Abstract
The Arabidopsis thaliana VERNALIZATION INDEPENDENCE (VIP) gene class has multiple functions in development, including repression of flowering through activation of the MADSbox gene FLC. Epigenetic silencing of FLC plays a substantial role in the promotion of flowering through cold (vernalization). To better understand how VIP genes influence development, we undertook a genetic and molecular study of the previously uncharacterized VIP5 and VIP6 genes. We found that loss of function of these genes also resulted in downregulation of other members of the FLC/MAF gene family, including the photoperiodic pathway regulator MAF1/FLM. We cloned VIP5 and VIP6 through mapping and transcriptional profiling. Both proteins are closely related to distinct components of budding yeast Paf1C, a transcription factor that assists in establishment and maintenance of transcription-promotive chromatin modifications such as ubiquitination of H2B by Bre1/Rad6 and methylation of histone H3 lysine-4 by the trithorax-related histone methylase Set1. Genetic analysis and coimmunoprecipitation experiments suggest that VIP5 and VIP6 function in the same mechanism as the previously described VIP3 and VIP4. Our findings suggest that an evolutionarily conserved transcriptional mechanism plays an essential role in the maintenance of gene expression in higher eukaryotes and has a central function in flowering.
INTRODUCTION
The activity of most eukaryotic genes results from the coordinated effort of a multitude of diverse factors that serve both to recognize the gene and to promote or repress initiation, elongation, and termination of transcription (Lee and Young, 2000). The access of the transcriptional machinery to gene regulatory regions, as well as its progression through transcribed regions, depends both on disruption of higher-order chromatin packaging and the accessibility of DNA at the nucleosomal level (Orphanides and Reinberg, 2000; Svejstrup, 2004). Recent attention in the field of transcription, originating predominately from studies in the budding yeast Saccharomyces cerevisiae, has turned to the astonishing array of factors that modify chromatin structure. These include chromatin-remodeling factors, which displace nucleosomes along the DNA, and histone-modifying enzymes, which add or remove various posttranslational modifications including small chemical groups (acetylation, phosphorylation, and methylation) and proteins (ubiquitination and SUMOylation) on nucleosomal histones. The number and pattern of histone modifications have been hypothesized to play a key role in orchestrating gene activity, both by directly affecting chromatin architecture and by providing interaction sites for other chromatin-associated proteins (Jenuwein and Allis, 2001; Fischle et al., 2003).
Superimposed on the complexity of transcription in higher eukaryotes is the requirement to alter gene expression in response to developmental cues and faithfully maintain patterns of gene activity in related cell types in the mature organism. The so-called trithorax group (trxG) and Polycomb group (PcG) proteins have been implicated as having crucial roles in the maintenance of activity states of developmental regulatory genes (Francis and Kingston, 2001). In fruit flies and mammals, trxG and PcG proteins maintain activity or repression, respectively, of the homeotic Hox genes set up during embryogenesis. This activity is accomplished at least in part by the ability of these and associated proteins to carry out and recognize various histone modifications, most notably lysine methylation (Fischle et al., 2003). In plants, although the role of trxG genes has not been well defined, it is becoming increasingly evident that at least the PcG proteins play crucial roles in various developmental progressions through maintenance of the repression of homeotic-function MADSbox genes (Goodrich et al., 1997; Gendall et al., 2001; Kohler et al., 2003).
An excellent model to study the epigenetic dynamics of developmentally important genes in eukaryotes is the silencing of the Arabidopsis thaliana MADS box floral repressor gene FLC, and associated initiation of flowering, after extended growth of the plant in the cold. Promotion of flowering by long periods of cold, a phenomenon known as vernalization, is an ecologically and agriculturally important response common to many plants and long recognized as having an epigenetic component (Lang, 1965). FLC is one member of a family of six closely related MADSbox proteins in Arabidopsis (Ratcliffe et al., 2001). FLC has been the most extensively studied gene of this family, both because of its substantial effect on the vernalization response and because genetic variation at FLC and its activator FRI are responsible for the natural diversity in flowering habit among Arabidopsis ecotypes (Lee et al., 1993; Johanson et al., 2000; Michaels et al., 2003). The other members of the FLC gene family, designated MAF1-MAF5, can act as floral repressors when expressed constitutively to high levels in transgenic plants, and at least MAF1, MAF2, MAF3, and MAF4 have been shown to be downregulated in vernalized plants (Ratcliffe et al., 2001, 2003). This suggests a conserved function for this clade of MADS box genes in mediating the vernalization response.
Genetic approaches have identified several factors required for silencing of FLC in vernalized plants (Surridge, 2004). These include VRN2, a homolog of the fly PcG protein Su(z)12 (Gendall et al., 2001), VRN1, a putative DNA binding protein (Levy et al., 2002), and VIN3, a plant homeodomain-containing protein (Sung and Amasino, 2004). Examination of vernalization-associated changes in histone modifications of FLC chromatin in wild-type and mutant plants is leading to a framework of a model for the involvement of chromatin changes in FLC silencing (Bastow et al., 2004; Sung and Amasino, 2004). The activity of VIN3, which accumulates during the cold and is associated with deacetylation of histone H3 within FLC promoter and intronic regions, may create favorable conditions for subsequent methylation of H3 at Lys residues K27 and K9 within these regions mediated by VRN2 and VRN1. Although information in plants is limited, studies in animals and fission yeast suggest that methylation at H3K9 within euchromatic regions promotes the formation of heterochromatin and long-term gene silencing, suggesting a precedence for the stable repression of FLC in vernalized plants.
To better understand the dynamics of FLC expression at the molecular level, we have used genetic screens to identify genes required for the maintenance of FLC activity in nonvernalized plants. To date, our group has identified at least 20 loci (Zhang et al., 2003), including seven that comprise the VERNALIZATION INDEPENDENCE (VIP) gene class (Zhang and van Nocker, 2002; Zhang et al., 2003). FLC expression is not detectable in strong vip mutants, indicating a critical function for these genes. VIP3 encodes a protein composed of so-called WD repeats. WD repeat proteins are well represented in eukaryotes, and are believed to coordinate dynamic protein assemblies (van Nocker and Ludwig, 2003). VIP4 encodes a highly charged protein closely related to budding yeast Leo1. Subsequent to our identification of VIP4, Leo1 was identified as a component of a ∼1.7 mD transcriptional complex called Paf1C (Mueller and Jaehning, 2002; see below).
Phenotypic analysis of vip mutants suggests that the VIP genes likely have additional roles unrelated to their activation of FLC. For example, vip mutants flower earlier than flc null mutants, suggesting that other flowering-time genes are targeted (Zhang et al., 2003). In addition, strong vip mutants exhibit mild developmental pleiotropy, which is not seen in an flc null mutant, suggesting that the VIP genes also target mechanisms unrelated to flowering (Zhang et al., 2003). The objectives of this research were to further characterize the mechanism by which the VIP genes activate FLC, through the identification of the VIP5 and VIP6 genes, and to investigate the role of these VIP genes in FLC-independent flowering and other developmental processes.
RESULTS
VIP5 and VIP6 Function in Concert with VIP3 and VIP4
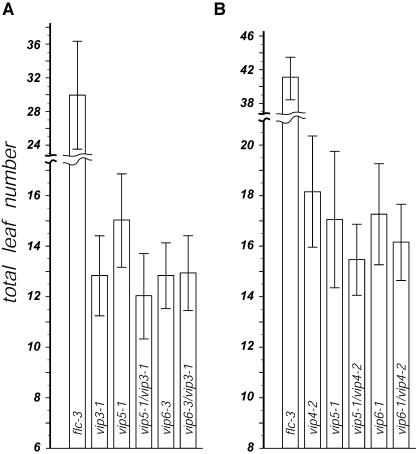
Based on phenotypic similarity among mutants at the seven VIP loci reported previously, we proposed that the respective genes work in concert in a common mechanism or pathway (Zhang and van Nocker, 2002; Zhang et al., 2003). To explore this idea further, we evaluated the phenotypic effects of combining strong vip3 and vip4 mutations with strong vip5 and vip6 mutations. In short-day photoperiods, where the promotive effects of extended daylengths are minimized, and under a variety of growth temperatures and light intensities, vip5 and vip6 single mutants flowered with a similar number of leaves to either vip3 or vip4 (Figure 1). As previously observed (Zhang et al., 2003), these mutants flowered significantly earlier than an flc null mutant (Figure 1). There was no significant difference in flowering time between any single mutant and any of the derived double mutant combinations evaluated. In addition, in the double mutants, we did not observe phenotypic effects that were more severe than those exhibited by any single mutant (data not shown). The lack of synergistic effects of coincident inactivity of these genes is consistent with our hypothesis that these genes are closely related in function, possibly as components of a protein complex or molecular pathway.
Figure 1.
Flowering Time of vip3, vip4, vip5, and vip6 Single and Double Mutants.
Flowering time (measured as the total number of rosette and cauline leaves produced) is indicated for (A) vip3-1, vip5-1, vip6-3, and derived double mutants and (B) vip4-2, vip5-1, vip6-1, and derived double mutants. Plants were grown under noninductive (8 h light/16 h dark) photoperiods. Results from independent experiments are shown; flowering time of the flc null mutant flc-3 in each experiment is shown for comparison. Values represent the mean and standard deviation for at least 20 plants of each genotype.
VIP5 and VIP6 Participate in the Regulation of a Heterogeneous Subset of Genes Including Other Members of the FLC/MAF Gene Family
The observation that strong vip3, vip4, vip5, and vip6 mutants flower earlier than an flc null mutant suggested that these genes participate in the regulation of flowering-time genes in addition to FLC. The unique (nonredundant) function of the FLC gene appears to be limited to flowering time, because flc null mutants do not exhibit gross defects beyond timing of flowering. In contrast, the developmental pleiotropy seen in strong vip mutants suggests that these genes participate in the regulation of a subset of genes that include, but are not limited to, FLC (Zhang et al., 2003).
To assist in the identification of these genes, and to evaluate similarity in molecular phenotype between vip5 and vip6 mutants, we performed transcriptional profiling experiments using Affymetrix ATH1 microarrays representing ∼22,700 Arabidopsis genes (Figure 2). To eliminate indirect effects on gene expression because of differential activity of FLC and its effects on flowering, we related transcriptional profiles of the strong vip5-1 or vip6-3 mutant plants, in which FLC transcripts are not detectable, with those of the flc-3 null mutant, which produces a dysfunctional transcript (Michaels and Amasino, 2001). All of these mutants were derived from the same parental genotype, and, under the long-day conditions in which these experiments were performed, flowered at approximately the same time and developmental stage (Zhang et al., 2003).
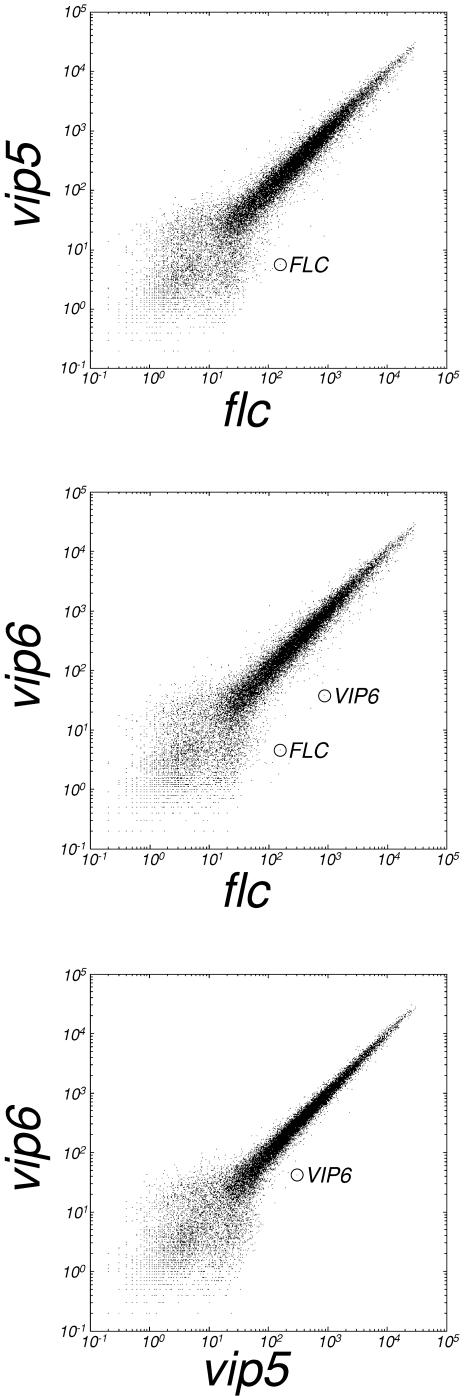
Figure 2.
Characteristics of Microarray Data Derived from flc, vip5, and vip6 Mutants.
Signal intensity was plotted to compare single replicates of flc with vip5 (top), flc with vip6 (middle), or vip5 with vip6 (bottom). The signal positions for FLC and/or VIP6 are indicated. For each comparison, representative data are shown.
We employed pairwise comparisons of the replicates and standard statistical analyses (see Methods) to define subsets of represented genes with expression affected in the vip5-1 mutant, the vip6-3 mutant, or in both the vip5-1 and vip6-3 mutants, relative to the flc-3 null mutant. Confirming the efficacy of this approach, and consistent with previous results based on RNA gel blotting (Zhang et al., 2003), we found that FLC transcripts were easily detectable in the flc-3 mutant, but undetectable (statistically absent) in the vip5 and vip6 mutants (Figure 2 and data not shown). In addition to FLC, ∼40 other genes showed a strong decrease in expression in the vip5 or vip6 mutants relative to flc-3; we also identified ∼20 genes that were strongly upregulated in the vip5 or vip6 mutants relative to flc-3 (see Supplemental Table 1 online). The genes that were misregulated in vip5 or vip6 were not obviously related with respect to structure, genomic location, or potential function (data not shown).
The essentially indistinguishable phenotype conferred by strong mutation at each vip locus (Zhang et al., 2003) suggested that a similar subset of genes would be affected in each mutant. This was indeed the case with vip5 and vip6. The data for these two mutants revealed a high degree of overlap (Figure 2); the subset of genes that showed a strong decrease in both mutants represented 79% of the strongly decreased genes in vip5, and 77% of the strongly decreased genes in vip6 (see Supplemental Table 1 online). The degree of overlap was nearly complete when the subset was defined by slightly relaxed criteria for either of the pairwise comparisons (see Methods). For example, all of the 42 genes exhibiting a strong decrease in vip5, as defined by the more stringent criteria, also met the relaxed criteria for a decrease in vip6 (see Supplemental Table 1 online). The overlap was also apparent when the data for vip5 and vip6 were compared directly using the more stringent criteria; for example, only two genes showed a strong decrease in vip6 relative to vip5, and one of these was subsequently identified as the VIP6 gene itself (Figure 2 and data not shown).
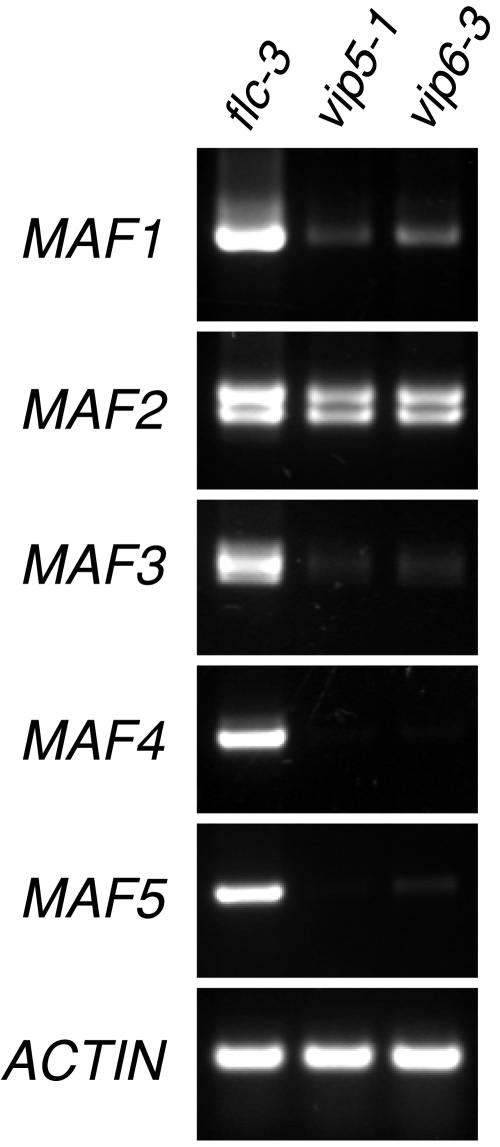
Interestingly, among the genes showing decreased expression in both vip5 and vip6 was the FLC paralog MAF1 (Ratcliffe et al., 2001; also known as FLM [Scortecci et al., 2001]). We confirmed this result through RT-PCR analysis (Figure 3). Like FLC, MAF1 acts as a repressor of flowering, and at least in the Columbia (Col) background is downregulated in vernalized plants (Ratcliffe et al., 2001; our unpublished results). The FLC/FLM MADS box clade in Arabidopsis is represented by four additional genes, designated MAF2-MAF5, that also can act as floral repressors (Ratcliffe et al., 2001, 2003). We considered whether VIP5 and VIP6 also participate in the regulation of these genes. MAF2, MAF4, and MAF5 were also represented on the microarrays, but their expression was statistically undetectable (MAF2 and MAF4) or did not exhibit a significant change (MAF5) in our microarray data. However, RT-PCR analysis indicated a modest but reproducible decrease in MAF2, and a marked silencing of the remaining MAF genes, in both vip5 and vip6 mutants, relative to the flc null (Figure 3). Notably, the involvement of VIP5 or VIP6 in the activation of the MAF genes did not depend on FLC activity, because this experiment was performed in an flc null genetic background. This suggests that the MAF gene family members represent additional regulatory targets of VIP5 and VIP6.
Figure 3.
Expression of the FLC-Related MAF Genes in flc, vip5, and vip6 Mutants.
Expression was monitored in flc-3, vip5-1, and vip6-3 plants by RT-PCR as described in Methods. Results shown are representative of two independent biological replicates.
VIP6 Encodes a Plant Homolog of the Paf1C Component Ctr9
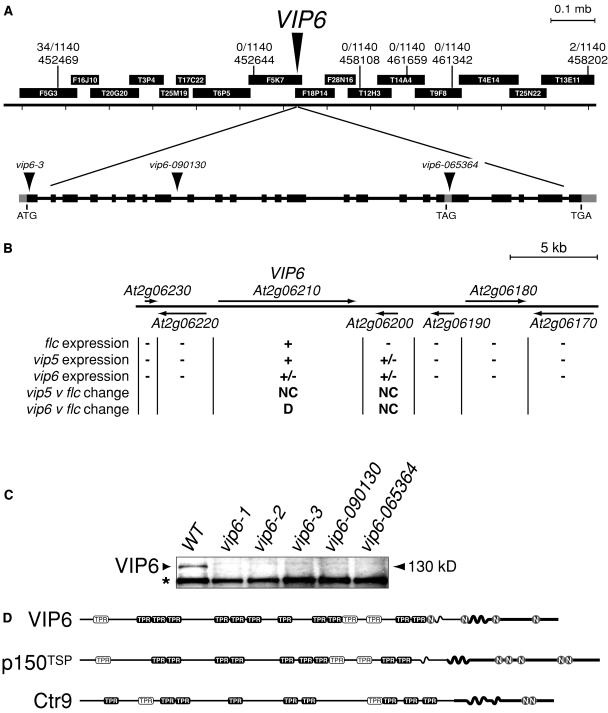
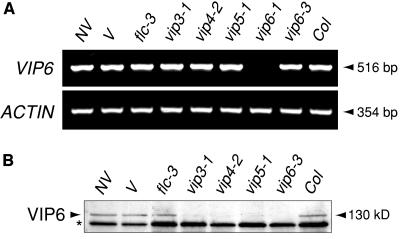
The VIP6 gene was represented by three alleles derived from fast-neutron mutagenesis (vip6-1) and T-DNA mutagenesis (vip6-2 and vip6-3) that, based on phenotypic similarity of the respective mutants, were of equivalent severity (data not shown). Initial attempts to identify VIP6 by characterizing genomic DNA flanking the T-DNA insertion site in the vip6-2 or vip6-3 mutants were not successful. Therefore, we used a positional cloning approach, and localized VIP6 within a ∼1.2-mb region of chromosome II (Figure 4A). Because no recombination was detected in the immediate region of VIP6, we analyzed the activity of the majority of genes within the ∼1.2-mb region in the vip6-3 mutant using data derived from microarray hybridizations (above). A single analyzed gene within this region, designated At2g06210, showed a statistically significant decreased expression in the vip6-3 mutant as compared with the flc-3 mutant (Figures 2 and 4B; data not shown). We were not able to detect At2g06210 transcripts in wild-type plants by RNA gel blotting, even using phosphorimaging and extended exposures. However, analysis of the At2g06210 gene by RT-PCR in wild-type plants and in the strong vip6-1 mutant revealed a strong decrease in mRNA accumulation in the mutant (Figure 5A). PCR analysis using T-DNA-specific primers and sets of overlapping primers encompassing the At2g06210 genomic region revealed the presence of T-DNA within the At2g06210 predicted transcribed region in the vip6-3 mutant (Figure 4A and data not shown). We were unable to amplify any region of At2g06210 genomic DNA from vip6-1 mutant plants, suggesting that the At2g06210 gene was deleted, and that vip6-1 represents a true null allele. As further evidence that At2g06210 represents the VIP6 gene, we analyzed the phenotype of two additional At2g06210 T-DNA insertion mutants from a collection developed at the Salk Institute Genomic Analysis Laboratory (SIGnAL; Alonso et al., 2003). For both lines, plants homozygous for the mutations exhibited a pleiotropic phenotype that was essentially indistinguishable from that of the three previously described vip6 mutants. The SIGnAL mutant alleles were isolated in the Col background, which does not strongly express FLC and flowers soon after germination; when introduced into the synthetic, winter-annual Col:FRISF2 background, these alleles conferred early flowering and loss of FLC expression (data not shown). Finally, antibodies raised against a portion of the At2g06210 protein recognized a ∼130-kD species in wild-type plants that was absent in plants carrying strong vip6 alleles (Figure 4C). Based on these observations, we concluded that At2g06210 is VIP6.
Figure 4.
Map Position, Structure, and Expression of the VIP6 Gene and Protein.
(A) Region of chromosome II containing the VIP6 gene. Molecular markers used in mapping are shown, with genetic distance (recombinations/chromosomes analyzed) between the vip6 mutation and marker indicated. Relevant BAC clones are shown. In the depiction of the VIP6 transcriptional unit, exons are shown as black (translated region) or gray (untranslated region) boxes, and an alternative exonic region detected in some cDNAs is shown as a gray box. The position of the start codon (ATG) and termination codons (TAG and TGA; the TAG termination codon is within the alternative exonic region) are shown. The positions of the T-DNA insertions in the vip6-3 allele and the SIGnAL alleles 090130 and 065364 are indicated.
(B) Expression of VIP6 and adjacent genes in the flc-3, vip5-1, and vip6-3 mutants. Predicted transcriptional units are indicated by arrows. Expression data were derived as described in Methods (+, detected in both replicates; +/− detected in one replicate; -, not detected; NC, no significant change in expression relative to flc mutant; D, significant decrease in expression relative to flc mutant). All predicted transcriptional units within ∼15 kb of the VIP6 gene that were represented on the microarray are shown.
(C) Expression of the VIP6 protein in wild-type plants (WT) and in five vip6 mutant backgrounds. An unrelated, immunoreactive protein species is indicated (*).
(D) Domain structure of the VIP6 protein and related murine p150TSP and budding yeast Ctr9. TPR motifs are indicated, with those most strongly resembling the canonical TPR motif shown in black. Potential nuclear localization signal sequences are also shown (N). Coiled-coil regions are depicted as sinuous segments. The highly charged regions of the C termini are represented with increased stroke weight.
Figure 5.
Analysis of VIP6 mRNA and Protein Abundance in Various Genetic Backgrounds and in Response to Vernalization.
(A) RT-PCR was used to compare VIP6 transcript levels in nonvernalized wild-type plants (NV); wild-type plants subjected to a 70-d cold treatment (V); the flc-3 null mutant; strong vip3, vip4, and vip5 mutants; the strong vip6-1 and vip6-3 mutants; and in the Col ecotype. For each sample, a portion of the ACTIN gene was amplified in parallel to demonstrate relative quantity and quality of the cDNA template.
(B) VIP6 protein was monitored by immunoblotting of extracts from nonvernalized wild-type plants (NV); wild-type plants subjected to a 70-d cold treatment (V); the flc-3 null mutant; strong vip3, vip4, and vip5 mutants; the strong vip6-3 mutant; and in the Col ecotype. An unrelated, immunoreactive protein species is indicated (*) to indicate the total amount of protein present in each lane.
Transgenic antisense expression of a VIP6 cDNA in a wild-type (Col:FRISF2) background conferred a broad degree of acceleration of flowering time, with approximately one-half of initial transformants (T1 plants) flowering during the course of the experiment, and the earliest flowering plants (∼10% of the population) flowering at approximately the same time as vernalized wild-type plants (see Supplemental Figure 1 online). Interestingly, only a minor fraction of VIP6 antisense plants exhibited developmental pleiotropy. As with flowering time, a range of pleiotropy was seen, with the most severe effects limited to the earliest-flowering T1 individuals. However, several of the earliest-flowering T1 plants did not exhibit obvious phenotypic defects other than flowering timing (data not shown).
We found that VIP6 transcript and protein levels were similar in vernalized and nonvernalized plants (Figure 5), suggesting that vernalization-mediated silencing of FLC does not directly involve modulation of VIP6 expression. Also, VIP6 mRNA and protein were expressed at wild-type levels in the Col genetic background, which lacks a functional FRI allele (Figure 5), suggesting that VIP6 does not regulate FLC downstream from FRI. Immunoblot analysis of dissected whole plants indicated that the VIP6 protein is ubiquitously expressed, with the strongest accumulation in apical tissues (data not shown).
We also detected VIP6 mRNA expression at wild-type levels in strong vip3, vip4, and vip5 mutants (Figure 5A), suggesting that the VIP6 gene was not subject to regulation by these other VIP genes. Interestingly, however, in contrast with wild-type plants, the VIP6 protein was not easily detectable in strong vip3, vip4, or vip5 genetic backgrounds (Figure 5B). We also observed this effect on VIP6 protein levels in vip1 and vip2 mutants (data not shown). This observation suggests a posttranslational role for these other VIP genes in maintaining VIP6 protein levels.
Based on sequence analysis of several cDNAs, VIP6 could encode two proteins of 1091 and 740 amino acids that would originate from alternative processing of a common precursor RNA (Figures 4A and 4D; see Supplemental Figure 2 online). These putative proteins differ only in the extent of their C termini, and contain so-called tetratricopeptide repeats (TPRs) throughout much of their length. TPRs are ∼34-amino acid domains found in proteins of diverse function, and are generally considered to mediate protein-protein interactions and/or assembly of protein complexes (D'Andrea and Regan, 2003). The larger form of the VIP6 protein contains predicted coiled-coil domains near its C terminus, and the C-terminal ∼200 amino acid region is highly enriched in charged amino acids such as Glu, Asp, Arg, and Lys (Figure 4D, see Supplemental Figure 2 online). This C-terminal region also contains four potential nuclear localization motifs (Figure 4D), suggesting compartmentalization in the nucleus. Immunoblot analysis using antibodies directed against the N-terminal region of the VIP6 protein recognized only a single, ∼130-kD species in wild-type plant extracts, suggesting that the longer form of the protein is relatively more abundant.
A query of public sequence databases identified known and hypothetical VIP6-related proteins in various divergent eukaryotes, including human, fruit fly, frog, slime mold, rice, and yeasts (see Supplemental Figure 2 online; data not shown). Of these, only the Ctr9 protein from budding yeast has been functionally characterized. This protein has been described as a component of Paf1C (Mueller and Jaehning, 2002), a transcription factor required for specific transcription-promotive covalent modifications of chromatin-associated histones: ubiquitination of H2B within its C-terminal domain, and methylation of H3 at residues K4, K36, and K79 (Ng et al., 2003a; Wood et al., 2003b). Paf1C is associated with the initiating and elongating forms of RNA polymerase II (Pol II; Mueller and Jaehning, 2002) and during elongation may serve as a platform for the association of specific histone methylases with chromatin (Hampsey and Reinberg, 2003). It has been postulated that Paf1C provides a mechanism for the memory of recent gene transcription, potentially by antagonizing the activity of silencing proteins and thus reinforcing the active state of genes (Ng et al., 2003b).
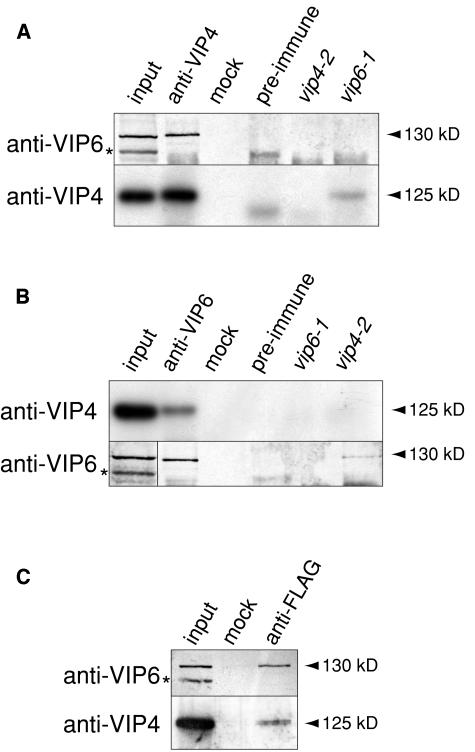
The VIP6 Protein Physically Interacts with VIP3 and VIP4 in Vivo
The observation that VIP6 encodes a Paf1C subunit homolog was especially intriguing in light of the previous identification of VIP4 as homologous to yeast Leo1 (Zhang and van Nocker, 2002). Leo1 copurified from yeast cells with Ctr9 and other Paf1C proteins (Mueller and Jaehning, 2002; Krogan et al., 2002; Squazzo et al., 2002), and so is probably an integral subunit of Paf1C. We performed coimmunoprecipitation experiments to determine if, like their yeast counterparts, VIP6 and VIP4 interact in vivo. Indeed, antisera generated against recombinant VIP4 protein (see Supplemental Figure 3A online) specifically immunoprecipitated a ∼130-kD protein from wild-type plant extracts that was strongly immunoreactive with anti-VIP6 antibodies (Figure 6A). This protein was absent from parallel immunoprecipitates using extracts from the strong vip6-1 mutant (Figure 6A). Conversely, anti-VIP6 antibodies immunoprecipitated an anti-VIP4 immunoreactive, ∼125-kD protein from wild-type extracts that was absent from immunoprecipitates from the strong vip4-2 mutant (Figure 6B). Only a marginally detectable amount of VIP6-immunoreactive protein was immunoprecipitated from vip4-2 extracts with anti-VIP6 IgGs (Figure 6B), consistent with the previous observation that VIP6 protein accumulation is dependent on functional VIP4.
Figure 6.
Coimmunoprecipitation of VIP3, VIP4, VIP5, and VIP6 in Vivo.
(A) and (B) Interaction between VIP4 and VIP6. Total protein from wild-type inflorescence apices (four lanes on left in each panel) was subjected to immunoprecipitation using anti-VIP4 IgGs (A) or anti-VIP6 IgGs (B). Immunoprecipitates were analyzed by protein gel blotting using anti-VIP6 or anti-VIP4 serum as indicated at left. No immunoreactive protein was detected when immunoprecipitations were performed in the absence of IgGs (mock) or using the respective preimmune sera. Parallel immunoprecipitations were performed using extracts from the strong vip4-2 and vip6-1 mutants (two lanes on right in each panel).
(C) Interaction between VIP3, and VIP4 and VIP6. Total inflorescence apex protein from vip3-1 plants expressing a transgenic copy of FLAG-epitope-tagged VIP3 (see Supplemental Figure 3B online) was subjected to immunoprecipitation using anti-FLAG antibody. Immunoprecipitates were analyzed by protein gel blotting using anti-VIP6 or anti-VIP4 serum as indicated at left. No immunoreactive protein was detected when immunoprecipitations were performed in the absence of antibody (mock). In each panel, an unrelated, VIP6-immunoreactive protein species present in total protein extracts is indicated (*). Immunoblots were developed using colorimetric detection (anti-VIP6) or enhanced chemiluminescence and autoradiography (anti-VIP4).
To determine if the VIP6 and VIP4 proteins also interact with the previously described VIP3 in vivo, we constructed and expressed a FLAG-epitope-tagged copy of the VIP3 protein in vip3-1 mutant plants (see Supplemental Figure 3B online). This epitope-tagged VIP3 protein fully complemented the vip3 mutant phenotype, indicating that it is functional (data not shown). Anti-FLAG antibodies specifically immunoprecipitated anti-VIP4- and anti-VIP6-immunoreactive proteins of the molecular masses expected for VIP4 and VIP6 (Figure 6C). Based on these observations, we conclude that VIP4, VIP6, and VIP3 interact in a protein complex in vivo.
VIP5 Encodes an Additional Paf1C Subunit Homolog
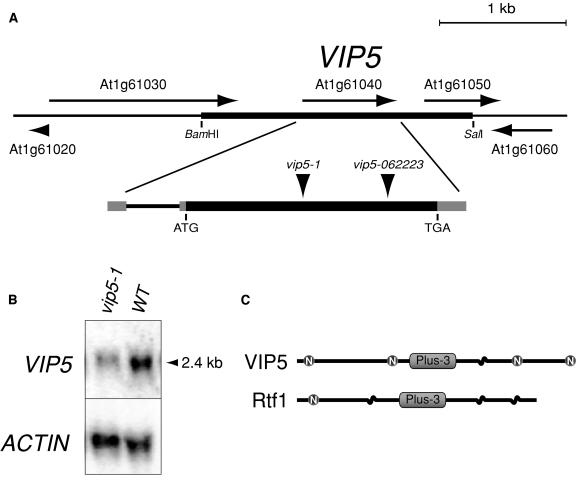
The homology of VIP4 and VIP6 with yeast Paf1C components brought up the possibility that other VIP genes encode plant homologs of additional Paf1C subunits. Besides Ctr9 and Leo1, the Paf1C complex includes at least three other proteins: Rtf1, Paf1, and Cdc73. Rtf1 is represented by a single Arabidopsis homolog, designated At1g61040 (data not shown). This gene is located within the likely genetic interval determined for both VIP2 and VIP5 (Zhang et al., 2003), and therefore we explored the possibility that At1g61040 was one of these genes. We sequenced the At1g61040 gene from the vip5-1 mutant and found a small insertion-deletion mutation that would terminate the reading frame after amino acid 319 of the predicted 643-amino acid protein (Figure 7A; see also Supplemental Figure 4 online). RNA gel blotting indicated a ∼2.4-kb species that was present at reduced levels in the vip5-1 mutant relative to wild-type plants, suggesting that this mutation affects mRNA accumulation in addition to protein sequence (Figure 7B). We also found that the SIGnAL T-DNA line 062223, which has an insertion within the open reading frame of At1g61040 (Figure 7A), exhibited a phenotype superficially indistinguishable from that of vip5-1 (data not shown). As further evidence that At1g61040 is VIP5, transgenic introduction of a ∼5.5-kb DNA containing the At1g61040 transcriptional unit into the vip5-1 mutant background fully complemented the vip5-1 phenotypes (Figure 7A and data not shown). Antisense expression of At1g61040 in a wild-type background conferred a varying degree of early flowering to a majority of primary (T1) transformants. Similar to the effect seen with VIP6-antisense plants, only a fraction of early-flowering plants exhibited strong developmental pleiotropy as seen in the vip5-1 mutant (see Supplemental Figure 1 online). Ectopic expression of VIP5 in transgenic wild-type plants did not confer obvious phenotypic consequences (data not shown).
Figure 7.
Structure and Expression of VIP5.
(A) VIP5 genomic region and transcriptional unit. Exons are shown as black (translated region) or gray (untranslated region) boxes. The positions of the start codon (ATG) and termination codon (TGA) are shown. The positions of the insertion/deletion in the vip5-1 allele and of the T-DNA insertion in the SIGnAL allele vip5-062223 are indicated. The BamHI/SalI fragment utilized in molecular complementation of the vip5 mutation is depicted as a thickened line encompassing the VIP5 gene.
(B) Gel blot analysis of VIP5 mRNA abundance in the strong vip5-1 mutant and in wild-type plants. The migration position of a ∼2.4-kb RNA size marker is indicated. The blot was subsequently hybridized with an ACTIN probe to indicate relative quantity and quality of mRNA in each lane.
(C) Domain structure of the VIP5 protein and related budding yeast Rtf1. The Plus-3 motif is indicated. Potential nuclear localization signal sequences are also shown (N). Coiled-coil regions are depicted as sinuous segments.
We found that VIP5 mRNAs were expressed to similar levels in strong vip3, vip4, and vip6 mutants, and, similar to VIP6, were expressed ubiquitously throughout the plant, with strongest accumulation in apices, and were unchanged in vernalized plants or in the Col background (data not shown). Based on current genomic annotation and sequence analysis of several cDNAs, At1g61040/VIP5 encodes a protein containing coiled-coil regions, four potential nuclear localization signal sequences, and a so-called Plus-3 domain (Figure 7C). The Plus-3 domain (PFam accession number PF03126), so named because of the presence of three conserved positively charged residues, has no recognized function, but is found in several other Arabidopsis and eukaryotic proteins (data not shown). The homologous yeast Rtf1 protein also contains these structural features (Figure 7C).
VIP Genes Are Not Required for Global Methylation of Histone H3
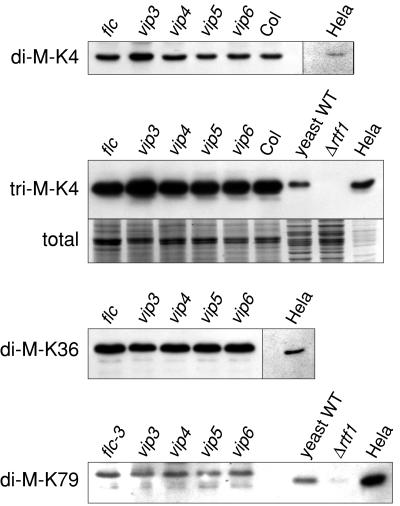
The conservation of the VIP4/Leo1, VIP5/Rtf1, and VIP6/Ctr9 proteins, and the role of the Paf1C complex in histone methylation in yeast, suggested that these proteins may be involved in histone methylation in plants. To address this, we examined global histone methylation profiles in strong vip4, vip5, and vip6 mutants. Chromatin histone-enriched proteins were extracted and analyzed by immunoblotting using antisera specific for histone H3 methylated at K4, K36, or K79. In each case, the antibodies reacted strongly with a single species of the predicted appropriate molecular mass (Figure 8), suggesting that these histone modifications are conserved in plants. However, there was no discernible difference in apparent abundance of modified histones in the vip4, vip5, or vip6 mutants when compared with control (flc-3 or Col) extracts. Similar results were obtained with a strong vip3 mutant (Figure 8). Thus, these VIP proteins do not appear to be essential for these histone modifications in Arabidopsis, at least when assayed in total plant tissues at the whole-genome level.
Figure 8.
Immunoblot Analysis of Histone H3 Methylation in Strong vip3, vip4, vip5, and vip6 Mutants, the flc-3 Null Mutant, and the Col Ecotype.
Histone-enriched extracts were resolved by SDS-PAGE and subjected to immunoblotting using antibodies directed against dimethylated Lys-4 (di-M-K4), trimethylated Lys-4 (tri-M-K4), dimethylated Lys-36 (di-M-K36), or dimethylated Lys-79 (di-M-K79). Histone-enriched extracts from human Hela cells, or total protein extracts from wild-type yeast and a yeast rtf1 deletion strain (Δrtf1) are included as controls. The separate images of Hela cell extract results were taken from the same immunoblot. A portion of a representative SDS-PAGE gel (stained with Coomassie blue) is shown to indicate relative quality and quantity of proteins in each lane (total).
DISCUSSION
Through the work reported here, we show that proteins related to a transcriptional complex from budding yeast are conserved in higher eukaryotes, and in Arabidopsis play a role in the expression of a diverse subset of genes including members of the FLC/MAF family of flowering-time regulators. Our findings add to the increasing complexity of mechanisms of both epigenetic gene regulation and flowering time.
The VIP Genes Have a Central Role in Flowering through Activation of the FLC/MAF Gene Family
Our previous observations that vip mutants flower earlier than flc null mutants suggested that other flowering-time genes in addition to FLC are targeted (Zhang et al., 2003). In accordance with this, here we found that loss of VIP5 or VIP6 function led to downregulation of not only FLC, but also other members of the FLC/MAF MADS-box gene family, all of which have the capacity to act as floral repressors (Ratcliffe et al., 2001, 2003; Scortecci et al., 2001). MADS box genes are commonly involved in regulatory cascades, and we considered the possibility that the downregulation of the MAF genes in vip5 and vip6 was mediated through silencing of FLC. However, these experiments were performed in an flc null genetic background and in strong vip mutants where FLC expression was not detected, suggesting that the regulation of the MAF genes occurred independently of FLC activity. Conversely, we considered the possibility that the observed silencing of FLC in the vip5 and vip6 backgrounds was an indirect result of downregulation of MAF genes. However, Ratcliffe et al. (2001, 2003) formerly demonstrated that FLC mRNA abundance was not affected by enhanced, constitutive expression of MAF1 or MAF2, or by mutation in MAF2, suggesting that FLC is normally not subject to regulation by at least these two genes. Therefore VIP5 and VIP6 likely regulate members of the FLC/MAF gene family independently.
The observed common regulation of distinct members of the FLC/MAF gene family by VIP5 and VIP6 is surprising because genetic and molecular analyses have identified clear differences in regulation and function among at least some of these genes. For example, mutation in FLC abrogated the late flowering conferred by functional FRI alleles and loss of function of autonomous pathway genes such as FVE and FCA (Sanda and Amasino, 1996), whereas having little effect on the photoperiodic response (Michaels and Amasino, 2001). In accordance with this, FLC gene expression was found to be strongly activated by FRI and repressed by the autonomous pathway genes, but relatively insensitive to regulation by genes intimately involved in photoperiodic flowering (Sheldon et al., 1999). By contrast, a strong maf1/flm mutation led to substantial loss of the photoperiodic flowering response and abrogated late flowering conferred by mutations in photoperiodic pathway genes (Scortecci et al., 2001, 2003). Also, MAF1/FLM is apparently not subject to appreciable regulation by FRI or autonomous pathway genes (Ratcliffe et al., 2001; Scortecci et al., 2001). Interestingly, however, like FLC, at least the MAF1-MAF4 genes have been reported to be downregulated after growth in the cold, albeit to different degrees and with different kinetics (Ratcliffe et al., 2001, 2003; our unpublished results).
The regulation of these FLC/MAF genes by both cold and the VIP genes might suggest a link between vernalization and VIP gene function. One possibility is that vernalization attenuates VIP activity, potentially through modifying abundance or activity of one or more VIP genes/proteins, thus resulting in FLC/MAF gene downregulation and silencing. However, we have not observed vernalization-associated changes in mRNA or protein levels for any of the VIPs tested, including VIP5 or VIP6. Also arguing against this possibility is the apparent requirement for VIP gene activity in unrelated developmental events in vernalized plants, as evidenced by the fact that the molecular and developmental pleiotropy of the respective vip mutants is not observed in vernalized wild-type plants (Zhang et al., 2003). Therefore, it is most likely that vernalization and the VIP genes regulate FLC/MAF genes through independent mechanisms.
VIP5 and VIP6 Define Important Pleiotropic Regulators of Development
Our transcriptional profiling experiments identified FLC as one of the genes most severely affected in the vip mutants (Figure 2), suggesting a special dependence on VIP activity. However, we also observed misregulation of a subset of genes not obviously related to FLC, and this was expected given the developmental pleiotropy conferred by the vip mutations. The subset of genes regulated by VIP5 and VIP6 appears diverse in structure, function, and genomic location (data not shown), and the common features that confer dependence on VIP5/VIP6 are not known. A trivial explanation is that expression of these genes may be localized to flowers or shoot and inflorescence apices, where the VIP5 and VIP6 genes are preferentially expressed. However, our microarray data indicated that expression of a variety of genes formerly reported to be localized to flowers or shoot/inflorescence apices, including VIP3 and VIP4, was independent of VIP5/VIP6 (data not shown).
The VIP Genes Cooperatively Regulate Gene Expression through a Mechanism Related to the Yeast Transcriptional Regulator Paf1C
The seven defined VIP genes carry out a common function in plant growth and development, based on their indistinguishable developmental pleiotropy (Zhang et al., 2003). Consistent with this, we did not observe enhanced effects on flowering time or development when vip3 or vip4 mutations were combined with vip5 or vip6 mutations. A common function for VIP5 and VIP6 was also reflected by the high degree of overlap among misregulated genes in the respective mutants. Because the VIP6 protein is not effectively expressed in the vip5 mutant background, the limited differences that we observed in transcriptional profiles between the vip5 and vip6 mutants could reflect roles for VIP5 that are independent of VIP6. Alternatively, although the vip5 and vip6 mutants were backcrossed to wild-type plants extensively before analysis, these distinctions could also have resulted from genetic lesions unrelated to the vip mutations that were sustained in the original mutagenesis and are still harbored by either vip5 or vip6.
Our observation that the abundance of VIP6 protein, but not mRNA, is dependent on functional VIP3, VIP4, and VIP5 could be explained by reduced posttranslational stability of VIP6 in the absence of participation in a protein complex, presumably involving VIP3, VIP4, and VIP5. The finding that at least VIP3, VIP4, and VIP6 physically interact in vivo is also consistent with the hypothesis that these proteins comprise a protein complex. We formerly reported that VIP4 is a plant homolog of budding yeast Leo1 (Zhang and van Nocker, 2002). Subsequently, it was revealed that Leo1 is a component of the Paf1C transcriptional complex (Mueller and Jaehning, 2002). Here, we identified VIP5 and VIP6 as homologous to the Paf1C components Rtf1 and Ctr9, respectively. These cumulative findings suggest that the Arabidopsis VIP proteins define a plant counterpart of Paf1C. Consistent with this, we have determined that the At1g79730 gene, which encodes a protein weakly related to the Paf1 component of the Paf1C complex, is likely VIP2 (M.J. Ek-Ramos and S. van Nocker, unpublished results). The WD-repeat protein VIP3 has obvious homologs in animals but not budding yeast (Zhang et al., 2003). WD-repeat proteins are common constituents of large chromatin-associated complexes (van Nocker and Ludwig, 2003), and it is tempting to speculate that VIP3 represents an elaboration of the Paf1C mechanism not relevant for the relatively simple chromatin of yeast.
Although the core components of yeast Paf1C are conserved in higher eukaryotes, their cellular and organismal role has not been explored. The VIP6/Ctr9 protein exhibits homology with murine p150TSP. This protein was previously isolated based on its in vitro affinity for an isolated Src homology (SH2) domain, a conserved, ∼100 amino acid, phosphopeptide binding module that has been best characterized as a component of proteins with roles in cellular signaling pathways including signal transducer and activator of transcription and suppressor of cytokine signaling proteins, janus kinases, and other tyrosine kinases (Pawson, 2004). Although these signaling pathways are generally not tightly conserved in plants, it remains a possibility that VIP6 couples transcription with plant-specific signaling pathways. In support of this, in vitro binding of p150TSP protein to SH2 is dependent on phosphorylation of Ser/Thr residues and the highly charged C-terminal region of p150TSP (Malek et al., 1996), features that are conserved in VIP6.
Paf1C plays a central role in transcription in yeast and, although not essential for viability, is required for full expression of a variety of yeast genes (Porter et al., 2002). Paf1C components assist in the ubiquitination of the C-terminal domain of histone H2B by the ubiquitin conjugating/ligase proteins Rad6/Bre1 (Ng et al., 2003a; Wood et al., 2003b). Ubiquitination of H2B within promoter regions, as well as ensuing deubiquitination by the SAGA histone acetyltransferase-associated Ubp8, is required for efficient activation of many genes in yeast (Henry et al., 2003). At least in yeast, H2B ubiquitination is also a prerequisite for methylation of histone H3 at lysines 4 and 79 by the histone methylases Set1/COMPASS and Dot1, respectively, within open reading frames (Wood et al., 2003b). These histone modifications have most often been associated with actively transcribed genes (Hampsey and Reinberg, 2003). At least the Rtf1 subunit of Paf1C is also required for efficient, locus-specific H3K36 methylation by an additional histone methylase, Set2, an activity that is apparently independent of H2B ubiquitination (Ng et al., 2003a). Unlike yeast strains deleted for components of Paf1C, Arabidopsis vip mutants did not exhibit detectable defects in methylation at H3K4, K36, or K79 when assayed on a bulk chromatin and total plant tissue basis. Potentially, such an activity is redundant in plants, or occurs in a tissue-specific or locus-specific manner.
Paf1C subunits associate with the initiating and elongating forms of Pol II (Mueller and Jaehning, 2002), and are bound within 5′ regions and open reading frames of various genes (Krogan et al., 2002; Simic et al., 2003). Given the association of Paf1C with elongating Pol II, the capacity of Paf1C to promote H3K4 methylation, and the observation that trimethylation at H3K4 is uniquely associated with actively transcribed loci, together with the apparent lack of enzymes that could demethylate histones, it has been hypothesized that H3K4 trimethylation comprises a molecular memory of recent gene transcription (Ng et al., 2003b). How this memory mechanism is manifested at the molecular level remains mostly unknown. However, methylation of H3K4 can recruit the Isw1 chromatin-remodeling ATPase (Santos-Rosa et al., 2003), which generates specific chromatin changes at the 5′ end of genes needed for correct distribution of Pol II throughout the transcribed region and assembly of the cleavage and polyadenylation machinery (Morillon et al., 2003).
The prospect that the homologous plant mechanism also participates in FLC transcription through generating active patterns of histone modification is intriguing. In animals, epigenetic maintenance of homeotic gene activity involving the trxG proteins also involves histone methylation at residues including H3K4. For example, the human trx-related MLL (mixed lineage leukemia) protein carries out H3K4 methylation at Hox loci in vivo (Milne et al., 2002). Similarly, in flies, H3K4 methylation by the epigenetic activator Ash1 is essential to maintain activity of homeotic genes in the developing embryo (Tripoulas et al., 1994; Beisel et al., 2002). Similar to the observed recruitment of Isw1 by methylated H3K4 in yeast, Ash1 activity involves the recruitment of the trxG chromatin-remodeling ATPase Brahma (Beisel et al., 2002). We hypothesize that the role of the VIP proteins in promoting FLC activity in nonvernalized plants is to provide a transcription-associated platform for the modification of FLC chromatin by a trxG-like mechanism, that would antagonize repression by a VRN2-associated PcG mechanism. This effect could involve the recruitment of chromatin-remodeling factors such as PIE1, an ISWI-family protein formerly shown to be required for full expression of FLC (Noh and Amasino, 2003). The only Arabidopsis trx-like protein to have been characterized to date, ATX1, functions in the activation of homeotic genes, and can methylate a synthetic peptide corresponding to the H3 amino-terminus on K4 in vitro (Alvarez-Venegas et al., 2003). A strong atx1 mutation conferred mildly delayed flowering and floral abnormalities that were seemingly distinct from that of the vip mutants (Alvarez-Venegas et al., 2003), suggesting that the activity of ATX1 is not closely tied to that of the VIP genes. However, the Arabidopsis genome encodes for several additional trx-related proteins that could promote FLC expression (Baumbusch et al., 2001).
A very recent study indicates that Paf1C also has transcriptional roles seemingly distinct from chromatin modification (Mueller et al., 2004). Loss of Rtf1 or Cdc73, which dissociated remaining Paf1C proteins from chromatin, conferred only mild phenotypes relative to those resulting from loss of other Paf1C proteins such as Paf1 or Ctr9. Similarly, loss of Paf1 or Ctr9 affected growth to a greater extent than loss of Bre1 or histone methylation (Wood et al., 2003a; Mueller et al., 2004). Moreover, loss of Paf1 or Rtf1 led to global defects in mRNA polyadenylation, suggesting an important function for Paf1C in posttranscriptional events. The increasingly diverse repertoire of activities attributed to Paf1C and its subunits allows wide latitude for speculation on the means by which the related plant VIP mechanism participates in FLC expression, and provides numerous avenues for further exploration. The identification of additional factors required to maintain FLC expression through genetic and biochemical methods holds exceptional promise and may illuminate many more unanticipated connections between basic transcription and development in higher eukaryotes.
METHODS
Plant and Yeast Material and Manipulations
The late-flowering, winter-annual introgression lines Col:FRISF2 (used here as the wild type) and Ler:FRISF2:FLCSF2 were as described previously (Zhang et al., 2003). The null flc-3 mutant in the Col:FRISF2 background was a gift from R. Amasino (University of Wisconsin). Populations derived from introgression line Col:FRISF2 mutagenized by fast-neutron radiation, ethyl methanesulfonate, or T-DNA insertion were described previously (Zhang et al., 2003). Mutant lines were backcrossed three times in succession to wild-type plants before phenotypic analysis. The vip5-1 and vip6-3 lines were backcrossed to wild-type plants an additional two times before microarray analysis. Sequence-indexed T-DNA-mutagenized lines developed at SIGnAL were obtained from the Arabidopsis Biological Resource Center at The Ohio State University (Columbus, OH). Standard genetic techniques were used in the production of double mutants. For vernalizing cold treatments, seeds were allowed to germinate on sterile media (Zhang and van Nocker, 2002) at 5°C in an 8-h-light/16-h-dark photoperiod for various lengths of time.
The Escherichia coli strain harboring BAC T7P1 was obtained from the Arabidopsis Biological Resource Center. Yeast strains YJJ662 (wild type) and its derivative YJJ1303 (rtf1 deletion) were obtained from T. Washburn and J. Jaehning, University of Colorado Health Science Center, and are described in Betz et al. (2002).
Cloning of VIP6
Positional cloning of the VIP6 gene utilized early-flowering F2 progeny of a single F1 individual derived from a cross between vip6-1 and introgression line Ler:FRISF2:FLCSF2. Bulked-segregant analysis was performed with 24 F2 individuals and molecular markers described by Lukowitz et al. (2000). Fine mapping was done entirely using molecular markers based on small insertion-deletion polymorphisms as characterized and cataloged by Cereon (Cambridge, MA) (http://www.arabidopsis.org/Cereon/index.jsp) and noted in Figure 4A.
Molecular Techniques
For production of VIP6 antisense plants, a 2.6-kb fragment corresponding to a portion of the translated region was amplified from apex cDNA using the primers VIP6FBam (5′-AAAGGATCCTATGGATTTGCAAGCAAATGATTG-3′) and VIP6RBam (5′-AAAGGATCCCTGTTGTTATGTATGAAATA-3′) and inserted into vector pPZP201:BAR:35S (Zhang and van Nocker, 2002). For production of VIP5 antisense plants, a 2-kb cDNA corresponding to the entire translated region was amplified from shoot apex-derived cDNA using primers VIP5FBam (5′-AAAGGATCCTATGGGTGATTTAGAGAACTTGC-3′) and VIP5RBam (5′-AAAGGATCCAAGAAGCAGTTTTCAGAAG-3′), and inserted into pPZP201:BAR:35S. For production of transgenic plants constitutively expressing VIP5 in the sense orientation, VIP5 coding and 3′ nontranslated region was amplified from genomic DNA using primers F35SVIP5 (5′-AAATCTAGACCTTAGAAGATTATGGGTGA-3′) and R35SVIP5 (5′-AAAGGATCCCCACGATCCATACACGAGCA-3′), and ligated into pPZP201:BAR:35S. For molecular complementation of the vip5 mutation, a ∼5.5-kb SalI/BamHI fragment DNA containing the At1g61040 transcriptional unit was excised from purified BAC T7P1 DNA and inserted into vector pPZP201:BAR (Zhang and van Nocker, 2002). Transgenic plants expressing FLAG-epitope-tagged VIP3 protein were engineered by ligating the VIP3 transcriptional unit, containing 1.2 kb of 5′/promoter DNA, into the plant expression vector pHuaFLAG (H. Zhang, unpublished results, available upon request), and introducing the resulting construction directly into the vip3-1 mutant background. The pHuaFLAG vector is based on pPZP201:BAR, and allows for a C-terminal translational fusion of two tandem copies of the FLAG epitope. The VIP3 transcriptional unit was amplified from wild-type plant DNA using the primers PstI-VIP3F (5′-AAACTGCAGTAACGCTCGAGCTTCTTCACCC-3′) and BamHI-VIP3R (5′-AAAGGATCCTGAGTAATCATAGAGCGATACA-3′).
RT-PCR Analysis
Relatively quantitative RT-PCR analysis of FLC/MAF gene family expression was performed using conditions and oligonucleotide primers as described by Ratcliffe et al. (2001, 2003). The number of cycles was varied for each primer set: MAF1, 35 cycles; FLC, MAF2, MAF3, and MAF4, 30 cycles; and ACTIN, 20 cycles. Similar conditions were used to analyze VIP6 expression, with primers VIP6FNcoI (5′-AAACCATGGATTTGCAAGCAAATGATTC-3′) and VIP6R2Bam (5′-AAAGGATCCAGTTATGTGGCCTTTCGCATGTACTC-3′) and 39 cycles.
Immunoblot Analysis
For use as antigen in rabbits, an N-terminal portion (amino acids 2 to 404) of the predicted VIP6 protein, and an N-terminal portion (amino acids 1 to 202) of the predicted VIP4 protein, were expressed in E. coli as hexahistidine fusions and purified using Ni2+-affinity chromatography. Anti-FLAG M2 monoclonal antibody was purchased from Sigma (St. Louis, MO; catalog no. F-3165).
Total protein extracts from aerial portions of 3-week-old plants were prepared as described previously (Zhang et al., 2003). Histone-enriched extracts were prepared as described by Moehs et al. (1988). Immunoblot analysis of histone-enriched extracts utilized the following antibodies: H3 di-M-K4 (Upstate, Lake Placid, NY; catalog no. 07-030), H3 di-M-K36 (Upstate; catalog no. 07-369), H3 di-M-K79 (Upstate; catalog no. 07-366), and H3 tri-M-K4 (Abcam, Cambridge, MA; catalog no. Ab 8580-50).
For immunoprecipitation experiments, anti-VIP4 and anti-VIP6 IgGs were purified by elution from Protein A-agarose (Roche, Indianapolis, IN) using a procedure described by the manufacturer. We used protein extracts from inflorescence apices, because VIP4 and VIP6 are strongly expressed in these tissues; ∼500 μg of protein extract, in a volume of 500 μL of extraction buffer (50 mM Tris-HCl, pH 8.0, 150 mM NaCl, 1 mM EDTA, 0.2% Triton X-100, containing 1 mM phenylmethylsulfonyl fluoride was incubated with 10 μL of IgGs, and mixed continuously for 2 h. Protein A-agarose beads (15 μL) were then added, and the mixture was incubated a further 1 h. Protein A-agarose beads were collected by centrifugation and washed with 1 mL ice-cold washing buffer (extraction buffer lacking Triton X-100) four times. After the final wash, the beads were resuspended in 30 μL of SDS-PAGE sample buffer. All immunoprecipitation procedures were performed at 4°C.
Immunoblotting was done as described by Harlow and Lane (1988), using polyvinylidene difluoride membranes (Bio-Rad, Hercules, CA) blocked with Tween-20 in phosphate-buffered saline, and alkaline-phosphatase-labeled, goat anti-rabbit IgGs (Bio-Rad), or enhanced chemiluminscence (Amersham Biosciences, Piscataway, NJ), using nitrocellulose membranes (Amersham) blocked with 3% skim milk in phosphate-buffered saline, and peroxidase-conjugated, anti-rabbit IgGs (Amersham).
Sequence Analyses
Motifs in the VIP5 and VIP6 proteins were identified using PFam version 13.0 on a Web server maintained by Washington University in St. Louis (http://pfam.wustl.edu/hmmsearch.shtml) or the PredictNLS server at the Columbia University Bioinformatics Center (http://cubic.bioc.columbia.edu/; Cokol et al., 2000). Other sequence analyses were performed using BLAST on Web servers maintained by the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov) or The Arabidopsis Information Resource (http://www.arabidopsis.org), and programs of the Genetics Computer Group (Madison, WI).
Microarray Analysis
Each of the two replicates for each genotype was composed of three independently derived samples. Each sample included between 12 and 20 plants. Total aerial tissues were harvested when the first flower was fully opened. Total RNA was isolated using Qiagen RNAeasy columns (Qiagen, Valencia, CA). Synthesis of cDNA employed the SuperScript double-stranded cDNA synthesis kit (Invitrogen, Carlsbad, CA) and 100 pmol of oligo(dT)24 primer (Proligo, Boulder, CO), following the manufacturers' instructions. Synthesis of biotinylated cDNA utilized the BioArray high yield RNA transcript labeling kit (Enzo Diagnostics, Farmingdale, NY). The Arabidopsis ATH1 Genome Array (Affymetrix, Santa Clara, CA) was used for hybridization. Hybridization and scanning of microarrays was performed at the Genomics Technology Support Facility at Michigan State University.
Microarray data were analyzed using the statistical algorithms within the Affymetrix Microarray Suite (MAS) 5.0 software. We employed pairwise comparisons of the independent biological replicates to identify genes that exhibited a marked change in transcript abundance according to an arbitrary stringent or relaxed definition. For the stringent definition, the gene must have been detected at a statistically significant level (i.e., called present or marginal by the MAS software) in both replicates of either the experiment (vip5 or vip6) or the baseline (flc). In addition, the MAS software must have observed a statistically significant change in expression (i.e., called decrease, marginal decrease, increase, or marginal increase) in at least three of the four comparisons, and the mean difference in signal intensity must have been threefold or greater. The number of genes that were detected and exhibited a significant change in expression of at least threefold in any one comparison of replicate sample pairs (i.e., flc versus flc, vip5 versus vip5, or vip6 versus vip6) was, at most, 80 (0.35% of microarrayed genes). For the relaxed definition, the gene must have been detected at a statistically significant level in either replicate of either the experiment or the baseline, the MAS software must have observed a statistically significant change in expression in at least two of the four comparisons, and the mean change in signal intensity must have been twofold or greater. The number of genes that met this relaxed criteria for any one comparison of replicate sample pairs was, at most, 512 (2.25% of microarrayed genes).
Microarray data discussed here have been deposited with the Gene Expression Omnibus database at the NCBI (http://www.ncbi.nlm.nih.gov/geo/; series no. GSE1516).
Supplementary Material
Acknowledgments
We acknowledge the gift of the vip6-3 and flc-3 mutants from Richard Amasino (University of Wisconsin), and yeast strains from Taylor Washburn and Judith Jaehning (University of Colorado Health Sciences Center). We thank Annette Thelen and staff of the Michigan State University Genomics Technology Support Facility for assistance with statistical analysis of microarray data, Ying Yan for assistance with plotting microarray data, Steve Rounsley (formerly of Cereon) for making the Cereon Arabidopsis Polymorphism Collection available, and Paolo Struffi for providing DNA for construction of pHuaFLAG. This work was supported by the Michigan Agricultural Experiment Station and a USDA National Research Initiative Competitive Grant (79-35304-5108) to S.V.N.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Steven van Nocker (vannocke@msu.edu).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026062.
References
- Alonso, J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657. [DOI] [PubMed] [Google Scholar]
- Alvarez-Venegas, R., Pien, S., Sadder, M., Witmer, X., Grossniklaus, U., and Avramova, Z. (2003). ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr. Biol. 13, 627–637. [DOI] [PubMed] [Google Scholar]
- Bastow, R., Mylne, J.S., Lister, C., Lippman, Z., Martienssen, R.A., and Dean, C. (2004). Vernalization requires epigenetic silencing of FLC by histone methylation. Nature 427, 164–167. [DOI] [PubMed] [Google Scholar]
- Baumbusch, L.O., Thorstensen, T., Krauss, V., Fischer, A., Naumann, K., Assalkhou, R., Schulz, I., Reuter, G., and Aalen, R.B. (2001). The Arabidopsis thaliana genome contains at least 29 active genes encoding SET domain proteins that can be assigned to four evolutionarily conserved classes. Nucleic Acids Res. 29, 4319–4333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beisel, C., Imhof, A., Greene, J., Kremmer, E., and Sauer, F. (2002). Histone methylation by the Drosophila epigenetic transcriptional regulator Ash1. Nature 419, 857–862. [DOI] [PubMed] [Google Scholar]
- Betz, J.L., Chang, M., Washburn, T.M., Porter, S.E., Mueller, C.L., and Jaehning, J.A. (2002). Phenotypic analysis of Paf1/RNA polymerase II complex mutations reveals connections to cell cycle regulation, protein synthesis, and lipid and nucleic acid metabolism. Mol. Genet. Genomics 268, 272–285. [DOI] [PubMed] [Google Scholar]
- Cokol, M., Nair, R., and Rost, B. (2000). Finding nuclear localization signals. EMBO Rep. 1, 411–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Andrea, L.D., and Regan, L. (2003). TPR proteins: The versatile helix. Trends Biochem. Sci. 28, 655–662. [DOI] [PubMed] [Google Scholar]
- Fischle, W., Wang, Y., and Allis, C.D. (2003). Binary switches and modification cassettes in histone biology and beyond. Nature 425, 475–479. [DOI] [PubMed] [Google Scholar]
- Francis, N.J., and Kingston, R.E. (2001). Mechanisms of transcriptional memory. Nat. Rev. Mol. Cell Biol. 2, 409–421. [DOI] [PubMed] [Google Scholar]
- Gendall, A.R., Levy, Y.Y., Wilson, A., and Dean, C. (2001). The VERNALIZATION 2 gene mediates the epigenetic regulation of vernalization in Arabidopsis. Cell 107, 525–535. [DOI] [PubMed] [Google Scholar]
- Goodrich, J., Puangsomlee, P., Martin, M., Long, D., Meyerowitz, E.M., and Coupland, G. (1997). A Polycomb-group gene regulates homeotic gene expression in Arabidopsis. Nature 386, 44–51. [DOI] [PubMed] [Google Scholar]
- Hampsey, M., and Reinberg, D. (2003). Tails of intrigue: Phosphorylation of RNA polymerase II mediates histone methylation. Cell 113, 429–432. [DOI] [PubMed] [Google Scholar]
- Harlow, E., and Lane, D. (1988). Antibodies: A Laboratory Manual. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press.)
- Henry, K.W., Wyce, A., Lo, W.S., Duggan, L.J., Emre, N.C., Kao, C.F., Pillus, L., Shilatifard, A., Osley, M.A., and Berger, S.L. (2003). Transcriptional activation via sequential histone H2B ubiquitylation and deubiquitylation, mediated by SAGA-associated Ubp8. Genes Dev. 17, 2648–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein, T., and Allis, C.D. (2001). Translating the histone code. Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- Johanson, U., West, J., Lister, C., Michaels, S., Amasino, R., and Dean, C. (2000). Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290, 344–347. [DOI] [PubMed] [Google Scholar]
- Kohler, C., Hennig, L., Spillane, C., Pien, S., Gruissem, W., and Grossniklaus, U. (2003). The Polycomb-group protein MEDEA regulates seed development by controlling expression of the MADS-box gene PHERES1. Genes Dev. 17, 1540–1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogan, N.J., Kim, M., Ahn, S.H., Zhong, G., Kobor, M.S., Cagney, G., Emili, A., Shilatifard, A., Buratowski, S., and Greenblatt, J.F. (2002). RNA polymerase II elongation factors of Saccharomyces cerevisiae: A targeted proteomics approach. Mol. Cell. Biol. 22, 6979–6992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, A. (1965). Physiology of flower initiation. In Encyclopedia of Plant Physiology, W. Ruhland, ed (Berlin: Springer-Verlag), pp. 1380–1536.
- Lee, I., Bleecker, A., and Amasino, R.M. (1993). Analysis of naturally occurring late flowering in Arabidopsis thaliana. Mol. Gen. Genet. 237, 171–176. [DOI] [PubMed] [Google Scholar]
- Lee, T.I., and Young, R.A. (2000). Transcription of eukaryotic protein-coding genes. Annu. Rev. Genet. 34, 77–137. [DOI] [PubMed] [Google Scholar]
- Levy, Y.Y., Mesnage, S., Mylne, J.S., Gendall, A.R., and Dean, C. (2002). Multiple roles of Arabidopsis VRN1 in vernalization and flowering time control. Science 297, 243–246. [DOI] [PubMed] [Google Scholar]
- Lukowitz, W., Gillmor, C.S., and Scheible, W.R. (2000). Positional cloning in Arabidopsis. Plant Physiol. 123, 795–805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malek, S.N., Yang, C.H., Earnshaw, W.C., Kozak, C.A., and Desiderio, S. (1996). p150TSP, a conserved nuclear phosphoprotein that contains multiple tetratricopeptide repeats and binds specifically to SH2 domains. J. Biol. Chem. 271, 6952–6962. [DOI] [PubMed] [Google Scholar]
- Michaels, S.D., and Amasino, R.M. (2001). Loss of FLOWERING LOCUS C activity eliminates the late-flowering phenotype of FRIGIDA and autonomous pathway mutations but not responsiveness to vernalization. Plant Cell 13, 935–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaels, S.D., He, Y., Scortecci, K.C., and Amasino, R.M. (2003). Attenuation of FLOWERING LOCUS C activity as a mechanism for the evolution of summer-annual flowering behavior in Arabidopsis. Proc. Natl. Acad. Sci. USA 100, 10102–10107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne, T.A., Briggs, S.D., Brock, H.W., Martin, M.E., Gibbs, D., Allis, C.D., and Hess, J.L. (2002). MLL targets SET domain methyltransferase activity to Hox gene promoters. Mol. Cell 10, 1107–1117. [DOI] [PubMed] [Google Scholar]
- Moehs, C.P., McElwain, E.F., and Spiker, S. (1988). Chromosomal proteins of Arabidopsis thaliana. Plant Mol. Biol. 11, 507–515. [DOI] [PubMed] [Google Scholar]
- Morillon, A., Karabetsou, N., O'Sullivan, J., Kent, N., Proudfoot, N., and Mellor, J. (2003). Isw1 chromatin remodeling ATPase coordinates transcription elongation and termination by RNA polymerase II. Cell 115, 425–435. [DOI] [PubMed] [Google Scholar]
- Mueller, C.L., and Jaehning, J.A. (2002). Ctr9, Rtf1, and Leo1 are components of the Paf1/RNA polymerase II complex. Mol. Cell. Biol. 22, 1971–1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller, C.L., Porter, S.E., Hoffman, M.G., and Jaehning, J.A. (2004). The Paf1 complex has functions independent of actively transcribing RNA polymerase II. Mol. Cell 14, 447–456. [DOI] [PubMed] [Google Scholar]
- Ng, H.H., Dole, S., and Struhl, K. (2003. a). The Rtf1 component of the Paf1 transcriptional elongation complex is required for ubiquitination of histone H2B. J. Biol. Chem. 278, 33625–33628. [DOI] [PubMed] [Google Scholar]
- Ng, H.H., Robert, F., Young, R.A., and Struhl, K. (2003. b). Targeted recruitment of Set1 histone methylase by elongating PolII provides a localized mark and memory of recent transcriptional activity. Mol. Cell 11, 709–719. [DOI] [PubMed] [Google Scholar]
- Noh, Y.S., and Amasino, R.M. (2003). PIE1, an ISWI family gene, is required for FLC activation and floral repression in Arabidopsis. Plant Cell 15, 1671–1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orphanides, G., and Reinberg, D. (2000). RNA polymerase II elongation through chromatin. Nature 407, 471–475. [DOI] [PubMed] [Google Scholar]
- Pawson, T. (2004). Specificity in signal transduction: From phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell 116, 191–203. [DOI] [PubMed] [Google Scholar]
- Porter, S.E., Washburn, T.M., Chang, M., and Jaehning, J.A. (2002). The yeast Paf1-RNA polymerase II complex is required for full expression of a subset of cell cycle-regulated genes. Eukaryot. Cell 1, 830–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Kumimoto, R.W., Wong, B.J., and Riechmann, J.L. (2003). Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15, 1159–1169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliffe, O.J., Nadzan, G.C., Reuber, T.L., and Riechmann, J.L. (2001). Regulation of flowering in Arabidopsis by an FLC homologue. Plant Physiol. 126, 122–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanda, S., and Amasino, R.M. (1996). Interaction of FLC and late-flowering mutations in Arabidopsis thaliana. Mol. Gen. Genet. 251, 69–74. [DOI] [PubMed] [Google Scholar]
- Santos-Rosa, H., Schneider, R., Bernstein, B.E., Karabetsou, N., Morillon, A., Weise, C., Schreiber, S.L., Mellor, J., and Kouzarides, T. (2003). Methylation of histone H3 K4 mediates association of the Isw1p ATPase with chromatin. Mol. Cell 12, 1325–1332. [DOI] [PubMed] [Google Scholar]
- Scortecci, K.C., Michaels, S.D., and Amasino, R.M. (2001). Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J. 26, 229–236. [DOI] [PubMed] [Google Scholar]
- Scortecci, K., Michaels, S.D., and Amasino, R.M. (2003). Genetic interactions between FLM and other flowering-time genes in Arabidopsis thaliana. Plant Mol. Biol. 52, 915–922. [DOI] [PubMed] [Google Scholar]
- Sheldon, C.C., Burn, J.E., Perez, P.P., Metzger, J., Edwards, J.A., Peacock, W.J., and Dennis, E.S. (1999). The FLF MADS box gene: A repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11, 445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simic, R., Lindstrom, D.L., Tran, H.G., Roinick, K.L., Costa, P.J., Johnson, A.D., Hartzog, G.A., and Arndt, K.M. (2003). Chromatin remodeling protein Chd1 interacts with transcription elongation factors and localizes to transcribed genes. EMBO J. 22, 1846–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Squazzo, S.L., Costa, P.J., Lindstrom, D.L., Kumer, K.E., Simic, R., Jennings, J.L., Link, A.J., Arndt, K.M., and Hartzog, G.A. (2002). The Paf1 complex physically and functionally associates with transcription elongation factors in vivo. EMBO J. 21, 1764–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, S., and Amasino, R.M. (2004). Vernalization in Arabidopsis thaliana is mediated by the PHD finger protein VIN3. Nature 427, 159–164. [DOI] [PubMed] [Google Scholar]
- Surridge, C. (2004). Plant development: The flowers that bloom in the spring. Nature 427, 112. [DOI] [PubMed] [Google Scholar]
- Svejstrup, J.Q. (2004). The RNA Pol II transcription cycle: Cycling through chromatin. Biochim. Biophys. Acta 1677, 64–73. [DOI] [PubMed] [Google Scholar]
- Tripoulas, N.A., Hersperger, E., LaJeunesse, D., and Shearn, A. (1994). Molecular genetic analysis of the Drosophila melanogaster gene absent, small or homeotic discs1 (ash1). Genetics 137, 1027–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nocker, S., and Ludwig, P. (2003). The WD-repeat protein superfamily in Arabidopsis: Conservation and divergence in structure and function. BMC Genomics 4, 50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood, A., Krogan, N.J., Dover, J., Schneider, J., Heidt, J., Boateng, M.A., Dean, K., Golshani, A., Zhang, Y., Greenblatt, J.F., Johnston, M., and Shilatifard, A. (2003. a). Bre1, an E3 ubiquitin ligase required for recruitment and substrate selection of Rad6 at a promoter. Mol. Cell 11, 267–274. [DOI] [PubMed] [Google Scholar]
- Wood, A., Schneider, J., Dover, J., Johnston, M., and Shilatifard, A. (2003. b). The Paf1 complex is essential for histone monoubiquitination by the Rad6-Bre1 complex, which signals for histone methylation by COMPASS and Dot1p. J. Biol. Chem. 278, 34739–34742. [DOI] [PubMed] [Google Scholar]
- Zhang, H., and van Nocker, S. (2002). The VERNALIZATION INDEPENDENCE 4 gene encodes a novel regulator of FLOWERING LOCUS C. Plant J. 31, 663–667. [DOI] [PubMed] [Google Scholar]
- Zhang, H., Ransom, C., Ludwig, P., and van Nocker, S. (2003). Genetic analysis of early flowering mutants in Arabidopsis defines a class of pleiotropic developmental regulator required for expression of the flowering-time switch FLOWERING LOCUS C. Genetics 164, 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.