Abstract
Resistance Gene Candidate2 (RGC2) genes belong to a large, highly duplicated family of nucleotide binding site–leucine rich repeat (NBS-LRR) encoding disease resistance genes located at a single locus in lettuce (Lactuca sativa). To investigate the genetic events occurring during the evolution of this locus, ∼1.5- to 2-kb 3′ fragments of 126 RGC2 genes from seven genotypes were sequenced from three species of Lactuca, and 107 additional RGC2 sequences were obtained from 40 wild accessions of Lactuca spp. The copy number of RGC2 genes varied from 12 to 32 per genome in the seven genotypes studied extensively. LRR number varied from 40 to 47; most of this variation had resulted from 13 events duplicating two to five LRRs because of unequal crossing-over within or between RGC2 genes at one of two recombination hot spots. Two types of RGC2 genes (Type I and Type II) were initially distinguished based on the pattern of sequence identities between their 3′ regions. The existence of two types of RGC2 genes was further supported by intron similarities, the frequency of sequence exchange, and their prevalence in natural populations. Type I genes are extensive chimeras caused by frequent sequence exchanges. Frequent sequence exchanges between Type I genes homogenized intron sequences, but not coding sequences, and obscured allelic/orthologous relationships. Sequencing of Type I genes from additional wild accessions confirmed the high frequency of sequence exchange and the presence of numerous chimeric RGC2 genes in nature. Unlike Type I genes, Type II genes exhibited infrequent sequence exchange between paralogous sequences. Type II genes from different genotype/species within the genus Lactuca showed obvious allelic/orthologous relationships. Trans-specific polymorphism was observed for different groups of orthologs, suggesting balancing selection. Unequal crossover, insertion/deletion, and point mutation events were distributed unequally through the gene. Different evolutionary forces have impacted different parts of the LRR.
INTRODUCTION
The majority of resistance genes (R-genes) in plants cloned so far encode nucleotide binding site–leucine rich repeat (NBS-LRR) proteins. The LRR region is hypothesized to form a series of β-sheets with solvent-exposed residues available for interaction with a variety of ligands (Jones and Jones, 1997). This is supported by significantly elevated nonsynonymous:synonymous nucleotide substitution ratios that are indicative of diversifying selection acting on the putative solvent-exposed residues for many of the resistance genes cloned (e.g., Parniske et al., 1997; McDowell et al., 1998; Meyers et al., 1998b; reviewed in Michelmore and Meyers, 1998). However, direct physical interaction between a NBS-LRR resistance protein and an avirulence gene product has only rarely been demonstrated (Jia et al., 2000). This in part led to the guard hypothesis in which NBS-LRR proteins function as members of multiprotein complexes and detect the binding of the avirulence protein to one or more members of the complex (Dangl and Jones, 2001).
Many resistance genes are clustered in plant genomes (reviewed in Michelmore and Meyers, 1998; Hulbert et al., 2001; Richly et al., 2002; Zhu et al., 2002; Meyers et al., 2003). These clusters often comprise tandem arrays of genes that determine resistance to multiple pathogens as well as to multiple variants of a single pathogen. The genes may be organized as tight clusters with little intervening sequence, for example, the RPP5 cluster in Arabidopsis thaliana spans 91 kb (Noël et al., 1999), or be spread over several megabases as is the Resistance Gene Candidate2 (RGC2) locus in lettuce (Lactuca sativa) (Meyers et al., 1998a). The functional and evolutionary significance of this clustered arrangement is unclear.
A variety of genetic events have been documented at clusters of resistance genes, but before our current study their relative importance to the evolution of new resistance specificities was poorly understood. Extensive sequence exchanges between paralogs within clusters of R-genes have been detected in tomato (Lycopersicon esculentum), lettuce, Arabidopsis, flax (Linum usitatissimum), rice (Oryza sativa), and maize (Zea mays) (Parniske et al., 1997; Song et al., 1997; McDowell et al., 1998; Meyers et al., 1998a; Caicedo et al., 1999; Ellis et al., 1999; Noël et al., 1999; Cooley et al., 2000; Dodds et al., 2001a; Van der Hoorn et al., 2001). Genetic analyses showed that recombination played a central role in the creation of genetic diversity at the Rp1 rust resistance complex of maize and could create genes with novel specificities (Hulbert, 1997). Spontaneous Rp1 mutants had deletions at this locus caused by unequal crossing-over between paralogs (Collins et al., 1999; Sun et al., 2001). Extensive work with the L, M, N, and P loci in flax demonstrated the role of recombination in the evolution of new specificities (Ellis et al., 1999; Luck et al., 2000; Dodds et al., 2001a, 2001b). Unequal crossing-over and gene conversion were the causes of spontaneous Dm3 mutants in lettuce (Chin et al., 2001). Recombination has also been important in the evolution of Cf-4/9 homologs in tomato (Parniske et al., 1997; Parniske and Jones, 1999). Because of the instability of some resistance loci, particularly the Rp1 genes, resistance genes were initially assumed to evolve rapidly in response to changes in the pathogen population.
However, molecular analysis of orthologs and paralogs of Pto in tomato and RGC2 paralogs in lettuce demonstrated that at least some resistance genes were evolving slowly (Michelmore, 1999). This led to the development of a birth-and-death model for resistance gene evolution, similar to that proposed for multigene families of the vertebrate immune system, in which gene duplication and unequal crossing-over followed by diversifying selection results in varying numbers of semi-independently evolving lineages of resistance genes (Nei et al., 1997; Michelmore and Meyers, 1998; Piontkivska and Nei, 2003). The recognition of the avirulence gene avrRpm1 predated the divergence of A. thaliana from A. lyrata (Stahl et al., 1999). Analysis of nucleotide polymorphisms flanking the RPM1 resistance gene in Arabidopsis spp subsequently indicated the long-term coexistence of resistance and susceptibility alleles consistent with the slow evolution of this single gene resistance locus (Stahl et al., 1999). No homoplasies were observed among RPS2 genes from 17 accessions of A. thaliana (Caicedo et al., 1999). Further support for the slow evolution of resistance genes comes from the ancient origin of pathogen recognition specificity; recognition of avrPto predated the divergence of Lycopersicon pimpinellifolium from L. hirsutum var glabratum (Riely and Martin, 2001).
Lettuce is an inbreeding cultivated species (2x = 2n = 18) in the Compositae family that is fully fertile with its likely progenitor L. serriola, a common weedy colonizer of disturbed habitats (Kesseli et al., 1991). L. serriola, L. saligna, and L. virosa exhibit increasing sexual incompatibility with L. sativa and represent the major wild species in the gene pool for cultivated lettuce. The center of diversity for these species is the Eastern Mediterranean (H. Kuang, E. Nevo, and R. Michelmore, unpublished data). L. serriola and to a lesser extent L. saligna have been important sources for the introgression of disease resistance, particularly to downy mildew (Bremia lactucae), into L. sativa (Crute, 1992).
The RGC2 locus in lettuce is one of the largest clusters of resistance gene candidates so far characterized in plants. At least eight Dm genes for resistance to downy mildew as well as a gene for resistance to root aphid have been mapped to this region (Kesseli et al., 1994). One RGC2 family member, Dm3 (RGC2B), is necessary and sufficient to confer resistance to isolates of B. lactucae that express the Avr3 avirulence gene (Meyers et al., 1998a; Shen et al., 2002). Dm3 is 13 kb long and encodes a large coiled-coil type NBS-LRR protein with ∼42 LRRs. More than 24 RGC2 genes (Dm3 paralogs) had been detected at this locus that covers at least 3 Mb in the cultivar Diana (Meyers et al., 1998a) but spanned <0.1 centimorgan in the crosses studied (Chin et al., 2001). The majority of these RGC2 genes are transcribed (Shen et al., 2002). Sequence exchange was detected between RGC2 paralogs and there has been significant diversifying selection on the putative solvent-exposed residues within the LRR region (Meyers et al., 1998b). Spontaneous losses of Dm3 specificity were shown to be attributable to deletions involving multiple RGC2 paralogs resulting from unequal crossing-over and, in one case, as the result of a gene conversion event in the 3′ end of Dm3 (Chin et al., 2001). Analysis of RGC2 diversity in wild and cultivated germplasm as well as within and between natural populations of L. serriola from Israel and California identified large numbers of haplotypes, indicating the presence of numerous resistance genes in L. serriola, L. saligna, and L. virosa (Sicard et al., 1999). However, there was little data on sequence diversity or the mechanisms generating this diversity.
To gain a detailed understanding of the genetic events occurring at the RGC2 locus in lettuce and their significance to the evolution of resistance gene candidates, we compared a large number of RGC2 sequences encoding fragments of the LRR region from seven genotypes of three Lactuca spp. This demonstrated that there are two distinct types of RGC2 genes (Type I and Type II) that differed in their patterns of sequence divergence. Frequent gene conversion events in the 3′ half of Type I genes have generated a large variety of RGC2 genes, homogenized intron sequences, and confounded allelic/orthologous relationships between RGC2 genes in different genotypes and species. By contrast, Type II genes evolved slowly and maintained obvious allelic/orthologous relationships in different genotypes and species. Different genetic events have occurred with different frequencies in Type I and Type II genes.
RESULTS
Characterization of RGC2 Genes from Genomic Libraries of cv Diana
RGC2 genes are members of a large cluster of paralogs that had been partially characterized previously; nine RGC2 genes had been completely sequenced and another 13 genes partially sequenced (Meyers et al., 1998a, 1998b). In this study, we sequenced additional RGC2 genes resulting in 29 to 31 completely or partially sequenced RGC2 genes from cv Diana (Figure 1). This included two new genes in cv Diana identified by sequencing of PCR amplification products (TDAE and TDAG; see Methods). The precise number is unclear because TDT or TDV could be from the same gene as TDAC, TDAE, or TDAG.
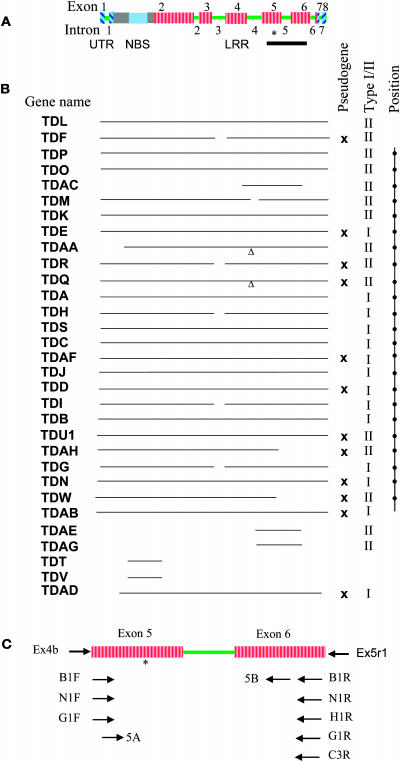
Figure 1.
Sequenced RGC2 Genes from Diana and Positions of Oligonucleotide Primers Used for Amplification Using PCR.
(A) Structure of RGC2 genes. The asterisk indicates the position of microsatellite MSATE6. The black bar below the gene indicates the region studied extensively in this article (Figure 1C).
(B) Regions of RGC2 paralogs sequenced from cv Diana. Pseudogenes are marked with an “x” in the third to last column. The last column shows the physical position of each RGC2 gene. TDL and TDF may be located at either end of the cluster. The position of the last six genes has not been determined. Small gaps in the sequences, mainly in introns, are indicated by “Δ.”
(C) Positions of primers used to amplify 3′ RGC2 fragments.
Three fragments in the MSATE6 profile of cv Diana (see below; Figure 2) were not present in the sequenced RGC2 genes. Therefore, there are at least 32 RGC2 genes in the major cluster resistance genes in cv Diana. The RGC2 genes are some of the largest genes known in plants, up to ∼15 kb with eight exons and seven introns and ∼5.8 kb of coding region. For all ∼30 sequenced RGC2 genes in Diana, the size of exons was highly conserved except for a few genes with large deletions and insertions (see below). The position of the introns was completely conserved; however, intron size varied greatly, particularly intron 3, which ranged from 363 to >6 kb.
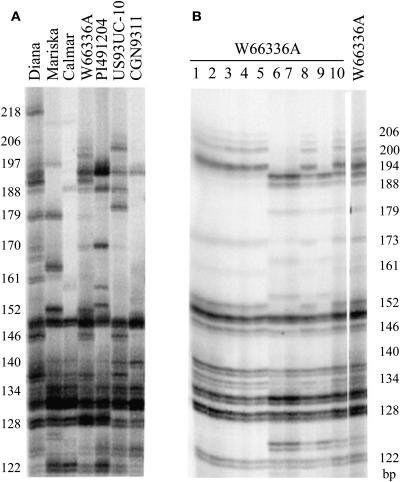
Figure 2.
Microsatellite MSATE6 Profiles of Lettuce Genomic DNA.
(A) MSATE6 profile of the seven genotypes sampled intensively in this study to estimate the minimum RGC2 copy number.
(B) MSATE6 profile of 10 individuals of L. serriola accession W66336A demonstrating the segregation of two RGC2 haplotypes. The last lane is a pool of ∼20 individuals of W66336A.
RGC2 genes encode NBS-LRR proteins of the non-TIR class of resistance genes. The N-terminal has ∼170 amino acids of unknown function, followed by an ∼300–amino acid putative NBS, followed by an ∼1300–amino acid LRR region (Meyers et al., 1998b). The number of LRRs with a consensus LxxLxxLxLxxCxx motif (where L refers to Leu or other aliphatic amino acid and x refers to any amino acid) varied from 40 to 47. Of the 29 to 31 RGC2 genes in cv Diana, at least 12 were apparently pseudogenes because they contained frameshift or nonsense mutations (Figure 1B). The overall nucleotide identities of RGC2 paralogs in Diana varied from 74 to 99% (mean = 80.4%). The most divergent genes included TDW and TDAH, which had frameshift mutations and therefore are probably pseudogenes. Sequence identity in the NBS region varied from 69.8 to 99.5% (mean = 77.7%) and 28 out of 392 pairwise nucleotide identities exceeded 95%. The mean nucleotide identity in the LRR region (82.4%) was slightly higher than that in the NBS and varied from 70.0 to 99.1% and varied over a similar range; however, only 1 out of 406 pairwise nucleotide identities exceeded 95%.
Isolation of RGC2 Fragments from Six Additional Genotypes of Lactuca spp
To amplify RGC2 fragments from other genotypes, oligonucleotide primers (Ex4b and Ex5r1) were designed based on conserved sites within the 3′ LRR region of the RGC2 genes in cv Diana (Figure 1, Table 1). This primer combination amplified RGC2 fragments from cv Diana with minimal PCR-induced errors (see Methods). Fragments of 1.5 to 2.4 kb from the 3′ region of RGC2 genes were amplified from an additional six genotypes of L. sativa (cultivars Mariska and Calmar), L. serriola (W66336A), and L. saligna (US93UC-10, CGN9311, and PI491204). We initially sequenced ∼40 clones from each of three independent PCR amplifications from each genotype. More than 90% of these clones contained sequences similar to RGC2 genes. Sequencing of the ∼120 clones/genotype represented 12, 10, 15, 34, 12, and 9 different RGC2 genes in Mariska, Calmar, US93UC-10, W66336A, CGN9311, and PI491204, respectively (Table 2). The number of PCR fragments amplified was close to saturated for all genotypes except W66336A because no or very few singletons were detected (Table 2). A total of 197 RGC2-containing clones were then sequenced for W66336A; this identified 39 distinct RGC2 sequences and decreased the number of singletons from 11 to 2. In all, 126 distinct RGC2 sequences were identified from the six genotypes plus Diana.
Table 1.
Sequences of Oligonucleotide Primers Used in PCR Amplification
| Use | Primer | Sequence 5′–3′ | Specific to |
|---|---|---|---|
| Nonresistance gene | 1007F | TGTGATTTCAACGAAGAAGCA | CL1007 |
| 1007R | AAAGAGTGAGAAACCCATGTC | CL1007 | |
| 1183F | GCACTCAGCAGAATCCGTAA | CL1183 | |
| 1183R | CTTCATCAGCATCACCACCT | CL1183 | |
| 1442F | TCCATCTCTCAATCCACCCAC | CL1442 | |
| 1442R | AATCAACACCACCGATACACC | CL1442 | |
| 368F | GGAGATCGGTTCTTGTTTCAG | CL368 | |
| 368R | AATGGTGTTGGAGAGAAAGC | CL368 | |
| 441F2 | CCCACAATACGCATCCCTAT | CL441 | |
| 441R2 | ATCACCACTTCCCATCAACG | CL441 | |
| 658F | GGCACCTCCTTCTGTCACTCA | CL658 | |
| 658R | GAAGACGACCCGTTAGTAGAA | CL658 | |
| Universal RGC2 primers | Ex4b | GCATTGTCAAGTGTGATTCCAT | Most RGC2 |
| Ex5r1 | GAATGAAAAATCCTCCTTCCC | Most RGC2 | |
| MSATE6 | 5E6 | AATGAAAGTGATWGTGAAG | Most RGC2 |
| 3E6 | TCWTCCCCAAGAAGAA | ||
| Type I genes | 5A | TTTATCGAATTAGATGTGGAAGGT | Type I |
| 5B | CAATCACCTCCTCCATCTCACT | Type I | |
| B1F | GAGAATAGAGTCTTGTGATGGCA | Type I | |
| B1R | CCCGTAAGACATGGAAGTTCTCT | Type I | |
| C3R | CCCGTAAGACTTTGAAGAAGTTG | Type I | |
| G1F | TTCAAGTGCTGAGAGTAATGGG | Type I | |
| G1R | GAAGACTTTCTAATATCAAGGAT | Type I | |
| H1R | CCCGTAAGACATGGAAGGTGTTT | Type I | |
| N1F | GCTTCAAGTGCTGACAGTAAAGTA | Type I | |
| Type II genes | F-for | GGGATGAAGGAGGTATTTGTTAG | Clade F |
| F-rev | CTGAAAACATCTAAGATAATAAAGAGA | Clade F | |
| K-for | CAAGTGCTGAATATATACAGGTG | Clade K | |
| K-rev | AAGGCGAGCAAGTGTTACAGTT | Clade K | |
| L-rev | TTGCCAATAGATTCTTCTTCC | Clade L | |
| M-for | CATGTTATGCAGCAGGACAAAG | Clade M | |
| M-rev | CCTTCCCCAAGCTAAACG | Clade M | |
| TC1 | CCTCAAGTAAGTTAGCTCTGTC | TC1 | |
| LC7 | GCCTGAGACGCTTTAGCCTTAG | LC7 | |
| RA7-for | CGCACTTGAAAGCCTGAGGAG | RA7 | |
| RA7-rev | AATCTCGTTCATTTTGCCATC | RA7 |
Table 2.
The Number of RGC2 Genes in the Seven Genotypes
| Genotype | Species | No. of Clones Sequenced | No. of Genes (Singletons)a | Error Rateb | Fragment Designation | No. of Bands in DNA Gel Blots | No. of Bands in MSATE6 | No. of Unaccounted Bandsc | Minimum Number of RGC2d |
|---|---|---|---|---|---|---|---|---|---|
| Calmar | L. sativa | 120 | 12 (2) | 1/14 kb | TC1-TC12 | 18 | 11 | 6 | 18 |
| Mariska | L. sativa | 107 | 10 (0) | 1/3 kb | TM1-TM10 | 16 | 12 | 3 | 13 |
| US93UC-10 | L. serriola | 104 | 15 (3) | 1/4 kb | LC1-LC15 RA1-RA12 | 13 | 16 | 5 | 20 |
| W66336A | L. serriola | 197 | 39 (2) | 1/10 kb | RB1-RB5 RAB1-RAB22 | 23 | 21 | 3 | 42 |
| CGN9311 | L. saligna | 104 | 12 (0) | 1/2 kb | LA1-LA12 | 11 | 14 | 1 | 13 |
| PI491204 | L. saligna | 96 | 9 (0) | 1/20 kb | LB1-LB9 | 11 | 9 | 3 | 12 |
| Diana | L. sativa | 158 | 22 | 1/10 kb | TD-e | 25 | 20 | 3 | 32 |
Total number of distinctive RGC2 fragments discovered.
Error rate was calculated by comparison of different clones of the same gene.
The number of bands that are present in MSATE6 but not encoded by any cloned RGC2 genes in a genotype.
The sum of the number of genes and the number of unaccounted bands.
TD followed by one or two letters.
Variation in the Number of RGC2 Genes in Different Genotypes
The number of genes amplified by PCR with primers Ex4b and Ex5r1 from the seven genotypes varied from 9 to 39 (Table 2). To determine whether this was because of differential amplification with primers designed from RGC2 sequences in cv Diana or reflected real variation in the number of RGC2 genes in the genome, copy number was assessed using several additional approaches. DNA gel blot hybridizations to genomic DNA were made using probes of pooled fragments that had been amplified by primers Ex4b and Ex5r1 from 20 diverse RGC2 genes in Diana. The number of bands varied from 11 in PI491204 and CGN9311 to 25 in Diana. There was a strong correlation between the number of bands in DNA gel blots and the estimated minimum number of RGC2 genes (see below) (r = 0.85, P < 0.01; Table 2).
Amplification of the microsatellite marker MSATE6 resulted in 20 distinct fragments from at least 29 of the 32 RGC2 genes in Diana (Figure 2A). The MSATE6 primers would be predicted to amplify from ∼90% of the sequences amplified using primers Ex4b and Ex5r1. The actual numbers of MSATE6 fragments amplified from genomic DNA of Mariska, Calmar, US93UC-10, W66336A, CGN9311, and PI491204 were 12, 11, 16, 21, 14, and 9, respectively (Figure 2A). The majority of bands in the MSATE6 profile were represented in the RGC2 sequences amplified by primers Ex4b and Ex5r1; however, a few previously undetected bands were identified (Table 2). There was a strong correlation between the number of MSATE6 fragments and number of fragments detected by DNA gel blot hybridization (r = 0.70, P < 0.05). The MSATE6 profile was therefore consistent with, but more informative than, DNA gel blot analysis. Therefore, these three approaches consistently confirmed significant variation in the copy number of RGC2 genes that was not correlated with species.
The minimum number of RGC2 genes in each genotype was calculated from the combined analysis of MSATE6 and the sequenced RGC2 fragments (Table 2). PI491204 (L. saligna) and cv Mariska had the lowest minimum copy numbers of 12 and 13, respectively; the minimum number of RGC2 genes in cv Calmar and US93UC-10 (L. saligna) was 20. However, the minimum number of RGC2 genes in cv Diana was 32.
Although W66336A (L. serriola) initially appeared to have more RGC2 genes than Diana (at least 42), we determined that W66336 is heterozygous, comprised of two distinct haplotypes. To check if W66336A was heterozygous at the RGC2 locus, 10 individuals each of W66336A and Diana were analyzed using microsatellite MSATE6. Segregation of two distinct MSATE6 profiles was observed in W66336A, including heterozygous individuals with a combination of the two profiles (Figure 2B). Therefore, the plants representing W66336A must have been segregating for the RGC2 locus, and the 39 RGC2 genes amplified from W66336A were derived from two L. serriola haplotypes (designated as A and B). Haplotype A has 14 bands in its MSATE6 profile, six of which were not present in haplotype B. Sequence analysis of the cloned fragments indicated that these six MSATE6 bands were derived from at least 12 RGC2 genes; therefore, these genes specific to haplotype A could be designated RA1 to RA12. On the other hand, haplotype B has 13 bands in its MSATE6 profile. There are five bands and five genes specific to haplotype B, which could be designated RB1 to RB5. The other 22 genes produced MSATE6 fragments common to both haplotypes and were therefore designated RAB1 to RAB22. Assuming that the 22 RAB genes and at least three unsequenced genes were equally distributed in the two haplotypes, the minimum copy number of RGC2 genes in haplotypes A and B within W66336A were estimated to be ∼25 and 18, respectively. By contrast, no segregation at the RGC2 locus was observed in cv Diana. Therefore, the copy number of RGC2 genes varied from ∼12 (PI491204, L. saligna) to >32 (cv Diana).
Large Insertions Encoding Multiple LRRs Resulted from Unequal Crossing-Over
The 126 distinct 3′ RGC2 sequences from the seven genotypes were aligned using ClustalX. The intron (intron 5) in this region was highly dissimilar between some genes and therefore could not be included in the alignment. Large insertions (>237 bp) had occurred in the exons of 32 fragments. Fragments with identical insertion sites and very similar inserted sequences (>97%) were considered to be derived from the same event. This indicated that the 32 large insertions were the results of 13 independent events (Table 3). Seven events were found in more than one gene; six of these were detected in multiple species and at least four events had occurred before speciation of L. serriola and L. saligna (Table 3).
Table 3.
Large Indels Predicted to Result from Unequal Crossing-Over
| Event ID | Fragment Names | Insertion Size | No. of LRRs | Insertion Positiona | Positions of Inserted Ends 5′–3′b | Recombination between | Event Time |
|---|---|---|---|---|---|---|---|
| BI5-1 | TDF, TC6, TM8, RAB7, RAB15 | 237 | 2.0 | 230 | 13–NA | Paralogs | Before speciation |
| BI5-2 | TDK, TC7, TM5, RA8, RAB6, RAB22, LA11, LB4 | 501 | 5.0 | 81 | 14–12 | Paralogs | Before speciation |
| BI5-3 | TDP, LA6 | 366 | 3.0 | 230 | 13–NA | Orthologs | Before speciation |
| BI5-4 | TM9, LB1 | 291 | 3.0 | 140 | 2–NA | Orthologs | Before speciation |
| BI6-1 | RAB2, LA5, LC1 | 285 | 3.0 | 35 | 16–16 | Paralogs | Before speciation |
| BI6-2 | TDU, TC4, RAB1, RAB17 | 285 | 3.0 | 35 | 16–16 | Paralogs | Before speciation |
| BI6-3 | LA10, LC2 | 258 | 2.5c | 53 | 25–10 | Paralogs | ND |
| BI6-4 | LC3 | 288 | 3.0 | 41 | 26–14 | Paralogs | ND |
| BI6-5 | TC2 | 291 | 3.0 | 17 | 25–22 | Paralogs | ND |
| BI6-6 | LC4 | 291 | 3.0 | 17 | 25–22 | Orthologs | ND |
| BI6-7 | RA1 | 291 | 3.0 | 17 | 25–22 | Paralogs | ND |
| BI6-8 | RA5 | 291 | 3.0 | 17 | 25–22 | Orthologs | ND |
| BI6-9 | TM7 | 291 | 3.0 | 17 | 25–22 | Paralogs | ND |
For BI5 events, numbers refer to base pairs from the first base of primer Ex4b; for BI6 events, numbers refer to base pairs from the first base of primer Ex5r1.
Positions of the ends of inserted sequences in consensus LRR motif. NA, not attempted because of poor sequence similarity; ND, not determined because of lack of information.
Includes two complete LRRs and 10 amino acids.
All of the 13 inserted sequences were direct repeats of LRR-encoding regions. This structure indicated that these large insertions were the result of unequal crossing-over within or between RGC2 genes. To investigate whether the unequal crossing-over had been intragenic or intergenic, sequence alignments were made for each of the 13 events. For each event, the fragments were divided at the insertion site, and the two sequences were independently aligned with all other RGC2 fragments. The two ends of the alignments were then trimmed and the duplicated regions analyzed. The duplicated sequences in nine of the 13 duplication events were more similar to other sequences than to each other (e.g., Figure 3); therefore, these duplications had resulted from unequal crossing-over between two different genes. For the other four duplication events, the duplicated sequences were most similar to each other (e.g., Figure 3B) but were not nearly identical, ranging from 89.7 to 95.8% nucleotide identity. These duplications could have been the result of an ancient intragenic unequal crossover event or the result of unequal crossing-over between closely related paralogs that were not identified in our analysis (Table 3).
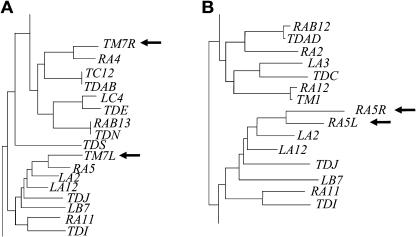
Figure 3.
Distance Trees Indicating the Origin of Duplications within the LRR.
Each duplicated region with large insertions was divided into two parts at the insertion sites (-L and -R, indicated by arrows). Then, the two fragments from all duplications were aligned with the same region of all RGC2 genes. Only parts of the distance tree indicating the origin of two duplications are shown.
(A) Duplications in fragment TM7 indicating a paralogous origin.
(B) Duplications in fragment RA5 indicating a potentially allelic origin.
The positions of the duplications were nonrandom. Four of the 13 duplication breakpoints occurred within a 150-bp region in exon 5. All of the other nine breakpoints occurred in exon 6 within a 35-bp region (Table 3). The duplications in genes TM7, RA5, RA1, LC4, and TC2 involved different sequences but occurred at exactly the same position. Therefore, the middle of exon 5 and the end of exon 6 have been hot spots for unequal crossing-over relative to the rest of the region analyzed (Figure 4).
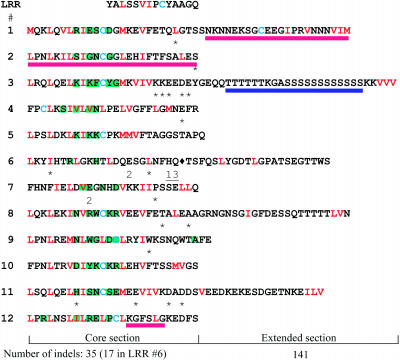
Figure 4.
Unequal Distribution of Point Mutation, Insertion/Deletion, and Recombination Events in RGC2 Genes from Lactuca spp.
Positions of these different events detected in all RGC2 sequences are indicated on the amino acid sequence of the 12 LRRs from TDB (RGC2B and Dm3). Aliphatic residues in the LRR consensus are shown in red. Position of intron 5 in LRR 6 is shown by a diamond. Hypervariable sites showing significant posterior probabilities using PAML indicative of diversifying selection are shaded in green. Hot spots for unequal crossing-over that resulted in duplications of one or more LRRs are underlined with a red bar. The positions of small, mostly in-frame, indels (<64 bp) in the core sections of each LRR are shown with asterisks; multiple occurrences of indels at the same position are indicated by numbers. The positions of indels in the extended sections of the LRR were too numerous to display and could not be positioned accurately because of sequence divergence between RGC2 sequences. The complex hyperpolymorphic microsatellite sequence in LRR 3 is shown by a blue bar.
The size of the inserted sequences varied from 237 to 501 bp, preserved the open reading frame, and encoded two to five LRRs. Most of the inserted fragments encoded complete LRRs and only one or two amino acids shorter than LRRs in TDB (Table 3). However, the inserted sequence in BI6-3 was 14 amino acids shorter and was missing 12 amino acids in the LRR consensus motif. It is unknown if sequences with a shortened LRR are functional. The majority of duplications preserved the predicted structure of the LRR with putative solvent-exposed β-sheets. The breakpoints of insertions were located outside the LRR consensus motif and in most cases resulted in the duplication of complete rather than truncated LRRs (Table 3).
Patterns of Sequence Diversity Identified Two Types of RGC2 Genes
A neighbor-joining distance tree was constructed for the 126 fragments using SUN7, an RGC2 fragment from sunflower (Helianthus annuus), as the outgroup (Figure 5). Two distinct types of genes (designated Type I and Type II) were recognizable based on the patterns of sequence diversity between the genes. The two types were separated by a node with a bootstrap value of 97%. A parsimony tree for the same set of fragments had similar topology to the neighbor-joining tree but a lower bootstrap value for the node separating Type I and Type II genes (data not shown).
Figure 5.
Distance Tree Derived from the 3′ Region of 126 RGC2 Genes from Seven Genotypes.
Genes from different genotypes are marked in different colors: L. sativa is shown as shades of blue, L. saligna as shades of red, and L. serriola as shades of green. SUN7 (an RGC2 homolog from sunflower) was used as the outgroup. Type I and Type II genes were distinguished by different tree topologies. The different Type II clades are designated at right.
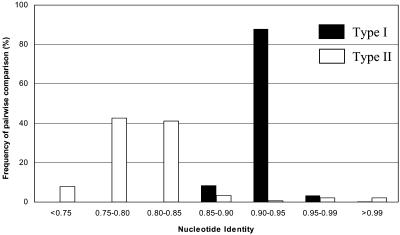
The Type I group contained 48 genes (38%) and usually had medium length branches (5 to10% nucleotide substitutions) in the distance tree. Most bootstrap values between Type I genes were <90% with the exception of 10 nodes. Their pairwise sequence identities varied from 86.9 to 100%, with the majority (87.9%) varying from 90 to 95%. Only four of the 1128 pairwise nucleotides identities within Type I genes were >99%. There was no clear relationship between taxonomy and position in the distance tree, and it was impossible to identify potentially allelic or orthologous relationships between genes in the Type I group except in a few cases, such as TC12 and TDAB.
In contrast with Type I, the branches linking the 78 Type II genes were either long (>10% nucleotide substitutions) or short (<5% nucleotide substitutions) (Figures 5 and 6). The bootstrap values between each tight clade in Type II were without exception 100%. Most (83.9%) pairwise sequence identities between genes in different Type II genes were <85%, whereas those between genes within each tight clade were >97%. Sequence identities of 90 to 95%, which were prevalent between Type I genes, were rare between Type II genes (Figure 6). Several of the tight clades had a single representative of each of the genotypes; six tight clades had two representatives from W66336A that contained two RGC2 haplotypes (see above). This distribution suggested that genes within each tight clade are alleles or orthologs.
Figure 6.
The Distribution of Pairwise Nucleotide Identities among 48 Type I and 78 Type II Genes.
The majority of Type I identities are between 90 and 95%; conversely, identities between Type II genes are either greater or less than that of Type I genes.
Sequence Exchanges Are Frequent between Type I Genes but Rare between Type II Genes
Besides differences in nucleotide similarity, another major difference between Type I and Type II RGC2 genes was the frequencies of sequence exchanges. A total of 79 independent sequence exchanges were detected between all 126 RGC2 fragments using Geneconv (P < 0.05). Seventy-six of them occurred between Type I genes; only two occurred between a Type I gene and a Type II gene and one between two Type II genes even though more Type II genes than Type I genes were represented in the 126 sequences. On average, each Type I gene had three sequence exchanges with other Type I genes in the 1.2-kb LRR-encoding region studied. The length of the sequence exchanged varied from 60 to 528 bp, with an average exchange length of 201 bp.
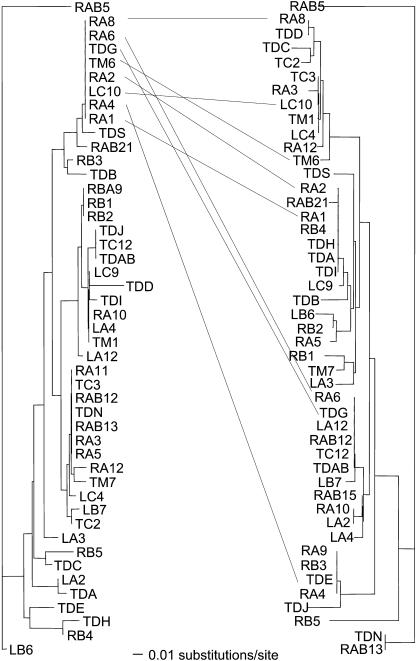
Sequence exchange among Type I genes was investigated in greater detail by comparing distance trees for 12 different sections of these sequences. First, the ∼1.2-kb sequences were divided into 12 LRR-encoding sections. Each section contained ∼100 bp, with one LxxLxxLxLxxCxx motif located in the middle. Trees were constructed for each of the 12 sections in the Type I genes. The topologies of these trees were considerably different from the tree constructed from the whole sequence of the Type I genes (Figure 7). Unlike the tree for the whole region that had medium branch lengths for Type I genes (Figure 5), these trees had several tight clades of identical or nearly identical sequences. The distribution of sequences within the trees varied; genes with high sequence similarity at the first LRR were not always similar at the last LRR (Figure 7). For example, one tight clade had eight (RA8, RA6, TDG, TM6, RA2, LC10, RA4, and RA1) sequences that were identical in the first 100 bp. However, these sequences varied by as much as 10% in the last 100 bp and were distributed throughout the tree for the last 100 bp (Figure 7).
Figure 7.
Distance Trees for LRR 1 (Left Tree) and LRR 12 (Right Tree) of Type I Genes Showing the Lack of Correlation between LRRs Derived from the Same Gene.
For illustration, genes with identical LRR 1, RA8, RA6, TDG, TM6, RA2, LC10, RA4, and RA1, are joined by lines to diversify positions in the tree for LRR 12.
Fragments encoding individual LRRs within each tight clade had >98% nucleotide identities and were often identical. Some tight clades had as many as 12 members representing all three species. The presence of identical fragments in the different species indicates that the rate of point mutation in Type I genes has been very low, just as it has been in Type II genes (see below). The diversity between Type I genes is therefore mainly attributable to exchanges of blocks of sequence encoding one or more LRRs rather than the accumulation of novel point mutations at the hypervariable sites within the consensus motif of LRR.
Individual Type I Sequences Are Rare in Natural Populations Because of Frequent Sequence Exchanges
To determine the prevalence of Type I sequences, either whole or in part, primers specific to a variety of Type I genes were used either together or in combination with conserved primers to amplify sequences from a panel of 40 additional wild accessions. These included 33 accessions of L. serriola, six accessions of L. saligna, and one accession of L. perennis. Oligonucleotide primers were designed to the hypervariable region within the consensus motif of first and last LRRs of Type I genes (Figure 1C, Tables 1 and 4). Combinations of one primer specific to a Type I gene with one conserved primer (either Ex4b or Ex5r1) were used to determine the frequency of each primer site in the panel of 40 accessions. The frequency of the most frequent specific primer sites ranged from 25 to >50% and was not correlated with their frequency in the initial seven accessions (Table 4).
Table 4.
The Relative Frequencies of Fragments Amplified Using Gene-Specific Primers from the Initial Seven Genotypes and the Frequencies Amplified from an Additional 40 Genotypes
| Seven Genotypesb | No. of Genesc | 40 Genotypesd
|
|||||
|---|---|---|---|---|---|---|---|
| Primers (Combination)a | Total | No. Sequenced | Fragment Name | Type I/II | |||
| N1F | Ex5r1 | 4 | 8 | 27 | 14 | _N | I |
| G1F | Ex5r1 | 4 | 9 | 10 | |||
| B1F | Ex5r1 | 1 | 1 | NAe | |||
| Ex4b | G1R | 5 | 13 | 11 | 11 | _H | I |
| Ex4b | C3R | 4 | 8 | 11 | 8 | _L | I |
| Ex4b | H1R | 3 | 7 | 14 | 5 | _W | I |
| Ex4b | B1R | 1 | 1 | NA | |||
| Ex4b | 5Bf | 3 | 4 | 19 | 7 | _M | I |
| Ex4b | 5Bf | 0 | 0 | 10 | 9 | _5B | II |
| N1F | C3R | 3 | 3 | 5 | 4 | _P | I |
| N1F | G1R | 2 | 2 | 4 | 3 | _D | I |
| N1F | 5B | 1 | 1 | 9 | 6 | _R | I |
| N1F | B1R | 0 | 0 | 0 | |||
| G1F | C3R | 1 | 1 | 2 | |||
| G1F | G1R | 2 | 2 | 3 | 2 | _Q | I |
| G1F | 5B | 1 | 1 | 0 | |||
| G1F | H1R | 1 | 1 | 10 | 3 | _V | I |
| B1F | B1R | 1 | 1 | 0 | |||
| B1F | 5B | 1 | 1 | 0 | |||
| 5A | 5B | 1 | 1 | 2 | |||
| Ex4b | K-rev | 7 | 8 | 36 | K_ | II | |
| K-for | Ex5r1 | 7 | 8 | 34 | 35 | K_ | II |
| K-for | K-rev | 7 | 8 | 34 | K_ | II | |
| Ex4b | L-rev | 7 | 7 | 34 | 34 | L- | II |
| AC-for | Ex5r1 | 6 | 6 | 32 | II | ||
| M-for | Ex5r1 | 6 | 7 | 24 | II | ||
| Ex4b | M-rev | 6 | 7 | 20 | II | ||
| Ex4b | F-rev | 4 | 5 | 21 | II | ||
| F-for | Ex5r1 | 4 | 5 | 17 | II | ||
| Q-for | Ex5r1 | 3 | 5 | 17 | II | ||
| Ex4b | LC7 | 1 | 1 | 3 | II | ||
| RA7-for | RA7-rev | 1 | 1 | 2 | II | ||
| Ex4b | TC1 | 1 | 1 | 3 | II | ||
Specific primers are underlined.
Number of the initial seven genotypes that have the primer sequences.
Number of genes in the seven genotypes with the primer sequences.
Number in the panel of 40 genotypes that have the primer sequences, as indicated by PCR amplifications.
NA, not available because of primer incompatibility.
The site of primer 5B was present only in Type I genes in the initial seven genotypes. However, primer combination Ex4b and 5B amplified both Type I and Type II genes.
Amplification products were only rarely obtained using pairs of Type I specific primers (Table 4). On average, the frequency of detecting amplification products from the 40 additional accessions was 8%. Primers specific to individual Type I genes could amplify products when combined with primers specific to different Type I genes (Table 4). Combinations of gene-specific primers that resulted in amplification products were as likely to have been derived from different Type I genes as from individual Type I genes present in the original seven genotypes (Table 4). The low frequencies of combinations of any two Type I specific sequences provided further evidence that Type I genes have undergone extensive shuffling, and many RGC2 genes encoding different combinations of individual LRR sequences may exist in nature. No PCR amplifications were observed when a Type I specific primer was used in combination with a Type II specific primer; this is consistent with minimal sequence exchange between Type I and Type II genes.
Type I Genes Comprise Diverse Chimeras in Natural Populations
To provide additional information on the evolution of Type I genes and to test whether the original amplification of the seven genotypes using the two universal primers (Ex4b and Ex5r1) had resulted in a biased sample, 72 of the fragments amplified using 10 different gene-specific primer combinations were cloned and sequenced (Table 4). These 72 additional sequences were aligned with the 48 Type I genes characterized previously from the seven genotypes, and a distance tree was constructed using RAB5, the Type II gene most similar to Type I, as the outgroup (data not shown).
Nine fragments, which were amplified by primer combination Ex4b and 5B, formed a tight clade with DNA identities of 99.1 to 100% and apparently belonged to a Type II clade that had not been detected in the initial seven genotypes. One of the nine fragments was from L. saligna; the other eight were from L. serriola. They were dissimilar to any other Type I or Type II gene (<85% nucleotide identities). These new Type II genes were 1.3 kb between primers Ex4b and 5B, whereas the Type I genes amplified by the same primer combination were 1.7 kb. They had a frequency of 25% in the 40 additional accessions but were apparently absent from the initial seven genotypes.
All of the other 63 sequences, together with the 48 Type I genes characterized earlier, exhibited similar sequence identities with each other. The distance tree for all 111 Type I genes had a similar topology to that for the Type I genes from the seven initial genotypes (data not shown). Sequences amplified by the same primer combinations were scattered throughout the tree, indicating that the original pair of primers (Ex4b and Ex5r1) had not provided a biased sample of sequences. Also, sequences amplified by the same primers were usually dispersed within the tree; conversely, genes that grouped together had often not been amplified using the same primer combination. Numerous sequence exchanges were observed between these 111 Type I genes (data not shown). These data provided further evidence of extensive shuffling of Type I sequences.
Highly Conserved Type II Alleles/Orthologs Occur at Different Frequencies in Natural Populations
The 78 Type II sequences were distributed in 18 tight clades (Figure 5). In addition, eight putative Type II genes had no obvious alleles or orthologs. The numbers of alleles/orthologs detected in each of the 18 Type II clades varied from two to eight. To investigate whether these differences in the frequencies of alleles/orthologs was because of sampling bias or reflected their actual prevalence in the seven haplotypes studied, the panel of 40 additional wild accessions were analyzed using 14 pairs of oligonucleotide primers specific to individual tight clades (Tables 1 and 4). In contrast with the results for Type I genes, RGC2 fragments were amplified from similar numbers of accessions by pairs of primers that were both specific to the clade and by pairs of primers consisting of a clade-specific and a conserved primer (e.g., K-for + K-rev compared with K-for + Ex5r1; Table 4). This is consistent with a low frequency of chimeric sequences. Subsequent sequencing of these PCR fragments showed that only alleles/orthologs were amplified using each primer combination. Fragments amplified using primers specific to clade K had 97.3 to 100% nucleotide identities. Similarly, fragments amplified using primers specific to L clade had 98.3 to 100% nucleotide identities with each other and were obvious alleles or orthologs of TDL (see below).
Some Type II clades were detected in many genotypes of different Lactuca species, whereas other clades were rare. The frequencies of K, L, and M alleles/orthologs were 38/40, 34/40, and 24/40, respectively, which was consistent with their prevalence in the initial seven genotypes (Table 4). Twenty-four of the 33 L. serriola genotypes but none of the six additional L. saligna genotypes had an F allele/ortholog; this was again consistent with the taxonomic distribution observed within the initial seven genotypes. Three primers (RA7, LC7, and TC1) specific to three Type II genes that were detected only once in the seven genotypes amplified products from only 2, 3, and 3 of the 40 additional genotypes, respectively (Table 4). Therefore, the variability in the prevalence of Type II alleles/orthologs in the initial seven genotypes was not an experimental artifact, and the different Type II clades varied in frequency from rare to almost ubiquitous.
Type II Orthologs Exhibit Trans-Specific Polymorphism
Fragments of six single-copy genes that were not resistance genes were chosen to serve as reference sequences for the analyses of the RGC2 genes and to confirm the taxonomic relationships of the seven Lactuca genotypes from which the RGC2 genes were cloned. Distance trees were constructed for the sequences of these six genes, and similar topologies were obtained for each tree (data not shown). Sequences from the same species always grouped tightly together and were often identical; intraspecific polymorphisms were rare (Table 5). Average KA:KS (nonsynonymous:synonymous nucleotide changes) ratios ranged from 0 to 0.23 for the six genes, consistent with purifying selection acting on these genes.
Table 5.
Single-Copy Nonresistance Genes from Lactuca spp
| BLASTX to GenBank
|
Linkage Groupa | Fragment Size (bp) | Point Mutations
|
||||
|---|---|---|---|---|---|---|---|
| Gene | Best Hit | E-Value | Between Speciesb | Within Speciesb | Total | ||
| CL368 | No hit | 8 | 310 | 2 (0.006) | 1 (0.001) | 3 | |
| CL441 | Far-red impaired response protein | 1e−30 | 6 | 681 | 19 (0.027) | 1 (0.0005) | 20 |
| CL658 | ZF-HD homeobox | 1e−38 | 6 | 393 | 8 (0.020) | 0 (0) | 8 |
| CL1007 | Acidic ribosomal protein | 1e−17 | 6 | 195 | 3 (0.015) | 0 (0) | 6 |
| CL1183 | 40S ribosomal protein S9 | 2e−68 | 6 | 429 | 1 (0.002) | 2 (0.002) | 3 |
| CL1442 | No hit | 9 | 789 | 10 (0.013) | 8 (0.021) | 18 | |
The rate (nucleotide changes per site) is shown in parentheses.
In contrast with the nonresistance genes, the distribution of RGC2 genes in the distance tree did not reflect the taxonomic relationships of their genotypes of origin. Different genotypes were dispersed throughout the tree rather than clustered in species-specific clades. A Templeton test (Templeton, 1983) indicated that the trees for the K and L Type II clades of RGC2 genes were significantly different from the trees based on nonresistance gene sequences (P < 0.0001). The relationships of alleles/orthologs from each accession varied within each Type II clade. Alleles from the same species were often not the most similar to each other; for example, in clade M, TM3 (an allele from cv Mariska) was the most divergent allele/ortholog, whereas the other two alleles from L. sativa (TDM and TC8) were identical or nearly identical to orthologs from L. saligna (LA8 and LB3). Such trans-specific polymorphisms may be indicative of balancing selection maintaining ancient polymorphism.
Point Mutations as the Most Common Polymorphism between Type II Alleles/Orthologs
Some RGC2 clades exhibited low frequencies of nucleotide ploymorphisms, even lower than single-copy nonresistance genes (Tables 5 and 6). The levels in nucleotide polymorphism observed between alleles/orthologs were clearly not consistent with markedly elevated rates of point mutation in the 3′ regions of RGC2 genes. Point mutations between alleles/orthologs were distributed throughout the LRR-encoding region rather than being concentrated in the hypervariable sites within each LRR, in contrast with polymorphisms between paralogs (see below). Point mutations were more prevalent than insertion/deletions (indels) and sequence exchanges between alleles/orthologs within individual Type II clades (Table 6). The total number of indels per clade was low, and only 23 indel polymorphisms were observed within clades (Table 6). The pairwise nucleotide identities between alleles/orthologs within a clade varied from 95 to 100% (mean = 97.8%). This was only slightly lower than nucleotide identities for the single-copy, nonresistance genes, which varied from 97.3 to 100% (mean = 98.9%).
Table 6.
Point Mutations and Sequence Exchanges Detected between Orthologs
| Number of Genes | Number of Polymorphic Sitesa | Number of Indelsa | Average Pairwise Nucleotide Identity (%)a | Sequence Exchangec
|
||||
|---|---|---|---|---|---|---|---|---|
| Clade | Size (bp)ab | Genecov | Visual Inspection | Four Gamete | ||||
| AE | 2 | 1266 (133) | 3 (0) | 0 (0) | 99.8 (100) | – | – | – |
| C5 | 2 | 1278 (406) | 1 (2) | 0 (1) | 99.9 (99.3) | – | – | – |
| AB8 | 2 | 1257 (373) | 5 (0) | 2 (1) | 99.4 (99.7) | – | – | – |
| DR | 2 | 1261 (310) | 6 (1) | 0 (0) | 99.5 (99.7) | – | – | – |
| C10 | 2 | 1263 (299) | 4 (0) | 0 (0) | 99.7 (100) | – | – | – |
| M9 | 2 | 1221 (200) | 1 (0) | 1 (1) | 99.9 (99.5) | – | – | – |
| LA10 | 2 | 1203 (255) | 31 (10) | 2 (2) | 97.4 (95.0) | – | – | – |
| LA7 | 2 | 1242 (316) | 8 (3) | 0 (0) | 99.4 (99.1) | – | – | – |
| RAB2 | 3 | 1194 (210) | 8 (3) | 0 (0) | 99.6 (99.0) | 0 | 0 | – |
| O | 3 | 1218 (295) | 20 (5) | 2 (2) | 98.9 (87.7) | 0 | 0 | – |
| P | 3 | 1278 (368) | 18 (12) | 3 (3) | 98.8 (97.3) | 0 | 0 | – |
| U | 4 | 1206 (227) | 42 (1) | 1 (1) | 98.2 (99.4) | 0 | 0 | 0 |
| Q | 5 | 1218 (289) | 17 (5) | 3 (0) | 99.4 (99.2) | 0 | 0 | 0 |
| F | 5 | 1237 (357) | 25 (11) | 2 (1) | 99.2 (98.8) | 0 | 0 | 0 |
| AC | 6 | 1239 (411) | 34 (10) | 3 (5) | 98.9 (98.5) | 1 | 2 | 3 |
| M | 7 | 1215 (272) | 25 (6) | 1 (0) | 99.4 (99.4) | 0 | 0 | 0 |
| L | 7 | 1263 (305) | 51 (19) | 3 (2) | 98.1 (96.4) | 2 | 3 | 4 |
| Kd | 43 | 1689 (322) | 155 (47) | 4 (10) | 98.4 (97.1) | 3 | 9 | 14 |
| 42 | 1683 (322) | 102 (33) | 3 (10) | 98.6 (97.3) | 3 | 9 | 13 | |
| 8 | 1683 (321) | 65 (21) | 1 (6) | 98.5 (97.1) | 1 | 4 | 6 | |
Numbers in front of parentheses refer to the coding regions between primers Ex4b and Ex5r1. Numbers in parentheses refer to intron 5.
The largest size among all orthologs within a clade is shown.
Sequence exchange was investigated using Geneconv (Sawyer, 1989; P < 0.05), visual inspection, and the four-gamete test (Hudson and Kaplan, 1985). –, not applicable.
Different subsets of K orthologs were analyzed: all 43 K orthologs cloned from Lactuca spp; 42 K orthologs, excluding the K ortholog from L. perennis; eight K orthologs from the seven genotypes analyzed initially.
Characterization of K and L Alleles/Orthologs from Additional Wild Accessions
To investigate allelic/orthologous variation and its genetic basis, K and L alleles/orthologs from the panel of 40 accessions were analyzed in detail. Primers that were specific to members in clade K were used in combination with the conserved primers (Ex4b or Ex5r1) to amplify K alleles/orthologs. Products were obtained from 38 of these genotypes using at least one of the primer combinations. We sequenced 35 of these products derived from 29 accessions of L. serriola plus five from L. saligna and one from L. perennis.
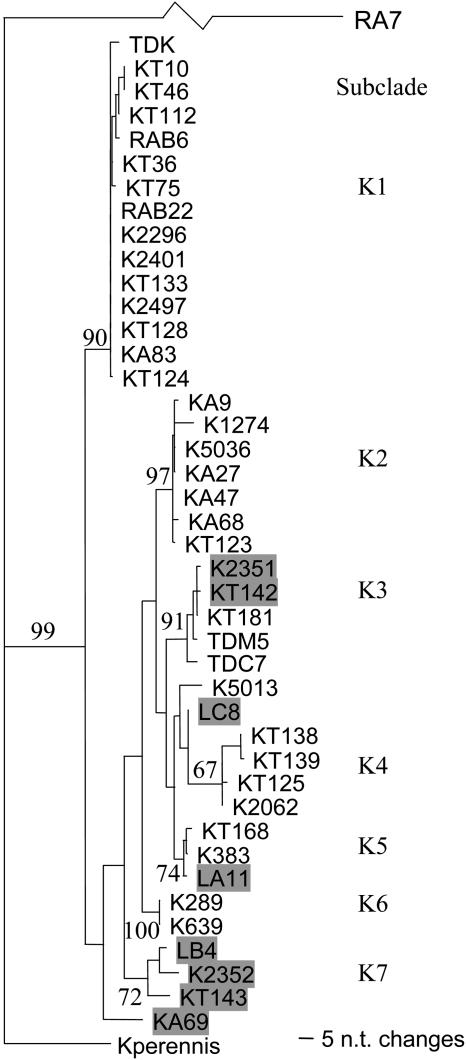
Sequences from 43 K alleles/orthologs, including the eight fragments from the seven genotypes characterized previously, were compared and a distance tree constructed using the closest paralog RA7 as the outgroup (Figure 8). The pairwise nucleotide identities between Kperennis (from L. perennis), the most divergent ortholog, and the other alleles/orthologs varied from 94.7 to 95.8%. The nucleotide identities between the other K alleles/orthologs were between 97.3 and 100%. These nucleotide identities were much higher than those between a K allele/ortholog and any paralog (<84.5%). K alleles/orthologs are therefore highly conserved in these four Lactuca spp, and they are clearly distinguishable from their closest paralog. Even the most divergent fragment from the sexually incompatible species, L. perennis, retained an obvious orthologous relationship with other alleles/orthologs in clade K.
Figure 8.
Neighbor-Joining Distance Tree of 43 K Alleles/Orthologs from 42 Genotypes of Lactuca spp.
Fragment RA7, the closest nonorthologous sequence, was used as outgroup; only half of the branch linking to RA7 is shown. Alleles from L. saligna are shaded. The values on the branches indicate the percentage of 1000 bootstrap replicates; values <60 are not shown.
Several subclades of K alleles/orthologs could be distinguished (Figure 8). Seven were separated by nodes with bootstrap values >67%; four of these nodes had bootstrap values >90%. These subclades may represent several allelic lineages. Each subclade frequently had representatives from multiple species. Subclade K3 includes two fragments from L. saligna and three fragments from L. serriola; K2351 from L. saligna differs from KT181 from L. serriola by only one base. There are three fragments in subclade K5, one from L. saligna and two from L. serriola that differed by only one or two bases. Such trans-specific polymorphism was consistent with previous observations and provides additional evidence that nucleotide differences were ancient and that balancing selection has maintained polymorphism within each Lactuca spp.
Only three, nine, and 14 sequence exchanges were detected among K alleles/orthologs using Geneconv, visual inspection, and the four-gamete method, respectively (Table 6). Fragments K289 and K639 in subclade K6 were extensive chimeras consisting of sequences from several different K alleles/orthologs (data not shown). K5013 was close to subclade K5 (Figure 8) but differed from K5 genes because of a sequence exchange in the first 700 bp between K5013 and KT10/KT46. The variation between KT125 and LC8 was caused by an ∼400 bp sequence exchange between KT125 and genes in subclade K3. The variation between TDC7 and the remaining subclade K3 genes was because of gene conversion with a gene from subclade K1. Therefore, detectable sequence exchanges between alleles/orthologs within clade K have occurred and may be important in generating diversity.
Similar results were obtained for the L clade. A total of 27 new sequences were obtained from the panel of 40 genotypes using primers specific to L alleles/orthologs. Excluding the MSATE6 sequence and fragment Lperennis from L. perennial, the 34 sequences (including seven from the initial seven genotypes) exhibited an average of 99.0% nucleotide identity and an obvious allelic/orthologous relationship.
Highly Conserved Introns in 3′ of Type I Genes but Not Type II Genes
Analysis of intron 5 sequences revealed very different characteristics for Type I and Type II genes. Four of the 111 Type I genes were excluded because three of them had a deletion of the entire intron 5 region and one had poor sequence. Of the remaining 107 Type I genes, the size of intron 5 ranged from 616 to 1037 bp, with an average of 679 ± 56 bp. The intron sequences could be readily aligned, and nucleotide identity varied from 94 to 100% with an average of 97%. This was greater than the nucleotide identities of the Type I flanking coding regions that ranged from 89 to 100% (average 92%). No hypervariable sites were evident within the intron. Intron variation was mainly attributable to evenly distributed point mutations and a few indels that were usually <20 bp. Insertions of 358 bp were observed in TDH and RB4. The inserted sequences were apparently derived from the same event because fragments TDH and RB4 were almost identical in the insertion and other regions. BLASTN analysis failed to detect significant sequence similarities to the inserted sequences, and there was no evidence that these sequences were related to retrotransposons or long terminal repeats.
Analysis of sequence exchanges in exons found that at least 22 sequence exchanges extended into the intron (see above). Sequence exchanges were also investigated by comparison of intron sequences of all Type I RGC2 genes. Analysis using Geneconv, visual inspection, and DnaSP detected 0, 6, and 18 exchanges, respectively, within the intron sequences. These were distributed evenly throughout the intron sequence. No elevated frequency of sequence exchange was observed relative to the flanking exon sequences.
To determine whether the high nucleotide identities between intron 5 sequences of Type I genes were unusual or whether this was typical of all introns in Type I RGC2 genes, nucleotide identities were calculated for all introns of sequenced Type I RGC2 genes, with the exception of intron 3, which varied dramatically in size (372 to >6 kb) (Table 7). High nucleotide identities were observed only in introns 4, 5, and 6 in the 3′ half of the gene; the nucleotide identities of these introns were higher than those of their flanking coding regions. By contrast, the nucleotide identities in introns 1 and 2 were lower than in their flanking coding regions. This variation in intron similarity in different regions may reflect the variation in frequency of sequence exchanges: sequence exchanges were frequent in the 3′ LRR region but rare in the 5′ half.
Table 7.
Average Nucleotide Identities (%) at Different Regions of RGC2 Genes in Diana and Mariska
| Type | All Coding | 5′ Enda | NBS | Exon 2 LRR | Intron 2 | Exon 3 | Intron 3b | Exon 4 | Intron 4 | Exon 5 | Intron 5 | Exon 6 | 3′ Endc |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| All | 80.4 ± 4.9 | 73.1 ± 9.3 | 77.7 ± 6.7 | 81.4 ± 6.0 | 81.1 ± 7.5 | 82.0 ± 7.1 | NAd | 84.4 ± 5.5 | 70.0 ± 18.4 | 82.8 ± 6.6 | 72.8 ± 17.0 | 83.2 ± 6.8 | 72.9 ± 16.7 |
| Type I | 86.9 ± 4.4 | 81.6 ± 12.2 | 81.6 ± 8.8 | 85.9 ± 6.6 | 85.8 ± 8.3 | 88.3 ± 5.7 | NA | 91.8 ± 1.7 | 94.4 ± 2.7 | 91.1 ± 2.4 | 97.3 ± 0.9 | 90.2 ± 2.7 | 92.5 ± 4.7 |
| Type II | 77.9 ± 4.7 | 71.0 ± 7.0 | 77.0 ± 5.9 | 78.3 ± 4.9 | 78.0 ± 6.2 | 77.2 ± 5.7 | NA | 80.4 ± 4.3 | <60 | 78.4 ± 4.9 | 63.0 ± 12.0 | 78.5 ± 6.7 | 62.1 ± 12.7 |
Including intron 1, 5′ untranslated region and ∼500 bp coding region 5′ to NBS, if available.
Intron 3 is too diverse to be aligned; nucleotide identities were not attempted.
Including intron 6, exon 7, intron 7, and exon 8, if available.
NA, not available because of large sequence differences.
By contrast, intron 5 of Type II genes varied greatly. Intron 5 of 78 Type II genes ranged in size from 133 to 788 bp, with an average of 317 bp. All intron 5 sequences except that in LC11 were <412 bp and were considerably shorter than intron 5 in Type I genes (679 ± 56 bp). Intron 5 in LC11 was 788 bp, but it had no sequence similarity with other RGC2 genes. BLAST analysis of the intron sequences did not reveal significant sequence similarity to transposon sequences. Introns from the different Type II clades were difficult to align because of low sequence identity. When alignments were possible for subsets of sequences, the highest nucleotide identity between intron 5 sequence of Type II genes from different clades was only 79%, and some were <50%, which was much lower than the average nucleotide identities between the exons of different Type II clades (∼80%).
Small Indels Occurred Mainly in Regions of Type II Genes Encoding Extended LRRs
The position and frequency of indel events were determined for the 126 RGC2 fragments from the seven genotypes. Indels at the same position and with the same size were considered as being potentially derived from the same event and, therefore, only counted once. The region containing the hypervariable microsatellite MSATE6 was considered separately.
A total of 141 independent indels were identified, excluding those involving the microsatellite MSATE6. Thirteen of them were large duplications resulting from unequal crossing-over as described above. There were also three large deletions, a 1.4-kb deletion in gene TDD, a 186-bp deletion in TDAD, and a 99-bp deletion in TM7. All of these 16 indels retained the open reading frame (ORF), except for the deletion in TDD, and changed the number of encoded LRRs by at least one. All of the other 125 indels were <64 bp, with an average of 8 ± 6 bp, and did not change the number of LRRs. The majority of these small indels also retained the ORF; only five resulted in frame-shift mutations.
The frequency of each small indel varied from 1 to 48 in the 126 RGC2 fragments. All 125 small indels detected were present in the 78 Type II genes, but only 14 were present in the 48 Type I genes (Table 8). The majority of the 141 indels were present in more than one species; five indels were present in more than one fragment but only from one species, and only 23 of the 141 indels were detected just once. The existence of frequent trans-specific polymorphism for these indel polymorphisms suggested that they may be under balancing selection.
Table 8.
Distribution of Indels in 12 LRRs of Type I and Type II RGC2 Genes
| Core Section
|
Extended Section
|
|||||
|---|---|---|---|---|---|---|
| LRR No. | Type I | Type II | Alla | Type I | Type II | Alla |
| 1 | 0 | 1 | 1 | 4 | 31 | 31 |
| 2b | 0 | 1 | 1 | |||
| 3 | 0 | 5 | 5 | 24c | 32 | 51 |
| 4b | 0 | 1 | 1 | |||
| 5b | 0 | 0 | 0 | |||
| 6d | 1 | 17 | 17 | 0 | 13 | 13 |
| 7b | 1 | 3 | 3 | |||
| 8 | 0 | 2 | 2 | 6 | 26 | 26 |
| 9b | 0 | 1 | 1 | |||
| 10b | 0 | 0 | 0 | |||
| 11 | 0 | 4 | 4 | 2 | 20 | 20 |
| 12b | 0 | 0 | 0 | |||
| Totale | 2 | 35 | 35 | 12 | 90 | 90 |
Number of indels in all 126 fragments. The numbers are not always additive because some indels are present in both Type I and Type II genes.
No extended section in these LRRs.
This includes indels involving microsatellite MSATE6.
LRR 6 includes the splicing sites for intron 5 at the boundary of the core and extended sections.
Total excludes the indels at the hyperpolymorphic microsatellite MSATE6 in LRR 3.
Diversifying Selection Acting on the LRR Region
The 12 conserved (unduplicated) LRRs in the 3′ fragments amplified from the seven genotypes were analyzed using multiple models within PAML (Yang, 1997; Yang et al., 2000) to examine selective forces acting on individual sites in the LRR-encoding region. The putative solvent-exposed residues of the LRR consensus motif had previously been shown to be under significant diversifying selection in RGC2 and other resistance genes (see references above). However, most of these earlier analyses had involved pooling of sequences encoding these sites along the gene and the analysis of KA:KS ratios between paralogs. In contrast with previous studies, the availability of many sequences allowed us to study individual sites within the LRR.
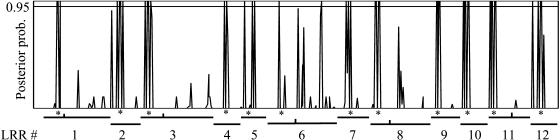
The M7 and M8 models of PAML were compared to investigate whether there were sites under diversifying selection. The likelihood-ratio test of M7 versus M8 was consistent with sites being under diversifying selection (χ2 = 1990, df = 2; P < 0.001). A total of 43 out of ∼493 sites were identified by model M8 as being under diversifying selection (P < 0.01; Figure 9). The 10th and 11th residues in most LRRs were clearly under diversifying selection (Figures 4 and 9). The 8th, 13th, and 14th residues in some LRRs, but not all, were also under diversifying selection. These inferences of diversifying selection have the caveat that recombination and gene conversion may have resulted in overestimations of positive selection (Anisimova et al., 2003).
Figure 9.
Distribution of Sites Identified as Being under Diversifying Selection within the 3′ LRR Region.
The positions of 12 LRRs amplified from the 3′ end of RGC2 genes from Lactuca spp are indicated below. The ticks in LRRs 1, 3, 6, 8, and 11 separate the core sections and extended sections. The positions of the hypervariable, putative solvent-exposed regions are indicated by asterisks.
The 3′ Half of Type I Genes Has Evolved Differently from the 5′ Half
To determine whether the 3′ half had undergone more frequent sequence exchanges relative to other regions of the gene, sequence exchanges and their distribution were analyzed between 26 RGC2 genes from cv Diana and four genes from Mariska for which we had full-length or near-full-length sequences (this article; T. Wroblewski and R. Michelmore, unpublished data). Of these, 17 (57%) were Type I, and 13 (43%) were Type II genes, based on the criteria described above. Because Geneconv ignores sites with indels, several alignments were examined to maximize the detection of sequence exchanges. Initially, 22 genes with no large indels were analyzed, and then genes with progressively larger indels were included to detect additional sequence exchanges. A total of 98 sequence exchange events were detected (P < 0.05). Ninety-one of the 98 (93%) sequence exchange events were between two Type I genes.
Eighty-two of the 98 sequence exchanges occurred in the 3′ half of the gene. The length of sequence exchange in the 3′ half was usually small, ranging from 72 to 392 bp with an average of 194 ± 68 bp, which is consistent with the average exchange length (203 bp) detected in the 126 RGC2 3′ fragments. By contrast, only 16 sequence exchanges were detected within the 5′ half of the gene, and most of them were large. Eight of the 16 sequence exchanges extended from the beginning of the gene to exon 4, and two of them were greater than 3 kb. If these eight events are considered to have resolved in the 3′ half, only eight (8%) sequence exchanges resolved within the 5′ half of the gene. Sequence exchange within the first 3 kb of seven genes (TDAB, TDAF, TDG, TDH, TDI, TDJ, and TDV) made these Type I genes highly conserved in the 5′ region, with nucleotide identities >96%. There were 3.7-kb sequence exchanges among genes TDA, TDE, and TDN at the 5′ end. Sequence exchanges between genes TDB and TDS and between genes TDC and TDD were 8.0 and 3.9 kb, respectively. Considering that these sequence exchanges are large and extend to the 5′ end, they might be the results of intergenic recombination. Alternatively, these genes might have been created by duplication followed by rapid divergence of their 3′ halves as a result of extensive gene conversions, whereas their 5′ half evolved slowly and remain highly conserved.
DISCUSSION
The distribution of variation in a genome is the result of an intricate interplay between mutation, recombination, selection, and demography and is influenced by the reproductive system and ecological constraints. Because of the complexity of their genetic structure and the potential diversity of selective forces acting on them, clusters of resistance genes represent both an opportunity and a challenge to determine the relative importance of each of these factors in the evolution of disease resistance.
This article describes the most comprehensive analysis to date of diversity at a cluster of resistance gene candidates in plants. Sequence analysis of 126 LRR sequences revealed a wide variety of genetic events that have resulted in a large number of diverse RGC2 genes. Variation occurred in gene copy number, LRR repeat number, and sequence. Point mutation, insertion, and recombination have all been important, but the position at which they occur in the gene and their impact has been different at different times in the evolution of RGC2 genes. Two distinct subsets of resistance gene candidates (designated Type I and Type II) within Lactuca spp exhibited different patterns of evolution in several respects (summarized in Table 9).
Table 9.
Summary of Differences between Type I and Type II RGC2 Genes
| Type I Genes | Type II Genes | |
|---|---|---|
| Location | Middle of Cluster | Edges of Cluster |
| Sequence identity | Medium | High within clade, but low between clades |
| Orthologous relationship | Not obvious | Obvious |
| Gene conversion | Frequent | Rare between different clades |
| Indels | Rare | Frequent between different clades |
| Selection | Diversifying | Purifying within clades |
| Frequency in nature | Low | Variable, mostly high |
| Intron nucleotide identity at 3′ end | Higher than flanking coding region | Very low between different clades |
| Rate of evolution | Rapid | Slow |
Methodological Considerations
The initial use of universal followed by more specific oligonucleotide primers resulted in the amplification and effective sampling of most but not all genes in a haplotype. Although not all RGC2 genes were amplified from all genotypes, there was no evidence of bias in our sampling. Multiple independent amplifications from each genotype provided robust data that identified single nucleotide differences with confidence. The isolation and comparison of large numbers of genes from multiple genotypes allowed more detailed analyses than were possible in previous comparisons that were usually between paralogs from one or a small number of haplotypes (e.g., Meyers et al., 1998b; Noël et al., 1999). In addition, previous studies have involved smaller clusters of R-genes; the large number of RGC2 genes within a haplotype made the different patterns of variation obvious.
There are several challenges to analyzing highly variable data from an inbreeding species. The reality analyzed here—a large locus of variable size with multiple alleles and paralogs potentially under selection, some of which exhibit high rates of gene conversion—is much more complicated than the situations modeled to date (Wiuf and Hein, 2000; Innan, 2002). Therefore, some of the specific values must be treated with caution. However, phylogenetic analysis was a useful tool to look for patterns of variation that revealed differences between two sets of sequences and allowed us to erect the hypothesis of two types of RGC2 genes that was subsequently validated by independent approaches.
Variation in Gene Copy Number
The numbers of RGC2 paralogs can vary greatly within and among these three species of Lactuca. Minimum estimated copy numbers varied from 12 to 32, with most genotypes having ∼20 RGC2 genes. The majority of copy number changes involved Type I genes. This is consistent with the involvement of Type I genes in frequent interparalog recombination. Resolution of such recombination complexes as a crossover would have resulted in changes of copy number, whereas noncrossover events would have resulted in gene conversions.
Lettuce cv Diana has at least 32 RGC2 genes. The large number of resistance gene candidates in Diana was most likely created by unequal crossing-over resulting in the amplification of only Type I genes in the center of the cluster because only one Type II clade (Q clade) has more than one copy from Diana. There are no obviously close pairs of Type I genes in Diana, implying that the Type I genes originated from different haplotypes or that the expansion and variation of Type I genes in cv Diana were created by unequal crossing-over followed by frequent gene conversions in the LRR region. Interestingly, the experimentally selected spontaneous losses of Dm3 specificity (Chin et al., 2001) involved deletions of Type I genes but not Type II genes, possibly indicating a reversal of the expansion process.
There are few data on the variation of R-gene copy number in other species. In Arabidopsis, deletions of resistance gene sequences have been documented at complex and single copy loci (McDowell et al., 1998; Stahl et al., 1999; Tian et al., 2002). Copy number variation (one to five copies) of Hcr9 homologs was observed at the Milky Way locus in three species of Cladosporium (Parniske and Jones, 1999). The copy numbers at the Rp1 locus varied from 1 to 15 in different maize cultivars (Collins et al., 1999; Sun et al., 2001; Ramakrishna et al., 2002). However, little variation of copy number has been observed in limited studies of several R-gene loci, such as RPP5 (Noël et al., 1999), N, and P in flax (Dodds et al., 2001a, 2001b).
Our observations of long-term variation in copy number are consistent with short-term evolutionary studies. Four of 167 recombination events at the RGC2 locus selected on the basis of flanking marker exchange occurred within the cluster of RGC2 genes and therefore resulted in haplotypes with nonparental copy numbers of RGC2 genes (Chin et al., 2001). Eleven out of 12 spontaneous mutants selected for the loss of Dm3 specificity carried deletions at the RGC2 locus (Chin et al., 2001). These were isolated from screens of 11,000 homozygous S2 families and 16,500 heterozygous F1 plants. Each lettuce plant can produce several thousand seeds. Therefore, it is likely that a few variant haplotypes are generated by every plant in each generation.
The observed variation in copy number is an integral component of the birth-and-death model of resistance gene evolution and is similar to that observed in vertebrate multigene families (Nei et al., 1997; Michelmore and Meyers, 1998; Piontkivska and Nei, 2003). Unequal crossing-over will generate variant haplotypes with deletions and duplications. The lack of representation of all genotypes in each of the Type II clades may be the consequence of stochastic processes or a lack of selective advantage. Changes in the number of R-genes maintained within a species may be caused by life histories that alter Ne, the effective population size, or by nonrandom loss of paralogs because of the heterogeneity of the fitness values of individual paralogs. Random duplications and losses of paralogs will be favored by situations that reduce Ne, such as those resulting from short-lived inbreeding populations inhabiting disturbed sites that are characteristic of Lactuca spp. Species that do not undergo genetic bottlenecks and retain a high Ne may be less susceptible to stochastic losses of individual genes. In such species, maintenance of individual genes will be more influenced by selective advantage or proximity to a paralog under selection.
Variation in LRR Number
Eighteen of the 28 full-length (or near-full-length) RGC2 genes from Diana and Mariska had 42 LRRs. This is a much larger number of repeats than present in R-genes in other species. The number of LRRs in both TIR and non-TIR R-genes in Arabidopsis averaged 14 with a range of 8 to 25 (Meyers et al., 2003). Most other R-genes are similar in size to the Arabidopsis genes; for example, Mi and I2 in tomato (14 and 17 LRR repeats; Milligan et al., 1998; Simons et al., 1998), P in flax (16 LRR repeats; Dodds et al., 2001b), and Rp1 in maize (24 LRR repeats; Collins et al., 1999). The second largest LRR reported to date is L in flax with 26 repeats. The significance of the numerous LRRs in RGC2 genes remains to be determined.
The remaining 10 full-length RGC2 sequences varied from 40 to 47 LRRs. Most of this variation occurred in the C-terminal half of these sequences. In the C-terminal region studied in detail from multiple genotypes, 91 (72%) had 12 LRRs, and the remaining 28% varied by one to five LRRs from the mode of 12. Most had additional LRRs as a result of unequal crossing-over with a paralogous gene. In nearly all cases, the changes in LRR number preserved the ORF and involved the exchange of complete LRR units such that the predicted three-dimensional structure of the region would be maintained; therefore, the predicted recombinant proteins are potentially functional. Consequently, although LRR number appears to be fairly stable and only varies within a narrow range, it is not fixed and variation in LRR number provides a mechanism for generating potentially radically different binding surfaces.
Variation as a Result of Small Indels
Two distinct size classes of indels were observed in RGC2 genes. Sixteen large indels altered the number of encoded LRRs as discussed above. The remaining 125 indels were small and encoded just a few amino acids. The majority of these small indels maintained the ORF; only five resulted in frame-shift mutations. The mechanism(s) that generated these indels is unclear. Similar in-frame (triplet) indels have been found in RPP13 homologs in Arabidopsis and Mla genes in barley (Hordeum vulgare) (Bittner-Eddy et al., 2000; Wei et al., 2002). The small RGC2 indels occurred preferentially in the extended section of the longer LRRs. This nonrandom distribution could be because of either tolerance of indels in these regions or selection. Several indel polymorphisms were trans-specific and occurred in multiple Type I and Type II genes, suggesting that some at least were ancient and may have been maintained by selection. The location of the small indels in the extended regions of the LRRs rather than the putative solvent-exposed surface implies that they may change the orientation of the binding surface(s) rather than primarily encoding residues that are involved in binding (Michelmore and Meyers, 1998; Mondragón-Palomino et al., 2002).
Variation Has Been Frequently Generated by Sequence Exchanges between Type I Genes
Although point mutation has been the ultimate source of variation, sequence exchange has been mainly responsible for the evolution of sequence diversity of RGC2 genes. Although gene conversion had been recognized within clusters of resistance genes in several species, its relative importance as a mechanism for generating variation and frequency was unclear (Michelmore and Meyers, 1998; Noël et al., 1999; Dodds et al., 2001a; Van der Hoorn et al., 2001). Our data demonstrates that gene conversion is a major driver for variation in RGC2 genes.
The most frequent genetic event generating variation in Type I RGC2 genes was sequence exchange between paralogs without alteration in the LRR number. Each Type I gene was a chimera of on average three segments over the ∼1.2-kb LRR region analyzed. Therefore, the LRRs were derived from numerous different genes, and the genealogy of these sequences is reticulate rather than monophyletic. Consequently, the topology of the tree shown in Figure 5 serves only to indicate their distinction from Type II sequences and does not reflect the evolutionary relationship between Type I genes. The length of the sequence exchanged varied from 60 to 528 bp, encoding one to three LRRs. Trees based on ∼100-bp sections (e.g., Figure 7) were consistent with a monophyletic origin of individual LRRs.
This combinatorial mechanism can readily generate a great variety of binding surfaces. If each LRR unit is considered as a quasi-independent evolutionary unit (as supported by the topologies of trees derived from individual LRRs; e.g., Figure 7), then there are potentially Σ = x1·x2…xn combinations, where x is the number of different sequences at each LRR position and n is the number of LRRs. Analysis of all 111 Type I genes showed that each LRR had more than 10 different sequences, any pair of which differed by at least 5% nucleotide substitutions and at least one amino acid change at the hypervariable sites. Therefore, more than 1012 different LRR combinations could be created by sequence exchange from this region alone, which represents only a fraction of the LRRs in RGC2 proteins.
Gene conversions in the LRR region may occur fairly frequently in nature. One of the spontaneous Dm3 mutations identified from a screen of 11,000 homozygous S2 families of Diana was a gene conversion of TDB (RGC2B) by a 1.5-kb sequence from the 3′ LRR of TDC (Chin et al., 2001). One individual plant of lettuce can produce thousands of seeds. Therefore, every three to four plants may produce one chimeric gene every generation.
The sequence exchanges detected in this study were most likely caused by gene conversions rather than crossovers. Some genes have identical or near identical sequences in several LRRs at the periphery of the amplified region but with distinct LRRs in the middle. Gene conversion may be more important than crossing-over in generating novel intragenic variation (Berry and Barbadilla, 2000). There is very little data for any organism on the sizes of gene conversion tracts and the relative frequencies at which recombination events are resolved as crossovers versus gene conversions. Sizes of gene conversion tracts at the bronze locus in maize are ∼1 kb (Dooner and Martinez-Ferez, 1997). Most of the higher eukaryotic data comes from Drosophila melanogaster, where gene conversion was estimated to be approximately four times more likely than crossing-over and the mean conversion tract length was 352 bp, similar to that observed within the 3′ region of Type I RGC2 genes (Hilliker et al., 1994). Conversion is five times more important than crossing-over for regions shorter than 352 bp and remains the main force disrupting linkage up to 1760 bp; only at longer distances was crossing-over more important (Hilliker et al., 1994; Haubold et al., 2002; Berry and Barbadilla, 2000). Therefore, gene conversion rather than crossing-over may be the predominant force influencing linkage of polymorphisms within RGC2 genes, whereas crossing-over may be almost exclusively responsible for breaking down the linkage disequilibrium between paralogs. This potential for decoupling of intragenic and intergenic rearrangements may have important evolutionary consequences. Genes under selective pressure for high variability, such as MHC loci (Hughes et al., 1993) and R-gene clusters, may undergo elevated rates of gene conversion relative to crossing-over. The elevated frequency of conversion relative to crossovers and the length of the conversion tract may have been selected at R-gene clusters to maximize the rate at which useful new combinations are generated.
Frequent Sequence Exchanges Homogenize Intron Sequences but Not Exon Sequences
Frequent sequence exchange tends to homogenize members in gene families (Ohta, 1983). Such concerted evolution is well documented for genes that encode abundant homogeneous proteins, for example, the vertebrate globin genes (Scott et al., 1984). However, the roles of unequal crossing-over and gene conversion in the evolution of other multigene families, such as at the MHC locus, are less clear (Ohta, 1991; Li, 1997; Nei et al., 1997; Takahata and Satta, 1998). The 3′ exons of Type I RGC2 genes are highly variable and exhibit no evidence for homogenization. The tendency of frequent sequence exchanges to homogenize sequences may be counterbalanced by diversifying selection, which has been indicated at hypervariable sites within the LRR consensus motif for RGC2 and many other R-genes.
Even though the frequent sequence exchanges between paralogs did not homogenize the coding region of Type I RGC2 genes, it did apparently homogenize their introns. The three introns in the 3′ half of the gene were more similar than the flanking exons. Homogenization of intron sequences has also been observed in the MHC and HLA gene families, which had frequent sequence exchanges and showed diversifying selection (Cereb et al., 1997; Hughes, 2000). Unlike the MHC and HLA families in mammals and RGC2 genes in lettuce, most genes typically have diverse introns in which the mutation rates are similar to Ks in their flanking exons (Hughes, 2000). Homogenization of RGC2 intron sequences probably occurred as a result of frequent sequence exchanges and genetic drift as well as a lack of selective constraint. High intron similarity, in turn, may facilitate sequence exchange.
Rates of Recombination Are Low and Heterogeneous within the RGC2 Cluster
Although gene conversion and unequal crossing-over are clearly important in generating variation at resistance loci, clusters of resistance genes are not hot spots for recombination. The absolute rates of recombination across clusters of resistance genes are low rather than elevated. The Dm3 region exhibited a recombination frequency 18-fold lower than the genome average, and no crossovers within RGC2 coding regions have been detected experimentally (Chin et al., 2001). Similarly, recombination rates across the Rp1 cluster in maize and the Mla cluster in barley were lower than the genome averages (Wei et al., 1999; Ramakrishna et al., 2002).
Mating system will also have a large impact on the pattern of recombination. In inbreeding species such as lettuce, haplotypes will rapidly become homozygous, reducing opportunities for recombination to rearrange alleles. Rare outcrossing and low recombination rates may provide few opportunities for the generation of novel haplotypes. Consequently, mis-pairing and sequence exchange between paralogs that seem to occur at a significant frequency within homozygotes (Chin et al., 2001) may be the predominant route for the generation of novel Type I genes. Conversely, in outbreeding species such as maize, novel haplotypes will rarely become homozygous when at low frequency; therefore, the pattern of recombination and distribution of polymorphism may be different.
Recombination events, crossovers, and gene conversions also seem to be distributed unequally within RGC2 genes. Higher rates of gene conversion were observed at the 3′ end rather than in the 5′ region. This could be because of structural or selective reasons. Differences in apparent recombination rates may actually reflect selection at linked sites. Strong selection resulting in a selective sweep and the rapid fixation of an allele or haplotype could result in an apparent decrease in recombination (Navarro and Barton, 2002). Also, background selection in which deleterious alleles are continuously removed from populations will decrease linked neutral variation (Tenaillon et al., 2002). Alternatively, sequence exchanges in the 3′ half may be tolerated, whereas recombinants in other parts of the gene may be selectively disadvantageous. If only certain recombinant molecules are functional or certain combinations of sequences are deleterious, only a subset of recombinant events will persist and, therefore, result in apparent recombinant hot spots. This may be the case with NBS-LRR encoding genes if some combinations of NBS and LRR domains are functional or lethal (Hwang et al., 2000; Luck et al., 2000). Conversely, balancing selection slows the coalescence of alleles and, hence, extends the time during which recombination can have an effect (Schierup et al., 2001). Therefore, even low rates of recombination can have a profound effect on allelic genealogy and recombination is expected to have happened with an inflated frequency close to a site(s) under balancing selection. Consequently, the selected position appears as an apparent recombination hot spot as may be the case with the 3′ ends of RGC2 genes.
Transitions between Type I and Type II Modes of Evolution
We hypothesize that Type I genes are stochastically converted into Type II genes and then Type II genes evolve independently. Type II genes will continue to evolve and be maintained if selectively advantageous or will disappear from the population because of purifying selection or drift if selectively neutral, consistent with the birth-and-death model (Michelmore and Meyers, 1998).
The events that cause the transition from Type I to Type II evolution are unknown but are likely to involve factors that reduce the tendency of paralogs to pair at meiosis. The frequency of pairing and sequence exchange between paralogs will be influenced by structural and sequence (dis)similarities. Therefore, the transitional event from Type I to Type II behavior may be an insertion or deletion event that reduces sequence exchange, which in turn allows more divergence that further represses mis-pairing and sequence exchange. Rapid divergence between repeated sequences has been observed and is believed to reduce illegitimate recombination between paralogs (Kricker et al., 1992). Transposon-mediated insertions or deletions in regions flanking RGC genes would also result in structural hemizygosity that could reduce pairing between paralogs. RGC2 genes are on average more than 100 kb apart, and many of the intervening sequences are retrotransposon related (Meyers et al., 1998a).
The Rate of Evolution
Previous claims of rapid evolution of resistance genes based on sequence analysis have relied on comparisons between paralogs within one or a few genotypes or comparisons between different genera. In contrast with these studies, our databases on many comparisons within closely related species show that many of the genetic events appear to be ancient as evidenced by trans-specific polymorphisms within Lactuca spp. Large and small indels as well as sequence polymorphisms were often present in multiple species and multiple clades. Six of the 13 large unequal crossing-over events that changed the numbers of LRR clearly occurred before speciation as did many of the small indels. It was impossible to determine the timing of the remaining seven large insertions because they were detected in single Type I genes. Sequence polymorphisms between the K alleles/orthologs were conserved across species. The rate of evolution therefore seemed to be slow, even at the hypervariable sites.
The most recent events seem to be changes in copy number and sequence exchange between Type I genes. The diversity of MSATE6 profiles of many genotypes and representation of genes in the genotypes studied here indicates that copy number of both Type I and Type II genes and, therefore, the composition of individual haplotypes is changing rapidly within each species (Sicard et al., 1999). The rarity of individual Type I genes and their obvious chimeric nature as well as the experimental detection of gene conversion indicates that sequence exchanges are occurring fairly frequently (Chin et al., 2001). Therefore, the RGC2 cluster is evolving rapidly in terms of its composition of Type I and Type II genes as well as the specific combinations of LRRs encoded by Type I genes.
Selective Forces Acting on R-Genes
A cluster of resistance gene candidates, such as the RGC2 locus, will be subject to a complex variety of overlapping selective forces. Purifying, diversifying, and frequency-dependent balancing selection are all likely to play a role in the evolution of an R-gene cluster. The consequences of their action are complicated by their effects on the whole haplotype rather than individual genes. The dynamics of specific paralogs and of individual haplotypes will be increasingly complex as the number of paralogs that are under selection increases. This is compounded further by heterogeneity in space and time of the biotic and abiotic environments, resulting in heterogeneities in the selection pressures.
Density-dependent balancing selection has been invoked as being responsible for maintaining diversity at several R-gene clusters (Michelmore and Meyers, 1998; Stahl et al., 1999; Tian et al., 2002). The presence of multiple polymorphisms in a population could be because of a lack of constraint and, therefore, tolerance of multiple residues, transitory polymorphism on its way to fixation, or balancing selection. Most tests for detecting balancing selection have been developed for large randomly mating populations, and population genetic theory is not developed well for inbred species (Li, 1997). However, the presence of numerous types of trans-specific polymorphisms, particularly at the putative solvent surface, is consistent with a significant impact of balancing selection on RGC2 haplotypes.
Balancing selection may be a result of heterogeneity in selection intensity because of a variety of factors. In the case of R-genes, it is likely that the effectiveness of a resistance specificity is directly related to the frequency of its corresponding avirulence factor in the pathogen population (McDonald and Linde, 2002). This will tend to set up cycles of density-dependent balancing selection because the efficacy of the resistance specificity decreases as its frequency increases, reflecting a decrease in the cognate avirulence allele. Balancing selection will also be caused by qualitative and quantitative heterogeneties in the pathogen over space and time. Different resistance specificities may be effective against different components of a single pathogen or against multiple pathogens. Several Dm specificities effective against B. lactucae as well as resistance to root aphid map to the RGC2 region (Farrara et al., 1987; Bonnier et al., 1994; T. Nakahara and R. Michelmore, unpublished data). Also, pathogens rarely exert a constant disease pressure because of stochastic, epidemiological, and climatic factors.
Haplotype Selection
The frequency of resistance specificities will be determined by the complex interaction of the above selective forces acting on the haplotype as a whole (Michelmore and Meyers, 1998). Selection will act on individual genes within a haplotype, but the frequency of the whole haplotype will depend on the sum of the effects of selection acting on all genes in the region. Linkage drag within genes and across the whole haplotype will result in linkage disequilibrium and in the maintenance of a reservoir of polymorphism within each haplotype that is not selected directly. Balancing selection acting on different members of a haplotype will maintain polymorphism within a species.
The rates of recombination across the cluster and the strength of selection will determine the amount of linkage disequilibrium within orthologs and between paralogs. The low rate of recombination across the RGC2 cluster is consistent with significant linkage disequilibrium across the locus (Sicard et al., 1999; Chin et al., 2001). The low rate recombination and linkage drag could explain the elevated KA:KS, even in Type II pseudogenes, observed in RGC2 and other R-genes (Noël et al., 1999). However, there has been some recombination between K alleles, and more detailed analysis of natural populations is required to determine the extent of linkage disequilibrium. Selection on only one gene at any one time will result in gradient of selection across the whole haplotype. However, different paralogs may be under selection by the same or different pathogen populations at different times, resulting in balancing selection on the haplotype as a whole. Therefore, the effect of selection is complicated when balancing selection acts to maintain alleles at two (multiple) loci.
Evidence for Heterogeneous Evolution in Other Species
There is little data on sequence diversity at R-gene clusters from multiple closely related genotypes in other genera. At RPP5 in Arabidopsis, one pair of fragments that were located at the periphery in Columbia-0 and Landsberg erecta showed obvious allelic relationships, whereas other homologs showed extensive sequence exchange (Noël et al., 1999). The best data for heterogeneous modes of evolution are from the N locus in flax, at which two paralogs appeared to be evolving independently, whereas two other paralogs had been involved in frequent sequence exchange (Dodds et al., 2001a). Comparable studies of other species in which clusters are characterized from multiple haplotypes are required to determine the prevalence of Type I and Type II genes.
Conclusions
Diversity in resistance gene candidates is shaped by the interaction of multiple normal genetic processes acting at a variety of times and in a variety of combinations to generate diversity rather than by novel genetic mechanisms specific to R-genes.
Gene copy number varies considerably between haplotypes consistent with a birth-and-death model of resistance gene evolution.
RGC genes within the same cluster can exhibit heterogeneous rates of evolution. One subset of genes evolves rapidly via sequence exchange between paralogs. A second subset evolves more slowly, independently of other paralogs.
There are two classes of indel events. Large indels involve one or more LRRs. Small indels involve a few amino acids and tend to occur in the backbone rather than the putative solvent exposed surface.
Rates of recombination and point mutation are not high in either type of RGC gene.
Rates of gene conversion appear to be elevated relative to crossovers. Conversion tracts approximately match regions encoding LRRs. The pattern and heterogeneity of variation suggests that the putative solvent exposed surfaces of β-sheet/β-turn of the LRRs are maintained, but their steric relationships may not be.
METHODS
Plant Materials
RGC2 genes were cloned from three lettuce cultivars of Lactuca sativa (Diana, Mariska, and Calmar), one wild accession of L. serriola (W66336A), and three wild accessions of L. saligna (US93UC-10, CGN9311, and PI491204). Details of the origins of each accession are described in Supplemental Table 1 online. DNA was extracted from a bulk of at least 20 plants for each accession using a modified CTAB procedure as described previously (Bernatzky and Tanksley, 1986). Cultivar Diana has resistance gene Dm3, and its RGC2 locus has been studied extensively (Meyers et al., 1998a, 1998b; Shen et al., 2002). Cultivar Mariska has Dm18, which is tightly linked to the RGC2 locus and has been introgressed from L. serriola (Maisonneuve et al., 1994). Cultivar Calmar contains no known resistance specificities encoded at the RGC2 locus, although it has known Dm genes elsewhere in the genome (Farrara et al., 1987). The L. serriola genotype and three L. saligna genotypes are resistant to all isolates of Bremia lactucae tested (at least 10 from diverse sources) and have been used in our breeding program as sources of resistance (O. Ochoa and R. Michelmore, unpublished data).
A panel of 40 additional wild accessions were also included in this study to investigate the frequencies of RGC2 genes/fragments. These included 33 accessions of L. serriola, six accessions of L. saligna, and one accession of L. perrenis. Nine of the 33 accessions of L. serriola were from a single population at Bolu, Turkey, and the others were randomly chosen from different populations. Details of the origins of each accession are described in Supplemental Table 1 online.
Sequencing of Single-Copy Nonresistance Genes
Six single-copy genes that were not resistance genes were chosen as reference genes for comparative purposes. A single copy of each gene was present in the genome, based on the DNA gel blot profiles of more than 30 genotypes of Lactuca spp after digestion with multiple endonucleases (Kesseli et al., 1994). These restiction fragment length polymorphism probes were derived from cDNA sequences from L. serriola (Tables 1 and 5; Kesseli et al., 1993). The six genes were at least 20 centimorgans away from the RGC2 locus and from each other (Kesseli et al., 1994). TBLASTX searches indicated that they encode proteins with a range of functions, none of which were obviously involved in plant–pathogen interactions (Table 5); therefore, these genes are likely to be under selective forces independent of those acting on resistance genes. PCR amplification and sequencing were performed as below.
Sequencing of RGC2 Paralogs in cv Diana
Before this study, numerous BAC and λ-phage clones containing RGC2 genes from L. sativa cv Diana had been identified, and nine RGC2 genes had been sequenced completely (Meyers et al., 1998a, 1998b). For our study, conserved oligonucleotide primers were designed throughout the RGC2 gene using the sequence information from these nine genes (GenBank accession numbers AF072267 to AF072275). These primers were used to amplify PCR products from additional RGC2 genes from cv Diana using DNA of the RGC2-containing BAC or λ-phage clones as template. The PCR products were treated with exonuclease I and shrimp alkaline phosphatase (U.S. Biochemical, Cleveland, OH) and then sequenced directly using the conserved primers. When necessary, additional sequencing was performed using primer walking. If more than one gene was present on a single BAC clone, gene-specific primers were used. To minimize sequencing errors and estimate error rates, all regions were sequenced at least twice from different PCR products.
Isolation of 3′ RGC2 Fragments from cv Diana Using PCR
To validate our PCR-based approach to assay variation, RGC2 fragments were amplified from cv Diana. Oligonucleotide primers, Ex4b and Ex5r1, were designed to amplify diverse RGC2 homologs based on conserved sites within the 3′ LRR region of the RGC2 genes in cv Diana (Figure 1, Table 1). PCR was performed with the Advantage HF 2-PCR kit following the manufacture's instructions (Clontech, Palo Alto, CA). Four independent reactions were performed for each genotype. The first three reactions were amplified in parallel with 2 min at 94°C, followed by 17 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min. The fourth reaction was amplified in parallel under the same conditions as above except that 25 cycles were performed. All PCR products were fractionated in 1% agarose gel. The lanes containing 25-cycle reactions were excised from the gel, visualized with ethidium bromide under UV light, and the positions of the products marked. Corresponding PCR products from the three 17-cycle reactions were excised from gel, purified using the gel purification kit (Qiagen, Hilden, Germany), and cloned into pCR2.1 vector using the TOPO TA cloning kit (Invitrogen, Carlsbad, CA).
PCR products from the three independent amplifications were cloned, and 182 clones were sequenced; 158 (87%) were RGC2 sequences representing 22 different RGC2 genes. Two genes were each represented by only one clone (singletons), whereas 28 clones were derived from one gene (TDD, a probable pseudogene with large deletion between Ex4b and Ex5r1). Sequences of the 22 RGC2 genes were compared with the sequences obtained previously from genomic libraries. Twenty of them had been identified previously, whereas two did not match any previously cloned RGC2 genes. To determine if these new sequences were genuine, previously unidentified RGC2 genes or PCR artifacts, primers specific to each were designed and found to successfully amplify genomic DNA from Diana.
The sequences of RGC2 genes obtained by PCR were compared with those obtained previously from genomic libraries to estimate the error rate. The overall average nucleotide mismatch rate was 1 per 10 kb. Mismatches occurred only in singletons and were not observed in different PCR reactions, suggesting that the mismatches were attributable to PCR and that the sequencing of RGC2 genes from genomic libraries was highly accurate. Furthermore, errors could be detected and corrected because three independent PCR amplifications were performed for each genotype. These data validated our approach as an accurate method for amplifying a fragment from the 3′ LRR region of the majority of RGC2 genes in cv Diana.
Cloning and Sequencing of 3′ Fragments of RGC2 Genes from Six Additional Lactuca Genotypes
The 3′ fragments of RGC2 genes were amplified from genomic DNA of six additional Lactuca genotypes using the conserved oligonucleotide primers Ex4b and Ex5r1. Approximately 40 random clones were initially sequenced from each of the three independent PCR reactions for each Lactuca genotype (∼120 total/genotype) using primer Ex4b. Sequences were compared using Sequencer (Genecodes, Ann Arbor, MI). Sequences were considered to be from the same gene if fewer than two nucleotide differences were present, unless these differences were observed from multiple PCR reactions. For each genotype, additional clones were sequenced until there are no or very few singletons. Three clones for each group of identical sequences in each genotype (if possible, one from each PCR amplification) were then sequenced completely in both directions.
Sequences amplified from different PCR reactions for the same genotypes were compared to estimate the error rate. This varied from 1 per 2 kb to 1 per 20 kb for the six genotypes (Table 2). The reason for this variation is unclear. Nevertheless, even the highest error rate of 1 per 2 kb in CGN9311 was tolerable for the purposes of our study. Furthermore, all errors occurred only in a single clone; therefore, in all but four cases, the errors could be detected and corrected by comparing the clones from the three independent PCR amplifications. In four cases, there were only two clones for that sequence; therefore, corrections could not be made.
Nomenclature of RGC2 Sequences
RGC2 sequences amplified from the seven initial genotypes were named to reflect their species and genotype of origin. The first letter indicates the species and the second the specific accession followed by a number (or letter). The names of the RGC2 genes from L. serriola W66336A were prefixed with RA, RB, or RAB because of heterozygosity (see Results for details); from L. saligna with LA for CGN9311, LB for PI491204, and LC for US93UC-10; from L. sativa with TM for cv Mariska and TC for cv Calmar. Genes from cv Diana were designated as TD followed by one or two letters rather than numbers. Previously, RGC2 genes from cv Diana were named with a RGC2 prefix followed by a letter (Meyers et al., 1998a; Chin et al., 2001). The letter after TD in this article corresponds to the letter after the RGC2 gene names used previously. For example, gene TDB in this article refers to the gene previously referred to as RGC2B.
When describing these genes in the context of other non-RGC2 sequences they should be referred to with a RGC2- prefix, for example, RGC2-RA21; for the purpose of brevity, the RGC2- prefix has been omitted throughout this article.
DNA Gel Blot Analysis
To estimate the number of RGC2 genes in a genome, DNA gel blot analysis was conducted according to the standard protocol (Sambrook et al., 1989). Genomic DNA was digested with HindIII endonuclease (New England Biolabs, Beverly, MA). Fragments were amplified using primers Ex4b and Ex5r1 from 20 diverse RGC2 genes in Diana and then pooled and labeled with 32P using the random-primer method (MultiPrime; Amersham, Buckinghamshire, UK) and used as the probe. Final washes were conducted at 65°C in 0.1% SDS and 1× SSPE (sodium sodium phosphate EDTA).
Microsatellite Analysis
Primers 5E6 and 3E6 flanking the complex trinucleotide repeat MSATE6 within exon 5 of RGC2 were used to amplify this microsatellite marker from the majority of RGC2 genes (Meyers et al., 1998a; Table 1). PCR was performed in 20 μL containing 2 mM MgCl2, 0.25 mM deoxynucleotide triphosphate, 50 mM KCl, 10 mM Tris, 1 unit of Taq, 50 ng of genomic DNA, 0.5 μM primer 3E6, and 0.05 μM primer 5E6 labeled with 1 μCi [γ33P]ATP. PCR products were resolved electrophoretically in a 5% polyacrylamide gel.
Sequence and Phylogenetic Analyses
DNA alignments were made using ClustalX (Thompson et al., 1994) and refined by eye using Genedoc (http://www.psc.edu/biomed/genedoc/). Neighbor-joining trees using Kimura's two-parameter model and maximum parsimony phylogenetic trees were constructed and bootstrap numbers were calculated using PAUP*4.0 (Sinauer Associates, Sunderland, MA). Nucleotide identity between two sequences was also calculated using PAUP*4.0. Diversifying selection was investigated using PAML (Yang, 1997; Yang et al., 2000). Models M7 and M8 in codeml of PAML were run for all RGC2 fragments obtained using the primer combination Ex4b and 5r1. Model M7 is a special case of Model 8 that assumes no selection, whereas Model 8 allows for positively selected sites (Yang et al., 2000). Diversifying selection was confirmed using a likelihood-ratio test by comparing the likelihood of models M8 and M7 (Yang et al., 2000).
Four methods were used to investigate sequence exchange between RGC2 genes because no one test is completely reliable under all genetic scenarios for detecting recombination (Posada et al., 2002). The first used the program Geneconv (Sawyer, 1989) that detects identical tracts within two otherwise divergent fragments. Analyzing all sequences simultaneously with Geneconv was not successful because there were too many indels in the alignment, which are ignored by Geneconv. Sequences with large deletions were therefore removed from the alignment before running Geneconv; no mismatch was allowed (gscal = 0), and only events with P < 0.05 were reported. Events detected by Geneconv were examined and confirmed visually. If one gene was detected as having gene conversions with several different genes at the same region, only the event with the largest conversion tract was reported. If several conversion events were detected between two sequences and the converted tracts differed by only one base, they were considered to be the result of a single gene conversion event followed by point mutations within the converted tract. The second method used the four-gamete method in DnaSP (Hudson and Kaplan, 1985; Rozas and Rozas, 1999). Recombination was indicated if four gametes for two polymorphic sites were present in four different sequences; this is an informative method but inevitably underestimates recombination as a result of the loss or lack of observation of one or more products of recombination (Berry and Barbadilla, 1999). The third method compared distance trees constructed using different sections of the sequences. The aligned sequences were divided into sections (such as different exons or regions encoding individual LRRs), and distance trees were constructed for each section. Large differences in position in different trees were indicative of sequence exchange. The fourth method relied on visual inspection. Sequence exchange was detected as divergent tracts between two otherwise conserved fragments or as a conserved tract between two otherwise divergent fragments. Only polymorphic sites were considered so that highly conserved regions were not falsely detected as sequence exchanges. Together, these methods provide robust tests for the occurrence of recombination; however, all methods tend to underestimate the extent of recombination that has occurred (Posada et al., 2002).
Sequence data from this article have been deposited with the EMBL/GenBank data banks under the following accession numbers: nonresistance genes from the seven genotypes, AY193417 to AY193458; RGC2 genes from cv Calmar, AY193503 to AY193514; from CGN9311, AY193515 to AY193526; from cv Diana, AY193527 to AY193556; from cv Mariska, AY193557 to AY193566; from PI491204, AY193567 to AY193575; from US93UC-10, AY193639 to AY193653; from W66336A, AY193654 to AY193692; RGC2 fragments from the panel of 40 genotypes amplified using Type I specific primers, AY193576 to AY193638; 5B alleles/orthologs, AY193459 to AY193467; K alleles/orthologs from the panel of 40 genotypes, AY193468 to AY193502.
Supplementary Material
Acknowledgments
We thank Barnaly Pande for critical reading of the manuscript and Dean Lavelle for help with sequencing. We also thank several anonymous reviewers for their useful suggestions. This research was supported by the U.S.-Israel Binational Agricultural Research and Development Fund program US-2547-95 and National Science Foundation Plant Genome Program DBI-0211923. E.N. also thanks the Ancell-Teicher Research Foundation for Molecular Genetics and Evolution for financial support.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Richard W. Michelmore (rwmichelmore@ucdavis.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025502.
References
- Anisimova, M., Nielsen, R., and Yang, Z. (2003). Effect of recombination on the accuracy of the likelihood method for detecting positive selection at amino acid sites. Genetics 164, 1229–1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatzky, R., and Tanksley, S.D. (1986). Genetics of actin-related sequences in tomato. Theor. Appl. Genet. 72, 314–321. [DOI] [PubMed] [Google Scholar]
- Berry, A., and Barbadilla, A. (2000). Gene conversion is a major determinant of genetic diversity at the DNA level. In Evolutionary Genetics: From Molecules to Morphology, R.S. Singh and C.B. Krimbas, eds (New York: Cambridge University Press), pp. 102–123.
- Bittner-Eddy, P.D., Crute, I.R., Holub, E.B., and Beynon, J.L. (2000). RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 21, 177–188. [DOI] [PubMed] [Google Scholar]
- Bonnier, J.F.M., Reinink, K., and Groenwald, R. (1994). Genetic analysis of Lactuca accession with the major gene resistance to lettuce downy mildew. Phytopathology 78, 462–468. [Google Scholar]
- Caicedo, A.L., Schaal, B.A., and Kunkel, B.N. (1999). Diversity and molecular evolution of the RPS2 resistance gene in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 96, 302–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cereb, N., Hughes, A.L., and Yang, S.Y. (1997). Locus-specific conservation of the HLA class I introns by intralocus recombination. Immunogenetics 47, 30–36. [DOI] [PubMed] [Google Scholar]
- Chin, D.B., Arroyo-Garcia, R., Ochoa, O., Kesseli, R.V., Lavelle, D.O., and Michelmore, R.W. (2001). Recombination and spontaneous mutation at the major cluster of resistance genes in lettuce (Lactuca sativa). Genetics 157, 831–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, N., Drake, J., Ayliffe, M., Sun, Q., Ellis, J., Hulbert, S., and Pryor, T. (1999). Molecular characterization of the maize Rp1-d rust resistance haplotype and its mutants. Plant Cell 11, 1365–1376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooley, M.B., Pathirana, S., Wu, H.J., Kachroo, P., and Klessig, D.F. (2000). Members of the Arabidopsis HRT/RPP8 family of resistance genes confer resistance to both viral and Oomycete pathogens. Plant Cell 12, 663–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crute, I.R. (1992). The role of resistance breeding in the integrated control of downy mildew (Bremia lactucae) in protected lettuce. Euphytica 63, 95–102. [Google Scholar]
- Dangl, J.L., and Jones, J.D.G. (2001). Plant pathogens and integrated defense responses to infection. Nature 411, 826–833. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001. a). Contrasting modes of evolution acting on the complex N locus for rust resistance in flax. Plant J. 27, 439–453. [DOI] [PubMed] [Google Scholar]
- Dodds, P.N., Lawrence, G.J., and Ellis, J.G. (2001. b). Six amino acid changes confined to the leucine-rich repeat β-strand/β-turn motif determine the difference between the p and p2 rust resistance specificities in flax. Plant Cell 13, 163–178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dooner, H.K., and Martinez-Ferez, I.M. (1997). Recombination occurs uniformly within the bronze gene, a meiotic recombination hotspot in the maize genome. Plant Cell 9, 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis, J.G., Lawrence, G.J., Luck, J.E., and Dodds, P.N. (1999). Identification of regions in alleles of the flax rust resistance gene L that determine differences in gene-for-gene specificity. Plant Cell 11, 495–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farrara, B., Illott, T.W., and Michelmore, R.W. (1987). Genetic analysis of factors for resistance to downy mildew (Bremia lactucae) in lettuce (Lactuca sativa). Plant Pathol. 36, 499–514. [Google Scholar]
- Haubold, B., Kroymann, J., Ratzka, A., Mitchell-Olds, T., and Wiehe, T. (2002). Recombination and gene conversion in a 170-kb genomic region of Arabidopsis thaliana. Genetics 161, 1269–1278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hilliker, A.J., Harauz, G., Reaume, A.G., Gray, M., Clark, S.H., and Chovnick, A. (1994). Meiotic gene conversion tract length distribution within the rosy locus of Drosophila melanogaster. Genetics 137, 1019–1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson, R.R., and Kaplan, N.L. (1985). Statistical properties of the number of recombination events in the history of a sample of DNA sequences. Genetics 111, 147–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, A.L. (2000). Evolution of introns and exons of class II MHC genes of vertebrates. Immunogenetics 51, 473–486. [DOI] [PubMed] [Google Scholar]
- Hughes, A.L., Hughes, M.K., and Watkins, D.I. (1993). Contrasting roles of interallelic recombination at the HLA-A and HLA-B loci. Genetics 133, 669–680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hulbert, S.H. (1997). Structure and evolution of the Rp1 complex conferring rust resistance in maize. Annu. Rev. Phytopathol. 35, 293–310. [DOI] [PubMed] [Google Scholar]
- Hulbert, S.H., Webb, C.A., Smith, S.M., and Sun, Q. (2001). Resistance gene complexes: Evolution and utilization. Annu. Rev. Phytopathol. 39, 285–312. [DOI] [PubMed] [Google Scholar]
- Hwang, C.F., Bhakta, A.V., Truesdell, G.M., Pudlo, W.M., and Williamson, V.M. (2000). Evidence for a role of the N terminus and leucine-rich repeat region of the Mi gene product in regulation of localized cell death. Plant Cell 12, 1319–1329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan, H. (2002). A method for estimating the mutation, gene conversion and recombination parameters in small multigene families. Genetics 161, 865–872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, Y., McAdams, S.A., Gregory, T.B., Hershey, H.P., and Valent, B. (2000). Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 19, 4004–4014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones, D., and Jones, J.D.G. (1997). The role of leucine-rich repeat proteins in plant defenses. Adv. Bot. Res. Adv. Plant Pathol. 24, 89–167. [Google Scholar]
- Kesseli, R.V., Ochoa, O., and Michelmore, R.W. (1991). Variation at RFLP loci in Lactuca spp. and origin of cultivated lettuce. Genome 34, 430–436. [Google Scholar]
- Kesseli, R.V., Paran, I., and Michelmore, R.W. (1994). Analysis of a detailed genetic map of Lactuca sativa constructed from RFLP and RAPD markers. Genetics 136, 1435–1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kesseli, R.V., Paran, I., Ochoa, O., Wang, W.-C., and Michelmore, R.W. (1993). Linkage map of lettuce (Lactuca sativa). In Genetic Maps, 6th ed., S.J. O'Brien, ed (Cold Spring Harbor, NY: Cold Spring Harbor Press), pp. 229–233.
- Kricker, M.C., Drake, J.W., and Radman, M. (1992). Duplication-targeted DNA methylation and mutagenesis in the evolution of eukaryotic chromosomes. Proc. Natl. Acad. Sci. USA 89, 1075–1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, W.H. (1997). Molecular Evolution. (Sunderland, MA: Sinauer Associates).
- Luck, J.E., Lawrence, G.J., Dodds, P.N., Shepherd, K.W., and Ellis, J.G. (2000). Regions outside of the leucine-rich repeats of flax rust resistance proteins play a role in specificity determination. Plant Cell 12, 1367–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maisonneuve, B., Bellec, Y., Anderson, P., and Michelmore, R.W. (1994). Rapid mapping of two genes for resistance to downy mildew from Lactuca serriola to existing clusters of resistance genes. Theor. Appl. Genet. 89, 96–104. [DOI] [PubMed] [Google Scholar]
- McDonald, B.A., and Linde, C. (2002). Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 40, 349–379. [DOI] [PubMed] [Google Scholar]
- McDowell, J.M., Dhandaydham, M., Long, T.A., Aarts, M.G., Goff, S., Holub, E.B., and Dangl, J.L. (1998). Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10, 1861–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B., Kozik, A., Griego, A., Kuang, H., and Michelmore, R.W. (2003). Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15, 809–834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Chin, D.B., Shen, K.A., Sivaramakrishnan, S., Lavelle, D.O., Zhang, Z., and Michelmore, R.W. (1998. a). The major resistance gene cluster in lettuce is highly duplicated and spans several megabases. Plant Cell 10, 1817–1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyers, B.C., Shen, K.A., Rohani, P., Gaut, B.S., and Michelmore, R.W. (1998. b). Receptor-like genes in the major resistance locus of lettuce are subject to divergent selection. Plant Cell 10, 1833–1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michelmore, R.W. (1999). Structure, function and evolution of resistance gene clusters in plants, particularly the Pto and Dm3 loci. In Proceedings of the 9th International Congress Molecular Plant Microbe Interaction, P.J.G.M. de Wit, T. Bisseling, and W. Stiekema, eds (St. Paul, MN: International Society for Plant-Microbe Interactions), pp. 232–237.
- Michelmore, R.W., and Meyers, B.C. (1998). Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res. 8, 1113–1130. [DOI] [PubMed] [Google Scholar]
- Milligan, S.B., Bodeau, J., Yaghoobi, J., Kaloshian, I., Zabel, P., and Williamson, V.M. (1998). The root knot nematode resistance gene Mi from tomato is a member of the leucine zipper, nucleotide binding, leucine-rich repeat family of plant genes. Plant Cell 10, 1307–1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mondragón-Palomino, M., Meyers, B.C., Michelmore, R.W., and Gaut, B.S. (2002). Patterns of positive selection in the complete NBS-LRR gene family of Arabidopsis thaliana. Genome Res. 12, 1305–1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Navarro, A., and Barton, N.H. (2002). The effects of multilocus balancing selection on neutral variability. Genetics 161, 849–863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei, M., Gu, X., and Sitnikova, T. (1997). Evolution by the birth-and-death process in multigene families of the vertebrate immune system. Proc. Natl. Acad. Sci. USA 94, 7799–7806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noël, L., Moores, T.L., van Der Biezen, E.A., Parniske, M., and Daniels, M.J. (1999). Pronounced intraspecific haplotype divergence at the RPP5 complex disease resistance locus of Arabidopsis. Plant Cell 11, 2099–2111. [PMC free article] [PubMed] [Google Scholar]
- Ohta, T. (1983). On the evolution of multigene families. Theor. Popul. Biol. 23, 216–240. [DOI] [PubMed] [Google Scholar]
- Ohta, T. (1991). Role of diversifying selection and gene conversion in evolution of major histocompatibility complex loci. Proc. Natl. Acad. Sci. USA 88, 6716–6720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parniske, M., Hammond-Kosack, K.E., Golstein, C., Thomas, C.M., Jones, D.A., Harrison, K., Wulff, B.B.H., and Jones, J.D.G. (1997). Novel disease resistance specificities result from sequence exchange between tandemly repeated genes at the Cf-4/9 locus of tomato. Cell 91, 821–832. [DOI] [PubMed] [Google Scholar]
- Parniske, M., and Jones, J.D.G. (1999). Recombination between diverged clusters of the tomato Cf-9 plant disease resistance gene family. Proc. Natl. Acad. Sci. USA 96, 5850–5855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piontkivska, H., and Nei, M. (2003). Birth-and-death evolution in primate MHC class I genes: Divergence time estimates. Mol. Biol. Evol. 20, 601–609. [DOI] [PubMed] [Google Scholar]
- Posada, D., Crandall, K.A., and Holmes, E.C. (2002). Recombination in evolutionary genomics. Annu. Rev. Genet. 36, 75–97. [DOI] [PubMed] [Google Scholar]
- Ramakrishna, W., Emberton, J., Ogden, M., SanMiguel, P., and Bennetzen, J.L. (2002). Structural analysis of the maize Rp1 complex reveals numerous sites and unexpected mechanisms of local rearrangement. Plant Cell 14, 3213–3223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richly, E., Kurth, J., and Leister, D. (2002). Mode of amplification and reorganization of resistance genes during recent Arabidopsis thaliana evolution. Mol. Biol. Evol. 19, 76–84. [DOI] [PubMed] [Google Scholar]
- Riely, B.K., and Martin, G.B. (2001). Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proc. Natl. Acad. Sci. USA 98, 2059–2064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozas, J., and Rozas, R. (1999). DnaSP version 3: An integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15, 174–175. [DOI] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press).
- Sawyer, S.A. (1989). Statistical tests for detecting gene conversion. Mol. Biol. Evol. 6, 526–538. [DOI] [PubMed] [Google Scholar]
- Schierup, M.H., Mikkelsen, A.M., and Hein, J. (2001). Recombination, balancing selection and phylogenies in MHC and self-incompatibility genes. Genetics 159, 1833–1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott, A.F., Heath, P., Trusko, S., Boyer, S.H., Prass, W., Goodman, M., Czelusniak, J., Chang, L.Y., and Slightom, J.L. (1984). The sequence of the gorilla fetal globin genes: Evidence for multiple gene conversions in human evolution. Mol. Biol. Evol. 1, 371–389. [DOI] [PubMed] [Google Scholar]
- Shen, K.A., Chin, D.B., Arroyo-Garcia, R., Ochoa, O.E., Lavelle, D.O., Wroblewski, T., Meyers, B.C., and Michelmore, R.W. (2002). Dm3 is one member of a large constitutively-expressed family of NBS-LRR encoding genes. Mol. Plant-Microbe Interact. 15, 251–261. [DOI] [PubMed] [Google Scholar]
- Sicard, D., Woo, S.S., Arroyo-Garcia, R., Ochoa, O., Nguyen, D., Korol, A., Nevo, E., and Michelmore, R.W. (1999). Molecular diversity at the major cluster of disease resistance genes in cultivated and wild Lactuca spp. Theor. Appl. Genet. 99, 405–418. [DOI] [PubMed] [Google Scholar]
- Simons, G., et al. (1998). Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell 10, 1055–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song, W.Y., Pi, L.Y., Wang, G.L., Gardner, J., Holsten, T., and Ronald, P.C. (1997). Evolution of the rice Xa21 disease resistance gene family. Plant Cell 9, 1279–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl, E.A., Dwyer, G., Mauricio, R., Kreitman, M., and Bergelson, J. (1999). Dynamics of disease resistance polymorphism at the Rpm1 loci of Arabidopsis. Nature 400, 667–671. [DOI] [PubMed] [Google Scholar]
- Sun, Q., Collins, N.C., Ayliffe, M., Smith, S.M., Drake, J., Pryor, T., and Hulbert, S.H. (2001). Recombination between paralogues at the Rp1 rust resistance locus in maize. Genetics 158, 423–438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahata, N., and Satta, Y. (1998). Improbable truth in human MHC diversity. Nat. Genet. 18, 204–206. [DOI] [PubMed] [Google Scholar]
- Templeton, A.R. (1983). Phylogenetic inference from restriction endonuclease cleavage site maps with particular reference to the evolution of humans and the apes. Evolution 37, 221–244. [DOI] [PubMed] [Google Scholar]
- Tenaillon, M.I., Sawkins, M.C., Anderson, L.K., Stack, S.M., Doebley, J., and Gaut, B.S. (2002). Patterns of diversity and recombination along chromosome 1 of maize (Zea mays ssp mays L.). Genetics 162, 1401–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian, D., Araki, H., Stahl, E., Bergelson, J., and Kreitman, M. (2002). Signature of balancing selection in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 11525–11530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson, J.D., Higgins, D.G., and Gibson, T.J. (1994). CLUSTAL W: Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoorn, R.A.L., Kruijt, M., Roth, R., Brandwagt, B.F., Joosten, M.H.A.J., and De Wit, P.J.G.M. (2001). Intragenic recombination generated two distinct Cf genes that mediate AVR9 recognition in the natural population of Lycopersicon pimpinellifolium. Proc. Natl. Acad. Sci. USA 98, 10493–10498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F., Gobelman-Werner, K., Morroll, S.M., Kurth, J., Mao, L., Wing, R., Leister, D., Schulze-Lefert, P., and Wise, R.P. (1999). The Mla (powdery mildew) resistance cluster is associated with three NBS-LRR gene families and suppressed recombination within a 240-kb DNA interval on chromosome 5S (1HS) of barley. Genetics 153, 1929–1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei, F.S., Wong, R.A., and Wise, R.P. (2002). Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14, 1903–1917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiuf, C., and Hein, J. (2000). The coalescent with gene conversion. Genetics 155, 451–462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Z. (1997). PAML: A program package for phylogenetic analysis by maximum likehood. Comput. Appl. Biosci. 13, 555–556. [DOI] [PubMed] [Google Scholar]
- Yang, Z., Nielsen, R., Goldman, N., and Pedersen, A.M.K. (2000). Codon-substitution models for heterogeneous selection pressure at amino acid sites. Genetics 155, 431–449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu, H.Y., Cannon, S.B., Young, N.D., and Cook, D.R. (2002). Phylogeny and genomic organization of the TIR and non-TIR NBS-LRR resistance gene family in Medicago truncatula. Mol. Plant-Microbe Interact. 15, 529–539. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.