Abstract
Puromycin-sensitive aminopeptidases (PSAs) participate in a variety of proteolytic events essential for cell growth and viability, and in fertility in a broad range of organisms. We have identified and characterized an Arabidopsis thaliana mutant (mpa1) from a pool of T-DNA tagged lines that lacks PSA activity. This line exhibits reduced fertility, producing shorter siliques (fruits) bearing a lower number of seeds compared with wild-type plants. Cytogenetic characterization of meiosis in the mutant line reveals that both male and female meiosis are defective. In mpa1, early prophase I appears normal, but after pachytene most of the homologous chromosomes are desynaptic, thus, by metaphase I a high level of univalence is observed subsequently leading to abnormal chromosome segregation. Wild-type plants treated with specific inhibitors of PSA show a very similar desynaptic phenotype to that of the mutant line. A fluorescent PSA-specific bioprobe, DAMPAQ-22, reveals that the protein is maximally expressed in wild-type meiocytes during prophase I and is absent in mpa1. Immunolocalization of meiotic proteins showed that the meiotic recombination pathway is disrupted in mpa1. Chromosome pairing and early recombination appears normal, but progression to later stages of recombination and complete synapsis of homologous chromosomes are blocked.
INTRODUCTION
The formation of haploid gametes during sexual reproduction in eukaryotes is dependent on the process of meiosis. It consists of a single round of replication followed in prophase I by the pairing and synapsis of homologous chromosomes to form bivalents. Recombination occurs between the nonsister chromatids within each bivalent. As prophase I finishes, the homologs condense taking up an equatorial position within the nucleus at metaphase I before undergoing a reductional division during anaphase I. A second equational division then occurs during which the sister chromatids of each chromosome separate to form the haploid gametes.
Numerous studies of meiosis in a variety of flowering plants have led to a thorough cytological description of the meiotic pathway. Nevertheless, it is only recently that significant progress has been made toward elucidating the molecular processes that underpin meiosis in plants (Mercier et al., 2001; Caryl et al., 2003). This has been because of the development of Arabidopsis as a model plant system for molecular studies, aided by the development of improved cytogenetic procedures for the study of the small chromosomes found in this species (Ross et al., 1996; Armstrong and Jones, 2001, 2003). As a result a considerable range of Arabidopsis thaliana meiotic genes have been identified affecting premeiosis, sister chromatid cohesion, synapsis, recombination, and chromosome segregation (reviewed in Bhatt et al., 2001; Mercier et al., 2001; Caryl et al., 2003).
A feature of meiotic development that has not been extensively investigated to date is the role of proteolytic enzymes. The selective degradation of proteins is a central feature of many intracellular processes. These include fundamentally important activities such as the regulation of cell cycle progression via the programmed degradation of cyclins (Koepp et al., 1999). Evidence is also emerging to suggest a meiotic role for proteolytic proteins. A recent study of microsporogenesis in Lilium longiflorium has revealed that proteinase activity is closely correlated with meiotic development. There is a temporal increase in general proteolytic activity commencing around midzygotene that persists throughout the remainder of prophase I (DeGuzman and Riggs, 2000). One class of proteases, the aminopeptidases cleave amino acids from the N terminus of oligopeptides. They are widely distributed in both eukaryotes and prokaryotes and are linked to a variety of cellular processes (Taylor, 1993). Recently activity of a leucine aminopeptidase, CoLAP, has been identified during meiotic prophase I in the meiocytes and the supporting cells that surround them in the basidiomycete Coprinus cinereus (Ishizaki et al., 2002). This led the authors to suggest that CoLAP may be required to control some of the biochemical events occurring during prophase I.
During a study aimed at the isolation of Arabidopsis mutants defective in meiotic chromosome synapsis we identified a meiotic mutant, from a population of 2000 T-DNA lines (Instituto Nacional de Investigaciones Agrarias (INIA), Madrid, Spain). Cytological analysis of meiosis in the mutant revealed that it exhibits a desynaptic phenotype. Molecular characterization of the line indicated that it contained a single T-DNA insertion in a gene annotated as a putative aminopeptidase. Further analysis of the mutant referred to hereafter as mpa1 (meiotic prophase aminopeptidase) revealed that MPA1 encodes a protein that is essential for homologous recombination during prophase I of meiosis in Arabidopsis. The protein has the characteristics of a puromycin-sensitive aminopeptidase and, thus, this study provides the first direct evidence that this class of enzyme performs an essential role in meiotic recombination.
RESULTS
Isolation and Preliminary Characterization of the Reduced Fertility Mutant mpa1
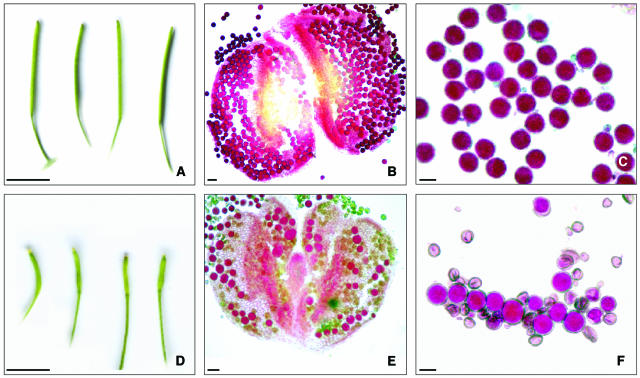
To identify mutants defective in meiosis, a population of 2000 T-DNA lines (INIA) was screened for reduced fertility. Putative meiotic mutants were identified on the basis of small silique size and a reduction in seed set compared with normal fertile plants. One of the lines, mpa1, produced siliques of approximately one-third normal size, generally containing only two to three seeds (Figure 1). Alexander staining (Alexander, 1969) revealed that, in contrast with the uniform round pollen from a normal fertile plant, that of the mutant was much more diverse in size and shape. In some instances the mpa1 pollen cytoplasm did not take up the Orange G from the Alexander stain, which is indicative of a lack of viability (Alexander, 1969).
Figure 1.
Comparative Fertility of Wild-Type Arabidopsis and the mpa1 Mutant.
(A) Wild-type siliques with a mean size of 12.78 mm (n = 500).
(B) Alexander staining of a wild-type anther.
(C) Alexander staining of wild-type pollen grains. Viable pollen grains are stained in red.
(D) mpa1 siliques with a mean size of 3.99 mm (n = 500).
(E) mpa1 anther.
(F) mpa1 pollen grains. Nonviable pollen grains do not stain red and have different size and morphology compared with the viable grains.
Bars in (A) and (D) = 5 mm; bars in (B) and (E) = 20 μm; and bars in (C) and (F) = 10 μm.
To investigate the nature of the T-DNA insertion in mpa1 total genomic DNA from the line was digested with both HindIII and EcoRV. DNA gel blot analysis was performed using the pGKB5 plasmid (Bouchez et al., 1993) used in the transformation as a probe. Single hybridizing bands of 10 and 6.5 kb were found in the HindIII and EcoRV digested DNA, respectively (see Supplemental Figure 1A online). Fluorescence in situ hybridization (FISH) of pGKB5 plasmid on pachytene chromosome spread preparations of mpa1 pollen mother cells revealed a single hybridization signal (see Supplemental Figure 1B online). Together, these results indicate that mpa1 has one T-DNA insertion site.
A cosegregation analysis was then performed to confirm that the mutant phenotype was genetically linked to the T-DNA insertion. Reciprocal crosses were performed between wild-type plants (Columbia) and mpa1 to generate two sets of F1 progeny. These were uniformly kanamycin resistant (Kmr) and fully fertile, indicating that mpa1 is a recessive character. F1 progeny were self-fertilized to produce an F2 generation. F2 seeds were germinated on kanamycin plates to score segregation of Kmr. A 3:1 segregation was obtained with both families ([Col × mpa1 F2; 182 Kmr:65 Kms; χ2 = 0.220, P > 0.5], [mpa1 × Col F2; 185 Kmr:62 Kms; χ2 = 0.001, P > 0.9]). The kanamycin-resistant plant seedlings were grown to maturity to test their fertility phenotype. This revealed a 2:1 fertile:semisterile segregation for both families ([Col × mpa1 F2; 87 fertile:45 semisterile. χ2 = 0.030; P > 0.5], [mpa1 × Col F2; 96 fertile:44 semisterile. χ2 = 0.226; P > 0.5]). Finally, 50 seeds from each semisterile plant (F3) were germinated on kanamycin plates to confirm that all the seeds were resistant to kanamycin ([Col × mpa1 F3; 50 × 45 = 2250 Kmr], [mpa1 × Col F3; 50 × 44 = 2200 Kmr]). Together these data provided a strong indication that the semisterile phenotype was the direct result of the T-DNA insertion in mpa1.
MPA1 Gene Structure and Localization of the T-DNA Insertion Site in mpa1
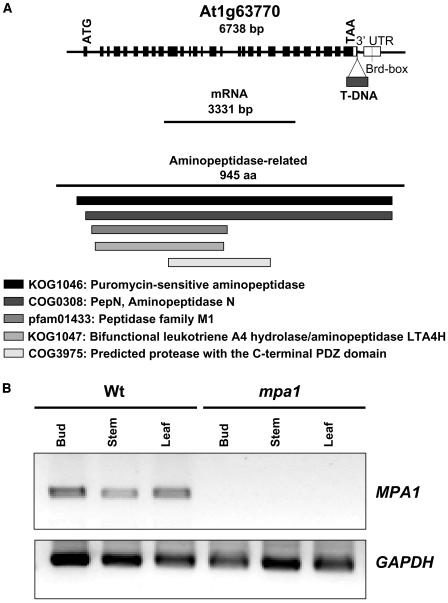
Inverse polymerase chain reaction (IPCR) was performed to isolate the plant genomic DNA flanking the T-DNA in mpa1. A fragment of 300 bp was amplified from mpa1 DNA digested with HindIII. This was cloned and sequenced to reveal that the T-DNA was inserted in the gene At1g63770 on chromosome 1. The sequence is predicted to encode a putative aminopeptidase, similar to the aminopeptidase N (α-hydrolase) from Escherichia coli. The sequencing indicated that the T-DNA was inserted in the 3′ untranslated region (nucleotide position 23,667,376) of At1g63770, subsequently referred to as MPA1 (Figure 2A). This was confirmed by PCR using specific primers for the appropriate region of the gene.
Figure 2.
Molecular Characterization of mpa1.
(A) The MPA1 gene structure with exons (black boxes) and introns, the cDNA, and the corresponding protein. The triangle indicates the position of the T-DNA insertion (nucleotide 23,667,376) and a line indicates the Brd box in the 3′untranslated region (UTR) (white boxes). The protein domains are listed.
(B) Expression analysis of MPA1 locus. RT-PCR products were amplified from RNA derived from flower buds at meiosis, stem, and leaf of both wild-type and mpa1 mutant plants. The GAPDH gene was used as a control.
The Entrez (NCBI) and The Arabidopsis Information Resource (TAIR) Web sites predict two splice transcript variants for At1g63770 (NC 003070). One transcript of 3229 bp (NM 105053) encoding a putative protein of 918 amino acids (NP 176563) and a longer transcript of 3331 bp (NM 202354) encoding a product of 945 amino acids (NP 974083). However, using 3′ rapid amplification of cDNA ends (RACE)-PCR, no evidence of the shorter transcript in bud tissue was found, whereas the longer transcript was readily isolated. Hence, it appears that the longer product is expressed during meiosis.
Expression of MPA1 in Wild-Type and Mutant Plants
The expression pattern of MPA1 was investigated using RT-PCR. Transcription of MPA1 in wild-type plants was detected in both reproductive and vegetative tissues with no discernible difference between them (Figure 2B). By contrast, there was no evidence of residual transcription in mpa1. This clearly indicated that despite the T-DNA insert lying downstream from the MPA1 stop codon the insertion had resulted in a null allele.
Meiosis Is Defective in mpa1
Although previous studies had hinted at a role for aminopeptidases in meiosis a direct link had not been demonstrated. We therefore performed a detailed cytological analysis of meiosis in male meiocytes from mpa1 compared with that in wild-type Arabidopsis (Figure 3). Early stages of prophase I in mpa1 were apparently normal, with leptotene and zygotene indistinguishable from the wild-type meiosis. At late pachytene the homologous chromosomes appeared (see later) to be synapsed, although there was some suggestion that the spacing between the homologs was less uniform than normal (see below) (Figure 3F). Differences between the wild type and the mutant became most apparent as the chromatin condensed at diplotene/diakinesis. It became clear that the majority of homologous chromosome pairs were not linked by chiasmata (Figure 3G). At metaphase I most of the chromosomes appeared as univalents (Figure 3H), such that overall the mean chiasma frequency in mpa1 was 0.9 per cell (n = 60) compared with 9.01 (n = 60) in wild-type plants (Sanchez-Moran et al., 2002). At the first meiotic division the few remaining bivalents segregated normally, however, the unsynapsed univalent chromosomes segregated aberrantly to produce unbalanced dyads and in some instances additional micronuclei. Equational segregation of the univalents was also detected, with the result that sister chromatids were detected at different poles at anaphase I (see Supplemental Figure 2 online). The defects in the first meiotic division led to second-division anomalies, such that at metaphase II the two nuclei rarely presented five chromosomes. Finally, at anaphase II more than four nuclei were frequently observed (Figure 3J).
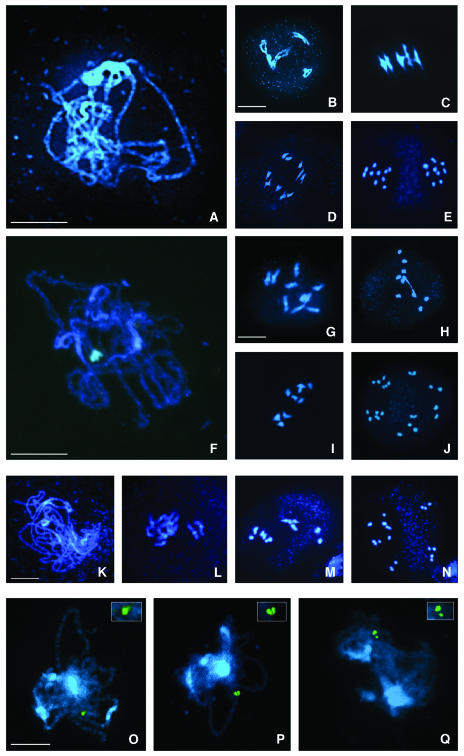
Figure 3.
Cytogenetical Analysis of Wild-Type and mpa1 Meiosis.
DAPI-stained meiotic stages.
(A) to (E) Wild-type male meiosis.
(A) Pachytene. Five fully synapsed bivalents.
(B) Diakinesis. The bivalents are clearly distinguished.
(C) Metaphase I. The five bivalents are coorientated on the spindle equator. Four ring bivalents (having chiasmata in both arms) and one rod bivalent (chiasmata in one arm only) can be distinguished.
(D) Anaphase I. Segregation of five chromosomes to each pole.
(E) Anaphase II. Separation of five chromatids toward each spindle pole.
(F) to (J) mpa1 male meiosis.
(F) Pachytene. The spacing between the homologous chromosomes appears greater than wild type, suggesting some abnormality in synapsis.
(G) Diakinesis. Eight univalents and one bivalent.
(H) Metaphase I. Only one rod bivalent and eight univalents.
(I) Anaphase I. Chromosome missegregation, with seven chromosomes moving to one pole and three to the other one.
(J) Anaphase II. Six groups with different numbers of chromosomes.
(K) to (N) mpa1 female meiosis.
(K) Pachytene. Abnormal synapsis.
(L) Diakinesis. Ten univalents.
(M) Metaphase I. One rod bivalent.
(N) Anaphase II. Four groups containing 3, 4, 3, and 10 chromatids, respectively.
(O) to (Q) Abnormal chromosome synapsis in mpa1.
(O) Fully synapsed pachytene chromosomes, the BAC F24J10 on both homologs is visualized as a single FISH signal.
(P) Abnormal synapsis with two discernable FISH signals, although still closely paired.
(Q) Abnormal synapsis with two clearly separated FISH signals. The homologous chromosomes are broadly spaced although fully aligned.
Bar = 10 μm.
The cytological analysis of the embryo sac mother cells of mpa1 showed that the female meiosis was similarly affected (Figures 3K to 3N). Although there were no apparent defects at early stages of prophase I, at diplotene some univalents were observed such that the number of metaphase I bivalents was lower than wild type and second division was abnormal (Figures 3K to 3N).
Chromosome Synapsis Appears Abnormal in mpa1
Although the pachytene nuclei from mpa1 appeared superficially normal, we noticed that the spacing of the fully aligned homologous chromosomes seemed more variable and in many places slightly greater than that in corresponding wild-type nuclei. From this we inferred that mpa1 may have a synaptic defect. To investigate further, FISH analysis with an Arabidopsis BAC F24J10 corresponding to an interstitial position on chromosome 1 was performed (Figures 3O to 3Q). The rationale was that if at pachytene, synapsis in the region of the BAC probe was complete this would give a single signal with FISH, whereas two discernible signals would suggest incomplete synapsis. In wild-type meiocytes at pachytene the vast majority of nuclei (89%; n = 75) gave a single FISH signal. By contrast, despite having fully aligned and apparently synapsed chromosomes, only 21% (n = 75) of the mpa1 pachytene nuclei gave a single FISH signal. Although this provides only a snapshot from one region of a single chromosome, it clearly suggests that in general, the homologous chromosomes in mpa1 tend to be spaced further apart than in the corresponding wild-type meiocytes and that normal synapsis has not been achieved.
Complementation of the mpa1 Mutant Restores Fertility
To confirm that the T-DNA insertion in mpa1 was responsible for the meiotic phenotype a 9396-bp genomic fragment corresponding to the coding region of the gene and 1508 bp of upstream and 1150 bp of downstream sequences, was cloned into the vector pCAMBIA1302. The DNA was then transformed into three different mpa1 plants. Three independent transformed lines were examined. In each case fertility was restored (see Supplemental Figure 3 online), such that average seed set was at wild-type levels (∼50 seeds per silique). Moreover, cytological examination of meiotic chromosome spreads prepared from meiocytes isolated from the complemented lines revealed that introduction of the wild-type MPA1 gene had restored normal meiosis (see Supplemental Figures 3B and 3C online).
MPA1 Encodes a Member of the M1 Family of Metallopeptidases
The amino acid sequence derived from MPA1 was used to search for conserved domains (NCBI) and protein homologs (BLASTP.2.2.1; NCBI). Analysis of the predicted amino acid sequence revealed the presence of the domain HEXXH(X)18E between residues 393 and 416 (see Supplemental Figure 4 online). This identifies MPA1 as a member of the M1 family of metaloproteases. Furthermore, the BLAST search indicated that MPA1 has homology to a subgroup of the M1 family, the puromycin-sensitive aminopeptidases previously found in Drosophila, mouse, and human (see Supplemental Figure 4 and Table 1 online).
Aminopeptidase Inhibitors Phenocopy the mpa1 Mutation
PSAs are specifically inhibited by puromycin (Constam et al., 1995) and a specific noncompetitive inhibitor, DAMPAQ-22 (Kakuta et al., 2003). Thus, to explore the possibility that MPA1 was a member of the PSA family, the effect of these inhibitors on Arabidopsis meiosis was investigated.
Inflorescence stems were immersed and maintained in a solution of puromycin at 1 μM or DAMPAQ-22 at 10 μM. One week after commencing application, the inflorescences exhibited a semisterile phenotype with both inhibitors (Figures 4A and 4F). The siliques appeared smaller in size than the wild-type control. Flower buds were fixed at 24, 48, and 72 h into the inhibitor application to analyze the meiotic process. After 24 h meiosis seemed to be normal, but at 48 h meiosis was clearly affected. Spread preparations of chromosomes from inflorescences treated with either puromycin or DAMPAQ-22 revealed an apparently normal prophase I. However, at later stages a desynaptic phenotype very similar to that in mpa1 became obvious with a majority of meiocytes containing univalents at metaphase I, which in turn resulted in chromosome missegregation (Figures 4B to 4J). Thus, both inhibitor treatments resulted in a meiotic defect that was remarkably similar to that in the mpa1 mutant.
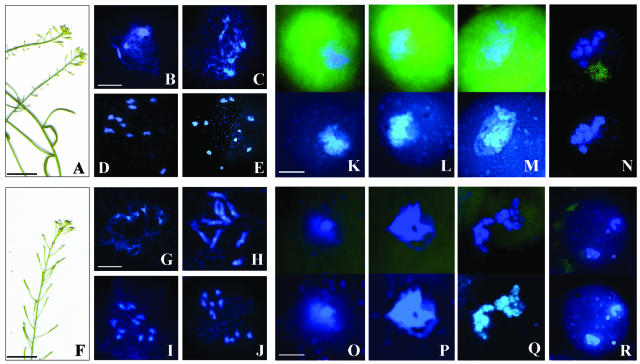
Figure 4.
Inhibitors of PSA Phenocopy the mpa1 Mutation.
(A) to (E) Puromycin (1 μM).
(A) Inflorescences exhibit a semisterile phenotype, with short siliques and a reduced number of seeds per silique.
(B) Pachytene. The homologous chromosomes exhibit slightly abnormal synapsis similar to mpa1.
(C) Late diplotene. The condensation of the chromatin appears more diffuse than the wild type.
(D) Metaphase I. Ten univalents.
(E) Telophase II. Five groups of cells.
(F) to (J) DAMPAQ-22 (10 μM).
(F) Semisterile phenotype of the inflorescences.
(G) Early diplotene.
(H) Diakinesis. Two bivalents and six univalents.
(I) Early anaphase I.
(J) Metaphase II. Two groups of six and four chromosomes.
(K) to (R) Fluorescent visualization of PSA with DAMPAQ-22 in the wild-type ([K] to [N]) and mpa1 ([O] to [R]) meiocytes. A fluorescent signal arising from binding of the bioprobe to MPA1 was detected during prophase I in the wild type.
(K) Leptotene. Showing a high level of fluorescence.
(L) Zygotene.
(M) Pachytene.
(N) Diakinesis. The fluorescent DAMPAQ-22 signal is absent in mpa1.
(O) Leptotene.
(P) Pachytene.
(Q) Diakinesis
(R) Telophase I. Four nuclei.
Bar = 10 μm.
DAMPAQ-22, a PSA-Specific Bioprobe, Localizes MPA1 in Prophase I Meiocytes
A feature of DAMPAQ-22 is that in addition to its use as a noncompetitive inhibitor of PSA, it may be used as a fluorescent bioprobe for visualization of the protein in living cells (Kakuta et al., 2003). We therefore used the inhibitor in conjunction with fluorescence microscopy of meiocyte spread preparations to investigate the expression of MPA1. Inflorescence stems from wild-type Arabidopsis and the mpa1 mutant were placed in DAMPAQ-22 solution (10 μM). After 48 h the meiocytes were analyzed. In wild-type plants a fluorescent signal was observed that was specific to meiocytes in prophase I (Figures 4K to 4N). The maximum signal was detected in meiocytes from zygotene through pachytene before decreasing as chromosome desynapsis took place during diplotene/diakinesis. That the meiocyte-associated fluorescent signal was because of the inhibitor binding to MPA1 was supported by the absence of a corresponding signal in prophase I meiocytes from the mpa1 mutant (Figures 4O to 4R). These observations indicate that MPA1 is present during meiotic prophase I and that activity of the protein is essential for homologous recombination to proceed.
The Meiotic Recombination Pathway Is Disrupted in mpa1
The desynaptic phenotype of mpa1 and presence of univalents at metaphase I strongly suggested that the recombination process in this mutant is disrupted. To investigate this further we performed immunolocalization studies with antibodies that detect key meiotic proteins (Figures 5 and 6). ASY1 is a protein that is expressed in late G2 and defines the onset of leptotene (Armstrong et al., 2003). The protein is essential for pairing and synapsis of homologous chromosomes. In wild-type plants at zygotene/pachytene ASY1 localizes to the bases of the chromatin loops that are in close association with the axial/lateral elements. Localization of the protein in mpa1 is indistinguishable from wild type, both in timing and distribution, suggesting that ASY1-mediated interaction between the chromatin and the chromosome axes is normal (Figures 5A and 5B). To analyze early recombination events in mpa1 the localization of the Rec A homolog, RAD51, was investigated using an anti-RAD51 Ab (Mercier et al., 2003). RAD51 is essential for meiotic recombination and in a majority of species is required for chromosome pairing (Zickler and Kleckner, 1999). Together with its meiotic homolog, DMC1, it forms complexes that localize on chromosomes during early prophase I as foci that correspond to sites of double-strand breaks initiated by the topoisomerase SPO11 (Moens et al., 2002).
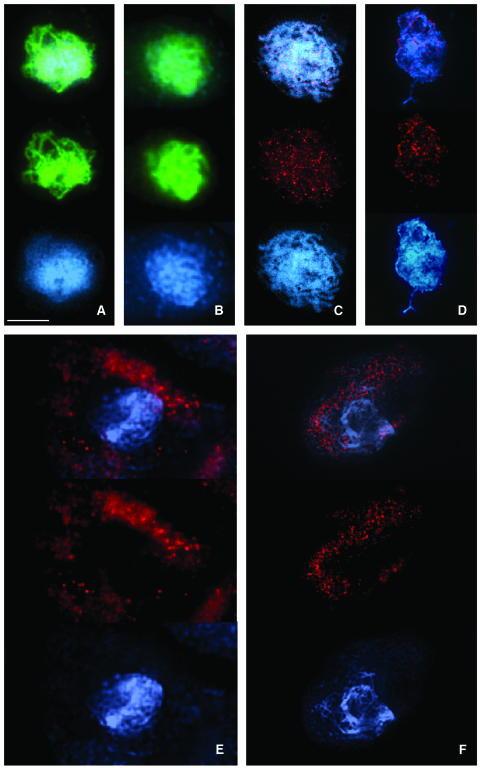
Figure 5.
Immunolocalization of Meiotic Proteins ASY1 and RAD51 in the Wild Type and mpa1.
(A) and (B) Immunolocalization of ASY1 (green).
(A) Wild-type pachytene. The axial/lateral elements are labeled.
(B) mpa1 pachytene. Identical localization to that in wild type, but the fluorescence signal seems to be slightly more diffuse, consistent with the mutant phenotype observed at diplotene.
(C) to (F) Immunolocalization of RAD51 (red).
(C) Wild-type zygotene. RAD51 appears as numerous foci on the chromosomes.
(D) mpa1 zygotene. In some cells RAD51 foci are detectable on the chromosomes.
(E) and (F) mpa1 zygotene. In ∼90% of cells RAD51 accumulates within the cytoplasm.
Bar = 10 μm.
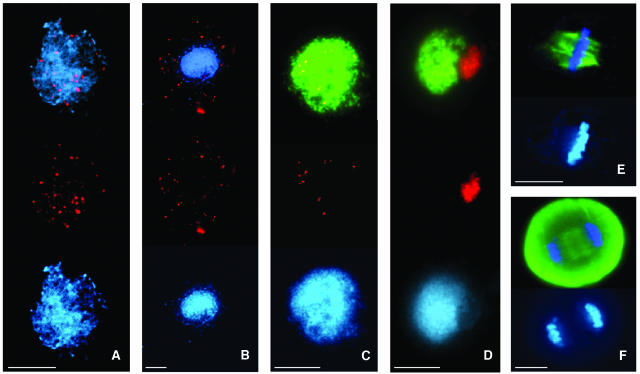
Figure 6.
Immunolocalization of MSH4, MLH1, and α-Tubulin in the Wild Type and mpa1.
(A) and (B) Immunolocalization of MSH4 (red).
(A) Wild-type leptotene/zygotene. MSH4 is distributed as multiple foci on the chromatin.
(B) mpa1 leptotene/zygotene. MSH4 is accumulated in the cytoplasm.
(C) and (D) Immunolocalization of MLH1 (red) and ASY1 (green).
(C) Wild-type zygotene. MLH1 foci are distributed on the chromosomes axes.
(D) mpa1 zygotene. MLH1 is found in association with the nucleolus.
(E) and (F) Localization of α-tubulin (green) in mpa1. Immunostaining of the mitotic spindle appears normal.
(E) Mitotic metaphase.
(F) Anaphase I. In all cases the slides were counterstained with DAPI (blue).
Bar = 10 μm.
In wild-type meiocytes RAD51 protein was detectable as numerous foci on chromosomes at late leptotene/zygotene (n = 30/30) (Figure 5C). A seemingly identical distribution of RAD51 was also detected in some mpa1 meiocytes at late leptotene/zygotene, but these were few in number (∼10% normal; n = 5/50) (Figure 5D). Instead, the majority of mpa1 meiocytes at this stage (∼90%) were found to have an accumulation of RAD51 within the cytoplasm. At zygotene this was fairly diffuse with some small foci, which became more pronounced toward the end of prophase I (Figures 5E and 5F). In addition the gradual decrease in RAD51 foci that is apparent in wild-type meiocytes as prophase I progresses (Mercier et al., 2003) was not observed in the mutant.
Next, we investigated the distribution of two proteins that act downstream of RAD51 in the recombination pathway. MSH4 is a eukaryotic homolog of the E. coli MutS mismatch repair protein (Eisen, 1998). It is thought to act during prophase I in conjunction with MSH5 on the subset of recombination initiations that are designated to form double Holliday junctions (DHJs) and subsequent homologous crossovers (Novak et al., 2001). Resolution of DHJs into crossovers is dependent on the E. coli MutL homolog, MLH1, a component of the late recombination nodules that associate to the sites of genetic crossovers (Moens et al., 2002). Antibodies raised against recombinant Arabidopsis homologs of the MSH4 and MLH1 proteins were used for immunolocalization studies with meiocytes from mpa1 and wild-type plants. In wild-type meiocytes MSH4 was initially detectable as multiple foci on the chromatin at leptotene and remained until late zygotene (Figure 6A; see Supplemental Figure 5A online). MLH1 foci were detectable on the chromosome axes at zygotene/pachytene (Figure 6C; see Supplemental Figure 5E online). Approximately 10 MLH1 foci per cell (n = 32) were observed, which corresponds with the number of crossovers previously determined by chiasma counts in wild-type Arabidopsis (Sanchez-Moran et al., 2002). However, in all the mpa1 meiocytes that were examined, distribution of both MSH4 and MLH1 was found to be abnormal. MSH4 appeared at late leptotene/zygotene, but rather than loading onto the chromatin it was found to accumulate in the cytoplasm (Figure 6B; see Supplemental Figure 5D online). In contrast with the wild type, MLH1 foci were not detected on the chromosomes axes; instead the protein was found in association with the nucleolus during zygotene and pachytene (Figure 6D).
Based on these results it would appear that in the absence of MPA1, recombination is apparently initiated as normal, but progression to crossovers is disrupted at an early stage resulting in an almost-total failure of recombination.
DISCUSSION
Regulated turnover of cellular proteins by an extensive family of proteolytic enzymes is a general feature of cellular metabolism. Meiosis requires the orchestrated, sequential assembly and disassembly of a range of protein complexes that participate in pairing, synapsis, and recombination of homologous chromosomes, suggesting that protease activity may play a crucial role in meiotic progression. In this study we have shown that this is indeed the case. Evidence has been presented that directly demonstrates an essential role for a proteolytic enzyme, MPA1, a member of the puromycin-sensitive family of aminopeptidases, during prophase I of meiosis.
MPA1 Is a Puromycin-Sensitive Aminopeptidase
Aminopeptidases hydrolyze individual amino acids localized at the N terminus of oligopeptides. They are distributed throughout different animal and plant tissues, reflecting important roles in a variety of biological process, including protein processing, regulation of peptide hormone action, viral infection, and cancer cell proliferation (reviewed in Taylor, 1993). Sequence analysis indicates the protein encoded by MPA1 belongs to the M1 metallopeptidase family of aminopeptidases that are characterized by a HEXXH(X)18E metal ion–coordination site and the GAMEN motif (Laustsen et al., 2001). MPA1 exhibits particular homology to puromycin-sensitive soluble aminopeptidase (EC 3.4.11.14), which is a member of the M1 family. PSA can be distinguished from other aminopeptidases on the basis of its specific sensitivity to inhibition by the antibiotic puromycin. Puromycin can have an effect on general protein biosynthesis, but when applied at a level of 1 μM it has been shown to be highly specific for PSA (Constam et al., 1995). Treatment of Arabidopsis inflorescences with either puromycin or DAMPAQ-22, a specific noncompetitive inhibitor of PSA (Kakuta et al., 2003), resulted in a phenocopy of the mpa1 mutation. Although the effect of the inhibitors on general protein synthesis was not directly assessed during these studies, the fact that the inflorescences remained viable throughout the period of inhibitor application and developed siliques suggests that any such effect was negligible. Thus, the inhibitor studies provide strong additional evidence to indicate that MPA1 is a PSA homolog.
Previous studies have implicated PSA in mitosis and possibly meiosis. Puromycin has been shown to block the cell cycle of COS cells and 3T3 fibroblasts in G2/M phase and induce apoptosis (Constam et al., 1995). A noncompetitive inhibitor of PSA, PIQ22, also reduces tumor cell invasion (Kagechika et al., 1999; Komoda et al., 2001). Studies suggest that PSA is associated with microtubules of the spindle apparatus during mitosis (Constam et al., 1995). Although this could also occur in Arabidopsis, immunostaining of mitotic and meiotic spindles in mpa1 with antitubulin antibody did not reveal any differences in organization from the wild type (Figures 6E and 6F). This suggests that if MPA1 is associated with the microtubules of the mitotic and meiotic spindle its role is not crucial.
Several studies have hinted at a possible link between aminopeptidase activity and meiosis. PSA-deficient male and female mice have been found to be infertile (Osada et al., 2001a, 2001b). Similarly, expression of a puromycin-sensitive aminopeptidase (dPSA) has been detected in Drosophila spermatocytes, and flies deficient in the protein exhibit reduced male fertility (Schulz et al., 2001). The levels of dPSA expression were reported to vary at different spermatocyte stages although no cytological analysis was performed to establish how this related to meiotic stage. An interesting feature of the dPSA transcript is the presence of two Brd boxes in the 3′ UTR (Schulz et al., 2001). This motif has been associated with posttranscriptional regulation (Lai and Posakony, 1997). An identical sequence is also present in the MPA1 transcript (3157 to 3163 residues). Recently, a C. elegans ortholog of PSA (PAM1) has been reported to be involved in embryogenesis and reproduction (Brooks et al., 2003). Furthermore, a microarray analysis of C. elegans mutants defective in germ line development suggest that PAM1 is expressed in both male and female germ lines (Reinke et al., 2000). Although these studies have not directly investigated the meiotic role of PSA it would seem likely based on our study of Arabidopsis that PSA activity is an important feature of meiosis in a broad range of organisms.
MPA1 Is Expressed during Prophase I of Meiosis
Two lines of evidence suggested that MPA1 functions during prophase I. First, cytological analysis of the mpa1 mutant revealed that despite apparently normal pairing of homologous chromosomes during early prophase I, complete synapsis was not achieved. Careful inspection of the homologs in mpa1 at pachytene suggested that the separation of the chromosomes was less uniform than normal. This was substantiated with FISH using a BAC corresponding to an interstitial site on chromosome 1. This revealed that synapsis was incomplete in more than four times as many nuclei in the mutant as compared with the wild type at the same stage. This observation could suggest a failure of synapsis or a delay in the process. Although further investigation is required to resolve this issue, we favor the latter possibility, as despite the lack of uniformity in spacing of the homologs, many regions looked to be normal. Subsequent desynapsis revealed that the frequency of crossovers was dramatically reduced in mpa1. Second, in good agreement with the cytological analysis of mpa1 the studies using the PSA-specific fluorescent bioprobe DAMPAQ-22 (Kakuta et al., 2003) clearly places expression during prophase 1 starting at leptotene and disappearing at late diplotene.
Although RT-PCR expression analysis demonstrated that MPA1 mRNA transcript is present in wild-type bud tissue, it was also detectable in leaf and stem. Discounting genes such as RAD51 that have a dual function in DNA repair and meiosis, this observation has been made for other genes that are solely meiotic, for example ASY1 and SWI1 (Caryl et al., 2000; Mercier et al., 2003). However, the proteins encoded by these genes are restricted to meiotic cells. At present we are unable to categorically state that MPA1 protein is not found in vegetative cells. Imaging with DAMPAQ-22 suggested that the protein was not in anther cells other than meiocytes, but other somatic cell types were not investigated. The finding that some meiotic genes are transcribed in somatic cells has led to the speculation that they may be subject to posttranscriptional regulation (Caryl et al., 2000; Mercier et al., 2003). In this respect the presence of a Brd box in the 3′ UTR of MPA1, a motif that has been shown to negatively regulate expression (Lai and Posakony, 1997) may be significant. However, further studies will be required to establish if this is indeed the case.
MPA1 Is Required for Progression of the Recombination Pathway in Prophase I
To define the activity and timing of action of MPA1 in more detail we investigated the distribution of several key meiotic proteins, namely ASY1, RAD51, MLH1, and MSH4 in mpa1. ASY1 was loaded onto the chromatin as normal in G2 and organized into the thread-like structures that are associated with the axial elements at leptotene and remain throughout prophase I (Armstrong et al., 2002). This indicated that the establishment of the chromosome axes and early pairing of homologous chromosomes is not perturbed in the absence of MPA1. This was substantiated by the BAC-FISH experiment that revealed that the homologs were accurately aligned, but synapsis might be abnormal or delayed.
In wild-type meiocytes numerous RAD51 foci accumulated during late leptotene corresponding to the positions of early recombination nodules (Anderson et al., 1997; Barlow et al., 1997; Moens et al., 1997). This was followed by their progressive disappearance at later stages of prophase I. Early recombination nodules are thought to coincide with the sites of recombination initiation, however, in the majority of cases they do not progress to form homologous crossovers. Recent studies indicate that in most instances they are resolved as noncrossovers possibly via the synthesis dependent strand annealing pathway that operates at the D-loop stage after RAD51/DMC1-catalyzed strand invasion of the homologous DNA (Allers and Lichten, 2001; de Massy, 2003). In most of the mpa1 meiocytes that were examined at late leptotene/zygotene, the protein was found in the cytoplasm rather than in association with the chromatin. Significantly, however, a few meiocytes were detected in which the number and distribution of the foci appeared normal. We interpret this as an indication that RAD51 does associate with the chromatin, presumably at SPO11-catalyzed double-strand breaks in the DNA (Mazin et al., 2000), but this association is possibly more transient than in wild-type plants. This interpretation would account for the fact that pairing of homologous chromosomes occurs as normal in mpa1 because studies in a range of species including plants indicate that this requires RAD51-mediated interactions between homologous chromosomes during early prophase I (Zickler and Kleckner, 1999; Pawlowski et al., 2003). Moreover, it would also explain why in contrast with recent findings in an Arabidopsis rad51 mutant, chromosome fragmentation does not occur in mpa1 (Li et al., 2004). Because the short-lived interaxis interactions that facilitate pairing would still occur and the associated double-strand breaks, some ∼90 to 95% of the total, would then be repaired via the synthesis dependent strand annealing pathway (Franklin et al., 1999; Allers and Lichten, 2001; Moens et al., 2002; Börner et al., 2004). This would restore the integrity of the chromosomes sufficiently to prevent the fragmentation found in the rad51 mutant. However, the subclass of 5 to 10% of recombination initiations that normally progress to stable strand invasion events and subsequently form crossovers require MSH4 localization and further recombination proteins such as MLH1, which as we have found does not occur in the mpa1 mutant. Thus, based on these observations it appears that RAD51 activity is largely unaffected, but the failure of MSH4 to localize to the recombination nodules appears to destabilize the interaction of the RAD51 with the chromatin such that it accumulates in the cytoplasm. Although the basis of such a destabilization is open to speculation, it is worth noting that RAD51 together with its catalytic partner, DMC1, has been shown to colocalize with a member of the RecQ helicase family, BLM (Moens et al., 2000, 2002). It has been proposed that BLM and the equivalent yeast protein Sgs1 may serve an antirecombination function removing excess recombination complexes (Moens et al., 2000; Rockmill et al., 2003). Thus, it is possible that in the event of disruption of the MSH4-dependent pathway all the recombination initiations that were destined for this route are removed by BLM resulting in the RAD51 accumulation. Although further studies will be required to establish the events in detail the existing data strongly indicate that MPA1 activity is directly or indirectly critical for the loading of MSH4 with subsequent effects on later stages of meiotic progression.
Despite the dramatic effect of the mpa1 mutation on homologous crossovers that are reduced by ∼90%, a residual level of recombination persists. Recent studies in yeast indicate that ∼85% of homologous crossovers are dependent on MSH4, with the remainder occurring via an alternative MUS81/MMS4-dependent route (de los Santos et al., 2003). Evidence is emerging to suggest that a comparable situation may also exist in Arabidopsis (Copenhaver et al., 2002; J. Higgins, unpublished data). Studies estimate that the proportion of crossovers in Arabidopsis that are MSH4 independent is between 0.1 and 0.2 (Copenhaver et al., 2002). This could account for the residual crossovers detected in the mpa1 mutant. It would also seem highly likely that the failure to achieve complete synapsis in mpa1 is also attributable to the effect of the mutation on MSH4 activity. This is based on studies in yeast, where mutation of MSH4 has been shown to result in a delay of synapsis such that only ∼50% of nuclei achieve full synapsis (Novak et al., 2001).
The immunolocalization studies indicate that both RAD51 and MSH4 accumulate within the cytoplasm in mpa1, something that is not found in wild-type meiocytes. At late prophase I both proteins are found to be associated with foci suggesting that they may be components of a protein complex or aggregate. Previously aggregates containing RAD51 have been reported late in meiosis in an Arabidopsis swi1 mutant (Mercier et al., 2003). Ultrastructural protein aggregates referred to as polycomplexes have been widely described in meiotic mutants, including those affecting recombination proteins. They arise as a result of disruption of synaptonemal complex (SC) morphogenesis and are comprised of proteins that are normally associated with the structure (Zickler and Kleckner, 1999). Although there is no direct evidence that the RAD51 and MSH4 are associated with polycomplexes in the mpa1 mutant, it is interesting to note that recent studies of synapsis initiation have demonstrated a close association of recombination proteins such as MSH4 with SC proteins (Börner et al., 2004; Fung et al., 2004). Hence it is conceivable that they may form aggregates in some mutant backgrounds.
Unlike MSH4, which in the mpa1 mutant remains distributed throughout the cytoplasm, MLH1 is found in association with the nucleolus. Although the significance of this is unknown, it is interesting to note that there is increasing evidence for a role of the nucleolus in regulating protein function and or availability (Visintin and Amon, 2000). We have previously shown that the meiotic protein SWI1 that is linked with the establishment of sister chromatid cohesion associates and accumulates to an extent in the nucleolus (Mercier et al., 2003). In budding yeast nucleolar localization of the meiosis-specific protein PCH2 seems to be important for checkpoint function. PCH2 is required to prevent meiotic interhomolog recombination within the repeated rRNA genes present in the nucleolus (San-Segundo and Roeder, 1999). Furthermore, it has been suggested that PCH2 perhaps sequesters within the nucleolus a protein required for the exit from pachytene (Roeder and Bailis, 2000). This proposal is based on analogy with CDC14 in yeast that is sequestered in the nucleolus until it is required to allow mitotic cell cycle exit (Murgia et al., 1998).
In summary, we have demonstrated that MPA1 plays an important role in meiosis. This evidence suggests that it is required to enable normal levels of homologous recombination to occur. Based on this investigation it appears that the window of MPA1 activity occurs at an early stage in the recombination pathway, soon after RAD51 is loaded onto the chromatin and before the loading of MSH4. Whether either protein is a direct substrate for MPA1 has yet to be established. The complexity of the recombination process is such that the apparent destabilization of RAD51 and disruption of MSH4 localization may reflect the effect of MPA1 on one or more other proteins in the process. Nevertheless, it is of interest to note that a direct effect of peptidases on DNA repair proteins is not without precedent as nucleotide excision repair activity of RAD23 is stimulated by activity of the proteosome in human cells (Walters et al., 2003).
METHODS
Material
The plant material employed for this study was the ecotype Columbia from Arabidopsis thaliana (2n = 10). For the isolation of a meiotic mutant, we analyzed 2000 T-DNA insertion lines obtained by J.M. Martínez-Zapater and J. Salinas (INIA, Madrid, Spain). The genomic background of these lines was also the ecotype Columbia. The in planta transformation method was the vacuum infiltration by the binary vector pGKB5 (Bouchez et al., 1993). This vector contains the genes conferring kanamycin and Basta resistance as plant selection markers.
The plants were grown in a glasshouse at 18 to 20°C, with supplementary lighting when necessary, using a 16-h daylength.
Cytogenetics
Pollen viability was assayed by Alexander staining (1969). Stained anthers were examined with a light microscope.
The bud flowers were fixed in Carnoy (absolute ethanol:chloroform:glacial acetic acid 6:3:1) and stored at −20°C. The method of Sanchez-Moran et al. (2001) was used for slide preparation of pollen mother cells. The female meiosis was studied in embryo-sac mother cells as described by Armstrong and Jones (2001). The slides were DAPI stained (10 μg/mL) in Vectashield (Vector Laboratories, Burlingame, CA) antifade mounting medium or kept at 4°C for FISH.
The FISH technique used was that previously described by Sanchez-Moran et al. (2001). The centromeric DNA probe, PAL1, and the BAC F24J10 were directly labeled with Spectrum Green (Amersham, Buckinghamshire, UK) by nick translation following the manufacturer's instructions (Roche, Basel, Switzerland). The binary vector pGKB5 containing the T-DNA was labeled with Biotin dUTP by nick translation (Roche).
The spreads and immunolocalization procedures were performed as previously described by Armstrong et al. (2002). The preimmune serum was used as a negative control. The following antibodies were used: the polyclonal rat anti-ASY1 and RAD51 antibodies were used as described by Armstrong et al. (2002) and Mercier et al. (2003), respectively. The commercial monoclonal rat anti-α-tubulin (Oxford Biotechnology, Oxfordshire, UK) antibody was used at a 1:20 dilution. MLH1 and MSH4 rat polyclonal antiserums were produced from the purified, recombinant proteins, containing 258 and 611 amino acids from each protein, respectively (J. Higgins, G.H. Jones, and F.C.H. Franklin, unpublished data).
The slides were studied by fluorescence microscopy using a Nikon Eclipse T300 microscope (Tokyo, Japan). Capture and analysis of images was done using an image analysis system, SmartCapture 2 (Digital Scientific, Cambridge, UK).
Genetic Analysis
The genetic segregation of mpa1 was tested by reciprocal crossing to wild type (Columbia) and characterizing the segregation of fertile versus reduced fertility phenotypes in the F2 generation. Cosegregation of the reduced fertility phenotype with T-DNA insertion of the gene was monitored by resistance of seedlings to kanamycin (50 μg/mL) on agar medium and by PCR with specific primers for the MPA1 gene and the T-DNA. Using the primers LB2 (5′-tgccaggtgcccacggaatag-3′) for the left border T-DNA and IFw (5′-gttctgcggttccccagtga-3′) and IRev (5′-acgatggcgggttcagcaga-3′) designed from the mpa1 gene sequence.
A 9396-bp XbaI/SphI genomic fragment was cloned in pCAMBIA1302 vector (Cambia GPO) to perform the complementation test. The genomic fragment contains the coding region of MPA1 (6738 bp), upstream sequence (1508 bp), and downstream sequence (1150 bp). The Arabidopsis transformation protocol was followed as described by Clough and Bent (1998). Three independent transformed lines were examined for fertility restoration and cytological analysis of meiosis.
Molecular Genetics
Different molecular approaches were performed to analyze the number of T-DNA inserts and isolate the gene affected.
Southern hybridization was performed as described by Caryl et al. (2000) to analyze the number of T-DNA insertions in mpa1 using the pGKB5 plasmid as a probe. Five micrograms of digested genomic DNA (HindIII and EcoRV) were transferred onto a Hybond N+ membrane (Amersham Life Sciences) by capillary blotting.
The plant genomic DNA flanking the T-DNA in mpa1 was isolated by inverse polymerase chain reaction. The IPCR protocol followed was similar to the one described by Ochman et al. (1998) with some modifications. Two micrograms of mpa1 genomic DNA were digested with different restriction enzymes: HindIII, BglII, PstI, SalI, and BamHI. The digested DNA products (0.1 to 0.5 μg/mL) were circularized using T4 DNA ligase (Invitrogen, Carlsbad, CA) at 15°C overnight. The IPCR was performed with circularized DNA (0.1 μg), primers LB1 (5′-cccagtgtcctattaccaatagc-3′) and LB2 (5′-tgccaggtgcccacggaatag-3′) (100 ng/μL) (MWG-Biotech, Ebersberg, Germany) and dNTPs (0.1 mM). A program of 1 min at 93°C, 1 min at 50°C, and 1 min at 72°C for 30 cycles was used for amplification. The IPCR products were cloned in the plasmid pCR2.1 and sequenced.
RT-PCR was performed to study the expression of MPA1 using the method of Agashe et al. (2002). Total RNA was isolated using Rneasy kit (Qiagen USA, Valencia, CA) from flower buds, leaves, and stems in mpa1 and the wild type. The one-step RT-PCR kit (Qiagen) was used following the manufacturer's instructions with the following primers: rtpcr1f (5′-aggtctcggcagtgtgctttc-3′) and rtpcr1r (5′-acaatgtcacccaagaacttg-3′) that amplified a region comprising residues 2115 to 2760 of the MPA1 cDNA. As a reference, GAPDH (present in all tissues) was used in identical conditions with the following primers: GAPD-N (5′-cttgaagggtggtgccaagaagg-3′) and GAPD-C (5′cctgttgtcgccaacgaagtcag-3′). The products were cloned in pCR2.1 and sequenced.
The 3′ RACE was performed using the following specific primers: 3′ RACE1f (5′-caagatggccaccaatttgac-3′) position 2439 and 3′ RACE1r (5′-gcttctgagaaataacattgccg-3′) position 3218 of the MPA1 cDNA.
Aminopeptidase-Inhibitor Assays and in Vivo Expression of PSA
The inflorescence stems of wild type were cut in water and submerged in different solutions with puromycin-sensitive aminopeptidase inhibitors: puromycin (0.75 μM) or DAMPAQ-22 (10 μM) following the method previously described for BrdU labeling of meiocytes (Armstrong et al., 2003). After 48 h the flower buds were fixed and the pollen mother cells were analyzed cytologically as reported above.
DAMPAQ-22 has also been used to study the in vivo expression of the PSA. DAMPAQ-22 is a fluorescent bioprobe designed from a noncompetitive inhibitor of PSA, MPAQ-22: 1-N-methyl-3-(2,6-diethylphenyil)-2,4(1H,3H)-quinazolinedione), that has been structurally modified to afford fluorescent visualization (Kakuta et al., 2003). Inflorescences of wild type were cut in water and submerged in a solution with DAMPAQ-22 (10 μM) for 48 h. The anthers were extracted out of the flowers using a dissecting microscope. The anthers were transferred onto a slide and incubated in enzyme mixture, 0.3% w/v pectolyase, 0.3% w/v cytohelicase, and 0.3% w/v cellulose (all Sigma-Aldrich, St. Louis, MO) in citrate buffer, pH 4.5, at 37°C for 20 min. Afterwards, the anthers were squashed in 7 μL of DAPI (10 μg/mL) in Vectashield antifade mounting medium (Vector). The slides were excited with a 360-nm filter and the emitted fluorescence was detected with a 405-nm filter as described by Kakuta et al. (2003). Water controls were used in each case.
Supplementary Material
Acknowledgments
This work was partially supported by the project BMC2002-01171 awarded by Ministerio de Ciencia y Tecnología (Madrid, Spain). Eugenio Sánchez-Morán was funded for part of this work by a European Union Marie Curie Grant QLK3-CT-2001-50690. Work in the laboratory of F.C.H.F and G.H.J. is funded by the Biotechnology and Biological Science Research Council. The authors also thank A. C. Jones (Functional Genomics and Proteomics Laboratories, University of Birmingham, Birmingham, UK) for technical support. Collaboration between the Madrid and Birmingham groups is supported by a Royal Society Research exchange grant. We are indebted to Yuichi Hashimoto for providing the DAMPAQ-22 inhibitor.
The authors responsible for distribution of materials integral to the finding presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) are: F. Christopher H. Franklin (f.c.h.franklin@bham.ac.uk) and Juan Luis Santos (jlsc53@bio.ucm.es).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.024992.
References
- Agashe, B., Prasad, C.K., and Siddiqi, I. (2002). Identification and analysis of DYAD: A gene required for meiotic chromosome organisation and female meiotic progression in Arabidopsis. Development 129, 3935–3943. [DOI] [PubMed] [Google Scholar]
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Allers, T., and Lichten, M. (2001). Differential timing and control of noncrossover and crossover recombination during meiosis. Cell 106, 47–57. [DOI] [PubMed] [Google Scholar]
- Anderson, L.K., Offenberg, H.H., Verkuijlen, W.M., and Heyting, C. (1997). RecA-like proteins are components of early meiotic nodules in lily. Proc. Natl. Acad. Sci. USA 94, 6868–6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Armstrong, S.J., Caryl, A.P., Jones, G.H., and Franklin, F.C.H. (2002). Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115, 3645–3655. [DOI] [PubMed] [Google Scholar]
- Armstrong, S.J., Franklin, F.C.H., and Jones, G.H. (2003). A meiotic time-course for Arabidopsis thaliana. Sex. Plant Reprod. 16, 141–149. [Google Scholar]
- Armstrong, S.J., and Jones, G.H. (2001). Female meiosis in wild-type Arabidopsis thaliana and in two meiotic mutants. Sex. Plant Reprod. 13, 177–183. [Google Scholar]
- Armstrong, S.J., and Jones, G.H. (2003). Meiotic cytology and chromosome behaviour in wild-type Arabidopsis thaliana. J. Exp. Bot. 54, 1–10. [DOI] [PubMed] [Google Scholar]
- Barlow, A.L., Benson, F.E., West, S.C., and Hulten, M.A. (1997). Distribution of the Rad51 recombinase in human and mouse spermatocytes. EMBO J. 16, 5207–5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhatt, A.M., Canales, C., and Dickinson, H.G. (2001). Plant meiosis: The means to 1N. Trends Plant Sci. 6, 114–121. [DOI] [PubMed] [Google Scholar]
- Bouchez, D., Camilleri, C., and Caboche, M. (1993). A binary vector based on Basta resistance for in planta transformation of Arabidopsis thaliana. C.R. Acad. Sci. Paris 316, 1188–1193. [Google Scholar]
- Börner, G.V., Kleckner, N., and Hunter, N. (2004). Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 117, 29–45. [DOI] [PubMed] [Google Scholar]
- Brooks, D.R., Hooper, N.M., and Isaac, R.E. (2003). The Caenorhabditis elegans orthologue of mammalian puromycin-sensitive aminopeptidase has roles in embryogenesis and reproduction. J. Biol. Chem. 278, 42795–42801. [DOI] [PubMed] [Google Scholar]
- Caryl, A.P., Armstrong, S.J., Jones, G.H., and Franklin, F.C.H. (2000). A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109, 62–71. [DOI] [PubMed] [Google Scholar]
- Caryl, A.P., Jones, G.H., and Franklin, F.C.H. (2003). Dissecting plant meiosis using Arabidopsis thaliana mutants. J. Exp. Bot. 54, 25–38. [DOI] [PubMed] [Google Scholar]
- Clough, S.J., and Bent, A.F. (1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Constam, D.B., Tobler, A.R., Rensing-Ehl, A., Kemler, I., Hersh, L.B., and Fontana, A. (1995). Puromycin-sensitive aminopeptidase. Sequence analysis, expression, and functional characterization. J. Biol. Chem. 270, 26931–26939. [DOI] [PubMed] [Google Scholar]
- Copenhaver, G.P., Housworth, E.A., and Stahl, F.W. (2002). Crossover interference in Arabidopsis. Genetics 160, 1631–1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGuzman, R., and Riggs, C.D. (2000). A survey of proteinases active during meiotic development. Planta 210, 921–924. [DOI] [PubMed] [Google Scholar]
- de los Santos, T., Hunter, N., Lee, C., Larkin, B., Loidl, J., and Hollingsworth, N.M. (2003). The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics 164, 81–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Massy, B. (2003). Distribution of meiotic recombination sites. Trends Genet. 19, 514–522. [DOI] [PubMed] [Google Scholar]
- Eisen, J.A. (1998). A phylogenomic study of the MutS family of proteins. Nucleic Acids Res. 26, 4291–4300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin, A.E., McElver, J., Sunjevaric, I., Rothstein, R., Bowen, B., and Cande, W.Z. (1999). Three-dimensional microscopy of the Rad51 recombination protein during meiotic prophase. Plant Cell 11, 809–824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fung, J.C., Rockmill, B., Odell, M., and Roeder, G.S. (2004). Imposition of crossover interference through non-random distribution of synapsis initiation complexes. Cell 116, 795–802. [DOI] [PubMed] [Google Scholar]
- Ishizaki, T., Tosaka, A., Nara, T., Aoshima, N., Namekawa, S., Watanabe, K., Hamada, F., Omori, A., and Sakaguchi, K. (2002). Leucine aminopeptidase during meiotic development. Eur. J. Biochem. 269, 826–832. [DOI] [PubMed] [Google Scholar]
- Kagechika, H., Komoda, M., Fujimoto, Y., Koiso, Y., Takayama, H., Kadoya, S., Miyata, K., Kato, F., Kato, M., and Hashimoto, Y. (1999). Potent homophthalimide-type inhibitors of B16F10/L5 mouse melanoma cell invasion. Biol. Pharm. Bull. 22, 1010–1012. [DOI] [PubMed] [Google Scholar]
- Kakuta, H., Koiso, Y., Nagasawa, K., and Hashimoto, Y. (2003). Fluorescent bioprobes for visualization of puromycin-sensitive aminopeptidase in living cells. Bioorg. Med. Chem. Lett. 13, 83–86. [DOI] [PubMed] [Google Scholar]
- Koepp, D.M., Harper, J.W., and Elledge, S.J. (1999). How the cyclins became a cyclin: Regulated proteolysis in the cell cycle. Cell 97, 431–434. [DOI] [PubMed] [Google Scholar]
- Komoda, M., Kakuta, H., Takahashi, H., Fujimoto, Y., Kadoya, S., Kato, F., and Hashimoto, Y. (2001). Specific inhibitor of puromycin-sensitive aminopeptidase with a homophthalimide skeleton: Identification of the target molecule and a structure-activity relationship study. Bioorg. Med. Chem. 9, 121–131. [DOI] [PubMed] [Google Scholar]
- Lai, E.C., and Posakony, J.W. (1997). The Bearded box, a novel 3′ UTR sequence motif, mediates negative post-transcriptional regulation of bearded and enhancer of split complex gene expression. Development 124, 4847–4856. [DOI] [PubMed] [Google Scholar]
- Laustsen, P.G., Vang, S., and Kristensen, T. (2001). Mutational analysis of the active site of human insulin-regulated aminopeptidase. Eur. J. Biochem. 268, 98–104. [DOI] [PubMed] [Google Scholar]
- Li, W., Chen, C., Markmann-Mulisch, U., Timofejeva, L., Schmelzer, E., Ma, H., and Reiss, B. (2004). The Arabidopsis AtRAD51 gene is dispensable for vegetative development but required for meiosis. Proc. Natl. Acad. Sci. USA 101, 10596–10601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazin, A.V., Zaitseva, E., Sung, P., and Kowalczykowski, S.C. (2000). Tailed duplex DNA is the preferred substrate for Rad51 protein-mediated homologous pairing. EMBO J. 19, 1148–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercier, R., Armstrong, S.J., Horlow, C., Jackson, N.P., Makaroff, C.A., Vezon, D., Pelletier, G., Jones, G.H., and Franklin, F.C.H. (2003). The meiotic protein SWI1 is required for axial element formation and recombination initiation in Arabidopsis. Development 130, 3309–3318. [DOI] [PubMed] [Google Scholar]
- Mercier, R., Grelon, M., Vezon, D., Horlow, C., and Pelletier, G. (2001). How to characterize meiotic functions in plants? Biochimie 83, 1023–1028. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Chen, D.J., Shen, Z., Kolas, N., Tarsounas, M., Heng, H.H., and Spyropoulos, B. (1997). Rad51 immunocytology in rat and mouse spermatocytes and oocytes. Chromosoma 106, 207–215. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Freire, R., Tarsounas, M., Spyropoulos, B., and Jackson, S.P. (2000). Expression and nuclear localization of BLM, a chromosome stability protein mutated in Bloom's syndrome, suggest a role in recombination during meiotic prophase. J. Cell Sci. 113, 663–672. [DOI] [PubMed] [Google Scholar]
- Moens, P.B., Kolas, N.K., Tarsounas, M., Marcon, E., Cohen, P.E., and Spyropoulos, B. (2002). The time course and chromosomal localization of recombination-related proteins at meiosis in the mouse are compatible with models that can resolve the early DNA-DNA interactions without reciprocal recombination. J. Cell Sci. 115, 1611–1622. [DOI] [PubMed] [Google Scholar]
- Murgia, C., Blaikie, P., Kim, N., Dans, M., Petrie, H.T., and Giancotti, F.G. (1998). Cell cycle and adhesion defects in mice carrying a targeted deletion of the integrin β4 cytoplasmic domain. EMBO J. 17, 3940–3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak, J.E., Ross-Macdonald, P.B., and Roeder, G.S. (2001). The budding yeast Msh4 protein functions in chromosome synapsis and the regulation of crossover distribution. Genetics 158, 1013–1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman, H., Gerber, A.S., and Hartl, D.L. (1998). Genetic applications of an inverse polymerase chain reaction. Genetics 120, 621–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osada, T., Watanabe, G., Kondo, S., Toyoda, M., Sakaki, Y., and Takeuchi, T. (2001. a). Male reproductive defects caused by puromycin-sensitive aminopeptidase deficiency in mice. Mol. Endocrinol. 15, 960–971. [DOI] [PubMed] [Google Scholar]
- Osada, T., Watanabe, G., Sakaki, Y., and Takeuchi, T. (2001. b). Puromycin-sensitive aminopeptidase is essential for the maternal recognition of pregnancy in mice. Mol. Endocrinol. 15, 882–893. [DOI] [PubMed] [Google Scholar]
- Pawlowski, W.P., Golubovskaya, I.N., and Cande, W.Z. (2003). Altered nuclear distribution of recombination protein RAD51 in maize mutants suggests the involvement of RAD51 in meiotic homology recognition. Plant Cell 15, 1807–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinke, V., Smith, H.E., Nance, J., Wang, J., Van Doren, C., Begley, R., Jones, S.J., Davis, E.B., Scherer, S., Ward, S., and Kim, S.K. (2000). A global profile of germline gene expression in C. elegans. Mol. Cell 6, 605–616. [DOI] [PubMed] [Google Scholar]
- Rockmill, B., Fung, J.C., Branda, S.S., and Roeder, G.S. (2003). The Sgs1 helicase regulates chromosome synapsis and meiotic crossing over. Curr. Biol. 13, 1954–1962. [DOI] [PubMed] [Google Scholar]
- Roeder, G.S., and Bailis, J.M. (2000). The pachytene checkpoint. Trends Genet. 16, 395–403. [DOI] [PubMed] [Google Scholar]
- Ross, K.J., Fransz, P., and Jones, G.H. (1996). A light microscopic atlas of meiosis in Arabidopsis thaliana. Chromosome Res. 4, 507–516. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran, E., Armstrong, S.J., Santos, J.L., Franklin, F.C.H., and Jones, G.H. (2001). Chiasma formation in Arabidopsis thaliana accession Wassileskija and in two meiotic mutants. Chromosome Res. 9, 121–128. [DOI] [PubMed] [Google Scholar]
- Sanchez-Moran, E., Armstrong, S.J., Santos, J.L., Franklin, F.C.H., and Jones, G.H. (2002). Variation in chiasma frequency among eight accessions of Arabidopsis thaliana. Genetics 162, 1415–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- San-Segundo, P.A., and Roeder, G.S. (1999). Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97, 313–324. [DOI] [PubMed] [Google Scholar]
- Schulz, C., Perezgasga, L., and Fuller, M.T. (2001). Genetic analysis of dPsa, the Drosophila orthologue of puromycin-sensitive aminopeptidase, suggests redundancy of aminopeptidases. Dev. Genes Evol. 211, 581–588. [DOI] [PubMed] [Google Scholar]
- Taylor, A. (1993). Aminopeptidases: structure and function. FASEB J. 7, 290–298. [DOI] [PubMed] [Google Scholar]
- Visintin, R., and Amon, A. (2000). The nucleolus: The magician's hat for cell cycle tricks. Curr. Opin. Cell Biol. 12, 752. [DOI] [PubMed] [Google Scholar]
- Walters, K.J., Lech, P.J., Goh, A.M., Wang, Q., and Howley, P.M. (2003). DNA-repair protein hHR23a alters its protein structure upon binding proteasomal subunit S5a. Proc. Natl. Acad. Sci. USA 100, 12694–12699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler, D., and Kleckner, N. (1999). Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33, 603–754. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.