Abstract
The BM niche comprises a tightly controlled microenvironment formed by specific tissue and cells that regulates the behavior of hematopoietic stem cells (HSCs). Here, we have provided a 3D model that is tunable in different BM niche components and useful, both in vitro and in vivo, for studying the maintenance of normal and malignant hematopoiesis. Using scaffolds, we tested the capacity of different stromal cell types to support human HSCs. Scaffolds coated with human mesenchymal stromal cells (hMSCs) proved to be superior in terms of HSC engraftment and long-term maintenance when implanted in vivo. Moreover, we found that hMSC-coated scaffolds can be modulated to form humanized bone tissue, which was also able to support human HSC engraftment. Importantly, hMSC-coated humanized scaffolds were able to support the growth of leukemia patient cells in vivo, including the growth of samples that would not engraft the BM of immunodeficient mice. These results demonstrate that an s.c. implantation approach in a 3D carrier scaffold seeded with stromal cells is an effective in vivo niche model for studying human hematopoiesis. The various niche components of this model can be changed depending on the context to improve the engraftment of nonengrafting acute myeloid leukemia (AML) samples.
Introduction
The BM niche comprises a tightly controlled microenvironment formed by various cell types that regulate the behavior of hematopoietic stem cells (HSCs). Currently, i.v. injection of cells in immunodeficient mouse models, followed by cellular studies of murine BM tissue, is the most common assay for studying normal and malignant human hematopoiesis. Although successful engraftment of primary acute myeloid leukemia (AML) is achievable, our own lab and others have shown that not all AML samples are able to engraft immunodeficient mice and that the engraftment ability of these samples is related to their clinical outcome (1–3). Over the past few years, this has prompted the development of new mouse strains encoding human cytokines and has also opened the door to a novel approach. By merging knowledge from biomaterials, tissue engineering, and cell-implantation fields, investigators have generated new models to mimic the native human hematopoietic microenvironment within s.c. 3D structures (4–5). Using human mesenchymal stromal cells (hMSCs) as stromal cell support within bone-forming implants, researchers have studied normal (6–10) and malignant (11–16) hematopoiesis.
In this context, we focused on the development of an implantable tool whereby different niche components could be tested, both in vitro and in vivo, in order to study both normal and malignant primary human hematopoietic cells.
Results and Discussion
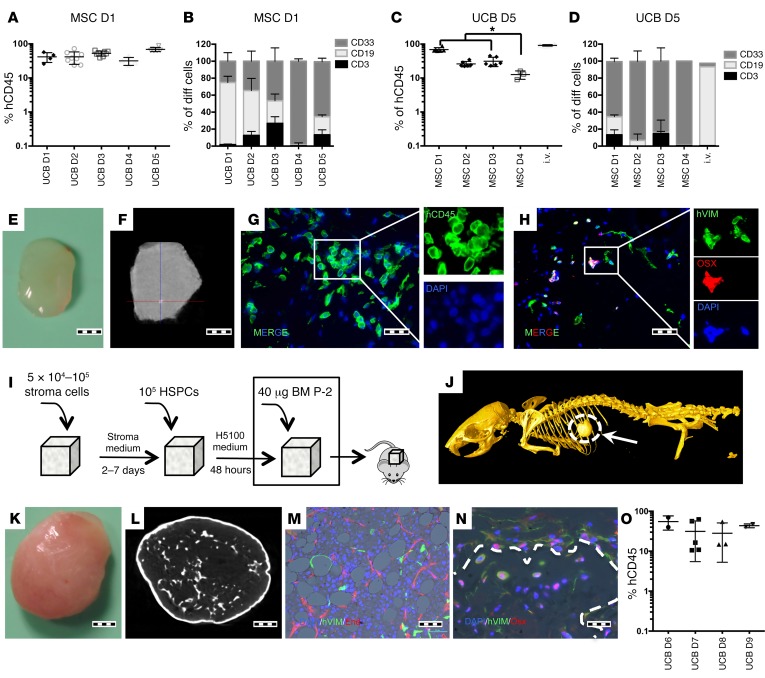
Preliminary assays were performed using different scaffold materials and various cell-seeding methods (data not shown). We found that injecting a stromal cell suspension into the center of a partially dehydrated gelatin-based porous scaffold (Gelfoam) provided a consistent coverage throughout the material (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI89364DS1). Further seeding of human hematopoietic cells using the same technique allowed adherence to preseeded stromal cells (Supplemental Figure 1), suggesting that this approach could be useful for hematopoietic cell studies. Importantly, the flexible nature of the selected biocompatible cell-carrier scaffold facilitates sectioning to the desired size via simple cutting while also allowing effective digestion using collagenase, meaning easy access to the cells for further studies. Next, we evaluated the supportiveness of mesenchymal, endothelial, or osteoblastic niche cells for the maintenance of human cord blood–derived hematopoietic stem and progenitor cells (CB-HSPCs) within the 3D model both in vitro and in vivo (Supplemental Figure 2). In vitro, most of the tested stromal cells promoted robust growth of all primary hematopoietic colony lineages (Supplemental Figure 2, B and C). In vivo, despite the detection of multilineage human engraftment in most stroma-seeded scaffolds, the hMSC-coated scaffolds had a significantly (P < 0.0001) (Supplemental Figure 2E, Left panel) higher capacity for maintaining human hematopoietic cell engraftment in primary mice. Using a secondary transplantation assay, we observed engraftment only with cells recovered from human osteoblast-, endothelial- and MSC-seeded scaffolds (Supplemental Figure 2E). Therefore, based on primary and secondary transplant assays, we decided to use hMSCs for further studies. Single-donor–derived hMSCs were used to test CB-HSPC interdonor variability in vivo (Figure 1). Although we found no significant differences in human hematopoietic cell engraftment, we did observe disparity in myeloid and lymphoid lineage distribution outcome between cord bloods (Figure 1, A and B respectively). Furthermore, hMSC intradonor variability was also tested measuring the engraftment of single-donor CB-HSPCs in scaffolds coated with hMSCs from different donors (Figure 1, C and D) and comparing it with that of i.v.-injected CB-HSPCs. In this case, we observed differences in the human hematopoietic hCD45+ cell-engraftment level (Figure 1C), indicating interdonor hMSC variability, while lineage distribution was similar for all tested cells, which in this case was mainly myeloid (Figure 1D). Interestingly, we observed that mouse hematopoietic cells migrated into the implanted scaffolds (Supplemental Figure 3), while vascularization of the scaffolds was observed using intravital microscopy (Supplemental Figure 4A). Additionally, histological characterization revealed some hCD45-positive cells next to vascular structures within the scaffolds (Supplemental Figure 4B). We also looked for the presence of hMSCs in the recovered scaffold (Supplemental Figure 5). Histology showed Vimentin expressing cells in scaffolds, while we were able to recover cells with hMSC phenotype from scaffolds (Supplemental Figure 3 and 5). None of the implanted scaffolds showed evidence of calcification (Figure 1, E and F), whereas a small portion of implanted hMSCs had differentiated to osteolineage (osterix+, Figure 1H, or osteocalcin+, Supplemental Figure 5B). We questioned whether bone formation within the implanted scaffolds would affect hematopoietic cell engraftment and therefore added BMP-2, an osteoinductive growth factor, to the implanted material (see Figure 1I). This approach yielded s.c. bone formation with vascularized BM and hMSCs, which had differentiated into adipose and osteoblastic lineages (Figure 1, J–N). Moreover, human hematopoietic cells had also engrafted in the newly formed mature BM tissue (Figure 1O).
Figure 1. UCB-derived HSPC engraftment in s.c. scaffolds.
(A and B) Comparative assay using a single hMSC donor and multiple UCB donors. Each point represents 1 mouse seeded with 6 scaffolds. For each UCB donor, 3 to 8 mice were transplanted. (A) hCD45 scaffold-engraftment level. D1-D5: different UCB Donors. (B) Lineage distribution of engrafted cells. (C and D) A comparative assay using a single UCB donor and multiple hMSC donors. Each point represents 1 mouse seeded with 6 scaffolds. For each hMSC donor, 3 to 6 mice were transplanted. (C) hCD45 scaffold–engraftment levels. For comparative purposes, the engraftment levels in BM of i.v.-injected mice are provided. Tukey’s test for multiple comparisons was applied. *P < 0.05. (D) Lineage distribution of engrafted cells. (E) Gross morphology of harvested scaffold. (F) microCT study of a harvested scaffold. The scaffold is mainly soft tissue, with sporadic calcification spots. (G) Immunofluorescence image showing hCD45 cells in the scaffold and (H) osterix-positive (OSX+) and osterix-negative human stroma cells. (I) Schematic of preparation and implantation of bone-forming scaffolds. (J) Whole-mouse microCT showing the s.c. ossicle formation. (K) Gross morphology of the harvested ossicle. (L) microCT study of a harvested ossicle. (M and N) Immunofluorescence images showing (M) human vimentin+ (hVIM) mesenchymal cells and adipocytes and mature neovascularized (endomucin+, End) BM. (N) Trabecular bone formed inside ossicles. Dotted line shows delimited trabecular bone area. hVIM+/Osx+ cells are osteocytes or osteoblasts in the bone surface areas. (O) hCD45+ engraftment levels in the ossicle model tested with multiple UCB donors. Each point represents 1 mouse with 2 scaffolds. For each UCB donor, 2 to 4 mice were transplanted. Scale bars: 1 mm (E, F, K, and L); 20 μm (G, H and N); 50 μm (M). All data were harvested at 12 weeks after implantation.
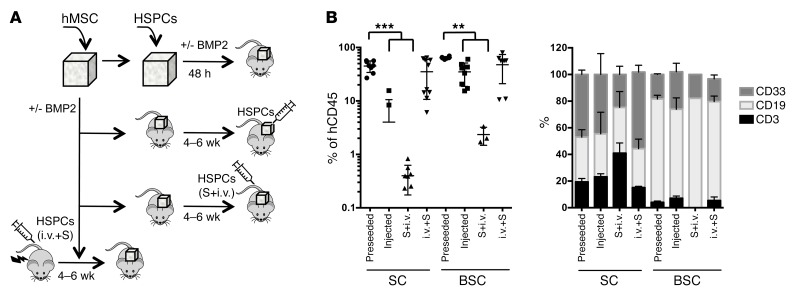
Following this, we evaluated different methods of seeding hHSPCs into the scaffold: either (a) preseeding hHSPCs before implantation; (b and c) first implanting the hMSC scaffolds and 4 to 6 weeks later injecting hHSPCs (b) intrascaffold; or (c) i.v. (S+i.v.); or (d) implanting hMSCs-scaffolds 4 to 6 weeks after transplantation of hHSPCs injected i.v. in sublethally irradiated mice (i.v.+S) (see schematic, Figure 2A). We found that preseeding hHSPCs within the scaffold before implantation not only gave the best engraftment level, but was also the quickest model to allow establishment of engraftment (only 10 to 12 weeks after implantation compared with 16 to 18 weeks for the other approaches) (Figure 2B). Additionally, in this model, we observed that human hematopoietic cells barely engraft the host BM (Supplemental Figure 6). We hypothesize that this could be related to the favorable microenvironment within the scaffold, but is most likely due to the fact that scaffolds were implanted in nonirradiated mice. Indeed, it is well known that NSG mice require a preconditioning regimen using either sublethal irradiation or busulfan pretreatment to engraft human HSPCs (17). Thus, the fact that low or no engraftment of host BM tissue was observed was expected. Moreover, the procedure named (d) i.v.+S showed a high level of engraftment in both the BM and scaffolds, indicating that hHSPCs were able to circulate to the neovasculature formed in the scaffolds and engraft there.
Figure 2. Different modality of transplantation of HSPCs into scaffold.
(A) Schematic of hMSC seeding and the different tested methods for CB-HSPCs inclusion in the scaffold: preseeded in the scaffolds before implantation in nonconditioned mice (preseeded); intrascaffold injection of hHSPCs in nonconditioned mice; i.v. injection of hHSPCs in scaffold-implanted nonconditioned mice (S+i.v.); and finally, irradiated mice injected i.v. with hHSPCs and 4 to 6 weeks later implanted with hMSC-seeded scaffolds (i.v.+S). All assays were performed without BMP2 (SC) or with BMP2 injection (BSC). All assays were performed with pools of HSPCs from different donors. (B) Analysis of human cells in the scaffolds. Percentage of hCD45+ cells (left panel) and proportion of hCD3+, hCD19+, and hCD33+ cells in the different scaffolds 12 weeks after implantation of HSPCs. Each point represents 1 mouse implanted with 2 to 6 scaffolds, which were pooled before the analysis. For each condition, 3 to 10 mice were transplanted. Preseeded approach shows higher engraftment levels than injected and S+i.v. approaches. Tukey’s multiple comparison test was applied: **P ≤ 0.005; ***P ≤ 0.0005.
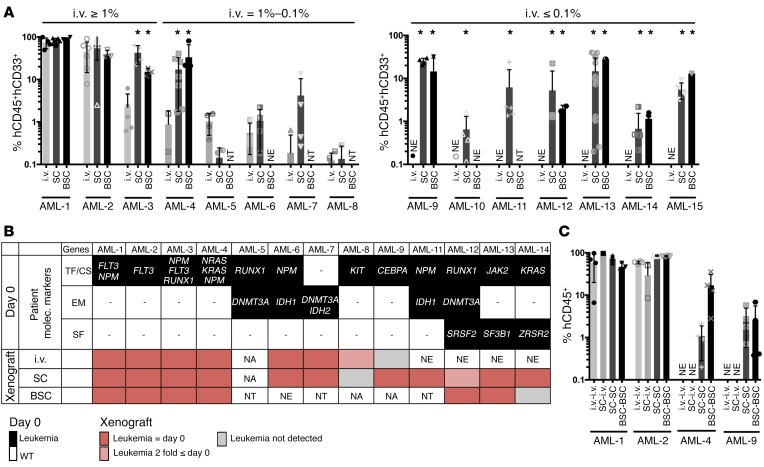
Next, we chose the preseeded protocol to test whether the “humanized” scaffolds were able to maintain AML patient samples in vivo. We selected a cohort of 15 AML patients (Supplemental Table 1), which included patients previously tested and classified as high engrafters (≥1%), low engrafters (1%–0.1%), and nonengrafters (≤0.1%) within the conventional i.v. transplantation model using NSG mice.
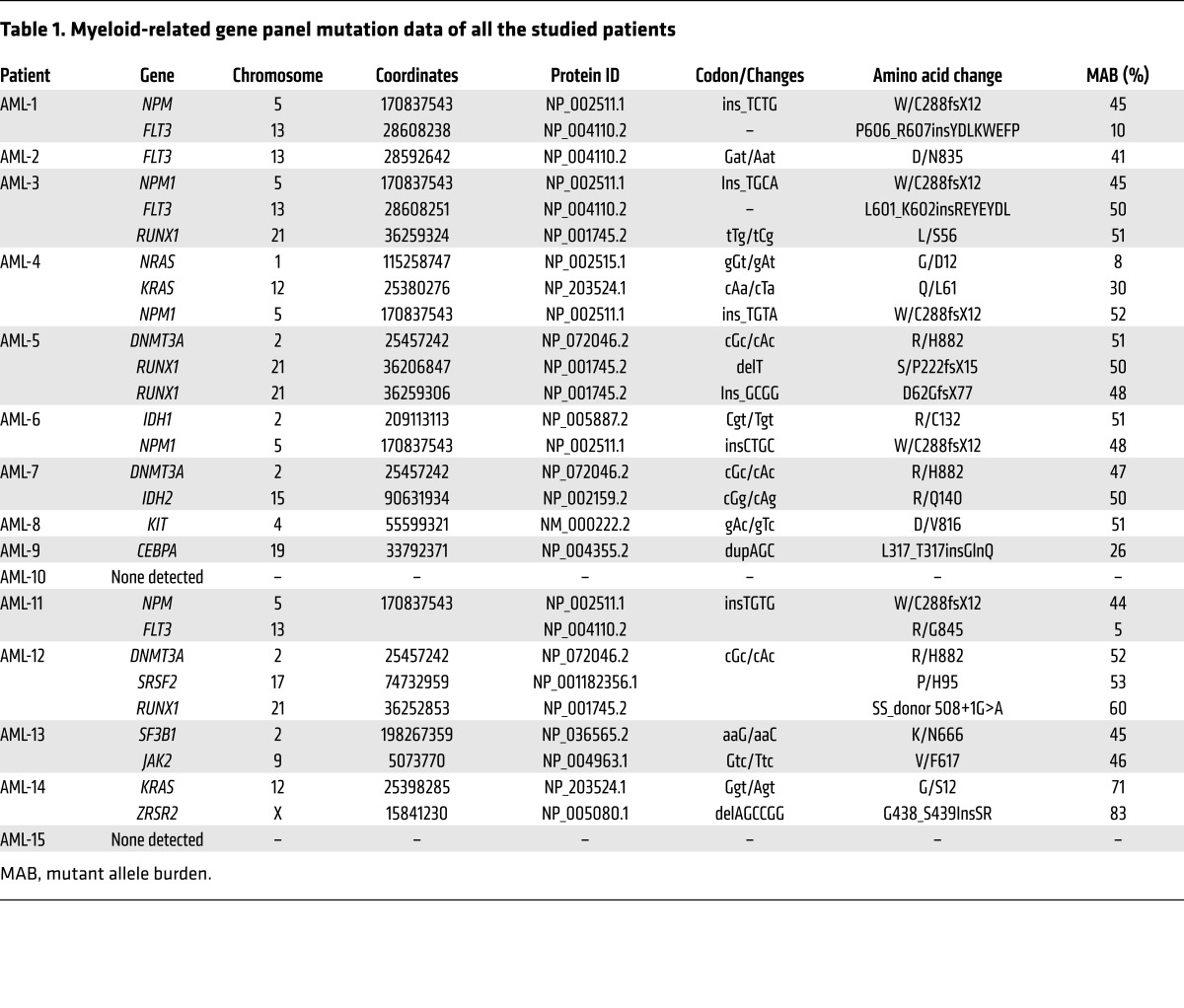
We generally observed a comparable or superior level of engraftment in the 2-scaffold–implanted models (SC and BSC) compared with conventional i.v. injection in all high- or low-engrafted samples tested (8 out of 8 in the scaffold-implanted model versus 4 out of 5 in the bone scaffold–implanted model) (Figure 3A), with mainly no to little engraftment in host BM (Supplemental Figure 7), except when higher AML cell doses were seeded in the scaffold (see Supplemental Figure 7, B and C). Importantly, leukemic engraftment of the nonengrafter samples was exclusively observed in the scaffold models (7/7 cases in the scaffold-implanted model versus 5 out of 6 in the bone scaffold–implanted model) (Figure 3A). In order to compare and track the AML burden in the xenotransplantation models, a panel of 24 genes, commonly mutated in myeloid malignancies, was used to screen hCD33+ pretransplanted cells obtained at day 0 from all patients (Table 1). Following on, patient-specific targeted mutation screen was performed on the posttransplanted cells. Analysis showed that in the implanted scaffolds, leukemic burden was largely maintained compared to the preimplanted patient cells, indicating that the scaffold model retained the original clonal architecture (Figure 3B and Supplemental Table 2). Finally, we tested to determine whether the leukemic cells present in these scaffolds (see absolute number of cells retrieved/mouse, Supplemental Figure 8) had leukemic stem cell capacity by performing secondary transplantation. For patients 1 and 2, which were high engrafters, we observed similar secondary engraftment whatever model was used, whereas for patients 4 and 9 (low engrafter and nonengrafter, respectively), secondary engraftment was only successful in the scaffold models. Interestingly, in this case, samples from primary scaffolds were not able to engraft the BM of secondary mice when transplanted i.v., suggesting their dependence on a humanized environment for their growth (Figure 3C).
Figure 3. AML engraftment in different implantation models.
(A) Percentage of hCD45+hCD33+ cells recovered from each xenograft implantation model compared with conventional i.v. injection in NSG mice 12 weeks after transplant for patients classified as high engrafters (≥ 1% engraftment), low engrafters (between 1 to 0.1%), and nonengrafters (< 0.1%) using conventional i.v. injection. i.v., i.v. injection; SC, scaffold-implantation; BSC, bone scaffold-implantation. Comparison between i.v. and SC or BSC was applied using Dunnett’s test. *P < 0.05. Each point represents 1 mouse implanted with 2 to 6 scaffolds. For each patient, 2 to 8 mice were transplanted per condition. (B) Graphical representation of gene mutations detected in CD33+ pretransplant and hCD45+hCD33+ postxenografted cells. Mutations are grouped in transcription factor and cell signaling (TF/CS) genes, epigenetic modifier (EM) genes, and splicing factor (SF) genes. NA, sample not available for testing; NE, no engraftment was detected in mice; NT, not tested. Red boxes, variant allele frequencies comparable to day 0; pink boxes, ≤ 2-fold day 0; gray boxes, not detected. Variant allele frequencies for xenotransplanted samples are the average between ≥1, where applicable. (C) Percentage of hCD45+ cells recovered from secondary recipients. Primary cells from i.v. mice were transplanted i.v. into secondary mice (i.v.-i.v.). Primary cells from SC were transplanted i.v. (SC-i.v.) or implanted into scaffold (SC-SC) in secondary mice. Primary cells from BSC were implanted into BSC in secondary mice (BSC-BSC). Each point represents 1 mouse. For each patient, 2 to 4 mice were transplanted per condition.
Table 1. Myeloid-related gene panel mutation data of all the studied patients.
Overall, our work describes a versatile humanized niche model for studying both normal and leukemic human hematopoietic cell biology, particularly for the less aggressive subtypes that fail to engraft in NSG mice. Recent reports demonstrate the use of humanized bone ossicles for the study of malignant hematopoiesis (11–16). Nevertheless, in all of these models, hMSCs were first induced to form bone ossicles before malignant hematopoietic cells were injected intrascaffold, making the duration of the whole experiment between 20 and 34 weeks compared with only 10 to 12 weeks in our study. Importantly, our data demonstrate that bone formation is not mandatory for human normal and leukemic engraftment, contrary to what was previously described (18, 19). Moreover, the flexibility of our reported approach should also help to decipher the role of different human stromal cell types (normal or malignant hMSCs or even comparing in vivo hMSC versus human endothelial cells versus osteoblasts) as well as the importance of particular signaling pathways within the hematopoietic niche, using genetically modified stromal cells to dissect the crosstalk between normal and/or leukemic human hematopoietic cells and their stroma. This model could also be used to screen drugs, specifically those that target stromal components.
Methods
Additional methods and associated references are available in the Supplemental Methods.
Study approval.
All animal experiments were performed under license PPL 70/8904 approved by the Home Office of the United Kingdom and in accordance with Cancer Research UK guidelines. The use of umbilical cord blood (UCB) and AML samples was approved by the East London Ethical Committee and carried out in accordance with the Declaration of Helsinki.
Statistics.
Prism Version 6 software (GraphPad) was used for statistical analysis. Data are presented as mean ± SEM. To determine the level of significance, statistical analysis was performed using either a Dunnett’s test for comparison of 2 groups or using a Tukey’s ANOVA test for multiple comparisons. A P value of less than 0.05 was considered significant.
Author contributions
AA, KF, and AH performed the research, analyzed data, and wrote the manuscript. SAM performed molecular analysis and analyzed results. JG provided AML samples and relevant patient information. GM provided the sequencing analysis before and after xenotransplantation. DB directed the research, analyzed data, and wrote the manuscript. DP performed research and analyzed data. All authors approved the manuscript.
Supplementary Material
Acknowledgments
We thank staff of the core facilities at the Francis Crick Institute (Biological Research Facility, Flow Cytometry, Experimental Histopathology, In Vivo Imaging, Electron Microscopy, and Cryo-Electron Microscopy) for their valuable help. This work was supported by The Francis Crick Institute, which receives its core funding from Cancer Research UK (FC001045), the UK Medical Research Council (FC001045), and the Wellcome Trust (FC001045).
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(2):543–548. https://doi.org/10.1172/JCI89364.
Contributor Information
Ander Abarrategi, Email: ander.abarrategi@crick.ac.uk.
Katie Foster, Email: katie.x.foster@gsk.com.
Ashley Hamilton, Email: ashley.x.hamilton@gsk.com.
Diana Passaro, Email: diana.passaro@crick.ac.uk.
John Gribben, Email: j.gribben@qmul.ac.uk.
Ghulam Mufti, Email: ghulam.mufti@kcl.ac.uk.
Dominique Bonnet, Email: dominique.bonnet@crick.ac.uk.
References
- 1.Pearce DJ, et al. AML engraftment in the NOD/SCID assay reflects the outcome ofAML: implications for our understanding of the heterogeneity of AML. Blood. 2006;107(3):1166–1173. doi: 10.1182/blood-2005-06-2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vargaftig J, et al. Frequency of leukemic initiating cells does not depend on the xenotransplantation model used. Leukemia. 2012;26(4):858–860. doi: 10.1038/leu.2011.250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanchez PV, et al. A robust xenotransplantation model for acute myeloid leukemia. Leukemia. 2009;23(11):2109–2117. doi: 10.1038/leu.2009.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Theocharides AP, Rongvaux A, Fritsch K, Flavell RA, Manz MG. Humanized hemato-lymphoid system mice. Haematologica. 2016;101(1):5–19. doi: 10.3324/haematol.2014.115212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lee J, et al. Implantable microenvironments to attract hematopoietic stem/cancer cells. Proc Natl Acad Sci USA. 2012;109(48):19638–19643. doi: 10.1073/pnas.1208384109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song J, et al. An in vivo model to study and manipulate the hematopoietic stem cell niche. Blood. 2010;115(13):2592–2600. doi: 10.1182/blood-2009-01-200071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Holzapfel BM, et al. Tissue engineered humanized bone supports human hematopoiesis in vivo. Biomaterials. 2015;61:103–114. doi: 10.1016/j.biomaterials.2015.04.057. [DOI] [PubMed] [Google Scholar]
- 8.Reinisch A, et al. Epigenetic and in vivo comparison of diverse MSC sources reveals an endochondral signature for human hematopoietic niche formation. Blood. 2015;125(2):249–260. doi: 10.1182/blood-2014-04-572255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torisawa YS, et al. Bone marrow-on-a-chip replicates hematopoietic niche physiology in vitro. Nat Methods. 2014;11(6):663–669. doi: 10.1038/nmeth.2938. [DOI] [PubMed] [Google Scholar]
- 10.Abarrategi A, et al. In vivo ectopic implantation model to assess human mesenchymal progenitor cell potential. Stem Cell Rev. 2013;9(6):833–846. doi: 10.1007/s12015-013-9464-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaiselbuh SR, Edelman M, Lipton JM, Liu JM. Ectopic human mesenchymal stem cell-coated scaffolds in NOD/SCID mice: an in vivo model of the leukemia niche. Tissue Eng Part C Methods. 2010;16(6):1523–1531. doi: 10.1089/ten.tec.2010.0179. [DOI] [PubMed] [Google Scholar]
- 12.Groen RW, et al. Reconstructing the human hematopoietic niche in immunodeficient mice: opportunities for studying primary multiple myeloma. Blood. 2012;120(3):e9–e16. doi: 10.1182/blood-2012-03-414920. [DOI] [PubMed] [Google Scholar]
- 13.Chen Y, et al. Human extramedullary bone marrow in mice: a novel in vivo model of genetically controlled hematopoietic microenvironment. Blood. 2012;119(21):4971–4980. doi: 10.1182/blood-2011-11-389957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reinisch A, et al. A humanized bone marrow ossicle xenotransplantation model enables improved engraftment of healthy and leukemic human hematopoietic cells. Nat Med. 2016;22(7):812–821. doi: 10.1038/nm.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sontakke P, et al. Modeling BCR-ABL and MLL-AF9 leukemia in a human bone marrow-like scaffold-based xenograft model. Leukemia. 2016;30(10):2064–2073. doi: 10.1038/leu.2016.108. [DOI] [PubMed] [Google Scholar]
- 16.Antonelli A, et al. Establishing human leukemia xenograft mouse models by implanting human bone marrow-like scaffold-based niches. Blood. 2016;128(25):2949–2959. doi: 10.1182/blood-2016-05-719021. [DOI] [PubMed] [Google Scholar]
- 17.Robert-Richard E, et al. Human cell engraftment after busulfan or irradiation conditioning of NOD/SCID mice. Haematologica. 2006;91(10):1384. [PubMed] [Google Scholar]
- 18.Chan CK, et al. Endochondral ossification is required for haematopoietic stem-cell niche formation. Nature. 2009;457(7228):490–494. doi: 10.1038/nature07547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scotti C, et al. Engineering of a functional bone organ through endochondral ossification. Proc Natl Acad Sci USA. 2013;110(10):3997–4002. doi: 10.1073/pnas.1220108110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.