ingersoll, J., Stern, D.B., and Kindle, K.L. (1997). Generation and maintenance of tandemly repeated extrachromosomal plasmid DNA in Chlamydomonas chloroplasts. Plant J. 11, 635–648. [DOI] [PubMed] [Google Scholar]
- Symmons, M.F., Jones, G.H., and Luisi, B.F. (2000). A duplicated fold is the structural basis for polynucleotide phosphorylase catalytic activity, processivity, and regulation. Structure Fold. Des. 8, 1215–1226. [DOI] [PubMed] [Google Scholar]
- Umen, J.G., and Goodenough, U.W. (2001). Chloroplast DNA methylation and inheritance in Chlamydomonas. Genes Dev. 15, 2585–2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Boudreau, E., Lemaire, S.D., Goldschmidt-Clermont, M., and Rochaix, J.D. (2000. b). Characterization of Mbb1, a nucleus-encoded tetratricopeptide-like repeat protein required for expression of the chloroplast psbB/psbT/psbH gene cluster in Chlamydomonas reinhardtii. Proc. Natl. Acad. Sci. USA 97, 14813–14818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaistij, F.E., Goldschmidt-Clermont, M., Wostrikoff, K., and Rochaix, J.D. (2000. a). Stability determinants in the chloroplast psbB/T/H mRNAs of Chlamydomonas reinhardtii. Plant J. 21, 469–482. [DOI] [PubMed] [Google Scholar]
- Van Dyck, E., and Clayton, D.A. (1998). Transcription-dependent DNA transactions in the mitochondrial genome of a yeast hypersuppressive petite mutant. Mol. Cell. Biol. 18, 2976–2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter, M., Kilian, J., and Kudla, J. (2002). PNPase activity determines the efficiency of mRNA 3′-end processing, the degradation of tRNA and the extent of polyadenylation in chloroplasts. EMBO J. 21, 6905–6914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse, P.M., Wang, M.B., and Lough, T. (2001). Gene silencing as an adaptive defence against viruses. Nature 411, 834–842. [DOI] [PubMed] [Google Scholar]
- Werner, R., and Dieter, M. (1998). Mating type determination of Chlamydomonas reinhardtii by PCR. Plant Mol. Biol. Rep. 16, 295–299. [Google Scholar]
- Westhoff, P., Alt, J., and Herrmann, R.G. (1983). Localization of the genes for the 2 Chlorophyll-a-conjugated polypeptides (Mol Wt 51 and 44 KD) of the photosystem-II reaction center on the spinach plastid chromosome. EMBO J. 2, 2229–2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westhoff, P., Farchaus, J.W., and Herrmann, R.C. (1986). The gene for the Mr 10,000 phosphoprotein associated with photosystem II is part of the psbB operon of the spinach plastid chromosome. Curr. Genet. 11, 165–169. [DOI] [PubMed] [Google Scholar]
- Westhoff, P., and Herrmann, R.G. (1988). Complex RNA maturation in chloroplasts: The psbB operon from spinach. Eur. J. Biochem. 171, 551–564. [DOI] [PubMed] [Google Scholar]
- Xu, F., Lin Chao, S., and Cohen, S.N. (1993). The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc. Natl. Acad. Sci. USA 90, 6756–6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Hirsh, M., and Schuster, G. (2001). Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell. Biol. 21, 5408–5416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yehudai-Resheff, S., Portnoy, V., Yogev, S., Adir, N., and Schuster, G. (2003). Domain analysis of the chloroplast polynucleotide phosphorylase reveals discrete functions in RNA degradation, polyadenylation, and sequence homology with exosome proteins. Plant Cell 15, 2003–2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, W., and Spreitzer, R.J. (1992). Chloroplast heteroplasmicity is stabilized by an amber-suppressor tryptophan tRNA (CUA). Proc. Natl. Acad. Sci. USA 89, 3904–3907. [DOI] [PMC free article] [PubMed] [Google Scholar]