Abstract
Cyclin-dependent kinases (CDKs) play essential roles in coordinate control of cell cycle progression. Activation of CDKs requires interaction with specific cyclin partners and phosphorylation of their T-loops by CDK-activating kinases (CAKs). The Arabidopsis thaliana genome encodes four potential CAKs. CAK2At (CDKD;3) and CAK4At (CDKD;2) are closely related to the vertebrate CAK, CDK7/p40MO15; they interact with cyclin H and phosphorylate CDKs, as well as the C-terminal domain (CTD) of the largest subunit of RNA polymerase II. CAK1At (CDKF;1) shows cyclin H-independent CDK-kinase activity and can activate a heterologous CAK, Mcs6, in fission yeast. In Arabidopsis, CAK1At is a subunit of a protein complex of 130 kD, which phosphorylates the T-loop of CAK2At and CAK4At and activates the CTD-kinase activity of CAK4At in vitro and in root protoplasts. These results suggest that CAK1At is a novel CAK-activating kinase that modulates the activity of CAK2At and CAK4At, thereby controlling CDK activities and basal transcription in Arabidopsis.
INTRODUCTION
The activation of cell division and transitions between different phases of the cell cycle are controlled by a family of cyclin-dependent Ser/Thr protein kinases (CDKs). The activity and substrate specificity of CDKs depend on their association with specific cyclin partners that act as regulatory subunits (Morgan, 1997). Activation of CDKs also requires phosphorylation of conserved Thr residues within their T-loops, such as residues Thr161 and Thr160 in human Cdc2 and CDK2, respectively. This activating phosphorylation is catalyzed by CDK-activating kinases (CAKs; Kaldis, 1999). Crystal structure analyses of monomeric CDK2 showed that the T-loop region blocks access of the potential substrates to the catalytic site (Morgan and De Bondt, 1994). Upon phosphorylation of Thr160 by CAK, the T-loop moves away and the cyclin A-CDK2 complex becomes fully active (Jeffrey et al., 1995; Russo et al., 1996).
CAK in fission yeast (Schizosaccharomyces pombe) phosphorylates Cdc2 and consists of catalytic Mcs6/Mop1/Crk1 and regulatory cyclin Mcs2 subunits (Damagnez et al., 1995). The Mcs2-Mcs6 CAK kinase is part of the general transcription factor IIH (TFIIH). In addition to Cdc2, it phosphorylates the C-terminal domain (CTD) of the largest subunit of RNA polymerase II thereby controlling both basal transcription and cell cycle progression (Buck et al., 1995; Damagnez et al., 1995; Lee et al., 1999). Recently, a monomeric kinase, Csk1, has been identified that phosphorylates the T-loops of both Mcs6 and Cdc2. This leads to the activation of these kinases (Hermand et al., 1998). Csk1 is thus defined as CAK-activating kinase (CAKAK), an upstream activating kinase of CAK/Mcs2-Mcs6 in fission yeast (Hermand et al., 2001; Saiz and Fisher, 2002).
Budding yeast (Saccharomyces cerevisiae) possesses a monomeric CAK, Cak1p, which shows low sequence similarity to Mcs6 and is required for the activation of Cdc28p through T-loop phosphorylation in vivo (Thuret et al., 1996; Cross and Levine, 2000; Ross et al., 2000). Unlike Mcs6, Cak1p lacks CTD-kinase activity (Kaldis et al., 1996). By contrast, a second budding yeast kinase, Kin28p, which is more closely related to Mcs6, is associated with TFIIH (Feaver et al., 1997; Keogh et al., 2002) and involved in phosphorylation of the CTD, but does not display Cdc28p phosphorylating activity (Cismowski et al., 1995). Recent reports indicate that Cak1p phosphorylates the T-loop of Kin28p and thereby stimulates its CTD-kinase activity (Kimmelman et al., 1999). This suggests that, despite their low sequence similarity, budding yeast Cak1p and fission yeast Csk1 perform similar functions by phosphorylating the CTD kinases, Kin28p and Mcs6.
One of the best characterized animal CAKs, CDK7/p40MO15, occurs in complex with cyclin H and phosphorylates both CDK and the CTD of RNA polymerase II (Kaldis, 1999; Wallenfang and Seydoux, 2002). CDK7 and cyclin H are close homologs of Mcs6 and Mcs2, respectively (Damagnez et al., 1995). A third subunit that stabilizes the cyclin H-CDK7 complex is MAT1 (Devault et al., 1995; Tassan et al., 1995), an assembly factor that has been identified in TFIIH complexes of mammals (Serizawa et al., 1995; Shiekhattar et al., 1995). MAT1 homologs, Tfb3/Rig2 and Pmh1, are also present in CAK complexes of budding and fission yeasts, respectively (Kaldis, 1999). However, no ortholog of either budding yeast Cak1p or fission yeast Csk1 was identified so far in vertebrates (Liu and Kipreos, 2002; Tsakraklides and Solomon, 2002).
The first plant CAK ortholog was identified as a Cdc2-related protein kinase named R2 in rice (Oryza sativa; Hata, 1991). R2 belongs to the CDK7 family, binds cyclin H, and phosphorylates both CDK and RNA polymerase II CTD (Yamaguchi et al., 1998, 2000). The sequenced Arabidopsis thaliana genome encodes four potential CAK orthologs, among which CAK2At, CAK3At, and CAK4At are closely related to CDK7 (Umeda, 2002). CAK2At and CAK4At phosphorylate both CDK and CTD, albeit with different preferences (Shimotohno et al., 2003). By contrast, CAK1At displays similarity to CDK7 only in restricted domains and unlike vertebrate-type CAKs it has CDK-activating kinase activity but no CTD-kinase activity (Umeda et al., 1998). Arabidopsis CAK1At-CAK4At were later designated CDKF;1, CDKD;3, CDKD;1, and CDKD;2, respectively, according to the nomenclature rules for CDKs proposed by Vandepoele et al. (2002). To better define the functions of Arabidopsis CAKs, we have identified a cyclin H homolog, At;CycH;1, and tested its interaction with CAK1At–CAK4At by monitoring CDK- and CTD-kinase activities and resolution of CAK complexes by gel filtration. The results show that CAK1At is active as a monomeric kinase and phosphorylates the T-loops of CAK2At and CAK4At. CAK1At activates the CTD-kinase activity of CAK4At in vitro and in Arabidopsis root protoplasts. The results suggest that phosphorylation of CDKs and RNA polymerase II CTD is mediated by a cascade of multiple CAKs in higher plants.
RESULTS
Identification of Arabidopsis Cyclin H At;CycH;1 That Interacts with CAK2At and CAK4At in Yeast Cells
In the Arabidopsis genome, a single gene, AT5G27620, codes for a cyclin H ortholog. We PCR-amplified the full-length cDNA derived from AT5G27620 from a cDNA library. The encoded protein (336 amino acids; 38 kD) was designated At;CycH;1, according to the nomenclature proposed by Renaudin et al. (1996). It shares 47.5 and 54.8% sequence identity with rice and poplar cyclin H homologs, respectively. It also shows 26 and 22.6% identity with human cyclin H and fission yeast Ccl1 within the conserved cyclin box domain that is indispensable for binding to CAKs (Nobel et al., 1997).
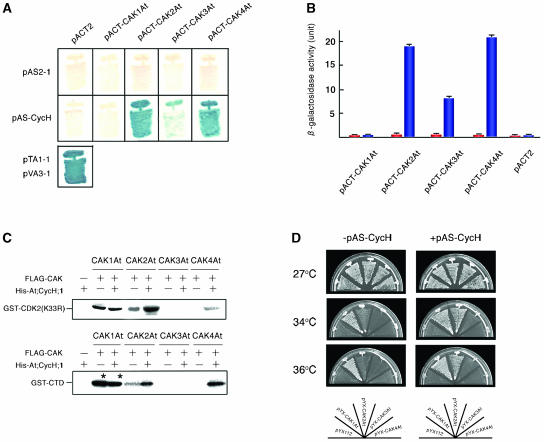
The interaction of cyclin H with all four putative Arabidopsis CAKs was tested in the yeast two-hybrid system. The coding regions of At;CycH;1 and CAK1At–CAK4At were cloned into the yeast expression vectors, pAS2-1 and pACT2, to test the interactions of fusion proteins carrying the GAL4-DNA binding and trans-activation domains using the His3 and lacZ reporter genes in the budding yeast strain Y190 (Harper et al., 1993). Whereas none of the constructs activated the lacZ reporter gene alone, cells expressing either CAK2At or CAK4At in combination with At;CycH;1 grew on a medium without His (data not shown) and showed lacZ activation (Figure 1A). A combination of CAK3At with At;CycH;1 conferred slower growth in the absence of His and a lower level of lacZ induction, whereas coexpression of CAK1At and At;CycH;1 did not reveal any interaction. Quantitative measurements of β-galactosidase activities confirmed the data observed by lacZ filter-lift assays. It showed that the interaction between CAK3At and At;CycH;1 conferred significantly lower level of lacZ activation in comparison with combinations of CAK2At and CAK4At with At;CycH;1 (Figure 1B). These results suggested that except for CAK1At, the other three Arabidopsis orthologs of vertebrate CAKs could bind At;CycH;1, although cyclin H binding capacity of CAK3At appeared to be lower.
Figure 1.
At;CycH;1 Binds and Activates CAK2At and CAK4At.
(A) Yeast two-hybrid assay. Budding yeast strain Y190 was transformed with Arabidopsis CAKs and At;CycH;1 cloned into pACT2 (pACT-CAK) and pAS2-1 (pAS-CycH), respectively, or with an empty vector. Transformants were grown on a minimal medium for 4 d at 30°C, and β-galactosidase activity of colonies was detected using the filter-lift assay. pTA1-1 and pVA3-1 were used as controls.
(B) Quantitative β-galactosidase assay. Y190 strain was transformed with pACT-CAK only (red bars) or with both pACT-CAK and pAS-CycH (blue bars). The values of three independent assays were averaged.
(C) CDK- and CTD-kinase activities of Arabidopsis CAKs expressed in insect cells. FLAG-tagged CAKs (FLAG-CAK) and His-tagged At;CycH;1 (His-At;CycH;1) were expressed in insect Sf9 cells, and 100 ng of protein extract was immunoprecipitated using anti-FLAG antibody. In the case of FLAG-CAK1At, 10 ng of protein was used for immunoprecipitation. Kinase assay was performed using GST-CDK2 (K33R) or GST-CTD as substrate. Asterisks indicate bands of autophosphorylated FLAG-CAK1At.
(D) The cak1ts suppressor activity of CAKs in budding yeast. CAK cDNAs cloned into pYX112 (pYX-CAK) were introduced into budding yeast strain GF2351 with (+) or without (−) At;CycH;1 in pAS2-1 (pAS-CycH). Transformants were grown on minimal medium at indicated temperatures for 4 d.
CAK2At and CAK4At Form Active Kinase Complexes with At;CycH;1 in Insect Cells
To test whether binding of At;CycH;1 would enhance the CDK- and CTD-kinase activities of Arabidopsis CAKs, FLAG epitope-tagged forms of CAK1At–CAK4At and a His-tagged form of At;CycH;1 were expressed in baculovirus-infected Sf9 insect cells. We confirmed that almost the same amount of FLAG-CAK proteins was produced in insect cells (data not shown). Protein extracts were immunoprecipitated with anti-FLAG antibody and subjected to kinase assays using glutathione S-transferase (GST)-fusions of human CDK2 and Arabidopsis RNA polymerase II CTD as substrates. CDK2 can be a substrate of CAKs in the absence of a cyclin partner, thus it is generally used in CDK-kinase assays. To preclude the possibility of autophosphorylation, a kinase-inactive mutant form of CDK2 (carrying a K33R replacement) was used (Poon et al., 1993). Immunoprecipitates from cells expressing FLAG-CAK2At alone phosphorylated both CDK2 and CTD, but coexpression of His-At;CycH;1 elevated the kinase activity fourfold to fivefold (Figure 1C). When expressed alone, FLAG-CAK4At phosphorylated neither substrate, but its CDK- and CTD-kinase activities were activated by coexpression of His-At;CycH;1. By contrast, FLAG-CAK3At did not exhibit CDK2- and CTD-kinase activities even in the presence of His-At;CycH;1, suggesting that CAK3At is probably an inactive CAK variant or it has another substrate specificity. Remarkably, FLAG-CAK1At showed ∼10- to 15-fold higher CDK2-kinase activity as compared with CAK2At and CAK4At, and this activity was not stimulated by At;CycH;1 (Figure 1C). Because the molecular mass of autophosphorylated CAK1At was identical to that of GST-CTD, we were unable to assess CTD phosphorylation by CAK1At in these assays. However, we observed previously that CAK1At immunoprecipitated from plant cells did not show any CTD-kinase activity (Umeda et al., 1998).
Coexpression of At;CycH;1 Enhances the cak1ts Suppressor Activity of CAK2At in Budding Yeast
Budding yeast Cak1p has CDK-kinase activity, but fails to phosphorylate the CTD of RNA polymerase II in vivo (Kaldis et al., 1996; Thuret et al., 1996; Sutton and Freiman, 1997). We observed previously that CAK1At and CAK2At, but neither CAK3At nor CAK4At, were able to suppress the cak1ts mutation of budding yeast (Shimotohno et al., 2003). To test whether coexpression of At;CycH;1 would affect the cak1ts suppressor activity of Arabidopsis CAKs, we coexpressed the pAS2-At;CycH;1 construct with CAK cDNAs cloned in the expression vector pYX112, downstream of the constitutive promoter of triose-phosphate isomerase gene in the cak1ts mutant budding yeast strain GF2351 (Thuret et al., 1996). Cells expressing CAK2At alone grew slowly at 34°C, but at 36°C, they were unable to survive. By contrast, cells coexpressing CAK2At and At;CycH;1 grew normally at both 34°C and 36°C, indicating that expression of At;CycH;1 enhanced the cak1ts suppressor activity of CAK2At (Figure 1D). On the other hand, CAK3At and CAK4At expressed alone or in combination with At;CycH;1 did not show any cak1ts suppressor activity. This result was consistent with our previous observations, which indicated that in insect cells CAK3At was inactive, whereas CAK4At showed significantly lower CDK2-kinase activity than CAK2At. As expected, CAK1At acted as a strong cak1ts suppressor independent of At;CycH;1 coexpression (Figure 1D).
Resolution of CAK Complexes by Gel Filtration
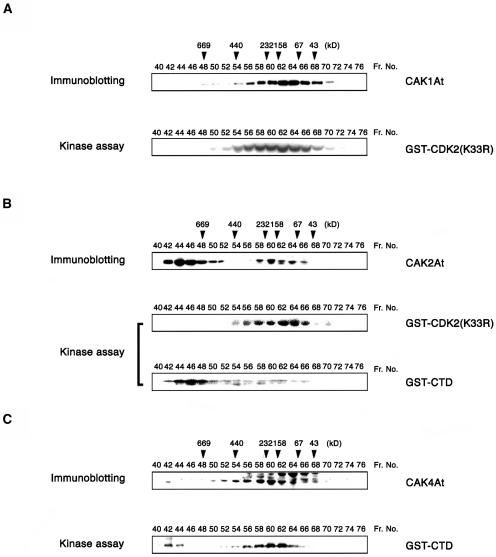
We have fractionated CAK1At, CAK2At, and CAK4At complexes from cultured Arabidopsis cells. In Sephacryl S300 gel exclusion chromatography, CAK1At eluted in a broad peak around 130 kD. Immunoprecipitation of chromatography fractions with an anti-CAK1At antibody indicated that the CAK1At complex had high CDK-kinase activity with GST-CDK2 (K33R) substrate (Figure 2A), but failed to phosphorylate GST-CTD (data not shown). CAK2At was detected in two different complexes with molecular masses of >700 and 130 kD, respectively. The larger complex phosphorylated GST-CTD but not GST-CDK2 (K33R), whereas the smaller one showed high CDK2-kinase activity but low CTD-kinase activity (Figure 2B). This fractionation pattern resembled those of CDK7-like kinases from other metazoa, which are known to form a TFIIH complex of over 700 kD and a heterotrimeric complex of 180 kD consisting of CDK7, cyclin H, and MAT1 subunits (Devault et al., 1995; Schultz et al., 2000). In comparison, CAK4At was detected in a complex of ∼200 kD, which phosphorylated the GST-CTD substrate (Figure 2C). A minor CAK4At complex of >700 kD was also observed, but displayed only low CTD-kinase activity. However, both CAK4At complexes showed only low CDK2-kinase activities (data not shown). These results, thus, confirmed our notion that CAK4At is a CTD kinase, whereas CAK2At is part of distinct CDK- and CTD-kinase complexes.
Figure 2.
Arabidopsis CAK Complexes in Suspension Cells.
Arabidopsis protein extract from suspension cells was fractionated by Sephacryl S300 gel exclusion chromatography, and 15 μL of each fraction was immunoblotted with (A) anti-CAK1At, (B) anti-CAK2At, or (C) anti-CAK4At antibody. Each fraction was immunoprecipitated with the antibody and assayed for kinase activity using GST-CDK2 (K33R) or GST-CTD as substrate. Arrowheads indicate the elution positions of marker proteins with their molecular masses in kD.
Identification of Functional Domains of CAK1At
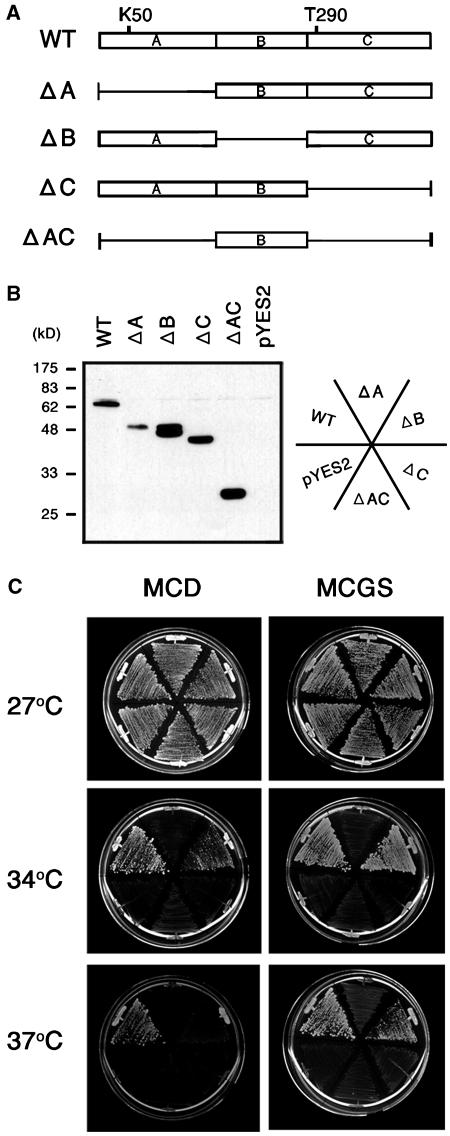
A unique structural feature distinguishing CAK1At from other Arabidopsis CAKs is that it carries an unusual insertion of 111 amino acids between the kinase active site and phosphoregulatory site, between amino acid positions 178 and 288 (Umeda et al., 1998). To determine whether this domain (named B-region in Figure 3A) is necessary for CAK1At activity, we expressed several His-tagged deletion derivatives of CAK1At using the galactose-inducible GAL1 promoter of pYES-DEST52 in budding yeast GF2351 cells. Immunoblotting with anti-His antibody showed that each mutant protein was stably produced in yeast cells (Figure 3B). Whereas removal of N- and C-terminal kinase domains fully abolished the cak1ts suppressor activity of CAK1At, cells carrying a deletion of B-region of CAK1At grew in galactose-containing minimal medium (MCGS), as well as somewhat slower in glucose-containing minimal medium (MCD), at restrictive temperature of 34°C (Figure 3C). At 37°C, cells harboring the wild-type and ΔB CAK1At constructs survived on MCGS, but only wild-type CAK1At rescued the cak1ts mutation on MCD medium. The suppression activity on MCD indicated a leakiness of GAL1 promoter and better complementation of cak1ts mutation by the wild-type CAK1At. In conclusion, these results indicated that domain B is not essential for the cak1ts suppressor activity of CAK1At.
Figure 3.
Complementation Test of Budding Yeast cak1ts Mutant with Deletion Derivatives of CAK1At.
(A) Schematic representation of wild-type (WT) and deletion mutants of CAK1At tentatively divided into three domains: A, B, and C. Domain B corresponds to the unusual insert of extra amino acids between positions 176 and 286. Lys50 (K50) at the kinase active site and Thr290 (T290) in the T-loop region are indicated. Solid bars represent truncated regions.
(B) Budding yeast strain GF2351 were transformed with each construct or the empty vector pYES2 and grown on MCGS. Yeast protein extracts were immunoblotted with anti-His antibody.
(C) Transformants were grown on MCD or on MCGS at 27, 34, or 37°C for 4 d.
CAK1At Functions as CAK-Activating Kinase in Fission Yeast
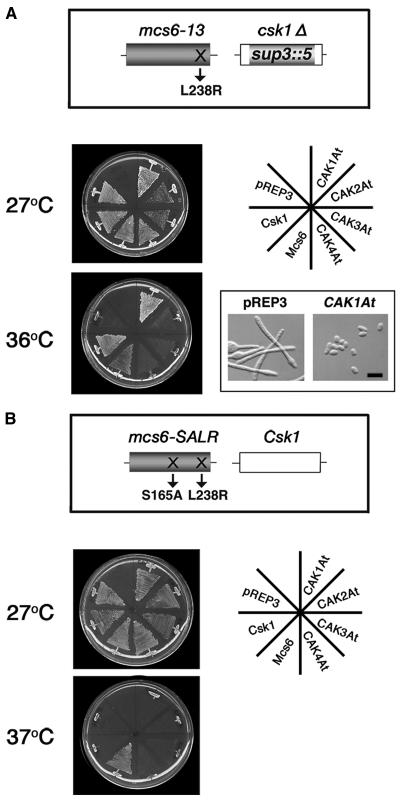
Budding yeast Cak1p is known to phosphorylate and activate Kin28p, which is a relative of CDK7-type CAKs in vertebrates (Espinoza et al., 1998; Kimmelman et al., 1999). Therefore, our cak1ts suppressor assay suggested that CAK1At may also have the activity to phosphorylate CDK7-related CAKs. To assay for CAKAK activity of CAK1At, we conducted a genetic complementation experiment using fission yeast mutants. In fission yeast, Mcs6 is a CDK7-related CAK, which is phosphorylated and activated by the monomeric CAKAK, Csk1 (Hermand et al., 1998). Fission yeast strain JS12 carries a mcs6-13 (L238R) mutation and a disrupted csk1Δ gene, which result in temperature-dependent cell cycle arrest (Hermand et al., 1998; Saiz and Fisher, 2002). We expressed Arabidopsis CAK1At-CAK4At under the control of a thiamine-repressible NMT1 promoter (Maundrell, 1990) of pREP3 in this strain, using fission yeast genes Csk1 or Mcs6 as controls. At the restrictive temperature of 36°C only transformants expressing CAK1At and the control Csk1 and Mcs6 constructs, but none of the other Arabidopsis CAKs, grew in minimal medium without thiamine (Figure 4A). Whereas JS12 transformants carrying the pREP3 vector alone displayed an elongated shape at 36°C, a short cell phenotype of CAK1At-expressing cells confirmed full suppression of cell cycle defect caused by the mcs6-13 and csk1Δ mutations. At the permissive temperature (27°C), JS12 cells carrying CAK2At or CAK3At grew slowly in the absence of thiamine (Figure 4A). We previously described similar observations when rice CAK R2 was overexpressed in fission yeast cells (Yamaguchi et al., 1998), suggesting that the extended C-terminal region of these CAKs might have an inhibitory effect on the yeast cells (Umeda, 2002).
Figure 4.
CAK1At Substitutes for the Function of Csk1 in Fission Yeast.
(A) Complementation of mcs6-13 and csk1Δ mutations. The open reading frame of Arabidopsis CAKs cloned into the vector pREP3 were introduced into fission yeast strain JS12 carrying a mcs6-13 (L238R) mutation and a disrupted csk1Δ gene. Transformants were grown on thiamine-free minimal media at 27 or 36°C for 5 d. Morphology of yeast cells carrying the empty vector or pREP3-CAK1At was observed after culturing without thiamine at 36°C. Bar = 10 μm.
(B) Complementation of mcs6-SALR mutation. Each plasmid was introduced into mcs6-SALR cells carrying the S165A and L238R mutations in Mcs6. Transformants were grown on thiamine-free minimal media at 27 or 37°C for 5 d. Fission yeast Csk1 and Mcs6 cloned into the pREP1 vector were used as controls.
Hermand et al. (2001) observed that activators of Cdc2, including Cdc13, Nim1, Cdc25, and Suc1, can also suppress the temperature-sensitive growth defect of strain JS12. Therefore, CAK2At that has CDK-kinase activity was expected to show suppressor activity in JS12. Our negative results with CAK2At suggested that either CAK2At has a significantly lower CDK-kinase activity than CAK1At, or in the JS12 suppressor assay, CAK1At substituted the function of fission yeast Csk1 by phosphorylation and activation of Mcs6 (L238R). To distinguish between these possibilities, we expressed all Arabidopsis CAKs in a mcs6-SALR mutant strain. This mutant strain, in addition to the L238R mutation, carried a S165A mutation in the T-loop region of Mcs6, thereby preventing its phosphorylation by wild-type Csk1 (Figure 4B). As shown by Hermand et al. (2001), only Mcs6, but not Csk1, suppressed the thermal sensitivity of mcs6-SALR, indicating that S165A mutation prevented the activation of Mcs6 by Csk1. Similar to Csk1, expression of CAK1At also failed to suppress the temperature-dependent cell cycle defect of mcs6-SALR mutant (Figure 4B). These results, thus, clearly demonstrated that CAK1At did not substitute Mcs6, rather it suppressed the mcs6-13 (L238R) mutation by acting as CAKAK and compensating for the csk1Δ defect in strain JS12.
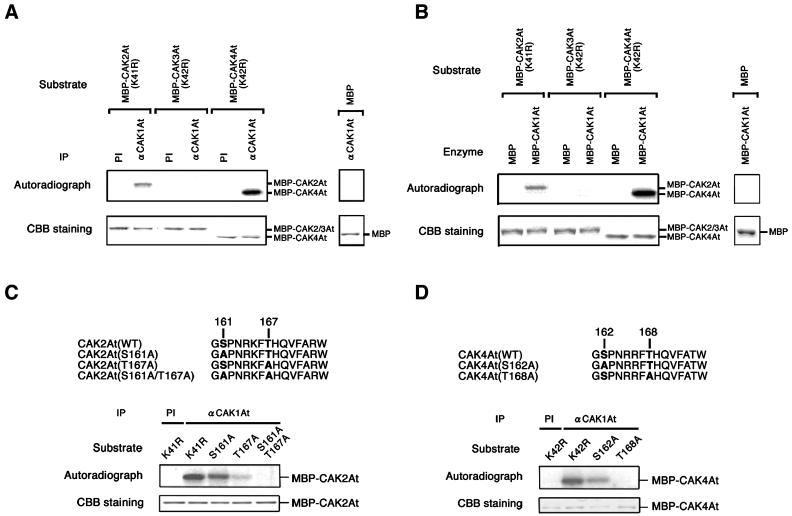
CAK1At Phosphorylates the T-Loop of CAK2At and CAK4At but Not CAK3At
As our suppressor assays with both budding and fission yeast mutants suggested that CAK1At is a CAK-activating kinase, we tested whether CAK1At could phosphorylate CAK2At, CAK3At, and CAK4At in vitro. The peak fraction of CAK1At complex, resolved by gel filtration (Figure 2A), was immunoprecipitated with anti-CAK1At antibody and assayed for CAKAK activity using maltose binding protein (MBP)-fused CAK2At, CAK3At, and CAK4At as substrates. To exclude autophosphorylation of substrates, a Lys residue in the catalytic cleft of each CAK was changed to Arg. The immunoprecipitated CAK1At complex efficiently phosphorylated MBP-CAK2At and MBP-CAK4At, but not MBP-CAK3At and the control MBP protein (Figure 5A). The same result was obtained when recombinant MBP-CAK1At produced in Escherichia coli was used instead of CAK1At purified from Arabidopsis cells (Figure 5B). This indicated that monomeric CAK1At could also phosphorylate both CAK2At and CAK4At. In both assays, CAK1At showed higher activity with CAK4At as compared with CAK2At.
Figure 5.
Phosphorylation of CAK2At and CAK4At by CAK1At.
(A) The CAK1At immunoprecipitates phosphorylate CAK2At and CAK4At. One hundred microliters of fraction No. 62 of gel exclusion chromatography (Figure 2) was immunoprecipitated with preimmune serum (PI) or with the anti-CAK1At antibody (αCAK1At), and the immunoprecipitates were subjected to kinase assays using each substrate.
(B) Recombinant CAK1At phosphorylates CAK2At and CAK4At. MBP-fused CAK2At, CAK3At, and CAK4At were incubated with 10 ng of MBP or MBP-CAK1At in the presence of [γ-32P]ATP. MBP was used as a control.
(C) CAK1At phosphorylates the T-loop of CAK2At. Bold letters in amino acid sequences represent potential phosphorylation sites within the T-loop. In each mutant, these sites were substituted to Ala. One hundred microliters of fraction 62 of gel exclusion chromatography (Figure 2) was immunoprecipitated with preimmune serum (PI) or with the anti-CAK1At antibody (αCAK1At), and the immunoprecipitates were subjected to kinase assays using each mutant protein fused to MBP as substrate.
(D) CAK1At phosphorylates the T-loop of CAK4At. Details are the same as for (C).
CDK7 enzymes are known to be phosphorylated at two major sites, corresponding to Ser164 and Thr170 in the T-loop region in humans (Labbe et al., 1994; Martinez et al., 1997; Akoulitchev and Reinberg, 1998). Comparison of amino acid sequences indicated that Arabidopsis CAK2At and CAK4At also contain conserved Ser and Thr residues within their T-loops. To determine whether these residues are targeted for phosphorylation by CAK1At, we have substituted Ser161 and Thr167 in CAK2At, and Ser162 and Thr168 in CAK4At with Ala, and used these mutant proteins as substrates for CAK1At kinase assays (Figures 5C and 5D). Phosphorylation activity of CAK1At with MBP-CAK2At (S161A) and MBP-CAK2At (T167A) was 80 and 20% as compared with that of the MBP-CAK2At (K41R) control, respectively. Combination of S161A and T167A mutations completely abolished the CAK1At phosphorylation activity, indicating that CAK1At indeed phosphorylated these residues in vitro (Figure 5C). Similarly, CAK1At phosphorylation of MBP-CAK4At was reduced to 30% by the S162A mutation and almost abolished by the T168A mutation (Figure 5D). These results demonstrated that CAK1At recognizes the activating phosphorylation sites within the T-loops of CAK2At and CAK4At.
CAK1At Phosphorylates CAK4At and Activates Its CTD-Kinase Activity
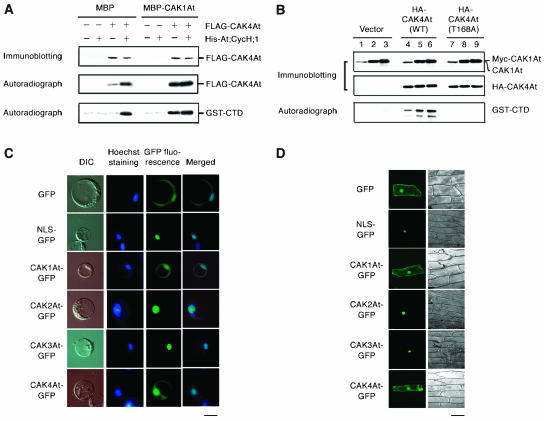
As CAK1At itself shows high CDK-kinase activity, we could not reliably determine whether CAK1At would activate the CDK-kinase activities of CAK2At and CAK4At. CAK1At does not phosphorylate the CTD of RNA polymerase II; we could, however, assay whether CAK1At would increase the CTD-kinase activity of CAK4At. A FLAG-tagged form of CAK4At with or without coexpression of His-tagged At;CycH;1 was immunoprecipitated from protein extracts of insect cells using an anti-FLAG antibody. Coexpression of His-At;CycH;1, as expected, resulted in a higher autophosphorylation activity of FLAG-CAK4At (Figure 6A). When MBP-CAK1At was added to the kinase reaction, both monomeric FLAG-CAK4At and the FLAG-CAK4At complex with His-At;CycH;1, showed higher autophosphorylation in comparison with control reactions performed with MBP (Figure 6A). Subsequently, MBP and MBP-CAK1At were removed by extensive washing and each FLAG-CAK4At sample was subjected to kinase assay using GST-CTD as substrate. Monomeric FLAG-CAK4At treated only with the control MBP protein showed no CTD-kinase activity, whereas FLAG-CAK4At complex with His-At;CycH;1 phosphorylated the GST-CTD substrate. Preincubation with MBP-CAK1At increased CTD-kinase activity of FLAG-CAK4At complexed with His-At;CycH;1, and also activated the monomeric FLAG-CAK4At kinase (Figure 6A). This indicates that CAK1At can phosphorylate CAK4At and activate its CTD-kinase activity independent of At;CycH;1.
Figure 6.
CAK1At activates the CTD-kinase activity of CAK4At.
(A) CAK4At activation in vitro. FLAG-CAK4At and/or His-At;CycH;1 were expressed in baculovirus-infected insect cells. Fifty nanograms of protein extract was immunoprecipitated with anti-FLAG antibody and immunoblotted with anti-CAK4 antibody (top). The immunoprecipitates were incubated with MBP or MBP-CAK1At in the presence of [γ-32P]ATP and autoradiographed (middle). After removal of MBP or MBP-CAK1At, the immunoprecipitates were subjected to kinase reaction using GST-CTD as substrate (bottom).
(B) CAK4At activation in Arabidopsis root protoplasts. The vector pMENCHU (lanes 1 to 3), pMENCHU-CAK4At (WT) (lanes 4 to 6) or pMENCHU-CAK4At (T168A) (lanes 7 to 9) was introduced into Arabidopsis root protoplasts. HA-CAK4At was coexpressed with 5 μg (lanes 2, 5, and 8) or 30 μg (lanes 3, 6, and 9) of pMESHI-CAK1At. Five micrograms of protein extract was immunoblotted with anti-CAK1At or anti-CAK4At antibody. Seventy micrograms of protein extract was immunoprecipitated with anti-HA antibody, followed by a kinase reaction using GST-CTD as substrate.
(C) Subcellular localization of GFP-fused CAKs in Arabidopsis protoplasts. Fluorescent microscopic images of CAK-GFP, and the corresponding images of DIC and Hoechst 33342 staining are shown. Merged images of Hoechst 33342 staining and GFP fluorescence are also shown. As a control, the nuclear localization signal (NLS) of Simian virus 40 fused to GFP was used. Bar = 10 μm.
(D) Subcellular localization of GFP-fused CAKs in onion epidermal cells. Fluorescent (left) and bright-field (right) images of cells are shown. Bar = 100 μm.
CAK1At Activates CAK4At in Arabidopsis Root Protoplasts
To confirm that CAK1At acts as CAK4At-activating kinase also in plant cells, we coexpressed c-Myc and hemagglutinin (HA) epitope-tagged forms of CAK1At and CAK4At, respectively, in Arabidopsis root protoplasts. HA-CAK4At was immunoprecipitated from protoplast protein extracts with anti-HA antibody and subjected to kinase assay with GST-CTD as substrate. The CTD-kinase activity of HA-CAK4At was elevated by coexpression with myc-CAK1At, and increasing the expression level of myc-CAK1At resulted in increasing phosphorylation of the CTD (Figure 6B). The T168A mutation in CAK4At caused a loss of CTD-kinase activity, which was not restored by CAK1At coexpression, indicating that the observed phosphorylation of the CTD was indeed dependent on CAK4At and its activation by coexpression of CAK1At. Consistent with their potential interactions in plant cells, green fluorescent protein (GFP) fusion proteins of CAK1At and CAK4At showed similar, overlapping cellular localization in the cytoplasm and nuclei of Arabidopsis protoplasts (Figure 6C) and onion (Allium cepa) epidermal cells (Figure 6D). By contrast, CAK2At and CAK3At displayed nearly exclusive nuclear localization, similar to that of a nuclearly targeted NLS-GFP control protein.
DISCUSSION
Is CAK1At a Cyclin-Independent CDK-Activating Kinase?
Our results indicate that CAK1At is a unique member of the Arabidopsis CDK-activating CAK kinase family that does not interact with the cyclin H homolog, At;CycH;1. Accordingly, the CDK-kinase activity of CAK1At is not enhanced by coexpression of At;CycH;1 in insect cells. We showed that recombinant CAK1At produced in E. coli is active without any other factor, indicating that CAK1At may not require a cyclin partner for its activity. Nevertheless, we observed that CAK1At in Arabidopsis cells occurs in a protein complex of 130 kD that shows high CDK-kinase but no CTD-kinase activity. It is, thus, likely that CAK1At is associated in vivo with other regulatory protein(s) that might control its activity in response to external or internal stimuli. Because monomeric CAK1At migrates as a protein of 62 kD in SDS-PAGE (Umeda et al., 1998), it is of course also possible that CAK1At forms a homodimer in vivo, which may account for the observed molecular mass of 130 kD. To distinguish between these possibilities, further analysis of the subunit composition of CAK1At complex is necessary.
A unique feature of CAK1At among other CAKs is that it carries an unusual insertion of 111 amino acids between its kinase active site and phosphoregulatory site. Our studies indicate that removal of this unique region does not affect the capability of CAK1At to suppress the budding yeast cak1ts mutation; hence, this region is not essential for the CDK-kinase activity of CAK1At. Nonetheless, a CAK1At mutant, which carries a deletion of this unique region, shows lower cak1ts suppressor activity than the wild-type kinase, as well as decreased CDK-kinase activity when expressed in Arabidopsis cells (data not shown). Genes encoding for CAK1At-related proteins also exist in other plant species. For example, CAK1At homologs in Euphorbia and rice, which show 49.7 and 35.9% sequence identity, respectively, also carry similar internal sequence domains with significant homology to that of CAK1At (data not shown). Therefore, we assume that this unique region may be involved in the control of interactions of CAK1At-related kinases with specific regulatory proteins or substrates acting in plant-specific signaling pathways.
CAK1At Is a Prototype of Plant-Specific CAK-Activating Kinases
CDK-activating kinases, such as Xenopus CDK7 and fission yeast Mcs6, are phosphorylated at Ser and Thr residues within their T-loops (Labbe et al., 1994; Martinez et al., 1997; Hermand et al., 1998). In Xenopus, phosphorylation of Thr170 is required for high affinity binding of CDK7 to cyclin H (Martinez et al., 1997). Recently, Liu et al. (2004) identified a novel human CAK homolog, p42, which shows sequence homology to both CDK7 and budding yeast Cak1p. It was responsible for CAK activity in vivo but had no CDK7-activating kinase activity, suggesting that the T-loop phosphorylation of CDK7 is likely performed by a yet unidentified human kinase. Here, we demonstrated that CAK1At directly phosphorylates CAK2At and CAK4At at conserved Ser and Thr residues within the T-loop and showed that this phosphorylation activates the CTD-kinase activity of CAK4At both in vitro and in plant cells. Mutations affecting Ser161 and Thr167 in the T-loop abolished the ability of CAK2At to suppress the cak1ts mutation in budding yeast (data not shown), which suggests that CAK2At requires phosphorylation of these invariant Ser and Thr residues to get activated. By contrast, we failed to detect CAK1At-mediated phosphorylation of CAK3At. The CAK3At has an altered T-loop sequence, GSPGRKFTHQVFARW, which has a substitution of Asn to Gly. This amino acid change could cause a conformational change of T-loop, and thus, prevent CAK3At from being a substrate of CAK1At. The nonphosphorylated form of CAK3At is probably incapable of forming a stable complex with cyclin H, and thus, has no enzyme activity and fails to suppress the budding yeast cak1ts mutation.
Our notion that CAK1At could function as an upstream kinase of Arabidopsis CAKs was supported by the fact that CAK1At suppressed the cell cycle defect caused by the mcs6 and csk1 mutations in fission yeast. Our results clearly show that CAK1At does not substitute the function of Mcs6, but it phosphorylates the Ser165 residue of Mcs6 T-loop region and thereby activates Mcs6's enzyme activity. Based on this feature, CAK1At is closely related to Csk1 that exhibits Mcs6-activating kinase activity (Hermand et al., 2001). Cak1p of budding yeast also shows functional similarity to CAK1At as it has the ability to phosphorylate the CDK7-family kinase Kin28p and thereby activate its CTD-kinase activity (Espinoza et al., 1998; Kimmelman et al., 1999). Recently, Tsakraklides and Solomon (2002) reported the comparison of biochemical properties of Csk1, Cak1p, and CAK1At. All these kinases were able to rescue the cak1ts mutation in a cyclin-independent manner and were insensitive to 5′-fluorosulfonylbenzoyladenosine. However, despite functional similarities, CAK1At shares no significant sequence similarity with Csk1 and Cak1p. Rather, it was classified into the CDK7 family by neighbor-joining and maximum likelihood methods, whereas Cak1p and Csk1 were included in the same family with significant bootstrap support, regardless of their low sequence homology (Liu and Kipreos, 2002). In particular, both Cak1p and Csk1 lack the glycine loop in the ATP binding region, which is highly conserved in CDK-related proteins, including CDK7 and CAK1At (Kaldis et al., 1996; Tsakraklides and Solomon, 2002). Cak1p and Csk1 orthologs have not been identified in species other than yeast. This leads to two possibilities: animals and plants may not require any ortholog; or they may possess functionally related kinases that are very divergent from Cak1p or Csk1. Our conclusion that Arabidopsis CAK1At functions as a CAKAK clearly supports the latter hypothesis. Using database searches, homologs of Arabidopsis CAK1At can only be identified in Euphorbia, rice, and soybean (Glycine max). Therefore, we propose that CAK1At-related kinases represent a distinct family of CAKAKs in the plant kingdom.
Multiple CAK Complexes Are Involved in CDK and CTD Phosphorylation in Arabidopsis
Our results show that recombinant CAK2At and CAK4At preferentially phosphorylate CDK2 and CTD, respectively, in association with At;CycH;1. This observation correlates well with the fact that overexpression of CAK2At, but not CAK4At, can suppress the budding yeast cak1ts mutation, which leads to reduced CDK-kinase activity (Shimotohno et al., 2003). However, neither CAK2At nor CAK4At suppressed the cell cycle defect caused by the mcs6-13 and Δcsk1 mutations in fission yeast. This discrepancy may be explained by the heterogeneity of regulatory cyclin H subunits. Namely, fission yeast Mcs2 is conserved in restricted domains of the cyclin box among various cyclin H homologs. This suggests that the association between CAK2At and Mcs2 in fission yeast cells might not be enough to rescue the temperature-sensitive mcs6-13 mutation. An association with Mcs2 could also be inhibited by a C-terminal extension of CAK2At that is specific to the plant CDK7 homologs (Umeda, 2002).
When Arabidopsis CAKs were fractionated by gel filtration, CAK2At was detected in two distinct protein complexes with molecular masses of >700 and 130 kD, which showed different CDK- and CTD-kinase activities. Although our anti-CAK2At antibody recognizes both recombinant CAK2At and CAK3At proteins (Shimotohno et al., 2003), the inactive nature of CAK3At indicates that in our assays we had monitored the enzyme activity of CAK2At. In comparison, the CTD-kinase activity of CAK4At was concentrated in a smaller complex of 200 kD, and only a small amount of CAK4At was recovered in a larger complex of over 700 kD. This result is surprising because CAK4At may be incorporated into TFIIH to exert higher CTD-kinase activity than CAK2At. In vertebrates, TFIIH complexes contain two DNA helicases, XPB and XPD. XPD, in particular, has a role in CAK anchoring and stabilizes the complex through interaction with MAT1 (Reardon et al., 1996; Busso et al., 2000). In Arabidopsis, putative DNA helicases are also encoded in the genome (Ribeiro et al., 1998; Costa et al., 2001), but no homolog of MAT1 has been identified so far. This suggests that another factor(s) may participate in weak anchoring of CAK4At to TFIIH, which could allow dissociation of the 200-kD complex from TFIIH during our protein extraction and fractionation processes.
In conclusion, our data show that CAK2At and CAK4At are vertebrate-type CAKs, which exhibit different phosphorylation activities on CDK and CTD, respectively. CAK1At has high CDK-kinase activity, but it is also involved in phosphorylation and activation of CAK2At and CAK4At. Therefore, CAK1At may be engaged not only in CDK activation, but also in the control of basal transcription, by modulating the CTD-kinase activity of CAK4At. The functional difference between CAK1At and CAK2At-CAK4At described in this article corresponds well to the classification of Arabidopsis CAKs into CDKF and CDKD classes (Joubès et al., 2000). To adopt the nomenclature rules for CDKs (Vandepoele et al., 2002), we shall hereafter use the terms of CDKF;1, CDKD;3, CDKD;1, and CDKD;2 for CAK1At-CAK4At, respectively. Unlike yeast Cak1p or Csk1, CAK1At (CDKF;1) has a conserved phosphoregulatory site at Thr290, which may be phosphorylated by a further upstream kinase to confer upregulation or downregulation of CAK1At (CDKF;1). Such a phosphorylation cascade is expected to link the CAKAK-CAK-CDK/CTD phosphorylation pathways to developmental and hormonal controls. Further studies on upstream signaling pathways could help us answer the question how this kinase cascade is involved in the control of cell cycle, transcription, and differentiation during plant development.
METHODS
Isolation of Cyclin H cDNA and Plasmid Constructions
The cDNA of At;CycH;1 was PCR-amplified from an Arabidopsis thaliana cDNA library made in the yeast expression vector pYX112 (Umeda et al., 1998). The open reading frame (ORF) of At;CycH;1 was amplified by PCR and cloned into the EcoRI site of the pAS2-1 vector (Clontech, Palo Alto, CA). The ORFs of CAK1At, CAK2At, CAK3At, and CAK4At were PCR-amplified and cloned into the XhoI, XhoI, SmaI, and BamHI sites of pACT2 (Clontech), respectively. CAK cDNAs cloned into the pYX112 or pREP3 vector (Shimotohno et al., 2003) were used for yeast complementation tests. For expression of deletion derivatives of CAK1At, each cDNA fragment was amplified by PCR and cloned into the Gateway entry vector pENTR/SD/D-TOPO (Invitrogen, San Diego, CA). Subsequently, a recombination reaction was performed between each entry clone and a destination vector pYES-DEST52 using LR Clonase (Invitrogen).
For expression in baculovirus-infected insect cells, the ORFs of CAK1At, CAK2At, CAK3At, and CAK4At were PCR-amplified and cloned into the pFAST-BAC-FLAG1 vector (Yamaguchi et al., 1998) at the sites of XhoI/SphI, EcoRI, EcoRI, and XhoI/SphI, respectively, such that they were in-frame with the FLAG sequence. The ORF of At;CycH;1 was cloned into the EcoRI site of pFASTBAC HTa (Invitrogen). Nucleotide substitutions were introduced into CAK cDNAs by using a Mutan-Super Express Km kit (Takara, Tokyo, Japan). To express MBP-fused proteins in Escherichia coli, wild-type and mutant cDNAs were cloned into the pMALC2 vector (New England Biolabs, Beverly, MA) as described by Shimotohno et al. (2003). For expression in Arabidopsis root protoplasts, the ORFs of CAK1At and CAK4At were amplified by PCR and cloned into the SalI and BamHI sites of pMESHI and pMENCHU (Ferrando et al., 2000, 2001), respectively. The c-Myc and HA epitope-tagged proteins were produced under the 35S promoter of Cauliflower mosaic virus in pMESHI and pMENCHU, respectively. To observe CAK localization, the ORFs of CAK1At, CAK2At, CAK3At, and CAK4At were PCR-amplified and cloned into the SalI/NcoI, SalI/NcoI, SalI, and SalI/NcoI sites of KS-GFP (Chiu et al., 1996), respectively.
Yeast Two-Hybrid Assay and Complementation Test
Yeast two-hybrid assay was performed with budding yeast (Saccharomyces cerevisiae) strain Y190 according to the protocol provided by Clontech. For the quantitative assay, yeast cells were cultured in the minimal medium, which lacked Leu and Trp, at 30°C to reach the stationary phase and were then diluted fivefold with YPD medium and cultured for an additional 4 h. pVA3-1, which contains the murine p53 fused to GAL4 DNA binding domain, and pTD1-1, which contains the SV40 large T-antigen cDNA fused to GAL4 transactivating domain, were used as controls. Complementation test was conducted with budding yeast strain GF2351 (MATa, civ1-4, ura3, leu2, trp1, lys2, ade2, ade3) (Thuret et al., 1996). Yeast protein extracts were prepared as described (Vitaly, 2000). Fission yeast (Schizosaccharomyces pombe) strains used for the complementation test were JS12 (h− csk1 Δmcs6-13 leu1-32 ade6-704) (Hermand et al., 1998) and mcs6-SALR (Hermand et al., 2001). Transformants were incubated on minimal medium in the presence or absence of 2.0 μM thiamine.
Immunoblotting and Kinase Assays with Recombinant Proteins
MBP-fusion proteins were expressed in E. coli strain BL21 and purified on amylose resin (New England Biolabs) using the manufacturer's protocol. GST-CDK2 (K33R) and GST-CTD were purified as described (Umeda et al., 1998). Expression of FLAG-CAK and His-At;CycH;1 proteins in insect cells was conducted as described by Yamaguchi et al. (2000). Cells were resuspended in five volumes of phosphate-buffered saline containing protease inhibitors (10 μg/mL each of leupeptine, pepstatin, aprotinin, benzamidine, and soybean trypsin inhibitor), lysed by sonication, and the supernatant was used for assays. Immunoblotting was conducted using ECL protein gel blotting detection reagents (Amersham Biosciences, Piscataway, NJ).
For the kinase assays, protein extracts were immunoprecipitated with anti-FLAG M2 affinity gel (Sigma, St. Louis, MO) or anti-CAK1At antibody (Umeda et al., 1998), and the immunoprecipitates were subjected to kinase reaction using 1 μg each of substrate protein. Ten nanograms of MBP-CAK1At were also used as enzyme. A detailed protocol for kinase assay was described previously (Umeda et al., 1998). To detect CAKAK activity of CAK1At, FLAG-CAK4At was immunoprecipitated from 100 ng of insect crude extract, using anti-FLAG antibody. Half of the immunoprecipitates were immunoblotted with anti-FLAG antibody, and the other half were incubated with 10 ng of MBP-CAK1At in the presence of [γ-32P]ATP. To examine the CTD-kinase activity of CAK4At, the immunoprecipitates were incubated with MBP-CAK1At in the presence of cold ATP, washed five times, and subjected to kinase reaction using GST-CTD as substrate.
Fractionation of Arabidopsis Protein Extracts by Gel Exclusion Chromatography
Arabidopsis cell suspension culture (ecotype Columbia) was maintained as described previously (Shimotohno et al., 2003). A crude extract that contained 60 mg of protein extracted from a culture, 4 d after subculturing, was brought to 70% saturation using ammonium sulfate. Precipitated proteins were dissolved in 1 mL of column buffer (20 mM Tris-HCl, pH 7.8, 100 mM NaCl, 5 mM β-glycerophosphate, 2 mM EGTA, 2 mM MgCl2, 0.5 mM DTT, 1 mM NaF, 0.1 mM Na3VO4, and 10% [w/w] glycerol) and loaded onto a Sephacryl S300 column (Amersham Biosciences). Elution profiles of proteins in LMW and HMW gel filtration calibration kits (Amersham Biosciences) were used for estimations of molecular mass. One hundred microliters of each fraction was immunoprecipitated with CAK antibodies (Shimotohno et al., 2003), and the immunoprecipitates were subjected to kinase reaction using 2 μg of GST-CDK2 (K33R) or GST-CTD as substrate.
Transient Expression in Arabidopsis Protoplasts
The preparation and transfection of root protoplasts have been described previously (see http://genetics.mgh.harvard.edu/sheenweb/). Protoplasts (4 × 104) were transfected with 30 μg of plasmid DNA and incubated at 23°C for 15 h in W5 solution under continuous illumination. To assess the CTD-kinase activity of CAK4At, protein extract was immunoprecipitated with anti-HA (12CA5) monoclonal antibody (Roche, Indianapolis, IN), and the immunoprecipitates were subjected to a kinase reaction using 2 μg of GST-CTD as substrate. Assays were conducted three times to examine the reproducibility. GFP constructs were introduced into protoplasts from the suspension cells as described by Ueda et al. (2001). DNA staining was conducted with 100 ng/mL of Hoechst 33342.
Transient Expression in Onion Epidermal Cells
Onion (Allium cepa) transformation was performed essentially as described by van den Ackerveken et al. (1996). Inner epidermal layers obtained from onions were placed on MS basal medium (Wako, Osaka, Japan) with 2% agar. Gold particles (1.6 μm; Bio-Rad, Hercules, CA) coated by 5 μg of plasmid DNA were briefly vortexed and bombarded onto each sample. Bombardment parameters were the following: a pressure of 7600 kPa, a target distance of 12 cm, and a decompression vacuum of 95 kPa in Hg. After bombardment, the samples were incubated at 22°C under continuous illumination for 18 h, followed by microscopic observations with confocal laser scanning microscope system (MicroRadiance MR/AG-2; Bio-Rad).
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession number AB051072, corresponding to AGI number AT5G27620.
Acknowledgments
We thank Carl Mann (CEA/Saclay) for the GF2351 cells, Robert Fisher (Memorial Sloan Kettering Cancer Center) for JS12 cells and the pREP1-Csk1 plasmid and Tomi Mäkelä (University of Helsinki) for mcs6-SALR cells. We are also grateful to Csaba Koncz (Max-Planck-Institut für Züchtungsforschung) for the pMENCHU and pMESHI plasmids and critical reading of the manuscript. We appreciate Nobuhiro Tsutsumi and Shin-ichi Arimura for help with particle bombardment. This work was supported by Grant-in-Aid for Scientific Research on Priority Areas (Grant 15031210) and by Research for the Future from the Japan Society for the Promotion of Science.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Masaaki Umeda (mumeda@iam.u-tokyo.ac.jp).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.025601.
References
- Akoulitchev, S., and Reinberg, D. (1998). The molecular mechanism of mitotic inhibition of TFIIH is mediated by phosphorylation of CDK7. Genes Dev. 12, 3541–3550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck, V., Russell, P., and Millar, J.B. (1995). Identification of a cdk-activating kinase in fission yeast. EMBO J. 14, 6173–6183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busso, D., Keriel, A., Sandrock, B., Poterszman, A., Gileadi, O., and Egly, J.M. (2000). Distinct regions of MAT1 regulate cdk7 kinase and TFIIH transcription activities. J. Biol. Chem. 275, 22815–22823. [DOI] [PubMed] [Google Scholar]
- Chiu, W.L., Niwa, Y., Zeng, W., Hirano, T., Kobayashi, H., and Sheen, J. (1996). Engineered GFP as a vital reporter in plants. Curr. Biol. 6, 325–330. [DOI] [PubMed] [Google Scholar]
- Cismowski, M.J., Laff, G.M., Solomon, M.J., and Reed, S.I. (1995). KIN28 encodes a C-terminal domain kinase that controls mRNA transcription in Saccharomyces cerevisiae but lacks cyclin-dependent kinase-activating kinase (CAK) activity. Mol. Cell. Biol. 15, 2983–2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa, R.M., Morgante, P.G., Berra, C.M., Nakabashi, M., Bruneau, D., Bouchez, D., Sweder, K.S., Van Sluys, M.A., and Menck, C.F. (2001). The participation of AtXPB/RAD25 homologue gene from Arabidopsis thaliana, in DNA repair and plant development. Plant J. 28, 385–395. [DOI] [PubMed] [Google Scholar]
- Cross, F.R., and Levine, K. (2000). Genetic analysis of the relationship between activation loop phosphorylation and cyclin binding in the activation of the Saccharomyces cerevisiae Cdc28p cyclin-dependent kinase. Genetics 154, 1549–1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damagnez, V., Mäkelä, T., and Cottarel, G. (1995). Schizosaccharomyces pombe Mop1-Mcs2 is related to mammalian CAK. EMBO J. 14, 6164–6172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devault, A., Martinez, A.M., Fesquet, D., Labbe, J.C., Morin, N., Tassan, J.P., Nigg, E.A., Cavadore, J.C., and Doree, M. (1995). MAT1 (‘menage a trois’) a new RING finger protein subunit stabilizing cyclin H-cdk7 complexes in starfish and Xenopus CAK. EMBO J. 14, 5027–5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinoza, F.H., Farrell, A., Nourse, J.L., Chamberlin, H.M., Gileadi, O., and Morgan, D.O. (1998). Cak1 is required for Kin28 phosphorylation and activation in vivo. Mol. Cell. Biol. 18, 6365–6373, Erratum. Mol. Cell Biol. 20, 1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feaver, W.J., Henry, N.L., Wang, Z., Wu, X., Svejstrup, J.Q., Bushnell, D.A., Friedberg, E.C., and Kornberg, R.D. (1997). Genes for Tfb2, Tfb3, and Tfb4 subunits of yeast transcription/repair factor IIH. Homology to human cyclin-dependent kinase activating kinase and IIH subunits. J. Biol. Chem. 272, 19319–19327. [DOI] [PubMed] [Google Scholar]
- Ferrando, A., Farràs, R., Jásik, J., Schell, J., and Koncz, C. (2000). Intron-tagged epitope: A tool for facile detection and purification of proteins expressed in Agrobacterium-transformed plant cells. Plant J. 22, 553–560. [DOI] [PubMed] [Google Scholar]
- Ferrando, A., Koncz, K.Z., Farràs, R., Tiburcio, A., Schell, J., and Koncz, C. (2001). Detection of in vivo protein interactions between Snf1-related kinase subunits with intron-tagged epitope-labelling in plants cells. Nucleic Acids Res. 29, 3685–3693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper, J.W., Adami, G.R., Wei, N., Keyomarski, K., and Elledge, S.J. (1993). The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell 75, 805–816. [DOI] [PubMed] [Google Scholar]
- Hata, S. (1991). cDNA cloning of a novel cdc2+/CDC28-related protein kinase from rice. FEBS Lett. 279, 149–152. [DOI] [PubMed] [Google Scholar]
- Hermand, D., Pihlak, A., Westerling, T., Damagnez, V., Vandenhaute, J., Cottarel, G., and Mäkelä, T.P. (1998). Fission yeast Csk1 is a CAK-activating kinase (CAKAK). EMBO J. 17, 7230–7238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermand, D., Westerling, T., Pihlak, A., Thuret, J.Y., Vallenius, T., Tiainen, M., Vandenhaute, J., Cottarel, G., Mann, C., and Mäkelä, T.P. (2001). Specificity of Cdk activation in vivo by the two Caks Mcs6 and Csk1 in fission yeast. EMBO J. 20, 82–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey, P.D., Russo, A.A., Polyak, K., Gibbs, E., Hurwitz, J., Massague, J., and Pavletich, N.P. (1995). Mechanism of CDK activation revealed by the structure of a cyclin A-CDK2 complex. Nature 376, 313–320. [DOI] [PubMed] [Google Scholar]
- Joubès, J., Chevalier, C., Dudits, D., Heberle-Bors, E., Inzé, D., Umeda, M., and Renaudin, J.-P. (2000). CDK-related protein kinases in plants. Plant Mol. Biol. 43, 607–620. [DOI] [PubMed] [Google Scholar]
- Kaldis, P. (1999). The cdk-activating kinase (CAK): From yeast to mammals. Cell. Mol. Life Sci. 55, 284–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaldis, P., Sutton, A., and Solomon, M.J. (1996). The Cdk-activating kinase (CAK) from budding yeast. Cell 86, 553–564. [DOI] [PubMed] [Google Scholar]
- Keogh, M.C., Cho, E.J., Podolny, V., and Buratowski, S. (2002). Kin28 is found within TFIIH and a Kin28-Ccl1-Tfb3 trimer complex with differential sensitivities to T-loop phosphorylation. Mol. Cell. Biol. 22, 1288–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimmelman, J., Kaldis, P., Hengartner, C.J., Laff, G.M., Koh, S.S., Young, R.A., and Solomon, M.J. (1999). Activating phosphorylation of the Kin28p subunit of yeast TFIIH by Cak1p. Mol. Cell. Biol. 19, 4774–4787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe, J.C., Martinez, A.M., Fesquet, D., Capony, J.P., Darbon, J.M., Derancourt, J., Devault, A., Morin, N., Cavadore, J.C., and Doree, M. (1994). p40MO15 associates with a p36 subunit and requires both nuclear translocation and Thr176 phosphorylation to generate cdk-activating kinase activity in Xenopus oocytes. EMBO J. 13, 5155–5164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, K.M., Saiz, J.E., Barton, W.A., and Fisher, R.P. (1999). Cdc2 activation in fission yeast depends on Mcs6 and Csk1, two partially redundant Cdk-activating kinases (CAKs). Curr. Biol. 9, 441–444. [DOI] [PubMed] [Google Scholar]
- Liu, J., and Kipreos, E.T. (2002). The evolution of CDK-activating kinases in CDK-activating kinase (CAK). In CDK-Activating Kinase (CAK), P. Kaldis, ed (Georgetown: Landes Bioscience), pp. 99–111.
- Liu, Y., Wu, C., and Galaktionov, K. (2004). p42, a novel cyclin-dependent kinase activating kinase in mammalian cells. J. Biol. Chem. 279, 4507–4514. [DOI] [PubMed] [Google Scholar]
- Martinez, A.M., Afshar, M., Martin, F., Cavadore, J.C., Labbe, J.C., and Doree, M. (1997). Dual phosphorylation of the T-loop in cdk7: Its role in controlling cyclin H binding and CAK activity. EMBO J. 16, 343–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maundrell, K. (1990). nmt1 of fission yeast: A highly transcribed gene completely repressed by thiamine. J. Biol. Chem. 265, 10857–10864. [PubMed] [Google Scholar]
- Morgan, D.O. (1997). Cyclin-dependent kinases: engines, clocks, and microprocessors. Annu. Rev. Cell Dev. Biol. 13, 261–291. [DOI] [PubMed] [Google Scholar]
- Morgan, D.O., and De Bondt, H.L. (1994). Protein kinase regulation: Insights from crystal structure analysis. Curr. Opin. Cell Biol. 6, 239–246. [DOI] [PubMed] [Google Scholar]
- Nobel, M.E., Endicott, J.A., Brown, N.R., and Johnson, L.N. (1997). The cyclin box fold: Protein recognition in cell-cycle and transcription control. Trends Biochem. Sci. 22, 482–487. [DOI] [PubMed] [Google Scholar]
- Poon, R.Y., Yamashita, K., Adamczewski, J.P., Hunt, T., and Shuttleworth, J. (1993). The cdc2-related protein p40MO15 is the catalytic subunit of a protein kinase that can activate p33cdk2 and p34cdc2. EMBO J. 12, 3123–3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reardon, J.T., Ge, H., Gibbs, E., Sancar, A., Hurwitz, J., and Pan, Z.Q. (1996). Isolation and characterization of two human transcription factor IIH (TFIIH)-related complexes: ERCC2/CAK and TFIIH. Proc. Natl. Acad. Sci. USA 93, 6482–6487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Renaudin, J.P., et al. (1996). Plant cyclins: A unified nomenclature for plant A-, B- and D-type cyclins based on sequence organization. Plant Mol. Biol. 32, 1003–1018. [DOI] [PubMed] [Google Scholar]
- Ribeiro, D.T., Machado, C.R., Costa, R.M., Praekelt, U.M., Van Sluys, M.A., and Menck, C.F. (1998). Cloning of a cDNA from Arabidopsis thaliana homologous to the human XPB gene. Gene 208, 207–213. [DOI] [PubMed] [Google Scholar]
- Ross, K.E., Kaldis, P., and Solomon, M.J. (2000). Activating phosphorylation of the Saccharomyces cerevisiae cyclin-dependent kinase, cdc28p, precedes cyclin binding. Mol. Biol. Cell 11, 1597–1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russo, A.A., Jeffrey, P.D., and Pavletich, N.P. (1996). Structural basis of cyclin-dependent kinase activation by phosphorylation. Nat. Struct. Biol. 3, 696–700. [DOI] [PubMed] [Google Scholar]
- Saiz, J.E., and Fisher, R.P. (2002). A CDK-activating kinase network is required in cell cycle control and transcription in fission yeast. Curr. Biol. 12, 1100–1105. [DOI] [PubMed] [Google Scholar]
- Schultz, P., Fribourg, S., Poterszman, A., Mallouh, V., Moras, D., and Egly, J.M. (2000). Molecular structure of human TFIIH. Cell 102, 599–607. [DOI] [PubMed] [Google Scholar]
- Serizawa, H., Mäkelä, T.P., Conaway, J.W., Conaway, R.C., Weinberg, R.A., and Young, R.A. (1995). Association of Cdk-activating kinase subunits with transcription factor TFIIH. Nature 374, 280–282. [DOI] [PubMed] [Google Scholar]
- Shiekhattar, R., Mermelstein, F., Fisher, R.P., Drapkin, R., Dynlacht, B., Wessling, H.C., Morgan, D.O., and Reinberg, D. (1995). Cdk-activating kinase complex is a component of human transcription factor TFIIH. Nature 374, 283–287. [DOI] [PubMed] [Google Scholar]
- Shimotohno, A., Matsubayashi, S., Yamaguchi, M., Uchimiya, H., and Umeda, M. (2003). Differential phosphorylation activities of CDK-activating kinases in Arabidopsis thaliana. FEBS Lett. 534, 69–74. [DOI] [PubMed] [Google Scholar]
- Sutton, A., and Freiman, R. (1997). The Cak1p protein kinase is required at G1/S and G2/M in the budding yeast cell cycle. Genetics 147, 57–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tassan, J.P., Jaquenoud, M., Fry, A.M., Frutiger, S., Hughes, G.J., and Nigg, E.A. (1995). In vitro assembly of a functional human CDK7-cyclin H complex requires MAT1, a novel 36 kDa RING finger protein. EMBO J. 14, 5608–5617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thuret, J.Y., Valay, J.G., Faye, G., and Mann, C. (1996). Civ1 (CAK in vivo), a novel Cdk-activating kinase. Cell 86, 565–576. [DOI] [PubMed] [Google Scholar]
- Tsakraklides, V., and Solomon, M.J. (2002). Comparison of Cak1p-like cyclin-dependent kinase-activating kinases. J. Biol. Chem. 277, 33482–33489. [DOI] [PubMed] [Google Scholar]
- Ueda, T., Yamaguchi, M., Uchimiya, H., and Nakano, A. (2001). Ara6, a plant-unique novel type Rab GTPase, functions in the endocytic pathway of Arabidopsis thaliana. EMBO J. 20, 4730–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umeda, M. (2002). CDK-activating kinases in higher plants in CDK-activating kinase (CAK). In CDK-Activating Kinase (CAK), P. Kaldis, ed (Georgetown, TX: Landes Bioscience), pp. 55–64.
- Umeda, M., Bhalerao, R.P., Schell, J., Uchimiya, H., and Koncz, C. (1998). A distinct cyclin-dependent kinase-activating kinase of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 95, 5021–5026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Ackerveken, G., Marois, E., and Bonas, U. (1996). Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87, 1307–1316. [DOI] [PubMed] [Google Scholar]
- Vandepoele, K., Raes, J., De Veylder, L., Rouzé, P., Rombauts, S., and Inzé, D. (2002). Genome-wide analysis of core cell cycle genes in Arabidopsis. Plant Cell 14, 903–916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitaly, V.K. (2000). Rapid and reliable protein extraction from yeast. Yeast 16, 857–860. [DOI] [PubMed] [Google Scholar]
- Wallenfang, M.R., and Seydoux, G. (2002). cdk-7 is required for mRNA transcription and cell cycle progression in Caenorhabditis elegans embryos. Proc. Natl. Acad. Sci. USA 99, 5527–5532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, M., Fabian, T., Sauter, M., Bhalerao, R.P., Schrader, J., Sandberg, G., Umeda, M., and Uchimiya, H. (2000). Activation of CDK-activating kinase is dependent on interaction with H-type cyclins in plants. Plant J. 24, 11–20 Erratum. Plant J. 25, 473. [DOI] [PubMed] [Google Scholar]
- Yamaguchi, M., Umeda, M., and Uchimiya, H. (1998). A rice homolog of Cdk7/MO15 phosphorylates both cyclin-dependent protein kinases and the carboxy-terminal domain of RNA polymerase II. Plant J. 16, 613–619. [DOI] [PubMed] [Google Scholar]