Abstract
Compromised apoptotic signaling is a prerequisite for tumorigenesis. The design of effective therapies for cancer treatment depends on a comprehensive understanding of the mechanisms that govern cell survival. The antiapoptotic proteins of the BCL-2 family are key regulators of cell survival and are frequently overexpressed in malignancies, leading to increased cancer cell survival. Unlike BCL-2 and BCL-XL, the closest antiapoptotic relative BCL-W is required for spermatogenesis, but was considered dispensable for all other cell types. Here, however, we have exposed a critical role for BCL-W in B cell survival and lymphomagenesis. Loss of Bcl-w conferred sensitivity to growth factor deprivation–induced B cell apoptosis. Moreover, Bcl-w loss profoundly delayed MYC-mediated B cell lymphoma development due to increased MYC-induced B cell apoptosis. We also determined that MYC regulates BCL-W expression through its transcriptional regulation of specific miR. BCL-W expression was highly selected for in patient samples of Burkitt lymphoma (BL), with 88.5% expressing BCL-W. BCL-W knockdown in BL cell lines induced apoptosis, and its overexpression conferred resistance to BCL-2 family–targeting BH3 mimetics. Additionally, BCL-W was overexpressed in diffuse large B cell lymphoma and correlated with decreased patient survival. Collectively, our results reveal that BCL-W profoundly contributes to B cell lymphoma, and its expression could serve as a biomarker for diagnosis and aid in the development of better targeted therapies.
Introduction
Apoptosis is a well-orchestrated process regulated by multiple genes, particularly those belonging to the BCL-2 family. The inherent ability of cells to undergo apoptosis in response to cellular stresses, including activation of oncogenes such as MYC, functions as a safeguard against tumorigenesis (1). Consequently, alterations resulting in aberrant expression of antiapoptotic BCL-2 family members and/or loss of expression of proapoptotic BCL-2 family members are well documented in most human cancers (2). The BCL-2 family has an established role in determining whether cells should live or die. However, knowledge about whether specific BCL-2 family members (other than BCL-2 itself) are dysregulated in or contribute to human malignancies and how this information can be exploited therapeutically remains incomplete.
Dysregulated MYC expression occurs in at least 70% of human malignancies and is a known driver of Burkitt lymphoma (BL) (3). The Eμ-MYC–Tg mouse model of MYC-induced B cell lymphomagenesis (4) has been instrumental in delineating the roles of BCL-2 family proteins in lymphoma, which has translated to many other human cancers (5). However, relative to other BCL-2 family members, little information is known about the role of the antiapoptotic protein BCL-W in cell survival and lymphomagenesis. This is likely due to early reports showing low expression of BCL-W in primary mouse hematopoietic cells and to observations that Bcl-w–deficient mice have no overt hematopoietic defects (6, 7). While BCL-W is recognized for its critical function in spermatogenesis (8, 9), there is emerging evidence to suggest that BCL-W may also have a role in tumorigenesis. Specifically, BCL-W overexpression cooperated with MYC to accelerate the development of myeloid leukemia (10). In addition, BCL-W overexpression was observed in patient samples of gastric and colorectal adenocarcinoma (11, 12) and in several human solid tumor cell lines (13). These data indicate that BCL-W may contribute to human cancers, but which malignancies and whether BCL-W contributes to tumorigenesis and cancer cell survival remain unclear.
Here, we report a previously unknown role for BCL-W in MYC-induced apoptosis and lymphomagenesis that we believe will have significant clinical implications. By evaluating BCL-W expression in human B cell lymphomas in combination with mouse modeling to directly assess the contribution of BCL-W in MYC-driven lymphomagenesis, we determined that BCL-W has a significant role in B cell lymphoma development and survival. Furthermore, we identified a mechanism by which MYC regulates BCL-W expression. Our data illuminate an unappreciated link between MYC and BCL-W that we believe significantly extends the knowledge of MYC and the role of BCL-W in tumorigenesis, both of which should aid in improving lymphoma prognostics, diagnostics, and therapeutics.
Results
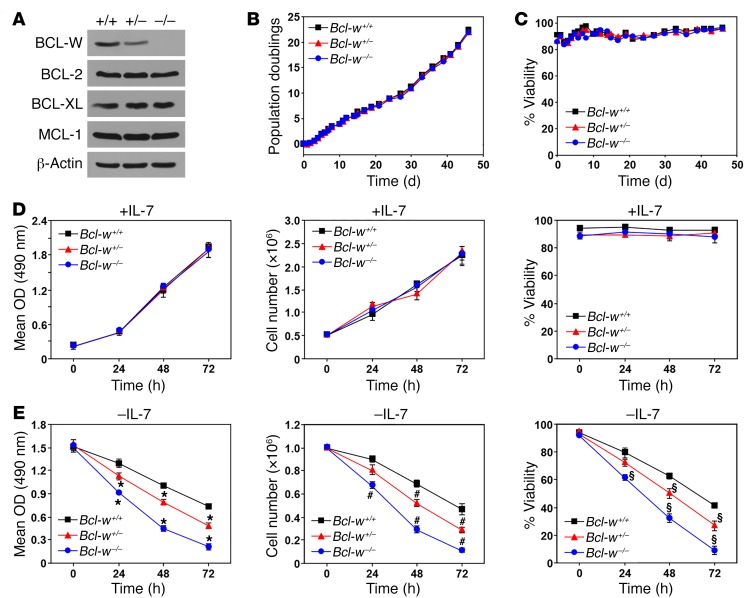
Cytokine deprivation–induced apoptosis is accelerated by loss of BCL-W.
BCL-W levels were reported to be low in lymphocytes and dispensable for lymphocyte survival due to a lack of defects in Bcl-w–null mice (6–8). However, we directly tested whether BCL-W contributed to the survival of B cells by generating cultures of primary pre–B cells isolated from BM of littermate-matched Bcl-w+/+, Bcl-w+/–, and Bcl-w–/– mice. BCL-W protein was readily detectable in WT pre–B cells (Figure 1A). BCL-W levels decreased by approximately half with loss of 1 allele of Bcl-w and were absent in the Bcl-w–null pre–B cells. The levels of other antiapoptotic BCL-2 family members were unaltered in the Bcl-w–deficient cells (Figure 1A). The growth rates and viability of the 3 genotypes of pre–B cells cultured under normal conditions were indistinguishable from each other (Figure 1, B–D). However, loss of Bcl-w affected cell survival following withdrawal of IL-7, an essential cytokine. Specifically, Bcl-w–null pre–B cells had significantly reduced cell expansion following IL-7 withdrawal, due to decreased cell numbers and viability (Figure 1E). Bcl-w heterozygous pre–B cells showed an intermediate reduction in growth and survival compared with WT and Bcl-w–/– pre–B cells after IL-7 withdrawal (Figure 1E). These results indicate that a loss of Bcl-w potentiates the negative consequences of cytokine deprivation in B cells and that BCL-W contributes to B cell survival.
Figure 1. Cytokine deprivation–induced pre–B cell death is accelerated by loss of Bcl-w.
BM from Bcl-w+/+, Bcl-w+/–, and Bcl-w–/– littermates was placed into IL-7–containing media on day 0. Western blotting was performed on pre–B cells that grew out of the cultures (A). Cells were counted at intervals, population doublings were calculated (B), and viability (C) was determined by trypan blue dye exclusion. (D and E) Equal numbers of pre–B cells of each genotype were plated with (D) or without (E) IL-7. MTS assays were performed in quadruplicate at intervals (left, D and E). Total viable cell numbers (middle, D and E) and viability (right, D and E) were measured with trypan blue in triplicate at the indicated intervals. Data shown are representative results of 2 to 4 independent experiments from cells isolated from 2 separate litters generated by different parents. Error bars indicate the SD. For E, *P < 0.0001, #P < 0.0004, and §P < 0.0009, by 1-way ANOVA.
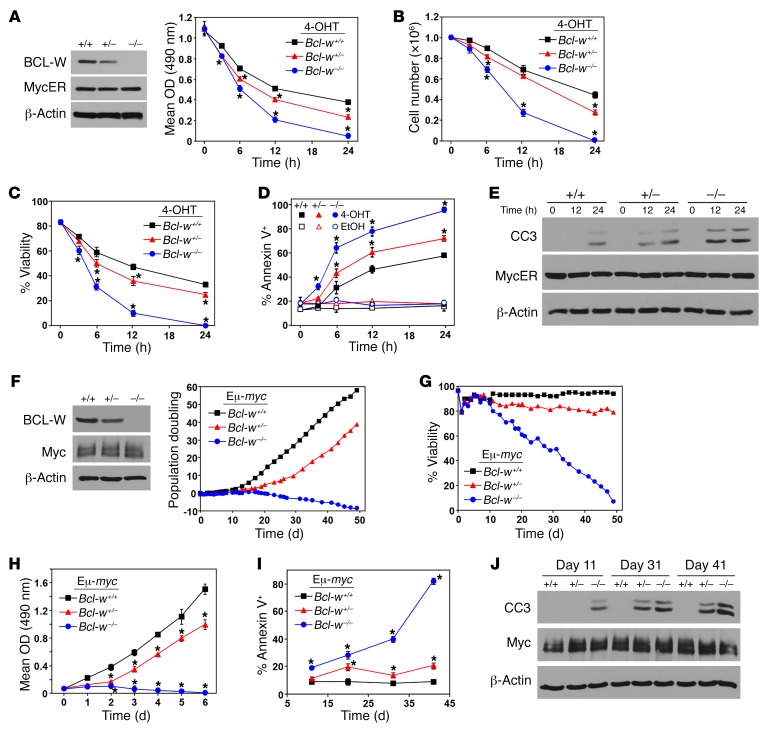
MYC-induced apoptosis is augmented by loss of BCL-W.
To determine whether loss of Bcl-w would confer sensitivity to other apoptotic stimuli such as hyperproliferative signals from oncogenes like MYC, we induced the expression of a 4-hydroxytamoxifen–inducible (4-OHT–inducible) form of MYC (MYCER) (14) in primary pre–B cell cultures from littermates (Figure 2A). Following activation of MYCER with 4-OHT, we observed a robust apoptotic response in pre–B cells lacking both alleles of Bcl-w. There was a decrease in cell growth (Figure 2A), total cell numbers (Figure 2B), and viability (Figure 2C) in the MYCER-activated Bcl-w–/– pre–B cells compared with WT pre–B cells. Moreover, the Bcl-w–null pre–B cells showed an increase in annexin V–positive cells (Figure 2D) and a more rapid appearance of cleaved caspase 3 (Figure 2E) following MYCER activation. We observed a modest, but significant, decrease in cell growth and survival in Bcl-w+/– pre–B cells compared with WT pre–B cells following MYCER activation (Figure 2, A–E). These data demonstrate that BCL-W is critical for B cell survival in the presence of MYC dysregulation.
Figure 2. MYC-induced apoptosis is augmented by loss of Bcl-w.
(A–E) BM-derived pre–B cells from Bcl-w+/+, Bcl-w+/–, and Bcl-w–/– littermates were infected with the 4-OHT–inducible MYCER. Western blot analysis was performed (A). Equal numbers of cells of each genotype were placed into culture. Following the addition of 4-OHT to activate MYCER, MTS assays (A) were performed in quadruplicate, total viable cell numbers (B) and viability (C) were determined using trypan blue dye (in triplicate), and annexin V positivity (in triplicate) (D) and caspase 3 cleavage (CC3) (E) were evaluated. (F–J) BM from Bcl-w+/+ Eμ-MYC, Bcl-w+/– Eμ-MYC, and Bcl-w–/– Eμ-MYC–Tg littermates prior to any detectable lymphoma was placed into culture on day 0, and cells were counted at intervals. Western blot analysis was performed on the pre–B cells that grew out of the cultures (F). Population doublings were calculated (F) and viability was determined by trypan blue dye exclusion (G). (H) Equal numbers of pre–B cells of each genotype were plated and MTS assays (quadruplicate) performed. (I and J) At the indicated intervals, cells were collected and annexin V positivity (I) and cleaved caspase 3 (J) assessed. Data shown are representative of 3 to 4 independent experiments using cells isolated from 3 separate litters generated by different parents. Error bars indicate the SD. *P < 0.0001 for A, *P < 0.029 for B, *P < 0.003 for C, *P < 0.028 for D (comparing 4-OHT–treated samples), *P < 0.0001 for H, and *P < 0.0002 for I, by 1-way ANOVA.
To further test the role of BCL-W in MYC-induced apoptosis in B cells, we derived pre–B cells from the BM of littermate-matched Bcl-w+/+, Bcl-w+/–, and Bcl-w–/– Eμ-MYC mice, which overexpress MYC specifically in B cells (4), prior to any sign of lymphoma development. Within 12 days after explantation into culture, pre–B cells (B220+CD19+CD43–IgM–) dominated the cultures for all 3 genotypes (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI89486DS1), indicating that loss of Bcl-w did not impair the ability of BM cells to differentiate into pre–B cells when MYC was overexpressed. However, when we evaluated pre–B cell growth by assessing the net population doublings beginning the day BM was isolated, we determined that Bcl-w–/– Eμ-MYC pre–B cells were unable to expand in culture (Figure 2F) due to decreased viability (Figure 2G). Even loss of 1 Bcl-w allele was sufficient to impair Eμ-MYC pre–B cell growth and survival (Figure 2, F and G). We obtained similar results when equal numbers of Eμ-MYC pre–B cells from each genotype were cultured and evaluated each day for 6 days (Figure 2H).
At intervals throughout the duration of these experiments, we assessed Eμ-MYC pre–B cells of each genotype for characteristics of apoptosis. We determined that loss of Bcl-w led to a significant increase in apoptosis over time compared with the WT counterpart, as evidenced by an increased number of annexin V–positive cells (Figure 2I) and the appearance of cleaved caspase 3 (Figure 2J). In addition, Bcl-w+/–Eμ-MYC pre–B cells also showed an elevated apoptotic index, as these cultures had increased annexin V staining (Figure 2I) and caspase 3 cleavage (Figure 2J) compared with Bcl-w+/+ Eμ-MYC pre–B cell cultures. These results further demonstrate that BCL-W has an essential role in B cell survival and inhibition of MYC-induced apoptosis.
To assess whether the effects of loss of Bcl-w following MYC dysregulation were B cell specific, we evaluated MYCER-expressing murine embryonic fibroblasts (MEFs). First, we assessed whether a deficiency in Bcl-w altered the response to MYCER activation under normal growth conditions (full serum). While WT MEFs had decreased growth and viability and increased apoptosis following MYCER activation, the effects were more robust in Bcl-w–null MEFs (Supplemental Figure 2, A–F). The apoptotic response was also accelerated in Bcl-w heterozygous MEFs following MYCER activation compared with the response observed in Bcl-w+/+ MEFs (Supplemental Figure 2, A–F). Under serum-free conditions, Bcl-w–null MEFs rapidly underwent apoptosis following MYCER activation (Supplemental Figure 2, A–C). After only 24 hours of MYCER activation, no viable cells remained in the Bcl-w–/– MEF cultures, whereas a third of their WT counterparts were still viable. Together, these results show that BCL-W is not only important for the survival of B cells, but also contributes significantly to the survival of nonhematopoietic cells in response to dysregulated MYC.
Loss of BCL-W profoundly delays MYC-driven lymphoma development.
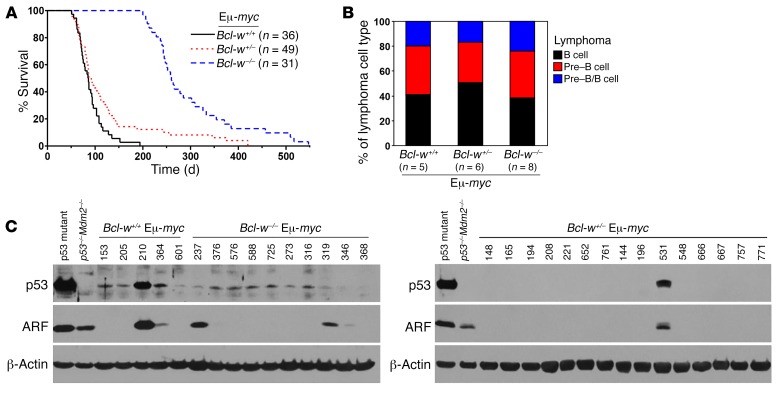
Given that loss of Bcl-w substantially increased the susceptibility of B cells to MYC-induced apoptosis, we postulated that a deficiency in Bcl-w would negatively affect MYC-driven B cell lymphoma development. To test this hypothesis, we generated a cohort of littermate-matched Eμ-MYC–Tg mice with 2, 1, or no Bcl-w alleles. Bcl-w–/– Eμ-MYC mice had an extremely long delay in lymphoma development (Figure 3A; P < 0.0001, log-rank test). This delay in MYC-induced lymphomagenesis resulted in a mean survival of 298.5 days for Bcl-w–/– Eμ-MYC mice, which was 3.3 times longer than the mean survival of their Bcl-w+/+ Eμ-MYC littermates (90.0 days). Of note, all of the Bcl-w+/+ Eμ-MYC mice developed lymphoma and were sacrificed before the first Bcl-w–/– Eμ-MYC mouse presented with lymphoma. Loss of just 1 Bcl-w allele also significantly delayed MYC-induced B cell lymphoma development (Figure 3A; P = 0.0338, log-rank test; mean survival of 120.7 days). Immunophenotyping revealed no difference in the type of B cell lymphomas that arose. Specifically, all 3 genotypes developed lymphomas characteristic of those that develop in Eμ-MYC–Tg mice (15), with a similar distribution of pre–B cell (B220+CD19+CD43–IgM–), B cell (B220+CD19+CD43–IgM+), and pre–B/B cell lymphomas (Figure 3B), including lymphomas that arose early or late. Therefore, a deficiency in Bcl-w caused a delay in B cell lymphoma development, but did not alter the type of lymphoma that emerged.
Figure 3. Loss of Bcl-w profoundly delays MYC-driven lymphomagenesis.
(A) Kaplan-Meier survival curves of Bcl-w+/+ Eμ-MYC, Bcl-w+/– Eμ-MYC, and Bcl-w–/– Eμ-MYC–Tg mice. P < 0.0001, comparing all 3 genotypes with each other; P < 0.0001, comparing Bcl-w–/– Eμ-MYC with Bcl-w+/+ Eμ-MYC; P = 0.0338, comparing Bcl-w+/– Eμ-MYC with Bcl-w+/+Eμ-MYC, by log-rank test. (B and C) Lymphomas of the indicated genotypes were subjected to (B) immunophenotyping to determine the type of lymphoma that emerged and (C) Western blotting to determine p53 and ARF status. Controls included a lymphoma-containing mutant p53 and p53–/– Mdm2–/– MEFs. Representative Western blots are shown; additional blots are shown in Supplemental Figure 4A. Summarized results of Western and Southern blotting are provided in Table 1.
To determine whether the protracted latency of lymphoma development in mice lacking Bcl-w was due to a deficiency in B cells, we performed immunophenotyping on 4 independent sets of littermate-matched precancerous Eμ-MYC mice to evaluate B cell populations. The data show no difference in the number of B cells in the spleens of Bcl-w–/– Eμ-MYC or Bcl-w+/– Eμ-MYC mice compared with their WT Eμ-MYC littermates (Supplemental Figure 3A). Similarly, there was no discernible difference in B cell numbers at all stages of development in the BM of Bcl-w–deficient Eμ-MYC mice (Supplemental Figure 3B). Evaluation of littermate-matched, non-Tg mice showed analogous results among the 3 genotypes (Supplemental Figure 3, A and B). Therefore, a deficiency in Bcl-w does not appear to alter B cell development in mice with normal or increased levels of MYC.
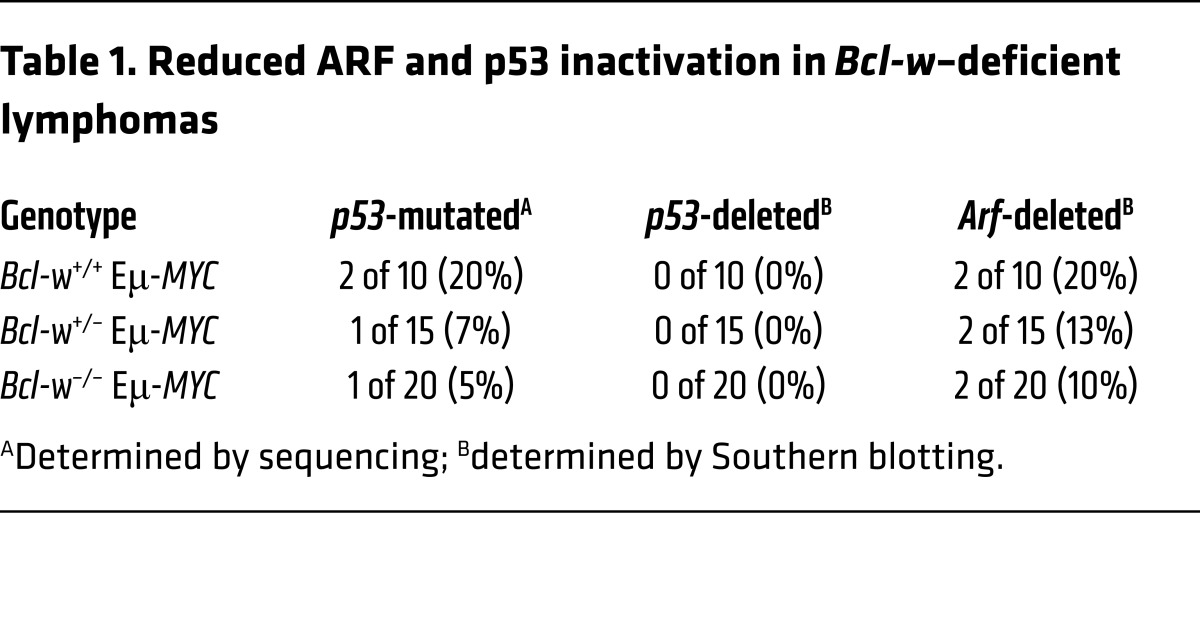
MYC overexpression selects for inactivation of the ARF/p53 tumor-suppressor pathway during lymphomagenesis, and alterations in proteins that influence apoptosis can change the frequency of ARF and/or p53 inactivation (16–18). To assess whether loss of Bcl-w affected the frequency of ARF and/or p53 inactivation, we performed Western (Figure 3C and Supplemental Figure 4A) and Southern (Supplemental Figure 4B) blot analyses of lymphomas from mice of all 3 genotypes. As mutations in p53 typically result in protein overexpression (16), we performed sequencing of p53 on samples that showed increased levels of p53 protein. We also sequenced p53 in lymphomas with increased ARF protein due to the negative feedback regulation of ARF by p53. Only lymphomas with elevated p53 protein harbored mutations in the DNA-binding domain of p53, the most frequently mutated region in p53 (19). Loss of Bcl-w resulted in less frequent mutations of p53 (Table 1). p53 deletions are rare in Eμ-MYC–Tg mice, and we detected no p53 deletions in any of the lymphomas from mice of the 3 genotypes (Table 1 and Supplemental Figure 4B). We observed a decrease in the frequency of Arf deletions in lymphomas that lacked Bcl-w (Table 1 and Supplemental Figure 4B). Together, these results suggest that, without Bcl-w, the selective pressure to inactivate the ARF/p53 pathway was reduced during lymphomagenesis.
Table 1. Reduced ARF and p53 inactivation in Bcl-w–deficient lymphomas.
MYC suppresses BCL-W expression.
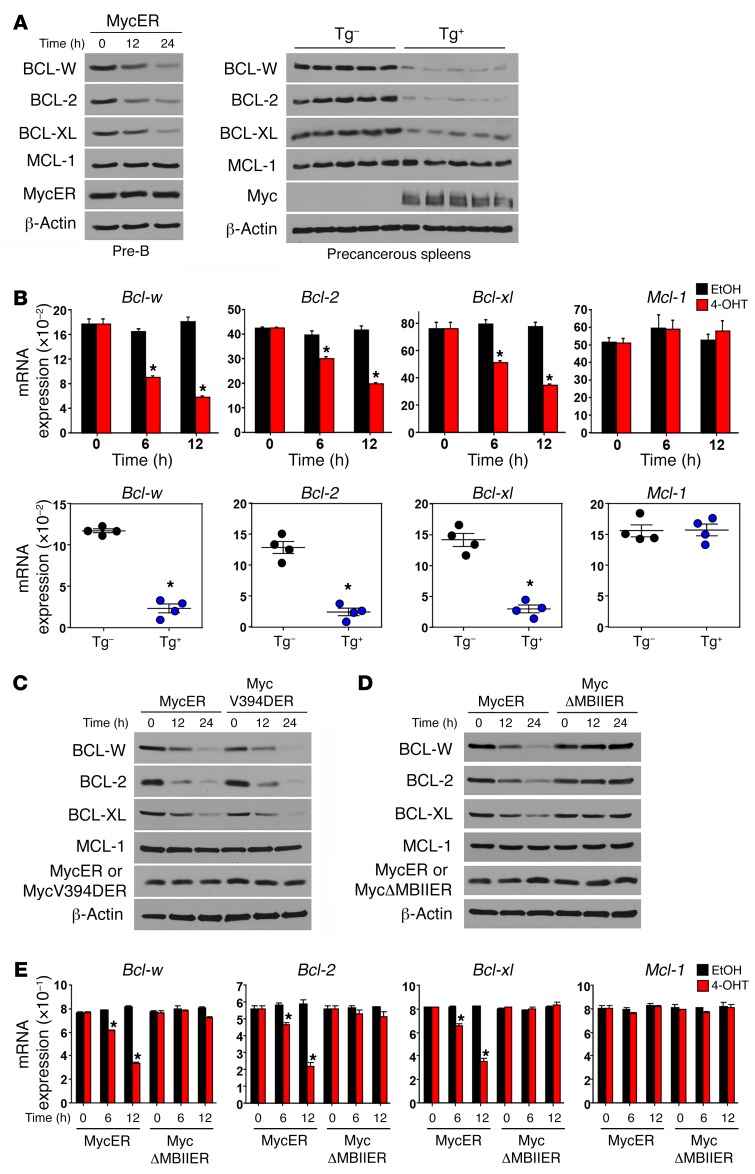
MYC activates apoptosis by activating p53, but also by suppressing the expression of BCL-2 and/or BCL-XL independently of p53 (20–23). We investigated whether BCL-W expression was regulated by MYC by expressing MYCER in pre–B cells. Activation of MYCER with the addition of 4-OHT decreased BCL-W as well as BCL-2 and BCL-XL protein levels (Figure 4A, left). Consistent with previous reports (20, 23), protein levels of the antiapoptotic BCL-2 family member MCL-1 did not change following MYCER activation (Figure 4A, left). As a separate, independent test of MYC regulation and to determine whether MYC regulates BCL-W expression in vivo, we evaluated spleens from precancerous Eμ-MYC mice and littermate-matched non-Tg spleens. Eμ-MYC spleens had reduced BCL-W protein levels as well as decreased BCL-2 and BCL-XL, but MCL-1 levels were unchanged compared with levels detected in the littermate-matched non-Tg spleens (Figure 4A, right). Evaluation of Bcl-w mRNA by quantitative real-time PCR (qRT-PCR) showed that, following MYCER activation in pre–B cells, Bcl-w mRNA levels decreased (Figure 4B, top panels). In addition, spleens from precancerous Eμ-MYC mice had reduced levels of Bcl-w mRNA compared with spleens from non-Tg littermates (Figure 4B, bottom panels). Taken together, MYC induces a decrease in BCL-W mRNA and protein levels in B cells.
Figure 4. MYC suppresses BCL-W expression in normal cells.
At intervals following the addition of 4-OHT, WT pre–B cells (A, left) and p53-null MEFs (C–E) expressing either WT MYCER (A, left, B, top, C, and D), MYCV394D-ER (C), or MYCΔMBII-ER (D and E) were Western blotted for the indicated proteins (A, C, and D), and mRNA levels were measured in triplicate by qRT-PCR (B, top, and E). Precancerous splenocytes from Eμ-MYC–Tg (Tg+) and non-Tg littermates (Tg–) were subjected to Western blotting (A, right) and qRT-PCR (in triplicate, B, bottom; n = 4 of each). mRNA expression was normalized to β-actin and presented as 2–ΔΔCt. Data shown are representative of 2 to 4 independent experiments. Error bars indicate the SEM. *P < 0.0071 (B) and *P < 0.005 (E), by t test.
To determine whether the MYC-induced decrease in BCL-W was cell-type specific, we also assessed WT MEFs expressing MYCER. Analogous to the results we obtained in pre–B cells, activation of MYCER in these nonhematopoietic cells decreased BCL-W levels (Supplemental Figure 5). Likewise, BCL-2 and BCL-XL levels were also reduced following MYCER activation (Supplemental Figure 5). Therefore, the MYC-induced decrease in BCL-W also occurred in primary nonhematopoietic cells, indicating that this was a generalizable consequence of MYC activation.
Suppression of BCL-W expression is independent of MIZ-1 and p53, but requires MYC transcriptional activity.
To gain insight into the mechanism by which MYC was modulating BCL-W expression, we performed a series of experiments using mutants of MYC. MYC can suppress the expression of some genes by inhibiting the transcriptional activator MIZ-1 (24). Using a mutant of MYCER containing a point mutation interrupting the MYC:MIZ-1 interaction (MYCV394D-ER) (25), we determined that BCL-W protein levels decreased in cells expressing MYCV394D-ER to an extent similar to that seen in cells with WT MYCER following activation with 4-OHT (Figure 4C). We also observed analogous reductions of BCL-2 and BCL-XL, but not of MCL-1 (Figure 4C). To evaluate whether a transcriptionally competent MYC was required to suppress BCL-W protein expression, we used a mutant of MYCER lacking the MYC box II domain, which is crucial for MYC transcriptional activity (MYCΔMBII-ER) (24). Upon addition of 4-OHT, we observed a decrease in BCL-W protein only in cells expressing the WT MYCER, but not in cells expressing the transcriptionally impaired MYCΔMBII-ER mutant (Figure 4D). The decrease in BCL-W protein expression was reflected at the mRNA level, as a reduction in Bcl-w mRNA only occurred in cells expressing WT MYCER, but not the transcriptionally impaired MYCΔMBII-ER (Figure 4E). These data demonstrate that MYC transcriptional activity is required to downregulate BCL-W expression. Furthermore, the experiments using MYCER mutants were performed in cells lacking p53 (Figure 4, C–E), indicating that the MYC-induced decrease in BCL-W expression occurs independently of p53.
BCL-W expression is modulated by the MYC-regulated miR-15 family.
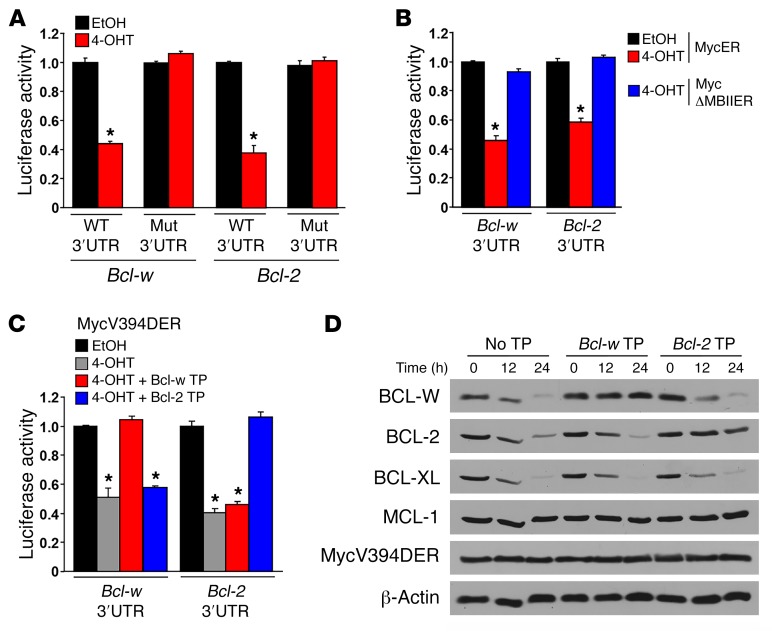
Upon evaluation of publically available ENCODE MYC ChIP-sequencing (ChIP-seq) data (26), MYC was not present at the Bcl-w promoter region in hematopoietic or nonhematopoietic cell lines (Supplemental Figure 6). Therefore, we postulated that MYC may be indirectly altering Bcl-w expression through miR. As we (Supplemental Figure 7) and others (27–29) have shown, Bcl-w mRNA is targeted and negatively regulated by the miR-15 family. Recently, we reported that MYC transcriptionally upregulates this miR family as a mechanism to downregulate Bcl-2 expression (22, 23). To test whether MYC activation leads to miR-15 family targeting of Bcl-w, we used a luciferase-based assay to detect direct binding of the miR-15 family to its predicted target site in the 3′-UTR of Bcl-w following MYC-induced transcriptional upregulation. Specifically, luciferase reporters harboring either a WT miR-15 family binding site of the Bcl-w 3′-UTR or a mutated version were introduced into p53–/– MEFs expressing the 4-OHT–inducible form of MYCER. Upon activation of MYCER, we detected a decrease in luciferase activity in cells containing the WT miR-15 family binding site, but not the mutated miR-15 family binding site (Figure 5A). As a positive control, we evaluated cells with WT or mutated miR-15 family binding sites in the 3′-UTR of Bcl-2 and obtained results analogous to those with the Bcl-w 3′-UTR (Figure 5A). These results indicate that MYC induces the expression of miR-15 family members, as we have recently reported (23), which then directly bind and negatively regulate Bcl-w expression.
Figure 5. BCL-W expression is regulated by MYC through transcriptional upregulation of the miR-15 family.
(A–C) A luciferase reporter containing the 3′-UTR of Bcl-w with a WT or mutated (Mut) miR-15 family binding site was transfected into p53–/– MEFs expressing MYCER (A), MYCΔMBII-ER (B), or MYCV394D-ER (C). Luciferase activity was measured in triplicate 48 hours after the addition of vehicle control (EtOH) or 4-OHT. A luciferase reporter containing either the WT or mutated miR-15 family binding site of the 3′-UTR of Bcl-2 served as a positive control for A–C. A β-gal reporter was cotransfected for normalization purposes. (C and D) miR-15 family binding sites in the 3′-UTR of Bcl-w and Bcl-2 were blocked with site-specific small molecules (Target Protectors [TP]) 24 hours prior to MYCV394D-ER activation. Following the addition of 4-OHT, luciferase activity (C) and protein levels (D) were assessed in the presence or absence of the Bcl-w and Bcl-2 Target Protectors. Data shown are representative of 2 to 4 independent experiments. Error bars indicate the SD. *P = 0.0017 (A), by t test; *P < 0.0001 (B and C), by 1-way ANOVA.
To further assess the mechanism by which MYC regulates the miR-15 family to target Bcl-w, we used the transcriptionally incompetent form of MYC. p53-null MEFs expressing either the WT MYCER or the deletion mutant MYCΔMBII-ER were transfected with a luciferase reporter containing the WT miR-15 family binding site of either the Bcl-w 3′-UTR or, as a positive control, the Bcl-2 3′-UTR. Following activation of WT MYCER, luciferase activity decreased in cells containing the Bcl-w or Bcl-2 3′-UTR (Figure 5B). In contrast, after addition of 4-OHT to the MYCΔMBII-ER–expressing cells, we detected no change in luciferase activity for either luciferase reporter (Figure 5B), indicating that a transcriptionally competent MYC is required for the miR-15 family–mediated decrease in Bcl-w expression.
We further validated this MYC-regulated miR-mediated mechanism by transfecting p53-null MEFs expressing the MYCER mutant unable to interact with MIZ-1 (MYCV394D-ER) with modified RNA molecules (Target Protectors) designed to block endogenous miR-15 family members from binding their target site in the 3′-UTR of Bcl-w. In addition to the Target Protectors, the MYCV394D-ER–expressing cells received luciferase reporters containing the miR-15 family binding site for either the 3′-UTR of Bcl-w or, as a control, Bcl-2. In the absence of any Target Protectors, MYCV394D-ER activation, which upregulates the miR-15 family (23), resulted in decreased luciferase activity in cells with either the Bcl-w or Bcl-2 luciferase reporters (Figure 5C). However, when the miR-15 family binding site of the Bcl-w or Bcl-2 3′-UTR was blocked by their respective Target Protectors, luciferase activity remained unchanged (Figure 5C). As an additional control to test the specificity of our system, cells containing the Bcl-w or Bcl-2 3′-UTR luciferase reporters were transfected with the opposite Target Protector. Following MYCV394D-ER activation with 4-OHT, luciferase activity decreased to an extent that was similar to that seen with no Target Protector present (Figure 5C). In addition, MYCV394D-ER activation decreased BCL-W and BCL-2 protein expression levels in the absence of any Target Protector (Figure 5D). However, BCL-W and BCL-2 levels remained unchanged when their respective Target Protectors were present to block the miR-15 family binding sites (Figure 5D). Together, these results provide direct mechanistic data showing that MYC transcriptionally upregulates the miR-15 family members that then target and decrease the expression of Bcl-w and that this occurs independently of both MIZ-1 and p53.
BCL-W is overexpressed in human BL.
We recently reported that the transformation status of the cell dictates whether MYC functions as a transcriptional activator or repressor of the miR-15 family (23). In normal, nontransformed cells, MYC transcriptionally upregulates the miR-15 family, which downregulates Bcl-2 and, as we show here, Bcl-w expression, leading to increased apoptosis. However, this apoptotic mechanism is inactivated in cancer cells, as MYC is converted from a transcriptional activator to a transcriptional repressor of the miR-15 family, which would allow Bcl-2 and, presumably, Bcl-w levels to be increased in cancer cells that overexpressed MYC (23).
We and others have reported that MYC-overexpressing murine lymphoma cells have reduced expression of the miR-15 family due to MYC-mediated repression (22, 23, 30). We evaluated miR-15a expression in formalin-fixed, paraffin-embedded BL patient samples, which constitutively overexpress MYC due to a MYC translocation, a defining feature of this lymphoma (31). As expected, miR-15a levels were significantly decreased in BL patient samples compared with levels in normal controls (Supplemental Figure 8A), whereas levels of miR-17-5p, which is induced by MYC in cancer cells, were increased in the BL samples (Supplemental Figure 8B). Therefore, as in murine B cell lymphoma, miR-15a levels are reduced in human B cell lymphoma.
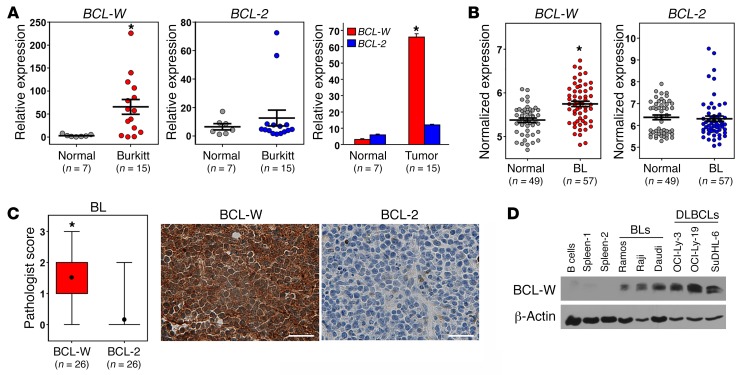
We next performed qRT-PCR to evaluate mRNA levels of BCL-W and BCL-2 from RNA isolated from formalin-fixed, paraffin-embedded BL patient samples and normal lymph node controls. Strikingly, BCL-W mRNA was overexpressed in the BL patient samples compared with expression levels detected in the normal tissue (Figure 6A and Supplemental Figure 9). Because low or undetectable BCL-2 levels are typical in BL and considered during diagnosis, BCL-2 levels were evaluated and showed no overexpression compared with normal tissue (Figure 6A and Supplemental Figure 9). As a separate, independent analysis of BCL-W mRNA expression in human BL, we evaluated publically available microarray gene expression–profiling data (http://www.ncbi.nlm.nih.gov/geo/). Consistent with our qRT-PCR data, BCL-W was significantly overexpressed in BL samples compared with either a mixed population of normal B cells or only normal germinal center B cells (Figure 6B and Supplemental Figure 10). We also assessed BCL-2 mRNA levels and determined that they were not elevated in BL cells compared with levels in normal B cells (Figure 6B). These data indicate that increased BCL-W mRNA expression is selected for in human BL, whereas BCL-2 is not.
Figure 6. BCL-W expression is selected for in human BL.
(A) qRT-PCR analysis (in triplicate) of samples from patients with BL (n = 15) compared with normal lymphoid control tissue (n = 7). mRNA levels were normalized to β-actin and presented as 2–ΔΔCt. Bars represent the mean ± SEM. *P = 9.1 × 10–4, by t test. (B) Microarray gene expression–profiling data for BCL-W and BCL-2 mRNA in BL (n = 57) compared with normal B cells (n = 49). Bars represent the mean ± SEM. Each circle represents 1 sample, and the y axis represents normalized expression of BCL-W and BCL-2. *P = 1.8 × 10–8, by t test. The data sets analyzed are listed in Supplemental Table 1. (C) Immunohistochemical analysis for BCL-W and BCL-2 protein was performed on samples from patients with BL (n = 26). Box and whisker plots of pathologist scores and representative images of BCL-W and BCL-2 staining from the same tumor sample are shown. Original magnification, ×40; scale bars: 200 μm. *P < 0.0001, by t test. For the box and whisker plots, the box represents the 25th and 75th percentiles, the line indicates the median, the circle indicates the mean, and the whiskers represent the maximum and minimum. (D) Protein expression was assessed by Western blotting for the indicated proteins in BL and DLBCL cell lines. Controls included tissue from 2 human spleens and purified B cells from human peripheral blood.
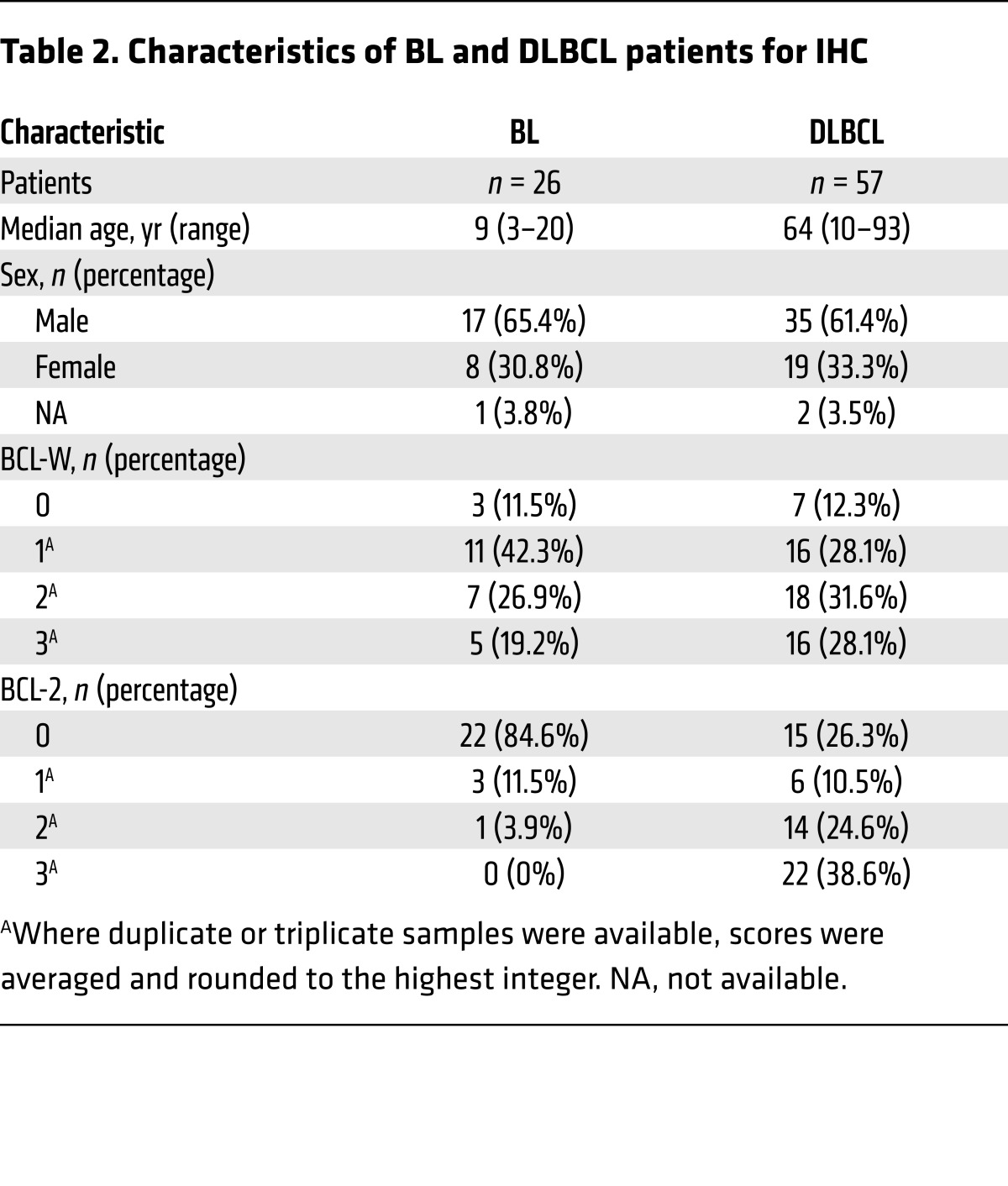
To determine whether the increase in BCL-W mRNA resulted in increased BCL-W protein expression, we performed immunohistochemical analyses of samples from patients with BL. Blinded scoring by a hematopathologist revealed that expression of BCL-W protein was a very frequent event in BL. Specifically, 88.5% (23 of 26) of the samples had positive staining (≥1 pathologist score) for BCL-W (Figure 6C, Table 2, Supplemental Figure 11). Only 15.4% (4 of 26) of the samples showed BCL-2 positivity, which was predominantly low. Additionally, BCL-W protein was overexpressed in BL cell lines compared with normal human splenic tissue and isolated B cells (Figure 6D). Evaluation of BCL-W levels in B cell lymphomas from Eμ-MYC–Tg mice showed a similarly high frequency of increased BCL-W protein expression (Supplemental Figure 12). These results indicate that, in addition to murine lymphoma, BCL-W may be a significant contributor to the pathogenesis of human BL.
Table 2. Characteristics of BL and DLBCL patients for IHC.
Targeting BCL-W in MYC-driven BL induces apoptosis.
Given that BCL-W expression is highly selected for in BL, we tested whether targeting BCL-W would affect BL cell survival. Because we have established a link between Bcl-w and the miR-15 family, which is repressed in BL, we hypothesized that increasing levels of the miR-15 family in these lymphoma cells would decrease BCL-W expression, leading to apoptosis. To test this, we ectopically expressed the miR-15a/16-1 cistron or empty vector as a control in 2 human BL cell lines. In both BL cell lines, ectopic expression of miR-15a/16-1 decreased BCL-W protein expression (Supplemental Figure 13A). Reduced levels of BCL-2 protein, which we previously reported occurred with ectopic expression of miR-15a/16-1 in murine lymphoma cells (23), were also detected and served as a positive control (Supplemental Figure 13A). The forced expression of miR-15a/16-1 reduced cell growth, total cell numbers, and viability (Supplemental Figure 13, B–D). Furthermore, ectopic miR-15a/16-1 expression induced apoptosis, as evidenced by the appearance of cleaved caspase 3 (Supplemental Figure 13A). Therefore, increasing expression of the miR-15 family members that target and downregulate BCL-W and BCL-2 induces apoptosis of BL cells.
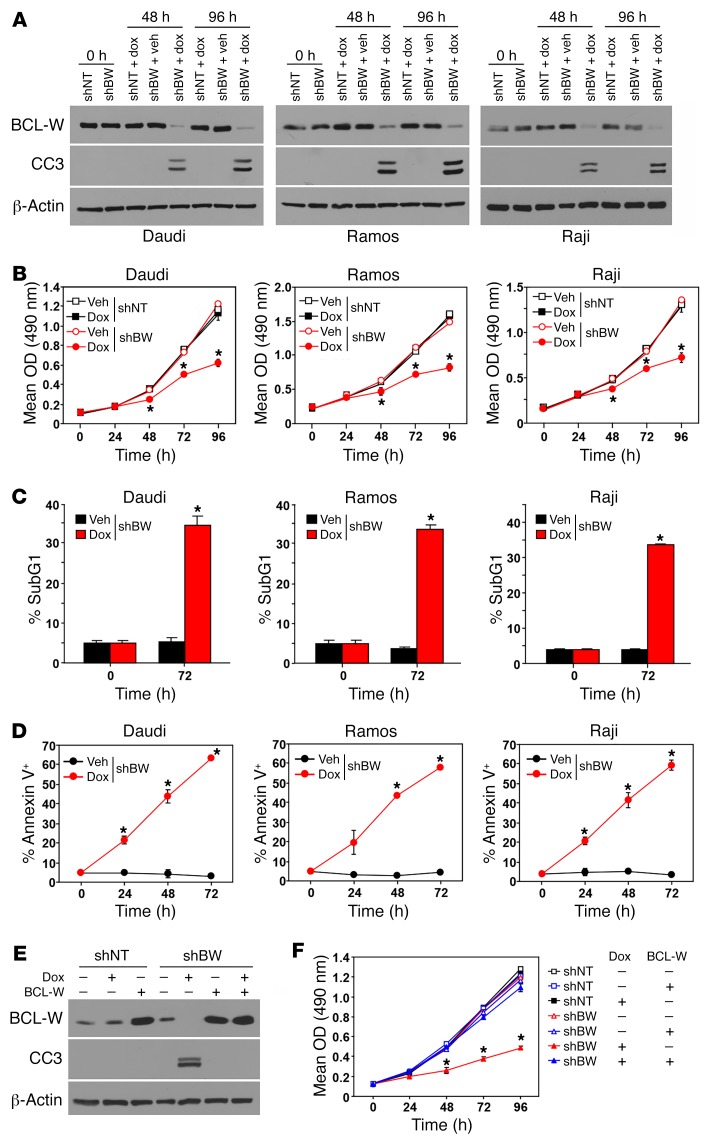
As an independent approach to specifically test the dependency of BL cells on BCL-W expression, we used a doxycycline-inducible (Dox-inducible) BCL-W shRNA in 3 human BL cell lines. Following the addition of Dox to activate expression of the BCL-W shRNA, BCL-W protein expression decreased (Figure 7A). This resulted in reduced cell growth in all 3 BL cell lines (Figure 7B). The reduction in cell growth was attributed to increased apoptosis, as indicated by the increased subG1 DNA content (Figure 7C), annexin V positivity (Figure 7D), and caspase 3 cleavage (Figure 7A). To test whether the negative consequences of BCL-W shRNA were specific and not due to off-target effects, BCL-W was ectopically expressed. BL cells with BCL-W shRNA and ectopic expression of BCL-W did not show a decrease in growth or an increase in caspase 3 cleavage after the addition of Dox, which was only evident in BL cells expressing the BCL-W shRNA alone following the addition of Dox (Figure 7, E and F). These data indicate that targeting BCL-W in BL, which selects for increased BCL-W expression, may be an effective therapeutic approach to treat BL.
Figure 7. Targeting BCL-W induces apoptosis in human BL.
(A–D) The indicated human BL cell lines were infected with a Dox-inducible lentivirus encoding either a BCL-W shRNA (shBW) or nontargeting (shNT) control shRNA. Following the addition of Dox or DMSO vehicle (Veh) control, Western blotting for cleaved caspase 3 (A), MTS assays (B, in quadruplicate), and staining for subG1 DNA content (C, in triplicate) and annexin V positivity (D, in triplicate) were performed. (E and F) Daudi cells were doubly infected with the Dox-inducible lentivirus expressing either a BCL-W shRNA (shBW) or nontargeting (shNT) control shRNA and a retrovirus expressing BCL-W (+) or empty vector control (–). Following the addition of Dox (+) or DMSO vehicle control (–), Western blotting for the indicated proteins (E) and MTS assays (F, in quadruplicate) were performed. Data shown are representative of 2 to 3 independent experiments. Error bars for B–D and F indicate the SD. *P < 6.4 × 10–4 (B), *P < 0.025 (C), *P < 0.034 (D), and *P < 0.0036 (F), by t test comparing shBW and Dox with shBW and vehicle.
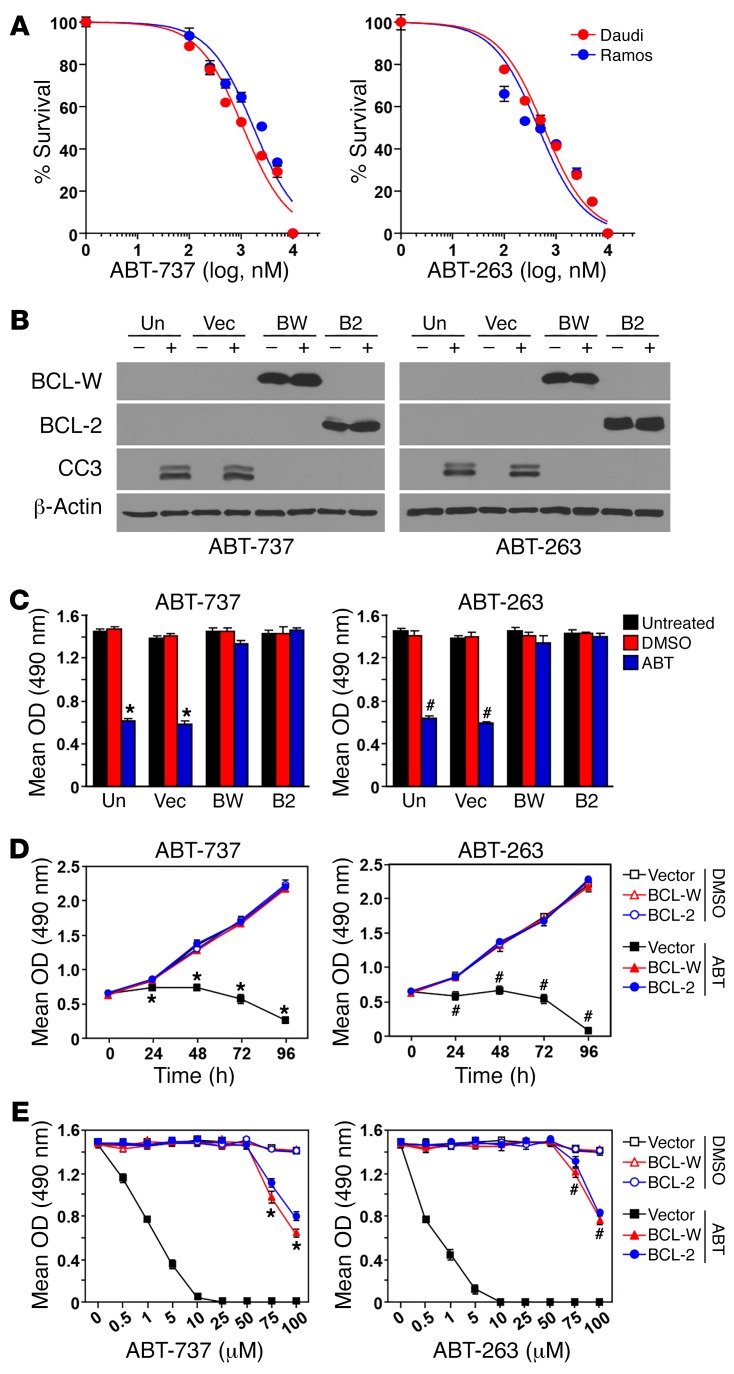
As an additional test of the requirements of BCL-W for BL survival, we took a pharmacological approach. The BH3 mimetic compounds ABT-737 and its analog ABT-263 directly activate apoptosis by binding and inhibiting the antiapoptotic BCL-2 proteins BCL-2, BCL-XL, and BCL-W (5). To evaluate the effect of ABT-737 and ABT-263 on BL, we first determined the IC50 of each in 2 human BL cell lines (Figure 8A). The IC50 for ABT-737 and ABT-263 was 500 nM and 1 μM, respectively, for Daudi cells and 250 nM and 2.5 μM, respectively, for Ramos cells. To determine whether BCL-W expression alone could protect the BL cells from ABT-737 and ABT-263, BCL-W was ectopically expressed (Figure 8B and Supplemental Figure 14A) and treated the BL cells with the IC50 of each. After 48 hours of treatment with the BH3 mimetics, the growth of BL cells expressing the control vector was significantly diminished (Figure 8C and Supplemental Figure 14B) as a result of increased apoptosis, as indicated by the appearance of cleaved caspase 3 (Figure 8B and Supplemental Figure 14A). However, cells overexpressing BCL-W were not sensitive to ABT-737 or ABT-263, as growth of these cells remained unaffected, and no caspase 3 cleavage was detected (Figure 8, B and C and Supplemental Figure 14, A and B). Additionally, 4-day kinetics experiments also showed that BL cells with forced overexpression of BCL-W were protected from the apoptosis-inducing properties of the BH3 mimetics (Figure 8D and Supplemental Figure 14C). Finally, increasing concentrations of ABT-737 and ABT-263 were administered to BL cell lines to determine the maximal dose at which the cells overexpressing BCL-W would no longer be protected. BL cells with the vector control quickly succumbed to BH3 mimetic treatment, with all cells dying following administration of 10 μM of either compound within 48 hours (Figure 8E and Supplemental Figure 14D). However, high concentrations (>50 μM) of either compound were necessary to kill the BL cells overexpressing BCL-W (Figure 8E and Supplemental Figure 14D). Results similar to those with BCL-W overexpression were obtained by overexpressing BCL-2 for all experiments with the BH3 mimetics (Figure 8 and Supplemental Figure 14). Collectively, these data show that BLs rely on BCL-W expression and that increased BCL-W levels in BL cells confer resistance to the BH3 mimetics ABT-737 and ABT-263.
Figure 8. Increased expression of BCL-W confers resistance to BL cells to BH3 mimetics.
(A) The IC50 of the BH3 mimetics ABT-737 and ABT-263 for 2 human BL cell lines (Daudi and Ramos) was determined using MTS assays (in quadruplicate, 48 h). (B–E) Daudi cells remained uninfected (Un) or were retrovirally infected to express empty vector (Vec), BCL-W (BW), or BCL-2 (B2). Following treatment with ABT-737 (+), ABT-263 (+), or DMSO vehicle control (–) at the determined IC50 (B–D) or the indicated concentrations (E) for 48 hours (B, C, and E) or the indicated intervals (D), Western blotting for cleaved caspase 3 (B) and MTS assays (C–E, in quadruplicate) were performed. Data shown are representative of 2 independent experiments for both inhibitors. For data in D and E, experiments with inhibitors were performed at the same time but graphed separately, thus the DMSO control data are the same. Error bars indicate the SD. *P < 6.67 × 10–6 ABT-737 and #P < 6.05 × 10–5 ABT-263 (ABT vs. DMSO) (C); *P < 2.69 × 10–4 ABT-737 and #P < 3.7 × 10–4 ABT-263 (vector with ABT vs. vector, BCL-W, or BCL-2 with DMSO) (D); *P < 7.33 × 10–4 ABT-737 and #P < 0.011 ABT-263 (BCL-W or BCL-2 with ABT vs. BCL-W or BCL-2 with DMSO) (E), by t test.
BCL-W overexpression in diffuse large B cell lymphoma correlates with worse survival.
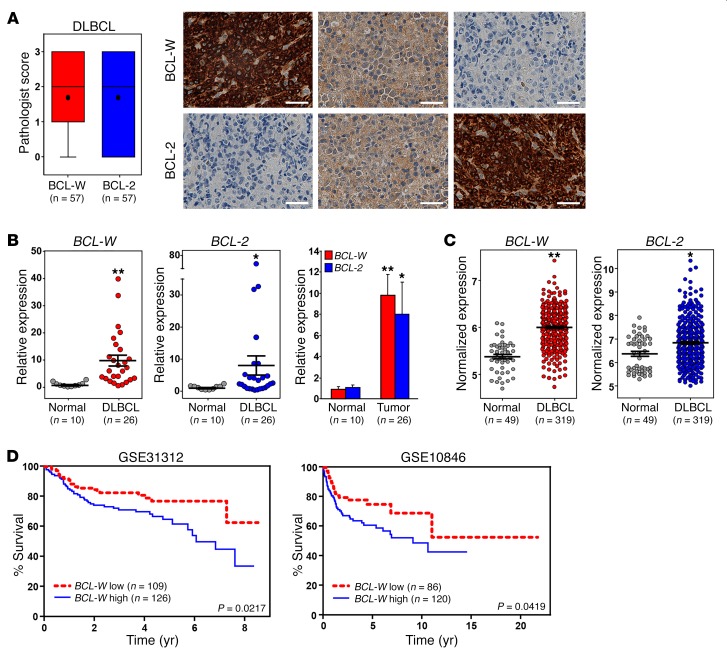
Given our results with BL, we questioned whether BCL-W overexpression was specific to BL or whether BCL-W may contribute to other human B cell lymphomas. We evaluated BCL-W protein levels in diffuse large B cell lymphoma (DLBCL) cell lines, which often have dysregulated MYC activity. Increased BCL-W protein levels were observed in the DLBCL cell lines compared with levels in the normal controls (Figure 6D). Additionally, immunohistochemical analysis of DLBCL patient samples showed that BCL-W protein was expressed in 87.7% (≥1 pathologist score) of the patient samples (Figure 9A, Table 2, Supplemental Figure 15). This frequency was consistent with the frequency (73.7%) we observed for elevated BCL-2 levels in DLBCL (Supplemental Figure 15), a known clinical feature of DLBCL (5). Additionally, while many DLBCL patient samples expressed both BCL-W and BCL-2 protein, approximately a quarter of the samples (26.3%) expressed only BCL-W protein and not BCL-2 protein (Supplemental Figure 15B). Of these samples, the vast majority (80%) had high (scoring a 2 or 3) BCL-W levels.
Figure 9. Increased expression of BCL-W in DLBCL correlates with poor patient survival.
(A) Immunohistochemical analysis for BCL-W and BCL-2 protein was performed on samples from patients with DLBCL (n = 57). Box and whisker plots of pathologist scores and representative images of BCL-W and BCL-2 staining from the same 3 tumor samples. Original magnification, ×40; scale bars: 200 μm. For the box and whisker plots, the box represents the 25th and 75th percentiles, the line indicates the median, the circle indicates the mean, and the whiskers represent the maximum and minimum. (B) qRT-PCR analysis in triplicate of samples from patients with DLBCL (n = 26) compared with normal lymphoid control tissue (n = 10). mRNA levels were normalized to β-actin and are presented as 2–ΔΔCt. Error bars represent the mean ± SEM. **P < 6.03 × 10–5 and *P = 0.014, by t test. (C) Microarray gene expression–profiling data for BCL-W and BCL-2 mRNA in DLBCL (n = 319) compared with normal B cells (n = 49). Lines represent the mean ± SEM. Each circle represents 1 sample, and the y axis represents normalized expression of BCL-W and BCL-2. **P < 2.2 × 10–16 and *P = 9.96 × 10–5, by t test. The data sets analyzed are listed in Supplemental Table 1. (D) Patient samples of DLBCL with low BCL-2 from the GSE31312 and GSE10846 data sets were each separated into 2 groups (BCL-W high and BCL-W low) on the basis of the median expression of BCL-W, and Kaplan-Meier analyses were performed. The P values in D were determined by log-rank test.
To determine whether the increase in BCL-W protein in DLBCL may be due to elevated mRNA levels, we performed qRT-PCR on patient samples of DLBCL, with normal lymph nodes and spleens serving as control lymphatic tissue. We determined that BCL-W was significantly overexpressed in DLBCL compared with levels detected in normal tissues (Figure 9B and Supplemental Figure 16). We obtained similar results for BCL-2 (Figure 9B and Supplemental Figure 16), which served as a positive control. Furthermore, we evaluated publically available microarray gene expression–profiling data on DLBCL to determine whether our mRNA results were reflected in larger patient populations (http://www.ncbi.nlm.nih.gov/geo/). Both BCL-W and BCL-2 were significantly overexpressed in DLBCL compared with mixed populations of normal B cells or only germinal center B cells (Figure 9C and Supplemental Figure 17). Together, our data show that BCL-W may also contribute to DLBCL.
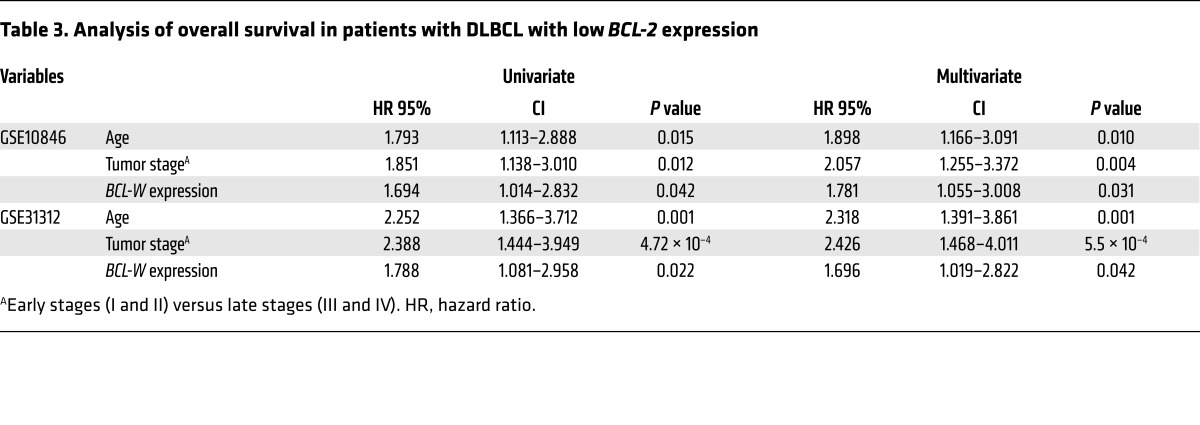
Next, we questioned whether there was a correlation between BCL-W and BCL-2 expression in DLBCL. Pearson’s correlations were calculated for BCL-W and BCL-2 expression for 2 independent data sets. Independently, both data sets showed a negative correlation between BCL-W and BCL-2 expression levels in DLBCL patient samples (for GSE10846, R = –0.1687 and P = 2.9 × 10–4; for GSE31312, R = –0.0948 and P = 3.99 × 10–2). Given that BCL-W was frequently overexpressed in DLBCL and survival rates of DLBCL are not high, we questioned whether increased expression of BCL-W correlated with DLBCL patient survival. To address this, the same 2 DLBCL data sets were evaluated, as both contained survival information. Because BCL-W expression was inversely correlated with BCL-2 expression, we assessed the contribution of increased BCL-W expression in samples with lower levels of BCL-2. Specifically, patient samples were simultaneously stratified on the basis of the median expression of BCL-2 and BCL-W into high or low expression groups for each gene. Then, using data on those patients with low BCL-2 expression, Kaplan-Meier survival curves were plotted for the high and low BCL-W expression groups. For both data sets, the patients with higher BCL-W levels and low BCL-2 levels had significantly worse overall survival than did those with lower levels of both BCL-W and BCL-2 (P = 0.0217 for GSE31312 and P = 0.0419 for GSE10846, log-rank tests, Figure 9D). Stratification of BCL-W expression for patients with high BCL-2 expression levels did not show a statistically significant difference in survival for those who overexpressed both. Next, we performed univariate and multivariate Cox regression analyses on both stratified data sets (with low BCL-2 expression), since additional prognostic variables (e.g., patient’s age and tumor stage) were available. From the univariate analysis, we determined that BCL-W expression, patient’s age, and tumor stage were significantly associated with overall survival (Table 3). Considering all 3 variables, multivariate Cox regression analysis indicated that BCL-W expression independently correlated with overall survival (Table 3). Together, these results indicate that BCL-W expression is a predictive indicator and independent determinant of overall survival in DLBCL patients with lower levels of BCL-2.
Table 3. Analysis of overall survival in patients with DLBCL with low BCL-2 expression.
Discussion
Prior to the current study, BCL-W was not believed to be an important contributor to hematopoietic cell survival. However, this conclusion was never tested and was based solely on expression levels and on the observation that no overt hematopoietic cell defects were detected in mice lacking Bcl-w (6–9). Here, our study has revealed an unexpected and, in our view, significant function for BCL-W in B cell survival and MYC-induced lymphomagenesis. We also provide important mechanistic insight into the regulation of BCL-W expression by MYC in both hematopoietic and nonhematopoietic cells. Most notably, our data provide striking evidence of a survival role for BCL-W in both human BL and DLBCL that we believe will have significant clinical implications.
Antiapoptotic BCL-2 family members inhibit death signals elicited by a number of cellular stressors, including cytokine withdrawal and activation of MYC (2). Given the dogma regarding BCL-W, we were initially surprised that in B cells, loss of Bcl-w conferred sensitivity to IL-7 withdrawal and MYC activation. Using multiple ex vivo and in vitro approaches, we demonstrated that BCL-W significantly contributes to inhibiting MYC-induced apoptosis in both hematopoietic and nonhematopoietic cells. Specifically, in the absence of BCL-W, primary pre–B cells and MEFs were considerably more susceptible to MYC-induced apoptosis than were cells that had WT BCL-W. Previously, it was reported that B cells lacking other antiapoptotic BCL-2 family members (BCL-2, BCL-XL, or MCL-1) were more sensitive to MYC-induced apoptosis (32–34), whereas loss of proapoptotic BCL-2 family members (BIM, BAX, PUMA, or NOXA) conferred resistance to MYC-induced apoptosis (17, 18, 35, 36). The p53 pathway also mediates apoptosis due to MYC dysregulation (16, 37). There was a decreased frequency of p53 pathway inactivation in the lymphomas that emerged in Eμ-MYC mice lacking Bcl-w, suggesting a decreased selective pressure to inactivate the p53 pathway during tumorigenesis. We previously reported that loss of the proapoptotic BCL-2 family member BAX altered the frequency of p53 inactivation in MYC-driven lymphomas (17). Together, our data highlight a previously unknown function of BCL-W in apoptosis triggered by deregulated MYC expression and suggest that BCL-W may have cross-talk with the p53 pathway.
To our knowledge, no link has yet been established between BCL-W expression and MYC activation. Here, we determined that MYC regulates BCL-W expression and identified the underlying mechanism through which this occurs. Specifically, we demonstrated that MYC suppressed the expression of BCL-W independently of its interaction with the transcriptional activator MIZ-1 and of p53, which has been reported to repress the expression of the related family members Bcl-2 and Bcl-xl (38, 39). Furthermore, our data revealed that a transcriptionally competent MYC was required for the reduction in BCL-W expression. However, there was no evidence of direct transcriptional activation by MYC, suggesting that MYC was indirectly regulating the expression of BCL-W. We (here) and others (27–29) have shown that Bcl-w is a target of the miR-15 family, which is known to be regulated by MYC (40). In addition, we recently demonstrated that MYC downregulates the expression of BCL-2 and BCL-XL by transcriptionally upregulating the levels of the miR-15 and let-7 families, respectively (22, 23). Therefore, we hypothesized that MYC was modulating the expression of BCL-W through the miR-15 family. Using luciferase reporter assays and site-specific small molecules designed to block the miR-15 family from binding the 3′-UTR of Bcl-w, we provided direct evidence that MYC, through the miR-15 family, modulates BCL-W expression. These data offer mechanistic evidence of a link between MYC, the miR-15 family, and BCL-W and, we believe, significantly expand our understanding of MYC-mediated apoptosis. Determining which transcriptional targets of MYC are responsible for the biological consequences of MYC dysregulation and how these are altered during cellular transformation provides important insights into lymphomagenesis. The data presented here, combined with previous findings (22, 23, 30, 40), indicate that MYC transcriptionally activates or represses miR to induce growth or apoptosis and that this is altered in cancers.
With our data showing that BCL-W is a critical mediator of MYC-induced apoptosis, we hypothesized that loss of Bcl-w would affect lymphomagenesis driven by dysregulated MYC. While Bcl-w heterozygosity delayed MYC-induced lymphoma development, remarkably, Bcl-w–/– Eμ-MYC–Tg mice developed lymphoma with a substantially extended latency compared with that of littermate-matched control Eμ-MYC mice. Specifically, Eμ-MYC mice lacking both Bcl-w alleles had a mean lifespan that was more than 3 times longer than that of their Bcl-w+/+ Eμ-MYC siblings. By the time all of the Bcl-w+/+ Eμ-MYC mice had succumbed to lymphoma, none of the Bcl-w–/– Eμ-MYC mice had developed lymphoma. Reports of the effects of deleting other prosurvival BCL-2 family members have shown differing results. For example, a Bcl-xl deficiency only slightly delayed Eμ-MYC lymphoma development (33), whereas deletion of Bcl-2 did not significantly alter lymphomagenesis (34). In contrast, loss of 1 allele of Mcl-1 profoundly delayed MYC-induced lymphoma development (41). Therefore, although there are multiple antiapoptotic BCL-2 family members, they do not appear to equally contribute to MYC-induced B cell lymphomagenesis.
While lymphoma development was profoundly extended in Bcl-w–/– Eμ-MYC mice, loss of Bcl-w did not impact the type of lymphoma that emerged. The lymphomas that arose, regardless of genotype or time of tumor incidence, were characteristic Eμ-MYC pre–B/B cell lymphomas. These observations suggested that the B cell populations in which transformation occurs to generate lymphomas in the Eμ-MYC mouse model (15) remained unaltered by loss of Bcl-w. Indeed, flow cytometric analysis revealed that the B cell populations in the spleen and BM of precancerous littermates were indistinguishable among the 3 genotypes. Previous reports using a transplant model with fetal liver–derived stem cells from Eμ-MYC Bcl-2–/– (34) or Eμ-MYC Bcl-xl–/– (33) mice documented reduced immature and mature or pro–B and pre–B cells, respectively, which is consistent with the cell types in which these proteins predominantly function (2). Despite having similar biochemical functions, our data support the concept that specific antiapoptotic BCL-2 family members are more important than other family members in different cell types or stages of differentiation (5). Knowing when and how individual prosurvival BCL-2 proteins are relied on during neoplastic transformation may provide additional strategies for treating MYC-driven lymphoma.
MYC is overexpressed and/or dysregulated in the majority of human cancers (3); therefore, we believe that our results demonstrating an essential role for BCL-W in MYC-induced lymphomagenesis have significant implications for cancer biology and therapy. Importantly, we exposed a previously unknown connection between BCL-W and the MYC-driven human BL, a lymphoma for which no other antiapoptotic BCL-2 family member has been linked. Using a combination of approaches, we showed that increased BCL-W expression is highly selected for in BL. Specifically, 88.5% of the patient samples expressed BCL-W protein, whereas only 15.4% expressed BCL-2. These data were supported by analyses of BCL-W and BCL-2 mRNA from BL patient samples, which showed that elevated levels of BCL-W mRNA were preferentially selected for, but BCL-2 was not. Furthermore, we demonstrate that knocking down BCL-W expression in human BL cell lines resulted in apoptosis, highlighting the importance of maintaining BCL-W levels in BL for continued cell survival. Additionally, elevated levels of BCL-W alone conferred resistance to BL cell lines to the apoptotic effects of the BH3 mimetic compounds ABT-737 and ABT-263, which target 3 antiapoptotic BCL-2 family members. Previously, it was reported that BCL-2 expression correlated with sensitivity to ABT-263 in a panel of human non-Hodgkin lymphoma cell lines (42) and sensitivity to ABT-737 in human chronic lymphocytic leukemia (43, 44), small-cell lung cancer cell lines (45), and Eμ-MYC lymphomas (46, 47). In addition, increased levels of other antiapoptotic family members, such as MCL-1 and BCL-XL, have also been shown to confer resistance to ABT-737 (42). Therefore, our data indicate that expression of BCL-W will likely need to be considered and monitored when using BH3 mimetics for the treatment of lymphomas. Moreover, our data suggest that BCL-W may be useful in the diagnosis of BL and aid in the development of targeted therapies for this malignancy.
We also investigated BCL-W in DLBCL with respect to BCL-2, a known contributor to DLBCL that, when overexpressed, correlates with reduced patient survival (5, 48). We determined that many samples from DLBCL patients expressed both BCL-W and BCL-2 protein, but approximately a quarter of the samples only expressed BCL-W protein, and of these, the vast majority had high BCL-W levels. Furthermore, gene expression profiling of large DLBCL data sets also showed an increase in BCL-W mRNA in DLBCL compared with normal B cells. We also determined that there was a negative correlation between BCL-W and BCL-2 levels in DLBCL, indicating that there may be a significant subset of DLBCL cells that preferentially selects for the overexpression of BCL-W rather than BCL-2. Importantly, those patients whose lymphoma did preferentially select for BCL-W overexpression while BCL-2 levels remained low had markedly worse survival rates, and in this context, the level of BCL-W was also an independent determinant of overall survival in patients with DLBCL. Of note, there was no additive negative affect on patient survival when both BCL-2 and BCL-W were overexpressed, suggesting that lymphoma cell survival is not further enhanced once a certain expression level of antiapoptotic BCL-2 family members is reached. In support of this concept, it has been hypothesized that a mechanism of resistance to the BH-3 mimetics involves the upregulation of other antiapoptotic BCL-2 family members that are not inhibited by the compounds (5). Therefore, our data show that BCL-W expression is an important contributor to and predictive indicator of DLBCL. Thus, BCL-W may serve as a novel biomarker for BL and more aggressive DLBCL. Collectively, our data indicate that BCL-W is an understudied contributor to lymphomas that could be added to the limited list of current predictive biomarkers to improve prognostication and the development of more effective individualized treatment strategies.
Methods
Mice.
C57Bl/6 Eμ-MYC-Tg mice (4) were mated with C57Bl/6 Bcl-w+/– mice (9) provided by Rosalind Segal (Harvard Medical School, Harvard University, Boston, Massachusetts, USA), and their offspring were intercrossed to obtain littermate-matched Bcl-w+/+, Bcl-w+/–, and Bcl-w–/– Eμ-MYC–Tg mice of both sexes. Only Eμ-MYC–Tg–positive males were mated to pass on the Tg. For survival studies, mice were sacrificed at humane endpoints, and tumors and tissues were harvested and analyzed.
Lymphoma phenotype and B cell development analysis.
Splenocytes and BM from littermates (prior to lymphoma development for Eμ-MYC–Tg mice) or lymphoma cells were analyzed by flow cytometry following incubation with fluorochrome-linked antibodies as previously reported (49, 50).
Patient samples and tissue acquisition.
Normal human B cells were purified from leukoreduction filters obtained from the Red Cross using the IMag Human B Lymphocyte Enrichment Set (BD Biosciences). Deidentified fresh spleens were obtained from the Cooperative Human Tissue Network at Vanderbilt University Medical Center (VUMC) (Nashville, Tennessee, USA). For immunohistochemical analysis of DLBCL, formalin-fixed, paraffin-embedded (FFPE) tissues were selected from archival pathology materials on the basis of a documented large B cell diagnosis. Two separate tissue microarrays (TMAs) were constructed using 2-mm diameter cores from 35 separate specimens represented in duplicate and 20 separate specimens represented in triplicate, respectively. In addition, 10 samples with high-grade morphologic features were examined as individual slides. Fifty-seven DLBCL samples were considered evaluable and included in the analyses. For qRT-PCR analysis of DLBCL, TissueScan cDNA arrays containing 26 samples from patients with DLBCL and 10 normal lymphoid tissue controls were obtained from OriGene. For immunohistochemical analysis of BL, slides from 28 deidentified patients diagnosed with BL were obtained from St. Jude Children’s Research Hospital (Memphis, Tennessee, USA). Twenty-six BL samples were considered evaluable and included in the analyses. Patient characteristics are listed in Table 2, and details on how the pathologist scored IHC are provided in Supplemental Figures 11 and 15. For qRT-PCR analysis of BL samples, 15 FFPE BL samples and 7 FFPE normal lymphoid control tissues were obtained from Thomas Jefferson University (Philadelphia, Pennsylvania, USA).
IHC.
Immunohistochemical analysis was performed on 4-μm sections from BM preparations with anti–BCL-W (sc-6172; Santa Cruz Biotechnology Inc.) and anti–BCL-2 (PA0117; Leica Biosystems). A Leica BOND-MAX stainer was used with the Bond Polymer Refine detection system for visualization (Leica Biosystems). Expression levels were scored by a practicing board-certified hematopathologist (ASK) on a 0–3 point scale in a blinded fashion and scoring only the neoplastic cells. After unblinding, scores for the replicate samples on the TMAs were averaged and a final score obtained.
Vectors.
MSCV-MYCER-IRES-GFP, MSCV-MYCΔMBIIER-IRES-GFP (MYC box II deletion mutant), and MSCV-MYCV394D-ER-IRES-GFP (MYC:MIZ-1 interaction mutant) bicistronic retroviral vectors have been previously described (23, 37). Luciferase reporter plasmids were constructed by cloning a 60-mer of the 3′-UTR of Bcl-w containing the predicted miR-15 family target sequence into pMIR-REPORT (Invitrogen, Thermo Fisher Scientific). Luciferase reporters containing mutated miR binding sites were similarly generated by cloning a 60-mer with a mutated (base substitutions) target sequence to prevent the miR-15 family from binding the 3′-UTR of Bcl-w into pMIR-REPORT. Both luciferase plasmids for the Bcl-2 3′-UTR have previously been reported (23). For luciferase assays, a reporter plasmid encoding β-gal was used for transfection normalization (Invitrogen, Thermo Fisher Scientific). For knockdown experiments, nontargeting and BCL-W shRNA sequences were cloned into the pInducer Dox-inducible shRNA system (51). To induce shRNA expression, cells were treated with 1 μg/ml Dox (Sigma-Aldrich) or vehicle control (DMSO). For overexpression and rescue experiments, retroviral vectors were generated to express BCL-W (MSCV-BCLW-IRES-RFP) or BCL-2 (MSCV-BCL2-IRES-RFP).
Cells, infection, and transfection.
Daudi, Ramos, Raji, Su-DHL-6, and NIH3T3 cells were cultured as described by the American Type Culture Collection (ATCC). OCI-Ly-19 and OCI-Ly-3 cells were cultured in RPMI-1640 containing 10% FBS. All cell lines were negative for MYCoplasma. MEFs were generated and cultured as previously described (37). Primary murine pre–B cell cultures were generated as previously described (16). Briefly, BM was harvested from 8- to 10-week-old Bcl-w+/+, Bcl-w+/–, and Bcl-w–/– littermate-matched mice or 4- to 6-week-old littermate-matched Eμ-MYC–Tg mice (prior to lymphoma development) with 1, 2, or no Bcl-w alleles. After red blood cell lysis, BM cells were cultured in RMPI supplemented with 10 ng/ml IL-7, 20% FBS, 55 μM β-mercaptoethanol, 2 mM glutamine, and penicillin-streptoMYCin. Primary pre–B cells and MEFs were infected with bicistronic retroviral vectors (described above) in the presence of 8 μg/ml polybrene as previously described (37). GFP-positive pre–B cells were isolated by fluorescence-activated cell sorting. Human BL cell lines were infected with Dox-inducible lentiviral vectors (see above) expressing either a nontargeting shRNA or BCL-W–targeting shRNA and/or retroviral vectors expressing either BCL-W or BCL-2 cDNA or empty vector control. p53–/– MEFs were transfected using Lipofectamine 2000 (Invitrogen, Thermo Fisher Scientific) with 200 nM miScript Target Protector for Bcl-w or Bcl-2 (QIAGEN) according to the manufacturers’ protocols.
Western blotting.
Whole-cell protein lysates were generated, and equal amounts of protein per lane were Western blotted as previously described (16). The following antibodies were used: p19ARF (GTX200780; GeneTex); p53 (Ab-7; Calbiochem); BCL-W (31H4, product no. 2724) and cleaved caspase 3 (Asp175, product no. 9661) (both from Cell Signaling Technology); BCL-2 (3F11) and BCL-XL (44/BCL-X) (both from BD Bioscience); MCL-1 (200-401-CR9; Rockland); MYC (9E10; EMD Millipore); and β-actin (AC-15; Sigma-Aldrich). p53 cDNA was sequenced as previously described (16).
Growth and apoptosis analyses.
For IL-7 deprivation experiments, BM-derived primary pre–B cells were washed twice with PBS and resuspended in complete media (described above) without IL-7. To activate MYCER, 1 μM 4-hydroxytamoxifen (4-OHT; Sigma-Aldrich) or vehicle control (EtOH) was added to culture media. Population doublings, cell numbers, and/or viability were determined by trypan blue dye exclusion (in triplicate) and proliferation by MTT (Sigma-Aldrich; 560 nm) or MTS (Promega; 490 nm) assays (in quadruplicate). Apoptosis was evaluated by Western blotting for cleaved caspase 3 (described above) and flow cytometry following propidium iodide (sub-G1/apoptotic DNA content) or annexin V/7-AAD staining (both in triplicate) as previously reported (52). The ABT-737 and ABT-263 compounds were obtained from Selleckchem and reconstituted with DMSO.
qRT-PCR.
Total RNA was isolated from cell lines using TRIzol (Invitrogen, Thermo Fisher Scientific) or the RecoverAll Total Nucleic Acid Isolation Kit for FFPE tissue (Thermo Fisher Scientific) according to the manufacturer’s protocols. cDNA was generated and SYBR Green (SABiosciences, QIAGEN) and TaqMan MicroRNA (Applied Biosystems) assays were performed in triplicate to measure mRNA and miR, respectively, as previously described (53, 54). qPCR of lymphoma TissueScan cDNA arrays (OriGene) was completed according to the manufacturer’s protocol. mRNA and miR expression was normalized to β-actin and RNU6b, respectively, and presented as 2–ΔΔCt. Primer sequences for mouse and human BCL-W, BCL-2, BCL-XL, MCL-1, and β-actin mRNA expression were obtained from the Harvard Medical School Primer Bank (55).
Luciferase assays.
p53–/– MEFs expressing the 4-OHT–inducible WT MYCER, MYCΔMBII-ER, or MYCV394D-ER were transfected in triplicate with luciferase reporters (described above), β-gal control plasmid, and/or 200 nM miScript Target Protectors (QIAGEN) designed to block miR-15 family binding sites in the 3′-UTR of Bcl-w and Bcl-2. Luciferase and β-gal activity was measured 24 hours after MYCER activation as previously described (23).
Microarray gene expression analysis.
Microarray gene expression profiling data for BL, DLBCL, and normal human B cells (germinal center centroblasts and centrocytes, naive B cells, and memory B cells) were generated on the Affymetrix Human Genome U133 Plus 2.0 platform following the standard Affymetrix protocol. Data sets (summarized in Supplemental Table 1) were downloaded from the NCBI’s Gene Expression Omnibus (GEO) database (56) or from the authors’ website. BL, DLBCL, and normal B cell CEL files were normalized using a robust multiarray average algorithm (57). Batch effects were adjusted using ComBat (58). Probe sets were averaged to obtain a single-expression intensity measure per gene per array. Differential expression was measured by an unpaired, 1-tailed t test. All analyses were carried out in R, version 3.2.3.
Patient survival and correlation analyses.
Two microarray gene expression–profiling data sets for DLBCLs (59, 60) were generated on the Affymetrix Human Genome U133 Plus 2.0 platform following the standard Affymetrix protocol. These data sets and clinicopathological information were downloaded from the NCBI’s GEO database (GSE10846 and GSE31312). For each data set, raw expression-profiling data (CEL files) were normalized using a robust multiarray average algorithm (57). Probe sets were averaged to obtain a single-expression intensity measure per gene per array. For survival analyses, patient samples were simultaneously stratified on the basis of the median expression of BCL-2 and BCL-W into high or low expression groups for each gene. Then, using data on those patients with low BCL-2 expression, Kaplan-Meier survival curves were plotted for the high and low BCL-W expression groups and compared using log-rank tests. Univariate and multivariate Cox proportional hazards regression analyses for overall DLBCL survival outcomes based on BCL-W expression when BCL-2 expression was low were also performed. Tumor stages were stratified into lower (I and II) and higher (III and IV) stages. Patients were divided into lower and higher age groups on the basis of the median age. Survival analyses were carried out using the R survival package, version 3.2.3.
Statistics.
A 1-tailed Student’s t test was used when comparing 2 groups. One-way ANOVA was performed with Bonferroni’s correction when comparing multiple groups. Survival curves were compared by log-rank tests. The figure legends indicate the statistical tests used, which groups were compared, and when values represent the mean ± SD or the mean ± SEM. Univariate and multivariate survival analyses were analyzed by Cox proportional hazards regression models. The Pearson’s correlation coefficient was used to determine the association between BCL-W and BCL-2 expression. A P value of less than 0.05 was considered statistically significant.
Study approval.
All studies using mice and deidentified human samples complied with state and federal guidelines and were approved by the IACUCs and/or IRBs of VUMC and Thomas Jefferson University. All deidentified human tissues were obtained from the Cooperative Human Tissue Network of VUMC, St. Jude Children’s Research Hospital, or Thomas Jefferson University.
Author contributions
CMA and CME designed the research studies. CMA performed the experiments. ASK performed the pathological analyses. RM completed the bioinformatics analyses. JKC and JZG provided samples from patients with BL for immunohistochemical and mRNA analyses, respectively. CMA, ASK, RM, and CME wrote the manuscript. All authors reviewed and edited the manuscript.
Supplementary Material
Acknowledgments
We thank Pia Arrate, Kelli Boyd, and Cindy Lowe for their technical assistance; Rosalind Segal for the Bcl-w mice; the Encyclopedia of DNA Elements (ENCODE) Consortium and production laboratories; and members of the Eischen laboratory for their helpful discussions. TMAs and IHC were performed by the Translational Pathology Shared Resource at VUMC. Support was provided by NCI for F31CA165728 (CMA), R01CA177786 (CME), R01CA148950 (CME), and the NCI Cancer Center support grant P30CA056036.
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(2):635–650https://doi.org/10.1172/JCI89486.
References
- 1.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 2.Hata AN, Engelman JA, Faber AC. The BCL2 family: Key mediators of the apoptotic response to targeted anticancer therapeutics. Cancer Discov. 2015;5(5):475–487. doi: 10.1158/2159-8290.CD-15-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5(10):1024–1039. doi: 10.1158/2159-8290.CD-15-0507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adams JM, et al. The c-MYC oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature. 1985;318(6046):533–538. doi: 10.1038/318533a0. [DOI] [PubMed] [Google Scholar]
- 5.Delbridge AR, Grabow S, Strasser A, Vaux DL. Thirty years of BCL-2: translating cell death discoveries into novel cancer therapies. Nat Rev Cancer. 2016;16(2):99–109. doi: 10.1038/nrc.2015.17. [DOI] [PubMed] [Google Scholar]
- 6.Gibson L, et al. bcl-w, a novel member of the bcl-2 family, promotes cell survival. Oncogene. 1996;13(4):665–675. [PubMed] [Google Scholar]
- 7.O’Reilly LA, et al. Tissue expression and subcellular localization of the pro-survival molecule Bcl-w. Cell Death Differ. 2001;8(5):486–494. doi: 10.1038/sj.cdd.4400835. [DOI] [PubMed] [Google Scholar]
- 8.Print CG, et al. Apoptosis regulator bcl-w is essential for spermatogenesis but appears otherwise redundant. Proc Natl Acad Sci USA. 1998;95(21):12424–12431. doi: 10.1073/pnas.95.21.12424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ross AJ, et al. Testicular degeneration in Bclw-deficient mice. Nat Genet. 1998;18(3):251–256. doi: 10.1038/ng0398-251. [DOI] [PubMed] [Google Scholar]
- 10.Beverly LJ, Varmus HE. MYC-induced myeloid leukemogenesis is accelerated by all six members of the antiapoptotic BCL family. Oncogene. 2009;28(9):1274–1279. doi: 10.1038/onc.2008.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lee HW, Lee SS, Lee SJ, Um HD. Bcl-w is expressed in a majority of infiltrative gastric adenocarcinomas and suppresses the cancer cell death by blocking stress-activated protein kinase/c-Jun NH2-terminal kinase activation. Cancer Res. 2003;63(5):1093–1100. [PubMed] [Google Scholar]
- 12.Wilson JW, et al. Bcl-w expression in colorectal adenocarcinoma. Br J Cancer. 2000;82(1):178–185. doi: 10.1054/bjoc.1999.0897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Placzek WJ, Wei J, Kitada S, Zhai D, Reed JC, Pellecchia M. A survey of the anti-apoptotic Bcl-2 subfamily expression in cancer types provides a platform to predict the efficacy of Bcl-2 antagonists in cancer therapy. Cell Death Dis. 2010;1:e40. doi: 10.1038/cddis.2010.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Littlewood TD, Hancock DC, Danielian PS, Parker MG, Evan GI. A modified oestrogen receptor ligand-binding domain as an improved switch for the regulation of heterologous proteins. Nucleic Acids Res. 1995;23(10):1686–1690. doi: 10.1093/nar/23.10.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harris AW, Pinkert CA, Crawford M, Langdon WY, Brinster RL, Adams JM. The E mu-MYC transgenic mouse. A model for high-incidence spontaneous lymphoma and leukemia of early B cells. J Exp Med. 1988;167(2):353–371. doi: 10.1084/jem.167.2.353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Eischen CM, Weber JD, Roussel MF, Sherr CJ, Cleveland JL. Disruption of the ARF-Mdm2-p53 tumor suppressor pathway in MYC-induced lymphomagenesis. Genes Dev. 1999;13(20):2658–2669. doi: 10.1101/gad.13.20.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eischen CM, Roussel MF, Korsmeyer SJ, Cleveland JL. Bax loss impairs MYC-induced apoptosis and circumvents the selection of p53 mutations during MYC-mediated lymphomagenesis. Mol Cell Biol. 2001;21(22):7653–7662. doi: 10.1128/MCB.21.22.7653-7662.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Egle A, Harris AW, Bouillet P, Cory S. Bim is a suppressor of MYC-induced mouse B cell leukemia. Proc Natl Acad Sci USA. 2004;101(16):6164–6169. doi: 10.1073/pnas.0401471101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller PA, Vousden KH. Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell. 2014;25(3):304–317. doi: 10.1016/j.ccr.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eischen CM, Woo D, Roussel MF, Cleveland JL. Apoptosis triggered by MYC-induced suppression of Bcl-X(L) or Bcl-2 is bypassed during lymphomagenesis. Mol Cell Biol. 2001;21(15):5063–5070. doi: 10.1128/MCB.21.15.5063-5070.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eischen CM, et al. Bcl-2 is an apoptotic target suppressed by both c-MYC and E2F-1. Oncogene. 2001;20(48):6983–6993. doi: 10.1038/sj.onc.1204892. [DOI] [PubMed] [Google Scholar]
- 22.Adams CM, Eischen CM. Histone deacetylase inhibition reveals a tumor-suppressive function of MYC-regulated miRNA in breast and lung carcinoma. Cell Death Differ. 2016;23(8):1312–1321. doi: 10.1038/cdd.2016.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Adams CM, Hiebert SW, Eischen CM. MYC induces miRNA-mediated apoptosis in response to HDAC inhibition in hematologic malignancies. Cancer Res. 2016;76(3):736–748. doi: 10.1158/0008-5472.CAN-15-1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eilers M, Eisenman RN. MYC’s broad reach. Genes Dev. 2008;22(20):2755–2766. doi: 10.1101/gad.1712408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Herold S, et al. Negative regulation of the mammalian UV response by MYC through association with Miz-1. Mol Cell. 2002;10(3):509–521. doi: 10.1016/S1097-2765(02)00633-0. [DOI] [PubMed] [Google Scholar]
- 26.ENCODE Project Consortium An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489(7414):57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang X, et al. miRNA-195 sensitizes human hepatocellular carcinoma cells to 5-FU by targeting BCL-w. Oncol Rep. 2012;27(1):250–257. doi: 10.3892/or.2011.1472. [DOI] [PubMed] [Google Scholar]
- 28.Yin KJ, et al. miR-497 regulates neuronal death in mouse brain after transient focal cerebral ischemia. Neurobiol Dis. 2010;38(1):17–26. doi: 10.1016/j.nbd.2009.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang T, et al. MicroRNA-15a induces cell apoptosis and inhibits metastasis by targeting BCL2L2 in non-small cell lung cancer. Tumour Biol. 2015;36(6):4357–4365. doi: 10.1007/s13277-015-3075-1. [DOI] [PubMed] [Google Scholar]
- 30.Chang TC, et al. Widespread microRNA repression by MYC contributes to tumorigenesis. Nat Genet. 2008;40(1):43–50. doi: 10.1038/ng.2007.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmitz R, Ceribelli M, Pittaluga S, Wright G, Staudt LM. Oncogenic mechanisms in Burkitt lymphoma. Cold Spring Harb Perspect Med. 2014;4(2):a014282 . doi: 10.1101/cshperspect.a014282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly GL, et al. Targeting of MCL-1 kills MYC-driven mouse and human lymphomas even when they bear mutations in p53. Genes Dev. 2014;28(1):58–70. doi: 10.1101/gad.232009.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kelly PN, Grabow S, Delbridge AR, Strasser A, Adams JM. Endogenous Bcl-xL is essential for MYC-driven lymphomagenesis in mice. Blood. 2011;118(24):6380–6386. doi: 10.1182/blood-2011-07-367672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly PN, Puthalakath H, Adams JM, Strasser A. Endogenous bcl-2 is not required for the development of Emu-MYC-induced B-cell lymphoma. Blood. 2007;109(11):4907–4913. doi: 10.1182/blood-2006-10-051847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Michalak EM, et al. Puma and to a lesser extent Noxa are suppressors of MYC-induced lymphomagenesis. Cell Death Differ. 2009;16(5):684–696. doi: 10.1038/cdd.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Garrison SP, et al. Selection against PUMA gene expression in MYC-driven B-cell lymphomagenesis. Mol Cell Biol. 2008;28(17):5391–5402. doi: 10.1128/MCB.00907-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zindy F, et al. MYC signaling via the ARF tumor suppressor regulates p53-dependent apoptosis and immortalization. Genes Dev. 1998;12(15):2424–2433. doi: 10.1101/gad.12.15.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Haldar S, Negrini M, Monne M, Sabbioni S, Croce CM. Down-regulation of bcl-2 by p53 in breast cancer cells. Cancer Res. 1994;54(8):2095–2097. [PubMed] [Google Scholar]
- 39.Sugars KL, Budhram-Mahadeo V, Packham G, Latchman DS. A minimal Bcl-x promoter is activated by Brn-3a and repressed by p53. Nucleic Acids Res. 2001;29(22):4530–4540. doi: 10.1093/nar/29.22.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bui TV, Mendell JT. MYC: Maestro of microRNAs. Genes Cancer. 2010;1(6):568–575. doi: 10.1177/1947601910377491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grabow S, Delbridge AR, Aubrey BJ, Vandenberg CJ, Strasser A. Loss of a single Mcl-1 allele inhibits MYC-driven lymphomagenesis by sensitizing pro-B cells to apoptosis. Cell Rep. 2016;14(10):2337–2347. doi: 10.1016/j.celrep.2016.02.039. [DOI] [PubMed] [Google Scholar]
- 42.Mérino D, et al. Bcl-2, Bcl-x(L), and Bcl-w are not equivalent targets of ABT-737 and navitoclax (ABT-263) in lymphoid and leukemic cells. Blood. 2012;119(24):5807–5816. doi: 10.1182/blood-2011-12-400929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Del Gaizo Moore V, Brown JR, Certo M, Love TM, Novina CD, Letai A. Chronic lymphocytic leukemia requires BCL2 to sequester prodeath BIM, explaining sensitivity to BCL2 antagonist ABT-737. J Clin Invest. 2007;117(1):112–121. doi: 10.1172/JCI28281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Al-Harbi S, et al. An antiapoptotic BCL-2 family expression index predicts the response of chronic lymphocytic leukemia to ABT-737. Blood. 2011;118(13):3579–3590. doi: 10.1182/blood-2011-03-340364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tahir SK, et al. Influence of Bcl-2 family members on the cellular response of small-cell lung cancer cell lines to ABT-737. Cancer Res. 2007;67(3):1176–1183. doi: 10.1158/0008-5472.CAN-06-2203. [DOI] [PubMed] [Google Scholar]
- 46.Whitecross KF, et al. Defining the target specificity of ABT-737 and synergistic antitumor activities in combination with histone deacetylase inhibitors. Blood. 2009;113(9):1982–1991. doi: 10.1182/blood-2008-05-156851. [DOI] [PubMed] [Google Scholar]
- 47.Mason KD, et al. In vivo efficacy of the Bcl-2 antagonist ABT-737 against aggressive MYC-driven lymphomas. Proc Natl Acad Sci USA. 2008;105(46):17961–17966. doi: 10.1073/pnas.0809957105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thieblemont C, Brière J. MYC, BCL2, BCL6 in DLBCL: impact for clinics in the future? Blood. 2013;121(12):2165–2166. doi: 10.1182/blood-2013-01-480392. [DOI] [PubMed] [Google Scholar]
- 49.Arrate MP, Vincent T, Odvody J, Kar R, Jones SN, Eischen CM. MicroRNA biogenesis is required for MYC-induced B-cell lymphoma development and survival. Cancer Res. 2010;70(14):6083–6092. doi: 10.1158/0008-5472.CAN-09-4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alt JR, Greiner TC, Cleveland JL, Eischen CM. Mdm2 haplo-insufficiency profoundly inhibits MYC-induced lymphomagenesis. EMBO J. 2003;22(6):1442–1450. doi: 10.1093/emboj/cdg133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Meerbrey KL, et al. The pINDUCER lentiviral toolkit for inducible RNA interference in vitro and in vivo. Proc Natl Acad Sci USA. 2011;108(9):3665–3670. doi: 10.1073/pnas.1019736108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adams CM, Eischen CM. Inactivation of p53 is insufficient to allow B cells and B-cell lymphomas to survive without Dicer. Cancer Res. 2014;74(14):3923–3934. doi: 10.1158/0008-5472.CAN-13-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang P, Lushnikova T, Odvody J, Greiner TC, Jones SN, Eischen CM. Elevated Mdm2 expression induces chromosomal instability and confers a survival and growth advantage to B cells. Oncogene. 2008;27(11):1590–1598. doi: 10.1038/sj.onc.1210788. [DOI] [PubMed] [Google Scholar]
- 54.McGirt LY, Adams CM, Baerenwald DA, Zwerner JP, Zic JA, Eischen CM. miR-223 regulates cell growth and targets proto-oncogenes in MYCosis fungoides/cutaneous T-cell lymphoma. J Invest Dermatol. 2014;134(4):1101–1107. doi: 10.1038/jid.2013.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang X, Spandidos A, Wang H, Seed B. PrimerBank: a PCR primer database for quantitative gene expression analysis, 2012 update. Nucleic Acids Res. 2012;40(Database issue):D1144–D1149. doi: 10.1093/nar/gkr1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Barrett T, Edgar R. Gene expression omnibus: microarray data storage, submission, retrieval, and analysis. Meth Enzymol. 2006;411:352–369. doi: 10.1016/S0076-6879(06)11019-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Irizarry RA, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 58.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8(1):118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 59.Lenz G, et al. Stromal gene signatures in large-B-cell lymphomas. N Engl J Med. 2008;359(22):2313–2323. doi: 10.1056/NEJMoa0802885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Visco C, et al. Comprehensive gene expression profiling and immunohistochemical studies support application of immunophenotypic algorithm for molecular subtype classification in diffuse large B cell lymphoma: a report from the International DLBCL Rituximab-CHOP Consortium Program Study. Leukemia. 2012;26(9):2103–2113. doi: 10.1038/leu.2012.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.