Abstract
Orphan GPCRs provide an opportunity to identify potential pharmacological targets, yet their expression patterns and physiological functions remain challenging to elucidate. Here, we have used a genetically engineered knockin reporter mouse to map the expression pattern of the Gpr182 during development and adulthood. We observed that Gpr182 is expressed at the crypt base throughout the small intestine, where it is enriched in crypt base columnar stem cells, one of the most active stem cell populations in the body. Gpr182 knockdown had no effect on homeostatic intestinal proliferation in vivo, but led to marked increases in proliferation during intestinal regeneration following irradiation-induced injury. In the ApcMin mouse model, which forms spontaneous intestinal adenomas, reductions in Gpr182 led to more adenomas and decreased survival. Loss of Gpr182 enhanced organoid growth efficiency ex vivo in an EGF-dependent manner. Gpr182 reduction led to increased activation of ERK1/2 in basal and challenge models, demonstrating a potential role for this orphan GPCR in regulating the proliferative capacity of the intestine. Importantly, GPR182 expression was profoundly reduced in numerous human carcinomas, including colon adenocarcinoma. Together, these results implicate Gpr182 as a negative regulator of intestinal MAPK signaling–induced proliferation, particularly during regeneration and adenoma formation.
Introduction
Deorphanization of GPCRs remains an active area of research, especially considering that approximately 40% of all approved drugs for humans target only a small fraction of the GPCRome (1, 2). In addition to elucidating the pharmacology of orphan GPCRs, it is crucial to characterize the anatomical locations and physiological functions of these receptors in vivo. G protein–coupled receptor 182 (GPR182, formerly known as G10D or adrenomedullin receptor [ADMR]) (3, 4), is a class A orphan GPCR with very little known about its expression, function, regulation, or pharmacology. GPR182 is grouped within the chemokine receptor family by phylogeny, with the atypical chemokine receptor 3 (ACKR3, formerly known as CXCR7 or RDC1) as its closest paralog, despite the two sharing a modest, less-than-30% sequence homology in mice and humans. GPR182 was previously considered a putative receptor for the multifunctional peptide adrenomedullin (4), however, these initial findings were not consistent among laboratories (5), and it was later shown that adrenomedullin signals through a different GPCR complex (6). Unfortunately, the former ADMR nomenclature is sometimes still used, which leads to confusion in the field. For example, GPR182 was reported to be expressed in numerous human pancreatic cancer cell lines, and knockdown of GPR182 in these cells decreased xenograft tumor growth, which the authors concluded was due to a loss of adrenomedullin signaling (7, 8). Anatomical expression profiling of the GPCRome demonstrated the relatively ubiquitous low expression of Gpr182 in most mouse tissues (9). More recently, Gpr182 was found to be highly expressed in developing murine and zebrafish endothelium and enriched in mammary tumor endothelium compared with normal mammary endothelium (10–12). Additionally, Gpr182 was identified among a group of factors that are significantly altered in a zebrafish model of myeloid leukemia (13). Thus, a significant advance of the current study is to map the expressional profile of Gpr182 using an in vivo mammalian reporter model, in which, in addition to the endothelium of numerous tissues, we observed expression within the gastrointestinal tract epithelia.
The epithelium of the gastrointestinal tract is one of the most dynamic tissues in the adult body and is primarily responsible for the absorption of dietary nutrients and also for fulfilling important endocrine, immune, and protective barrier functions. To maintain its proper functions, the intestinal epithelium must undergo continuous turnover, with the entire small intestinal epithelium renewing every week in humans and in mice. This constant renewal is driven by an active population of intestinal stem cells (ISCs) that are located at the base of the crypts of Lieberkühn, where they give rise to rapidly dividing daughter transit-amplifying progenitor cells that differentiate into the absorptive or secretory lineages responsible for functions of the intestine (14–17). Current views hold that 2 distinct pools of ISCs exist in the intestinal epithelium: the crypt base columnar (CBC) ISCs, which are positioned between differentiated Paneth cells and mediate normal homeostatic renewal, and “damage-resistant” ISCs, which act as reserve ISCs that are activated following injury (14, 15, 17, 18). With the discovery of numerous ISC-specific markers including leucine-rich repeat containing G protein–coupled receptor 5 (Lgr5), leucine-rich repeats and Ig-like domains 1 (Lrig1), achaete-scute family bHLH transcription factor 2 (Ascl2), olfactomedin 4 (Olfm4), HOP homeobox (Hopx), telomerase reverse transcriptase (Tert), BMI1 proto-oncogene, polycomb ring finger (Bmi1), and SRY (sex-determining region Y)-box 9 (Sox9), our understanding of both of these ISC populations has drastically expanded over the past decade (19–26). It is evident that the activity and proliferation of these ISCs must be tightly controlled by numerous signaling pathways and redundant mechanisms in order to maintain homeostasis in the dynamic gut microenvironment (14, 27). Furthermore, oncogenic mutations specifically in ISCs can drastically enhance adenoma formation in mice (20, 28). Thus, defining the factors that regulate ISC proliferation and survival is critical in order to better understand ISC function during homeostasis, damage repair, and cancer and, hence, to better therapeutically target these ISCs.
In this study, we aimed to identify novel functions of the orphan GPR182 in vivo by first mapping the murine Gpr182 expression pattern during development and in adulthood, and then next elucidating the effects of Gpr182 reduction on intestinal homeostasis, regeneration, and adenoma formation.
Results
Orphan GPR182 is widely expressed throughout development and adulthood.
The Gpr182tm2a(KOMP)Wtsi/+ mouse (hereafter referred to as the Gpr182lacZ/+ mouse) was generated and used to both map the murine Gpr182 expression pattern during development and adulthood as well as act as a loss-of-function model (29). A gene trap cassette bearing an En2 splice acceptor upstream of a lacZ reporter was knocked into the murine Gpr182 locus immediately downstream of exon 1, resulting in expression of lacZ instead of the endogenous protein–coding sequence in exon 2 (Figure 1A). When crossed to achieve homozygosity, the Gpr182lacZ/lacZ animals had a significant reduction of approximately 85% in endogenous Gpr182 expression in jejunum and negligible levels in other tissues compared with Gpr182+/+ mice (Supplemental Figure 1, A and B; supplemental material available online with this article; doi:10.1172/JCI87588DS1). When crossed with the ubiquitously expressed CMV-Cre mouse, the resulting Gpr182Δ/Δ CMV-Cre mice had undetectable Gpr182 expression in the jejunum compared with levels in Gpr182+/+ Cre controls (Supplemental Figure 1A). Like the Gpr182lacZ/lacZ mice, the Gpr182Δ/Δ CMV-Cre–KO mice were born at expected Mendelian ratios and lived to adulthood with no observed phenotypic abnormalities.
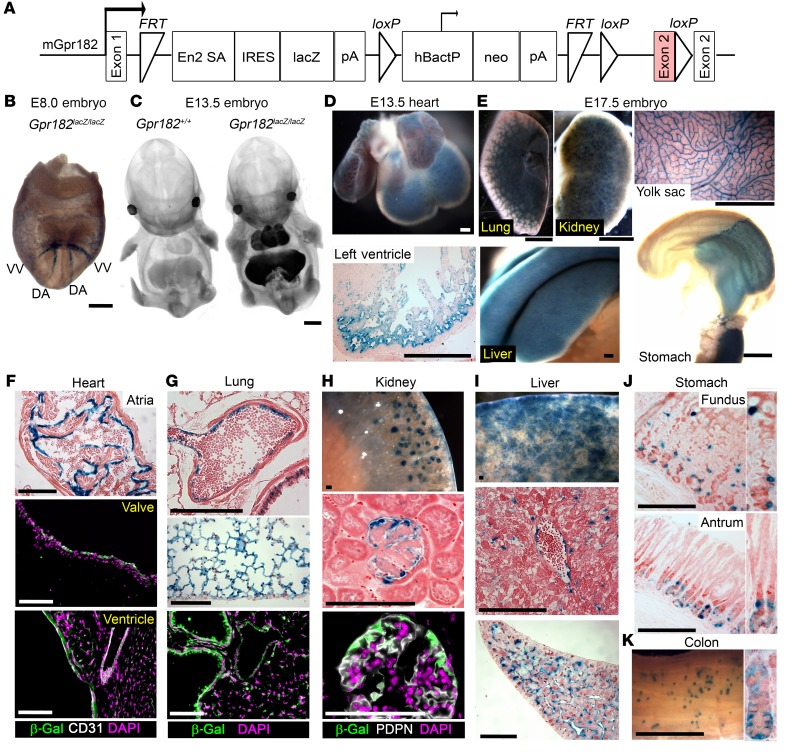
Figure 1. Murine Gpr182 expression profile during development and adulthood.
(A) Targeting vector of the Gpr182tm2a(KOMP)Wtsi/+ lacZ reporter mouse model (29, 62, 63). The GPR182 protein–coding region is shaded in pink. mGPR182, murine GPR182. (B) Whole-mount X-gal staining of E8 Gpr182lacZ/lacZ embryo. DA, dorsal aorta; VV, vitelline vein. (C) Optical projection tomography of whole-mount X-gal–stained WT and Gpr182lacZ/lacZ E13.5 embryos. X-Gal staining in Gpr182lacZ/lacZ E13.5 heart (D) and E17.5 lung, kidney, liver, yolk sac, and stomach, pancreas, and duodenum (E). Representative Gpr182 lacZ expression in adult heart (F), lung (G), kidney (H), liver (I), glandular stomach (J), and colon (K) stained with X-gal and/or β-gal (green). Sections were costained with DAPI (purple) and either the endothelial marker CD31 (F) or the podocyte marker podoplanin (PDPN) (H). X-Gal–stained sections were counterstained with eosin (F–H) or Neutral Red (D and I–K). Scale bars: 200 μm (B and D), 1 mm (C, E, and K), and 100 μm (F–J).
We detected expression of Gpr182 lacZ by β-gal activity as early as E8.0 in dorsal aorta and vitelline veins (Figure 1B and Supplemental Figure 2A), similar to previously published ISH observations (10). By E13.5, we observed Gpr182 expression in numerous organs including heart, lung, liver, aorta, and carotid arteries (Figure 1C and Supplemental Figure 2, B and C). Expression of Gpr182 in embryonic heart was detected in both atria and ventricles, particularly in the ventricular trabecular region (Figure 1D). At E17.5, Gpr182 localization remained prominent in the heart, lungs, and liver, but was also observed in embryonic kidneys, glandular stomach, intestine, spleen, and yolk sac vasculature (Figure 1E and Supplemental Figure 2D).
In adult mice Gpr182 was widely expressed throughout the body. Cardiac expression was localized to the endocardium of the atria and ventricles, as well as in the heart valves and coronary arteries (Figure 1F and Supplemental Figure 2E). Notably, we observed little to no staining in cardiomyocytes, fibroblasts, epicardium, or capillary endothelium. Lungs expressed high levels of Gpr182 primarily in endothelial cells (Figure 1G, Supplemental Figure 1C and Supplemental Figure 2F). Though Gpr182 was expressed in renal tubules during late gestation (Supplemental Figure 2D), we found that adult renal localization was primarily enriched in the glomeruli, where it appeared strikingly specific to podocytes (Figure 1H and Supplemental Figure 2G). Sinusoidal endothelial cells, rather than hepatocytes, were the primary cell type expressing Gpr182 in the liver (Figure 1I). We found that Gpr182 was expressed throughout the fundus and antrum (Figure 1J, Supplemental Figure 2H, and Supplemental Figure 3). Interestingly, this Gpr182 localization was detected in gastric epithelial cells primarily near the base of the glands in both fundus and antrum, although we observed a few lacZ+ cells higher up the gland in the fundus. We did not detect staining in acid-secreting parietal cells (Figure 1J and Supplemental Figure 3, B and C). We detected X-gal staining in the distal colon, also at the base of the crypts (Figure 1K and Supplemental Figure 3). Gpr182 X-gal staining was also robust in spermatocytes of the adult testis (Supplemental Figure 2I). Spleen and lymph nodes revealed endothelial Gpr182 localization, with little to no detectable staining in mature or developing hematopoietic and lymphoid lineages (Supplemental Figure 2, J and K). We observed little to no X-gal staining in skeletal muscle, pancreas, brain, spinal cord, or dorsal root ganglia of adult mice (Supplemental Figure 2, L–O).
Gpr182 is expressed throughout the intestine and is enriched in small ISCs.
Whole-mount X-gal staining and histology revealed an interesting pattern of staining in the crypt epithelial cells of the small intestine, with only a few positively stained secretory cells in the villi and relatively little staining in enterocytes or submucosal endothelial cells (Figure 2, A and B, and Supplemental Figure 3). We found that β-gal staining was most intense in the stem cell zone at the base of crypts of Lieberkühn compared with the transit-amplifying cells residing higher in the crypt/villus axis (Figure 2, C–E). More specifically, β-gal–stained thin cells with elongated nuclei were intercalated between Lysozyme+ Paneth cells (Figure 2E), closely resembling the pattern of other markers of the active CBC ISCs (19–22). The gastrointestinal lacZ expression pattern of Gpr182Δ/+ CMV-Cre and Gpr182Δ/Δ CMV-Cre mice was similar to that observed in Gpr182lacZ/+ mice (Supplemental Figure 3).
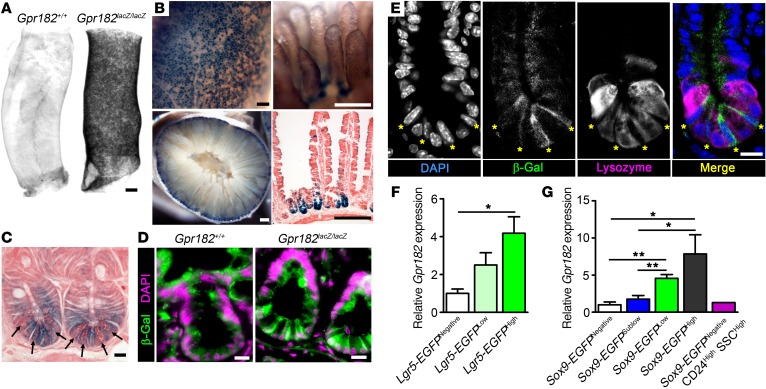
Figure 2. Gpr182 is enriched in CBC ISCs.
(A) Optical projection tomography of X-gal–stained optically cleared duodenum from Gpr182+/+ and Gpr182lacZ/lacZ adult mice. (B) Representative whole-mount X-gal–stained small intestine from adult Gpr182lacZ/lacZ mice. (C) Cross-sectional X-gal staining of adult Gpr182lacZ/lacZ small intestine crypts counterstained with eosin. Arrows indicate Paneth cells. (D) Immunofluorescence of β-gal (green) and DAPI (purple) of intestine from adult Gpr182+/+ and Gpr182lacZ/lacZ mice. (E) Crypt from Gpr182lacZ/lacZ jejunum costained with β-gal (green), the Paneth cell marker Lysozyme (magenta), and DAPI (blue). Yellow asterisks mark CBC ISCs as defined by morphologic position, elongated nuclei, and Lysozyme negativity. (F) Relative Gpr182 expression in isolated epithelial cell populations from (F) Lgr5-EGFP mice and (G) Sox9-EGFP–Tg mice. Differentiated epithelium (Lgr5-EGFPNegative); early progenitors (Lgr5-EGFPLow); CBC ISCs (Lgr5-EGFPHigh or Sox9-EGFPLow); enterocytes/goblet cells (Sox9-EGFPNegative); transit-amplifying cells (Sox9-EGFPSublow); enteroendocrine/Tuft/+4 ISCs (Sox9-EGFPHigh); and Paneth cells (Sox9-EGFPNegative CD24High SSCHigh). Expression was normalized to Lgr5-EGFPNegative or Sox9-EGFPNegative cell populations and Actb. Biological replicates: n = 2–5 mice per population (F–G). Scale bars: 500 μm (A), 200 μm (B), and 10 μm (C–E). *P < 0.05 and **P < 0.01, by 1-way ANOVA with Tukey’s multiple comparisons test.
To further verify the enrichment of Gpr182 in CBC ISCs in models without altered Gpr182 expression, the Lgr5-EGFP and Sox9-EGFP BAC–Tg mice were used to isolate different intestinal epithelial cell populations with distinct levels of EGFP, as previously described (18, 19, 25, 30–32). Gpr182 transcripts were significantly enriched in the Lgr5-EGFPHigh CBC ISC population compared with transcript expression of the differentiated Lgr5-EGFPNegative population, with intermediate expression in the Lgr5-EGFPLow progenitors (Figure 2F). Likewise, Gpr182 mRNA was enriched in the CBC ISC population (Sox9-EGFPLow) when compared with populations containing transit-amplifying cells (Sox9-EGFPSublow) and a mixed population of enterocytes and goblet cells (Sox9-EGFPNegative) (Figure 2G). Furthermore, Gpr182 expression was very low in isolated Paneth cells. Consistent with the previous observation of rare β-gal+ cells in the villus region, we found that Gpr182 also was enriched in the Sox9-EGFPHigh cell population, which is a mixed population of enteroendocrine cells, tuft cells, and activatable reserve ISCs (18, 31, 32). While Gpr182 is not exclusively expressed in CBC ISCs, like Lgr5, these data provide evidence that Gpr182 is enriched in CBC ISCs at the crypt base, as well as in the Sox9-EGFPHigh cell population that contains activatable reserve ISCs.
Gpr182 reduction does not alter intestinal proliferation during homeostasis in vivo.
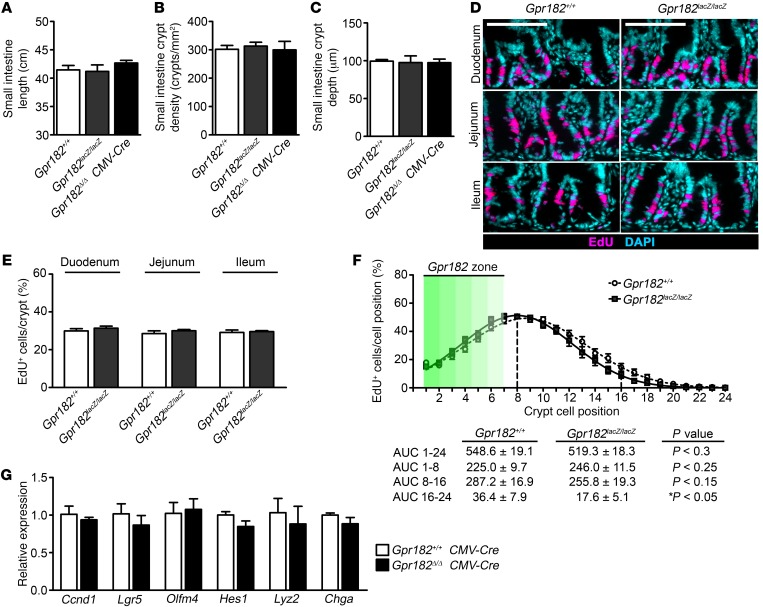
To evaluate the potential roles of GPR182 in ISCs, we first characterized intestinal proliferation during homeostasis in Gpr182lacZ/lacZ mice and, consistent with the highly regulated nature of the ISC niche, we found no significant effects of Gpr182 reduction on homeostatic intestinal proliferation. For example, small intestine and colon lengths were unchanged in the Gpr182lacZ/lacZ animals compared with that observed in controls (Figure 3A and Supplemental Figure 4A). Intestinal crypt density, as a proxy for crypt fission, was unaltered between the genotypes (Figure 3B). Crypt depth was also unchanged, consistent with no significant differences in basal proliferation, as evidenced by similar numbers of EdU+ cells per crypt, across the genotypes in both the small intestine and colon (Figure 3, C–E, and Supplemental Figure 4, B–D). Likewise, there was no difference in the number of phosphorylated histone H3+ (p–histone H3+) cells per crypt in the small intestine (Gpr182+/+: 1.26 ± 0.13 cells/crypt and Gpr182lacZ/lacZ: 1.33 ± 0.10 cells/crypt, n = 5). The β-gal+ cells were primarily located in the lowest 4 cell positions of the crypt, and there were no significant changes in the position of proliferating transit-amplifying zone cells between the genotypes (Figure 3F). Additionally, expression levels of cyclin D1 (Ccnd1) and Lgr5 were unchanged in the Gpr182Δ/Δ CMV-Cre mouse jejunum compared with levels detected in Gpr182+/+ CMV-Cre controls (Figure 3G). Expression of the Notch targets Olfm4 and Hes1 as well as the differentiation markers Chga and Lysozyme were not significantly altered in these mice. Collectively, these data demonstrate that genetic reduction of Gpr182 does not lead to altered intestinal proliferation in vivo, which is consistent with the highly regulated control of intestinal proliferation during homeostasis.
Figure 3. Reduced Gpr182 does not alter basal proliferation in vivo.
(A) Length of small intestines from adult Gpr182+/+, Gpr182lacZ/lacZ, and Gpr182Δ/Δ CMV-Cre mice. (B) Morphometric quantification of crypt density and (C) histological quantification of crypt depth among Gpr182 genotypes. (D) Representative images and (E) EdU incorporation quantification of intestinal proliferation in Gpr182+/+ and Gpr182lacZ/lacZ animals. (F) Analysis of the cellular position of EdU+ cells along the crypt axis expressed as a percentage of the total number of cells in that position in all crypts. n = 20–60 open crypts per region per mouse. The Gpr182 zone (green gradient) is the relative cellular position of β-gal+ cells along the crypt axis. (G) Relative expression of Ccnd1, Lgr5, Olfm4, hairy and enhancer of split 1 (Hes1), chromogranin A (Chga), and Lysozyme (Lyz2) in whole jejunum from Gpr182+/+ CMV-Cre and Gpr182Δ/Δ CMV-Cre mice. Expression was normalized to Gpr182+/+ and 18S. Biological replicates: n = 3–5 mice per genotype. Scale bars: 100 μm. Significance was determined by either 1-way ANOVA with Tukey’s multiple comparisons test.
Mice with Gpr182 knockdown show intestinal hyperproliferation during intestinal regeneration following irradiation injury.
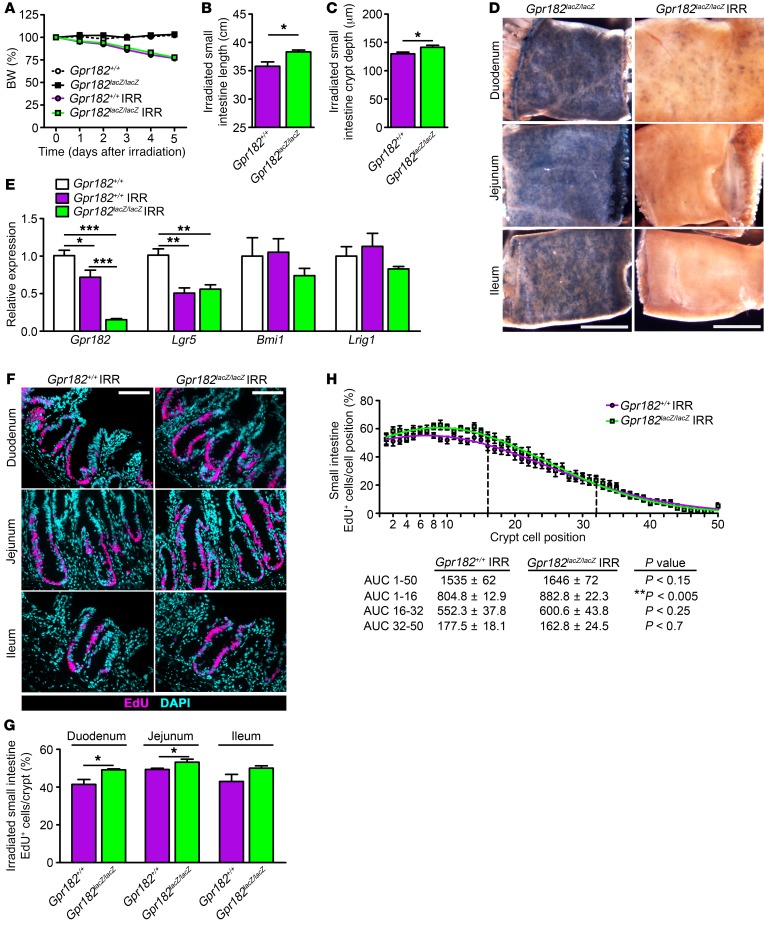
To test whether GPR182 may be involved with regulating proliferation during intestinal regeneration following damage, Gpr182+/+ and Gpr182lacZ/lacZ mice were exposed to a high dose (14 Gy) of ionizing radiation delivered to the abdomen. It is well established that intestinal irradiation (IRR) causes apoptosis of actively dividing cells, leading to the temporary loss of proliferative crypts and villi, with subsequent surviving ISCs expanding and regenerating the damaged intestinal epithelium and peak proliferation occurring 5 days after IRR (18, 32–35). Both Gpr182+/+ and Gpr182lacZ/lacZ IRR animals lost approximately 20% of their initial BW 5 days after IRR compared with the non-IRR controls (Figure 4A). Interestingly, the small intestine and colon were significantly longer in the Gpr182lacZ/lacZ IRR mice compared with controls (Figure 4B and Supplemental Figure 4A). During regeneration, Gpr182lacZ/lacZ IRR mouse small intestines had significantly increased crypt depth compared with that seen in IRR control mice (Figure 4C). While X-gal staining was observed in crypts throughout the small intestine in non-IRR Gpr182lacZ/lacZ mice (Figure 4D and Supplemental Figure 3A), the vast majority of regenerative crypts in the Gpr182lacZ/lacZ IRR mice were X-gal negative (Figure 4D). Gpr182 expression in whole jejunum was decreased at this phase of regeneration in Gpr182+/+ mice compared with expression in non-IRR controls (Figure 4E). Likewise, expression of the CBC ISC marker Lgr5 was decreased in IRR tissue, but unchanged between genotypes. Expression of the activatable ISC markers Bmi1 and Lrig1 was unchanged between the genotypes, although expression trended lower in the Gpr182lacZ/lacZ IRR mouse jejunum.
Figure 4. Decreased Gpr182 leads to hyperproliferation during the regeneration phase after irradiation-induced injury.
Adult Gpr182+/+ (purple) and Gpr182lacZ/lacZ (green) mice were challenged with a single 14-Gy dose of radiation (IRR) to the abdomen. (A) BW changes (percentage of pre-IRR weight) of Gpr182+/+ and Gpr182lacZ/lacZ mice for 5 days following IRR compared with BW of non-IRR control mice. Small intestine length (B) and regenerating crypt depth (C) in IRR-treated Gpr182+/+ and Gpr182lacZ/lacZ animals. (D) Whole-mount X-gal–stained small intestine from non-IRR–treated Gpr182lacZ/lacZ and IRR-treated Gpr182lacZ/lacZ animals 5 days after IRR. (E) Relative expression of Gpr182 and ISC markers Lgr5, Bmi1, and Lrig1 in whole jejunum from Gpr182+/+ IRR, Gpr182lacZ/lacZ IRR, and non-IRR Gpr182+/+ animals. Expression was normalized to non-IRR Gpr182+/+, Gapdh, and 18S. (F) Representative images and (G) EdU quantification of intestinal proliferation among IRR Gpr182+/+ and Gpr182lacZ/lacZ animals. (H) Analysis of the cellular position of EdU+ cells along the crypt axis expressed as a percentage of the total number of cells in that position in all regenerating crypts. n = 10–60 open crypts per region per mouse. Biological replicates: n = 4–5 mice per genotype. Scale bars: 5 mm (D) and 100 μm (F). *P < 0.05, **P < 0.01, and ***P < 0.001, by 2-tailed Student’s t test (B, C, and G), 1-way ANOVA with Tukey’s multiple comparisons test (E), or Mann-Whitney t test of the AUC (H).
There were significantly more EdU+ cells per crypt in the regenerating duodenum and jejunum of Gpr182lacZ/lacZ IRR mice compared with Gpr182+/+ IRR controls, with comparable, but not statistically significant, trends in the ileum and colon (Figure 4, F and G, and Supplemental Figure 4, C and D). The percentage of EdU+ cells was increased in the bottom third of the regenerating crypt (Figure 4H). The number of p–histone H3+ cells per crypt was also significantly increased in the regenerating Gpr182lacZ/lacZ duodenum (Gpr182+/+: 2.51 ± 0.39 cells/crypt and Gpr182lacZ/lacZ: 3.74 ± 0.18 cells/crypt, n = 4–5; *P <0.05). Together, these data indicate a role for GPR182 in limiting proliferation during intestinal epithelial regeneration.
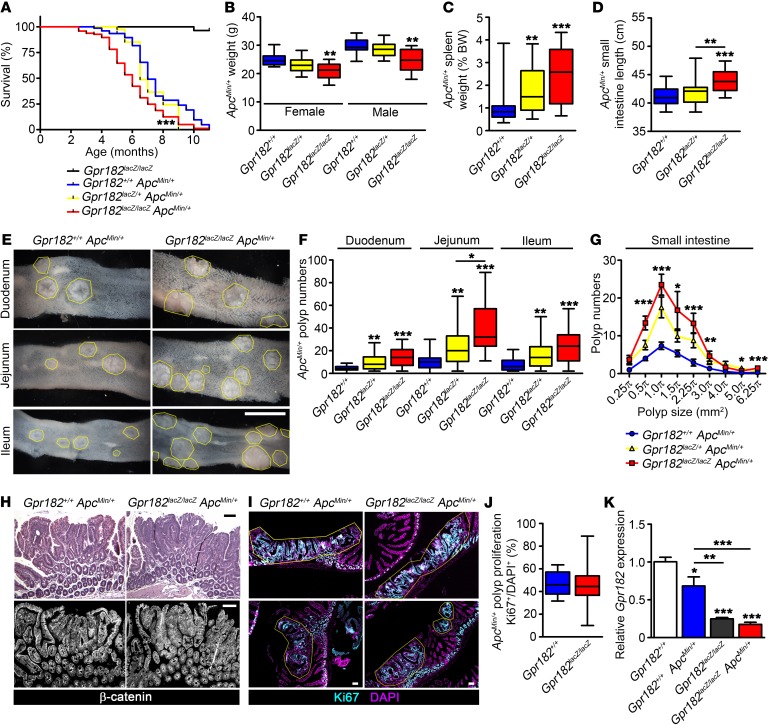
Mice with reduced Gpr182 that carry the ApcMin/+ allele have increased intestinal adenoma burden and decreased survival.
Since Gpr182 acts to inhibit proliferation during regeneration, we reasoned that Gpr182 levels may also be important in deregulated proliferation during disease, such as intestinal adenoma and carcinoma. Gpr182lacZ/lacZ adults up to 2 years of age did not develop spontaneous intestinal adenomas (n = 6; data not shown). Thus, to investigate whether Gpr182 is involved in intestinal adenoma initiation and progression, Gpr182lacZ/lacZ mice were crossed with the well-characterized ApcMin mouse model, which develops spontaneous intestinal and colonic adenomas caused by aberrant activation of the Wnt/β-catenin signaling pathway (36, 37). Both male and female 5-month-old Gpr182lacZ/+ ApcMin/+ and Gpr182lacZ/lacZ ApcMin/+ mice were characterized for adenoma formation and compared with Gpr182+/+ ApcMin/+ controls. The majority of Gpr182+/+ ApcMin/+ (71 of 76) and Gpr182lacZ/+ ApcMin/+ (37 of 39) animals survived past 5 months of age (Figure 5A). However, significantly fewer Gpr182lacZ/lacZ ApcMin/+ (62 of 96) mice survived past 5 months, indicating that genetic reduction of Gpr182 exacerbates the lethality of C57BL/6-ApcMin/+ mice. Furthermore, both Gpr182lacZ/lacZ ApcMin/+ male and female mice had significantly lower BW at 5 months compared with the BW of sex-matched controls (Figure 5B). The spleens from both Gpr182lacZ/+ and Gpr182lacZ/lacZ animals weighed significantly more when compared with spleen weights of controls (Figure 5C). The small intestines were also significantly longer in the Gpr182lacZ/lacZ ApcMin/+ animals compared with control small intestine lengths (Figure 5D), which was similar to the response in mice treated with IRR (Figure 4C). Together, these data suggest that attenuated Gpr182 expression is detrimental to the health and survival of ApcMin/+ animals. For both sexes, macroscopic polyp numbers throughout the intestine were 3- to 4-fold higher in Gpr182lacZ/lacZ ApcMin/+ mice compared with polyp numbers in Gpr182+/+ ApcMin/+ controls (Figure 5, E and F). Moreover, the haploinsufficient Gpr182lacZ/+ ApcMin/+ mice also showed significantly more polyps in the small intestine than did Gpr182+/+ ApcMin/+ controls, further supporting the idea of a genetic dose-dependent inverse correlation between Gpr182 levels and polyp formation. There were significantly more polyps of all sizes in the Gpr182lacZ/lacZ ApcMin/+ mice than in the Gpr182+/+ ApcMin/+ mice, but the relative distribution of small and large polyps was unchanged between these genotypes (Figure 5G).
Figure 5. Reduced Gpr182 increases lethality and polyp numbers in ApcMin/+ mice.
(A) Kaplan-Meier survival curve comparing Gpr182lacZ/lacZ Apc+/+ (black), Gpr182+/+ ApcMin/+ (blue), Gpr182lacZ/+ ApcMin/+ (yellow), and Gpr182lacZ/lacZ ApcMin/+ (red) animals. (B) BW of 5-month-old female and male ApcMin mice from different Gpr182 genotypes. (C) Spleen weight normalized to BW for Gpr182+/+ ApcMin/+, Gpr182lacZ/+ ApcMin/+, and Gpr182lacZ/lacZ ApcMin/+ mice. (D) Small intestine length from Gpr182lacZ/lacZ ApcMin/+ animals compared with controls. (E) Representative images of the number and size of adenomas from Gpr182+/+ ApcMin/+ and Gpr182lacZ/lacZ ApcMin/+ mouse intestines. (F) Quantification of polyp numbers throughout the small intestines of Gpr182+/+ ApcMin/+, Gpr182lacZ/+ ApcMin/+, and Gpr182lacZ/lacZ ApcMin/+ mice. (G) Distribution of the number of polyps throughout the small intestine based on approximate polyp size. (H) Histology for 5-month-old Gpr182+/+ ApcMin/+ and Gpr182lacZ/lacZ ApcMin/+ jejunum polyps labeled with H&E and β-catenin (white). (I and J) Polyp proliferation assessed by Ki67 (blue) staining normalized to DAPI (purple) in Gpr182+/+ ApcMin/+ and Gpr182lacZ/lacZ ApcMin/+ mice. (K) Gpr182 expression in polyps from jejunum of Gpr182+/+ ApcMin/+ and Gpr182lacZ/lacZ ApcMin/+ mice compared with expression levels in Apc+/+ jejunum. Expression was normalized to Gpr182+/+, Gapdh, and 18S. Scale bars: 5 mm (E) and 100 μm (H and I). Biological replicates: n = 25–100 mice per genotype (A); n = 10–20 mice per genotype per sex (B); n = 700–1,700 polyps from 20 to 35 mice per genotype (C, D, F, and G); n = 15–60 polyps from 5 mice (J); and n = 3–5 mice per genotype (K). *P < 0.05, **P < 0.01, and ***P < 0.001, by Mantel-Cox test (A), 1-way ANOVA Kruskal-Wallis test with Dunn’s multiple comparisons test (B–G), Mann-Whitney t test (J), or 1-way ANOVA with Tukey’s multiple comparisons test (K).
The ApcMin/+ model shows fewer and more sporadic polyps in the colon after 5 months. Consistently, colon polyps occurred in 66% of the Gpr182+/+ ApcMin/+ mice assessed, while the incidence of colon polyps was 84% in the Gpr182lacZ/lacZ ApcMin/+ mice. Moreover, we observed a 2-fold increase in the number of colon polyps in the Gpr182lacZ/lacZ ApcMin/+ animals compared with colon polyp numbers in the Gpr182+/+ ApcMin/+ controls (2.0 ± 0.3 versus 1.0 ± 0.2 polyps, respectively; *P < 0.05). Together, these data indicate that decreased Gpr182 expression is sufficient to exacerbate ApcMin/+ adenoma formation and burden throughout the intestine, further supporting the idea that Gpr182 has a role in negatively regulating intestinal proliferative capacity.
While adenoma formation was increased in the Gpr182lacZlacZ ApcMin/+ animals, no morphological differences were observed in Gpr182lacZ/lacZ polyps compared with Gpr182+/+ polyps (Figure 5H). Furthermore, there were no qualitative or quantitative differences in overall proliferation within individual polyp cross-sections between the Gpr182+/+ and Gpr182lacZ/lacZ animals (Figure 5, I and J), although polyp proliferation in the Gpr182lacZlacZ ApcMin/+ mice appeared more heterogeneous. Gpr182 expression was significantly decreased in ApcMin/+ polyps compared with normal WT jejunum (Figure 5K). Consistent with the highly mosaic nature of adenomas (38, 39), macroscopic and histologic analysis of Gpr182-associated X-gal and β-gal antibody staining revealed a mosaic staining pattern between polyps and within individual polyps (Supplemental Figure 5). We consistently observed X-gal staining in microadenomas (Supplemental Figure 5C), and polyp Ki67-labeled proliferation appeared inversely correlated with Gpr182 β-gal staining, supporting the notion of heterogeneous Gpr182 promoter activity (Supplemental Figure 5, D and E). Together, these data support the idea that GPR182 plays a role in preventing adenoma formation, but it does not appear to directly alter adenoma progression.
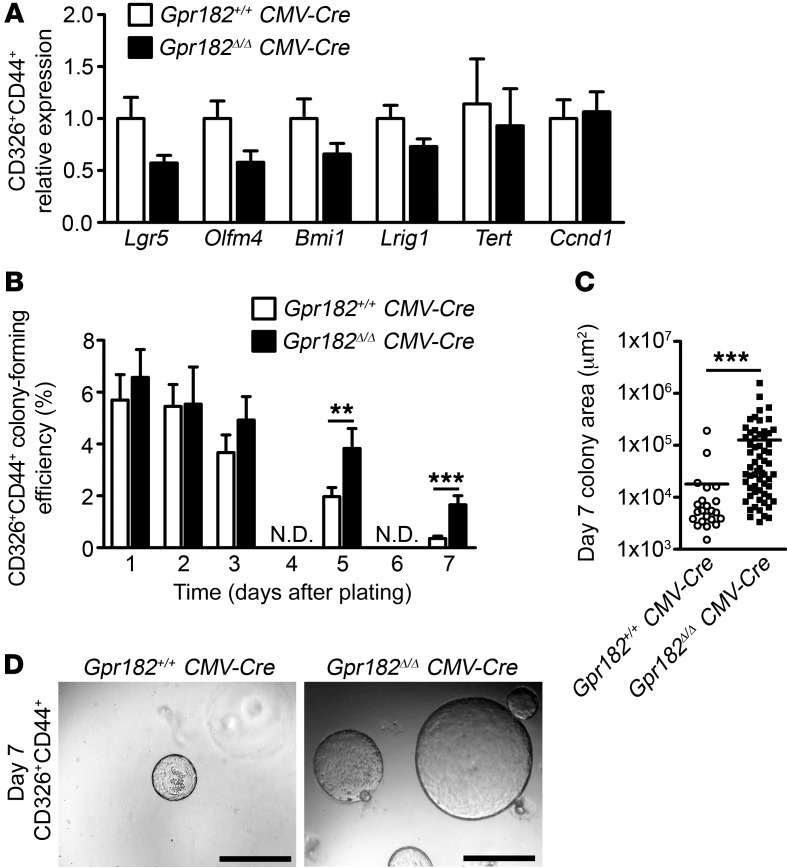
Loss of Gpr182 increased single-cell organoid growth efficiency.
Considering the proliferative phenotypes observed during regeneration and adenoma formation in Gpr182lacZ/lacZ animals, we sought to determine whether the loss of Gpr182 altered intestinal stem cell numbers and/or proliferative capacity. We isolated single crypt epithelial cells from Gpr182+/+ CMV-Cre and Gpr182Δ/Δ CMV-Cre jejunum using FACS to select CD326+CD44+ cells, as previously reported (40, 41). Neither the percentage of CD326+ epithelial cells nor the proportion of CD44– and CD44+ epithelial cells was altered with the loss of Gpr182 (Supplemental Figure 6, A and B). We observed no significant differences in the expression of either active (Lgr5 and Olfm4) or reserve (Bmi1, Tert, Lrig1) ISC markers in the CD326+CD44+ cell population from Gpr182+/+ and Gpr182Δ/Δ mice (Figure 6A). Single CD326+CD44+ and CD326+CD44– cells were seeded in Matrigel and cultured in supplemented media for 7 days to assay ISC numbers, survival, and proliferative capacity, as previously reported (40–42). As expected, CD326+CD44– cells formed very few colonies that were viable after 7 days, with no significant difference observed between Gpr182+/+ and Gpr182Δ/Δ mice (Supplemental Figure 6, C–E). Initially the efficiency of CD326+CD44+ cells in forming organoids was similar between Gpr182+/+ and Gpr182Δ/Δ mice (Figure 6B), suggesting a relatively equal distribution of ISCs and/or progenitors between the genotypes. However, after 5 days and 7 days in culture, when only organoids initiated and maintained by self-renewing ISCs are present, Gpr182Δ/Δ mice had significantly more viable and larger organoids than did Gpr182+/+ mice (Figure 6, B–D). A number of these Gpr182Δ/Δ organoids were X-gal+ and had an increased number of p–histone H3+ cells (Supplemental Figure 6, E and F). These data demonstrate that isolated ISCs from these animals have increased survival and proliferative potential ex vivo and further the conclusion that GPR182 acts to regulate the proliferative capacity of ISCs in the small intestine.
Figure 6. Loss of Gpr182 leads to increased growth efficiency of single-crypt epithelial cells ex vivo.
(A) Relative expression of ISC markers in isolated CD326+CD44+ cells from Gpr182+/+ CMV-Cre and Gpr182Δ/Δ CMV-Cre jejunum. Expression was normalized to Gpr182+/+ CMV-Cre, Actb, and 18S. (B) Growth efficiency of single CD326+CD44+ cells isolated from Gpr182+/+ CMV-Cre and Gpr182Δ/Δ CMV-Cre jejunum and cultured for 7 days ex vivo. One thousand CD326+CD44+ cells were initially plated in triplicate per mouse. (C) Area quantification of CD44+ colonies after 7 days in culture. (D) Representative images of organoid density and size after 7 days in culture. Scale bars: 500 μm. Biological replicates: n = 3 mice. Significance was determined by 2-tailed Student’s t test (A and B) or Mann-Whitney t test (C).
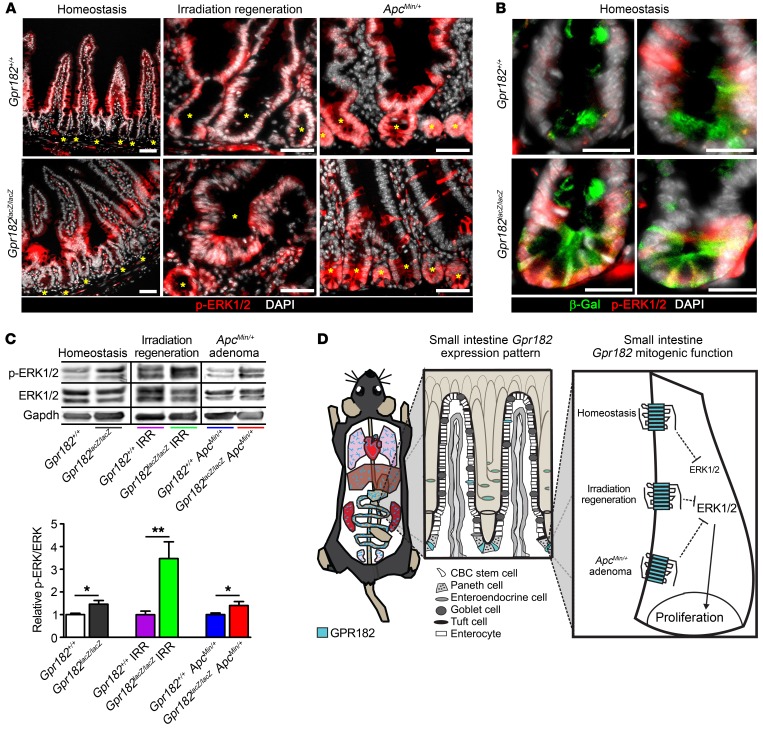
Decreased GPR182 is correlated with aberrant ERK1/2 activation during homeostasis, regeneration, and in ApcMin/+ mice.
Class A GPCRs commonly activate or inhibit the MAPK signaling cascade, which has numerous downstream effects on cell proliferation and survival, but the signal transduction pathways of the orphan GPR182 have not been characterized. ERK1/2 signaling was localized in both epithelial and nonepithelial cells in the small intestine. During homeostasis, epithelial p-ERK1/2 appeared highest in the transit-amplifying zone and right above the crypt, with relatively little activity in the crypt base ISCs (Figure 7, A and B) (43). However, during regeneration and in ApcMin/+ animals, p-ERK1/2 was much higher throughout the epithelium, especially at the base of crypts (Figure 7A). Mosaic p-ERK1/2 was also observed within adenomas from ApcMin/+ animals (Supplemental Figure 5F). We observed occasional β-gal+ p-ERK1/2+ ISCs in Gpr182lacZ/lacZ mice during homeostasis (Figure 7B). Significantly increased p-ERK1/2 was observed in Gpr182lacZ/lacZ jejunum compared with Gpr182+/+ controls, and this increase was observed during homeostasis, regeneration, and in ApcMin/+ polyps (Figure 7C). Interestingly, the increased organoid growth efficiency in Gpr182Δ/Δ CD326+CD44+ single cells was dependent on the addition of exogenous EGF (Supplemental Figure 7, A–C). The expression of Egfr was not altered in Gpr182Δ/Δ or Gpr182lacZ/lacZ IRR jejunum compared with expression in controls (Supplemental Figure 7D). These data suggest that EGF-mediated MAPK signaling is still required for the increased proliferative capacity of these Gpr182-null organoids.
Figure 7. Reduced Gpr182 leads to elevated ERK1/2 signaling upstream of the hyperproliferative intestinal crypt microenvironment.
(A) Immunofluorescence of p-ERK1/2 (red) staining in Gpr182+/+ and Gpr182lacZ/lacZ intestine during homeostasis, 5 days after IRR, and in ApcMin/+ animals. Yellow asterisks mark crypts. (B) Colocalization of β-gal+ (green) ISCs with p-ERK1/2 in Gpr182lacZ/lacZ compared with Gpr182+/+ crypts. Scale bars: 50 μm (A) and 20 μm (B). (C) Representative immunoblots and quantification of relative p-ERK1/2 expression in unchallenged, 5 days post-IRR, and ApcMin/+ polyps from whole jejunum lysates derived from Gpr182+/+ and Gpr182lacZ/lacZ mice. Samples were normalized to total ERK1/2 and Gpr182+/+. GAPDH was used as a loading control. Biological replicates: n = 5–10 mice per genotype per condition. *P < 0.05 and **P < 0.01, by unpaired t test. (B) Model summarizing GPR182 β-gal expression pattern (blue) in the whole mouse (heart, lungs, liver, stomach, small intestine, colon, kidney, and testis), with more specific expressional detail within the small intestine. GPR182 functions to inhibit ERK1/2 signaling to regulate the proliferative capacity of the intestine.
Unlike ERK1/2 signaling, the phosphorylation of other signaling kinases, including p38 MAPK, AKT, STAT3, and YAP, appeared relatively unaltered in whole jejunum in Gpr182+/+ and Gpr182lacZ/lacZ mice (Supplemental Figure 8). Therefore, the correlation between decreased Gpr182 and increased ERK1/2 signaling, along with the crypt and ISC localization of p-ERK1/2, especially during regeneration and in ApcMin/+ mice, suggests that GPR182 regulation of ERK1/2 occurs upstream of the increased proliferative capacity we observed in the Gpr182lacZ/lacZ mice (Figure 7D).
GPR182 expression is reduced in numerous human carcinomas, including colon adenocarcinomas.
Considering that reduced Gpr182 increased the intestinal proliferative capacity in mice, we sought to determine whether GPR182 expression levels are changed in human carcinomas. First, we found that GPR182 expression was low in numerous human colorectal carcinoma cell lines, including CaCo2, HT29, Colo205, and SW480 (Supplemental Figure 9). Second, RNA-sequencing (RNA-seq) data from The Cancer Genome Atlas (TCGA) (44, 45) confirmed that GPR182 expression was significantly lower in colon adenocarcinoma when compared with expression levels in patient-matched normal colon tissue (Figure 8) and nonmatched samples (normal colon: 0.86 ± 0.17, n = 41; colon adenocarcinoma: 0.24 ± 0.04, n = 285; ***P < 0.0001, by Mann-Whitney t test). GPR182 expression was also significantly downregulated in numerous carcinomas in tissues other than the colon, including bladder, breast, bile duct, kidney, liver, lung, pancreas, thyroid, and uterine carcinomas (Figure 8). While loss of GPR182 may not drive carcinoma initiation in mice, these data further support the idea that low GPR182 expression is inversely correlated with increased proliferative capacity in mice and humans.
Figure 8. Low GPR182 expression in human carcinomas.
Relative GPR182 expression determined by TCGA RNA-seq of patient-matched normal (N) and tumor (T) tissue. Bladder urothelial carcinoma (BLCA), n = 19; breast invasive carcinoma (BRCA), n = 98; cholangiocarcinoma (CHOL), n = 9; colon adenocarcinoma (COAD), n = 26; kidney chromophobe (KICH), n = 25; kidney renal clear cell carcinoma (KIRC), n = 71; kidney renal papillary cell carcinoma (KIRP), n = 32; liver hepatocellular carcinoma (LIHC), n = 50; lung adenocarcinoma (LUAD), n = 57; lung squamous cell carcinoma (LUSC), n = 50; pancreatic adenocarcinoma (PAAD), n = 58; rectum adenocarcinoma (READ), n = 6; stomach adenocarcinoma (STAD), n = 32; thyroid carcinoma (THCA), n = 57; uterine corpus endometrial carcinoma (UCEC), n = 7. *P < 0.05, **P < 0.01, and ***P < 0.001, by Wilcoxon matched-pairs, 2-tailed t test.
Discussion
We generated and used a genetic Gpr182-lacZ–knockin mouse model to gain insights into the tissue localization and physiological functions of GPR182. Although it is possible that genetic reduction of Gpr182 expression may influence its lacZ reporter localization, our characterization using the lacZ reporter is entirely consistent with previously published ISH and microarray data (9, 10), as well as expression analysis from GPR182-independent models, including Lgr5-EGFP and Sox9-EGFP mice. The intestinal expression pattern — particularly the crypt localization — piqued our interest and became the focus of this study. However, it is worth emphasizing that Gpr182 is expressed in a variety of tissues and cell types, particularly in the endothelium, which will certainly prompt additional physiological studies. Furthermore, these types of physiological in vivo studies may prove instructive with regard to the eventual “deorphanization,” or identification, of endogenous ligand(s) of GPR182, which currently remains unknown.
Our study shows that Gpr182 is expressed in the epithelium throughout the gastrointestinal tract and is particularly enriched in the CBC ISCs of the small intestine. Like LGR5, GPR182 is a GPCR enriched in ISCs but appears to be a less selective ISC marker. The genetic reduction of Gpr182 did not alter intestinal morphology or proliferation during homeostasis in vivo, which is consistent with the tightly regulated control of epithelial proliferation for maintaining intestinal health. However, under conditions of stress-induced proliferation, such as regeneration following injury, ApcMin mutation, or ex vivo culture in elevated Wnt signaling growth conditions, the reduction of Gpr182 caused hyperproliferative phenotypes, which were associated with elevated ERK1/2 signaling. Thus, we conclude that GPR182 normally functions to directly or indirectly inhibit MAPK-induced intestinal proliferation (Figure 7D). This inhibitory function is consistent with the finding that GPR182 expression is significantly reduced in human colon adenocarcinoma — a novel finding that expands the growing cadre of negative regulators of proliferation, including LRIG1, that have been shown to be downregulated during colorectal tumorigenesis (20, 46–49). Conversely, numerous pro-proliferative ISC markers, such as LGR5, BMI1, and SOX9, have all been shown to be upregulated in human colorectal carcinomas (50).
MAPK signaling plays a critical role during regeneration and tumorigenesis, which is evidenced by the increased p-ERK1/2 localization within the crypt and ISCs during these conditions. Intestinal epithelial conditional deletion of ERK1/2 leads to rapid malabsorption and lethality (43). ERK1/2 activation through surface receptors, such as TLR and/or EGFR, is required for adenoma formation in ApcMin/+ mice, which can be blocked by MEK/ERK inhibitors (51, 52). In addition, loss of the RTK negative regulator Lrig1 leads to constitutively active EGFR and other ERbB receptors, which lead to increased ERK1/2 signaling, hyperproliferation, and tumorigenesis (20, 53). Oncogenic mutations in KRAS activate numerous downstream kinases, including ERK1/2, which rarely initiate tumorigenesis alone but, when combined with Apc inactivation, lead to increased adenoma formation and progression (48, 54–58). GPCRs can activate ERK1/2 directly through both G protein– and β-arrestin–mediated signaling, as well as indirectly through modulation of EGF/EGFR expression and/or EGFR transactivation (59, 60). An activating mutation in Gsα (GNAS R201C) leads to increased cAMP and ERK1/2 activation, which increases adenoma formation when crossed with ApcMin/+ mice (61) to an extent similar to that observed in the Gpr182lacZ/lacZ ApcMin/+ mice. Alternatively, the trending decrease in Lrig1 expression in CD326+CD44+ cells and in Gpr182lacZ/lacZ IRR mice supports the notion that ErbB signaling may be enhanced in these mice and contributes to the hyperproliferative phenotypes. Additionally, organoids arising from single ISCs were dependent on the addition of exogenous EGF for the enhanced growth and survival observed in Gpr182Δ/Δ ISCs. Thus, it remains possible that GPR182 could directly inhibit ERK1/2 through the reduction of cAMP levels and/or indirectly through the negative regulation of the EGFR signaling pathway. Alternatively, considering that the closest paralog to GPR182 is ACKR3, a member of the “decoy” atypical chemokine receptor subfamily, it is possible that the effects of GPR182 are manifested by its function as a nonsignaling ligand sink or “decoy” receptor. Unfortunately, until a ligand (or ligands) is identified, it is challenging and premature to try to delineate the precise signal transduction pathways that are directly associated with GPR182 activity in different cell types.
To our knowledge, this is the first study to use a genetic mouse model to simultaneously map Gpr182 localization patterns and elucidate novel physiological functions for the negative regulation of intestinal proliferative capacity, especially during regeneration and adenoma formation. Future studies to identify the ligand for this exciting and physiologically relevant orphan GPCR will shed light on its tractability as a potential therapeutic target.
Methods
Experimental animals.
The Gpr182tm2a(KOMP)Wtsi/+ (knockout first/promoter driven) mice used in this study were created from an embryonic stem (ES) cell clone (EPD0365_4_C08) obtained from the National Center for Research Resources–NIH–supported Knockout Mouse Project (KOMP) repository and generated by the CHORI, Sanger Institute, and UC Davis (CSD) Consortium for the NIH-funded KOMP (29). The CSD-targeted allele has been previously published (62, 63). To achieve ubiquitous deletion, Gpr182tm2a(KOMP)Wtsi/+ mice were crossed with the B6.C-Tg(CMV-cre)1Cgn/J Tg mouse line (The Jackson Laboratory; stock no. 006054). For adenoma studies, Gpr182lacZ/+ mice were crossed with the C57BL/6J-ApcMin/+/J mouse line (The Jackson Laboratory; stock no. 002020). All Gpr182-associated mouse lines were maintained on an isogenic C57BL/6 background. Previously published Sox9-EGFP BAC–Tg mice on a CD-1 background and C57BL/6 Lgr5-EGFP mice (The Jackson Laboratory; stock no. 008875) were used for cell isolation and expression verification (18, 19, 25, 30–32). The genotyping primers used are listed in Supplemental Table 1.
Animal procedures and dissection.
For irradiation challenge, 10- to 14-week-old female Gpr182+/+ and Gpr182lacZ/lacZ mice were subjected to abdominal ionizing radiation as previously described (18). Briefly, under isoflurane anesthesia, mice received 14 Gy radiation (50 cm, F1 Filter; X-Rad 320; Precision X-Ray) to their abdomens. Experimental and control mice were irradiated at the same time to ensure equivalent radiation dosages and rates (~2.8 Gy/min for 300 s). Mice were observed and weighed daily to monitor the severity of irradiation and overall health. Five days after irradiation, mice were injected i.p. with 4 μg 5-ethynyl-2′-deoxyuridine (EdU) per 1 g BW. After 90 minutes, mice were euthanized, and gastrointestinal tissue was collected for histology and biochemistry. For adenoma studies, male and female mice were monitored for health (physical appearance, BW, bloody stool) and euthanized for tissue collection at 5 months of age. The small intestines and colons were removed and flushed with cold PBS. The entire intestinal length was measured and then separated into duodenum, jejunum and ileum. The small intestines and colons from all ApcMin/+ mice were opened along the entire length, flattened, and macroscopic polyp numbers and sizes were quantified. Tissues were then processed for histology, whole-mount X-gal staining, and biochemistry. For cryogenic sections, tissue was fixed in 4% paraformaldehyde, cryoprotected in 30% sucrose, and embedded in OCT before cryosectioning. For histology, tissues were fixed in either 4% paraformaldehyde or 10% zinc formalin and then paraffin embedded and sectioned.
Single-cell isolation and ex vivo culture.
Single cells from jejunum of 8-week-old female Gpr182+/+ CMV-Cre and Gpr182Δ/Δ CMV-Cre mice were isolated and cultured as previously described (31, 32, 40–42, 64). Briefly, jejuna were flushed and incubated in 3 mM EDTA (Corning) and 10 μM Y27632 (Selleck Chemicals) in Dulbecco’s PBS. Villi were gently scraped with a pipette tip, and the epithelium was separated from the submucosa by shaking. The epithelial crypts were further dissociated into single cells in 0.3 U/ml dispase (Thermo Fisher Scientific), 10 μg/ml DNase (Thermo Fisher Scientific), and Y27632 in HBSS. Single cells were stained with APC/Cy7-conjugated anti-CD326 (1:100; catalog 118218); Brilliant Violet 421–conjugated anti-CD44 (1:100; catalog 338810); and 7-AAD (catalog 420404) (all from BioLegend). FACS was conducted on an SH800Z Cell Sorter (Sony). Debris, doublets, and dead cells were excluded by size and 7-AAD. Viable CD326+CD44– and CD326+CD44+ cells (1,000 cells/well) were collected into Matrigel. Culture media (31) were supplemented every 2 days (unless otherwise noted) with 50 ng/ml EGF (Invitrogen, Thermo Fisher Scientific); 100 ng/ml Noggin (Peprotech); 100 nM valproic acid (Sigma-Aldrich); 3 μM CHIR99021 (Selleck Chemicals); and 25% conditioned R-spondin 2 media.
Whole-mount X-gal staining.
Whole-mount X-gal staining was adapted from previously published protocols (19). Briefly, whole tissue was fixed in 0.2% glutaraldehyde (Electron Microscopy Sciences) for 24 hours at 4°C. Tissue was washed and permeabilized with 0.1% triton and incubated in 1 mg/ml X-gal staining buffer (Bioline) for approximately 24 hours at room temperature in the dark. Tissue was washed and post-fixed in 4% paraformaldehyde for 24 hours at 4°C. Samples were washed, imaged, and paraffin embedded, and sections were counterstained with 1% Neutral Red solution (Sigma-Aldrich) or eosin.
Optical projection tomography.
Whole-mount X-gal–stained tissues were embedded in 1% agarose (catalog 105128; Thermo Fisher Scientific), dehydrated overnight in absolute methanol, and cleared in benzyl alcohol/benzyl benzoate (1:2) as previously described (65). Optically cleared specimens were mounted onto aluminum chucks and scanned with a 3001m Optical Projection Tomography Scanner (BiOPTonics) under bright-field illumination.
IHC.
Paraffin and cryogenic sections were rehydrated, permeabilized, and blocked with 5% normal donkey serum. When required, slides were boiled in citrate buffer for antigen retrieval. Slides were then stained overnight at room temperature with the following primary antibodies: chicken anti–β-gal (1:1,500; catalog BGL-1040; Aves Labs Inc.); rabbit anti-lysozyme (1:1,000; catalog PA0391; Leica Biosystems); rabbit anti–p-histone H3 (1:500; catalog 9701; Cell Signaling Technology); rat anti-Ki67 (1:30; M7249; DakoCytomation); rabbit anti–p-ERK1/2 (1:300; catalog 4370; Cell Signaling Technology); rabbit anti–β-catenin (1:500; catalog ab16051; Abcam); rabbit anti-DCLK1 (1:1,000; catalog ab31704; Abcam); rat anti-CD31 (1:100; catalog ab56299; Abcam); and Syrian hamster anti-podoplanin (1:200; 8.1.1; Developmental Studies Hybridoma Bank). Sections were rinsed, blocked, and incubated in the dark for 90 minutes at room temperature with the following secondary antibodies from Jackson ImmunoResearch (1:200): donkey anti-rabbit Cy2 (catalog 711-225-152), donkey anti-rabbit Alexa Fluor 594 (catalog 711-585-152), donkey anti-chicken Alexa Fluor 647 (catalog 703-605-155), donkey anti-chicken Alexa Fluor 594 (catalog 703-585-155), donkey anti-rat Cy3 (catalog 712-165-150), donkey anti-mouse Alexa Fluor 488 (catalog 715-545-151), and goat anti-Syrian hamster FITC (catalog 107-095-142); and Hoechst 33258 (1:1,000; catalog B1155; Sigma-Aldrich). The Click-iT EdU Alexa Fluor 594 Kit (catalog C10339; Invitrogen, Thermo Fisher Scientific) was used according to the manufacturer’s instructions.
Imaging and image processing.
Whole-mount tissue was imaged using a Leica MZ16FA dissecting stereoscope outfitted with a QImaging Micropublisher 5.0 RTV color CCD camera. Paraffin-embedded H&E- and X-gal–stained slides were imaged using a Leitz Dialux 20 with a QImaging Micropublisher 5.0 RTV color CCD camera or an Olympus BX61 microscope equipped with a QImaging RETIGA 4000R CCD color camera with Volocity software (Improvision). Fluorescence IHC images were acquired on a Nikon E800 fluorescence microscope with a Hamamatsu Orca CCD camera and MetaMorph software (Molecular Devices) or a Zeiss LSM 700 confocal microscope. Organoids were imaged on an Olympus IX81 fluorescence microscope with a Hamamatsu Orca camera and MetaMorph Basic software. Fluorescent images were pseudo-colored using ImageJ software (NIH). Image processing, including cropping, brightness, and contrast adjustments, were altered equally across comparable images using ImageJ and Photoshop CS4 (Adobe).
Morphometry and proliferation quantitation.
Crypt depth was quantified by measuring the distance from the bottom of nuclei at the base to the top of the crypt from 5 to 30 open crypts from each region of the intestine from 5 mice per genotype per treatment. Crypt density was assessed by whole-mount microscopy. Three fields per intestinal area per genotype were imaged, and crypt the area from 10 crypts per field was measured with ImageJ. The approximate number of crypts that would fit into a 1-mm2 area was calculated. Proliferation was assayed by EdU incorporation and the Click-iT Detection Kit (Thermo Fisher Scientific) and analyzed by counting the number of EdU+ cells per open crypt normalized to the total number of DAPI+ crypt cells and expressed as a percentage. Cell positions of EdU+ cells along the crypt/villus axis were recorded during quantification. Ten to sixty whole, open crypts were included from each area of the intestine (duodenum, jejunum, ileum, and distal colon) from 5 mice per genotype per treatment. Polyps were counted and dimensions were approximated from all polyps measuring 1 × 1 mm2 or larger throughout the small intestine in all experimental mice, and the polyp area was calculated assuming an elliptical shape. A total of 700 to 1,700 polyps were counted from 20 to 35 mice per genotype. Proliferation of individual polyps was determined by threshold quantification of Ki67+ signal as a ratio of DAPI+ signal in cross-sections using ImageJ software. Viable organoids with defined cell borders were counted on days 1, 2, 3, 5, and 7 when they were imaged. Researchers were blinded to the mouse genotype and treatment.
Real-time PCR analysis.
Tissues were either snap-frozen in liquid nitrogen or stored in RNAlater (Ambion, Thermo Fisher Scientific). Enriched cell populations labeled with CD326 and CD44 or differential Sox9-EGFP or Lgr5-EGFP levels as previously described (18, 30, 40–42) were isolated by FACS and collected into RNAqueous-Micro Lysis Solution (Ambion, Thermo Fisher Scientific). Likewise, the Paneth cell–enriched population was isolated using a previously published protocol (31). Human colorectal carcinoma cell lines (ATCC), HUVECs (Lonza), and human intestinal epithelial cells, provided by J.F. Beaulieu (University of Sherbrooke, Quebec, Canada), were cultured and collected as previously reported (66). RNA was extracted using either TRIzol Reagent (Invitrogen, Thermo Fisher Scientific) and a Percellys bead homogenizer or RNAqueous-Micro Kit (Ambion, Thermo Fisher Scientific) according to standard procedures and then subsequently treated with DNase1 (RQ1; Promega) and reverse transcribed with M-MLV (Invitrogen, Thermo Fisher Scientific) or iScript (Bio-Rad Laboratories). Quantitative gene expression was assayed with 2× TaqMan Master Mix (Applied Biosystems or Bio Basic Inc.) and run on a StepOne Plus Real-Time PCR System (Applied Biosystems, Thermo Fisher Scientific). Primers and probes for real-time PCR (RT-PCR) are listed in Supplemental Table 1. Relative expression levels were determined by the ΔΔCt method and normalized to reference gene expression of Gapdh, Actb, or 18S.
Western blotting and analysis.
Protein was extracted using Lens Homogenization Buffer with DTT and cOmplete Protease Inhibitor Cocktail Tablets (Roche) and a bead homogenizer (Percellys) according to standard procedures. Concentrations were determined using a Pierce bicinchoninic acid (BCA) protein assay kit (Thermo Fisher Scientific) read on a Mithras LB 940 Multimode Microplate Reader (Berthold Technologies). Protein was run on Mini-Protean TGX SDS-PAGE Gel (Bio-Rad Laboratories) and transferred onto a nitrocellulose (GE Healthcare) membrane. Blots were blocked and stained in 5% BSA diluted in TBS with 0.1% Tween-20. Blots were incubated overnight at 4°C with the following primary antibodies: rabbit anti-p42/44 MAPK (ERK1/2) (1:1,000; catalog 9102; Cell Signaling Technology); rabbit anti–p-p42/44 MAPK (ERK1/2) (Thr202/Tyr204) (1:1,000; catalog 9101; Cell Signaling Technology); rabbit anti-AKT (1:1,000; catalog 4691; Cell Signaling Technology); rabbit anti–p-AKT (Ser473) (1:2,000; catalog 4060; Cell Signaling Technology); mouse anti–β-catenin (1:200; catalog 610153; BD Biosciences); rabbit anti-p38 MAPK (1:1,000; catalog 8690; Cell Signaling Technology); rabbit anti–p-p38 MAPK (Thr180/Tyr182) (1:1,000; catalog 9211; Cell Signaling Technology); mouse anti-STAT3 (1:1,000; catalog 9139; Cell Signaling Technology); rabbit anti–p-STAT3 (Tyr705) (1:2,000; catalog 9145; Cell Signaling Technology); rabbit anti-YAP (1:1,000; catalog 14074; Cell Signaling Technology); rabbit anti–p-YAP (Ser127) (1:1,000; catalog 13008; Cell Signaling Technology); and mouse anti-GAPDH (1:5,000; catalog NB300-285; Novus Biologicals). Blots were rinsed, blocked in 5% nonfat milk, and incubated in the dark for 60 minutes at room temperature with the following secondary antibodies: goat anti-rabbit DyLight 680 (1:12,000; catalog 35568; Thermo Fisher Scientific) and goat anti-mouse DyLight 800 (1:12,000; catalog SA5-10176; Thermo Fisher Scientific). An Odyssey CLx (LI-COR Biosciences) was used for imaging. Image brightness and contrast changes were made equally across all comparable images using Image Studio and Adobe Photoshop CS4. Phosphorylated and total protein quantification was analyzed by densitometry using ImageJ and normalized to Gpr182+/+ controls. GAPDH was used to ensure equal loading.
RNA-seq analysis.
RNA-seq data were downloaded from the TCGA Data Portal (44, 45). RSEM upper-quantile–normalized values from Illumina HiSeq_RNASeqV2 from patient-matched tumor and normal tissue were log2 transformed. Samples with an expression value of 3 or lower were indistinguishable from background values and were thus considered a 0 value.
Statistics.
Statistical analysis was performed with GraphPad Prism 5.0 (GraphPad Software), and all data are represented as the mean ± SEM. The unpaired or Mann-Whitney t test was used for data analysis of 2 groups with biological replicate numbers of less than 10 or more than 10, respectively. For parametric data with more than 2 comparisons and biological replicate numbers of less than 10, a 1-way ANOVA with Tukey’s multiple comparisons test were used. For nonparametric data analyzing more than 2 groups and biological replicate numbers of more than 10, a 1-way ANOVA with Kruskal-Wallis and Dunn’s multiple comparisons tests were used. The Mantel-Cox test was used to analyze Kaplan-Meier survival data. Patient-matched tumor and normal tissues were compared with a Wilcoxon matched-pairs t test and nonmatched tumors were compared using an unpaired Mann-Whitney t test. All t tests were 2 tailed. A P value of less than 0.05 was considered statistically significant.
Study approval.
All animal studies were conducted in accordance with protocols approved by the IACUC of the University of North Carolina at Chapel Hill.
Author contributions
DOK designed research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript. REB conducted mouse irradiation procedures and provided technical and intellectual support. BZ conducted ex vivo culture experiments and provided technical and intellectual support. STE acquired data, provided technical and intellectual support, and edited the manuscript. ATM acquired data, provided technical and intellectual support, and edited the manuscript. MBS and CMP conducted bioinformatics analysis and provided technical support. SD provided reagents and intellectual support. STM designed research studies, acquired data, and provided reagents and intellectual support. PKL designed research studies and provided reagents and technical and intellectual support. KMC designed research studies, provided intellectual support, and edited the manuscript.
Supplementary Material
Acknowledgments
We thank the University of North Carolina Lineberger Animal Histopathology Core (NIH CA16086); the Center for Gastrointestinal Biology and Disease (NIH P30-DK34987); the Microscopy Services Laboratory; as well as Kirk McNaughton and Ashley Ezzell of the Histology Research Core. We also thank the current and past members of the Caron and Lund laboratories for discussions. This work was supported by NIH grants RO1-HD060860 and RO1-DK099156 (to KMC); NIH grant F31-CA174194 and a University of North Carolina Cell and Molecular Physiology Fellner Fellowship (to DOK); NIH grant F31-DK107137 (to BZ); NIH grant 2-T32-DK07686 (to ATM); NIH grants 1-F30-CA200345 and T32-CA071341-17 (to MBS); NIH grant P50-CA58223-09A1 and a grant from the Breast Cancer Research Foundation (to CMP); NIH grant RO1-DK091427 (to STM); and NIH grant RO1-DK040247-19 (to PKL).
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(2):593–607. https://doi.org/10.1172/JCI87588.
Contributor Information
R. Eric Blue, Email: eric_blue@med.unc.edu.
Bailey Zwarycz, Email: bzwarycz@email.unc.edu.
Scott T. Espenschied, Email: scott.espenschied@duke.edu.
Amanda T. Mah, Email: atmah@stanford.edu.
Shengli Ding, Email: shengli_ding@med.unc.edu.
Kathleen M. Caron, Email: kathleen_caron@med.unc.edu.
References
- 1.Rask-Andersen M, Almen MS, Schioth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10(8):579–590. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- 2.Civelli O, Reinscheid RK, Zhang Y, Wang Z, Fredriksson R, Schioth HB. G protein-coupled receptor deorphanizations. Annu Rev Pharmacol Toxicol. 2013;53:127–146. doi: 10.1146/annurev-pharmtox-010611-134548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harrison JK, Barber CM, Lynch KR. Molecular cloning of a novel rat G-protein-coupled receptor gene expressed prominently in lung, adrenal, and liver. FEBS Lett. 1993;318(1):17–22. doi: 10.1016/0014-5793(93)81318-T. [DOI] [PubMed] [Google Scholar]
- 4.Kapas S, Catt KJ, Clark AJ. Cloning and expression of cDNA encoding a rat adrenomedullin receptor. J Biol Chem. 1995;270(43):25344–25347. doi: 10.1074/jbc.270.43.25344. [DOI] [PubMed] [Google Scholar]
- 5.Kennedy SP, Sun D, Oleynek JJ, Hoth CF, Kong J, Hill RJ. Expression of the rat adrenomedullin receptor or a putative human adrenomedullin receptor does not correlate with adrenomedullin binding or functional response. Biochem Biophys Res Commun. 1998;244(3):832–837. doi: 10.1006/bbrc.1998.8349. [DOI] [PubMed] [Google Scholar]
- 6.McLatchie LM, et al. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393(6683):333–339. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 7.Ramachandran V, Arumugam T, Hwang RF, Greenson JK, Simeone DM, Logsdon CD. Adrenomedullin is expressed in pancreatic cancer and stimulates cell proliferation and invasion in an autocrine manner via the adrenomedullin receptor, ADMR. Cancer Res. 2007;67(6):2666–2675. doi: 10.1158/0008-5472.CAN-06-3362. [DOI] [PubMed] [Google Scholar]
- 8.Ramachandran V, et al. The ADMR receptor mediates the effects of adrenomedullin on pancreatic cancer cells and on cells of the tumor microenvironment. PLoS One. 2009;4(10):e7502. doi: 10.1371/journal.pone.0007502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Regard JB, Sato IT, Coughlin SR. Anatomical profiling of G protein-coupled receptor expression. Cell. 2008;135(3):561–571. doi: 10.1016/j.cell.2008.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takase H, et al. Genome-wide identification of endothelial cell-enriched genes in the mouse embryo. Blood. 2012;120(4):914–923. doi: 10.1182/blood-2011-12-398156. [DOI] [PubMed] [Google Scholar]
- 11.Xiao L, Harrell JC, Perou CM, Dudley AC. Identification of a stable molecular signature in mammary tumor endothelial cells that persists in vitro. Angiogenesis. 2014;17(3):511–518. doi: 10.1007/s10456-013-9409-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sumanas S, Jorniak T, Lin S. Identification of novel vascular endothelial-specific genes by the microarray analysis of the zebrafish cloche mutants. Blood. 2005;106(2):534–541. doi: 10.1182/blood-2004-12-4653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alghisi E, et al. Targeting oncogene expression to endothelial cells induces proliferation of the myelo-erythroid lineage by repressing the Notch pathway. Leukemia. 2013;27(11):2229–2241. doi: 10.1038/leu.2013.132. [DOI] [PubMed] [Google Scholar]
- 14.Clevers H. The intestinal crypt, a prototype stem cell compartment. Cell. 2013;154(2):274–284. doi: 10.1016/j.cell.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 15.Barker N. Adult intestinal stem cells: critical drivers of epithelial homeostasis and regeneration. Nat Rev Mol Cell Biol. 2014;15(1):19–33. doi: 10.1038/nrm3721. [DOI] [PubMed] [Google Scholar]
- 16.Gracz AD, Magness ST. Defining hierarchies of stemness in the intestine: evidence from biomarkers and regulatory pathways. Am J Physiol Gastrointest Liver Physiol. 2014;307(3):G260–273. doi: 10.1152/ajpgi.00066.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chia LA, Kuo CJ. The intestinal stem cell. Prog Mol Biol Transl Sci. 2010;96:157–173. doi: 10.1016/B978-0-12-381280-3.00007-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Landeghem L, et al. Activation of two distinct Sox9-EGFP-expressing intestinal stem cell populations during crypt regeneration after irradiation. Am J Physiol Gastrointest Liver Physiol. 2012;302(10):G1111–1132. doi: 10.1152/ajpgi.00519.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker N, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449(7165):1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 20.Powell AE, et al. The pan-ErbB negative regulator Lrig1 is an intestinal stem cell marker that functions as a tumor suppressor. Cell. 2012;149(1):146–158. doi: 10.1016/j.cell.2012.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van der Flier LG, et al. Transcription factor achaete scute-like 2 controls intestinal stem cell fate. Cell. 2009;136(5):903–912. doi: 10.1016/j.cell.2009.01.031. [DOI] [PubMed] [Google Scholar]
- 22.van der Flier LG, Haegebarth A, Stange DE, van de Wetering M, Clevers H. OLFM4 is a robust marker for stem cells in human intestine and marks a subset of colorectal cancer cells. Gastroenterology. 2009;137(1):15–17. doi: 10.1053/j.gastro.2009.05.035. [DOI] [PubMed] [Google Scholar]
- 23.Takeda N, Jain R, LeBoeuf MR, Wang Q, Lu MM, Epstein JA. Interconversion between intestinal stem cell populations in distinct niches. Science. 2011;334(6061):1420–1424. doi: 10.1126/science.1213214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sangiorgi E, Capecchi MR. Bmi1 is expressed in vivo in intestinal stem cells. Nat Genet. 2008;40(7):915–920. doi: 10.1038/ng.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Formeister EJ, Sionas AL, Lorance DK, Barkley CL, Lee GH, Magness ST. Distinct SOX9 levels differentially mark stem/progenitor populations and enteroendocrine cells of the small intestine epithelium. Am J Physiol Gastrointest Liver Physiol. 2009;296(5):G1108–1118. doi: 10.1152/ajpgi.00004.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montgomery RK, et al. Mouse telomerase reverse transcriptase (mTert) expression marks slowly cycling intestinal stem cells. Proc Natl Acad Sci U S A. 2011;108(1):179–184. doi: 10.1073/pnas.1013004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scoville DH, Sato T, He XC, Li L. Current view: intestinal stem cells and signaling. Gastroenterology. 2008;134(3):849–864. doi: 10.1053/j.gastro.2008.01.079. [DOI] [PubMed] [Google Scholar]
- 28.Barker N, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457(7229):608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 29. UC Davis. Knockout Mouse Project (KOMP) Respository. https://www.komp.org Accessed November 30, 2016.
- 30.Gracz AD, Ramalingam S, Magness ST. Sox9 expression marks a subset of CD24-expressing small intestine epithelial stem cells that form organoids in vitro. Am J Physiol Gastrointest Liver Physiol. 2010;298(5):G590–G600. doi: 10.1152/ajpgi.00470.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gracz AD, et al. A high-throughput platform for stem cell niche co-cultures and downstream gene expression analysis. Nat Cell Biol. 2015;17(3):340–349. doi: 10.1038/ncb3104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roche KC, Gracz AD, Liu XF, Newton V, Akiyama H, Magness ST. SOX9 maintains reserve stem cells and preserves radioresistance in mouse small intestine. Gastroenterology. 2015;149(6):1553–1563.e10. doi: 10.1053/j.gastro.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan KS, et al. The intestinal stem cell markers Bmi1 and Lgr5 identify two functionally distinct populations. Proc Natl Acad Sci U S A. 2012;109(2):466–471. doi: 10.1073/pnas.1118857109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hua G, et al. Crypt base columnar stem cells in small intestines of mice are radioresistant. Gastroenterology. 2012;143(5):1266–1276. doi: 10.1053/j.gastro.2012.07.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Metcalfe C, Kljavin NM, Ybarra R, de Sauvage FJ. Lgr5+ stem cells are indispensable for radiation-induced intestinal regeneration. Cell Stem Cell. 2014;14(2):149–159. doi: 10.1016/j.stem.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 36.Moser AR, Pitot HC, Dove WF. A dominant mutation that predisposes to multiple intestinal neoplasia in the mouse. Science. 1990;247(4940):322–324. doi: 10.1126/science.2296722. [DOI] [PubMed] [Google Scholar]
- 37.Morin PJ, et al. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 38.Kozar S, et al. Continuous clonal labeling reveals small numbers of functional stem cells in intestinal crypts and adenomas. Cell Stem Cell. 2013;13(5):626–633. doi: 10.1016/j.stem.2013.08.001. [DOI] [PubMed] [Google Scholar]
- 39.Nakanishi Y, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45(1):98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- 40.Gracz AD, et al. Brief report: CD24 and CD44 mark human intestinal epithelial cell populations with characteristics of active and facultative stem cells. Stem Cells. 2013;31(9):2024–2030. doi: 10.1002/stem.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wang F, et al. Isolation and characterization of intestinal stem cells based on surface marker combinations and colony-formation assay. Gastroenterology. 2013;145(2):383–95.e1. doi: 10.1053/j.gastro.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sato T, et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature. 2009;459(7244):262–265. doi: 10.1038/nature07935. [DOI] [PubMed] [Google Scholar]
- 43.de Jong PR, et al. ERK5 signalling rescues intestinal epithelial turnover and tumour cell proliferation upon ERK1/2 abrogation. Nat Commun. 2016;7:11551. doi: 10.1038/ncomms11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoadley KA, et al. Multiplatform analysis of 12 cancer types reveals molecular classification within and across tissues of origin. Cell. 2014;158(4):929–944. doi: 10.1016/j.cell.2014.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. National Institute of Health National Cancer Institute National Human Genome Research Institute. The Cancer Genome Atlas (TCGA) Data Portal. https://tcga-data.nci.nih.gov/docs/publications/tcga/? Accessed November 30, 2016.
- 46.Noah TK, et al. SPDEF functions as a colorectal tumor suppressor by inhibiting β-catenin activity. Gastroenterology. 2013;144(5):1012–1023.e6. doi: 10.1053/j.gastro.2013.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bossuyt W, et al. Atonal homolog 1 is a tumor suppressor gene. PLoS Biol. 2009;7(2):e39. doi: 10.1371/journal.pbio.1000039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Depeille P, et al. RasGRP1 opposes proliferative EGFR-SOS1-Ras signals and restricts intestinal epithelial cell growth. Nat Cell Biol. 2015;17(6):804–815. doi: 10.1038/ncb3175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nam KT, et al. Loss of Rab25 promotes the development of intestinal neoplasia in mice and is associated with human colorectal adenocarcinomas. J Clin Invest. 2010;120(3):840–849. doi: 10.1172/JCI40728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Espersen ML, Olsen J, Linnemann D, Høgdall E, Troelsen JT. Clinical implications of intestinal stem cell markers in colorectal cancer. Clin Colorectal Cancer. 2015;14(2):63–71. doi: 10.1016/j.clcc.2014.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Lee SH, et al. ERK activation drives intestinal tumorigenesis in Apc(min/+) mice. Nat Med. 2010;16(6):665–670. doi: 10.1038/nm.2143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Roberts RB, et al. Importance of epidermal growth factor receptor signaling in establishment of adenomas and maintenance of carcinomas during intestinal tumorigenesis. Proc Natl Acad Sci U S A. 2002;99(3):1521–1526. doi: 10.1073/pnas.032678499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wong VW, et al. Lrig1 controls intestinal stem-cell homeostasis by negative regulation of ErbB signalling. Nat Cell Biol. 2012;14(4):401–408. doi: 10.1038/ncb2464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sansom OJ, et al. Loss of Apc allows phenotypic manifestation of the transforming properties of an endogenous K-ras oncogene in vivo. Proc Natl Acad Sci USA. 2006;103(38):14122–14127. doi: 10.1073/pnas.0604130103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dow LE, et al. Apc restoration promotes cellular differentiation and reestablishes crypt homeostasis in colorectal cancer. Cell. 2015;161(7):1539–1552. doi: 10.1016/j.cell.2015.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Phelps RA, et al. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell. 2009;137(4):623–634. doi: 10.1016/j.cell.2009.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haigis KM, et al. Differential effects of oncogenic K-Ras and N-Ras on proliferation, differentiation and tumor progression in the colon. Nat Genet. 2008;40(5):600–608. doi: 10.1038/ng.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Janssen KP, et al. APC and oncogenic KRAS are synergistic in enhancing Wnt signaling in intestinal tumor formation and progression. Gastroenterology. 2006;131(4):1096–1109. doi: 10.1053/j.gastro.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 59.Yu FX, et al. Regulation of the Hippo-YAP pathway by G-protein-coupled receptor signaling. Cell. 2012;150(4):780–791. doi: 10.1016/j.cell.2012.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Prenzel N, et al. EGF receptor transactivation by G-protein-coupled receptors requires metalloproteinase cleavage of proHB-EGF. Nature. 1999;402(6764):884–888. doi: 10.1038/47260. [DOI] [PubMed] [Google Scholar]
- 61.Wilson CH, McIntyre RE, Arends MJ, Adams DJ. The activating mutation R201C in GNAS promotes intestinal tumourigenesis in Apc(Min/+) mice through activation of Wnt and ERK1/2 MAPK pathways. Oncogene. 2010;29(32):4567–4575. doi: 10.1038/onc.2010.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Testa G, et al. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38(3):151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]
- 63.Bradley A, et al. The mammalian gene function resource: the International Knockout Mouse Consortium. Mamm Genome. 2012;23(9-10):580–586. doi: 10.1007/s00335-012-9422-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yin X, Farin HF, van Es JH, Clevers H, Langer R, Karp JM. Niche-independent high-purity cultures of Lgr5+ intestinal stem cells and their progeny. Nat Methods. 2014;11(1):106–112. doi: 10.1038/nmeth.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Karpinich NO, Kechele DO, Espenschied ST, Willcockson HH, Fedoriw Y, Caron KM. Adrenomedullin gene dosage correlates with tumor and lymph node lymphangiogenesis. FASEB J. 2013;27(2):590–600. doi: 10.1096/fj.12-214080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Andres SF, Simmons JG, Mah AT, Santoro MA, Van Landeghem L, Lund PK. Insulin receptor isoform switching in intestinal stem cells, progenitors, differentiated lineages and tumors: evidence that IR-B limits proliferation. J Cell Sci. 2013;126(Pt 24):5645–5656. doi: 10.1242/jcs.132985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.