Abstract
Myotonic dystrophy type I (DM1) is a disabling multisystemic disease that predominantly affects skeletal muscle. It is caused by expanded CTG repeats in the 3′-UTR of the dystrophia myotonica protein kinase (DMPK) gene. RNA hairpins formed by elongated DMPK transcripts sequester RNA-binding proteins, leading to mis-splicing of numerous pre-mRNAs. Here, we have investigated whether DM1-associated muscle pathology is related to deregulation of central metabolic pathways, which may identify potential therapeutic targets for the disease. In a well-characterized mouse model for DM1 (HSALR mice), activation of AMPK signaling in muscle was impaired under starved conditions, while mTORC1 signaling remained active. In parallel, autophagic flux was perturbed in HSALR muscle and in cultured human DM1 myotubes. Pharmacological approaches targeting AMPK/mTORC1 signaling greatly ameliorated muscle function in HSALR mice. AICAR, an AMPK activator, led to a strong reduction of myotonia, which was accompanied by partial correction of misregulated alternative splicing. Rapamycin, an mTORC1 inhibitor, improved muscle relaxation and increased muscle force in HSALR mice without affecting splicing. These findings highlight the involvement of AMPK/mTORC1 deregulation in DM1 muscle pathophysiology and may open potential avenues for the treatment of this disease.
Introduction
Myotonic dystrophy type I (DM1; OMIM #160900) is a multisystemic neuromuscular disorder, which represents the most common form of muscular dystrophy in adults (1). In particular, DM1 patients suffer from muscle wasting, weakness, and myotonia. DM1 is an autosomal dominant disease caused by an expansion of unstable CTG repeats located within the 3′-UTR of the dystrophia myotonica protein kinase (DMPK) gene. Toxic expanded transcripts containing RNA hairpins formed by the triplet repeats accumulate as RNA foci in the nuclei of affected cells (2, 3). These mutant transcripts are thought to sequester RNA-binding proteins, such as muscleblind-like 1 (MBNL1), and to increase CUG triplet repeat RNA-binding protein 1 (CUGBP1) levels. The resulting splicing defects are considered the primary cause of DM1 symptoms (4–6). HSALR mice, which carry a CTG repeat expansion in the human skeletal actin (HSA) gene, constitute a well-characterized mouse model for DM1 (5). These mice express (CUG)n-expanded transcripts specifically in skeletal muscle and reiterate the dystrophic phenotype and myotonic discharges observed in muscle of patients. HSALR mice also recapitulate DM1 molecular characteristics such as foci accumulation, MBNL1 sequestration, and splicing abnormalities (5, 7, 8). Therapeutic strategies have mainly focused on targeting DM1-associated mis-splicing and mRNA toxicity (9–11), although a more complete understanding of pathogenic pathways would clearly be of interest for the development of alternative or additional therapeutic options.
Recently, deregulation of cellular processes and signaling pathways important for maintaining proper muscle homeostasis has been reported in DM1. This includes abnormal activation of the ubiquitin-proteasome system and increased autophagic flux, which were both related to muscle atrophy and weakness in DM1 (12–14). In parallel, perturbation in the PKB/Akt pathway may arise from altered expression of the insulin receptor, which correlates with glucose intolerance in DM1 patients (15). Although PKB/Akt deregulation has been reported in Dmpk-deficient mice (16), in DM1 flies (13), and in DM1 human neural stem cells (17), contradictory results have been obtained in human muscle cells (18, 19).
To obtain further insight into the pathomechanisms associated with the disease, we investigated whether deregulated metabolic pathways may be involved in muscle alterations in DM1. We uncovered that muscles from HSALR mice do not efficiently respond to fasting by displaying impaired activation of AMPK and delayed inhibition of the mTOR complex 1 (mTORC1) pathway. Moreover, we observed mild perturbations of the autophagic flux in both HSALR muscle and myotubes from DM1 patients, which may arise from AMPK/mTORC1 deregulation. Importantly, we established that treatments normalizing these pathways improved skeletal muscle strength and strongly reduced the myotonia in HSALR mice. Our data provide evidence for the pathological role of metabolic pathways in DM1 and may open interesting avenues for alternative therapeutic strategies for the disease.
Results
AMPK and mTORC1 pathways are deregulated in HSALR muscle.
To identify pathomechanisms involved in DM1-related muscle alterations, we examined the potential deregulation of metabolic pathways in HSALR mice (Supplemental Figure 1A; supplemental material available online with this article; doi:10.1172/JCI89616DS1). To this purpose, the activation state of key proteins was compared in muscle from 2-month-old mice analyzed in fed conditions or subjected to a physiological stimulus like fasting (20). No major difference was observed in the activation state of AMPK, PKB/Akt, and mTORC1 pathways in muscle from fed mutant and control mice, as reflected by the similar phosphorylation levels of AMPK (AMPKP172), PKB/Akt (AktP473), and the mTORC1 targets ribosomal protein S6 kinase (p70S6KP389) and S6 ribosomal protein (S6P235/6 and S6P240/4) (Figure 1, A and B). After 24 hours of starvation, HSALR mice showed impaired activation of the AMPK pathway, as revealed by the reduced levels of AMPKP172 in tibialis anterior (TA) mutant muscle (Figure 1A). Regardless of the nutritional status, protein expression of the known AMPK regulatory kinases liver kinase B1 (LKB1) and TGF-β–activated kinase 1 (TAK1) was unchanged in HSALR muscle compared with control (Figure 1A). In contrast, mutant muscle displayed an altered expression profile for Ca2+-calmodulin–dependent kinase II (CaMKII) isoforms, with marked reduction in levels of the CaMKIIβM muscle-specific form and of its phosphorylated, active form (Figure 1A). Such deregulation was consistent with splicing defects in the Camk2 genes previously described in tissues from DM1 patients and mouse models (11, 21–23). We confirmed by quantitative PCR that splicing of Camk2b was altered in muscle from HSALR mice (exon 13 exclusion; Supplemental Figure 1B), while overall expression of Camk2 transcripts was unchanged in comparison with controls (Supplemental Figure 1C). As CaMKII regulates AMPK (24–26), these results suggest that impaired AMPK activation in HSALR muscle may rely on mis-splicing–dependent CaMKII deficiency.
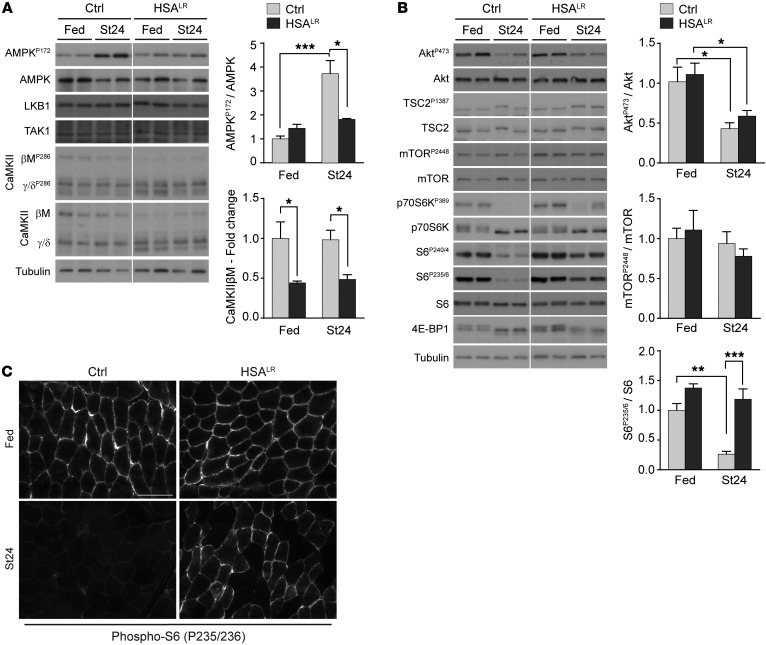
Figure 1. AMPK and mTORC1 pathways do not respond to starvation in HSALR muscle.
(A and B) Two-month-old HSALR and control (Ctrl) mice were examined in fed conditions and after 24 hours of starvation (St24). Immunoblots for phospho- (P) and total proteins of the AMPK (A) and mTORC1 (B) pathways reveal reduced AMPK activation and increased phosphorylation of some mTORC1 targets upon starvation in mutant muscle. Samples were run on the same gel but were noncontiguous. Protein quantification is given for AMPKP172 (n = 4 Ctrl and 3 HSALR), CaMKIIβM, AktP473, mTORP2448 (Fed, n = 3; St24, n = 4), and S6P235/6 (Fed, n = 3; St24, n = 7 Ctrl and 6 HSALR). Data are relative to fed control mice and are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, 2-way ANOVA with Tukey’s multiple comparisons test correction. (C) Immunostaining on muscle cross sections from fed and starved (St24) HSALR and control (Ctrl) mice shows high levels of phospho-S6 in mutant muscle upon starvation. Scale bar: 100 μm.
In parallel, higher phosphorylation of p70S6K and S6 was detected upon starvation in HSALR muscle compared with control muscle (Figure 1B). Accumulation of phosphorylated S6 in muscle from starved mutant mice was further confirmed by immunostaining (Figure 1C), suggesting an abnormal activation of the mTORC1 signaling in HSALR mice. The specificity of the staining was confirmed by use of the S6P235/6 blocking peptide and by immunostaining of sections from muscles with a constant activation (TSCmKO; ref. 27) or depletion (RAmKO; ref. 28) of mTORC1 (Supplemental Figure 1D). Notably, no major change in the phosphorylation of mTOR was observed in mutant and control muscles from fed versus starved mice (Figure 1B). Moreover, upon starvation, changes in 4E-BP1 levels were similar between HSALR and control muscles (Figure 1B), consistent with previous reports indicating differential regulation of mTORC1 targets (29). Interestingly, mTORC1 deregulation was not related to abnormal activity of PKB/Akt, since levels of the active phosphorylated form of PKB/Akt were efficiently decreased upon starvation in mutant mice (Figure 1B). Accordingly, we did not detect any changes in the splicing (exon 11, mis-spliced in DM1 patients) or expression of the gene encoding insulin receptor (Insr) in TA muscle from 2-month-old HSALR mice (Supplemental Figure 1E). Moreover, mTORC1 and AMPK activation state in nonmuscle tissue, such as liver, was similar in control and mutant mice (Supplemental Figure 1F), indicating that deregulation of these pathways is confined to skeletal muscles, which specifically express (CUG)n-expanded transcripts.
In an attempt to normalize mTORC1/AMPK pathways, control and HSALR mice were subjected to starvation for 45 hours. Mutant mice lost less weight than controls after prolonged starvation (Supplemental Figure 2). Moreover, upon 45 hours of starvation, 4E-BP1 and phospho-S6 levels were similar in mutant and control muscles, while AMPK phosphorylation showed only a trend toward increase in HSALR muscle (Figure 2A). We next addressed whether pharmacological treatments would be sufficient to modulate AMPK/mTORC1 pathways in HSALR mice. Control and mutant mice were treated for 5 days with metformin, a drug known to induce AMPK signaling. The treatment slightly activated AMPK in muscle from starved HSALR mice, which was accompanied by a decrease in phospho-S6 levels (Figure 2B). Conversely, a single injection of rapamycin, a canonical inhibitor of mTORC1, strongly reduced S6P235/6 levels in muscle from HSALR mice, to levels similar to those in controls. This further confirmed that S6 deregulation is dependent on mTORC1/p70S6K and does not involve ERK/p90S6K, which can also phosphorylate S6 at Ser235/6 (30). Levels of AMPKP172 remained unchanged in rapamycin-treated mice (Figure 2C). Although no obvious change was detected in AMPK-dependent phosphorylation of TSC2 (TSC2P1387), an upstream inhibitor of mTORC1 (Figure 1B), these results suggest that AMPK deregulation may primarily be responsible for the defective response to starvation and for mTORC1 signaling perturbation in HSALR muscle.
Figure 2. AMPK and mTORC1 pathways can be modulated by caloric and pharmacological treatments in HSALR muscle.
Immunoblots for phospho- (P) and total AMPK and S6 proteins reveal efficient inhibition of mTORC1 signaling upon 45 hours of starvation (St45, A) and with metformin (MetF, B) or rapamycin (Rapa, C) treatment in muscle from HSALR mice. AMPK activation shows a trend toward increase in mutant muscle with metformin treatment (B). Samples were run on the same gel but were noncontiguous. Protein quantification is shown for AMPKP172 and S6P235/6 (Fed, n = 3; St45, n = 4 Ctrl and 3 HSALR; Veh [B], n = 3; MetF, n = 4; Veh [C], n = 4 Ctrl and 3 HSALR; Rapa, n = 3 per genotype). Data are relative to fed (A) or vehicle-treated (B and C) control mice and are mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, 2-way ANOVA with Tukey’s multiple comparisons test correction.
Autophagic flux is perturbed in HSALR muscle.
It is well established that mTORC1 and AMPK are key regulators of autophagy and that perturbation of their activities can lead to severe tissue alterations, especially in skeletal muscle (31–33). To determine whether the expression of the CUG repeats impairs the autophagy process, we assessed the ability of HSALR muscle to induce autophagy when the mice were subjected to starvation. First, we evaluated levels of the soluble (LC3I) and autophagosome-associated (LC3II) forms of the widely used LC3B (MAP1LC3 for microtubule-associated protein light chain 3) autophagy marker. The amount of LC3II correlates with the intracellular accumulation of autophagic vesicles (34). Under fed conditions, LC3II levels were increased, although not significantly, in mutant muscle, which reflects either a slight increase in autophagy induction or a mild defect in the degradation steps (Figure 3A and Supplemental Figure 3A). After 24 hours of starvation, a clear switch from LC3I to LC3II occurred in control muscle, while HSALR muscle displayed reduced changes in LC3 levels and LC3II/LC3I ratio (Figure 3A and Supplemental Figure 3A). To confirm these results, we starved HSALR and control mice expressing the GFP-LC3 fusion protein for 24 hours. In control muscle, a striking increase in the number of GFP-LC3–positive puncta, representing autophagic vesicles, was observed upon starvation (Figure 3B and Supplemental Figure 3B). In HSALR mice, the number of puncta was higher under fed conditions, but was significantly less increased upon starvation as compared with control muscle (Figure 3B and Supplemental Figure 3B). These results confirmed that autophagy is perturbed in HSALR muscle, which may rely on impaired induction and/or degradation steps.
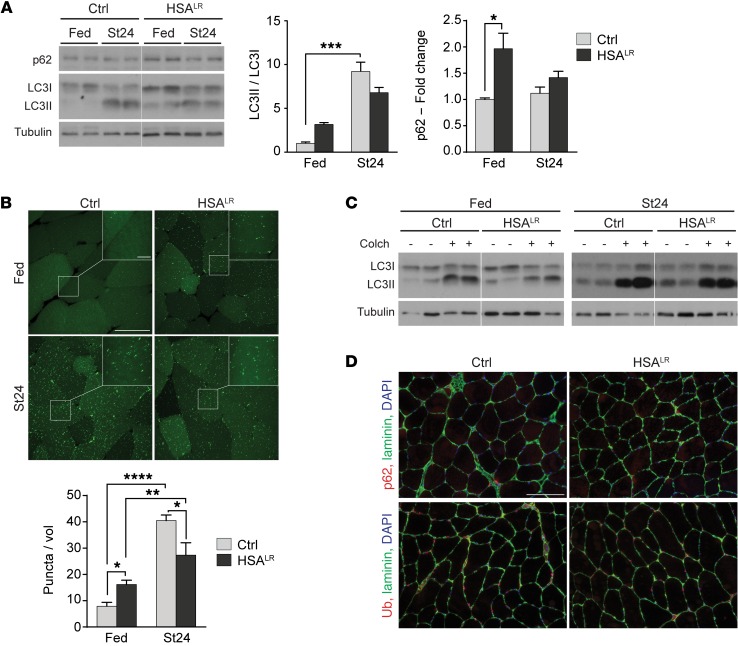
Figure 3. HSALR muscles show mild deregulation of the autophagic flux.
(A) Immunoblots for autophagy-related proteins show accumulation of autophagic substrates in HSALR TA muscle in fed conditions. A reduced LC3I-to-LC3II switch is observed in mutant muscle upon 24 hours of starvation (St24), compared with control (Ctrl). Samples were run on the same gel but were noncontiguous. (Fed, n = 3; St24, n = 7 Ctrl and 6 HSALR for LC3 ratio, n = 4 for p62.) For LC3I and LC3II levels, see Supplemental Figure 3A. (B) HSALR mice expressing GFP-LC3 display increased number of GFP-positive puncta in TA muscle compared with control (Ctrl) in fed conditions (n = 3 Ctrl and 4 HSALR), but reduced accumulation after 24 hours of starvation (St24, n = 3). Scale bar: 50 μm; 10 μm for insets. A volume unit (vol) corresponds to 2.8 × 103 μm3. (C) Treatment with colchicine (Colch) leads to milder changes in LC3II levels in TA muscle from fed and starved HSALR mice, compared with control (Ctrl) mice. For LC3II/LC3I quantification, see Supplemental Figure 3C. (D) Immunostaining of muscle sections from starved control (Ctrl) and HSALR mice reveals no major accumulation of p62 or ubiquitinated proteins in mutant muscle. Scale bar: 100 μm. Data are relative to control fed mice and represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, 2-way ANOVA with Tukey’s multiple comparisons test correction.
To assess the status of the autophagic flux, mice were treated for 2 days with colchicine, a drug preventing degradation of the autophagic content. Under both fed and starved conditions, colchicine induced a major switch from LC3I to LC3II in control muscle (Figure 3C). Comparing colchicine-treated and untreated mice, we observed that the fold change in the LC3II/LC3I ratio was less in HSALR muscle compared with controls, in both fed and starved conditions (Figure 3C and Supplemental Figure 3C). This result ruled out that elevated LC3II levels in fed conditions were due to increased autophagy induction in HSALR muscle; accumulation of autophagic vesicles was therefore likely related to restricted degradation. Consistently, levels of the autophagosome cargo protein p62 were higher in muscle from fed mutant mice than in controls (Figure 3A). However, we detected neither p62 aggregates nor accumulation of ubiquitinated proteins in muscle from fed and starved HSALR mice (Figure 3D), indicating that autophagy is only mildly affected. Similarly, distribution of lysosomal vesicles, visualized by LAMP1 immunostaining, was unchanged between 2-month-old mutant and control mice (Supplemental Figure 3D). Together, these results suggest that autophagy is slightly deregulated in DM1 muscle, which results from reduced degradation in combination with attenuated autophagy induction upon starvation.
Under fed conditions, increased amounts of p62 and LC3II were not due to abnormal transcript expression in HSALR muscle (Figure 4A). Moreover, expression of the Map1lc3b, Sqstm (encoding p62), and Ctsl (encoding cathepsin L) genes was unchanged after 24 hours of fasting, but we observed an efficient induction of the genes upon 45 hours of starvation in both mutant and control muscles (Figure 4A and Supplemental Figure 4A). It should be noted that following prolonged starvation, autophagy induction remained weaker in mutant muscle compared with control muscle (Figure 4B and Supplemental Figure 4B). To gain further insight into autophagy deregulation, we assessed the phosphorylation state of Unc-51–like kinase 1 (ULK1), as mTORC1 and AMPK phosphorylate and thereby inhibit or activate ULK1, respectively (31). Upon starvation, levels of the inactive form of ULK1 (ULK1P757) remained slightly higher in mutant muscle as compared with control muscle, while no major difference was observed for its active form (ULK1P317; Figure 4C and Supplemental Figure 4C). Interestingly, rapamycin and metformin treatments were both sufficient to increase LC3II levels in control muscle, but did not induce autophagy in HSALR muscle (Figure 4, D and E, and Supplemental Figure 4, D and E). Hence, mTORC1/AMPK deregulation in conjunction with mTORC1/AMPK–independent mechanisms likely contributes to autophagy perturbation in HSALR muscle.
Figure 4. HSALR muscles display perturbed response of autophagy to caloric and pharmacological treatments.
(A) Expression of autophagy-related genes is efficiently upregulated after 45 hours of starvation (St45) in HSALR TA muscle. Data are normalized to Actn2 levels (Fed, n = 4; St45, n = 4 Ctrl and 3 HSALR). Data are relative to control fed mice and represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, 2-way ANOVA with Tukey’s multiple comparisons test correction. (B) Immunoblots reveal limited switch from LC3I to LC3II in HSALR muscle upon 45 hours of starvation (St45) compared with controls (Ctrl). Samples were run on the same gel but were noncontiguous. For LC3II/LC3I quantification, see Supplemental Figure 4B. (C) Levels of the inhibited phosphorylated form of ULK1 (SerP757) remain slightly higher upon starvation in HSALR muscle, compared with control (Ctrl) muscle. For quantification, see Supplemental Figure 4C. (D and E) Immunoblots for LC3 show blunted induction of LC3II upon rapamycin (Rapa, D) or metformin (MetF, E) treatments, compared with controls (Ctrl). For LC3II/LC3I quantification, see Supplemental Figure 4, D and E. Samples were run on the same gel but were noncontiguous.
Lastly, upon starvation, expression of the atrogenes Trim63 and Fbxo32 was similarly induced in HSALR and control muscles (Supplemental Figure 4F). However, caspase- and trypsin-like activities associated with the proteasome system were increased in muscle from fed and starved mutant mice, compared with control animals (Supplemental Figure 4G). This is consistent with a previous report showing higher proteasome activity in muscle from a DM1 mouse model expressing 550 CTG triplets (12).
Involvement of AMPK/mTORC1 deregulation in DM1 pathology.
To ascertain the relevance of the changes observed in HSALR mice for DM1 pathology, we evaluated the activation state of AMPK/mTORC1 signaling in muscle biopsies from 3 DM1 patients (P1–3) of 33, 34, and 49 years of age. There was no major difference in total and phosphorylated levels of PKB/Akt and AMPK proteins in muscle from DM1 patients compared with age-matched control individuals (C1/2 and C3–5 aged 30 and 50 years, respectively) (Figure 5A). Notwithstanding, we noticed that levels of the active phosphorylated forms of p70S6K and S6 were increased in muscle biopsies from DM1 patients compared with control individuals (Figure 5A). However, the nutritive status of the patients at the time of the biopsy could not be ascertained and may have influenced the results. For this reason, we next tested the ability of DM1 human muscle cells to modulate mTORC1/AMPK signaling in response to energy and nutrient deprivation. Fibroblasts of 3 DM1 patients (DM-L1–3) were transduced with MyoD and differentiated for 10 days into myotubes, before being subjected to growth medium or to amino acid– and glucose-deprived conditions (i.e., PBS) for 3 hours. Upon deprivation, levels of the active phosphorylated forms of PKB/Akt and S6 were strongly reduced in control muscle cells; there was no major activation of AMPK in comparison with enriched conditions (Figure 5B). A similar response was observed in DM1 muscle cells, although they retained higher phosphorylation of S6 in deprived conditions compared with control cells (Figure 5B). In parallel, a major switch from LC3I to LC3II occurred in control muscle cells upon deprivation. LC3II levels were further increased in control cells treated with chloroquine, consistent with high autophagy induction in deprived cells (Figure 5C). In contrast, LC3II levels were only slightly changed when DM1 myotubes were subjected to deprivation, even in the presence of chloroquine, indicating that the autophagic flux is blocked at the induction steps (Figure 5C). Together, these data indicate that DM1 human muscle cells do not efficiently respond to nutrient/energy deprivation and display deregulation of the autophagy process.
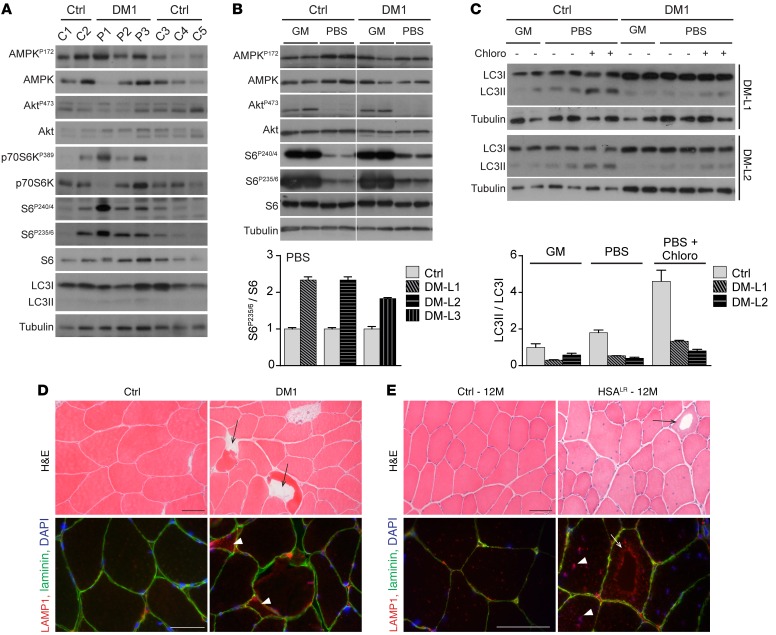
Figure 5. Autophagy perturbation contributes to muscle alterations in DM1.
(A) Protein lysates from muscle biopsies of control individuals (C1–5) and DM1 patients (P1–3) were analyzed for phospho- (P) and total proteins of the AMPK and PKB/Akt–mTORC1 pathways. (B) MyoD-transduced fibroblasts from controls (Ctrl) and DM1 patients were differentiated to myotubes and subjected to growth medium (GM) or deprived conditions (PBS) for 3 hours. Immunoblots for phospho- (P) and total proteins reveal increased phospho-S6 levels upon deprivation in the 3 cell lines of DM1 patients (DM-L1–3), compared with controls. Samples were run on the same gel but were noncontiguous. Quantification is given for deprived conditions; values are mean ± SEM of technical replicates. (C) Immunoblots for LC3 marker show defective accumulation of LC3II in DM1 myotubes upon energy and amino acid deprivation (PBS) as well as with deprived conditions and chloroquine treatment (Chloro), compared with control cells (Ctrl). Quantification of LC3II/LC3I ratio is shown for 2 DM1 cell lines (DM-L1/2) in enriched (GM) and deprived conditions; values are mean ± SEM of technical replicates. (D) H&E stain reveals the presence of vacuolated fibers (arrows) in muscle biopsy from 1 DM1 patient, together with lysosomal accumulation (arrowheads) observed by immunostaining in some affected muscle fibers (red, bottom panel). Scale bars: 50 μm. (E) Vacuoles (arrows) are observed in muscle from aging HSALR mice; the periphery of the vacuoles is strongly reactive with anti-LAMP1 antibodies (red, bottom panel), indicating accumulation of lysosomal structures in these regions. High density of lysosomes is also observed in nonvacuolated muscle fibers from 12-month-old (12M) mutant mice (arrowheads), compared with muscle from age-matched control mice (Ctrl). Scale bars: 50 μm.
To test the relevance of autophagy changes in DM1, we looked for muscle alterations related to autophagy defects in muscle biopsies from DM1 patients and in muscle from aged HSALR mice. As previously reported (14, 35–37), vacuolated fibers were observed in muscle biopsy of 1 DM1 patient, out of the 3 examined (Figure 5D). Lysosome accumulation was also detected in affected fibers from DM1 muscle (Figure 5D). However, in contrast to biopsies from an inclusion body myositis (IBM) patient, there was no accumulation of LC3, ubiquitinated proteins, or p62 in DM1 patient muscles (Supplemental Figure 5A). Consistently, LC3 levels detected by Western blot were similar in DM1 and control biopsies (Figure 5A). Interestingly, we observed some intracellular vacuoles in muscle from 12-month-old HSALR mice, as well as accumulation of the lysosomal marker LAMP1 near the vacuolar structures and myonuclei (Figure 5E). Secondary antibodies alone did not react with the vacuoles, and we did not observe such features in muscle from age-matched control mice (Figure 5E and Supplemental Figure 5B). Electron microscopy confirmed the presence of vacuoles in mutant mouse muscle: they were surrounded by dense, disorganized areas of contractile elements and usually limited by a single, discontinuous membrane (Supplemental Figure 5C). Together with the lysosome staining, these features argue for the presence of autophagic vacuoles in muscle from older HSALR mice. These results suggest that autophagy perturbation may contribute to the alteration of muscle tissue in DM1, but is unlikely to be a predominant feature of the disease.
AICAR, an AMPK agonist, abrogates myotonia in HSALR mice.
In light of the deregulation of AMPK signaling in HSALR muscle, we investigated whether AMPK normalization would have a beneficial effect on muscle function in mutant mice. As readout of the disease, we evaluated myotonia by measuring the late relaxation time of skeletal muscle (i.e., time to reduce the maximal force from 50% to 10%) after ex vivo tetanic stimulation (8). As reported previously, we observed no change in the late relaxation time of soleus muscle from HSALR mice compared with controls (Supplemental Figure 6A), whereas this parameter was strongly increased in extensor digitorum longus (EDL) mutant muscle (Figure 6A). Since AMPK activation by metformin has recently been shown to correct mis-splicing in human DM1 cells in vitro (38), 4- and 12-month-old control and HSALR mice were treated with metformin for 10 days. Despite using high doses of metformin, we observed only a limited and nonreproducible effect of the treatment on AMPK/S6 activation state in these groups of mice analyzed under basal nutritive conditions (Supplemental Figure 6B). Besides, the treatment failed to reduce the late relaxation time of EDL muscle in mutant mice (Figure 6B), and it did not modify the expression and splicing of genes affected in DM1, including Clcn1 (encoding CLC-1, chloride channel protein 1; inclusion of exon 7a) and Atp2a1 (encoding sarcoplasmic/endoplasmic reticulum Ca2+-ATPase 1; exclusion of exon 22) (Figure 6C and Supplemental Figure 6C).
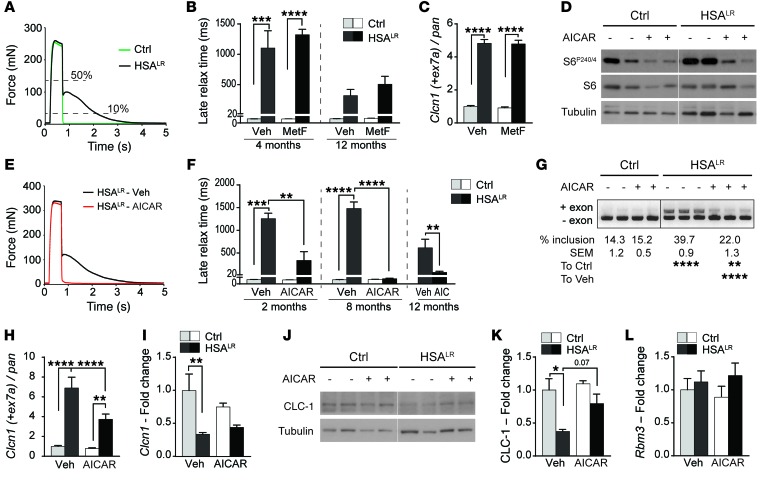
Figure 6. AICAR markedly decreases myotonia in HSALR mice and reduces mis-splicing in mutant muscle.
(A) In vitro tetanic stimulation of EDL muscle reveals strongly increased relaxation time in HSALR muscle. (B) Metformin (MetF) treatment does not reduce muscle late relaxation time in 4-month-old (Ctrl, n = 5; HSALR, n = 6 Veh and 8 MetF) and 12-month-old (Ctrl, n = 3; HSALR, n = 7 Veh and 8 MetF) HSALR mice, as compared with vehicle-treated mutant mice. (C) Inclusion of exon 7a of the Clcn1 gene is not changed in muscle from metformin-treated (MetF) HSALR mice, compared with vehicle-treated mice (n = 3). (D) Immunoblots for phospho- and total S6 protein reveal efficient inhibition of indirect AMPK target in muscle from control (Ctrl) and mutant mice treated with AICAR. Samples were run on the same gel but were noncontiguous. (E) AICAR treatment normalizes the time to relax of HSALR muscle upon tetanic stimulation, compared with muscle from vehicle-treated (Veh) mutant mice. (F) Late relaxation time is significantly reduced in EDL muscle from 2-month-old (n = 3 Ctrl and 4 HSALR), 8-month-old (Ctrl, n = 3; HSALR, n = 6 Veh and 7 AICAR), and 12-month-old (n = 4 Veh and 5 AICAR) HSALR mice that were treated with AICAR, as compared with age-matched vehicle-treated (Veh) mutant mice. (G–K) End-point PCR (G) and quantitative PCR (H and I) reveal strong reduction in exon 7a inclusion of the Clcn1 gene in muscle from HSALR mice treated with AICAR, compared with vehicle-treated (Veh) mutant mice (Ctrl, n = 3; HSALR, n = 5 Veh and 4 AICAR [G], n = 5 [I]). Protein levels of CLC-1 are also increased in mutant muscle from AICAR-treated mice (J and K, n = 3 Ctrl and 4 HSALR). (L) Quantitative PCR shows similar transcript levels of Rbm3 in muscle from AICAR-treated and untreated mice (n = 3 Ctrl and 4 HSALR). Data represent mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, 2-way ANOVA with Tukey’s multiple comparisons test correction (except 12M AICAR, unpaired 2-tailed Student’s t test).
As we did not detect any effect of metformin on HSALR muscle, we tested whether 5-aminoimidazole-4-carboxamide ribonucleotide (AICAR), an agonist of AMPK, may constitute an alternative strategy to target AMPK activation in muscle. Following a 7-day treatment with AICAR, control and mutant muscles showed a clear reduction in phospho-S6 levels (Figure 6D). We further confirmed that AICAR increased phosphorylated levels of AMPK and those of its direct target acetyl-CoA carboxylase shortly after the last injection (30 minutes), while inhibition of the indirect AMPK target S6 was detected only after 2 hours in control muscle (Supplemental Figure 6D). Importantly, following tetanic stimuli, a strong and significant reduction in the late relaxation time of EDL muscle was detected in 2- and 12-month-old AICAR-treated mutant mice, and myotonia was completely abrogated with AICAR in the group of 8-month-old HSALR mice (Figure 6, E and F). Normalization of the half-relaxation time (i.e., time to reduce the maximal force from 100% to 50%) of mutant muscle was also observed upon treatment (Supplemental Figure 6E). As mis-splicing of the Clcn1 gene is thought to be the primary cause of myotonia in DM1 (7, 39–41), we investigated whether the effect of AICAR was related to changes in Clcn1 splicing. By end-point PCR, a significant improvement of the misregulated Clcn1 splicing was detected upon AICAR treatment (Figure 6G). We confirmed by quantitative PCR that the expression of the mis-spliced Clcn1 transcript (containing exon 7a) was strongly reduced in muscle from AICAR-treated HSALR mice, while overall transcript levels of Clcn1 were unchanged compared with those in untreated mutant mice (Figure 6, H and I). Furthermore, AICAR led to a slight increase in CLC-1 protein levels in mutant muscle (Figure 6, J and K). It is worth noting that AICAR did not change splicing of the Atp2a1 and Camk2b genes in HSALR muscle (Supplemental Figure 6, F and G). Moreover, we did not detect any reduction in transcript levels of Rbm3, encoding RNA-binding protein 3 (Figure 6L), previously suggested to mediate the effect of AMPK activation on splicing (38).
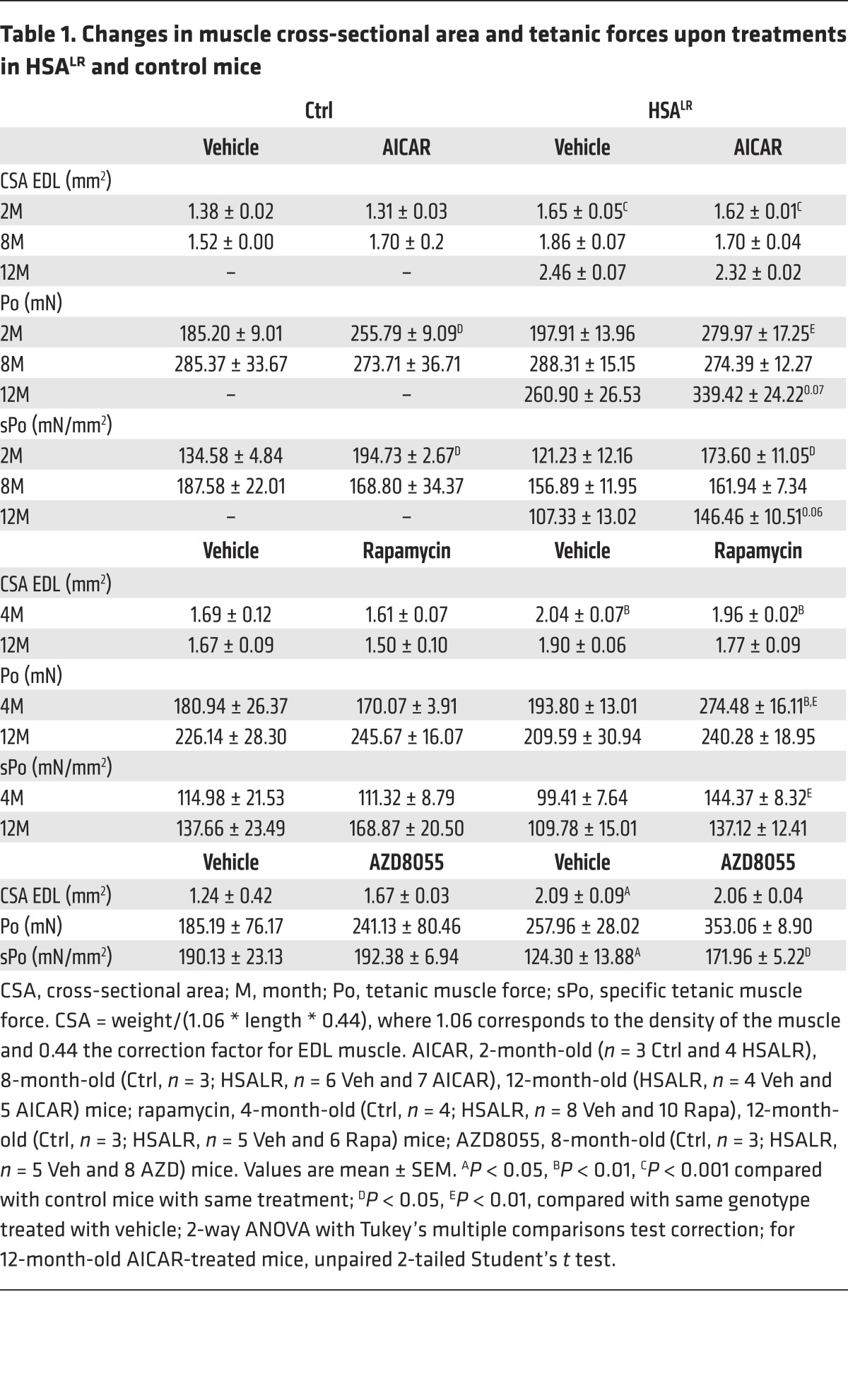
Besides its effect on myotonia, we wondered whether AICAR treatment would change muscle force in mutant mice. As initially reported (5), we detected neither muscle wasting nor reduction in total twitch (Pt) and tetanic (Po) muscle forces in HSALR mice compared with controls (Table 1 and Supplemental Table 1). Nonetheless, as cross-sectional area [CSA: mass/(density * length * correction factor)] of EDL muscle was increased in mutant mice, specific muscle forces (sPt and sPo), representative of the contractile capacity of the myofibers, were reduced in HSALR mice compared with control animals (Table 1 and Supplemental Table 1). Upon AICAR treatment, we observed that both total and specific forces of EDL muscle were increased in 2-month-old HSALR and control mice, but not in older mice (Table 1 and Supplemental Table 1). Altogether, these results indicate that targeting AMPK activation by AICAR improves muscle function in HSALR mice by reducing myotonia and potentially increasing muscle force, at least in part, through splicing correction.
Table 1. Changes in muscle cross-sectional area and tetanic forces upon treatments in HSALR and control mice.
Rapamycin treatment improves muscle function in HSALR mice.
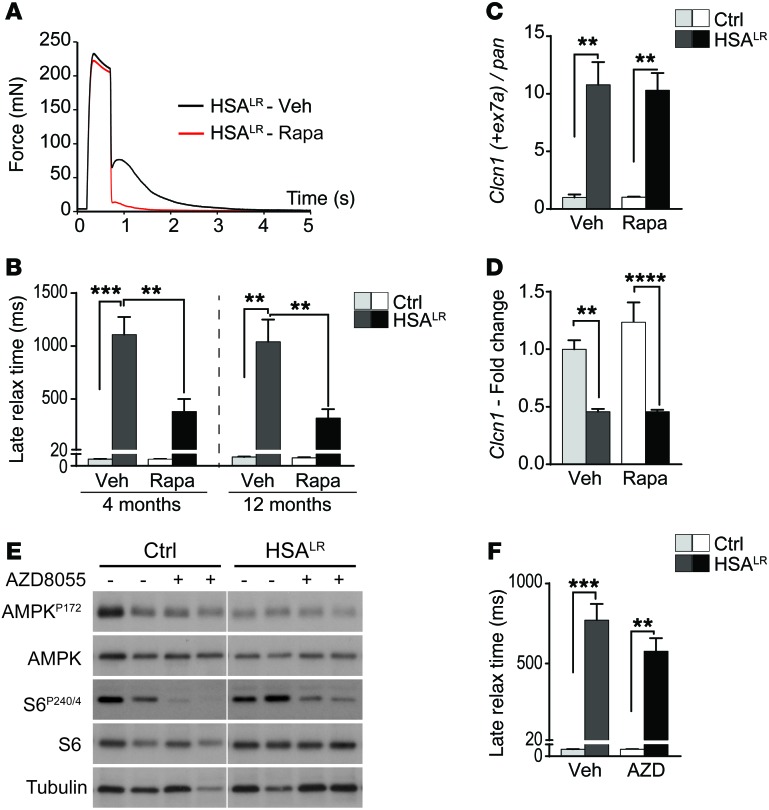
Based on the abnormal activation of mTORC1 signaling detected in HSALR muscle, we wondered whether indirect mTORC1 inhibition was part of the effect of AICAR and whether direct mTORC1 inhibition would improve muscle function in mutant mice. To this purpose, we subjected 4- and 12-month-old mice to rapamycin treatment for 7 and 10 days, respectively. Rapamycin treatment efficiently inhibited mTORC1 signaling, as shown by reduced phospho-S6 levels in muscle from control and HSALR mice (Supplemental Figure 7A). Rapamycin did not affect muscle half-relaxation time (Supplemental Figure 7B), but significantly reduced the late relaxation time of EDL muscle from 4- and 12-month-old HSALR mice (Figure 7, A and B). Moreover, we detected a significant increase in total and specific muscle forces in young rapamycin-treated mutant mice compared with vehicle-treated animals. Forces remained unchanged upon treatment in older mice (Table 1 and Supplemental Table 1).
Figure 7. Rapamycin improves muscle function in HSALR mice via splicing-independent mechanisms.
(A) Rapamycin treatment strongly reduces the time to relax of HSALR muscle upon tetanic stimulation, compared with muscle from vehicle-treated (Veh) mutant mice. (B) Rapamycin (Rapa) treatment significantly reduces late relaxation time of muscle from 4-month-old (Ctrl, n = 4; HSALR, n = 8 Veh and 10 Rapa) and 12-month-old (Ctrl, n = 3; HSALR, n = 5 Veh and 6 Rapa) HSALR mice, as compared with age-matched, vehicle-treated mutant mice. (C and D) Splicing (C) and overall transcript expression (D) of the Clcn1 gene are not modified in muscle from rapamycin-treated (Rapa) HSALR mice, compared with vehicle-treated (Veh) mutant mice. Values are relative to vehicle-treated control mice (n = 3 Ctrl and 4 Veh-treated and 5 Rapa-treated HSALR). (E) Treatment with AZD8055 for 10 days efficiently reduces phosphorylation of mTORC1 target, S6, in control (Ctrl) and HSALR muscle, but does not change AMPK activation. Samples were run on the same gel but were noncontiguous. (F) AZD8055 (AZD) does not reduce late relaxation time of EDL mutant muscle, compared with vehicle-treated (Veh) mutant mice. (n = 3 Ctrl and 5 Veh and 8 AZD HSALR mice.) Data represent mean ± SEM. **P < 0.01, ***P < 0.001, ****P < 0.0001, 2-way ANOVA with Tukey’s multiple comparisons test correction.
To test whether the effect of rapamycin on muscle function relied on splicing improvement, we assessed Clcn1 mis-splicing (i.e., exon 7a inclusion) by quantitative PCR. Interestingly, rapamycin affected neither Clcn1 splicing (Figure 7C) nor the overall transcript expression of the gene (Figure 7D) in mutant muscle. Consistently, splicing of the Atp2a1 gene was also not restored in rapamycin-treated HSALR mice (Supplemental Figure 7C). Together, these data suggest that mTORC1 inhibition by rapamycin is sufficient to improve muscle function in HSALR mice likely through splicing-independent mechanisms.
Since rapamycin has been shown to impact on channel function (e.g., ryanodine receptor 1, RyR1) via its binding to FKBP12, we wondered whether mTORC1 inhibition or the drug itself mediated the effect of the treatment on myotonia. Hence, control and HSALR mice were treated for 10 days with AZD8055, an ATP-competitive inhibitor of mTORC1. We confirmed that AZD8055 strongly decreased phospho-S6 levels in control and mutant muscle, while the activation state of AMPK remained unchanged (Figure 7E). In contrast to rapamycin, AZD8055 had no effect on late relaxation time of mutant muscle (Figure 7F). Nonetheless, total and specific forces of EDL muscle were increased in AZD8055-treated mutant mice, as observed with rapamycin (Table 1 and Supplemental Table 1). Together, these results indicate that mTORC1 inhibition may ameliorate the contractile capacity of muscle in HSALR mice and suggest that improvement of muscle relaxation upon rapamycin and AICAR treatments may be independent of mTORC1.
AICAR, but not rapamycin, leads to nuclear foci dispersion in HSALR muscle.
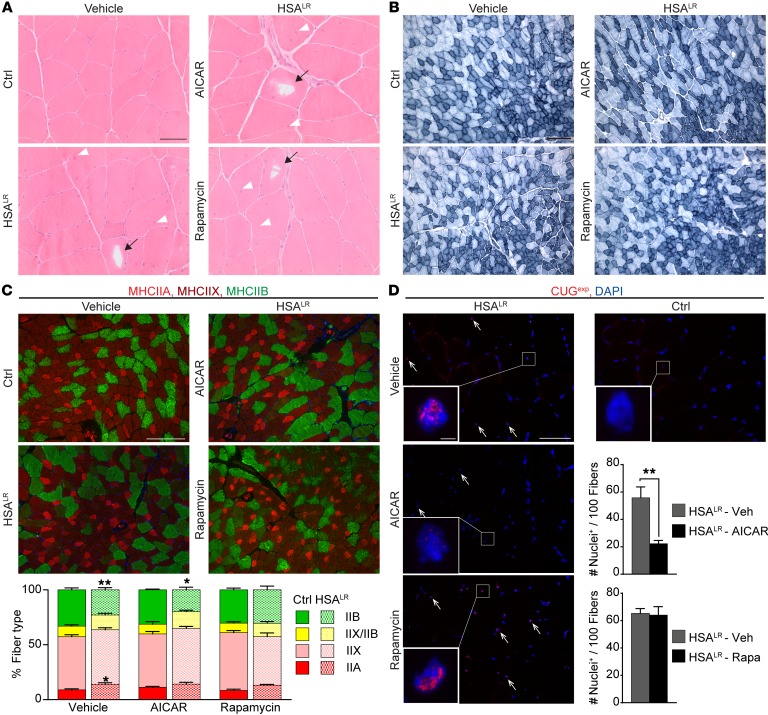
To further understand the beneficial effect of AICAR and rapamycin in HSALR mice, we investigated whether the treatments improved muscle function by affecting the properties of the diseased muscle. First, we did not observe major changes in the histopathology of HSALR muscle upon 7-day AICAR or 10-day rapamycin treatment, compared with untreated conditions (Figure 8A). Notably, vacuoles remained present in muscle fibers from 12-month-old AICAR- or rapamycin-treated HSALR mice (Figure 8A), indicating that the treatments were not sufficient to reverse muscle alterations related to impaired autophagy in aging mice. As no myotonia was detected in the slow, soleus muscle of HSALR mice and as AICAR and rapamycin were previously shown to alter muscle fiber types (42, 43), we tested whether changes in muscle function upon treatments were related to modification of muscle metabolic and contractile capacities. By reduced nicotinamide adenine dinucleotide (NADH) staining, we first observed that the overall oxidative property of TA muscle was unchanged in AICAR- and rapamycin-treated mutant mice, compared with untreated animals (Figure 8B). Immunostaining against type I, IIA/X, and IIB myosin heavy chains (MHCs) was then conducted in TA muscle from HSALR and control mice to identify changes in muscle contractile properties upon treatment. Few type I fibers were present in all the muscles analyzed (data not shown). Mutant muscle displayed a switch to slower fibers (i.e., increased and reduced proportion of IIA and IIB fibers, respectively) compared with control muscle (Figure 8C). However, upon AICAR or rapamycin, there was no significant change in the proportion of the different fiber types in comparison with vehicle-treated mice (Figure 8C).
Figure 8. AMPK activation by AICAR leads to nuclear foci dispersion in HSALR muscle.
(A and B) H&E (A) and NADH (B) stains reveal no major change in muscle histopathology and oxidative capacity upon AICAR or rapamycin treatment in HSALR mice. Arrowheads and arrows show internalized nuclei and vacuoles, respectively. Scale bars: 50 μm (A), 200 μm (B). (C) Immunostaining for type IIA (bright red), IIX (dark red), and IIB (green) myosin heavy chains (MHC) reveals no significant change in the respective proportion of fiber types in TA mutant muscle upon AICAR (n = 3 Ctrl and 4 HSALR) or rapamycin (n = 4) treatment, compared with vehicle-treated HSALR mice (n = 6 Ctrl and 7 HSALR). Scale bar: 200 μm. (D) FISH on TA muscle sections with a Cy3-CAG10 DNA probe shows accumulation of nuclear foci in HSALR muscle (arrows). The number of stained nuclei is significantly decreased upon AICAR treatment (n = 4), but not with rapamycin (Rapa, n = 3), compared with vehicle-treated (Veh) mutant mice. Foci are not detected in control (Ctrl) muscle. Scale bar: 50 μm; 2 μm for insets. Data in C and D represent mean ± SEM. *P < 0.05, **P < 0.01, 2-way ANOVA with Tukey’s multiple comparisons test correction.
Since aggregation of (CUG)n-expanded RNA in nuclear foci is a histological hallmark in DM1 diseased muscle, we next wondered whether the treatments affected their accumulation in HSALR muscle. To this purpose, we performed FISH using a CAG10 DNA probe on TA muscle sections from mutant untreated and treated mice. Numerous foci were observed in HSALR muscle, while none were detected in control muscle (Figure 8D). Interestingly, the number of nuclei showing foci was significantly decreased in muscle from AICAR-treated mutant mice, while no change was observed with rapamycin (Figure 8D). Moreover, foci appeared more diffuse in positive nuclei from mutant muscle upon AICAR treatment, compared with untreated conditions (Figure 8D). Altogether, these results indicate that changes in muscle metabolic and contractile properties do not account for the beneficial effect of the short treatments applied to HSALR mice, while reduced muscle pathology upon AICAR-mediated acute AMPK activation likely involves nuclear foci dispersion in the mutant muscle.
Discussion
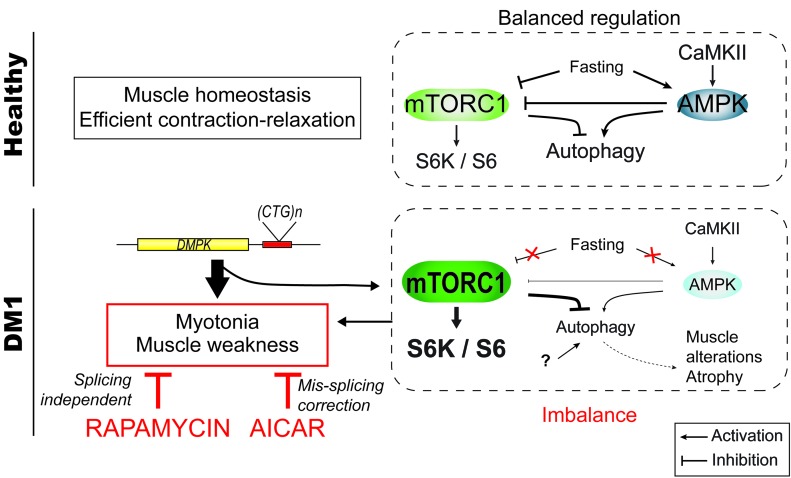
The pathogenic mechanisms underlying DM1 disease are still not well understood, and most investigations so far have focused on splicing defects caused by mRNA toxicity. In this study, we uncovered that in DM1, the AMPK and the mTORC1 pathways are deregulated and that the autophagic flux is perturbed in skeletal muscle. Most importantly, we established that AICAR and rapamycin, which interfere with AMPK/mTORC1 signaling, ameliorate DM1 muscle function (Figure 9).
Figure 9. Scheme depicting the deregulation of AMPK/mTORC1 signaling pathways in DM1 muscle.
In healthy muscle, the pathways are tightly regulated depending on external and internal stimuli (e.g., growth factors, energy, nutrients). Upon fasting, mTORC1 is inhibited, while AMPK is activated, leading to the induction of autophagy. In DM1, skeletal muscle does not respond to fasting conditions. Deregulation of the AMPK/mTORC1 signaling likely contributes to muscle dysfunction: rapamycin, an inhibitor of mTORC1, and AICAR, an AMPK agonist, both lead to marked reduction of myotonia and normalize muscle weakness in DM1 mice. The underlying mechanisms include RNA splicing–dependent and –independent mechanisms.
AMPK signaling and PKB/Akt–mTORC1 signaling are central metabolic pathways in muscle cells, and their deregulation has been related to muscle alterations and disease (27, 32, 44). We found that DM1 muscle shows an altered response to energy/nutrient-deprived conditions, with impaired AMPK activation and abnormal activation of mTORC1 signaling. Although we have not studied the upstream mechanisms involved in this deregulation, mis-splicing–dependent CaMKII deficiency could well account for the limited AMPK activation in DM1 muscle (21, 23–26). Interestingly, Jones et al. recently reported increased GSK3β levels and activity in HSALR muscle (45), which may also contribute to the perturbation of AMPK in DM1 muscle (46). In parallel, AMPK constitutes an upstream regulator of mTORC1 (47), and its deregulation could thus be responsible for abnormal mTORC1 activation in DM1 muscle cells. Nonetheless, as we did not detect changes in 4E-BP1 levels or mTOR phosphorylation in mutant muscle, evidence that the state of mTORC1 is modified in DM1 muscle and not only the activation of its downstream axis p70S6K/S6 is missing. Previous reports suggested that mTORC1 is inhibited in DM1 human neural and muscle cells (14, 17), although results were only obtained in vitro and the underlying mechanisms have not been investigated. On the basis of the abnormal splicing and protein trafficking of the insulin receptor in metabolic tissues in DM1 (15, 16), it has also been hypothesized that PKB/Akt–mTORC1 may be less responsive to insulin. However, results regarding the activation state of PKB/Akt signaling in DM1 human muscle biopsies or cells are conflicting (18, 19). In our experiments, we did not detect changes in PKB/Akt activation or in the expression of Insr in HSALR muscle, suggesting that AMPK/mTORC1 deregulation is independent of insulin receptor deficiency.
Autophagy as a major catabolic process essential for proteostasis has also been suggested to contribute to muscle alterations in DM1 (48). The involvement of autophagy in DM1 was largely deduced from the presence of autophagic vesicles and/or accumulation of autophagic markers in DM1 cells, but usually without dynamic measurement of the autophagic flux (13, 14, 17, 19, 36, 37, 49). In our study, we combined several methods to establish that mild changes in autophagic markers in muscle from HSALR mice are caused by autophagic flux limitation during the degradation steps. Further, we showed that even prolonged starvation did not fully induce autophagy in mutant muscle. We hypothesize that AMPK- and mTORC1-independent mechanisms contribute to autophagy perturbation, as rapamycin and metformin were not sufficient to normalize the flux in HSALR mice. Although this autophagy deregulation may contribute to muscle atrophy in DM1, it is unlikely to be the main pathogenic event, as autophagic features were scarce in DM1 muscle biopsies, compared with diseases primarily related to autophagy defects, such as vacuolar myopathies.
Importantly, we identified that AMPK/mTORC1 deregulation likely contributes to alteration of muscle function in DM1. Myotonia, which is due to membrane hyperexcitability, is thought to be caused primarily by mis-splicing and thereby deficiency in the CLC-1 (5, 7, 39, 41). In contrast to a recent report studying cultured human DM1 cells (38), we did not find any effect of metformin on the mis-splicing of DM1-affected genes or on the severe myotonia observed in HSALR mice. While we cannot rule out that changes in dosage and administration may lead to different results, much higher concentrations may be required to efficiently stimulate AMPK in rodent muscle tissue. Although metformin is commonly used to treat diabetes, clinical evaluation of its effect on muscle function in DM1 patients is lacking. In contrast to metformin, we found a profound effect of AICAR, a more potent AMPK agonist. AICAR led to a strong reduction of myotonia in HSALR mice, which correlated with improved splicing of the Clcn1 gene and increased protein levels of the channel. While AMPK activation was related to repression of the RNA-binding protein Rbm3 in vitro, Rbm3 expression was unchanged in HSALR muscle upon AICAR treatment. As we observed dispersion of the nuclear foci formed by the (CUG)n-expanded RNA aggregation after AICAR treatment, the effect of AMPK activation on splicing may be mediated by its interaction with other RNA-binding proteins, such as hnRNP H, which were implicated in foci stability in DM1 (50, 51). Hence, one may argue that AMPK deregulation likely contributes to pathogenesis in DM1 muscle by perturbing RNA-binding proteins and thereby accentuating foci stability and mis-splicing events (52–54). Notably, we cannot rule out that amelioration of muscle relaxation also relies on splicing-independent mechanisms. In particular, changes in sodium- and calcium-activated potassium channels or in Ca2+ homeostasis, which have also been suggested to contribute to myotonia in DM1 (55–60), may mediate some of the observed effect. Consistently, AMPK has been shown to modulate chloride and potassium channels in several cell types, including cardiomyocytes (61, 62). Such mechanisms may also contribute to the beneficial effect of rapamycin, as it occurs in the absence of splicing changes. Rapamycin could influence intracellular calcium mobilization by dissociating FKBPs (FK506-binding protein) from RyR1, thereby modifying channel activity (63–65). Although AZD8055-dependent mTORC1 inhibition did not improve muscle relaxation, further investigations are required to rule out the involvement of mTORC1 signaling in myotonia reduction, given the complexity of the signaling.
While muscle weakness is observed in DM1 patients, it was initially not reported in HSALR mice (5). Consistently, in our study, total muscle force was not affected in mutant mice but we observed reduced specific strength of EDL muscle in HSALR mice. Both specific and total muscle forces were increased upon AICAR, rapamycin, and AZD8055 treatments in young mutant mice, which may be mediated by mTORC1 inhibition as the signaling was shown to modulate Ca2+ homeostasis and excitation-contraction coupling in skeletal muscle (66). Myotonia reduction and increase in muscle force were not caused by modified metabolic and contractile properties of the mutant muscle upon the applied short-term treatments. However, one can hypothesize that the changes expected upon long-term administration of these drugs (i.e., switch toward slower fibers) may further positively affect DM1 muscle function.
In conclusion, we identified that deregulation of AMPK/mTORC1 signaling, together with mild autophagy perturbation, contributes to DM1-associated muscle alterations. We showed that treatments targeting the AMPK/mTORC1 imbalance are beneficial for muscle function, though to varying degrees. Whether alternative AMPK/mTORC1–targeting compounds, as well as changes in the dosage, administration mode, or treatment duration, may further improve muscle function remains to be investigated. As the drugs used in our study can target the pathways body-wide, they may also be beneficial in other tissues and thus may represent new treatment options for DM1.
Methods
Mice.
Homozygous mice of the mouse line LR20b carrying about 250 (CTG) repeats within the HSA transgene (HSALR) were obtained from Thornton and colleagues (University of Rochester Medical Center, Rochester, New York, USA) (5). Mice of the corresponding background strain (FVB/N) were used as control. GFP-LC3 HSALR and GFP-LC3 FVB/N mice were obtained by crossing of GFP-LC3–expressing mice (20) with HSALR or FVB/N mice. Mice were genotyped for HSALR transgenes by quantification of ACTA1 levels normalized to endogenous actin (mouse Acta1) in genomic DNA. Mice were maintained in a conventional specific-pathogen–free facility with a fixed light cycle (23°C, 12-hour dark-light cycle). Mice were injected i.p. with colchicine (Sigma-Aldrich; 0.4 mg/kg) for 2 days, rapamycin (LC Laboratories) for 1 day (4 mg/kg) or 7 or 10 days (2 mg/kg), AICAR (Toronto Research Chemicals; 500 mg/kg) for 7 days, or AZD8055 (LC Laboratories; 10 mg/kg) for 10 days. Mice were treated with metformin (Sigma-Aldrich; 300 mg/kg) by gavage for 5 or 10 days. For starvation experiments, mice were sacrificed after 12 hours of food deprivation followed by 4 hours of free access to food (fed), or after 24 or 45 hours of food deprivation but free access to water (starved). In vitro force measurement of EDL and soleus muscles was conducted as previously described (32). Half- and late relaxation times were calculated according to Moyer et al. (8).
Human muscle cells and biopsy samples.
Muscle biopsies frozen in nitrogen-cooled isopentane from 3 DM1 patients, aged 33, 34, and 49 years, and from an IBM patient were analyzed and compared with 5 control muscle samples from age-matched individuals showing no clinical signs of DM1 and normal muscle histology. MyoD-transduced fibroblasts from control individuals and patients were cultured in growth medium (DMEM, 10% FBS, 50 μg/ml gentamicin) at 37°C under 5% CO2. At confluence, transduction into myoblasts was induced by differentiation medium (DMEM, 50 μg/ml gentamicin, 3 μg/ml doxycycline hyclate, 10 μg/ml human recombinant insulin) (67). Myotubes obtained after 10 days were incubated for 3 hours in growth medium (refed), PBS (starved), or PBS supplemented with chloroquine (100 μM).
Western blotting.
Cell pellets and muscles powdered in liquid nitrogen were lysed in cold RIPA+ buffer (50 mM Tris-HCl pH 8, 150 mM NaCl, 1% NP-40, 0.5% sodium deoxycholate, 0.1% SDS, 1% Triton X, 10% glycerol, phosphatase and protease inhibitors). Following dosage (BCA Protein Assay, Sigma-Aldrich), proteins were separated on SDS-polyacrylamide gels and transferred to nitrocellulose membrane. Blots were blocked in TBS, 3% BSA, 0.1% Tween-20, and incubated overnight at 4°C with primary antibodies, then for 2 hours with HRP-labeled secondary antibodies. Immunoreactivity was detected using the ECL Western blot detection reagent LumiGLO (KPL) and exposed to Super RX-N films (Fujifilm). Protein expression was normalized to α-actinin, α-tubulin, or the total protein of the corresponding phosphorylated form. Antibodies used are listed in Supplemental Methods.
Polymerase chain reaction.
Total RNAs were extracted with the RNeasy Mini Kit (Qiagen), reverse transcribed with the SuperScript III First-Strand Synthesis System (Invitrogen), and amplified with the Power SYBR Green Master Mix (Applied Biosystems) or the Hot FirePol EvaGreen qPCR Mix (Solis BioDyne). Expression of specific spliced or pan transcripts was analyzed by end-point PCR and electrophoresis, or by quantitative PCR with Step One software and normalization to Actn2 expression. Primers are listed in Supplemental Table 2.
Histology and immunofluorescence.
Muscles were frozen in liquid nitrogen–cooled isopentane. Eight-micrometer muscle sections were stained with H&E or NADH, and observed with an upright microscope (DMR, Leica). For immunostaining, sections were unfixed or fixed with 4% paraformaldehyde (PFA), cold acetone, or methanol; for some, microwave antigen retrieval was used. Sections were then blocked in PBS, 3% BSA, incubated sequentially with primary and appropriate secondary fluorescent antibodies (Invitrogen), mounted with Vectashield medium (Vector), and observed with a Leica fluorescent microscope.
GFP-LC3 puncta analysis.
For GFP-LC3 detection, mice were perfused with 4% PFA, and muscles were incubated in 30% sucrose overnight. Cryosections were washed and mounted. Images were recorded using a Leica confocal microscope with ×63 or ×100 objectives. The number of GFP-LC3 puncta was counted on the 3D reconstructed images with Imaris (version 8.1.2) software. Seven to twelve image stacks were quantified for each muscle, and the average number of GFP-LC3 puncta per volume unit defined within a single fiber (20.8 × 20.8 × 6.5 μm3) was used for statistical analyses. All GFP quantifications were done in a blinded way.
Fluorescence in situ hybridization.
FISH was conducted on muscle cryosections as previously described by Batra et al. (68), using a Cy3-CAG10 DNA probe. Nuclear foci were observed with a Leica confocal microscope with ×40 and ×100 objectives.
Statistics.
Quantitative data are displayed as mean ± SEM of independent samples, with n (number of individual experiments) ≥ 3. Statistical analysis of values was performed using Student’s t test or 2-way ANOVA test with Tukey’s multiple comparisons test correction, with a 0.05 level of confidence accepted for statistical significance.
Study approval.
Muscle biopsies from DM1 patients were obtained from the Neuromuscular Tissue Bank (Department of Neurosciences, University of Padova, Padova, Italy) through the Telethon Network of Genetic Biobanks and the EuroBioBank, in accordance with European recommendation and Italian legislation on ethics. Control and IBM human muscle biopsies were from the Department of Pathology, University Hospital Basel (Basel, Switzerland); their use was approved by the Ethical Committee of the University Hospital Basel. Human fibroblast cell lines were obtained from the platform for immortalization of human cells at the Institut de Myologie (Paris, France). Fibroblasts derived from skin biopsies were obtained from the MyoBank-AFM bank of tissues for research at the Institut de Myologie, a partner in the European Union network EuroBioBank, in accordance with European recommendation and French legislation on ethics. All animal studies were performed in accordance with the European Union guidelines for animal care and approved by the Veterinary Office of the Canton of Basel city (application number 2601).
Author contributions
MB and PC performed most of the experiments, analyzed the data, and wrote the paper with input from all authors. NR, KC, TW, CE, and BE conducted quantitative PCR and Western blot analyses, GFP-LC3 quantification, muscle dissection, cryosections and inorganic staining, and electron microscopy analyses, respectively. SF and CA provided human muscle biopsies. DF developed and provided transduced fibroblasts from DM1 patients. MAR and MS helped conceive the project and edited the manuscript. MS secured funding. PC designed and directed the research project.
Supplementary Material
Acknowledgments
We thank C.A. Thornton for the generous gift of HSALR mice, and J. Hench and G. Schweighauser (Institute of Pathology, University of Basel, University Hospital Basel) for their help in preparing electron microscopy samples. We are grateful to S. Lin (Biozentrum, University of Basel) for his technical expertise on muscle force measurement and Imaris software, and to J. Kinter and L. Tintignac (Neuromuscular Research Group, University of Basel, University Hospital Basel) for their constructive comments on the project. We also thank the platform for immortalization of human cells from the Institut de Myologie (Paris, France) for providing human transduced fibroblasts. MHC antibodies, developed by H.M. Blau and S. Schiaffino, were obtained from the Developmental Studies Hybridoma Bank (University of Iowa, Iowa City, Iowa, USA). This work was supported by EuroBioBank and Telethon Network of Genetic Biobanks (grant GTB 12001D to CA), the University of Basel and University Hospital Basel (MS), the University of Basel–Stadt and Basel-Landschaft (MAR), the Neuromuscular Research Association Basel (MS, PC), the Swiss Foundation for Research on Muscle Diseases (MAR, MS), and the Swiss National Science Foundation (PC, MS).
Footnotes
Conflict of interest: M. Sinnreich owns shares of Novartis and is coinventor on a patent application for drug discovery in DM1 (EP 16/166212.7). M. Sinnreich’s institution (University Hospital Basel) has received research support from CSL Behring and Roche, not in relation to this study. C. Angelini is part of the European Board of Genzyme-Sanofi.
Reference information:J Clin Invest. 2017;127(2):549–563. https://doi.org/10.1172/JCI89616.
Contributor Information
Marielle Brockhoff, Email: marielle.brockhoff@unibas.ch.
Nathalie Rion, Email: nathalie.rion@stud.unibas.ch.
Kathrin Chojnowska, Email: kathrin.chojnowska@unibas.ch.
Tatiana Wiktorowicz, Email: tatiana.wiktorowicz@unibas.ch.
Christopher Eickhorst, Email: christopher.eickhorst@unibas.ch.
Beat Erne, Email: beat.erne@unibas.ch.
Stephan Frank, Email: Stephan.Frank@usb.ch.
Corrado Angelini, Email: corrado.angelini@unipd.it.
Denis Furling, Email: denis.furling@upmc.fr.
Michael Sinnreich, Email: michael.sinnreich@usb.ch.
Perrine Castets, Email: perrine.castets@unibas.ch.
References
- 1.Machuca-Tzili L, Brook D, Hilton-Jones D. Clinical and molecular aspects of the myotonic dystrophies: a review. Muscle Nerve. 2005;32(1):1–18. doi: 10.1002/mus.20301. [DOI] [PubMed] [Google Scholar]
- 2.Brook JD, et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3′ end of a transcript encoding a protein kinase family member. Cell. 1992;69(2):385. doi: 10.1016/0092-8674(92)90418-C. [DOI] [PubMed] [Google Scholar]
- 3.Mahadevan M, et al. Myotonic dystrophy mutation: an unstable CTG repeat in the 3′ untranslated region of the gene. Science. 1992;255(5049):1253–1255. doi: 10.1126/science.1546325. [DOI] [PubMed] [Google Scholar]
- 4.Lin X, et al. Failure of MBNL1-dependent post-natal splicing transitions in myotonic dystrophy. Hum Mol Genet. 2006;15(13):2087–2097. doi: 10.1093/hmg/ddl132. [DOI] [PubMed] [Google Scholar]
- 5.Mankodi A, et al. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289(5485):1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 6.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280(5364):737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 7.Mankodi A, et al. Expanded CUG repeats trigger aberrant splicing of ClC-1 chloride channel pre-mRNA and hyperexcitability of skeletal muscle in myotonic dystrophy. Mol Cell. 2002;10(1):35–44. doi: 10.1016/S1097-2765(02)00563-4. [DOI] [PubMed] [Google Scholar]
- 8.Moyer M, Berger DS, Ladd AN, Van Lunteren E. Differential susceptibility of muscles to myotonia and force impairment in a mouse model of myotonic dystrophy. Muscle Nerve. 2011;43(6):818–827. doi: 10.1002/mus.21988. [DOI] [PubMed] [Google Scholar]
- 9.Herrendorff R, et al. Identification of plant-derived alkaloids with therapeutic potential for myotonic dystrophy type I. J Biol Chem. 2016;291(33):17165–17177. doi: 10.1074/jbc.M115.710616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wheeler TM, et al. Reversal of RNA dominance by displacement of protein sequestered on triplet repeat RNA. Science. 2009;325(5938):336–339. doi: 10.1126/science.1173110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sobczak K, Wheeler TM, Wang W, Thornton CA. RNA interference targeting CUG repeats in a mouse model of myotonic dystrophy. Mol Ther. 2013;21(2):380–387. doi: 10.1038/mt.2012.222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vignaud A, et al. Progressive skeletal muscle weakness in transgenic mice expressing CTG expansions is associated with the activation of the ubiquitin-proteasome pathway. Neuromuscul Disord. 2010;20(5):319–325. doi: 10.1016/j.nmd.2010.03.006. [DOI] [PubMed] [Google Scholar]
- 13.Bargiela A, Cerro-Herreros E, Fernandez-Costa JM, Vilchez JJ, Llamusi B, Artero R. Increased autophagy and apoptosis contribute to muscle atrophy in a myotonic dystrophy type 1 Drosophila model. Dis Model Mech. 2015;8(7):679–690. doi: 10.1242/dmm.018127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beffy P, et al. Altered signal transduction pathways and induction of autophagy in human myotonic dystrophy type 1 myoblasts. Int J Biochem Cell Biol. 2010;42(12):1973–1983. doi: 10.1016/j.biocel.2010.08.010. [DOI] [PubMed] [Google Scholar]
- 15.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat Genet. 2001;29(1):40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 16.Llagostera E, et al. Role of myotonic dystrophy protein kinase (DMPK) in glucose homeostasis and muscle insulin action. PLoS One. 2007;2(11):e1134. doi: 10.1371/journal.pone.0001134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Denis JA, et al. mTOR-dependent proliferation defect in human ES-derived neural stem cells affected by myotonic dystrophy type 1. J Cell Sci. 2013;126(pt 8):1763–1772. doi: 10.1242/jcs.116285. [DOI] [PubMed] [Google Scholar]
- 18.Li X, Zhang W, Lv H, Wang ZX, Yuan Y. [Activities of Akt pathway and their correlation with pathological changes in myotonic dystrophy] Beijing Da Xue Xue Bao. 2010;42(5):526–529. [PubMed] [Google Scholar]
- 19.Loro E, et al. Normal myogenesis and increased apoptosis in myotonic dystrophy type-1 muscle cells. Cell Death Differ. 2010;17(8):1315–1324. doi: 10.1038/cdd.2010.33. [DOI] [PubMed] [Google Scholar]
- 20.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15(3):1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Suenaga K, et al. Muscleblind-like 1 knockout mice reveal novel splicing defects in the myotonic dystrophy brain. PLoS One. 2012;7(3):e33218. doi: 10.1371/journal.pone.0033218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Du H, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat Struct Mol Biol. 2010;17(2):187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamori M, et al. Splicing biomarkers of disease severity in myotonic dystrophy. Ann Neurol. 2013;74(6):862–872. doi: 10.1002/ana.23992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart PC, et al. MnSOD upregulation sustains the Warburg effect via mitochondrial ROS and AMPK-dependent signalling in cancer. Nat Commun. 2015;6:6053. doi: 10.1038/ncomms7053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee H, et al. NQO1-induced activation of AMPK contributes to cancer cell death by oxygen-glucose deprivation. Sci Rep. 2015;5:7769. doi: 10.1038/srep07769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raney MA, Turcotte LP. Evidence for the involvement of CaMKII and AMPK in Ca2+-dependent signaling pathways regulating FA uptake and oxidation in contracting rodent muscle. J Appl Physiol. 2008;104(5):1366–1373. doi: 10.1152/japplphysiol.01282.2007. [DOI] [PubMed] [Google Scholar]
- 27.Bentzinger CF, et al. Differential response of skeletal muscles to mTORC1 signaling during atrophy and hypertrophy. Skelet Muscle. 2013;3(1):6. doi: 10.1186/2044-5040-3-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bentzinger CF, et al. Skeletal muscle-specific ablation of raptor, but not of rictor, causes metabolic changes and results in muscle dystrophy. Cell Metab. 2008;8(5):411–424. doi: 10.1016/j.cmet.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 29.Kang SA, et al. mTORC1 phosphorylation sites encode their sensitivity to starvation and rapamycin. Science. 2013;341(6144):1236566. doi: 10.1126/science.1236566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Roux PP, et al. RAS/ERK signaling promotes site-specific ribosomal protein S6 phosphorylation via RSK and stimulates cap-dependent translation. J Biol Chem. 2007;282(19):14056–14064. doi: 10.1074/jbc.M700906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim J, Kundu M, Viollet B, Guan KL. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat Cell Biol. 2011;13(2):132–141. doi: 10.1038/ncb2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Castets P, et al. Sustained activation of mTORC1 in skeletal muscle inhibits constitutive and starvation-induced autophagy and causes a severe, late-onset myopathy. Cell Metab. 2013;17(5):731–744. doi: 10.1016/j.cmet.2013.03.015. [DOI] [PubMed] [Google Scholar]
- 33.Masiero E, et al. Autophagy is required to maintain muscle mass. Cell Metab. 2009;10(6):507–515. doi: 10.1016/j.cmet.2009.10.008. [DOI] [PubMed] [Google Scholar]
- 34.Klionsky DJ, et al. Guidelines for the use and interpretation of assays for monitoring autophagy. Autophagy. 2012;8(4):445–544. doi: 10.4161/auto.19496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ueda H, et al. Decreased expression of myotonic dystrophy protein kinase and disorganization of sarcoplasmic reticulum in skeletal muscle of myotonic dystrophy. J Neurol Sci. 1999;162(1):38–50. doi: 10.1016/S0022-510X(98)00290-1. [DOI] [PubMed] [Google Scholar]
- 36.Swash M, Fox KP. Abnormal intrafusal muscle fibres in myotonic dystrophy: a study using serial sections. J Neurol Neurosurg Psychiatr. 1975;38(1):91–99. doi: 10.1136/jnnp.38.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ludatscher RM, Kerner H, Amikam S, Gellei B. Myotonia dystrophica with heart involvement: an electron microscopic study of skeletal, cardiac, and smooth muscle. J Clin Pathol. 1978;31(11):1057–1064. doi: 10.1136/jcp.31.11.1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laustriat D, et al. In vitro and in vivo modulation of alternative splicing by the biguanide metformin. Mol Ther Nucleic Acids. 2015;4:e262. doi: 10.1038/mtna.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wheeler TM, Lueck JD, Swanson MS, Dirksen RT, Thornton CA. Correction of ClC-1 splicing eliminates chloride channelopathy and myotonia in mouse models of myotonic dystrophy. J Clin Invest. 2007;117(12):3952–3957. doi: 10.1172/JCI33355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lueck JD, et al. Chloride channelopathy in myotonic dystrophy resulting from loss of posttranscriptional regulation for CLCN1. Am J Physiol Cell Physiol. 2007;292(4):C1291–C1297. doi: 10.1152/ajpcell.00336.2006. [DOI] [PubMed] [Google Scholar]
- 41.Charlet-BN, Savkur RS, Singh G, Philips AV, Grice EA, Cooper TA. Loss of the muscle-specific chloride channel in type 1 myotonic dystrophy due to misregulated alternative splicing. Mol Cell. 2002;10(1):45–53. doi: 10.1016/S1097-2765(02)00572-5. [DOI] [PubMed] [Google Scholar]
- 42.Pauly M, et al. AMPK activation stimulates autophagy and ameliorates muscular dystrophy in the mdx mouse diaphragm. Am J Pathol. 2012;181(2):583–592. doi: 10.1016/j.ajpath.2012.04.004. [DOI] [PubMed] [Google Scholar]
- 43.Lee CS, et al. Ligands for FKBP12 increase Ca2+ influx and protein synthesis to improve skeletal muscle function. J Biol Chem. 2014;289(37):25556–25570. doi: 10.1074/jbc.M114.586289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grumati P, et al. Autophagy is defective in collagen VI muscular dystrophies, and its reactivation rescues myofiber degeneration. Nat Med. 2010;16(11):1313–1320. doi: 10.1038/nm.2247. [DOI] [PubMed] [Google Scholar]
- 45.Jones K, et al. GSK3β mediates muscle pathology in myotonic dystrophy. J Clin Invest. 2012;122(12):4461–4472. doi: 10.1172/JCI64081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Suzuki T, et al. Inhibition of AMPK catabolic action by GSK3. Mol Cell. 2013;50(3):407–419. doi: 10.1016/j.molcel.2013.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inoki K, Zhu T, Guan KL. TSC2 mediates cellular energy response to control cell growth and survival. Cell. 2003;115(5):577–590. doi: 10.1016/S0092-8674(03)00929-2. [DOI] [PubMed] [Google Scholar]
- 48.Castets P, Frank S, Sinnreich M, Rüegg MA. “Get the balance right”: pathological significance of autophagy perturbation in neuromuscular disorders. J Neuromuscul Dis. 2016;3(2):127–155. doi: 10.3233/JND-160153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dunne PW, Ma L, Casey DL, Harati Y, Epstein HF. Localization of myotonic dystrophy protein kinase in skeletal muscle and its alteration with disease. Cell Motil Cytoskeleton. 1996;33(1):52–63. doi: 10.1002/(SICI)1097-0169(1996)33:1<52::AID-CM6>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 50.Kim N, et al. AMPKα2 translocates into the nucleus and interacts with hnRNP H: implications in metformin-mediated glucose uptake. Cell Signal. 2014;26(9):1800–1806. doi: 10.1016/j.cellsig.2014.03.023. [DOI] [PubMed] [Google Scholar]
- 51.Kim DH, Langlois MA, Lee KB, Riggs AD, Puymirat J, Rossi JJ. HnRNP H inhibits nuclear export of mRNA containing expanded CUG repeats and a distal branch point sequence. Nucleic Acids Res. 2005;33(12):3866–3874. doi: 10.1093/nar/gki698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zou T, et al. Polyamines modulate the subcellular localization of RNA-binding protein HuR through AMP-activated protein kinase-regulated phosphorylation and acetylation of importin alpha1. Biochem J. 2008;409(2):389–398. doi: 10.1042/BJ20070860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang W, et al. AMP-activated kinase regulates cytoplasmic HuR. Mol Cell Biol. 2002;22(10):3425–3436. doi: 10.1128/MCB.22.10.3425-3436.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Finley J. Alteration of splice site selection in the LMNA gene and inhibition of progerin production via AMPK activation. Med Hypotheses. 2014;83(5):580–587. doi: 10.1016/j.mehy.2014.08.016. [DOI] [PubMed] [Google Scholar]
- 55.Santoro M, et al. Alternative splicing alterations of Ca2+ handling genes are associated with Ca2+ signal dysregulation in myotonic dystrophy type 1 (DM1) and type 2 (DM2) myotubes. Neuropathol Appl Neurobiol. 2014;40(4):464–476. doi: 10.1111/nan.12076. [DOI] [PubMed] [Google Scholar]
- 56.Vihola A, et al. Altered expression and splicing of Ca(2+) metabolism genes in myotonic dystrophies DM1 and DM2. Neuropathol Appl Neurobiol. 2013;39(4):390–405. doi: 10.1111/j.1365-2990.2012.01289.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kimura T, et al. Alternative splicing of RyR1 alters the efficacy of skeletal EC coupling. Cell Calcium. 2009;45(3):264–274. doi: 10.1016/j.ceca.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kimura T, et al. Altered mRNA splicing of the skeletal muscle ryanodine receptor and sarcoplasmic/endoplasmic reticulum Ca2+-ATPase in myotonic dystrophy type 1. Hum Mol Genet. 2005;14(15):2189–2200. doi: 10.1093/hmg/ddi223. [DOI] [PubMed] [Google Scholar]
- 59.Tang ZZ, et al. Muscle weakness in myotonic dystrophy associated with misregulated splicing and altered gating of Ca(V)1.1 calcium channel. Hum Mol Genet. 2012;21(6):1312–1324. doi: 10.1093/hmg/ddr568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Franke C, Hatt H, Iaizzo PA, Lehmann-Horn F. Characteristics of Na+ channels and Cl– conductance in resealed muscle fibre segments from patients with myotonic dystrophy. J Physiol (Lond) 1990;425:391–405. doi: 10.1113/jphysiol.1990.sp018110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Andersen MN, Rasmussen HB. AMPK: a regulator of ion channels. Commun Integr Biol. 2012;5(5):480–484. doi: 10.4161/cib.21200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sukhodub A, et al. AMP-activated protein kinase mediates preconditioning in cardiomyocytes by regulating activity and trafficking of sarcolemmal ATP-sensitive K(+) channels. J Cell Physiol. 2007;210(1):224–236. doi: 10.1002/jcp.20862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kaftan E, Marks AR, Ehrlich BE. Effects of rapamycin on ryanodine receptor/Ca(2+)-release channels from cardiac muscle. Circ Res. 1996;78(6):990–997. doi: 10.1161/01.RES.78.6.990. [DOI] [PubMed] [Google Scholar]
- 64.Avila G, Dirksen RT. Rapamycin and FK506 reduce skeletal muscle voltage sensor expression and function. Cell Calcium. 2005;38(1):35–44. doi: 10.1016/j.ceca.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 65.Brillantes AB, et al. Stabilization of calcium release channel (ryanodine receptor) function by FK506-binding protein. Cell. 1994;77(4):513–523. doi: 10.1016/0092-8674(94)90214-3. [DOI] [PubMed] [Google Scholar]
- 66.Lopez RJ, et al. Raptor ablation in skeletal muscle decreases Cav1.1 expression and affects the function of the excitation-contraction coupling supramolecular complex. Biochem J. 2015;466(1):123–135. doi: 10.1042/BJ20140935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chaouch S, et al. Immortalized skin fibroblasts expressing conditional MyoD as a renewable and reliable source of converted human muscle cells to assess therapeutic strategies for muscular dystrophies: validation of an exon-skipping approach to restore dystrophin in Duchenne muscular dystrophy cells. Hum Gene Ther. 2009;20(7):784–790. doi: 10.1089/hum.2008.163. [DOI] [PubMed] [Google Scholar]
- 68.Batra R, et al. Loss of MBNL leads to disruption of developmentally regulated alternative polyadenylation in RNA-mediated disease. Mol Cell. 2014;56(2):311–322. doi: 10.1016/j.molcel.2014.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.