Abstract
The blends of flavor compounds produced by fruits serve as biological perfumes used to attract living creatures, including humans. They include hundreds of metabolites and vary in their characteristic fruit flavor composition. The molecular mechanisms by which fruit flavor and aroma compounds are gained and lost during evolution and domestication are largely unknown. Here, we report on processes that may have been responsible for the evolution of diversity in strawberry (Fragaria spp) fruit flavor components. Whereas the terpenoid profile of cultivated strawberry species is dominated by the monoterpene linalool and the sesquiterpene nerolidol, fruit of wild strawberry species emit mainly olefinic monoterpenes and myrtenyl acetate, which are not found in the cultivated species. We used cDNA microarray analysis to identify the F. ananassa Nerolidol Synthase1 (FaNES1) gene in cultivated strawberry and showed that the recombinant FaNES1 enzyme produced in Escherichia coli cells is capable of generating both linalool and nerolidol when supplied with geranyl diphosphate (GPP) or farnesyl diphosphate (FPP), respectively. Characterization of additional genes that are very similar to FaNES1 from both the wild and cultivated strawberry species (FaNES2 and F. vesca NES1) showed that only FaNES1 is exclusively present and highly expressed in the fruit of cultivated (octaploid) varieties. It encodes a protein truncated at its N terminus. Green fluorescent protein localization experiments suggest that a change in subcellular localization led to the FaNES1 enzyme encountering both GPP and FPP, allowing it to produce linalool and nerolidol. Conversely, an insertional mutation affected the expression of a terpene synthase gene that differs from that in the cultivated species (termed F. ananassa Pinene Synthase). It encodes an enzyme capable of catalyzing the biosynthesis of the typical wild species monoterpenes, such as α-pinene and β-myrcene, and caused the loss of these compounds in the cultivated strawberries. The loss of α-pinene also further influenced the fruit flavor profile because it was no longer available as a substrate for the production of the downstream compounds myrtenol and myrtenyl acetate. This phenomenon was demonstrated by cloning and characterizing a cytochrome P450 gene (Pinene Hydroxylase) that encodes the enzyme catalyzing the C10 hydroxylation of α-pinene to myrtenol. The findings shed light on the molecular evolutionary mechanisms resulting in different flavor profiles that are eventually selected for in domesticated species.
INTRODUCTION
Strawberry is a member of the rose family (Rosaceae, subfamily Rosoideae, tribe Potentilleae) in the genus Fragaria. There are four basic fertility groups in Fragaria, distinguished by their ploidy level or chromosome number. F. vesca is the most common native species, contains 14 chromosomes, and is a diploid (Hancock, 1999). The cultivated varieties of commercial strawberries, usually designated as F. × ananassa, are almost all octaploids, containing 56 chromosomes, and derive mainly from the octaploids F. chiloensis (native to South America) and F. virginiana (native to the eastern United States). Most other evolutionary relationships within the genus are not clear. F. vesca may be the ancestor of the other Fragaria species because it occurs in most areas where these other species also grow. F. vesca chromosomes are able to pair with those of many of these other Fragaria species, including the octaploids. The first strawberry species were domesticated ∼2000 years ago, and the first commercial strawberry was introduced 250 years ago (Hancock, 1999). A remarkable difference exists between the fruit of the diploid wild species and the modern, cultivated species, not only in terms of fruit size and yield but also in flavor and aroma profile (Pyysalo et al., 1979; Honkanen and Hirvi, 1990).
The flavor of fruits is generally determined by tens if not hundreds of constituents, most of them generated during the ripening phase and typically in concentrations of 10 to 100 ppm of the fruit fresh weight (Maarse, 1991). Nearly all flavor compounds are formed from nonvolatile precursors (e.g., amino acids and lipids), and in some fruit, such as citrus, they accumulate in specialized structures adapted to contain high levels (Turner et al., 1998). Just as in other fruit, a complex mixture of more than 300 compounds has been detected in ripening strawberry (Zabetakis and Holden, 1997). These compounds can be grouped into more than a dozen chemical classes, including organic acids, aldehydes, ketones, alcohols, esters, lactones, sulfur compounds, acetals, furans, phenols, terpenes, and epoxides. Individual members of these groups, although often present in minute quantities, may have a significant impact on the overall aroma of the strawberry. Volatile flavors may also be glycoconjugated and, thus, stored in the fruit as nonvolatile compounds (Perez et al., 1997).
Early research on fruit flavor first focused on identifying flavor components present in the different fruit species and later on characterizing the volatiles that convey the characteristic odor unique to a particular fruit and unraveling their biogenesis. To date, only a few genes that directly influence fruit flavor biogenesis have been reported. These include the tomato (Lycopersicon esculentum) alcohol dehydrogenase (ADH2) (Bicsak et al., 1982; Longhurst et al., 1994; Speirs et al., 1998), alcohol acyltransferases from ripe wild and cultivated strawberry, melon (Cucumis melo), and banana fruit (Musa spp) (Aharoni et al., 2000; Yahyaoui et al., 2002; Beekwilder et al., 2004), an O-methyltransferase from the cultivated strawberry (Wein et al., 2002), and terpene synthases from citrus species (Lucker et al., 2002; Sharon-Asa et al., 2003) and apple (Malus domestica) (Pechous and Whitaker, 2004).
Terpenoids, and mainly the C10 (monoterpenes) and C15 (sesquiterpenes) members of this family, have been identified at varying levels in the flavor profiles of most if not all soft fruit (Maarse, 1991). In some species, they are of great importance for the characteristic flavor and aroma. For example, most citrus species are rich in various terpenoid components (Weiss, 1997). Another example is mango (Mangifera indica), in which terpenes comprise the main volatiles of most cultivars studied to date (MacLeod and Pieris, 1984).
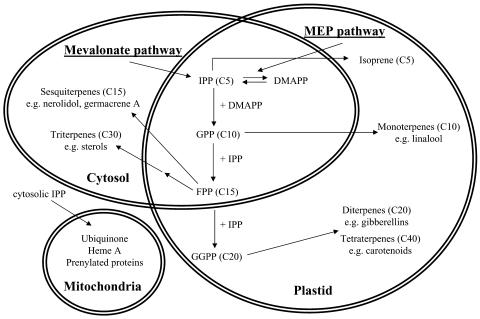
All terpenoids are derived from either the mevalonate pathway active in the cytosol or the plastidial 2-C-methyl-d-erythritol 4-phosphate pathway (Rodriguez-Concepcion and Boronat, 2002) (Figure 1). Both pathways lead to the formation of isopentenyl diphosphate (IPP) and its allylic isomer dimethylallyl diphosphate (DMAPP), the basic terpenoid biosynthesis building blocks. In both compartments, IPP and DMAPP are used by prenyltransferases in condensation reactions producing prenyl diphosphates. Condensation of IPP and DMAPP catalyzed by the prenyltransferase geranyl diphosphate (GPP) synthase yields GPP, the immediate precursor of monoterpenes. The condensation of two IPP units with one DMAPP by the action of farnesyl diphosphate (FPP) synthase generates the precursor for sesquiterpene biosynthesis. After the formation of these precursors, the various monoterpenes and sesquiterpenes are generated through the action of terpenoid synthases (TPSs; Trapp and Croteau, 2001). Primary terpene skeletons formed by TPSs might be further modified by hydroxylation, oxidation, double bond reduction, acylation, glycosylation, and methylation (Lange and Croteau, 1999). The complexity of terpenoid biosynthesis is further increased by subcellular compartmentation of the enzymes involved (Cunillera et al., 1997). For example, multiple isoforms of the gene for FPP synthase have been detected in Arabidopsis thaliana, with FPS1S and FPS2 encoding cytosol-targeted proteins, whereas FPS1L encodes a mitochondrially targeted protein (Cunillera et al., 1997). The FPS1 gene is bifunctional and uses alternative transcription start sites or selection of alternative translation initiation codons to generate either the cytosolic isoform (FPS1S) or the mitochondrial isoform (FPS1L).
Figure 1.
Compartmentation of Isoprenoid Biosynthesis in the Plant Cell.
The mevalonate pathway is active in the cytosol (and supplies IPP to mitochondria), whereas the methylerythritol 4-phosphate (MEP) pathway is active in plastids. Enzymatic steps similar in both the cytosolic and plastidic pathways are represented in the common area. GGPP, geranylgeranyl diphosphate.
The ability of plant species to produce one set of compounds and not another is commonly attributed to the evolution of new genes encoding enzymes with different characteristics or to altered gene expression. Although other molecular mechanisms exist that allow plants to alter their metabolic profiles during evolution, our information on such processes, especially in relation to plant secondary metabolism, is limited. Several scenarios have been proposed, including changes in the localization of enzymes or regulatory proteins by mutations in coding parts of genes that specify subcellular localization or changes in the localization of substrate biosynthesis and the transportation of the substrate to a different subcellular location (for a review, see Pichersky and Gang, 2000). An interesting example is provided by the biosynthesis of quinolizidine alkaloids in Lupinus (Suzuki et al., 1996; Wink and Roberts, 1998), where the quinolizidine skeleton is formed in chloroplasts, whereas acylation occurs after intracellular transportation to the cytosol and mitochondria. It was suggested that the fact that the acyl donor, tigloyl-CoA (derived from Ile), is produced within the mitochondria has caused the tigloylation reaction to occur within the mitochondria. Thus, primary metabolic processes and the organelles in which they are active are attuned to achieve metabolic diversity (De Luca and St. Pierre, 2000), and as a result the chances of creating beneficial metabolites or of stopping to produce others are enhanced.
Here, we present a detailed molecular and biochemical analysis of the biosynthesis of the major terpenoids produced during ripening of cultivated and wild strawberry species. Identification of marked differences in the production of specific monoterpenes and sesquiterpenes between the two species provided us with an opportunity to examine the mechanisms by which metabolic diversity in fruit flavor composition may have evolved.
RESULTS
Terpenoids of Wild and Cultivated Strawberry Species Differ
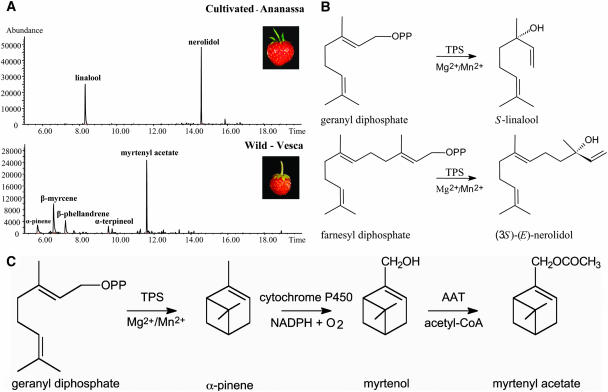
In the headspace of cultivated strawberry, only the monoterpene linalool and the sesquiterpene nerolidol were detected, whereas wild strawberry fruit emitted only monoterpenes (α-pinene, β-myrcene, α-terpineol, and β-phellandrene), myrtenyl acetate, and low levels of myrtenol, which were not detected in the cultivated species (Figure 2A). The biosynthesis of the terpenoids detected in the headspace of both species requires the action of monoterpene and sesquiterpene synthases using GPP or FPP as their substrate, respectively (Figures 2B and 2C). Isolation and characterization of the genes encoding these terpene synthases was therefore our first step in trying to explain the differences in the terpenoid profiles between the two strawberry species.
Figure 2.
Terpenoid Production in Wild and Cultivated Strawberry Species.
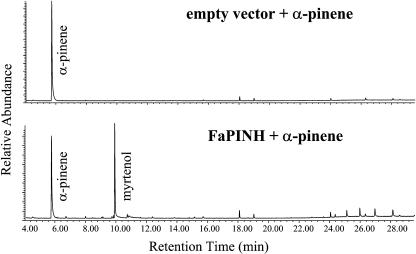
(A) Terpenoids detected by headspace analysis of ripe fruits. GC-MS chromatograms (selected mass-to-charge ratio 93) after headspace Tenax trapping (see Methods) showing the different terpenes emitted by cultivated (top) and wild (bottom) ripe strawberry fruit. A trace of the monoterpene alcohol myrtenol was also detected in wild strawberry (data not shown).
(B) Reactions catalyzed by terpene synthases (TPS) for the formation of the monoterpene alcohol linalool and the sesquiterpene alcohol nerolidol.
(C) Reactions catalyzed by a terpene synthase (TPS) enzyme for the formation of the monoterpene α-pinene, a cytochrome P450 enzyme catalyzing a subsequent hydroxylation step at C10 forming myrtenol and an alcohol acyltransferase (AAT) forming myrtenyl acetate.
Cloning and Characterization of the Terpene Synthase Gene Responsible for the Production of the Volatiles Linalool and Nerolidol by Cultivated Strawberry and Its Wild Strawberry Counterpart
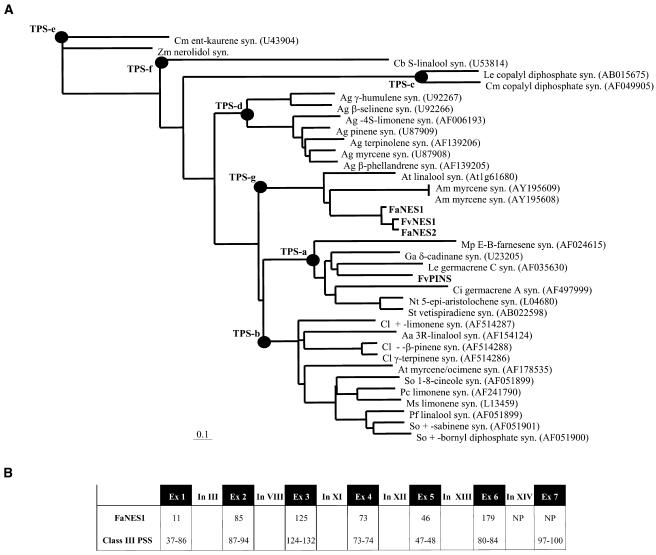
The Strawberry Nerolidol Synthase Proteins Are Members of a Recently Defined Seventh Subfamily of TPS (TPS-g)
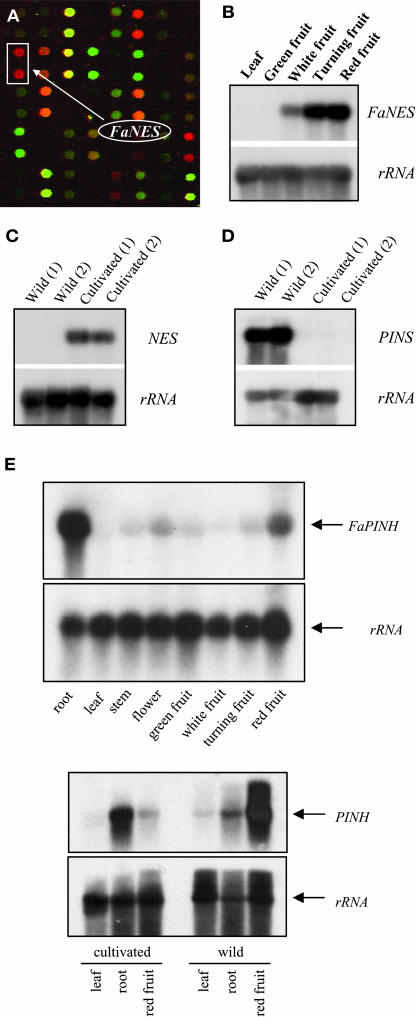
Analysis of microarray data (see Aharoni and O'Connell, 2002) comparing gene expression among various developmental stages and fruit tissues revealed strong upregulated expression of a putative terpene synthase gene homolog (F. ananassa Nerolidol Synthase1 [FaNES1]) in the receptacle tissue during ripening (Figure 3A). An RNA gel blot study confirmed the increase in FaNES1 transcript levels during fruit ripening, whereas no expression could be detected in leaf tissue (Figure 3B). It also revealed that FaNES1 was strongly expressed in cultivated strawberry varieties but hardly expressed at all in wild strawberry species (Figure 3C).
Figure 3.
Expression of the Terpene Synthase and Hydroxylase Genes in Wild and Cultivated Strawberry Species.
(A) Detection of FaNES1 expression in ripe strawberry receptacle tissue using cDNA microarrays. Red and green signals represent higher gene expression in the receptacle and achene (seeds) tissues, respectively (yellow signal indicates similar levels in both tissues).
(B) FaNES expression in tissues of the cultivated strawberry detected by RNA gel blots.
(C) and (D) Mirror images of NES and Pinene Synthase (PINS) expression in ripe fruits of wild and cultivated strawberry species detected by RNA gel blots. The numbers (1) and (2) mark two different wild species or cultivated varieties of strawberry (see Methods).
(E) Expression analysis of the F. ananassa Pinene Hydroxylase (FaPINH) gene in different tissues of cultivated (top pair of blots) and the wild strawberry in leaf, root, and red ripe fruit tissues (bottom pair of blots). The entire FaNES1, FvPINS, and FaPINH cDNAs were used to hybridize the RNA gel blots. An rRNA probe was used as a control for equal loading.
Sequence analysis showed that the predicted protein exhibited sequence similarities to both monoterpene and sesquiterpene synthases from other plant species, but was relatively short: 511 amino acid residues compared with 550 to 620 residues in known monoterpene and sesquiterpene synthases (Bohlmann et al., 1998; Figure 4). The predicted FaNES1 protein showed the greatest sequence homology to a recently described (+)-3S-linalool synthase from Arabidopsis (Chen et al., 2003; At1g61680, 51.3% at the amino acid level).
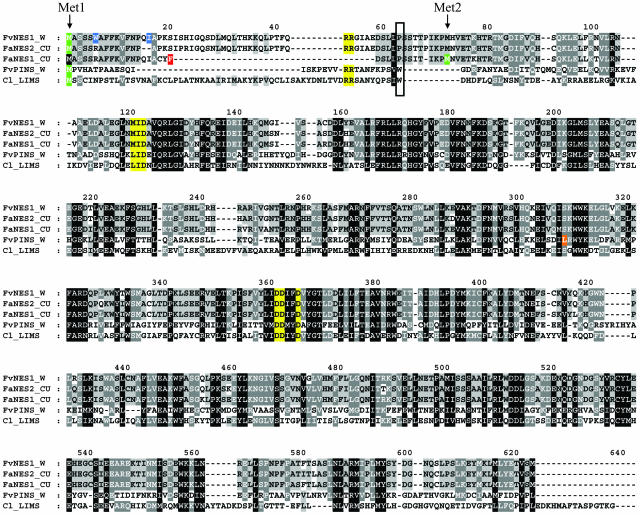
Figure 4.
Protein Sequence Alignment of the Strawberry Terpene Synthases.
The protein sequences of FaNES1, FaNES2, FvNES1, and FvPINS derived from cultivated (CU) and wild (W) strawberry species were aligned to the sequences of the Citrus limon limonene synthase (Cl_LIMS; GenBank accession number AF514287). The stop codon in the FaNES1 gene sequence, between Met1 (shaded green in all sequences except FaNES1) and Met2 (shaded green only in FaNES1) immediately follows the red shaded Phe residue (F). The RR, L/M-I-D, and DDXXD motifs conserved in monoterpene synthases are shaded yellow. Black, gray, and light gray shading represent 100, 80, and 60% conserved identity between residues, respectively. A 2-bp insertion (CC insertion; see also Figure 11) in the cultivated strawberry species results in a frameshift and an immediate stop codon in the middle of the FaPINS gene-coding region (instead of the orange shaded Leu residue [L] in FvPINS). Substitution (W6R) and removal (I16Δ) of the residues shaded blue in FvNES1 resulted in a change of targeting from plastid localization to localization in mitochondria (mainly) as well as to plastids (see also C12 in Figure 9). The residues present at the ninth position succeeding the twin Arg of the RR(x)8W motif normally found in the N-terminal part of class III TPS proteins are boxed. The motif is not entirely conserved in FaNES2 and FvNES1 because it contains a Pro (P) instead of a Trp (W) residue.
The plant TPS gene family has been classified into six subfamilies (designated TPS-a through TPS-f), with each subfamily sharing a minimum of 40% identity at the amino acid level (Bohlmann et al., 1998; Figure 5A). Recently, Dudareva et al. (2003) defined a seventh TPS subfamily (TPS-g), and in view of the criteria described above, FaNES1 is a member of this subfamily (Figure 5A). TPS genes have also been grouped into three classes based on their genomic structure (Trapp and Croteau, 2001): class I (12 to 14 introns), class II (9 introns), and class III (6 introns). Compared with class III TPS genes, FaNES1 contains a shorter first exon (encoding 11 amino acid residues compared with 37 to 86 in other genes) and, interestingly, has the lowest number of introns detected to date in TPS genes because it lacks intron XIV (Figure 5B).
Figure 5.
Strawberry NES Genes as Members of a New Family of Terpene Synthases (TPS-g).
(A) Phylogenetic analysis after ClustalX alignment of terpene synthases representing the seven different TPS families described to date (TPS-a to TPS-g). The alignment used PAM 350 and the neighbor-joining method. The ent-kaurene synthase (syn.) protein was defined as an out-group when rooting the tree. In the scale, bar 0.1 is equal to 10% sequence divergence. So, Salvia officinalis; Pf, Perilla frutescens; Pc, Perilla citridora; Ms, Mentha spicata; Cl, Citrus limon; At, Arabidopsis thaliana; Aa, Artemisia annua; Ga, Gossypium arboreum; Le, Lycopersicon esculentum; St, Solanum tuberosum; Sc, Solidago canadensis; Mp, Mentha piperita; Ag, Abies grandis; Cb, Clarkia breweri; Cm, Cucurbita maxima; Zm, Zea mays; Am, Antirrhinum majus; Ci, Cichorium intybus; Nt, Nicotiana tabacum. GenBank accession numbers are shown in brackets.
(B) FaNES1 contains only five introns, compared with the six normally present in class III terpene synthases. Numbers of amino acid residues encoded by each exon are depicted. Intron numbers were derived from Trapp and Croteau (2001). Ex, exon; In, intron; NP, not present.
The FaNES1 Gene Is Exclusively Present and Highly Expressed in the Cultivated Varieties and Encodes a Protein Truncated at Its N Terminus
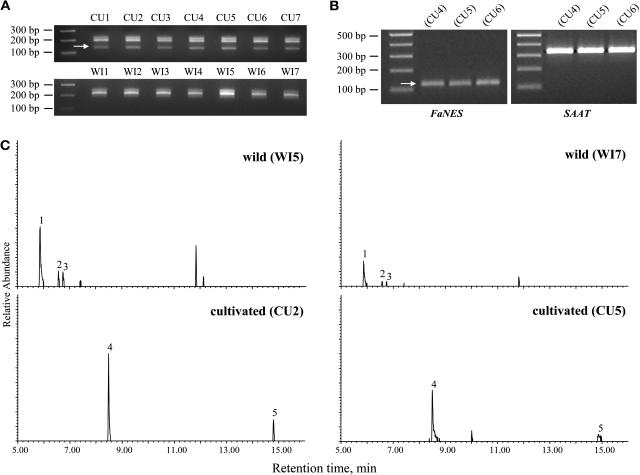
In view of the unusually short length of the predicted protein from FaNES1, we searched its gene sequence for potential translation start sites upstream of the ATG codon initially identified (termed Met2, see Figure 4). Such a codon was detected 96 bp upstream of the original ATG codon. However, the sequence between the two ATG codons would not be properly translated because of a stop codon halfway between the two (Figure 4). PCR on genomic DNA using oligonucleotides flanking the two ATG codons revealed that both the wild and the cultivated strawberry species also contain genes with a longer and uninterrupted sequence in between the two translation start sites (Figure 6A). However, none of the wild species tested (>12, seven of them shown in Figure 6A) contained the short fragment corresponding to the FaNES1 gene. By contrast, all cultivated species contained such a fragment (>70 tested, seven of them shown in Figure 6A).
Figure 6.
Correlation of the Presence and Expression of NES Genes and Volatile Product Formation in Wild and Cultivated Strawberry Species.
(A) PCR on genomic DNA of seven cultivated (CU1 to CU7) and seven wild strawberry species (WI1 to WI7) using oligonucleotides flanking the Met1 to Met2 regions, which should amplify all known gene fragments (see Figure 4). The arrow indicates a fragment of ∼150 bp corresponding to the FaNES1 gene. The larger fragments correspond to other NES genes, including those with a proper targeting signal.
(B) RT-PCR (left) using RNA derived from red, ripe fruit of cultivated varieties using the same oligonucleotides used in (A). The single band corresponds to the 150-bp fragment detected in (A). Larger fragments corresponding to the other NES transcripts were not detected under these RT-PCR conditions. A larger amount of cDNA and more cycles of amplification were used to clone the fragment corresponding to the FaNES2 gene because of its low abundance. The strawberry alcohol acyltransferase gene (SAAT) (Aharoni et al., 2000) was used as a control (right).
(C) Headspace analysis of fruits from two wild and two cultivated strawberry lines (tested molecularly in [A] and [B]) showing the presence or absence of monoterpenes (1, α-pinene; 2, sabinene; 3, β-myrcene; 4, linalool) and the sesquiterpene nerolidol (5).
We subsequently isolated the corresponding full-length cDNAs from the cultivated and wild strawberries (FaNES2 and F. vesca NES1, respectively) using specific oligonucleotides and rapid amplification of cDNA ends PCR (Figure 4). The cloning of FaNES2 and FvNES1 cDNAs was hampered by the fact that both showed very low levels of expression. Indeed, RT-PCR experiments (using the same oligonucleotides used previously for amplification on genomic DNA; see Methods) demonstrated that FaNES1 is the dominantly expressed gene in ripe cultivated strawberry fruit, whereas expression of FaNES2 and other NES-related genes was barely detectable (Figure 6B). Because the different NES cDNAs share very high homology at the nucleotide level (97% identity starting from the second Met), the results of the RNA gel blots probed with the full-length FaNES1 demonstrate that none of the NES-related genes in the cultivated species (including FaNES2) are expressed at detectable levels in leaf or green fruit tissue (Figure 3B) and that in the wild species none of them (including FvNES1) are expressed at detectable levels in red stage fruit tissue (Figure 3C). The molecular results presented above for the FaNES1 gene at the genomic and gene expression levels correlated in all cases with the presence or absence of nerolidol and linalool in the different cultivated and wild strawberry lines tested (Figure 6C).
It is evident that the FaNES1 N terminus is a truncated version of the corresponding regions in FaNES2 and FvNES1 because stretches of amino acids from both sides of the deleted region in FaNES1 are identical in the three proteins (Figure 4). Despite the clear differences at the N terminus, the three full-length strawberry proteins show high sequence conservation (95% identity). The putative FaNES2 and FvNES1 proteins are longer than FaNES1 (578 and 580 residues, respectively, versus 511 residues) and contain N-terminal regions (between the two Met residues; see Figure 4) of 59 and 61 amino acids, respectively. Interestingly, FaNES2 and FvNES1 contain a motif, RR(X)8P, that is similar to the RR(X)8W motif normally found in the N-terminal part (40 to 80 residues from the initial Met) of class III TPS proteins (Aubourg et al., 2002) (Figure 4). This small change in the motif has also been reported for other putative TPSs (e.g., in Arabidopsis; Aubourg et al., 2002). The RR(X)8W/P motif is not present in the FaNES1 protein, nor, surprisingly, in its closest homologs, the (+)-3S-linalool synthase from Arabidopsis (At1g61680), the (+)-3S-linalool synthase from Clarkia breweri (Dudareva et al., 1996), and the three monoterpene synthases reported recently from snapdragon (Antirrhinum majus) (Dudareva et al., 2003).
Recombinant NES Enzymes Generate (S)-Linalool and (3S)-(E)-Nerolidol from GPP and FPP, Respectively
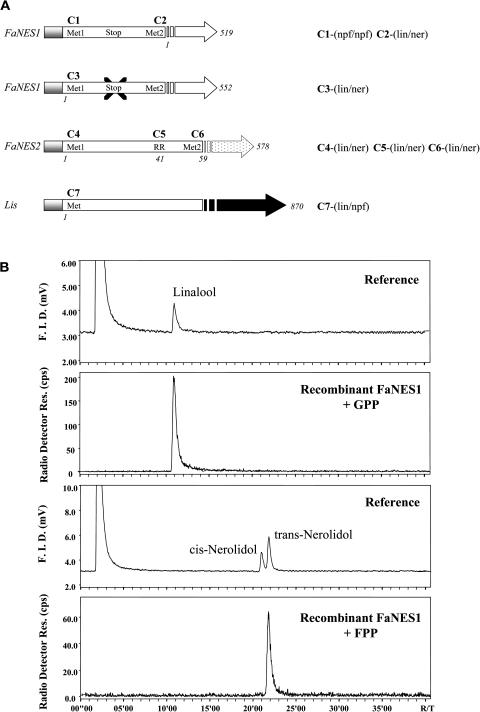
To determine the activity of the NES enzymes and to analyze whether this is influenced by the difference in their N termini, we conducted enzyme assays using the FaNES1 and FaNES2 recombinant proteins produced in Escherichia coli cells. We analyzed six different constructs (Figure 7, C1 to C6), harboring different parts of the proteins, either from Met1 (constructs C1 and C3, with the stop codon removed to allow translation from Met1, and C4), the twin Arg (RR and C5), or from Met2 (constructs C2 and C6) for activity with both GPP and FPP (Figure 7A). Using a seventh construct (Figure 7, C7), we also examined the activity with both substrates of the C. breweri linalool synthase recombinant enzyme (Lis; Dudareva et al., 1996). All variants of FaNES1 and FaNES2 efficiently converted GPP and FPP into the monoterpene and sesquiterpene alcohols linalool and nerolidol, respectively (Figure 7B).
Figure 7.
Enzymatic Activity Assays Using the FaNES1 and FaNES2 Recombinant Enzymes Produced in E. coli Cells.
(A) Seven different constructs (C1 to C7) used for the production of recombinant proteins and tested for enzyme activity. In C3, the stop codon present between the two Met residues (Met1 and Met2) in FaNES1 was removed by site-directed mutagenesis, allowing translation from Met1. All proteins were produced as fusions with a His-tag at their N termini (marked in gray). Results of enzyme activity assays with GPP and FPP are shown on the right-hand side of the construct schemes. lin, linalool; ner, nerolidol; npf, no product formed.
(B) Radio-GC analyses of radiolabeled products formed from 20 μM [3H]-geranyl diphosphate (top two panels) or [3H]-farnesyl diphosphate (bottom two panels) by heterologously produced, His-tag purified FaNES1 protein. Panel 1, flame ionization detector (F.I.D.) signal of coinjected, unlabeled linalool. Panel 3, unlabeled standards of (Z)-nerolidol and (E)-nerolidol. Panels 2 and 4, radio traces showing radiolabeled products. For further details, see Methods.
The results obtained with radio–gas chromatography (radio-GC) (Figure 7B) were further confirmed using gas chromatography–mass spectrometry (GC-MS) by comparing retention time and mass spectrum with authentic standards (data not shown). With both substrates, side products were undetectable by either radio-GC (Figure 7B) or GC-MS. In contrast with the strawberry FaNES1 and FaNES2 proteins, the recombinant Lis protein only converted GPP into linalool, but not FPP into nerolidol. Recombinant His-tag purified FaNES1 protein was used to determine enzyme kinetics. Assays over a range of GPP and FPP concentrations from 2 to 100 μM yielded the typical hyperbolic saturation curves for both substrates, but with slightly different parameters. The Km for FPP was 8.1 μM, whereas for GPP it was 29.0 μM. The Vmax with both substrates also differed slightly, being 2.3 nmol product h−1·μg−1 protein for GPP and 3.0 nmol product h−1·μg−1 protein for FPP.
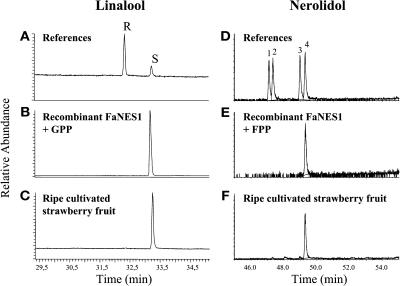
Enantioselective GC-MS analysis was performed to determine the enantiomeric composition of both products formed by the recombinant FaNES1 enzyme (construct C2 in Figure 7A) as well as the linalool and nerolidol emitted by ripe strawberry fruit (Figure 8). The recombinant FaNES1 enzyme converted GPP (with ∼100% efficiency) to S-linalool and FPP to (3S)-E-nerolidol. The results corresponded to the conformation of linalool and nerolidol emitted by the cultivated strawberry fruit during ripening.
Figure 8.
Chiral Analysis of Linalool and Nerolidol Produced by the Recombinant FaNES1 Protein and Ripe Fruit of the Cultivated Strawberry.
(A) and (D) References of R and S linalool enantiomers and of nerolidol enantiomers; 1 and 2, (3R) and (3S) (Z)-nerolidol (elution order not known); 3, (3R)(E)-nerolidol; 4, (3S)(E)-nerolidol.
(B) and (E) Enzyme activity assays with the recombinant FaNES1 enzyme and either GPP or FPP as substrate.
(C) and (F) Linalool and nerolidol produced by ripe, cultivated strawberry fruit.
GFP Localization Experiments Show That FaNES1 Localizes to the Cytosol, whereas FaNES2 and FvNES1 Are Localized to Mitochondria and/or Plastids
Starting from Met1, the putative FaNES2 and FvNES1 proteins and the short region in between Met1 and Met2 in FaNES1 (when translated after removal of the stop codon) contain a relatively large number of Ser and Ala residues (Figure 4). This is characteristic of mitochondrial and plastidic localization signals (Small et al., 1998). Various prediction programs (e.g., http://www.inra.fr/predotar/) suggested, with high probability, that the N termini of FaNES2 and FvNES1 serve as a targeting signal to mitochondria and plastids, respectively. By contrast, the FaNES1 protein was not predicted to contain such a targeting signal.
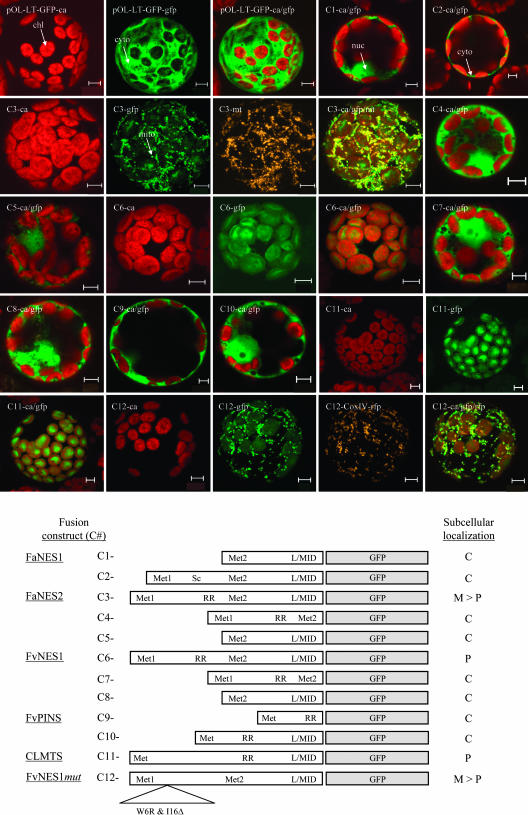
To test the targeting predictions, we analyzed the targeting capacity of the different N-terminal portions of the NES proteins in vivo. The 5′ regions of the various cDNAs were fused to a green fluorescent protein (GFP) reporter gene and transferred to tobacco (Nicotiana tabacum) protoplasts, which were subsequently analyzed for transient GFP expression using confocal laser scanning microscopy (Figure 9). Fusions of the 5′ region of FaNES1 to GFP showed that the deletion and the stop codon between Met1 and Met2 still allow translation, which starts from Met2 and results in a cytosolic localization of the protein (Figure 9, C1 and C2). The results of the GFP fusion studies further showed a dual targeting to both mitochondria and chloroplasts (mainly to mitochondria) for the FaNES2 fragment (Figure 9, C3), whereas GFP fluorescence was localized to the chloroplasts when it was fused to the FvNES1 5′ region (Figure 9, C6). In both cases, the region between Met1 and Met2 alone was not sufficient for localization to the plastids or mitochondria and led to GFP fluorescence in the cytosol (Figure 9, C4/C5 and C7/C8).
Figure 9.
Transient Expression of GFP Fusions in Tobacco Protoplasts Detected by Confocal Laser Scanning Microscopy.
pOL-LT-GFP is the original vector used to fuse the different strawberry gene fragments to GFP, which directs GFP expression to the cytosol and nucleoplasm (see Methods). Chloroplasts (chl; ∼5 μm in size), mitochondria (mito; ∼1 μm in size), cytosol (cyto), and nucleoplasm (nuc) are indicated by arrows. If several images are shown for a single construct, they are derived from the same protoplast. ca, chlorophyll autofluorescence detected in the red channel; gfp, green fluorescent protein fluorescence detected in the green channel; ca/gfp, combined red and green channels; mt, MitoTracker mitochondrial stain detected in the orange channel or merged with the green and red channels (ca/gfp/mt). Bottom panel, schematic and localization results for each fusion construct. Met1 and Met2, Met residues at the N termini of the proteins (Figure 4); L/MID, conserved motif in terpene synthases (Figure 4); Sc, stop codon; C, cytosol; P, plastids; M > P, more in mitochondria than in plastids. CLMTS, GFP fusion of the 5′ region of a typical monoterpene synthase from Citrus limon (Lucker et al., 2002).
The cytosolic, mitochondrial, and plastidic patterns of GFP expression observed with the different NES protein fusions were identical to those obtained with several positive controls: the pOL-LT-GFP empty vector (cytosol and nucleus), Mitotracker stain, and red fluorescent protein (RFP) expression directed by the cytochrome oxidase IV mitochondrial targeting signal (mitochondria) and RpoT;3 (data not shown) for plastidic localization. A comparison of the nearly identical targeting signals of FaNES2 and FvNES1 and the use of Predotar (http://www.inra.fr/predotar/) allowed us to identify two amino acid residues that, when modified (Figure 4, W6R substitution and I16Δ deletion), were able to change the plastidic targeting of FvNES1 to the dual mitochondrial and plastidic targeting of FaNES2 (Figure 9, C12). These results confirm that the N termini of the proteins encoded by FaNES2 and FvNES1 are mitochondrial and/or plastidic targeting signals. In FaNES1, the deletions and the introduction of a premature stop codon produce a transcript in which the targeting sequence is excluded from the coding region. The outcome of these events is cytosolic localization of the FaNES1 encoded protein in cultivated strawberries.
Cloning and Characterization of Genes Responsible for the Production of the Major Volatiles Produced by Wild Strawberry and Their Counterparts from Cultivated Strawberry
The FvPINS Gene Encodes the Enzyme Catalyzing the Biosynthesis of Multiple Monoterpenes in the Wild Species, whereas Its Cultivated Counterpart FaPINS Is Nonfunctional
As demonstrated above (Figure 2), wild strawberry species produce a range of monoterpenes that are not produced by the cultivated species. Nam et al. (1999) reported on the isolation of a cDNA fragment from wild strawberry fruit (clone 6.1.V1) that corresponded to a putative sesquiterpene synthase. They showed that the corresponding gene expression increased during fruit ripening and was not detectable in vegetative tissues. Interestingly, the mRNA could not be detected either in various tissues of a cultivated variety.
We used the sequence information of clone 6.1.V1 to isolate the full-length cDNA from wild strawberry (FvPINS). Analyzing FvPINS expression in wild and cultivated strawberries showed an inverse correlation with FaNES1 gene expression because the former was exclusively expressed in the fruit of wild strawberries (cf. Figures 3C and 3D). The FvPINS protein sequence (555 amino acids long) was most closely related to other sesquiterpene synthases present in the public databases, with high homology to a δ-cadinene synthase from cotton (Gossypium hirsutum) (GenBank accession number U23205; see Figure 5). The FvPINS and FaNES1 proteins share only 30% identity, but in both proteins the N-terminal region (up to the RR motif) is relatively short (Figure 4). In contrast with the NES proteins, FvPINS contains the complete RR(x)8W motif normally found in the N-terminal part of class III TPS proteins.
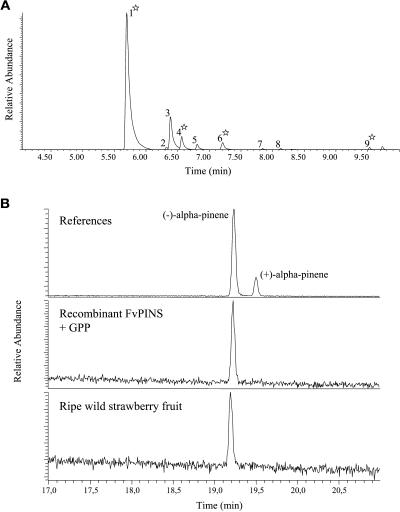
Heterologous expression in E. coli cells showed that FvPINS is a genuine monoterpene synthase, forming multiple monoterpenes, such as the major products α-pinene, β-phellandrene, and β-myrcene from GPP (Figure 10A). Because the protein sequence of FvPINS showed its highest homology to sesquiterpene synthases, we also analyzed the capacity of the recombinant enzyme to use FPP. However, we were unable to detect activity when FPP was used as a substrate. Finally, the enantiomeric composition of the product formed by the recombinant FvPINS was analyzed and compared with that in the headspace of the wild strawberry species. Both the heterologous enzyme product and the α-pinene in the headspace of wild strawberry showed an enantiomeric excess of >99% for (−)-α-pinene (Figure 10B).
Figure 10.
Analysis of the Monoterpene Products Formed from GPP in Assays with the Recombinant FvPINS Enzyme.
(A) Compounds detected in the volatile profile of the wild strawberry species are marked by a star (cf. Figure 2A). 1, α-pinene; 2, β-pinene; 3, sabinene; 4, β-myrcene; 5, α-phellandrene; 6, β-phellandrene; 7, dihydromyrcenol (tentative); 8, α-terpinolene (tentative); 9, α-terpineol (tentative).
(B) Multidimensional GC-MS analysis of the enantiomeric composition of the α-pinene formed from GPP in an assay with the recombinant FvPINS enzyme and the α-pinene present in the headspace of wild strawberry.
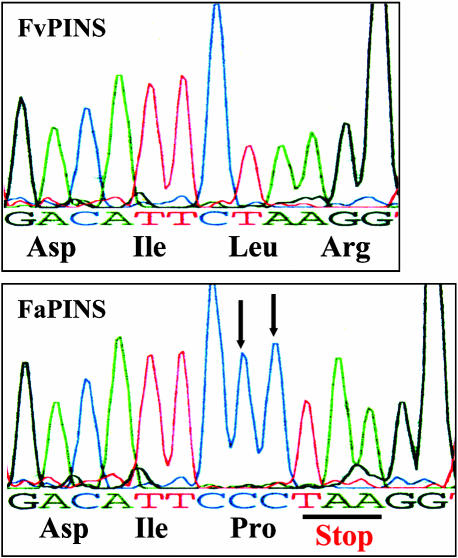
Although hampered by low expression, we succeeded in cloning the cDNA corresponding to FvPINS from the cultivated species (FaPINS). Nucleic acid alignment of FvPINS and FaPINS revealed only seven nucleic acid differences. Interestingly, an insertion of two tandem cytosine nucleotides in the middle of the coding region of FaPINS caused a frameshift that was followed directly by a UAA stop codon (Figure 11). To confirm that the CC insertion in FaPINS was not a PCR artifact, we cloned the cDNA twice again from different RNA samples and obtained the same result. In addition, we performed PCR on genomic DNA derived from both strawberry species. Using oligonucleotides flanking the CC insertion region, 20 clones were isolated and sequenced from each species. Whereas none of the fragments derived from the wild species contained a CC insertion, six of the 20 fragments isolated from the cultivated strawberry did contain it. Thus, only some of the FaPINS alleles in the genome of the octaploid cultivated species contain the insertion and, hence, the frameshift and subsequent stop codon.
Figure 11.
Insertion in the Middle of the FaPINS Gene-Coding Region.
A 2-bp insertion (CC, indicated by arrows) results in a frameshift and an immediate stop codon in the middle of the cultivated strawberry FaPINS gene-coding region (see also Figure 4). The insertion was not detected in the FvPINS gene derived from the wild strawberry.
The Monoterpene Synthase, FvPINS, Does Not Contain a Targeting Signal at Its N Terminus
In light of our previous results with the FaNES1 protein, we decided to analyze the targeting of the FvPINS protein. Targeting prediction programs (e.g., Predotar) were unable to identify a possible targeting signal in the FvPINS protein. Indeed, when the two regions derived from the 5′ end of the FvPINS gene (encoding 20 and 70 amino acid residues) were tested for their targeting capacity, the typical cytosolic GFP expression was observed (Figure 9, C9 and C10). By contrast, GFP fusion proteins of the 5′ region of a typical monoterpene synthase active in young green lemon peel tissue (Citrus limon) (Lucker et al., 2002) showed a clear plastidic subcellular localization, as expected (Figure 9, C11).
Cloning and Characterization of a Cytochrome P450 Gene from Wild and Cultivated Strawberry Encoding the Enzyme Catalyzing the Hydroxylation of α-Pinene to Myrtenol
Similar to a vast number of plant secondary metabolites, flavor components are very often further derivatized by the activity of modifying enzymes (Wein et al., 2002). Myrtenol, a monoterpene alcohol formed by the C10 hydroxylation of α-pinene (Figures 2A and 2C), has previously been described as one of the few compounds that may contribute to the typical aroma of the wild strawberry species (Honkanen and Hirvi, 1990). By contrast, cultivated strawberry varieties have never been reported to produce or emit myrtenol (Pyysalo et al., 1979; Hirvi and Honkanen, 1982; Honkanen and Hirvi, 1990; Wintoch et al., 1991; Zabetakis and Holden, 1997).
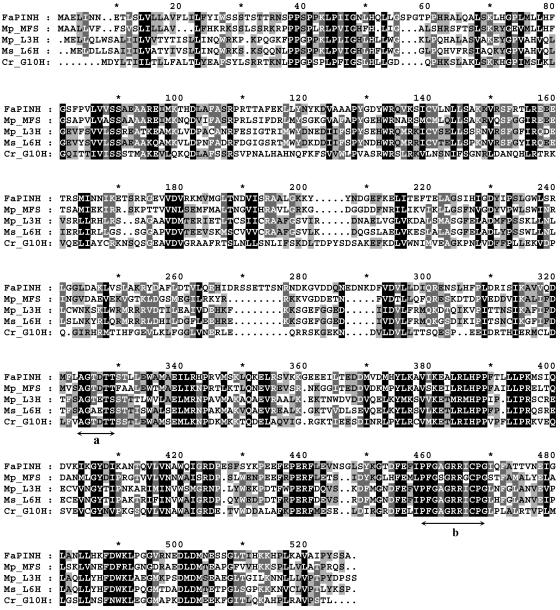
As described above, α-pinene is one of the monoterpenes we detected in wild species but not in cultivated species (Figure 2), and it was also the primary product of the recombinant FvPINS enzyme. The most likely enzymes to catalyze the oxidation of α-pinene to myrtenol are cytochrome P450 monooxygenases. By mining an EST sequence collection generated by random sequencing of a cultivated strawberry ripe fruit cDNA library (Aharoni and O'Connell, 2002), we identified five different EST clones that showed homology to cytochrome P450 genes cloned from a vast number of other organisms. Protein sequence alignment to other published cytochrome P450s showed that, of these five, the D59 clone is related to the CYP71 family (Figure 12). This family of cytochrome P450 proteins has previously been shown to be associated with monoterpene metabolism (Hallahan et al., 1994; Lupien et al., 1999; Bertea et al., 2001).
Figure 12.
Protein Sequence Alignment of the Strawberry FaPINH and Cytochrome P450s from Mint (Mentha) and Catharanthus roseus.
Mint (Mp, Mentha × piperita; Ms, Mentha spicata) proteins are menthofuran synthase (MFS) (GenBank accession number AF346833; Bertea et al., 2001), (−)-4S-limonene-3-hydroxylase (L3H; GenBank accession number AF124817), and (−)-4S-limonene-6-hydroxylase (L6H; GenBank accession number AF124815) (Lupien et al., 1999); the Catharanthus roseus (Cr) protein is geraniol 10 hydroxylase (G10H) (GenBank accession number AJ251269; Collu et al., 2001). The conserved oxygen binding and heme binding domains (Schuler, 1996) are marked a and b, respectively. Black, gray, and light gray shading represent 100, 80, and 60% conserved identity between residues, respectively.
Detailed gene expression analysis using the five different fragments as probes for RNA gel blot hybridizations revealed that clone D59 showed increased expression in the ripe red strawberry fruit but was also expressed, to even higher levels, in roots (FaPINH in Figure 3E). It has been reported that myrtenol glycoside is a component of cultivated strawberry roots (Wintoch, 1993), and this could support the view that the enzyme corresponding to the D59 gene is indeed the α-pinene hydroxylase. The protein putatively encoded by the D59 gene showed the highest homology (49 to 50% identity) to three Arabidopsis proteins with unknown functions (CYP71A26, CYP71A25, and CYP71A22).
We then analyzed the production of myrtenol in fruit and roots of various wild and cultivated strawberry species. The free form of myrtenol was detected in ripe fruit of four wild species, but not in any of the eight cultivated species examined (Table 1). The same pattern was detected for glycosylated myrtenol and myrtenyl acetate. On the other hand, relatively high levels of free and glycosylated myrtenol (more than in ripe fruit tissue of the wild species) were detected in the roots of both species. Enzyme assays with the recombinant D59 protein (termed FaPINH) produced in yeast microsomes showed that the substrate α-pinene was hydroxylated at C-10 to form myrtenol (Figure 13). The (−)-α-pinene form was preferred to (+)-α-pinene as a substrate. A dozen other monoterpenes were also tested as substrates for the recombinant FaPINH protein, and in several cases they could also be hydroxylated at the position corresponding to C-10 in α-pinene. For example, (+)- and (−)-limonene were hydroxylated at C10 to yield perilla alcohol with only slightly lower efficiency than α-pinene (data not shown). Cloning of the corresponding gene from the wild species (termed FvPINH) showed that the proteins from the wild and the cultivated species differed by only three amino acid residues (data not shown).
Table 1.
Presence of Free Myrtenol, Glycosidically Bound Myrtenol, and Myrtenyl Acetate in Ripe Fruit and Roots of Various Wild and Cultivated Strawberries
| Species | Name | Tissue | Free Form (mg/kg) Myrtenol | Glycosidically Bound (mg/kg) Myrtenol | Free Form (mg/kg) Myrtenyl Acetate |
|---|---|---|---|---|---|
| Wild | PRI line 92189 | Ripe fruit | 0.109 | 0.040 | 0.155 |
| Wild | PRI line H1 | Ripe fruit | 0.608 | 0.090 | 0.392 |
| Wild | PRI line 92190 | Ripe fruit | 0.352 | 0.050 | 0.604 |
| Wild | Yellow wonder | Ripe fruit | 0.227 | NDa | 0.418 |
| Cultivated | Elsanta | Ripe fruit | ND | ND | ND |
| Cultivated | Calypso | Ripe fruit | ND | ND | ND |
| Cultivated | Camerosa | Ripe fruit | ND | ND | ND |
| Cultivated | Gorrela | Ripe fruit | ND | ND | ND |
| Cultivated | Sure crop | Ripe fruit | ND | ND | ND |
| Cultivated | Senga sengana | Ripe fruit | ND | ND | ND |
| Cultivated | Virginiana 352 | Ripe fruit | ND | ND | ND |
| Cultivated | Elsanta | Roots | 7.230 | 0.239 | ND |
| Wild | PRI line 92189 | Roots | 4.634 | 0.271 | ND |
ND, not detected (<0.001 mg/kg).
Figure 13.
Production of Myrtenol by the Recombinant FaPINH Enzyme.
Recombinant protein extracted from yeast microsomes and harboring either the empty vector or the vector containing the FaPINH coding region was used for enzymatic assays using (−)-α-pinene as a substrate. Total ion chromatograms from the GC-MS analysis of the products are shown.
We further analyzed PINH gene expression in leaf, root, and ripe fruit tissues of wild and cultivated species (Figure 3E). The results show that PINH is expressed at high levels in ripe fruit of the wild species, higher than in ripe fruit of the cultivated species. Expression of PINH was detected in roots of both strawberry species (though at higher levels in the cultivated species), whereas only very low levels could be detected in leaves of both species.
DISCUSSION
Many authors have described the large diversity of secondary metabolites that are produced by plants and the differences observed between wild and cultivated species (Hanson et al., 1996; Hartmann, 1996; Kliebenstein et al., 2001; Hadacek, 2002; Schwab, 2003). Fruit volatile flavors serve as a useful model for studying such variations, in view of the large number of different compounds associated with it and the variability in its composition across different fruit varieties and between cultivated fruit varieties and their wild ancestors (Maarse, 1991).
Data on the molecular mechanisms that direct changes in fruit flavors during evolution and domestication are very limited. Recently, Tadmor et al. (2002) identified a wild species allele in tomato that negatively affects tomato fruit aroma and that was selected against during domestication. They suggested that this loss of the ability to produce high levels of phenylacetaldehyde had contributed to the development of the desirable aroma of the cultivated tomato. Future identification of genes from both wild and cultivated tomato species that influence phenylacetaldehyde biosynthesis might explain how this compound has been lost from the flavor profile during breeding.
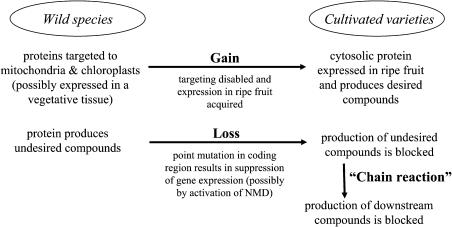
In this investigation, we identified a marked difference between wild and cultivated strawberry species in the production of specific volatile terpenoid flavor components. We used a combination of molecular and biochemical tools to examine the differences observed between the two species. The results suggest that domestication of strawberry has involved selection for specific alleles in the cultivated species, which in turn has contributed to a strongly modified flavor profile (Figures 2 and 14).
Figure 14.
Gain and Loss of Terpenoids in Ripe Strawberry Fruit.
Schematic depiction of mechanisms causing the gain and loss of flavor and aroma compounds in strawberry during evolution, which may have been specifically selected for in domesticated species.
The FaNES1 and FvPINS Proteins Account for the Production of the Dominant Terpenes Produced by Ripe Fruit of Cultivated and Wild Strawberry Species, Respectively
To understand the change in terpene composition in strawberry, we isolated putative terpene synthase genes from the wild and cultivated species. Expression of the two genes isolated (FaNES1 and FvPINS) and the catalytic activity of their corresponding enzymes correlated with the terpenoid products generated during fruit ripening: linalool and nerolidol in cultivated and olefinic monoterpenes in wild strawberry. FaNES1 was only detected in the genomes of octaploid species, including the octaploid strawberry parents F. chiloensis and F. virginiana. This gene was not found in any of the diploid species or in the genome of the hexaploid F. moschata (B. Denoyes-Rothan and D. Sargent, unpublished data). This observation suggests that the FaNES1 gene evolved close to or during the octaploid stage in strawberry. Expression of FaNES1 was high in cultivated strawberry varieties, whereas FvPINS expression was virtually absent, and the reverse was true in the wild species.
Enzymatic activity assays using the recombinant proteins produced in E. coli demonstrated that the protein encoded by FaNES1 could generate with similar efficiency both S-linalool and (3S)-(E)-nerolidol (depending on whether the monoterpene precursor GPP or the sesquiterpene precursor FPP was used). The enzyme kinetics data seem to support a genuine dual monoterpene and sesquiterpene synthase activity, with a Km for FPP of 8.1 μM, which is well within the range of Km values reported for several other sesquiterpene synthases (Bouwmeester et al., 2002). Although the Km for GPP of 29.0 μM is relatively high for a monoterpene synthase, it is comparable to and even lower than the Km values reported for two other linalool synthases from Mentha citrata (56 μM for GPP; Crowell et al., 2002) and Artemisia annua (64 μM for GPP; Jia et al., 1999). Moreover, the Vmax values for the two substrates (GPP and FPP) hardly differ. This is supported by the radio-GC and GC-MS analyses. These not only show similar levels of product formation from the two substrates, but the absence of any major side products also suggests tight control of the enzymatic catalysis. Our in vivo experiments also showed that the protein encoded by FaNES1 (when targeted to the plastids) in transgenic Arabidopsis plants generated both linalool (at high levels) and nerolidol (Aharoni et al., 2003). Despite the very low level of expression of the FaNES2 gene, we cannot rule out the possibility that plastidic FaNES2 enzyme activity could make a minor contribution to the production of linalool in the cultivated strawberry species.
In contrast with FaNES1, the FvPINS recombinant enzyme was only able to use GPP to generate α-pinene as the major product and various other monoterpene side products. For both proteins, compounds produced by the recombinant enzymes in vitro fully matched the enantiomers detected in planta. The fact that the major monoterpene in the headspace of wild strawberry was β-myrcene, whereas the major product of recombinant FvPINS was α-pinene, might be explained by the fact that, in vivo, α-pinene is further converted to myrtenol and its derivatives (myrtenol glycoside and myrtenyl acetate) (Figure 2, see also Table 1). Myrtenol can be converted to its corresponding acetate ester by a wild strawberry alcohol acyltransferase (VAAT) described recently by Beekwilder et al. (2004).
The ability of both terpene synthases from strawberry to generate more than one product (when tested in vitro), each of which could be detected in the headspace of the fruit, further strengthens the idea that a large proportion of the diversity in secondary metabolite composition, for example, in the flavor blend of a fruit, is directed by the ability of single enzymes to generate multiple products (Schwab, 2003). For example, the formation of most volatile esters in the wild and cultivated strawberry (more than 100 have been reported) could be explained by the action of just a few alcohol acyl transferases (Aharoni et al., 2000; Beekwilder et al., 2004).
Altered Gene Expression and Subcellular Localization of the NES Genes and Proteins Mediate the Production of Linalool and Nerolidol by the Cultivated Species
The unusually short length of the putative protein encoded by FaNES1 prompted us to investigate in detail the N termini of the various proteins encoded by the genes cloned from both wild and cultivated species. In doing so, we discovered that the FaNES1 protein lacks the conserved RRx8W motif, previously described for other monoterpene synthases, and predicted to be important for the catalysis of monoterpene formation (Williams et al., 1998). Only recently, Dudareva et al. (2003) distinguished a new subfamily of terpene synthases (TPS-g) containing monoterpene synthases from Arabidopsis (At1g61680; Chen et al., 2003) and snapdragon (ama1e20, ama0c15, and ama0a23; Dudareva et al., 2003). The members of this family all lack the RRx8W motif, share a high sequence identity, show little relatedness with terpene synthases in other subfamilies, produce acyclic monoterpenes, and, interestingly, were all associated with floral scent. The FaNES1 protein could also be included in this family, on the basis of its sequence similarity and its function in volatile emission from reproductive organs. However, it represents the first member of this new family that can efficiently generate a sesquiterpene (e.g., nerolidol).
We have also discovered a deletion in the N-terminal part of FaNES1 compared with the same region in FvNES1 and FaNES2 (Figure 4). All the monoterpene synthases functionally characterized to date possess an N-terminal transit peptide that directs their import into the plastid, after which this transit peptide is cleaved (Williams et al., 1998). Indeed, our GFP localization experiments showed that the N-terminal part of a lemon limonene synthase directs the GFP protein to the plastids. Similar analyses of the targeting properties of the various NES proteins detected GFP expression either in both plastids and mitochondria (FaNES2 and FvNES) or only in the cytosol (FaNES1). These findings were supported by the results obtained from various targeting prediction programs (e.g., http://www.inra.fr/predotar/). The localization of the highly expressed FaNES1 to the cytosol in cultivated strawberry, its in vitro activity with the two substrates GPP and FPP (producing linalool and nerolidol, respectively, with about equal efficiency), and the fact that these two terpene alcohols are the major headspace volatiles of cultivated strawberry suggest that both linalool and nerolidol are produced in the cytosol in strawberry.
Even stronger evidence that both monoterpene and sesquiterpene biosynthesis occurs in the cytosol of ripe strawberry fruit cells was provided by the analysis of a genuine monoterpene synthase (FvPINS) from wild strawberry, which is distantly related to FaNES1 in sequence and activity. Just as for FaNES1, GFP localization assays showed that the FvPINS N-terminal part lacks targeting ability and directs GFP expression to the cytosol (Figure 9). This suggests that in strawberry fruit, the monoterpene precursor GPP and the sesquiterpene precursor FPP are both present in sufficient amounts in the cytosol to support the production of monoterpenoids and sesquiterpenoids, respectively.
Biosynthesis of monoterpenes in the cytosol, although surprising, is not a completely new concept because Bouvier et al. (2000) have suggested that monoterpenes may be produced in the cytosol in nonphotosynthetic tissues such as fruit, glands, or other secretory tissues. These authors also suggested that the Arabidopsis GPP synthase encoded two isoforms, one targeted to the plastids and the other to the cytosol. Similarly, the formation of shikonin, a monoterpene derivative, was reported to occur in the cytosol of Lithospermum erythrorhizon cells (Heide and Berger, 1989; Sommer et al., 1995).
Preliminary feeding experiments demonstrated that application of labeled mevalonic acid to ripe strawberry resulted in incorporation of the label to linalool. By contrast, feeding labeled 1-deoxy-d-xylulose did not result in labeled linalool. Because biosynthesis of mevalonic acid is expected to occur in the cytosol, the feeding experiments suggest that the formation of linalool in ripe strawberry fruit proceeds in the cytoplasm rather than plastids (Matthias Wuest, personal communication).
With regard to the evolutionary accession of linalool and nerolidol production, we suggest that ancestral genes most likely encoded proteins with mitochondrial and/or plastidic targeting signals (both known sites for terpene formation in plants; Bouvier et al., 2000), such as those present in the FaNES2 and FvNES1 proteins. These proteins may have been similar to the higher terpene synthases, such as diterpenes synthases involved in primary metabolism, which were proposed by Trapp and Croteau (2001) to serve as the ancestors of terpene synthase genes responsible for natural product biosynthesis. According to these authors, duplication and divergence in structural and functional specialization of these ancestral genes was accompanied by sequential intron loss (from 14 down to six introns). FaNES1 is unusual in this respect because it contains only five introns, the smallest number detected so far for terpene synthases. The absence, or low levels, of terpenoid precursors in the mitochondria or plastids of strawberry may have resulted in the absence of selection pressure to maintain active organellar NES proteins. Deletions and insertions of stop codons in the coding regions of ancestral proteins such as FaNES2 disabled the targeting signal region (which in FaNES2 targeted predominantly to mitochondria) and led to a cytosolic localization (as in the protein encoded by FaNES1). The change in localization (and a sufficiently high expression) exposed the enzyme to a new or larger pool of substrates (both GPP and FPP) that enabled it to form both linalool and nerolidol.
Several other examples of relocation of isozymes performing analogous functions at different intracellular locations and involving either single or multiple genes have been reported for plants and other organisms (Danpure, 1995). For example, in humans, rabbits, and guinea pigs, the ability to target the metabolic enzyme alanine:glyoxalate aminotransferase (AGAT) to the mitochondria was lost because of mutation of the translation start site (the most upstream of the two possible ones) from ATG to ATA in the human gene, to ACA in the rabbit gene, and to GTT in the guinea pig gene (Danpure, 1997). The loss of the mitochondrial targeting signal at the N terminus allowed permanent targeting of AGAT to the peroxisomes. Peroxisomal targeting of AGAT in humans is most important because mislocalization of AGAT to mitochondria causes a decrease in glyoxylate detoxification and results in the hyperoxaluria type 1 disease.
In plants, several examples of molecular mechanisms resulting in the acquisition of a targeting signal have been provided by the study of gene transfer from organelles to the nucleus (Adams et al., 2002). Activation of a nuclear gene after transfer requires the rapid acquisition of several regulating and targeting elements before it becomes inactivated by random mutations (Daley et al., 2002). In the case of the soybean (Glycine max) nuclear-encoded cytochrome c oxidase subunit (GmCox2), a mitochondrial targeting presequence of 124 amino acids was acquired (Daley et al., 2002). The authors suggested that an intron located only 1 bp upstream of the start codon could have been involved in an exon shuffling-like process during presequence acquisition.
In recent years, numerous enzymes associated with secondary metabolism have been shown to be promiscuous with respect to substrate (e.g., various alcohols and acyl-CoAs used by SAAT for ester formation) (Aharoni et al., 2000). This phenomenon might be exploited by plants to enhance metabolic diversity by changing enzyme localization and, hence, substrate availability, as in the case of the strawberry NES enzymes. Of course, the change in FaNES protein localization was not yet sufficient for the biosynthesis of substantial amounts of products. High level, ripe fruit tissue specific gene expression was as important and required additional mutation, possibly in the promoter region of the gene. As a result of such molecular evolutionary events, FaNES1 acquired the capability to produce high levels of both linalool and nerolidol in ripe strawberry fruit. Thus, this study provides an example in which metabolic diversity is created by a change in the localization of enzymes and not, as is more commonly found, by mutations in genes resulting in functionally different enzymes.
Loss of Several Monoterpenes, Including Myrtenol, by the Cultivated Strawberry Species
This study also investigated the reverse situation, in which monoterpenes normally produced by wild strawberry species could not be detected in the volatile profile of cultivated varieties. We suggest that the presence of mutated alleles of FaPINS, containing a premature translational stop caused by a frameshift, has led to this loss of flavor components in the fruit (Figure 14). A probable explanation for the low steady state levels of FaPINS mRNA in the cultivated species might be the activity of RNA surveillance mechanisms such as nonsense-mediated decay (NMD). As a result of NMD, abnormal mRNAs containing premature translation termination codons are efficiently eliminated by degradation, so that production of undesirable truncated proteins is avoided (Isshiki et al., 2001).
Little is known about NMD in plants compared with what is currently known for yeast and mammals (Petracek et al., 2000), although it is clear that NMD does occur. Non-sense or frameshift mutations have been shown to reduce drastically the abundance of mRNAs in several cases, such as in the genes for soybean Kti3, trypsin inhibitor (Jofuku et al., 1989), bean (Phaseolus vulgaris) phytohemagglutinin (Voelker et al., 1990; Hoof and Green, 1996), pea (Pisum sativum) ferredoxin (Dickey et al., 1994; Petracek et al., 2000), and the rice (Oryza sativa) waxy gene (Isshiki et al., 2001). Because we also identified genomic fragments in the cultivated species that do not contain the insertion mutation, these fragments might correspond to a root-specific α-pinene synthase gene, whereas the fruit contains only truncated alleles that induce NMD. In this context, it is worth mentioning that three separate attempts to clone a nontruncated FaPINS gene from ripe fruit were unsuccessful and resulted in the isolation of only the mutated gene. This might also explain the formation of α-pinene in roots of the cultivated species and the low-level but detectable expression in roots of the cultivated strawberry when the entire FaPINS cDNA was used as a probe for RNA gel blot analysis (data not shown).
Further studies are needed to determine whether the non-sense mutation detected in the FaPINS gene is the only reason for the absence of its transcript in the fruit or whether other factors, such as changes in the promoter region or reduction in the activity of a regulatory protein, also play a role in this phenomenon. The loss of the ability to produce α-pinene also leads to the loss of other characteristic flavor components (myrtenol and its derivatives) of wild strawberry species (Wintoch et al., 1991) in cultivated strawberries, despite the fact that the FaPINH gene encoding the enzyme catalyzing the hydroxylation of α-pinene to myrtenol is still expressed in the ripe fruit of the cultivated species.
Gain and Loss of Fruit Flavors during Evolution and Domestication and Their Significance
The presence of the FaNES1 gene in the genome of certain strawberry species has apparently provided them with a strong selective advantage, which was systematically passed on to today's domesticated strawberries. Flavor and aroma are important selection criteria used by strawberry breeders. Both linalool, which imparts a sweet, floral, citrus-like note, and nerolidol, providing a rose, apple, green note, may have been specifically selected for because they positively influence the aroma of the cultivated varieties. Linalool in particular influences the odor of strawberries (Larsen and Poll, 1992), and its presence in large amounts in the cultivar Senga sengana (used extensively in the production of jams) contributes to the intense and pleasant character of this cultivar (Maarse, 1991). By contrast, the olefinic monoterpenes, namely, α-pinene, β-phellandrene, and β-myrcene, are major components of tree oleoresins (e.g., grand fir [Abies grandis]) (Steele et al., 1998; Martin et al., 2002) and contribute the turpentine-like, woody, resinous, and unpleasant odor of wild strawberry that was perhaps selected against by breeders. The production of nerolidol and linalool by the cultivated varieties may also have provided a strong selective advantage in terms of pathogen resistance because both compounds have been shown to be highly potent antimicrobial substances (Kubo et al., 1993; Carson and Riley, 1995).
In conclusion, we have demonstrated that independent molecular mechanisms have led to changes in the fruit flavor volatile blend in strawberry, under evolutionary and possibly breeding pressure. The remarkable diversity of more than 100,000 low molecular mass natural plant products (Dixon, 2001) suggests that many other, as yet unknown, molecular evolutionary processes may have occurred in plants, resulting in the production of particular metabolites.
METHODS
Plant Material
Greenhouse-grown strawberry (Fragaria spp) varieties and lines of wild species from the Plant Research International (PRI) breeding collection were used. Volatile analysis (Figure 2A) was conducted using Elsanta as the cultivated variety and PRI accession 92189 as the wild species. For RNA gel blots, we used the Elsanta cultivar (Figures 3B, 3E, and 3F), the PRI accessions H1 and 92189 as wild species (W), and Gorella and Holiday as cultivated forms (CU; in Figures 3C and 3D). PCR on genomic DNA and expression analysis using RT-PCR (Figure 6) were performed using CU1 (cv Sure crop), CU2 (cv Holiday), CU3 (cv Senga sengana), CU4 (cv Gorella), CU5 (cv Calypso), CU6 (cv Elsanta), CU7 (PRI accession 75169), and WI1 (PRI accession FA-1), WI2 (PRI accession FA-2), WI3 (PRI accession FA-3), WI4 (Yellow wonder), WI5 (Alexandria), WI6 (PRI accession 92189), and WI7 (PRI accession H2).
Cloning of Full-Length NES and PINS cDNAs
Full-length cDNAs were cloned using the Smart RACE cDNA amplification kit (Clontech, Palo Alto, CA) according to the manufacturer's instructions, with slight modifications to annealing temperatures (normally reduced by 5 to 10°C relative to recommended temperatures) or numbers of cycles (up to 35 cycles). The published fragment of the FvPINS gene (Nam et al., 1999) was used to design oligonucleotides and clone the entire FvPINS and FaPINS cDNAs. PCR, restriction digests, plasmid DNA isolation, and gel electrophoresis were performed using standard protocols. All fragments were purified from the gel using the GFX purification kit (Amersham, Buckinghamshire, UK). Cloning of PCR fragments was either done to the PCR Script (Stratagene, La Jolla, CA) or pCR 4Blunt-TOPO (Invitrogen, Carlsbad, CA) vectors (for blunt-end products generated when using Pfu DNA polymerase) or to the pGEM-T Easy (Promega, Madison, WI) vector (when A-tailed PCR products were generated by means of Taq DNA polymerase). Sequencing was done using the ABI 310 capillary sequencer according to the manufacturer's instructions (Applied Biosystems, Perkin-Elmer, Foster City, CA). Sequence analysis was conducted using the DNASTAR (DNASTAR, Madison, WI) and GeneDoc (http://www.psc.edu/biomed/genedoc) programs.
Heterologous Expression of NES and PINS Proteins in Escherichia coli Cells and Enzyme Kinetics
The modified pRSET B (Invitrogen) expression vector was used for expression in E. coli cells (Stratagene; BL21 Gold DE3 strain) as described previously (Aharoni et al., 2000). Briefly, the cloning of the various NES fragments to a modified pRSET B vector yielded a fusion protein at the N terminus with a peptide that included an ATG translation initiation codon, as well as a series of six His residues (His-tag) and Anti-Xpress epitope proteins, and eliminated the native Met ATG codon present in the strawberry cDNAs. For protein purification (50 mL of bacterial culture), we used nickel-nitrilotriacetic acid agarose spin columns under native conditions as described by the manufacturers (Qiagen, Valencia, CA). First eluates (200 μL) from the His-tag columns were used for enzyme activity assays. Heterologous proteins were characterized after His-tag purification by diluting 100 μL of the eluent to 1 mL with assay buffer containing 15 mM Mopso, pH 7.0, 10% (v/v) glycerol, 10 mM MgCl2, 1 mM MnCl2, 1 mM sodium ascorbate, and 2 mM DTT. We added 20 μM [3H]-GPP or [3H]-FPP to the assay. After the addition of a 1-mL redistilled pentane overlay, the tubes were carefully mixed and incubated for 1 h at 30°C. The assays were extracted as previously described (Bouwmeester et al., 2002) and analyzed using radio-GC on a Carlo-Erba 4160 series gas chromatograph, equipped with a RAGA-90 radioactivity detector (Raytest, Straubenhardt, Germany) and GC-MS (Bouwmeester et al., 1999, 2002). Enzyme kinetics were determined using His-tag purified enzyme, essentially as previously described (de Kraker et al., 1998).
Isolation of Nucleic Acids and Expression Analysis
Total cellular DNA isolation was performed as described by Marty et al. (2000). RNA isolation and expression analysis using either RNA gel blots or cDNA microarrays (different fruit tissues and leaves) were conducted as described previously (Aharoni et al., 2000). The microarray experiment comparing gene expression between achene and receptacle tissues was described previously (Aharoni and O'Connell, 2002). The entire FaNES1 and FvPINS cDNAs were used as probes for the RNA gel blot analysis. The PCR on genomic DNA and the RT-PCR of the various cultivated and wild species (Figure 6) both used the oligonucleotides AAP418 (forward, 5′-TGCTGATCATAGATCAGATGG-3′) and AAP456 [reverse, 5′-TGCT(C/T)GGTTTCAACGT(G/T)CAT-3′], flanking the Met1 and Met2, for the amplification of the NES fragments. The oligonucleotides AAP162 (forward, 5′-GGATGAACATGGAGACAGTG-3′) and AAP157 (reverse, 5′-TCCAATCCATGTCCTCC-3′) were used for the amplification of a 350-bp SAAT fragment as a control for the amount of cDNA. In the RT-PCR analysis, the first strand cDNA was synthesized from 1 μg of DNAase-treated (Invitrogen, Life Technologies kit) total RNA using the Superscript II RNase H reverse transcriptase kit as described by the manufacturer (Invitrogen, Life Technologies). We used standard PCR conditions for both PCR on genomic DNA and RT-PCR (55°C annealing temperature, 1-min extension; 35 cycles).
Protein Localization Experiments
The various fragments used for localization analysis were fused upstream of, and in frame with, the GFP gene in the cloning sites (SpeI and SalI) present in the cassette of pOL-LT-GFP-L64T65 (modified pOL-GFPS65C; Peeters et al., 2000), using XbaI and XhoI restriction sites (at the 5′ and 3′ ends, respectively) introduced by PCR. Tobacco (Nicotiana tabacum) protoplasts (cv SRI) were prepared and transformed as described earlier (Negrutiu et al., 1987) using 50 μg of plasmid DNA (of each construct, also when two constructs were cotransformed). MitoTracker Red CMXRos staining was conducted according to the manufacturer's instructions (Molecular Probes, Eugene, OR). Protoplasts were examined 24 h after transformation with a Carl Zeiss confocal laser scanning microscope (LSM 510; Jena, Germany) with an argon ion laser. The fluorescence of GFP (absorption, 488 nm; emission 507 nm) was obtained using the 488-nm laser line, and the emission signal was collected using a band-pass filter (band-pass 505 to 550). The ×40 objective was used for imaging, and the excitation intensity was set at 2 to 4% in most cases. The long-wavelength signal of the chlorophyll was collected using a long-pass filter (LP650). When the protoplasts were stained with MitoTracker Red (absorption 578 nm; emission 599 nm), images were obtained in the multiexcitation mode, using the 488- and 543-nm laser lines sequentially. Both chlorophyll autofluorescence and MitoTracker Red emission were collected via a long-pass filter (LP650). RFP (absorption, 558 nm; emission, 585 nm) was monitored with excitation at 543 nm, and the emission signal was detected with a band-pass filter (band-pass 560 to 615). Optical sections were taken along the optical axis and projected into one image with the Zeiss LSM image browser (Carl Zeiss).
Heterologous Expression of FaPINH Protein in Yeast
The construct for expression in Saccharomyces cerevisiae was generated by amplifying the entire FaPINH coding region by PCR with Pfu DNA polymerase using oligonucleotides that introduced a BglII restriction site upstream of the start codon (AAP113, 5′-CAGATCTATGGAAGCCACTTCTTGGGTTAC-3′) and an EcoRI site downstream of the stop codon (AAP114, 5′-CCTTAAGAGAAGCTAGTAGCTGGAACC-3′). PCR products digested with BglII and EcoRI were ligated to the pYeDP60 expression vector (Pompon et al., 1996), which was digested with BamHI and EcoRI in between the artificial promoter GAL10-CYC1 and the PGK terminator. Recombinant pYeDP60 plasmids were transferred into S. cerevisiae cells using the lithium acetate method (Ito et al., 1983). The yeast host cells employed, WAT11U and WAT21U (Pompon et al., 1996), were kindly provided by P. Urban. Both strains contain an insertion in the endogenous NADPH-cytochrome P450 reductase locus (CPR1) that is replaced by either ATR1 or ATR2, the two NADPH-cytochrome P450 reductase genes from Arabidopsis thaliana (Urban et al., 1997). Transformants were selected on SGI (7 g/L of yeast nitrogen base, 1 g/L of bactocasamino acid, 20 g/L of glucose, and 20 mg/L of dl-Trp) medium, and expression was performed in YPL medium (induction with 2% galactose) (Pompon et al., 1996). Samples of harvested yeast cells were assayed spectrophotometrically for cytochrome P450 content as detected by CO binding spectra (Omura and Sato, 1964). Intact cells were also harvested and used for an in vivo assay, by incubation in 50 mM Tris-HCl buffer, pH 7.4, containing 0.1 mM DTT and 1 mM EDTA in the presence of 500 μM (−)-α-pinene. The reaction was allowed to proceed for 2 h at 30°C with gentle shaking. Controls containing either no cofactors or substrate and cells containing vector without insert were also used for in vivo assays. The reaction was stopped by chilling the mixture on ice and extracting twice with 1.0-mL portions of pentane:diethyl ether (4:1). The combined extract was passed through a short column of silica gel and anhydrous MgSO4, after which the column was washed with 1.5 mL of diethyl ether to remove residual compounds. After concentration under N2, the samples were analyzed by GC-MS. Samples (2 μL) were analyzed by an automated injector on a Hewlett-Packard 5890 series II gas chromatograph equipped with a 30 m × 0.25 mm inner diameter, fused silica column coated with a 0.25-μm film of HP-5MS (Hewlett-Packard, Palo Alto, CA) and a Hewlett-Packard 5972A mass selective detector. GC oven temperature was programmed at an initial temperature of 45°C for 1 min, with a ramp of 10°C min−1 to 280°C, and a final time of 10 min. Full spectra were recorded for major reaction products, which were identified by comparing retention times with authentic standards and by comparing spectra with those of the NSB75K library, using the G1033A NIST probability-based matching algorithm. The identity of the product was confirmed by coincidence of retention time with the authentic standard. Transformants containing significant cytochrome P450 levels as well as control yeast cells harboring the expression vector without insert were also used to prepare microsomes by means of published procedures (Pompon et al., 1996). Microsome preparations were evaluated by CO-difference spectrum and were assayed for (−)-α-pinene hydroxylation.
The reaction mixture for the recombinant enzyme activity assay, in a final volume of 1 mL, contained 50 mM Tris-HCl, pH 7.4, 1 mM EDTA, 0.1 mM DTT, 0.8 units of glucose-6-phosphate dehydrogenase, 2 mM glucose-6-phosphate, 5 μM flavin adenine dinucleotide, 5 μM flavin mononucleotide, 1 mM NADPH, and 200 μL of microsomal preparation. We added 200 nmol (−)-α-pinene (in <10 μL, which has no detectable influence on the reaction) to the mixture to start the reaction, which was allowed to proceed for 2 h at 30°C with gentle shaking. Microsomes isolated from cells harboring the empty vector were employed as controls. The reaction was stopped by chilling the mixture on ice, and the reaction products were extracted and analyzed as described above for the in vivo assay.
Analysis of Fruit Volatiles
For the purpose of headspace analyses, red ripe strawberry fruits were enclosed in 0.7-liter glass jars fitted with a Teflon-lined lid equipped with an inlet and an outlet. A vacuum pump was used to draw air through the glass jar at ∼100 mL min−1, the incoming air being purified through a glass cartridge (140 × 4 mm) containing 150 mg Tenax TA (20/35 mesh; Alltech, Breda, the Netherlands). At the outlet, the volatiles emitted by the detached fruits were trapped on a similar Tenax cartridge. Volatiles were sampled over 24 h. Cartridges were eluted using 3 × 1 mL of redistilled pentane-diethyl ether (4:1). Of these samples, 2 μL was analyzed by GC-MS, using an HP 5890 series II gas chromatograph equipped with an HP-5MS column (30 m × 0.25 mm i.d., 0.25 μm film thickness) and an HP 5972A mass selective detector as described by Bouwmeester et al. (1999).
Solid-Phase Extraction
The XAD-2 column was purchased from Supelco (Bellefonte, PA). Myrtenol, phenol, methanol, and diethyl ether were obtained from Aldrich (Deisenhofen, Germany). Samples were stored at −20°C until work-up. Frozen samples were weighed and submerged in an equal volume of water, homogenized by means of an Ultra-Turrax, and centrifuged (2000g; 10 min). The pellets were washed twice, and the supernatants were combined (∼40 mL) and subjected to solid-phase extraction on XAD-2 (20 cm, 1 cm i.d). The XAD-2 column was preconditioned with 50 mL of methanol and 100 mL of water. After application of the sample, the XAD-2 was successively washed with 50 mL of water, 50 mL of diethyl ether, and 80 mL of methanol. The diethyl ether extract was dried over sodium sulfate and concentrated to ∼100 μL. Phenol (0.1 mg/mL) was added as an internal standard. The methanol extract was concentrated in vacuo to ∼1 mL.
Enzymatic Hydrolysis and GC-MS Analysis
Enzymatic hydrolysis was performed by dissolving an aliquot of the methanol extract, as described above, in 2 mL 0.2 M phosphate buffer, pH 5.5. The solution was extracted twice with the same volume of diethyl ether to remove free alcohols. Subsequently, we added 200 μL of Rohapect D5L (Röhm, Darmstadt, Germany), a pectinolytic enzyme preparation exhibiting glycosidase activity. After an incubation period of 24 h at 37°C, the liberated aglycons were extracted twice with 1 mL of diethyl ether. The combined organic layers were dried over sodium sulfate and concentrated. Capillary GC-MS analysis was performed with a Fisons GC 8000 series (Fisons Instruments, Engelsbach, Germany) coupled to a Fisons MD800 quadrupole mass detector fitted with a split injector (1:20) at 230°C. A DB-Wax fused silica capillary column (30 m × 0.25 mm i.d.; film thickness = 0.25 μm) (J and W, Folsom, CA) was used with a run program from 50°C for 3 min to 220°C for 10 min, with a temperature increase of 4°C min−1, using a 2 mL·min−1 helium gas flow rate. Xcalibur for Windows software was used for data acquisition. The significant MS operating parameters were as follows: ionization voltage 70 eV (electron impact ionization), ion source temperature 220°C, and interface temperature 250°C. Constituents were identified by comparing their mass spectra and retention indices with those of authentic reference compounds. The absolute configuration of (E)-nerolidol was determined as described by Bouwmeester et al. (1999). Multidimensional GC-MS on a Fisons 8160 GC connected to a Fisons 8130 GC and a Fisons MD 800 quadrupole mass spectrometer was used to analyze the absolute configuration of linalool and α-pinene as described by Lucker et al. (2002) for α-pinene or with the following modifications for linalool. The fused silica capillary column in GC1 was maintained at 60°C, then programmed to 240°C at 10°C min−1 with He gas flow at 3 mL min−1. The fused silica capillary column in GC2 was maintained at 60°C (15 min), then programmed to 200°C at 2°C min−1 with He gas flow at 3 mL min−1. Separation of linalool enantiomers was achieved with the second GC, using a 25 m × 0.25 mm i.d. fused silica capillary column coated with a 0.15-mm film of 2,3-di-O-ethyl-6-O-tert-butyl dimethylsilyl-β-cyclodextrin/PS086.
Sequence data from this article have been deposited with the EMBL/GenBank data libraries under accession numbers AX529067, AX528996, and AX529025.
Acknowledgments
We would like to thank Ian Small, Nemo Peeters, Maureen Hanson, and Andreas Weihe for the GFP and RFP vectors. We are grateful to Patrick Smit, Mark Hink, and Jan Willem Borst for assistance with confocal laser scanning microscopy, to Maarten Koornneef, Andy Pereira, Robert Hall, and Christopher Danpure for comments on the manuscript, and to Joost Lucker for providing the citrus limon monoterpene synthase cDNA and helpful discussions. We are also grateful to Philipe Urban for the yeast vectors and strains, to Beatrice Denoyes-Rothan and Dan Sargent for the PCR analysis of the different wild strawberry species, and to Matthias Wuest for sharing data before publication. We thank Bert Meulenbroek, Jos Kanne, and Eric Van de Weg for plant material and genomic DNA of the different strawberry genotypes and Eran Pichersky for the Lis DNA.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Asaph Aharoni (asaph.aharoni@weizmann.ac.il).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.023895.
References
- Adams, K.L., Daley, D.O., Whelan, J., and Palmer, J.D. (2002). Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell 14, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A., Giri, A.P., Deuerlein, S., Griepink, F., de-Kogel, W.J., Verstappen, F.W.A., Verhoeven, H.A., Jongsma, M.A., Schwab, W., and Bouwmeester, H.J. (2003). Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 15, 2866–2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A., et al. (2000). Identification of the SAAT gene involved in strawberry flavor biogenesis by use of DNA microarrays. Plant Cell 12, 647–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aharoni, A., and O'Connell, A.P. (2002). Gene expression analysis of strawberry achene and receptacle maturation using DNA microarrays. J. Exp. Bot. 53, 2073–2087. [DOI] [PubMed] [Google Scholar]
- Aubourg, S., Lecharny, A., and Bohlmann, J. (2002). Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics 267, 730–745. [DOI] [PubMed] [Google Scholar]
- Beekwilder, J., Alvarez-Huerta, M., Neef, E., Verstappen, F.W.A., Bouwmeester, H.J., and Aharoni, A. (2004). Functional characterization of enzymes forming volatile esters from strawberry and banana. Plant Physiol. 135, 1865–1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertea, C.M., Schalk, M., Karp, F., Maffei, M., and Croteau, R. (2001). Demonstration that menthofuran synthase of mint (Mentha) is a cytochrome P450 monooxygenase: Cloning, functional expression, and characterization of the responsible gene. Arch. Biochem. Biophys. 390, 279–286. [DOI] [PubMed] [Google Scholar]
- Bicsak, T.A., Kann, L.R., Reiter, A., and Chase, T., Jr. (1982). Tomato alcohol dehydrogenase: Purification and substrate specificity. Arch. Biochem. Biophys. 216, 605–615. [DOI] [PubMed] [Google Scholar]
- Bohlmann, J., Meyer Gauen, G., and Croteau, R. (1998). Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 95, 4126–4133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouvier, F., Suire, C., d'Harlingue, A., Backhaus, R.A., and Camara, B. (2000). Molecular cloning of geranyl diphosphate synthase and compartmentation of monoterpene synthesis in plant cells. Plant J. 24, 241–252. [DOI] [PubMed] [Google Scholar]
- Bouwmeester, H.J., Kodde, J., Verstappen, F.W.A., Altug, I.G., de Kraker, J.-W., and Wallaart, T.E. (2002). Isolation and characterization of two germacrene A synthase cDNA clones from chicory. Plant Physiol. 129, 134–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouwmeester, H.J., Verstappen, F.W.A., Posthumus, M.A., and Dicke, M. (1999). Spider mite-induced (3S)-(E)-nerolidol synthase activity in cucumber and lima bean. The first dedicated step in acyclic C11-homoterpene biosynthesis. Plant Physiol. 121, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson, C.F., and Riley, T.V. (1995). Antimicrobial activity of the major components of the essential oil of Melaleuca alternifolia. J. Appl. Bacteriol. 78, 264–269. [DOI] [PubMed] [Google Scholar]
- Chen, F., Tholl, D., D'Auria, J.C., Farooq, A., Pichersky, E., and Gershenzon, J. (2003). Biosynthesis and emission of terpenoid volatiles from Arabidopsis flowers. Plant Cell 15, 481–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collu, G., Unver, N., Peltenburg Looman, A.M.G., van der Heijden, R., Verpoorte, R., and Memelink, J. (2001). Geraniol 10-hydroxylase, a cytochrome P450 enzyme involved in terpenoid indole alkaloid biosynthesis. FEBS Lett. 508, 215–220. [DOI] [PubMed] [Google Scholar]
- Crowell, A.L., Williams, D.C., Davis, E.M., Wildung, M.R., and Croteau, R. (2002). Molecular cloning and characterization of a new linalool synthase. Arch. Biochem. Biophys. 405, 112–121. [DOI] [PubMed] [Google Scholar]
- Cunillera, N., Boronat, A., and Ferrer, A. (1997). The Arabidopsis thaliana FPS1 gene generates a novel mRNA that encodes a mitochondrial farnesyl-diphosphate synthase isoform. J. Biol. Chem. 272, 15381–15388. [DOI] [PubMed] [Google Scholar]
- Daley, D.O., Adams, K.L., Clifton, R., Qualmann, S., Millar, A.H., Palmer, J.D., Pratje, E., and Whelan, J. (2002). Gene transfer from mitochondrion to nucleus: Novel mechanisms for gene activation from Cox2. Plant J. 30, 11–21. [DOI] [PubMed] [Google Scholar]
- Danpure, C. (1995). How can the products of a single gene be localized to more than one intracellular compartment? Trends Cell Biol. 5, 230–238. [DOI] [PubMed] [Google Scholar]
- Danpure, C. (1997). Variable peroxisomal and mitochondrial targeting of alanine: Glyoxylate aminotransferase evolution and disease. Bioessays 19, 317–326. [DOI] [PubMed] [Google Scholar]
- de Kraker, J.W., Franssen, M.C.R., de Groot, A., König, W.A., and Bouwmeester, H.J. (1998). (+)-Germacrene A biosynthesis. The committed step in the biosynthesis of bitter sesquiterpene lactones in chicory. Plant Physiol. 117, 1381–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Luca, V., and St. Pierre, B. (2000). The cell and developmental biology of alkaloid biosynthesis. Trends Plant Sci. 5, 168–173. [DOI] [PubMed] [Google Scholar]
- Dickey, L.F., Nguyen, T.T., Allen, G.C., and Thompson, W.F. (1994). Light modulation of ferredoxin mRNA abundance requires an open reading frame. Plant Cell 6, 1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon, R.A. (2001). Natural products and plant disease resistance. Nature 411, 843–847. [DOI] [PubMed] [Google Scholar]
- Dudareva, N., Cseke, L., Blanc, V.M., and Pichersky, E. (1996). Evolution of floral scent in Clarkia: Novel patterns of S-linalool synthase gene expression in the C. breweri flower. Plant Cell 8, 1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudareva, N., Martin, D., Kish, C.M., Kolosova, N., Gorenstein, N., Faldt, J., Miller, B.M., and Bohlmann, J. (2003). (E)-beta-ocimene and myrcene synthase genes of floral scent biosynthesis in snapdragon: Function and expression of three terpene synthase genes of a new terpene synthase subfamily. Plant Cell 15, 1227–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadacek, F. (2002). Secondary metabolites as plant traits: Current assessment and future perspectives. Crit. Rev. Plant Sci. 21, 273–322. [Google Scholar]
- Hallahan, D.L., Lau, S.M.C., Harder, P.A., Smiley, D.W.M., Dawson, G.W., Pickett, J.A., Christoffersen, R.E., and O'Keefe, D.P. (1994). Cytochrome P-450-catalysed monoterpenoid oxidation in catmint (Nepeta racemosa) and avocado (Persea americana); evidence for related enzymes with different activities. Biochim. Biophys. Acta 1, 94–100. [DOI] [PubMed] [Google Scholar]
- Hancock, J.F. (1999). Strawberries. (Wallingford, UK: CABI Publishing).
- Hanson, M.A., Gaut, B.S., Stec, A.O., Fuerstenberg, S.I., Goodman, M.M., Coe, E.H., and Doebley, J.F. (1996). Evolution of anthocyanin biosynthesis in maize kernels: The role of regulatory and enzymatic loci. Genetics 143, 1395–1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, T. (1996). Diversity and variability of plant secondary metabolism: A mechanistic view. Entomol. Exp. Appl. 80, 177–188. [Google Scholar]
- Heide, L., and Berger, U. (1989). Partial purification and properties of geranyl pyrophosphate synthase from Lithospermum erythrorhizon cell cultures. Arch. Biochem. Biophys. 273, 331–338. [DOI] [PubMed] [Google Scholar]
- Hirvi, T., and Honkanen, E. (1982). The volatiles of two new strawberry cultivars, “Annelie” and “Alaska Pioneer”, obtained by backcrossing of cultivated strawberries with wild strawberries, Fragaria vesca, Rugen and Fragaria virginiana. Z. Lebensm. Unters. Forsch. 175, 113–116. [Google Scholar]
- Honkanen, E., and Hirvi, T. (1990). The flavour of berries. In Food Flavours, I.D. Morton and A.J. MacLeod, eds (Amsterdam: Elsevier Scientific Publications), pp. 125–193.
- Hoof, A.V., and Green, P.J. (1996). Premature nonsense codons decrease the stability of phytohemagglutinin mRNA in a position-dependent manner. Plant J. 10, 415–424. [DOI] [PubMed] [Google Scholar]
- Isshiki, M., Yamamoto, Y., Satoh, H., and Shimamoto, K. (2001). Nonsense-mediated decay of mutant waxy mRNA in rice. Plant Physiol. 125, 1388–1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito, H., Fukuda, Y., Murata, K., and Kimura, A. (1983). Transformation of intact yeast cells treated with alkali cations. J. Bacteriol. 153, 163–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia, J.W., Crock, J., Lu, S., Croteau, R., and Chen, X.Y. (1999). (3R)-linalool synthase from Artemisia annua L.: cDNA isolation, characterization, and wound induction. Arch. Biochem. Biophys. 372, 143–149. [DOI] [PubMed] [Google Scholar]
- Jofuku, K.D., Schipper, R.D., and Goldberg, R.B. (1989). A frameshift mutation prevents Kunitz trypsin inhibitor mRNA accumulation in soybean embryos. Plant Cell 1, 427–435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein, D.J., Lambrix, V.M., Reichelt, M., Gershenzon, J., and Mitchell-Olds, T. (2001). Gene duplication in the diversification of secondary metabolism: Tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13, 681–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo, I., Muroi, H., and Himejima, M. (1993). Antibacterial activity against Streptococcus mutans of mate tea flavor components. J. Agric. Food Chem. 41, 107–111. [Google Scholar]
- Lange, B.M., and Croteau, R. (1999). Genetic engineering of essential oil production in mint. Curr. Opin. Plant Biol. 2, 139–144. [DOI] [PubMed] [Google Scholar]
- Larsen, M., and Poll, L. (1992). Odour thresholds of some important aroma compounds in strawberries. Z. Lebensm. Unters. Forsch. 195, 120–123. [Google Scholar]
- Longhurst, T., Lee, E., Hinde, R., Brady, C., and Speirs, J. (1994). Structure of the tomato Adh2 gene and Adh2 pseudogenes, and a study of Adh2 gene expression in fruit. Plant Mol. Biol. 26, 1073–1084. [DOI] [PubMed] [Google Scholar]
- Lucker, J., El Tamer, M.K., Schwab, W., Verstappen, F.W.A., van der Plas, L.H.W., Bouwmeester, H.J., and Verhoeven, H.A. (2002). Monoterpene biosynthesis in lemon (Citrus limon) cDNA isolation and functional analysis of four monoterpene synthases. Eur. J. Biochem. 269, 3160–3171. [DOI] [PubMed] [Google Scholar]
- Lupien, S., Karp, F., Wildung, M., and Croteau, R. (1999). Regiospecific cytochrome P450 limonene hydroxylases from mint (Mentha) species: cDNA isolation, characterization, and functional expression of (-)-4S-limonene-3-hydroxylase and (-)-4S-limonene-6-hydroxylase. Arch. Biochem. Biophys. 368, 181–192. [DOI] [PubMed] [Google Scholar]
- Maarse, H. (1991). Volatile Compounds in Foods and Beverages. (New York: Marcel Dekker).
- MacLeod, A.J., and Pieris, N.M. (1984). Comparison of the volatile components of some mango cultivars. Phytochemistry 23, 361–366. [Google Scholar]
- Martin, D., Tholl, D., Gershenzon, J., and Bohlmann, J. (2002). Methyl jasmonate induces traumatic resin ducts, terpenoid resin biosynthesis, and terpenoid accumulation in developing xylem of Norway spruce stems. Plant Physiol. 129, 1003–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marty, I., Douat, C., Tichit, L., Jungsup, K., Leustek, T., and Albagnac, G. (2000). The cystathionine-gamma-synthase gene involved in methionine biosynthesis is highly expressed and auxin-repressed during wild strawberry (Fragaria vesca L.) fruit ripening. Theor. Appl. Genet. 100, 1129–1136. [Google Scholar]
- Nam, Y.W., Tichit, L., Leperlier, M., Cuerq, B., Marty, I., and Lelievre, J.M. (1999). Isolation and characterization of mRNAs differentially expressed during ripening of wild strawberry (Fragaria vesca L.) fruits. Plant Mol. Biol. 39, 629–636. [DOI] [PubMed] [Google Scholar]
- Negrutiu, I., Shillito, R., Potrykus, I., Biasini, G., and Sala, F. (1987). Hybrid genes in the analysis of transformation conditions. Plant Mol. Biol. 8, 363–373. [DOI] [PubMed] [Google Scholar]
- Omura, T., and Sato, R. (1964). The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J. Biol. Chem. 239, 2370–2378. [PubMed] [Google Scholar]
- Pechous, S.W., and Whitaker, B.D. (2004). Cloning and functional expression of an (E,E)-alpha-farnesene synthase cDNA from peel tissue of apple fruit. Planta 219, 84–94. [DOI] [PubMed] [Google Scholar]
- Peeters, N.M., Chapron, A., Giritch, A., Grandjean, O., Lancelin, D., Lhomme, T., Vivrel, A., and Small, I. (2000). Duplication and quadruplication of Arabidopsis thaliana cysteinyl- and asparaginyl-tRNA synthetase genes of organellar origin. J. Mol. Evol. 50, 413–423. [DOI] [PubMed] [Google Scholar]
- Perez, A.G., Cert, A., Rios, J.J., and Olias, J.M. (1997). Free and glycosidically bound volatile compounds from two banana cultivars Valery and Pequena Enana. J. Agric. Food Chem. 45, 4393–4397. [Google Scholar]
- Petracek, M.E., Tuyen, N., Thompson, W.F., and Dickey, L.F. (2000). Premature termination codons destabilize ferredoxin-1 mRNA when ferredoxin-1 is translated. Plant J. 21, 563–569. [DOI] [PubMed] [Google Scholar]
- Pichersky, E., and Gang, D.R. (2000). Genetics and biochemistry of secondary metabolites in plants: An evolutionary perspective. Trends Plant Sci. 5, 439–445. [DOI] [PubMed] [Google Scholar]
- Pompon, D., Louerat, B., Bronine, A., and Urban, P. (1996). Yeast expression of animal and plant P450s in optimized redox environments. Methods Enzymol. 272, 51–64. [DOI] [PubMed] [Google Scholar]
- Pyysalo, T., Honkanen, E., and Hirvi, T. (1979). Volatiles of wild strawberries, Fragaria vesca L., compared to those of cultivated berries, Fragaria X ananassa cv. Senga sengana. J. Agric. Food Chem. 27, 19–22. [Google Scholar]
- Rodriguez-Concepcion, M., and Boronat, A. (2002). Elucidation of the methylerythritol phosphate pathway for isoprenoid biosynthesis in bacteria and plastids. A metabolic milestone achieved through genomics. Plant Physiol. 130, 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schuler, M.A. (1996). Plant cytochrome P450 monooxygenases. Crit. Rev. Plant Sci. 15, 235–284. [Google Scholar]
- Schwab, W. (2003). Metabolome diversity: Too few genes, too many metabolites? Phytochemistry 62, 837–849. [DOI] [PubMed] [Google Scholar]
- Sharon-Asa, L., Shalit, M., Frydman, A., Bar, E., Holland, D., Or, E., Lavi, U., Lewinsohn, E., and Eyal, Y. (2003). Citrus fruit flavor and aroma biosynthesis: Isolation, functional characterization, and developmental regulation of Cstps1, a key gene in the production of the sesquiterpene aroma compound valencene. Plant J. 36, 664–674. [DOI] [PubMed] [Google Scholar]
- Small, I., Wintz, H., Akashi, K., and Mireau, H. (1998). Two birds with one stone: Genes that encode products targeted to two or more compartments. Plant Mol. Biol. 38, 265–277. [PubMed] [Google Scholar]
- Sommer, S., Severin, K., Camara, B., and Heide, L. (1995). Intracellular localization of geranylpyrophosphate synthase from cell cultures of Lithospermum erythrorhizon. Phytochemistry 38, 623–627. [Google Scholar]
- Speirs, J., Lee, E., Holt, K., Kim, Y., Scott, N.S., Loveys, B., Schuch, W., and Kim, Y.D. (1998). Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols. Plant Physiol. 117, 1047–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele, C.L., Katoh, S., Bohlmann, J., and Croteau, R. (1998). Regulation of oleoresins in grand fir (Abies grandis). Plant Physiol. 116, 1497–1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki, H., Koike, Y., Murakoshi, I., and Saito, K. (1996). Subcellular localization of acyltransferases for quinolizidine alkaloid biosynthesis in Lupinus. Phytochemistry 42, 1557–1562. [Google Scholar]
- Tadmor, Y., Fridman, E., Gur, A., Larkov, O., Lastochkin, E., Ravid, U., Zamir, D., and Lewinsohn, E. (2002). Identification of malodorous, a wild species allele affecting tomato aroma that was selected against during domestication. J. Agric. Food Chem. 50, 2005–2009. [DOI] [PubMed] [Google Scholar]
- Trapp, S.C., and Croteau, R.B. (2001). Genomic organization of plant terpene synthases and molecular evolutionary implications. Genetics 158, 811–832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, G.W., Berry, A.M., and Gifford, E.M. (1998). Schizogenous secretory cavities of Citrus limon (L.) Burm. f. and a reevaluation of the lysigenous gland concept. Int. J. Plant Sci. 159, 75–88. [Google Scholar]
- Urban, P., Mignotte, C., Kazmaier, M., Delorme, F., and Pompon, D. (1997). Cloning, yeast expression, and characterization of the coupling of two distantly related Arabidopsis thaliana NADH-cytochrome P450 reductases with P450 CYP73A5. J. Biol. Chem. 272, 19176–19186. [DOI] [PubMed] [Google Scholar]
- Voelker, T.A., Moreno, J., and Chrispeels, M.J. (1990). Expression analysis of a pseudogene in transgenic tobacco: A frameshift mutation prevents mRNA accumulation. Plant Cell 2, 255–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wein, M., Lavid, N., Lunkenbein, S., Lewinsohn, E., Schwab, W., and Kaldenhoff, R. (2002). Isolation, cloning and expression of a multifunctional O-methyltransferase capable of forming 2,5-dimethyl-4-methoxy-3(2H)-furanone, one of the key aroma compounds in strawberry fruits. Plant J. 31, 755–765. [DOI] [PubMed] [Google Scholar]
- Weiss, E.A. (1997). Essential Oil Crops. (Wallingford, UK: CABI Publishing).
- Williams, D.C., McGarvey, D.J., Katahira, E.J., and Croteau, R. (1998). Truncation of limonene synthase preprotein provides a fully active ‘pseudomature’ form of this monoterpene cyclase and reveals the function of the amino-terminal arginine pair. Biochemistry 37, 12213–12220. [DOI] [PubMed] [Google Scholar]
- Wink, M., and Roberts, M.F. (1998). Compartmentation of alkaloid synthesis, transport, and storage. In Alkaloids: Biochemistry, Ecology and Medicinal Applications, M.F. Roberts and M. Wink, eds (New York: Plenum Press), pp. 239–262.
- Wintoch, H. (1993). Studies on Glycosidically Bound Aroma Compounds from Plant Tissues. PhD dissertation (Wuerzburg, Germany: University of Wuerzburg).
- Wintoch, H., Krammer, G., and Schreier, P. (1991). Glycosidically bound aroma compounds from two strawberry fruit species, Fragaria vesca f. semperflorens and Fragaria X ananassa, cv. Korona. Flavour Fragrance J. 6, 209–215. [Google Scholar]
- Yahyaoui, F.E.L., Wongs-Aree, C., Latche, A., Hackett, R., Grierson, D., and Pech, J.C. (2002). Molecular and biochemical characteristics of a gene encoding an alcohol acyl-transferase involved in the generation of aroma volatile esters during melon ripening. Eur. J. Biochem. 269, 2359–2366. [DOI] [PubMed] [Google Scholar]
- Zabetakis, I., and Holden, M.A. (1997). Strawberry flavor: Analysis and biosynthesis. J. Sci. Food Agric. 74, 421–434. [Google Scholar]