Abstract
Transcript patterns elicited in response to attack reveal, at the molecular level, how plants respond to aggressors. These patterns are fashioned both by inflicted physical damage as well as by biological components displayed or released by the attacker. Different types of attacking organisms might therefore be expected to elicit different transcription programs in the host. Using a large-scale DNA microarray, we characterized gene expression in damaged as well as in distal Arabidopsis thaliana leaves in response to the specialist insect, Pieris rapae. More than 100 insect-responsive genes potentially involved in defense were identified, including genes involved in pathogenesis, indole glucosinolate metabolism, detoxification and cell survival, and signal transduction. Of these 114 genes, 111 were induced in Pieris feeding, and only three were repressed. Expression patterns in distal leaves were markedly similar to those of local leaves. Analysis of wild-type and jasmonate mutant plants, coupled with jasmonate treatment, showed that between 67 and 84% of Pieris-regulated gene expression was controlled, totally or in part, by the jasmonate pathway. This was correlated with increased larval performance on the coronatine insensitive1 glabrous1 (coi1-1 gl1) mutant. Independent mutations in COI1 and GL1 led to a faster larval weight gain, but the gl1 mutation had relatively little effect on the expression of the insect-responsive genes examined. Finally, we compared transcript patterns in Arabidopis in response to larvae of the specialist P. rapae and to a generalist insect, Spodoptera littoralis. Surprisingly, given the complex nature of insect salivary components and reported differences between species, almost identical transcript profiles were observed. This study also provides a robustly characterized gene set for the further investigation of plant–insect interaction.
INTRODUCTION
Herbivorous insects, together with their higher plant hosts, make up a massive proportion of the earth's biodiversity and biomass, and herbivory is responsible for the entry of most of the carbon into the second trophic level of the biosphere. During evolution, many plant and insect groups radiated simultaneously, and plant–insect interactions have been a powerful driving force in coevolution (Ehrlich and Raven, 1964; Mauricio and Rausher, 1997; Berenbaum, 2001; Gaunt and Miles, 2002; Nosil et al., 2002; Becerra, 2003). Given the constant pressure from herbivory, it is to be expected that counterresponses are both highly evolved and exquisitely regulated. This is clearly the case. First lines of defense are preformed chemical and physical barriers. These are important in nature, reducing access to or availability of plant resources to attackers (Hartmann and Ober, 2000). However, if these barriers are breached, active, inducible defenses are of central importance in reducing herbivory (Agrawal, 1998). Inducible defenses involve a broad range of proteins and other molecules whose synthesis is spatially and temporally controlled (Karban and Baldwin, 1997; Walling, 2000). Herbivores trigger at least two types of inducible defense responses: direct defenses that result in the inhibition of their growth and/or development and indirect defenses including the release of bouquets of plant volatiles that will be detrimental by attracting the herbivore's parasitoids and predators (Paré and Tumlinson, 1999; Walling, 2000). These defense mechanisms appear to be tightly regulated, permitting economy in times of peace and presenting a moving defense horizon to an attacker. It is therefore a priority to identify signal pathways regulating inducible defense responses and to quantitate their impact on gene expression.
Many herbivores are specialized to feed on a single plant species or family, whereas a minority are polyphagous (Bernays, 1998; Schoonhoven et al., 1998). Induced plant responses to insects are characterized by some level of specificity (Karban and Baldwin, 1997). For example, some induced defenses in wild radish (Raphanus sativus) protect against generalist, but not specialist, herbivores (Agrawal, 1999). Ethylene signaling was shown to affect resistance to a generalist herbivore but not to a specialist herbivore in Arabidopsis thaliana (Stotz et al., 2000). It is, however, not clear whether these responses arise from differences in plant metabolism or transcriptional activity or whether they reflect differences in insect physiology or susceptibility.
The recent development of genomic transcript profiling methods has allowed significant progress in the study of plant defense responses (Reymond, 2001; Wan et al., 2002), and genes differentially expressed during plant–insect interactions are being characterized (Korth, 2003). For example, the molecular responses of Nicotiana attenuata to folivory by the specialist herbivore Manduca sexta were studied by differential display of one-twentieth of the transcriptome and uncovered 16 upregulated and 9 downregulated cDNAs (Hermsmeier et al., 2001). An extension of this analysis reported 73 differentially regulated transcripts, of which several clones coding for proteins involved in photosynthesis and growth were reported to be downregulated (Hui et al., 2003). In a previous microarray study, we monitored the expression of ∼150 defense-related genes in Arabidopsis plants mechanically wounded or challenged with caterpillars of the crucifer specialist Pieris rapae (Reymond et al., 2000). The comparison of expression profiles revealed a difference between insect-attacked or wounded plants, particularly in the expression of dehydration-inducible genes. The use of a similar microarray showed that aphids feeding on Arabidopsis leaves induce genes involved in oxidative stress, calcium-dependent signaling, and pathogenesis-related responses (Moran et al., 2002). However, a global and comparative view of transcript changes during interaction with a specialist or a generalist herbivore is lacking.
Underlying the control of many inducible defenses to insects is a complex and crucially important regulatory network: the jasmonate pathway (Liechti and Farmer, 2002). Jasmonates are a family of lipid regulators derived from tri-unsaturated fatty acids. Several reports have shown that the jasmonate pathway is crucial for a protection against insect attack in both the laboratory and the natural environment (Orozco-Cardenas et al., 1993; Howe et al., 1996; McConn et al., 1997; Baldwin, 1998; Stintzi et al., 2001). Together, the jasmonate family of regulators exerts powerful control of the activation of downstream defense genes (Reymond et al., 2000; Schenk et al., 2000), participates in the developmental control of physical defense production (Heil et al., 2001), interacts with pathogen-activated defense signal pathways (Feys and Parker, 2000), and controls the production of volatiles that participate in indirect defense processes (Thaler et al., 2002; Van Poecke and Dicke, 2002; Schmelz et al., 2003). Other recent reports on induced defense against chewing insects have illustrated the importance of the jasmonate, salicylate, and ethylene pathways during interaction between Arabidopsis and the generalist Egyptian cotton worm, Spodoptera littoralis (Stotz et al., 2000, 2002). The octadecanoid and the salicylic acid pathways were also shown to be involved in indirect parasitoid attraction by Arabidopsis (Van Poecke and Dicke, 2002).
Which inputs lead to activation of the jasmonate pathway during herbivore feeding? Two components are proposed to activate this and other defense-related signal pathways: mechanical tissue damage and pattern recognition (Farmer, 2000). The pattern recognition component of some chewing insects often involves production of fatty acylated amino acids (Alborn et al., 1997; Halitschke et al., 2001) and/or enzymes (Mattiacci et al., 1995; Musser et al., 2002) that can powerfully influence host gene expression via the jasmonate pathway. Salivary components are highly complex and, when applied to mechanically wounded tissues, can have powerful effects on gene expression, by either inducing or repressing the level of some transcripts (Frey et al., 2000; Halitschke et al., 2001, 2003; Schittko et al., 2001). Moreover, differences in the biochemical composition of insect saliva between species has been reported (Alborn et al., 2003), potentially leading to differential host responses to particular herbivore species (Korth and Dixon, 1997; De Moraes et al., 1998; Dicke, 1999).
The first goal of this study was to identify target genes regulated by herbivory and to assess the importance of key signal pathways in their regulation. Genes regulated in response to larvae of the pierid butterfly P. rapae were identified using a cDNA microarray representing approximately one-quarter to one-third of the Arabidopsis genome. The importance of the jasmonate signal pathway in the control of gene expression during Pieris attack was investigated by determining the proportion of the genes regulated by this pathway in wild-type and mutant plants defective in jasmonate signaling. The jasmonate pathway proved to be quantitatively important in the global regulation of transcript induction by the specialist herbivore P. rapae and was found to impact larval development. Oxylipin signature analysis of levels of jasmonates in response to P. rapae feeding correlated well with gene expression data. We then tested whether a second herbivore of contrasting dietary strategy would induce different sets of genes. We employed larvae of the generalist noctuid moth S. littoralis. Results from experiments comparing transcriptional responses to Pieris and Spodoptera were surprising because host responses to both insects were highly similar.
RESULTS
Statistical Analysis of Genes Regulated in Response to P. rapae
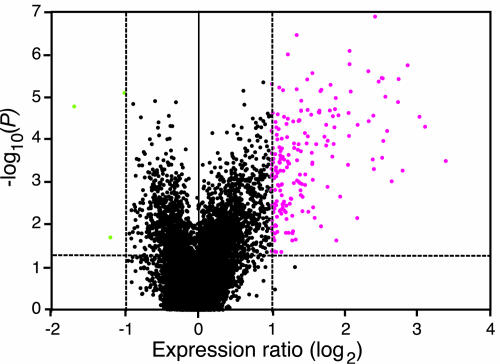
In this study, we used a microarray containing 12,135 Arabidopsis ESTs representing ∼7200 unique genes. Differentially expressed genes were selected using two criteria: expression ratio ≥2-fold or ≥-2-fold and Student's t test P value < 0.05 (see Methods). The choice of the twofold cutoff was determined from six independent control experiments showing that the addition of biological and technical variability produce a range of expression ratios between −1.5 and 1.5 (see Supplemental Figure 1 online). With these stringent criteria, we thus may have underestimated the extent of the upregulation or downregulation of genes. Figure 1 shows an example of the statistical analysis of data obtained from plants challenged with P. rapae. ESTs (195) representing 111 unique genes were induced above twofold (P < 0.05), whereas the levels of only three ESTs were repressed. Because of redundancy in the EST collection, the majority of the insect-induced genes were represented by more than one EST on the microarray (see Supplemental Table 1 online), and replicates displayed a similar expression pattern. If we had only used the commonly accepted threshold of twofold induction, only two ESTs with a P value > 0.05 would have been selected (Figure 1). On the other hand, many genes have a high probability of induction (P < 0.001), although their expression fold change is between 1.5 and 2. These latter genes are potentially interesting candidates that would require careful testing to determine whether changes in their expression have biological relevance. A minute change in transcript abundance of a transcription factor could lead to a substantial biological effect, whereas the same change for a storage protein might have less impact. For this study, using two selection criteria (fold change and P value) allowed us to identify strongly upregulated or downregulated genes with a high level of confidence. To estimate the false discovery rate (FDR), we calculated a q value for all significant genes (Storey and Tibshirani, 2003) and found that, for plants challenged with P. rapae, the FDR was 10.3% at P = 0.05, diminishing to 1.5% at P = 0.001 (see Supplemental Figure 2 and Table 1 online).
Figure 1.
Statistical Analysis of Arabidopsis Gene Induction by P. rapae.
Volcano plot where gene expression ratios (log2 fold change) are plotted against the negative log10-transformed P values from a t test calculation. Arabidopsis plants were challenged with P. rapae larvae for 3 to 5 h, and gene expression ratios between challenged and unchallenged plants were calculated from five independent replicate experiments. In parallel, expression ratios were obtained from six independent experiments of control versus control plants. Mean log2-transformed expression ratios for both types of comparisons were used for a t test conducted for each gene. The vertical dashed lines represent a twofold threshold in gene induction or repression. The horizontal dashed line represents a significance level of 0.05. Genes with statistically significant differential expression (P < 0.05) and fold change >2 and >−2 are shown in magenta and green, respectively.
Initial experiments concentrated on Arabidopsis leaves damaged by Pieris larvae. According to our criteria, we found 111 upregulated genes after 3 to 5 h of feeding, whereas only three genes were repressed (see Supplemental Table 1 online). Pieris-induced genes were classified based on their potential cellular function into 10 functional groups, and selected genes are shown in Table 1. Viewed broadly, they include genes involved in defense, indole glucosinolate metabolism, phenolic pathway, oxylipin synthesis, auxin and ethylene synthesis, detoxification processes, abiotic stress, reallocation of resources, signal transduction, and transcription factors. Moreover, 16 genes encoding proteins of unknown function are differentially regulated (see Supplemental Table 1 online).
Table 1.
Microarray Data for Selected Genes Induced by P. rapae
| Probe Identification and Putative Function | AGI Code | Wild Type | P Value | coi1-1 gl1 | P Value |
|---|---|---|---|---|---|
| Defense protein | |||||
| Lectin | At3g16400 | 9.14 ± 1.65 | <0.001 | 1.15 ± 0.15 | 0.615 |
| Cys proteinase | At4g11320 | 4.35 ± 0.72 | <0.001 | 1.07 ± 0.10 | 0.780 |
| β-1,3-glucanase (PR2) | At3g57260 | 3.96 ± 1.32 | 0.011 | 1.41 ± 0.23 | 0.093 |
| β-Glucosidase | At1g52400 | 5.78 ± 1.65 | 0.007 | 1.50 ± 0.58 | 0.581 |
| Hypersensitive response-induced protein | At5g62740 | 2.07 ± 0.17 | <0.001 | 1.05 ± 0.08 | 0.532 |
| Indole glucosinolate metabolism | |||||
| Anthranilate synthase | At5g05730 | 2.62 ± 0.20 | <0.001 | 1.08 ± 0.07 | 0.113 |
| Trp synthase α subunit | At3g54640 | 2.11 ± 0.07 | <0.001 | 1.10 ± 0.12 | 0.477 |
| Trp synthase β subunit | At5g54810 | 2.17 ± 0.14 | <0.001 | 1.19 ± 0.08 | 0.009 |
| Cytochrome P450 (CYP79B2) | At4g39950 | 3.37 ± 0.40 | <0.001 | 1.07 ± 0.13 | 0.285 |
| Cytochrome P450 (CYP83B1) | At4g31500 | 3.71 ± 0.39 | <0.001 | 1.55 ± 0.26 | 0.210 |
| Myrosinase-associated protein, putative | At1g54010 | 2.36 ± 0.25 | <0.001 | 1.05 ± 0.05 | 0.644 |
| Myrosinase binding protein, putative | At3g16420 | 2.66 ± 0.20 | <0.001 | 0.99 ± 0.08 | 0.411 |
| Phenolic metabolism | |||||
| Chorismate mutase | At5g22630 | 2.23 ± 0.28 | <0.001 | 1.37 ± 0.20 | 0.095 |
| Prephenate dehydratase | At3g44720 | 2.02 ± 0.28 | 0.003 | 1.65 ± 0.39 | 0.069 |
| Phe ammonia lyase (PAL1) | At2g37040 | 2.92 ± 0.33 | 0.002 | 1.41 ± 0.17 | 0.772 |
| Cinnamoyl CoA reductase | At1g15950 | 2.06 ± 0.16 | 0.005 | 1.09 ± 0.06 | 0.371 |
| Caffeic acid O-methyltransferase | At5g54160 | 2.83 ± 0.50 | 0.005 | 1.21 ± 0.09 | 0.295 |
| Flavonol 4′-sulfotransferase | At1g74100 | 2.35 ± 0.14 | <0.001 | 1.24 ± 0.03 | 0.011 |
| Tyr aminotransferase, putative | At2g24850 | 4.58 ± 1.10 | <0.001 | 1.19 ± 0.12 | 0.290 |
| Oxylipin metabolism | |||||
| Lipoxygenase (LOX2) | At3g45140 | 3.21 ± 0.62 | <0.001 | 1.01 ± 0.04 | 0.560 |
| Lipoxygenase (LOX3) | At1g17420 | 5.67 ± 1.14 | <0.001 | 1.25 ± 0.18 | 0.939 |
| Allene oxide synthase (AOS) | At5g42650 | 5.99 ± 0.93 | <0.001 | 1.05 ± 0.06 | 0.334 |
| 12-Oxophytodieonate reductase (OPR3) | At2g06050 | 4.31 ± 0.56 | <0.001 | 1.23 ± 0.09 | 0.482 |
| Hydroxyjasmonate sulfotransferase | At5g07010 | 3.81 ± 0.44 | 0.023 | 0.91 ± 0.11 | 0.446 |
| JA amino-synthetase (JAR1) | At2g46370 | 2.05 ± 0.18 | <0.001 | 1.08 ± 0.08 | 0.599 |
| Hydroperoxide lyase (HPL) | At4g15440 | 2.30 ± 0.21 | 0.004 | 1.01 ± 0.03 | 0.443 |
| Metabolite/hormone biosynthesis | |||||
| Nitrilase (NIT3) | At3g44320 | 2.15 ± 0.19 | <0.001 | 1.01 ± 0.06 | 0.932 |
| IAA-Ala hydrolase (IAR3) | At1g51760 | 6.97 ± 0.98 | <0.001 | 1.13 ± 0.05 | 0.283 |
| S-Adenosylmethionine synthase | At4g01850 | 2.25 ± 0.11 | <0.001 | 1.26 ± 0.11 | 0.341 |
| Detoxification, redox processes | |||||
| Glutathione S-transferase (GST1) | At1g02930 | 4.23 ± 0.92 | 0.002 | 3.91 ± 1.39 | 0.016 |
| Glutathione S-transferase (GST5) | At2g29450 | 5.70 ± 1.18 | <0.001 | 1.13 ± 0.08 | 0.345 |
| GSH-dependent dehydroascorbate reductase | At1g19570 | 6.01 ± 0.92 | <0.001 | 1.20 ± 0.24 | 0.369 |
| Thioredoxin | At1g45145 | 2.17 ± 0.33 | 0.004 | 1.23 ± 0.09 | 0.234 |
| Cytochrome b5 | At2g46650 | 2.25 ± 0.37 | 0.001 | 0.86 ± 0.12 | 0.133 |
| Abiotic stress | |||||
| Aquaporin | At2g37180 | 2.81 ± 0.57 | 0.001 | 1.31 ± 0.08 | 0.147 |
| Tonoplast integral potein | At3g16240 | 2.83 ± 0.23 | <0.001 | 1.20 ± 0.17 | 0.425 |
| Dehydrin (ERD10) | At1g20450 | 2.97 ± 0.66 | <0.001 | 3.27 ± 0.50 | <0.001 |
| Heat-shock cognate protein (HSC70-3) | At3g09440 | 2.33 ± 0.29 | 0.003 | 1.98 ± 0.14 | 0.006 |
| Reallocation of resources | |||||
| Vegetative storage protein (VSP2) | At5g24770 | 6.36 ± 2.12 | <0.001 | 0.94 ± 0.05 | 0.639 |
| Hexose transporter | At5g26340 | 2.32 ± 0.42 | 0.005 | 1.64 ± 0.25 | 0.085 |
| Galactinol synthase | At2g47180 | 2.81 ± 0.29 | <0.001 | 0.94 ± 0.09 | 0.605 |
| β-Fructosidase | At1g12240 | 2.46 ± 0.44 | <0.001 | 0.93 ± 0.07 | 0.689 |
| Signal transduction | |||||
| Calmodulin-related (TCH3) | At2g41100 | 2.26 ± 0.51 | 0.014 | 2.68 ± 0.62 | 0.004 |
| Transducin (WD40 repeat protein) | At1g04140 | 2.05 ± 0.24 | 0.001 | 1.16 ± 0.15 | 0.389 |
| PIP kinase-like protein | At1g01470 | 2.67 ± 0.44 | <0.001 | 2.74 ± 0.40 | <0.001 |
| Transcription factors | |||||
| bHLH protein (AtMYC2) | At1g32640 | 3.59 ± 0.40 | <0.001 | 1.54 ± 0.14 | 0.009 |
| NAC-domain transciption factor (NAM-like) | At1g52890 | 3.16 ± 0.61 | <0.001 | 1.18 ± 0.07 | 0.162 |
| Zinc-finger-like protein | At3g52800 | 2.58 ± 0.47 | 0.002 | 2.41 ± 0.66 | 0.022 |
| MYB-related protein | At5g67300 | 2.84 ± 0.48 | 0.001 | 1.70 ± 0.16 | 0.015 |
| Transcription factor II homolog | At4g31720 | 2.42 ± 0.25 | <0.001 | 1.04 ± 0.01 | 0.715 |
Relative changes in gene expression after challenge with P. rapae were measured in wild-type and coi1-1 gl1 mutant plants. Mean expression ratios (± se) are calculated from five (wild type) and four (coi1-1 gl1) biologically independent experiments. The P values denote the significant difference of the mean log-transformed ratios of challenged over unchallenged plants. Only representatives of functional classes are shown. The complete list of induced genes is given in Supplemental Table 1 online. bHLH, basic helix-loop-helix; PIP, phosphatidyl inositol phosphate.
Global Importance of the Jasmonate Pathway
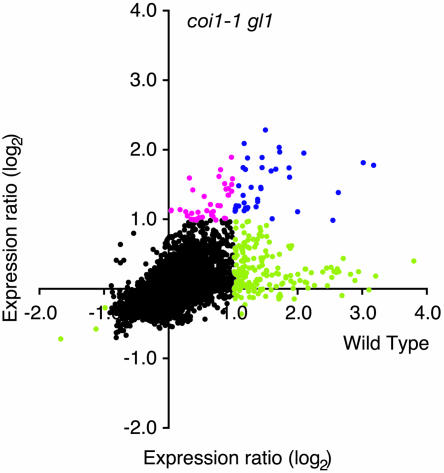
Many studies have provided information on the role of the jasmonate pathway in resistance to insects (Howe et al., 1996; McConn et al., 1997; Stintzi et al., 2001; Thaler et al., 2002; Halitschke and Baldwin, 2003). To assess the importance of the jasmonate signal pathway in transcriptional reprogramming during the response to insect herbivores, we employed the coronatine insensitive1 glabrous1 (coi1-1 gl1) mutant, which is defective in jasmonate perception (Feys et al., 1994). P. rapae caterpillars were allowed to feed on wild-type and coi1-1 gl1 mutant leaves, and gene expression profiles were compared after 3 to 5 h. Figure 2 shows that a large group of ESTs are only induced in wild-type plants, whereas some others are induced in both genotypes. Interestingly, some ESTs were predominantly or only induced in coi1-1 gl1 plants. We found that 74 (67%) of the 111 Pieris-inducible genes had an expression ratio below 1.5-fold in the mutant plant, indicating a strong COI1 dependence (Table 1; see Supplemental Table 1 online). Conversely, there were 16 transcripts that were significantly induced in coi1-1 gl1 plants (ratio > 2, P < 0.05). These showed very similar expression ratios and P values in wild-type and mutant plants, although the estimated FDR was noticeably higher in coi1-1 gl1 (FDR 47.7%, P = 0.05; see Supplemental Table 1 online). For 19 genes (17%), the induction level varied between 1.5-fold and twofold in coi1-1 gl1, showing potentially a partial need for a functional COI1 protein. Among the three repressed genes, two clones showed COI1 dependence, and one had an expression ratio of 0.61 in the mutant, showing again a partial effect of the mutation. Thus, the regulation of a large proportion (67 to 84%) of the Pieris-activated genes seems to involve, totally or partially, the jasmonate pathway. Because coi1-1 gl1 plants are in a genetic background containing a mutation in the GL1 gene (Feys et al., 1994), we wanted to verify that this mutation was not responsible for the observed differential gene expression. Using a dedicated small-scale microarray, we found that Pieris larvae reproducibly induced a very similar set of genes in wild-type and in gl1 plants. However, for 10% of the genes, the expression ratio was bigger than 2 in the wild type and smaller than 1.75 in gl1 (see Supplemental Table 5 online).
Figure 2.
Contribution of the Jasmonate Pathway to Insect-Inducible Gene Expression.
Relative changes in gene expression after challenge with P. rapae were measured in wild-type and coi1-1 gl1 mutant plants. Expression ratios calculated from experiments comparing challenged and unchallenged wild-type plants (five biologically independent replicates) are plotted against expression ratios between challenged and unchallenged coi1-1 gl1 plants (four biologically independent replicates). Black dots represent genes that showed no changes in gene expression (see Methods). Blue dots represent genes that were induced in both genotypes. Magenta dots represent genes only induced in coi1-1 gl1 plants, and green dots represent genes only induced in wild-type plants.
In a complementary experiment, we treated wild-type plants with methyl jasmonate (MeJA) and found 52 induced transcripts (see Supplemental Table 1 online; ratio > 2, FDR 33.6%, P = 0.05). As expected, the majority of the COI1-dependent genes were significantly upregulated by MeJA (39/74). However, some COI1-dependent genes (15/74) were only weakly induced by the treatment (1.5-fold to twofold), whereas the expression ratios of 18 genes were below 1.5. This indicates that the regulation of some transcripts is under the control of COI1 but that these genes are partially or not induced by a treatment with MeJA alone. The jasmonate-inducible genes also included a few transcripts that were not induced by P. rapae (see Supplemental Table 1 online).
Effect of the Jasmonate Pathway on Larval Performance
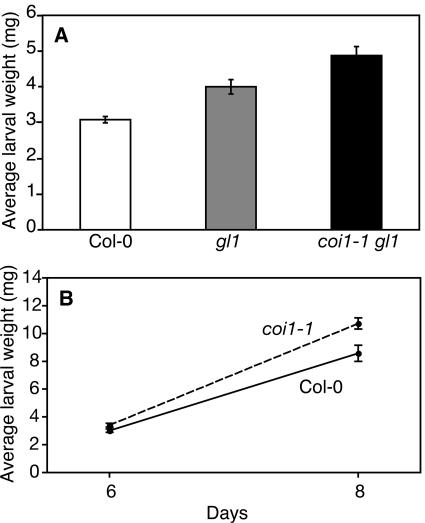
Because several genes induced by Pieris were not induced in the jasmonate-insensitive mutant coi1-1 gl1, we tested whether these genes contribute to plant resistance against this herbivore. We developed a bioassay where freshly hatched P. rapae larvae were placed on plants and their weight measured after 6 or 8 d of feeding. This assay proved to be more reliable and quantitative than estimating leaf area consumption. We verified that first instar larvae induced the same set of genes as older larvae used for the other experiments (data not shown). Larvae feeding on coi1-1 gl1 plants were significantly heavier than those feeding on wild-type plants (Student's t test, P < 0.001) (Figure 3A). We included the gl1 mutant as a control in the assay and observed that larvae feeding on coi1-1 gl1 were also significantly heavier than larvae feeding on gl1 plants (P = 0.009). Interestingly, we observed that Pieris larvae grew significantly more on gl1 plants than on Columbia-0 (Col-0) (P < 0.001). More detailed experiments were undertaken to assess the impact of the coi1-1 mutation (in the absence of gl1) on larval feeding. The results show clearly that insects were significantly heavier after feeding on coi1-1 than on wild-type plants (P = 0.038 at 6 d, P = 0.005 at 8 d; Figure 3B). Thus, we demonstrate here the functional importance of the jasmonate pathway for resistance to Pieris rapae.
Figure 3.
Influence of the Jasmonate Pathway on P. rapae Larval Performance.
(A) The growth of P. rapae larvae was tested on wild-type, gl1, and coi1-1 gl1 mutant plants. Freshly hatched Pieris larvae were placed simultaneously on each Arabidopsis genotype, and larval weight (mean ± se) was measured after 6 d of feeding.
(B) Pieris larvae were placed on wild-type plants and on the non-glabrous coi1-1 mutant, and larval weight (mean ± se) was measured after 6 and 8 d of feeding.
Levels of Jasmonate Family Members Increase after Insect Feeding
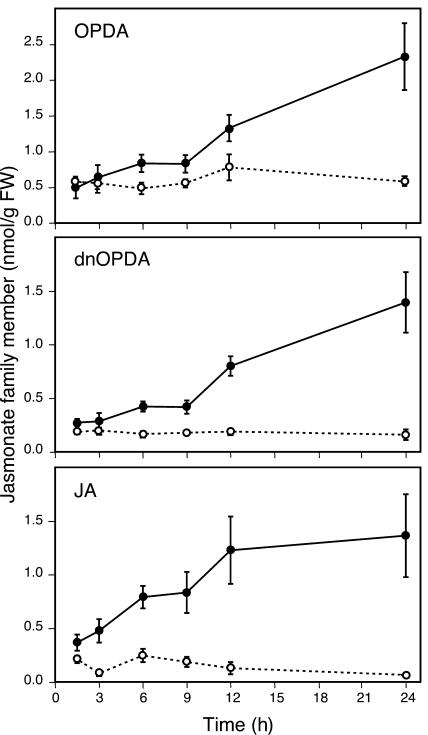
Having observed that several enzymes involved in the biosynthesis of JA were upregulated by Pieris and that the jasmonate pathway controlled a large proportion of the insect-induced genes through COI1, we decided to quantify jasmonate family members in plants challenged with P. rapae during a 24-h feeding period. In addition to JA, we chose to monitor the jasmonate precursor 12-oxo-phytodienoic acid (OPDA) because it has been shown to play a critical role in resistance of Arabidopsis against an insect herbivore (Stintzi et al., 2001). We also measured dinor oxo-phytodienoic acid (dnOPDA), a 16-carbon structural homolog of OPDA, which accumulates after wounding (Stintzi et al., 2001). As shown in Figure 4, levels of OPDA, dnOPDA, and JA steadily increased during feeding by Pieris larvae over the 24-h time course. OPDA reached a level of 2.5 nmol/g of fresh weight after 24 h, whereas JA and dnOPDA levels were just below 1.5 nmol/g of fresh weight. In undamaged control plants, levels of the three oxylipins stayed constant over the 24-h period and were <0.8 nmol/g of fresh weight for OPDA and <0.2 nmol/g of fresh weight for JA or dnOPDA.
Figure 4.
Accumulation of Jasmonate Family Members after Challenge with P. rapae.
Oxylipins were extracted from leaves at different times after challenge with first-instar P. rapae larvae. Kinetics of OPDA, dnOPDA, and JA accumulation were followed in control (open symbols and dashed lines) and infested (closed symbols and solid lines) plants. Data are the mean ± se of four independent determinations.
Contribution of Insect-Derived Cues to Transcript Regulation
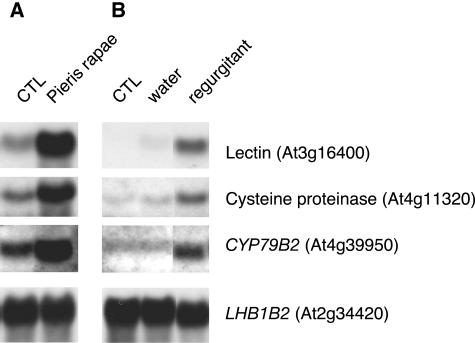
Several studies have shown that responses to insect feeding are greater or different to responses to mechanical damage alone. This is the case for induced volatiles (Paré and Tumlinson, 1997), jasmonate synthesis (McCloud and Baldwin, 1997), or for the accumulation of wound-induced transcripts (Korth and Dixon, 1997). We compared the gene expression response of leaves eaten by P. rapae with that of leaves that had been wounded using a cork borer. As expected, the two treatments resulted in different transcript profiles. We found 36 genes induced by wounding (ratio > 2, FDR 24.2%, P = 0.05; see Supplemental Table 1 online). These data indicated many genes that were Pieris inducible but not activated, or activated very weakly, by a puncture wound. To investigate the role of insect-specific factors on gene expression, we selected three of these genes. Regurgitant of Pieris caterpillars that had been feeding on wild-type Arabidopsis plants was collected and applied to leaf punctures. After 6 h, we measured the transcript levels of Pieris-inducible genes by gel blot analysis. As shown in Figure 5, application of insect regurgitant enhanced the transcript level of a putative lectin (At3g16400), a Cys proteinase (At4g11320), and a cytochrome P450 (CYP79B2; At4g39950) gene. Application of water alone had no or little effect, indicating that the wound caused by making the punctures was not sufficient to powerfully induce the insect-specific transcripts. Furthermore, experiments applying frequent physical wounds meant to mimic the speed and duration of insect feeding increased the expression of these genes but not to the level caused by insect feeding or regurgitant application (data not shown).
Figure 5.
Insect Regurgitant Stimulates the Expression of Pieris-Inducible Genes.
The expression of three defense-related genes was analyzed in plants that were either challenged with P. rapae larvae for 3 to 5 h (A) or treated with oral regurgitant for 6 h (B). For regurgitant treatment, three 1-mm holes per leaf were made and 1 μL of regurgitant obtained from fourth- or fifth-instar larvae was added to each hole. A control with water alone showed very little induction. LHB1B2 was used as a loading control.
Because jasmonate family members accumulate during Pieris feeding (Figure 4) and are likely to be ingested by caterpillars, we speculated that these molecules could be found in insect regurgitant and that they could be responsible for the enhanced transcript level observed after regurgitant application. To test this hypothesis, we fed Pieris larvae with allene oxide synthase (aos; Park et al., 2002) or fatty acid desaturase (fad3-2 fad7-2 fad8; McConn et al., 1997) mutant plants that do not accumulate JA, OPDA, or dnOPDA. When regurgitant collected from mutant plants was applied to wild-type plants, the same induction of insect-specific genes was observed (data not shown), indicating that the elicitor activity of the regurgitant was not attributable to a member of the jasmonate family.
Overlapping Gene Expression Patterns Local and Distal to the Feeding Site
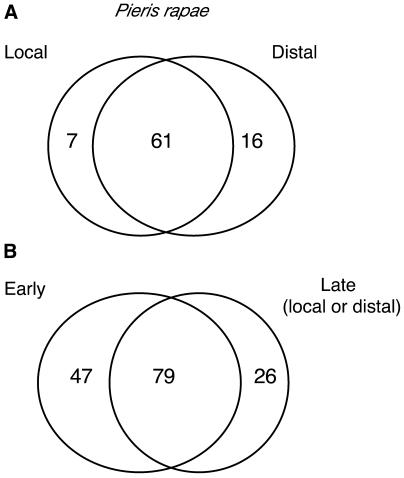
During attack it is important for the plant to mount defenses in undamaged distal leaves. Assessing gene expression 24 h after a 3- to 5-h period of Pieris attack revealed 66 genes that are upregulated in distal leaves (ratio > 2, FDR 67%, P = 0.05). Among the 66 induced transcripts, those for 54 genes had an expression ratio > 1.5 in the insect-damaged leaves, indicating a strong overlap between local and systemic responses to herbivory at this time point (Figure 6; see Supplemental Table 2 online). A few genes (12) are classed as being expressed in the distal undamaged leaves and not in the insect damaged leaves. Inspection of the data reveals that several of these genes encode calcium response–related proteins or glutathione S-transferases (see Supplemental Table 2 online). Furthermore, when we compared the genes activated after the initial 3- to 5-h feeding phase to those activated at the 24-h time point, we found that 79 genes (52% of the total) upregulated early were still up at a later time point, illustrating a relatively long lasting upregulation for the majority of the transcripts (Figure 6; see Supplemental Table 2 online).
Figure 6.
Comparison of Local and Distal Gene Expression after Insect Feeding.
(A) Venn diagram representing the distribution of induced transcripts between damaged (local) and undamaged (distal) leaves. P. rapae larvae were allowed to feed for 3 to 5 h then removed from plants. Plants were left in a growth cabinet under constant illumination for another 24 h until harvesting and microarray analysis. This experiment was repeated three times.
(B) Venn diagram representing the distribution of early (3 to 5 h challenge, five independent replicates) and late (3 to 5 h challenge followed by 24 h without larvae, three independent replicates) transcripts.
The numbers in the overlapping area indicate the shared number of genes in the comparisons and include genes with an average expression ratio ≥ 2 in one experiment and ≥ 1.5 in the other. Numbers outside the overlapping area represent genes specific for one experiment with an expression ratio ≥ 2 in one experiment and ≤ 1.5 in the other.
A Specialist and a Generalist Insect Trigger Similar Transcript Profiles
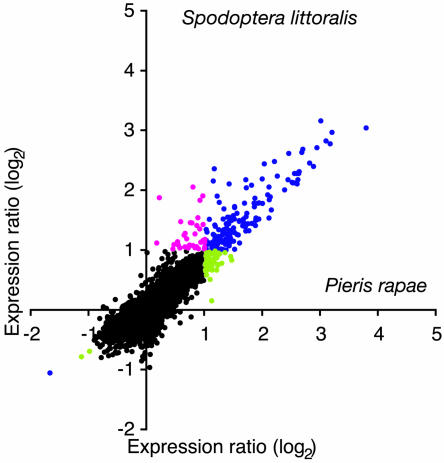
Direct comparison of gene expression levels after feeding by the specialist P. rapae or the generalist S. littoralis revealed remarkably similar patterns between the two insects after a short (3 to 5 h) feeding phase (Figure 7). Challenge with Spodoptera larvae induced 88 transcripts and repressed 1 (ratio > 2, FDR 23.8%, P = 0.05; see Supplemental Table 1 online). The vast majority of differentially regulated transcripts behaved similarly in response to both insects, and there were no clear examples of genes that were upregulated by one insect and whose expression was not altered by the second insect. We found 80 genes that were significantly upregulated by both insects and only one common gene that was repressed after 3 to 5 h. (P < 0.05; see Supplemental Table 1 online). Moreover, 36 genes were upregulated more than twofold with one insect and >1.5-fold with the second insect. Only genes homologous to ASPARAGINE SYNTHETASE (At3g47340) and the protein phosphatase gene ABSCISSIC ACID INSENSITIVE1 (At4g26080) were specifically induced by Pieris (see Supplemental Table 1 online). When we compared the expression profile of Pieris-damaged plants with that of Spodoptera-damaged plants 24 h after an initial feeding phase of 3 to 5 h, we again found a very similar pattern both in local and distal leaves (data not shown).
Figure 7.
Comparison of Transcript Profiles between a Specialist and a Generalist Herbivore.
Relative changes in gene expression were measured after 3 to 5 h challenge with the specialist P. rapae or the generalist S. littoralis. Expression ratios calculated from experiments comparing Pieris-challenged and unchallenged plants (five biologically independent replicates) are plotted against expression ratios from ratios between Spodoptera-challenged and unchallenged plants (five biologically independent replicates). Black dots represent genes that showed no changes in gene expression (see Methods). Blue dots represent genes that were induced by both insects. Magenta dots represent genes only induced by S. littoralis, and green dots represent genes only induced by P. rapae.
Cluster Analysis Reveals Distinct Responses to an Insect, to Wounding, and to MeJA
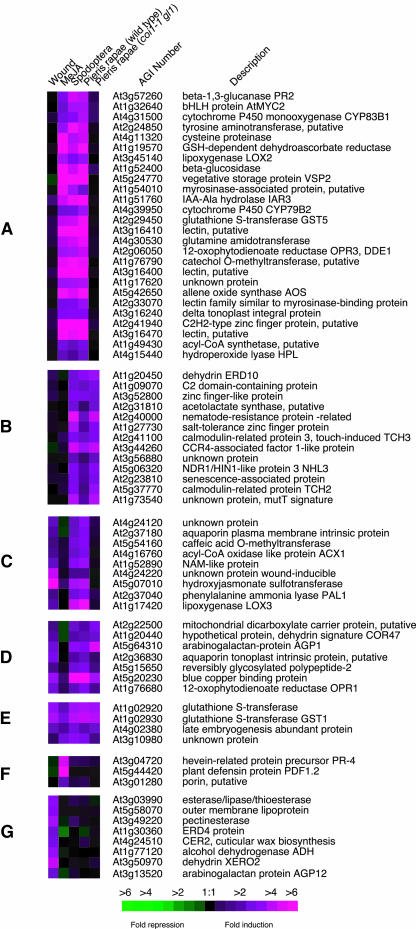
Data from different treatments (Pieris feeding, Spodoptera feeding, MeJA treatment, and mechanical wounding) were analyzed by hierarchical clustering and provided different categories of transcriptional responses. Seven clusters of genes are presented in Figure 8. Genes preferentially responding to insects, but not to the punctual wounding we inflicted, are shown in Figure 8A; they require the presence of COI1 for induction by Pieris and are induced by MeJA treatment. This cluster is enriched in genes typical of biotic stress responses, including, for example, pathogenesis-related genes and genes involved in oxylipin and indole glucosinolate metabolism. Figure 8B shows several genes that are also preferentially induced by insects and not by wounding; however, they do not depend on COI1 and are not upregulated by MeJA. This group contains elements mainly related to abiotic stress responses, such as dehydrins, touch genes, and salt- and senescence-related genes. Figure 8C is comprised of genes induced by wounding and P. rapae. Interestingly, the Pieris-induced accumulation of these transcripts requires COI1, but the genes are not induced by MeJA treatment. These genes do not represent a defined functional category, but their regulation might illustrate a new signaling pathway that is independent of JA but needs COI1. Figures 8D and 8E contain genes upregulated in all treatments except that genes in Figure 8D are not MeJA inducible. This group includes general stress-response genes such as GLUTATHIONE S-TRANSFERASE1 (At1g02930) and OXOPHYTODIENOATE REDUCTASE1 (OPR1; At1g76680). Genes in Figure 8F are mainly induced by MeJA and consist of defense-related genes, HEVEIN-LIKE (At3g04720) and PLANT DEFENSIN1.2 (PDF1.2; At5g44420), that are typical markers of MeJA treatment. Finally wound-specific genes (Figure 8G) include, for example LIPOPROTEIN (At5g58070), DEHYDRIN (XERO2, At3g50970), and PECTINESTERASE (At3g49220); Pieris did not significantly activate these genes.
Figure 8.
Clustered Display of Genes Responding to Insect Feeding, Mechanical Damage, and MeJA Treatment.
Hierarchical clusters of selected genes are shown to illustrate differential induction patterns after insect feeding, mechanical damage, and treatment with MeJA. The experiment with coi1-1 gl1 plants challenged with P. rapae is included to visualize the importance of the jasmonate pathway in response to chewing insects. The clusters in (A) to (G) are discussed in the text. Representative genes of each cluster are displayed here.
DISCUSSION
Insect-Regulated Genes
Although the majority of insect herbivores are specialists, there are also a considerable number of generalists, raising the question whether wild-type plants respond differently when attacked by insects representing these two classes. To compare transcriptome reprogramming in response to two different chewing insect species that differ in the degree of specialization, we used a cDNA microarray containing ∼12,000 ESTs and concentrated primarily on the Arabidopsis–P. rapae interaction. Based on rigorous selection criteria, five biologically independent replicates, and thorough statistical analyses, we identified 111 genes that were upregulated by feeding larvae of the specialist P. rapae on wild-type Arabidopsis plants (Table 1; see Supplemental Table 1 online). Among genes likely to be directly involved in defense against insects, we find several putative lectins (At3g16400, At3g15356, At3g16460, and At2g33070). Lectins are carbohydrate-binding proteins, many of which have insecticidal activities (Peumans and Vandamme, 1995) thought to be based on specific binding to glycoproteins in the insect gut. We also observed the accumulation of a Cys proteinase (At4g11320). This gene is homologous to a maize (Zea mays) gene whose product was shown previously to reduce caterpillar growth by disrupting the peritrophic matrix of the midgut (Pechan et al., 2002).
We identified several induced genes potentially involved in indole glucosinolate metabolism. Glucosinolates are well-characterized defense compounds that function against herbivores. Upon tissue damage, glucosinolates are hydrolyzed by specific thioglucosidases called myrosinases, and this reaction releases an array of toxic compounds, such as nitriles, isothiocyanates, epithionitriles, and thiocyanates (Rask et al., 2000). Indole glucosinolates have a core structure comprising a β-d-thioglucose group linked to a sulfonated aldoxime moiety and a Trp side chain. These compounds are thought to be part of a constitutive defense mechanism (Jander et al., 2001; Lambrix et al., 2001), but levels of specific indole glucosinolates can also increase upon elicitor, MeJA, or salicylic acid (SA) treatment (Doughty et al., 1995; Brader et al., 2001; Mikkelsen et al., 2003). Several genes involved in the biosynthesis of indole glucosinolates were shown to be transcriptionally regulated by MeJA (Brader et al., 2001; Mikkelsen et al., 2003). Recently, it was shown that Arabidopsis lyrata plants accumulate indole glucosinolates upon damage by P. rapae and that these compounds, when incorporated in Pieris diet, reduced their survival and growth (Agrawal and Kurashige, 2003). We show here that several genes likely to be involved in the myrosinase-glucosinolate system are activated in response to P. rapae feeding on A. thaliana plants (Table 1). Indeed, genes responsible for the biosynthesis of Trp (anthranilate synthase, At5g05730; Trp synthase α subunit, At3g54640; Trp synthase β subunit, At5g54810) and for the subsequent oxidation of Trp to form indole-3-acetaldoxime (cytochrome P450 [CYP79B2], At4g39950; cytochrome P450 [CYP83B1], At4g31500) were coordinately upregulated by Pieris. Two genes with homology to myrosinase-associated proteins were also upregulated (Table 1).
Phenolic secondary metabolites have been proposed to play a variety of roles in defense as phytoalexins, radical scavengers, or structural barriers (e.g., lignin, cell wall cross-linking). We observed the activation of several genes involved in the shikimic-chorismic acid pathway. Chorismate mutase (At5g22630) and prephenate dehydratase (At3g44720) are enzymes required for the synthesis of arogenate, the precursor of both Phe and Tyr. Tyr aminotransferase (At2g24850) forms 4-hydroxyphenylpyruvate, which could either lead to the synthesis of phenolic cross-linkers in the cell wall or to the production of prenylquinones with radical scavenging properties (Lopukhina et al., 2001). Upregulated members of the phenylpropanoid pathway included PHENYLALANINE AMMONIA LYASE1 (At2g37040), CINNAMOYL-CoA REDUCTASE (At1g15950), CAFFEIC ACID O-METHYL TRANSFERASE (At5g54160), or FLAVONOL 4′-SULFOTRANSFERASE (At1g74100).
Several genes participating in the biosynthesis of JA were induced by Pieris, including LIPOXYGENASE2 (LOX2, At3g45140), AOS (At5g42650), and OPR3 (At2g06050) (Table 1). Two genes implicated in the modification of JA were upregulated, the JA amino-synthetase JASMONATE RESISTANT1 (JAR1, At2g46370) and HYDROXYJASMONATE SULFOTRANSFERASE (At5g07010). These two genes could be responsible for the formation of biologically active oxylipins (Staswick et al., 2002; Gidda et al., 2003). We also found an induction for HYDROPEROXIDE LYASE (HPL, At4g15440), a key enzyme in the synthesis of small defense-related aldehydes that has been shown to affect aphid performance (Vancanneyt et al., 2001). A putative 13-LIPOXYGENASE of unknown function (LOX3, At1g17420) was also upregulated. Ethylene is a modulator in defense signal transduction (Reymond and Farmer, 1998). Genes encoding two enzymes involved in ethylene synthesis were upregulated (S-ADENOSYLMETHIONINE SYNTHASE2, At4g01850; 1-AMINOCYCLOPROPANE-1-CARBOXYLATE [ACC]-OXIDASE, At1g05010), although in the case of ACC OXIDASE, the induction was just below the cutoff value of twofold (Table 1; see Supplemental Table 1 online). Additionally, the transcript abundance of NITRILASE3 (NIT3, At3g44320), and IAA-ALANINE RESISTANT3 coding for an IAA-Ala hydrolase (IAR3, At1g51760) implicated in the production and release of auxin, were enhanced by Pieris.
Glutathione S-transferases (GSTs) are a group of stress response proteins that contribute to cellular survival after oxidative damage. Several GST genes (At1g02920, At1g02930, and At2g29450) were activated as well as those encoding other enzymes involved in the antioxidant defense systems, including monodehydroascorbate reductase (At3g09940), dehydroascorbate reductase (At1g19570), thioredoxin (At1g45145), and cytochrome b5 (At2g46650). Taken together, the induced production of these cell protectants suggests that oxidative stress is caused by the damage imposed by herbivore feeding. The transcript abundance of other general stress-responsive proteins was also increased in Pieris-challenged plants, including, for example, water stress–related genes like AQUAPORINS (At2g36830, At2g37180, and At3g16240), DEHYDRINS (ERD10, At1g20450; COR47, At1g20440), or RIBONUCLEASE1 (At2g02990) (Table 1).
Plants move and sequester resources during attack and remobilize them when conditions are more favorable (Strauss and Agrawal, 1999). We identified several candidate genes that might play a role in resource reallocation: VEGETATIVE STORAGE PROTEIN2 (VSP2, At5g24770), HEXOSE TRANSPORTER (At5g26340), GALACTINOL SYNTHASE (At2g47180), β-FRUCTOSIDASE (At1g12240), and INVERTASE (At3g13790) (Table 1; see Supplemental Table 1 online). Finally, insect feeding activated a series of genes involved in signal transduction and transcriptional regulation (Table 1), including members of different families of transcription factors, like the basic helix-loop-helix ATMYC2 (At1g32640), a NAC-domain protein (At1g52890), a zinc-finger-like protein (At3g52800), or a MYB-related protein (At5g67300).
Our results demonstrate that an overriding feature of the response of wild-type plants to Pieris feeding is the induction of transcription. Only three genes were downregulated by Pieris and, to a lesser extent, by Spodoptera. They encode two proteins of unknown function (At5g44680 and At5g11420) and a protein regulated by gibberellin (At1g74670) (see Supplemental Table 1 online). In recent analyses of the interaction between the specialist herbivore M. sexta and its host plant N. attenuata (Hermsmeier et al., 2001; Hui et al., 2003), the downregulation of several photosynthetic-related transcripts was reported. We did not observe a repression of photosynthetic-related transcripts, and this might illustrate differences between plant species or might also reflect technical differences between small- and large-scale microarrays. By comparison with other plant/pathogen interactions, large-scale study of Arabidopsis responses after infection with the bacterial pathogen Pseudomonas syringae revealed an induction of 950 transcripts and a downregulation of 1005 transcripts 7 h after infection, this representing ∼15% of the genome (Scheideler et al., 2002). Early responses of Arabidopsis to the fungal pathogen Alternaria brassicicola resulted in the induction of ∼645 genes (equivalent to 8% of the genome), but very little repression was observed (Van Wees et al., 2003). Similarly, infection by diverse viruses induced 114 genes common to all inoculations (1.4% of the genome), whereas only a few genes were repressed (Whitham et al., 2003). In this study, we find that the molecular response to insects results in the induction of many genes and the repression of very few genes. By extrapolation of the data to genomic scale, we can estimate that the short-term response to herbivory results in the upregulation of ∼1.3% of the transcriptome in the damaged leaves. This figure is a minimum value because expression ratios we used below the twofold threshold might still be biologically relevant and because low abundance transcripts might not be detectable by conventional microarray analyses. In the Arabidopsis–P. rapae interaction, the activation of host gene expression is thus the dominant transcriptional response.
In undamaged distal leaves 24 h after an initial feeding phase of 3 to 5 h, a strikingly similar pattern of gene expression was observed. Figure 6 reveals that there is a large overlap between gene expression programs in damaged leaves and in distal leaves. We conclude that there are few differences in transcript patterns between local and distal leaves at this time. Because late instar larvae feed rapidly, they might consume leaf material before it has reached a sufficiently high level of resistance. It is thus important for the plant to activate defense responses distal to the attack site and, thus, to prepare for further aggression. Together, the data reveal that the response of Arabidopsis to insect feeding is active and consistent throughout damaged and undamaged leaves.
Quantitative Importance of the Jasmonate Pathway
A goal of this study was to provide a quantitative estimate of the number of herbivore-responsive genes regulated via the jasmonate pathway. This was accomplished using fully replicated and biologically independent experiments involving well-defined mutants. The results for the Pieris–Arabidopsis interaction showed that 67 to 84% of insect-inducible genes are under the control of the jasmonate pathway, attesting to the global nature of jasmonate signaling in this interaction. By comparison, in the study of plant responses to A. brassicicola, only 40% of the induced genes were found to be COI1 dependent (Van Wees et al., 2003). Expression profiling of sorghum plants colonized by phloem-feeding aphids revealed that only a weak induction of MeJA-regulated defense genes occurs and that a strong induction of SA-dependent pathogenesis-related genes is observed (Zhu-Salzman et al., 2004). These findings indicate clearly that the contribution of the JA pathway to the control of gene expression may vary depending on the type of plant–aggressor interaction.
The large reduction of Pieris-induced gene expression in coi1-1 gl1 plants suggested that this mutant might be less resistant to herbivore attack. Indeed, we showed that Pieris larvae grew better on coi1-1 gl1 than on wild-type or gl1 plants (Figure 3A). Similar findings were reported for the interaction between S. littoralis and coi1-1 gl1 plants where larvae inflicted significantly more damage to coi1-1 gl1 than to wild-type Col-0 plants (Stotz et al., 2002).
Two caveats to the interpretation of the data on gene expression or insect feeding remain. First, we assume that the coi1-1 allele is both a null mutant and that it regulates all jasmonate responses without affecting other signaling processes. This latter point is not yet demonstrated in Arabidopsis. However, complementary experiments using MeJA treatment to activate gene expression showed that the majority of COI1-dependent genes are activated by MeJA (Figure 8A; see Supplemental Table 1 online). Second, we have employed a coi1-1 gl1 double mutant and we cannot formally rule out epistatic interactions between the two mutations. However, detailed investigation of the impact of the jasmonate pathway showed that the independent coi1-1 mutation (in the absence of gl1) permitted increased weight gain of P. rapae larvae (Figure 3B). This showed that, alone, coi1-1 mutation affects insect performance. Furthermore, 90% of genes induced by Pieris in wild-type plants were also upregulated in the gl1 mutant (see Supplemental Table 5 online), indicating that the contribution of the gl1 mutation to the transcript pattern observed in coi1-1 gl1 plants is relatively low.
Relative Impact of Trichomes on Insect Resistance
We observed that larvae grew better on glabrous (gl1) than on Col-0 plants (Figure 3A). This effect seemed to be dependent on the developmental stage of the larvae with slightly less effect of the gl1 mutation observed with larger larvae (N. Bodenhausen, P. Reymond, and E.E. Farmer, unpublished results). In tomato (Lycopersicon esculentum), a mutation in a gene homologous to COI1 rendered mutant plants more susceptible to spider mites (Li et al., 2004b). In this example, and in the study with Spodoptera (Stotz et al., 2002), the mutant plants were either glabrous or had an abnormal development of glandular trichomes, and the relative contribution of trichomes to the response was not assayed. It is known that trichomes, which are constitutive structural barriers, play an important role in resistance to insects and that plants respond to herbivory by increasing the density or number of these structures (Mauricio and Rausher, 1997; Agrawal, 1998). Artificial damage and JA treatment elevate trichome production in Arabidopsis (Traw and Bergelson, 2003), and it was shown that black mustard (Brassica nigra) damaged with P. rapae had an increased trichome density (Traw and Dawson, 2002). Our data illustrate the potential effect of trichomes on larval performance and also underscore the importance of genetic background when analyzing mutant responses with insects (many Arabidopsis mutants are in a gl1 genetic background). However, Pieris larvae grew better on coi1-1 gl1 than on gl1 plants, indicating that jasmonate-dependent gene induction is necessary for a full response to herbivory even in the absence of trichomes. Further work will be necessary to exactly identify which of the COI1-dependent genes are responsible for this resistance.
Jasmonate Levels during Feeding
Another indication that the JA pathway is involved in plant–insect interactions is the finding that levels of jasmonates increase constantly in the damaged leaves during Pieris feeding. Quantitation of the levels of three jasmonates (JA, dnOPDA, and OPDA) revealed similar accumulation for each family member. Interestingly, the ratio of JA to OPDA was found to be <1 throughout the 24-h time course. This contrasts with jasmonate accumulation in response to mechanical wounding where the JA to OPDA ratio can exceed 2 (Reymond et al., 2000; Stintzi et al., 2001). OPDA levels are, thus, proportionally higher in Pieris damaged leaves than in mechanically wounded leaves. This may be one of many factors explaining differences in gene expression in mechanically wounded and insect-damaged leaves.
We have recently shown that OPDA can be an important signaling molecule per se. An OPDA reductase 3 (opr3) null mutant that lacks JA but can produce OPDA had near wild-type resistance to the dipteran Bradysia impatiens, whereas plants lacking all jasmonates were susceptible (Stintzi et al., 2001). We attributed this resistance to the presence of OPDA and showed that exogenous OPDA treatment powerfully upregulated several defense-related genes, many of which are also sensitive to JA. This indicated the presence of a signal input in opr3 requiring OPDA and COI1, but not JA, for the regulation of some defense gene expression (Stintzi et al., 2001). Interestingly, in this study we observed that several transcripts induced in Pieris-damaged wild-type plants (and not induced in the coi1-1 gl1 mutant) were not enhanced after MeJA treatment (Figure 8C; see Supplemental Table 1 online). They could represent a group of genes regulated more powerfully by OPDA than by JA, or, alternatively, they might require JA and another wound-inducible signal for correct regulation. For example, two ethylene biosynthesis genes are induced by Pieris, and this hormone has been shown to act concomitantly with JA to control defense gene expression (Penninckx et al., 1998). Another explanation could be that those genes depend on GL1, although as already mentioned, we find that only a small percentage of insect-regulated genes are potentially under the control of this gene (see Supplemental Table 5 online). Additionally, we observed that a few transcripts, for example PDF1.2 (see Supplemental Table 1 online), were induced after MeJA treatment but not after insect feeding (Figure 8F). Because challenge with Pieris larvae causes an increase of endogenous JA (Figure 4), this suggests that an insect-mediated signal interferes with the jasmonate pathway to suppress the induction of some specific genes. This phenomenon was found in N. attenuata where insect-induced ethylene was able to reduce jasmonate-induced nicotine accumulation at the transcription level (Winz and Baldwin, 2001). We also showed that PDF1.2 is not induced after wounding, although there is a large increase in JA levels (Reymond et al., 2000). Alternatively, it is possible that these genes respond to MeJA levels greater than those produced naturally.
Other Signal Inputs
For genes whose induction by Pieris was COI1 independent, other signaling pathways must operate to control their expression. In some cases, these genes were also wound inducible, and we assume that some abiotic responses like dehydration or touch could be involved. A recent report indicated that UV-B and caterpillar herbivory induced some genes in common (Izaguirre et al., 2003). Further work will be necessary to identify the exact signals responsible for the upregulation of genes not controlled directly by the jasmonate pathway. Interestingly, we found very few SA-responsive genes after feeding with P. rapae. SA could nevertheless be important because the antagonistic relationship between the SA and JA pathways is well documented (Kunkel and Brooks, 2002). It has been shown that the SA pathway might play a role in regulating direct defense against S. littoralis via the inhibition of the octadecanoid acid pathway (Stotz et al., 2002), and in a similar study, Arabidopsis mutants compromised in SA-dependent defense responses exhibited reduced levels of feeding by Trichoplusia ni caterpillars (Cui et al., 2002). Recently, a WRKY transcription factor was found to be an essential component of the cross talk between JA and SA, acting as an activator of SA-induced genes and a repressor of JA-responsive genes (Li et al., 2004a). A role for SA in indirect defense is also documented. Methyl salicylate is a component of the blend emitted by caterpillar-damaged Arabidopsis (Van Poecke et al., 2001) that elicits an electrophysiological response in two parasitoids that attack the caterpillars (Smid et al., 2002) and attracts predatory mites (De Boer and Dicke, 2004).
The largest cluster in Figure 8A contains genes that are insect and MeJA inducible. These genes were not wound inducible, although this might relate to the fact that we used single wounds rather than prolonged wounding in the experiments. Pieris salivary factors, which are absent in mechanical wounding experiments, might also help induce these genes, perhaps by stimulating high endogenous rise in JA synthesis. Alternatively, the insect might be reapplying ingested jasmonates to wounded tissues. To test this, we raised larvae on fad triple mutant plants that lack jasmonates and their precursors, tri-unsaturated fatty acids (McConn and Browse, 1996). The regurgitant from insects that had fed on the mutant plants still elicited the expression of selected insect-inducible defense genes (data not shown), as was the case for regurgitant from wild-type plants (Figure 5). We can thus rule out, at least in this system, insect-borne jasmonates in elicitation. Jasmonate-regulated gene expression is attributable to changes in levels of endogenous jasmonates induced by a combination of wounding and salivary factors. Theses factors in saliva could include, for example, β-glucosidase (Mattiacci et al., 1995) or fatty acid–amino acid conjugates (FACs). FACs have been previously described to be present in insect regurgitant and cause the accumulation of defense-related transcripts (Halitschke et al., 2001). However, because the fad triple mutant lacks linolenic acid, which is the precursor of many FACs (Paré et al., 1998), and because we could not detect linolenic acid in the regurgitant of Pieris larvae fed with fad triple mutant plants (see Supplemental Figure 3 online), our data suggest that 18:3-derived FACs are not among the biologically active components of Pieris saliva responsible for the induction of insect-inducible genes. Moreover, the observation that mechanical wounding activates several genes that are not activated by P. rapae raises the possibility that components of the regurgitant suppress the induction of wound-responsive genes. Such a mechanism has been observed in the tobacco (Nicotiana tabacum)–Helicoverpa zea interaction where a glucose oxidase contained in the insect's saliva suppressed nicotine production (Musser et al., 2002). Finally, the identification of insect-responsive genes allows a search for conserved elements in their promoters. The precise analysis of the activation of such elements, for example, by purified or synthetic elicitors, could be useful for the development of new and targeted approaches for reducing insect damage in cultivated crops.
Gene Expression Responses to a Specialist and to a Generalist
Most species of phytophagous insects have narrow diets, feeding on plants from one or two taxonomic groups or even from one single species (Bernays, 1998; Schoonhoven et al., 1998). There are several hypotheses to explain this abundance of specialists in nature (Ehrlich and Raven, 1964; Bernays and Graham, 1988; Jaenike, 1990; Fry, 1996), and specialization clearly provides selective advantages. Specialist herbivores have long been predicted to be more tolerant to defense substances of their hosts compared to generalists (Blau et al., 1978; Van der Meijden, 1996; Bernays, 1998). In accordance with this is a report looking at variation in the glucosinolate-myrosinase chemical defense system in Arabidopsis (Kliebenstein et al., 2002). Using recombinant inbred lines, the authors identified several quantitative trait loci controlling insect feeding that were correlated with the glucosinolate-myrosinase system. They showed that glucosinolates and myrosinase have larger effects on generalist than on specialist herbivores. Moreover, the crucifer specialist insect Plutella xylostella contains a sulfatase activity that modifies glucosinolates and prevents the formation of toxic hydrolysis products (e.g., isothiocyanates) arising from the glucosinolate-myrosinase complex (Ratzka et al., 2002). This enzyme activity is absent from P. rapae, but it was shown recently that P. rapae larvae contain a larval gut protein that leads to the formation of less toxic nitriles instead of isothiocyanates (Wittstock et al., 2004). Even though Pieris seems to have adapted to defensive glucosinolates, to the point that it uses them to select host plants for oviposition (Du et al., 1995), induced production of these same chemicals plays a defensive role by reducing Pieris larval development (Agrawal and Kurashige, 2003).
Very little is known about the differential molecular responses of the plant when confronted with either specialists or generalists. We reasoned that one explanation for the difference in dietary strategy was that specialists might have evolved mechanisms with which to minimize or even suppress inducible host defense gene expression. Such mechanisms would probably be too difficult to evolve for generalists that face the disparate defensive strategies of many different plants. If this were the case, we would have expected to see less induced gene expression in the Arabidopsis–specialist interaction than in the Arabidopsis–generalist interaction. A converse hypothesis was that plants might be able to better recognize specialist herbivores that they encounter more frequently. This interaction might lead to stronger and more diverse defense gene expression in response to an adapted specialist than to an infrequent generalist attacker. Specialization would then be the result of an enhanced tolerance of specialists to the induced defense gene products. We directly addressed these questions by comparing P. rapae, a crucifer specialist, and S. littoralis, a generalist herbivore. In contrast with our expectations, the two insects elicited remarkably similar gene expression profiles in Arabidopsis. Thus, despite the coevolution between an adapted specialist and its plant host, the plant recognizes the specialist as strongly as the generalist. Supporting this is the report that Cotesia parasitoids do not discriminate between volatiles induced by their host, P. rapae, and by the non-host, Spodoptera exigua, feeding on Arabidopsis (Van Poecke et al., 2003). Both findings imply that plants trigger similar defenses when confronted by insects that cause similar damage and that dietary strategy seems not to be governed by the differential activation of a defense response.
In conclusion, Pieris and Spodoptera, two insects with strongly different dietary strategies, clearly elicit highly similar molecular responses in the host. These responses are active with relatively little transcriptional repression observed. Using rigorous criteria, we have identified a list of inducible genes that respond to a specialist and a generalist chewing herbivore and have a quantitative estimate of the importance of the jasmonate pathway in this response. Directly and indirectly the jasmonate pathway controls or influences the expression of the majority of genes identified as being insect-activated, and these genes may have a significant effect on insect performance. The gene set we defined in this study will now permit the rigorous comparison of gene expression programs in response to widely divergent herbivores that differ not only in dietary strategy but also in feeding behavior.
METHODS
Plant and Insect Growth Conditions
Growth of wild-type (Col-0) and mutant Arabidopsis thaliana plants was described previously (Reymond et al., 2000). The coi1-1 gl1 mutant was supplied by J.G. Turner, the non-glabrous coi1-1 mutant was a gift from Jane Glazebrook, and the gl1 mutant was obtained from S. Somerville. Spodoptera littoralis (Egyptian cotton worm) eggs were obtained from Syngenta (Stein, Switzerland) and raised on cabbage (Brassica capitata) plants in a phytotron (25°C, 65% relative humidity, 14-h light period, 100 μmol m−2 s−1) until larvae reached the fourth or fifth instar stage. Cultivation and handling of Pieris rapae larvae were described previously (Reymond et al., 2000).
Plant Treatments
Plants were 6 to 7 weeks old at the time of treatment. All experiments were biologically independent; they were conducted with several plantings done at intervals of several weeks. For experiments aimed at estimating individual variability in gene expression, 24 untreated Arabidopsis plants were split into two groups of 12 plants, and rosette leaves from each group were pooled, harvested, and immediately stored in liquid nitrogen. This experiment was repeated six times.
For insect challenge, two fourth- or fifth-instar larvae were allowed to feed on a plant for 3 to 5 h in a growth chamber (20°C, 60% relative humidity, 100 μmol m−2 s−1) until ∼20% of leaf area was removed. For each experiment, damaged leaf tissue from 12 challenged plants was harvested and immediately stored in liquid nitrogen. Leaves from 12 control unchallenged plants were collected at the same time. Experiments were repeated five times for challenge with P. rapae or S. littoralis on wild-type plants and four times for P. rapae on coi1-1 gl1 plants. For gene expression analyses in local and distal leaves, larvae were removed manually after 3 to 5 h of feeding, and plants were left in a phytotron under constant illumination for another 24 h until harvesting. Local (damaged) and distal (undamaged) leaves were collected separately. Unchallenged plants of the same age were used as controls. Three independent experiments were conducted.
Mechanical damage was performed with a cork borer, and ∼20% of leaf area was removed from each rosette leaf, roughly mimicking the amount of tissue removed by insect feeding. Plants were then placed in a phytotron for 3 to 5 h until harvesting. Four independent experiments were done. For MeJA treatment, 12 plants were exposed for 6 h to 4 μmol racemic MeJA (Aldrich, Buchs, Switzerland; in 10 μL EtOH deposited on a cotton swab) in an 11-liter hermetic Plexiglas box. Control plants were exposed to ethanol only. Expression ratios were obtained from three biologically independent replicates.
For treatment with insect regurgitant, three 1-mm holes per leaf were made with a cork borer and 1 μL of regurgitant obtained from fourth- or fifth-instar P. rapae larvae was added to each hole. In control plants, 1 μL of water was added to each hole. After 6 h, three to four leaves were harvested for RNA gel blot analyses. The experiment was repeated three times.
Insect Feeding Trials
The effect of plant genotype on insect herbivory was studied using freshly hatched P. rapae larvae. Arabidopsis Col-0, gl1, coi1-1, and coi1-1 gl1 plants were grown as mentioned above, except that there was only one plant per pot. Plants were 5 weeks old at the time of treatment and were placed in a growth chamber (20°C, 65% relative humidity, 100 μmol m−2 s−1). One newly hatched larva was placed on each plant (approximately 16 to 24 plants per genotype). Larvae were not caged because they do not move from plant to plant as long as the leaves of adjacent plants do not touch each other. After 6 or 8 d of feeding, larvae were removed and weighed to the nearest 0.01 mg using a Mettler-Toledo MT5 balance (Mettler-Toledo, Greifensee, Switzerland). Caterpillar weights at egg hatch are assumed to be equal; thus, only final weights are measured. These experiments were repeated with similar results.
Microarray Experiments and Data Analysis
A set of 12,135 ESTs was obtained from the Arabidopsis Biological Resource Center (Columbus, OH). The cDNA inserts were amplified with universal primers M13 forward 5′-GTTTTCCCAGTCACGTTG-3′ and M13 reverse 5′-TGAGCGGATAACAATTTCACACAG-3′ using PCR. The PCR products were checked for quality by gel electrophoresis, purified with a QIAquick-96 PCR purification kit (Qiagen, Basel, Switzerland). Printing of DNA and processing of aldehyde slides (TeleChem, San Jose, CA) was performed as described previously (Reymond et al., 2000). mRNA isolation and preparation of fluorescent probes was done according to published protocols (Reymond et al., 2000; Stintzi et al., 2001) and www.unil.ch/ibpv/microarrays.htm. Microarrays were scanned with a ScanArray 4000 (Packard BioScience, Zurich, Switzerland). Photomultiplier and laser power settings were adjusted so that the expression ratio of the majority of genes was as close to 1.0 as possible and signal intensities were below saturation of the scanner. The average fluorescence intensity for each fluor and for each gene was determined using the ImaGene program (BioDiscovery, Los Angeles, CA). Median background fluorescence around each gene spot was calculated and subtracted from each spot. A measure of spot quality was used as follows: spots where the difference between spot and background mean intensity was below two times the background standard deviation were flagged and removed from the analyses. Signal values <1000 (two to three times the average background intensity) were manually raised to 1000 to avoid extreme expression ratios. To avoid spatial bias and the effect of systematic differences between print tips, signal intensities were normalized so that the distribution of log ratios within each subgrid had a median of zero (Yang et al., 2002).
Normalized signal intensities were used to calculate expression ratios. To estimate the individual variability of gene expression between untreated plants, we performed a control hybridization using two independent samples obtained from plants harvested the same day. This experiment was replicated six times on six different days. We observed that all but seven ESTs had expression ratios between −1.5 and 1.5 (see Supplemental Figure 1 online). A Student's t test (one sample hypothesis) on log2-transformed expression ratios indicated that 677 ESTs (5%) had an expression ratio different from 1.0 (P < 0.05). To identify differentially expressed genes in treated samples, a Student's t test (two sample hypothesis, equal variance) was conducted between log2-transformed expression ratios from control versus control experiments and log2-transformed ratios from control versus treated experiments. Based on the control experiments, we thus decided to use a threshold of twofold for the expression ratio to increase the chance of identifying strongly induced genes. For all of the experiments, we thus only considered genes with an expression ratio ≥2 or ≥−2 and a P value < 0.05.
To address the issue of multiple comparisons and identify the proportion of false positives among all the genes identified as being differentially expressed, we calculated an FDR using a method developed for genome-wide studies (Storey and Tibshirani, 2003). This method computes a q value for each gene using the distribution of P values of all measurements. The q value for a particular gene reflects the proportion of false positives incurred when calling this gene significant. This calculation of FDR is supposed to give a fair estimation of the number of false positives and is considered as appropriate for microarray analyses (Cui and Churchill, 2003; Storey and Tibshirani, 2003). We noticed that high FDR values are estimated when the number of induced genes is relatively small. For example, a set of genes that were induced with the same intensity and with the same statistics in two separate experiments (challenge with P. rapae in wild-type or coi1-1 gl1 plants) had a much higher q value in the second experiment (see Supplemental Table 1 online). This might be because of the fact that the coi1-1 gl1 mutation has a very strong effect on the observed response and that this drastically reduces the number of induced genes. Thus interexperiment comparison adds to data interpretation, and FDR calculations might be too conservative in some cases.
Hierarchical gene clustering was done with the program Cluster (Eisen et al., 1998). Most data processing and analysis were done using a relational database (Nomad, University of California, San Francisco, http://ucsf-nomad.sourceforge.net) installed locally and adapted using custom Perl scripts. All genes presented in this work were resequenced to confirm their identity.
RNA Extraction and RNA Gel Blot Analyses
For RNA gel blot analysis, we used probes for a lectin gene (EST accession number N96505), a Cys proteinase gene (EST accession number T22938), a cytochrome P450 gene CYP79B2 (EST accession number T42902), and as a loading control a probe for LHB1B2 (EST accession number R89981). Probes were labeled with digoxigenin (Roche Biochemicals, Rotkreuz, Switzerland) using universal primers located on the cloning vector.
Quantitative Analysis of Jasmonate Family Members
Six-week-old plants were either infested with P. rapae caterpillars or left untreated. Caterpillar-infested plants were obtained by placing 20 first instar P. rapae caterpillars on two Arabidopsis plants (10 caterpillars per plant). At several time points after infestation (0 to 24 h), caterpillars were removed and the leaf material of the two plants was harvested, pooled, and immediately frozen in liquid nitrogen before storage. At the same time points, leaf material from two control plants was harvested, pooled, frozen, and stored. For each time point, new sets of infested and control plants were used. Extraction and quantification of JA, OPDA, and dnOPDA in the leaf samples were performed according to published protocols (Weber et al., 1997; Stintzi et al., 2001).
Microarray data from this article have been deposited with the Array-Express database (http://www.ebi.ac.uk/arrayexpress) under accession numbers E-MEXP-158 to E-MEXP-164.
Supplementary Material
Acknowledgments
We thank Martine Damond for the preparation of PCR products, Boris Kunstner for maintenance of the plants, and Robin Liechti for installing and adapting the Nomad database. Roland Reist (Syngenta, Stein, Switzerland) kindly provided S. littoralis eggs. We thank Darlene Goldstein and Jerome Goudet for their help with the statistical analysis and Dawn Little for critically reading the manuscript. This work was supported by the Leenaards Foundation, the Swiss National Science Foundation Grants 3100-054942 and 3100AO-101711, and by a genomics grant from the University of Lausanne.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantcell.org) is: Edward E. Farmer (edward.farmer@unil.ch).
Online version contains Web-only data.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.104.026120.
References
- Agrawal, A.A. (1998). Induced responses to herbivory and increased plant performance. Science 279, 1201–1202. [DOI] [PubMed] [Google Scholar]
- Agrawal, A.A. (1999). Induced responses to herbivory in wild radish: Effects on several herbivores and plant fitness. Ecology 80, 1713–1723. [Google Scholar]
- Agrawal, A.A., and Kurashige, N.S. (2003). A role for isothiocyanates in plant resistance against the specialist herbivore Pieris rapae. J. Chem. Ecol. 29, 1403–1415. [DOI] [PubMed] [Google Scholar]
- Alborn, H.T., Brennan, M.M., and Tumlinson, J.H. (2003). Differential activity and degradation of plant volatile elicitors in regurgitant of tobacco hornworm (Manduca sexta) larvae. J. Chem. Ecol. 29, 1357–1372. [DOI] [PubMed] [Google Scholar]
- Alborn, H.T., Turlings, T.C.J., Jones, T.H., Stenhagen, G., Loughrin, J.H., and Tumlinson, J.H. (1997). An elicitor of plant volatiles from beet armyworm oral secretion. Science 276, 945–949. [Google Scholar]
- Baldwin, I.T. (1998). Jasmonate-induced responses are costly but benefit plants under attack in native populations. Proc. Natl. Acad. Sci. USA 95, 8113–8118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becerra, J.X. (2003). Synchronous coadaptation in an ancient case of herbivory. Proc. Natl. Acad. Sci. USA 100, 12804–12807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berenbaum, M.R. (2001). Chemical mediation of coevolution: Phylogenetic evidence for Apiaceae and associates. Ann. Mo. Bot. Gard. 88, 45–59. [Google Scholar]
- Bernays, E., and Graham, M. (1988). On the evolution of host specificity in phytophagous arthropods. Ecology 69, 886–892. [Google Scholar]
- Bernays, E.A. (1998). Evolution of feeding behavior in insect herbivores. Bioscience 48, 35–44. [Google Scholar]
- Blau, P.A., Feeny, P., Contardo, L., and Robson, D.S. (1978). Allylglucosinolate and herbivorous caterpillars: Contrast in toxicity and tolerance. Science 200, 1296–1298. [DOI] [PubMed] [Google Scholar]
- Brader, G., Tas, E., and Palva, E.T. (2001). Jasmonate-dependent induction of indole glucosinolates in Arabidopsis by culture filtrates of the nonspecific pathogen Erwinia carotovora. Plant Physiol. 126, 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, J.P., Jander, G., Racki, L.R., Kim, P.D., Pierce, N.E., and Ausubel, F.M. (2002). Signals involved in Arabidopsis resistance to Trichoplusia ni caterpillars induced by virulent and avirulent strains of the phytopathogen Pseudomonas syringae. Plant Physiol. 129, 551–564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, X.Q., and Churchill, G.A. (2003). Statistical tests for differential expression in cDNA microarray experiments. Genome Biol. 4, 210.1–210.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer, J.G., and Dicke, M. (2004). The role of methyl salicylate in prey searching behavior of the predatory mite Phytoseiulus persimilis. J. Chem. Ecol. 30, 255–271. [DOI] [PubMed] [Google Scholar]
- De Moraes, C.M., Lewis, W.J., Pare, P.W., Alborn, H.T., and Tumlinson, J.H. (1998). Herbivore-infested plants selectively attract parasitoids. Nature 393, 570–573. [Google Scholar]
- Dicke, M. (1999). Are herbivore-induced plant volatiles reliable indicators of herbivore identity to foraging carnivorous arthropods? Entomol. Exp. Appl. 91, 131–142. [Google Scholar]
- Doughty, K.J., Kiddle, G.A., Pye, B.J., Wallsgrove, R.M., and Pickett, J.A. (1995). Selective induction of glucosinolates in oilseed rape leaves by methyl jasmonate. Phytochem. 38, 347–350. [Google Scholar]
- Du, Y.J., Vanloon, J.J.A., and Renwick, J.A.A. (1995). Contact chemoreception of oviposition-stimulating glucosinolates and an oviposition-deterrent cardenolide in 2 subspecies of Pieris napi. Physiol. Entomol. 20, 164–174. [Google Scholar]
- Ehrlich, P.R., and Raven, P.H. (1964). Butterflies and plants: A study in coevolution. Evolution 18, 586–608. [Google Scholar]
- Eisen, M.B., Spellman, P.T., Brown, P.O., and Botstein, D. (1998). Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. USA 95, 14863–14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farmer, E.E. (2000). Adding injury to insult: Pathogen detection and responses. Genome Biol. 1, 1012.1011–1012.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feys, B.J., and Parker, J.E. (2000). Interplay of signaling pathways in plant disease resistance. Trends Genet. 16, 449–455. [DOI] [PubMed] [Google Scholar]
- Feys, B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frey, M., Stettner, C., Pare, P.W., Schmelz, E.A., Tumlinson, J.H., and Gierl, A. (2000). An herbivore elicitor activates the gene for indole emission in maize. Proc. Natl. Acad. Sci. USA 97, 14801–14806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry, J.D. (1996). The evolution of host specialization: Are trade-offs overrated? Am. Nat. 148 (suppl.), S84–S107. [Google Scholar]
- Gaunt, M.W., and Miles, M.A. (2002). An insect molecular clock dates the origin of the insects and accords with palaeontological and biogeographic landmarks. Mol. Biol. Evol. 19, 748–761. [DOI] [PubMed] [Google Scholar]
- Gidda, S.K., Miersch, O., Levitin, A., Schmidt, J., Wasternack, C., and Varin, L. (2003). Biochemical and molecular characterization of a hydroxyjasmonate sulfotransferase from Arabidopsis thaliana. J. Biol. Chem. 278, 17895–17900. [DOI] [PubMed] [Google Scholar]
- Halitschke, R., and Baldwin, I.T. (2003). Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuata. Plant J. 36, 794–807. [DOI] [PubMed] [Google Scholar]
- Halitschke, R., Gase, K., Hui, D., Schmidt, D.D., and Baldwin, I.T. (2003). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. VI. Microarray analysis reveals that most herbivore-specific transcriptional changes are mediated by fatty acid-amino acid conjugates. Plant Physiol. 131, 1894–1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halitschke, R., Schittko, U., Pohnert, G., Boland, W., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. III. Fatty acid-amino acid conjugates in herbivore oral secretions are necessary and sufficient for herbivore-specific plant responses. Plant Physiol. 125, 711–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartmann, T., and Ober, D. (2000). Biosynthesis and metabolism of pyrrolizidine alkaloids in plants and specialized insect herbivores. Top. Curr. Chem. 209, 207–243. [Google Scholar]
- Heil, M., Koch, T., Hilpert, A., Fiala, B., Boland, W., and Linsenmair, K.E. (2001). Extrafloral nectar production of the ant-associated plant, Macaranga tanarius, is an induced, indirect, defensive response elicited by jasmonic acid. Proc. Natl. Acad. Sci. USA 98, 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermsmeier, D., Schittko, U., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. I. Large-scale changes in the accumulation of growth- and defense-related plant mRNAs. Plant Physiol. 125, 683–700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., Lightner, J., Browse, J., and Ryan, C.A. (1996). An octadecanoid pathway mutant (JL5) of tomato is compromised in signaling for defense against insect attack. Plant Cell 8, 2067–2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hui, D., Iqbal, J., Lehmann, K., Gase, K., Saluz, H.P., and Baldwin, I.T. (2003). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. V. Microarray analysis and further characterization of large-scale changes in herbivore-induced mRNAs. Plant Physiol. 131, 1877–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izaguirre, M.M., Scopel, A.L., Baldwin, I.T., and Ballare, C.L. (2003). Convergent responses to stress. Solar ultraviolet-B radiation and Manduca sexta herbivory elicit overlapping transcriptional responses in field-grown plants of Nicotiana longiflora. Plant Physiol. 132, 1755–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaenike, J. (1990). Host specialization in phytophagous insects. Annu. Rev. Ecol. Syst. 21, 243–273. [Google Scholar]
- Jander, G., Cui, J.P., Nhan, B., Pierce, N.E., and Ausubel, F.M. (2001). The TASTY locus on chromosome 1 of Arabidopsis affects feeding of the insect herbivore Trichoplusia ni. Plant Physiol. 126, 890–898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karban, R., and Baldwin, I.T. (1997). Induced Responses to Herbivory. (Chicago: University of Chicago Press).
- Kliebenstein, D., Pedersen, D., Barker, B., and Mitchell-Olds, T. (2002). Comparative analysis of quantitative trait loci controlling glucosinolates, myrosinase and insect resistance in Arabidapsis thaliana. Genetics 161, 325–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth, K.L. (2003). Profiling the response of plants to herbivorous insects. Genome Biol. 4, 221.1–221.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korth, K.L., and Dixon, R.A. (1997). Evidence for chewing insect-specific molecular events distinct from a general wound response in leaves. Plant Physiol. 115, 1299–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel, B.N., and Brooks, D.M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. [DOI] [PubMed] [Google Scholar]
- Lambrix, V., Reichelt, M., Mitchell-Olds, T., Kliebenstein, D.J., and Gershenzon, J. (2001). The Arabidopsis epithiospecifier protein promotes the hydrolysis of glucosinolates to nitriles and influences Trichoplusia ni herbivory. Plant Cell 13, 2793–2807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, J., Brader, G., and Palva, E.T. (2004. a). The WRKY70 transcription factor: A node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., Zhao, Y.F., McCaig, B.C., Wingerd, B.A., Wang, J.H., Whalon, M.E., Pichersky, E., and Howe, G.A. (2004. b). The tomato homolog of CORONATINE-INSENSITIVE1 is required for the maternal control of seed maturation, jasmonate-signaled defense responses, and glandular trichome development. Plant Cell 16, 126–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liechti, R., and Farmer, E.E. (2002). The jasmonate pathway. Science 296, 1649–1650. [DOI] [PubMed] [Google Scholar]
- Lopukhina, A., Dettenberg, M., Weiler, E.W., and Hollander-Czytko, H. (2001). Cloning and characterization of a coronatine-regulated tyrosine aminotransferase from Arabidopsis. Plant Physiol. 126, 1678–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattiacci, L., Dicke, M., and Posthumus, M.A. (1995). Beta-glucosidase: An elicitor of herbivore-induced plant odor that attracts host-searching parasitic wasps. Proc. Natl. Acad. Sci. USA 92, 2036–2040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mauricio, R., and Rausher, M.D. (1997). Experimental manipulation of putative selective agents provides evidence for the role of natural enemies in the evolution of plant defense. Evolution 51, 1435–1444. [DOI] [PubMed] [Google Scholar]
- McCloud, E.S., and Baldwin, I.T. (1997). Herbivory and caterpillar regurgitants amplify the wound-induced increases in jasmonic acid but not nicotine in Nicotiana sylvestris. Planta 203, 430–435. [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., Creelman, R.A., Bell, E., Mullet, J.E., and Browse, J. (1997). Jasmonate is essential for insect defense in Arabidopsis. Proc. Natl. Acad. Sci. USA 94, 5473–5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikkelsen, M.D., Petersen, B.L., Glawischnig, E., Jensen, A.B., Andreasson, E., and Halkier, B.A. (2003). Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol. 131, 298–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran, P.J., Cheng, Y.F., Cassell, J.L., and Thompson, G.A. (2002). Gene expression profiling of Arabidopsis thaliana in compatible plant-aphid interactions. Arch. Insect Biochem. Physiol. 51, 182–203. [DOI] [PubMed] [Google Scholar]
- Musser, R.O., Hum-Musser, S.M., Eichenseer, H., Peiffer, M., Ervin, G., Murphy, J.B., and Felton, G.W. (2002). Herbivory: Caterpillar saliva beats plant defences. Nature 416, 599–600. [DOI] [PubMed] [Google Scholar]
- Nosil, P., Crespi, B.J., and Sandoval, C.P. (2002). Host-plant adaptation drives the parallel evolution of reproductive isolation. Nature 417, 440–443. [DOI] [PubMed] [Google Scholar]
- Orozco-Cardenas, M., McGurl, B., and Ryan, C.A. (1993). Expression of an antisense prosystemin gene in tomato plants reduces resistance toward Manduca sexta larvae. Proc. Natl. Acad. Sci. USA 90, 8273–8276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré, P.W., Alborn, H.T., and Tumlinson, J.H. (1998). Concerted biosynthesis of an insect elicitor of plant volatiles. Proc. Natl. Acad. Sci. USA 95, 13971–13975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré, P.W., and Tumlinson, J.H. (1997). De novo biosynthesis of volatiles induced by insect herbivory in cotton plants. Plant Physiol. 114, 1161–1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paré, P.W., and Tumlinson, J.H. (1999). Plant volatiles as a defense against insect herbivores. Plant Physiol. 121, 325–331. [PMC free article] [PubMed] [Google Scholar]
- Park, J.H., Halitschke, R., Kim, H.B., Baldwin, I.T., Feldmann, K.A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. [DOI] [PubMed] [Google Scholar]
- Pechan, T., Cohen, A., Williams, W.P., and Luthe, D.S. (2002). Insect feeding mobilizes a unique plant defense protease that disrupts the peritrophic matrix of caterpillars. Proc. Natl. Acad. Sci. USA 99, 13319–13323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninckx, I.A., Thomma, B.P., Buchala, A., Metraux, J.P., and Broekaert, W.F. (1998). Concomitant activation of jasmonate and ethylene response pathways is required for induction of a plant defensin gene in Arabidopsis. Plant Cell 10, 2103–2113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peumans, W.J., and Vandamme, E.J.M. (1995). Lectins as plant defense proteins. Plant Physiol. 109, 347–352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rask, L., Andreasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). Myrosinase: Gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113. [PubMed] [Google Scholar]
- Ratzka, A., Vogel, H., Kliebenstein, D.J., Mitchell-Olds, T., and Kroymann, J. (2002). Disarming the mustard oil bomb. Proc. Natl. Acad. Sci. USA 99, 11223–11228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond, P. (2001). DNA microarrays and plant defence. Plant Physiol. Biochem. 39, 313–321. [Google Scholar]
- Reymond, P., and Farmer, E.E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. [DOI] [PubMed] [Google Scholar]
- Reymond, P., Weber, H., Damond, M., and Farmer, E.E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheideler, M., Schlaich, N.L., Fellenberg, K., Beissbarth, T., Hauser, N.C., Vingron, M., Slusarenko, A.J., and Hoheisel, J.D. (2002). Monitoring the switch from housekeeping to pathogen defense metabolism in Arabidopsis thaliana using cDNA arrays. J. Biol. Chem. 277, 10555–10561. [DOI] [PubMed] [Google Scholar]
- Schenk, P.M., Kazan, K., Wilson, I., Anderson, J.P., Richmond, T., Somerville, S.C., and Manners, J.M. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. USA 97, 11655–11660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schittko, U., Hermsmeier, D., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. II. Accumulation of plant mRNAs in response to insect-derived cues. Plant Physiol. 125, 701–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz, E.A., Alborn, H.T., Banchio, E., and Tumlinson, J.H. (2003). Quantitative relationships between induced jasmonic acid levels and volatile emission in Zea mays during Spodoptera exigua herbivory. Planta 216, 665–673. [DOI] [PubMed] [Google Scholar]
- Schoonhoven, L.M., Jermy, T., and Van Loon, J.J.A. (1998). Insect-Plant Biology: From Physiology to Evolution. (London: Chapman and Hall).
- Smid, H.A., van Loon, J.J.A., Posthumus, M.A., and Vet, L.E.M. (2002). GC-EAG-analysis of volatiles from Brussels sprouts plants damaged by two species of Pieris caterpillars: Olfactory receptive range of a specialist and a generalist parasitoid wasp species. Chemoecology 12, 169–176. [Google Scholar]
- Staswick, P.E., Tiryaki, I., and Rowe, M.L. (2002). Jasmonate response locus JAR1 and several related Arabidopsis genes encode enzymes of the firefly luciferase superfamily that show activity on jasmonic, salicylic, and indole-3-acetic acids in an assay for adenylation. Plant Cell 14, 1405–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E.E. (2001). Plant defense in the absence of jasmonic acid: The role of cyclopentenones. Proc. Natl. Acad. Sci. USA 98, 12837–12842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey, J.D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Genome Biol. 100, 9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stotz, H.U., Koch, T., Biedermann, A., Weniger, K., Boland, W., and Mitchell-Olds, T. (2002). Evidence for regulation of resistance in Arabidopsis to Egyptian cotton worm by salicylic and jasmonic acid signaling pathways. Planta 214, 648–652. [DOI] [PubMed] [Google Scholar]
- Stotz, H.U., Pittendrigh, B.R., Kroymann, J., Weniger, K., Fritsche, J., Bauke, A., and Mitchell-Olds, T. (2000). Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against egyptian cotton worm but not diamondback moth. Plant Physiol. 124, 1007–1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strauss, S.Y., and Agrawal, A.A. (1999). The ecology and evolution of plant tolerance to herbivory. Trends Ecol. Evol. 14, 179–185. [DOI] [PubMed] [Google Scholar]
- Thaler, J.S., Farag, M.A., Pare, P.W., and Dicke, M. (2002). Jasmonate-deficient plants have reduced direct and indirect defences against herbivores. Ecol. Lett. 5, 764–774. [Google Scholar]
- Traw, M.B., and Bergelson, J. (2003). Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133, 1367–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traw, M.B., and Dawson, T.E. (2002). Differential induction of trichomes by three herbivores of black mustard. Oecologia 131, 526–532. [DOI] [PubMed] [Google Scholar]
- Vancanneyt, G., Sanz, C., Farmaki, T., Paneque, M., Ortego, F., Castanera, P., and Sanchez-Serrano, J.J. (2001). Hydroperoxide lyase depletion in transgenic potato plants leads to an increase in aphid performance. Proc. Natl. Acad. Sci. USA 98, 8139–8144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Meijden, E. (1996). Plant defence, an evolutionary dilemma: Contrasting effects of (specialist and generalist) herbivores and natural enemies. Entomol. Exp. Appl. 80, 307–310. [Google Scholar]
- Van Poecke, R.M.P., and Dicke, M. (2002). Induced parasitoid attraction by Arabidopsis thaliana: Involvement of the octadecanoid and the salicylic acid pathway. J. Exp. Bot. 53, 1793–1799. [DOI] [PubMed] [Google Scholar]
- Van Poecke, R.M.P., Posthumus, M.A., and Dicke, M. (2001). Herbivore-induced volatile production by Arabidopsis thaliana leads to attraction of the parasitoid Cotesia rubecula: Chemical, behavioral, and gene-expression analysis. J. Chem. Ecol. 27, 1911–1928. [DOI] [PubMed] [Google Scholar]
- Van Poecke, R.M.P., Roosjen, M., Pumarino, L., and Dicke, M. (2003). Attraction of the specialist parasitoid Cotesia rubecula to Arabidopsis thaliana infested by host or non-host herbivore species. Entomol. Exp. Appl. 107, 229–236. [Google Scholar]
- Van Wees, S.C., Chang, H.S., Zhu, T., and Glazebrook, J. (2003). Characterization of the early response of Arabidopsis to Alternaria brassicicola infection using expression profiling. Plant Physiol. 132, 606–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walling, L.L. (2000). The myriad plant responses to herbivores. J. Plant Growth Regul. 19, 195–216. [DOI] [PubMed] [Google Scholar]
- Wan, J., Dunning, F.M., and Bent, A.F. (2002). Probing plant-pathogen interactions and downstream defense signaling using DNA microarrays. Funct. Integr. Genomics 2, 259–273. [DOI] [PubMed] [Google Scholar]
- Weber, H., Vick, B.A., and Farmer, E.E. (1997). Dinor-oxo-phytodienoic acid: A new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. USA 94, 10473–10478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitham, S.A., Quan, S., Chang, H.S., Cooper, B., Estes, B., Zhu, T., Wang, X., and Hou, Y.M. (2003). Diverse RNA viruses elicit the expression of common sets of genes in susceptible Arabidopsis thaliana plants. Plant J. 33, 271–283. [DOI] [PubMed] [Google Scholar]
- Winz, R.A., and Baldwin, I.T. (2001). Molecular interactions between the specialist herbivore Manduca sexta (Lepidoptera, Sphingidae) and its natural host Nicotiana attenuata. IV. Insect-induced ethylene reduces jasmonate- induced nicotine accumulation by regulating putrescine N- methyltransferase transcripts. Plant Physiol. 125, 2189–2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittstock, U., Agerbirk, N., Stauber, E.J., Olsen, C.E., Hippler, M., Mitchell-Olds, T., Gershenzon, J., and Vogel, H. (2004). Successful herbivore attack due to metabolic diversion of a plant chemical defense. Proc. Natl. Acad. Sci. USA 101, 4859–4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, Y.H., Dudoit, S., Luu, P., Lin, D.M., Peng, V., Ngai, J., and Speed, T.P. (2002). Normalization for cDNA microarray data: A robust composite method addressing single and multiple slide systematic variation. Nucleic Acids Res. 30, e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu-Salzman, K., Salzman, R.A., Ahn, J.-E., and Koiwa, H. (2004). Transcriptional regulation of sorghum defense determinants against a phloem-feeding aphid. Plant Physiol. 134, 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.