Abstract
We recently demonstrated that selective expression of the Rho GTPase-activating protein ARHGAP42 in smooth muscle cells (SMCs) controls blood pressure by inhibiting RhoA-dependent contractility, providing a mechanism for the blood pressure–associated locus within the ARHGAP42 gene. The goals of the current study were to identify polymorphisms that affect ARHGAP42 expression and to better assess ARHGAP42’s role in the development of hypertension. Using DNase I hypersensitivity methods and ENCODE data, we have identified a regulatory element encompassing the ARHGAP42 SNP rs604723 that exhibits strong SMC-selective, allele-specific activity. Importantly, CRISPR/Cas9–mediated deletion of this element in cultured human SMCs markedly reduced endogenous ARHGAP42 expression. DNA binding and transcription assays demonstrated that the minor T allele variation at rs604723 increased the activity of this fragment by promoting serum response transcription factor binding to a cryptic cis-element. ARHGAP42 expression was increased by cell stretch and sphingosine 1-phosphate in a RhoA-dependent manner, and deletion of ARHGAP42 enhanced the progression of hypertension in mice treated with DOCA-salt. Our analysis of a well-characterized cohort of untreated borderline hypertensive patients suggested that ARHGAP42 genotype has important implications in regard to hypertension risk. Taken together, our data add insight into the genetic mechanisms that control blood pressure and provide a potential target for individualized antihypertensive therapies.
Introduction
Although hypertension is a major risk factor for stroke, myocardial infarction, and kidney failure (1), surprisingly little is known about the genetic mechanisms that contribute to its development. GWAS have begun to identify common genetic variants that contribute to variations in blood pressure (BP) (see refs. 2, 3 for reviews). While some of these loci have been fairly well characterized, others are found within genes that have no known connection to the control of BP. One such locus was identified on chromosome 11 within the GTPase-activating protein (GAP) ARHGAP42 (4–6). The minor allele at this locus was associated with a decrease in BP of about 0.5 mmHg per allele and had a minor allele frequency (MAF) of 0.27 in the European population in which it was identified (5).
ARHGAP42, also known as GRAF3, is a member of the GRAF (GAP for Rho associated with focal adhesion kinase) family of Rho-specific GAPs previously characterized by our group (7–10). Based on our demonstration that ARHGAP42 was highly and selectively expressed in smooth muscle in mouse and human tissues (11) and the fact that Rho GTPase RhoA signaling controls smooth muscle cell (SMC) contractility (12), we hypothesized that ARHGAP42’s association with BP was mediated by its ability to modulate vascular resistance. Indeed, Arhgap42-deficient mice exhibited significant hypertension and increased pressor responses to ANGII and endothelin 1, and these effects were prevented by treatment with the Rho kinase (ROCK) inhibitor Y-27632 (11). Further supporting this idea, we showed that large and small arteries from Arhgap42-deficient mice exhibited increased contractility in vitro and in vivo, while kidney structure and function were unchanged.
According to the HaploReg v4 database from the Broad Institute, the BP-associated allele in the European population is currently defined by 4 SNPs (rs604723, rs633185, rs607562, and rs667575) in high linkage disequilibrium (LD) (r2 > 0.8). Importantly, all of these SNPs are present within the noncoding 80-kb first intron of ARHGAP42. Thus, we hypothesized that minor allele variations within the BP-associated locus increase ARHGAP42 expression by enhancing the transcriptional activity of a yet-to-be-identified regulatory element. The goals of the present study were to identify the transcription mechanisms that drive ARHGAP42 expression in SMCs, to test whether variations within the ARHGAP42 BP-associated locus affect ARHGAP42 transcription, and to further characterize the role of ARHGAP42 expression in the regulation of BP and the development of human hypertension.
Results
Allele-specific differences of ARHGAP42 expression in SMCs.
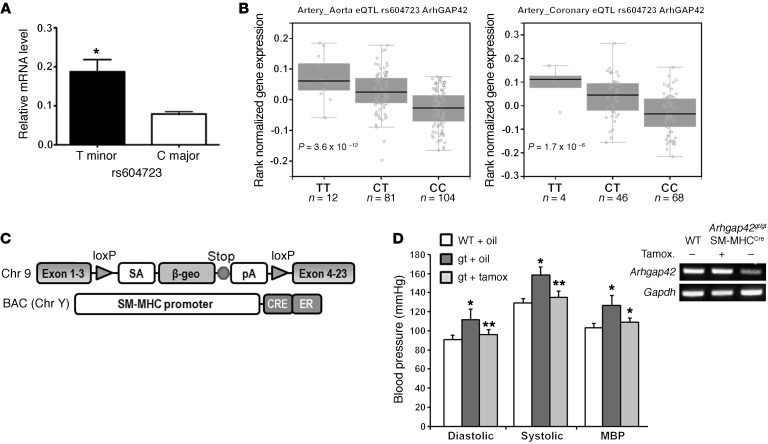
To begin to test our hypothesis that BP-associated variations in the ARHGAP42 gene have allele-specific effects on ARHGAP42 expression in SMCs, we developed PCR primers that can distinguish between the major C and minor T alleles at the rs604723 SNP and used quantitative PCR to measure ARHGAP42 mRNA levels in human aortic SMC (HuAoSMC) cultures that are heterozygous at the ARHGAP42 BP-associated locus. One caveat to this approach is that it can only measure unprocessed transcripts that contain the first intron. As shown in Figure 1A, ARHGAP42 transcripts containing the minor allele were significantly higher than those containing the major allele, and control experiments with and without DNase and with and without reverse transcriptase confirmed that these results were not influenced by contaminating genomic DNA (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/JCI88899DS1). In strong support of these findings, the NIH-sponsored Genotype-Tissue Expression (GTEx) Consortium identified an expression quantitative trait locus (eQTL) at rs604723 by correlating ARHGAP42 mRNA levels in human tibial artery samples with ARHGAP42 genotype (13). As shown in Figure 1B, our more recent analysis of GTEx data revealed that ARHGAP42 mRNA levels in aorta and coronary artery samples were approximately 3-fold higher in samples taken from individuals homozygous for the minor allele at rs604723 than from individuals homozygous for the major allele. Aside from a relatively small difference in ARHGAP42 expression in subcutaneous adipose (a highly vascularized tissue), ARHGAP42 eQTLs were not detected in other organs, strongly supporting the idea that the BP effects of this locus are mediated by changes in ARHGAP42 expression in SMCs.
Figure 1. ARHGAP42 expression in SMCs is regulated by allele-specific mechanisms and controls BP.
(A) Total RNA isolated from HuAoSMCs heterozygous at the rs604723 SNP (C/T) was subjected to first-strand cDNA synthesis using reverse transcriptase. Reaction products were then subjected to a TaqMan-based PCR assay using allele-specific primers to the ARHGAP42 rs604723 variation. Data represent mean ± SEM of n = 4 experiments; *P < 0.01 vs. the major C allele (Student’s t test). (B) ARHGAP42 mRNA levels were measured by the Genotype-Tissue Expression (GTEx) Consortium. The minor T ARHGAP42 allele at the rs604723 polymorphism was significantly associated with increased ARHGAP42 expression in aortic and coronary artery samples. (C) Schematic of the Arhgap42 gene-trap and SM-MHCCreERT2 mice used for SMC-specific ARHGAP42 rescue experiments. (D) WT and Arhgap42gt/gt SM-MHCCreERT2 mice were injected i.p. with vehicle (corn oil) or tamoxifen (100 mg/kg) for 5 consecutive days as indicated. Two weeks after the last injection, BP was measured by tail cuff method, and Arhgap42 mRNA levels in the aorta were measured by semiquantitative RT-PCR analysis using primers to exons 1 and 4. Data are expressed as mean ± SEM; n = 6 for WT and Arhgap42gt/gt SM-MHCCreERT2 mice with vehicle treatment, n = 5 for Arhgap42gt/gt SM-MHCCreERT2 mice with tamoxifen treatment. *P < 0.05 vs. WT; **P < 0.05 vs. corn oil–treated (ANOVA). Note that tamoxifen treatment restored Arhgap42 expression and reduced BP to WT levels (representative of 3 separate experiments).
ARHGAP42 expression in SMCs controls BP.
To provide additional direct evidence that expression of ARHGAP42 in SMCs is critical for BP regulation, we rescued its expression in our hypertensive global Arhgap42-deficient gene-trap mice (Arhgap42gt/gt) by crossing them to an SM-MHCCreERT2 line that expresses a tamoxifen-inducible Cre under the control of the smooth muscle myosin heavy chain promoter (14). As shown in Figure 1C, the inhibitory Arhgap42gt/gt gene-trap cassette is flanked by loxP sites, and treatment of Arhgap42gt/gt SM-MHCCreERT2 mice with tamoxifen permanently restored ARHGAP42 expression in aortic SMCs, and completely reversed the hypertensive phenotype in this model by 2 weeks after tamoxifen treatment (Figure 1D and Supplemental Figure 2). These data provide conclusive evidence that ARHGAP42 levels in SMCs control BP homeostasis and that the hypertensive phenotype in Arhgap42gt/gt mice was reversible and most likely due to an increase in SMC contractility.
ARHGAP42 genotype and human hypertension.
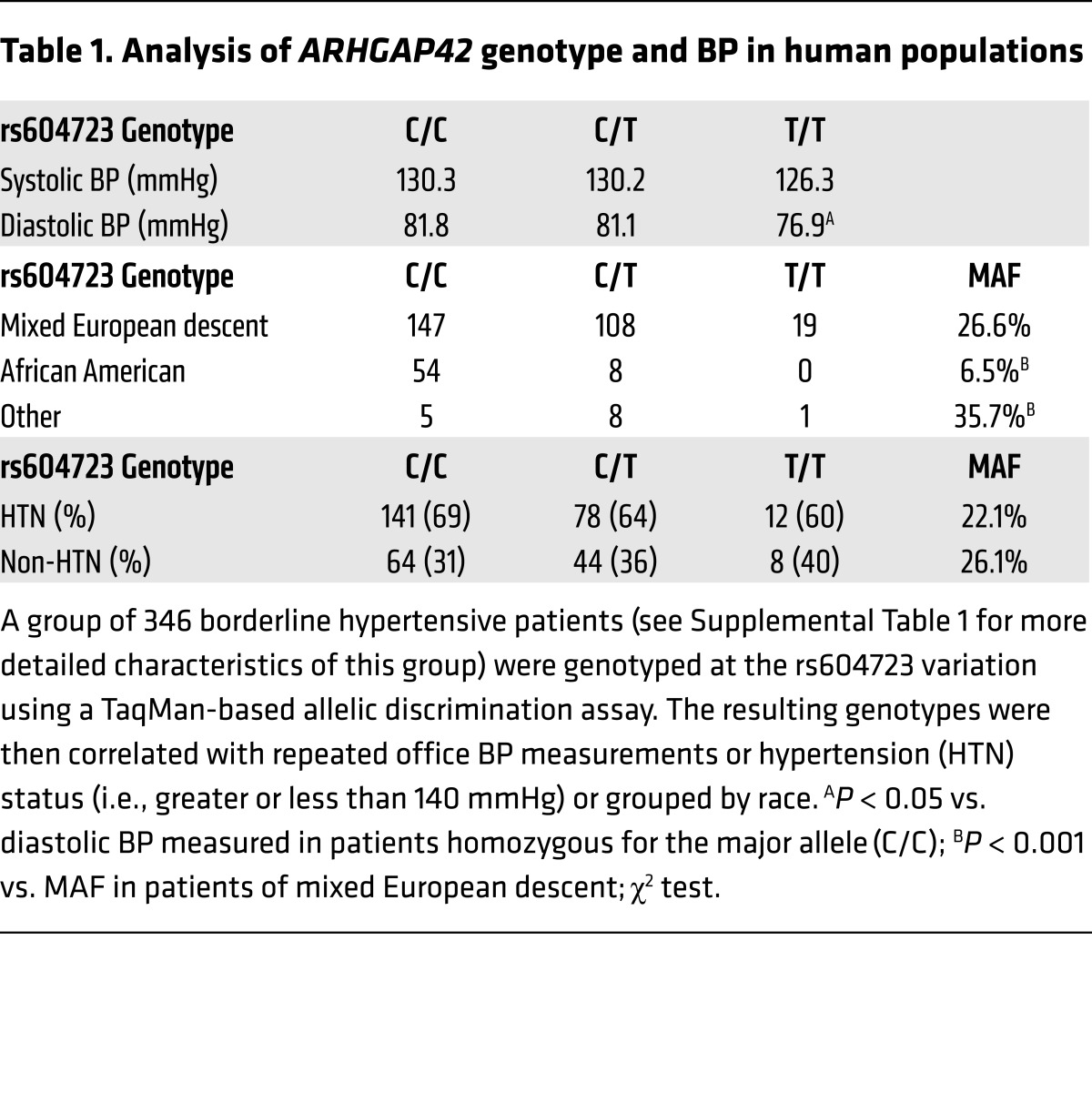
To better assess ARHGAP42’s role in the development of human hypertension, we genotyped a group of 346 borderline hypertensive patients who were part of a clinical study comparing the effectiveness of BP monitoring protocols at the University of North Carolina (refs. 15, 16, and Supplemental Table 1). Importantly, BPs in this group were extremely well characterized by repeated office measurements, and none of these individuals were being treated with antihypertensive therapies. Several findings illustrated in Table 1 are worth noting. First, we observed an age-independent decrease in BP in subjects homozygous for the minor allele compared with subjects homozygous for the major allele (76.9 mmHg vs. 81.8 mmHg; P = 0.028). The relative magnitude of this decrease (5 mmHg vs. 1 mmHg measured in GWAS) suggested that ARHGAP42’s effects on BP may be greater than originally estimated. Second, the MAF (as defined by rs604723) trended lower in the group of patients who were categorized as hypertensive as defined by American Heart Association guidelines (systolic BP > 140 mmHg), and the percentage of subjects that exhibited hypertension trended lower in subjects who had copies of the minor allele (69%, 64%, and 60% for the C/C, C/T, and T/T genotypes, respectively). Finally, the MAF of the protective T allele in this cohort was dramatically lower in African Americans than in subjects of mixed European descent (6.5% vs. 26.6%; P < 0.001), and sequencing of over 1,000 individuals from this and additional University of North Carolina cardiovascular cohorts (17–20) confirmed this result (Supplemental Table 2).
Table 1. Analysis of ARHGAP42 genotype and BP in human populations.
Identification of regulatory elements within the ARHGAP42 gene.
It is clear that gene regulatory regions exhibit distinct chromatin signatures, and we and others have shown that SMC-specific gene expression is regulated by alterations in chromatin structure (21–24). Thus, to prioritize our search for regulatory regions that drive ARHGAP42 expression, we performed DNase hypersensitivity measurements in HuAoSMCs to identify regions of open chromatin. As shown in the ARHGAP42 gene schematic in Figure 2A, we identified 2 approximately 600-bp DNase hypersensitivity sites (DHS1 and DHS2) within the first intron and a larger DHS that covered about 2 kb of the transcription start site (TSS). Additional analyses of Encyclopedia of DNA Elements (ENCODE) data sets revealed that all 3 regions were marked by histone modifications known to be associated with transcriptionally active regions (i.e., H3K4 methylation and H3K27 acetylation) and contained stretches of highly conserved sequence. Of particular interest, the rs604723 SNP was in the middle of a highly conserved 100-bp region at the center of the DHS2. All 3 regions were PCR-amplified from human genomic DNA and cloned into the pGL3 luciferase vector. It is important to note that the DHS1 region was extended to include the nearby SNP rs633185, so that the functional effects of this variation could be tested. Also, because the other 2 SNPs within the BP-associated LD block (rs607562 and rs667575) were not near open chromatin regions or conserved DNA sequences, we did not further examine their contributions to ARHGAP42 expression.
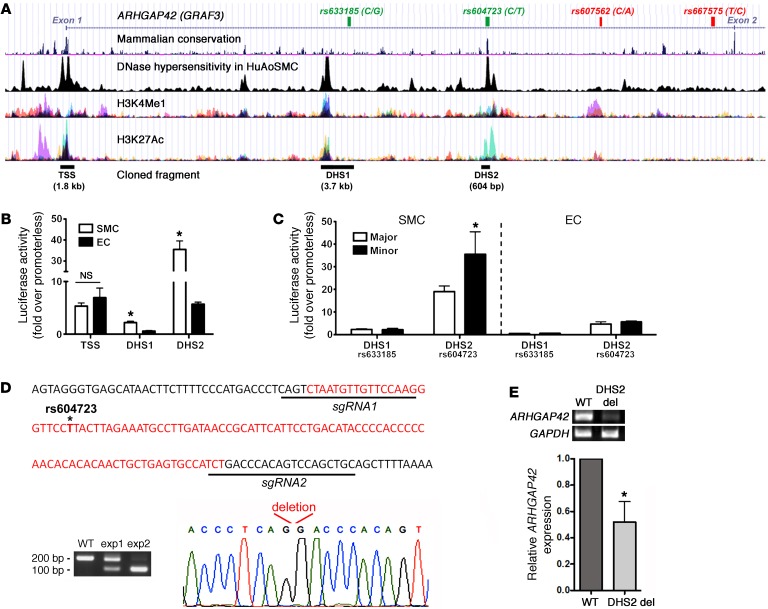
Figure 2. An enhancer within the ARHGAP42 first intron displays strong SMC-specific and allele-specific activity and is required for endogenous ARHGAP42 expression.
(A) Map of the chromatin determinations used to characterize potential regulatory elements near the ARHGAP42 BP-associated locus. The SNPs that define the BP-associated allele (r2 > 0.8) are shown at the top. (B) The indicated DNase-hypersensitive (DHS) regions were cloned into the pGL3 luciferase vector and transfected into primary human bronchial SMCs and mouse ECs. Luciferase activity in cell lysates was measured 2 days later and is expressed as fold over the promoterless pGL3 vector. Data represent mean ± SEM of n = 6 experiments; *P < 0.001 vs. in ECs (Student’s t test). (C) Site-directed mutagenesis was used to test the effects of the major/minor alleles on DHS1 and DHS2 enhancer activity. Data represent mean ± SEM of n = 6 experiments; *P < 0.01 vs. the major allele (Student’s t test). (D) Schematic of the 102-bp deletion (in red) generated by our CRISPR/Cas9–mediated gene editing protocol. (E) ARHGAP42 message was measured by semiquantitative RT-PCR in human bronchial SMC cultures transfected with expression plasmids encoding Cas9 and the guide RNAs shown in D (n = 5). The reduction in ARHGAP42 expression was normalized to the efficiency of DHS2 deletion, which ranged from 45% to 95%. Data represent mean ± SEM of n = 5 separate experiments; *P < 0.05 vs. cells transfected with empty guide RNA expression plasmid (Student’s t test).
As shown in Figure 2B, the DHS2 exhibited very strong transcriptional activity (38-fold over the promoterless pGL3 vector) that was significantly higher in SMCs than in endothelial cells (ECs), suggesting that this element is an important driver of SMC-selective ARHGAP42 expression. The TSS exhibited moderate activity in SMCs and in ECs, consistent with the idea that it functions as more of a basal promoter. We next used site-directed mutagenesis to generate allelic series for the rs633185 and rs604723 SNPs within the context of the DHS1 and DHS2 regulatory elements, respectively. As shown in Figure 2C, the DHS2 containing the minor T allele exhibited significantly higher activity than the DHS2 containing the major C allele, while the variation at rs633185 had no effect on the relatively low SMC-selective activity of the DHS1 region. Interestingly, the rs604723 variation did not affect DHS2 activity in ECs.
To test whether the DHS2 element was required for expression of the endogenous ARHGAP42 gene, we used CRISPR/Cas9–mediated gene editing to delete the 100-bp conserved element within the DHS2 in human bronchial SMC (HuBrSMC) cultures (Figure 2D). Although we observed somewhat variable deletion efficacy (from 45% to 95% in 5 separate experiments), our deletion protocol resulted in a significant decrease in ARHGAP42 mRNA expression (Figure 2E). These results strongly support our conclusion that the DHS2 region regulates ARHGAP42 expression and that rs604723 is the functional SNP in this BP-associated LD block.
The minor allele sequence at rs604723 binds serum response factor.
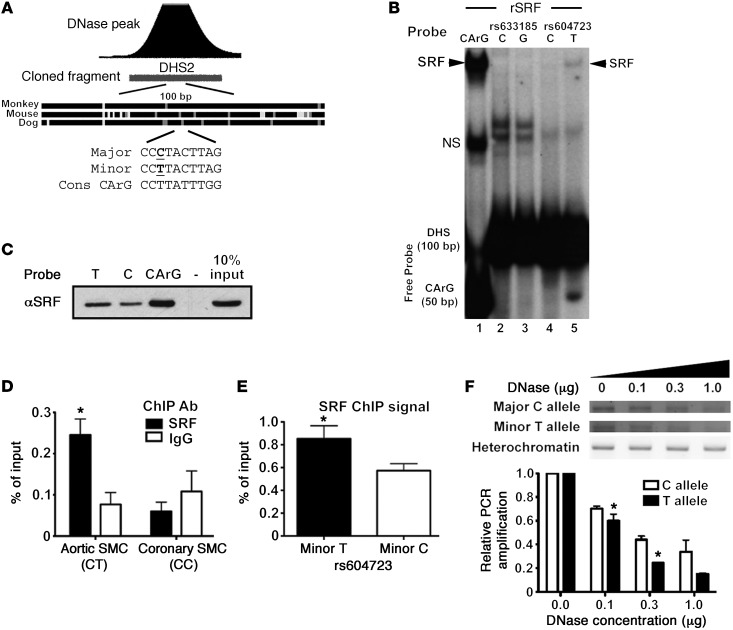
To begin to identify the mechanisms that mediate the transcriptional activity of the DHS2 fragment and the effects of the minor T allele variation, we used gel shift assays to compare protein binding to 100-bp probes that encompassed the highly conserved region at the center of the DHS2. As shown in Supplemental Figure 3, we observed 2 relatively weak T allele–specific binding complexes, 1 of which had a mobility similar to that of serum response factor (SRF), a transcription factor known to be critical for SMC-specific gene expression (see ref. 25 for a review). Interestingly, the presence of the minor T allele at rs604723 results in a DHS2 sequence that conforms to a consensus SRF-binding CArG element at 8 out of 10 residues, while the presence of the major C allele within the A/T-rich region would be predicted to inhibit SRF binding to this sequence (Figure 3A). To test the involvement of SRF, we performed additional gel shift assays using recombinant SRF protein. Our results clearly show that the minor T allele sequence interacted with SRF while the major C allele sequence did not (Figure 3B). Similar results were obtained when biotin-labeled T and C allele oligonucleotide probes were conjugated to avidin-coated beads and used to precipitate SRF from SMC lysates (Figure 3C). We next used ChIP assays to test whether SRF bound to the DHS2 in the context of the endogenous ARHGAP42 gene. As shown in Figure 3D, we observed significant SRF binding in our HuAoSMCs that are heterozygous (C/T) at rs604723 but no binding when ChIP assays were performed in human coronary SMCs that are homozygous major (C/C) at this sequence. Moreover, the increased presence of T allele–containing DNA in SRF immunoprecipitates (Figure 3E) strongly suggested that SRF interacts more readily with this sequence in the context of the endogenous gene, and targeted allele-specific DNase hypersensitivity assays revealed that the DHS2 region containing the T allele sequence was in a more active chromatin conformation (Figure 3F).
Figure 3. The minor T allele at rs604723 promotes SRF binding.
(A) Schematic of sequence conservation at the center of the DHS2 region and of CArG homology at the rs604723 SNP. (B) Gel shift assays were performed by combining recombinant SRF (rSRF) with radiolabeled 100-bp oligonucleotide probes containing the major or minor alleles at rs633185 and rs604723. Representative image shown from n = 2. (C) Biotin-labeled 20-bp oligonucleotides containing the major C or minor T allele at rs604723 or a consensus CArG element were conjugated to streptavidin beads and incubated with HuAoSMC nuclear extracts. Washed immunoprecipitates were analyzed for the presence of SRF by Western blotting. Data are representative of 2 separate experiments. (D) ChIP assays were used to measure SRF binding to the DHS2 region in cultured HuAoSMCs and HuCoSMCs that are heterozygous (CT) and homozygous major (CC) at the rs604723 SNP, respectively. Data represent mean ± SEM of n = 4 experiments; *P < 0.05 vs. IgG in HuAoSMCs (Student’s t test). (E) SRF-ChIP immunoprecipitates from HuAoSMCs were subjected to a TaqMan-based assay that discriminates between the major and minor alleles at the rs604723 SNP. Data represent mean ± SEM of n = 4 experiments; *P < 0.01 vs. major allele (Student’s t test). (F) Increasing amounts of DNase I (0–1 μg) were added to permeabilized nuclei isolated from HuAoSMCs. Following genomic DNA isolation, allele-specific primers were used to amplify a 300-bp region containing the rs604723 SNP. Data represent mean ± SEM of n = 3 experiments; *P < 0.05 (Student’s t test).
SRF is required for ARHGAP42 expression and for the effects of the rs604723 variation.
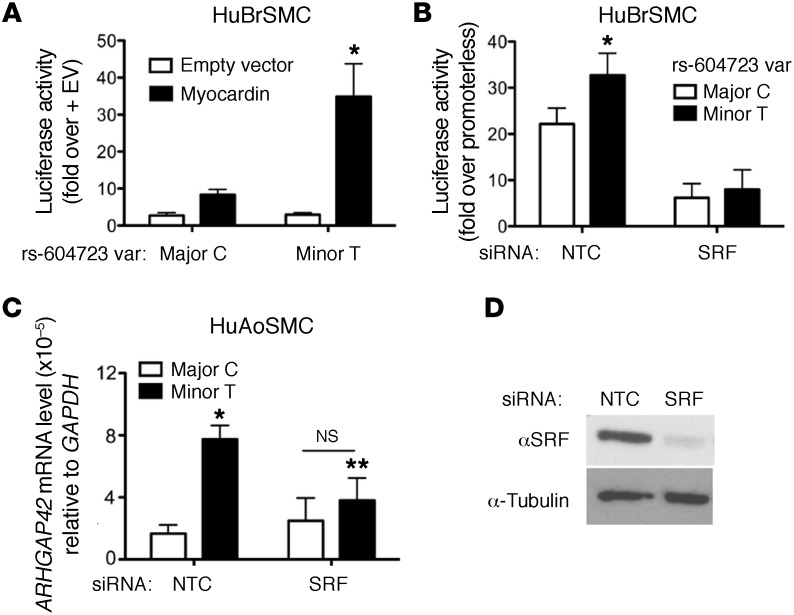
To test whether ARHGAP42, like most other SMC-specific markers, is regulated by SRF, we used several gain/loss-of-function approaches. As shown in Figure 4A, overexpression of the SRF cofactor myocardin in HuBrSMCs transactivated the DHS2-luciferase fragment containing the minor T allele 12-fold, but had significantly less of an effect on the DHS2 fragment containing the major C allele. Moreover, knockdown of SRF in these cells by siRNA decreased the transcriptional activity of the DHS2 minor allele to a level that was similar to that of the major allele (Figure 4B). Importantly, SRF knockdown in our HuAoSMC line also decreased the level of endogenous ARHGAP42 message containing the minor allele as measured by our allele-specific reverse transcription PCR (RT-PCR) methods (Figure 4C).
Figure 4. The allele-specific activity of the DHS2 enhancer is SRF-dependent.
(A) Major and minor DHS2 luciferase constructs were transfected into HuBrSMCs along with myocardin or empty expression vector. Data represent mean ± SEM of n = 5 experiments; *P < 0.05 vs. major allele plus myocardin (Student’s t test). (B) DHS2-luciferase activity was measured in HuBrSMCs treated with control (NTC) or SRF siRNA. Data represent mean ± SEM of n = 6 experiments; *P < 0.01 vs. the minor allele (Student’s t test). (C) Allele-specific GRAF3 mRNA levels were measured by semiquantitative RT-PCR in control and SRF knockdown HuAoSMCs. Data represent mean ± SEM of n = 3 experiments; *P < 0.05 vs. the major C allele in control cells; **P < 0.05 vs. minor T allele in control cells (Student’s t test). (D) Confirmation of SRF knockdown in SMCs treated with control or SRF siRNAs. Data are representative of 3 separate experiments.
ARHGAP42 expression is upregulated by RhoA signaling.
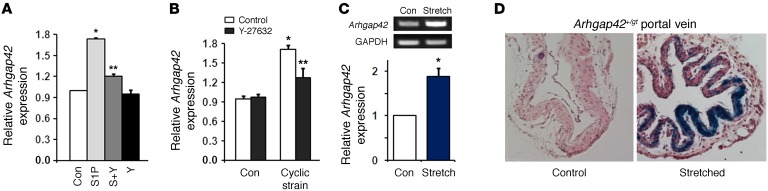
We and others have shown that RhoA signaling enhances SMC-specific gene expression by promoting the nuclear localization of the myocardin-related transcription factors (MRTFs) (26–28). In support of this mechanism, Arhgap42 expression in primary rat aortic SMCs was upregulated by sphingosine 1-phosphate, a strong activator of RhoA signaling in many cell types, including SMCs (29), and this effect was abolished by pretreatment with the ROCK inhibitor Y-27632 (Figure 5A). Physical forces such as cell stretch and tension are also well-known activators of RhoA signaling and are known to be increased in the vessel wall under hypertensive conditions (30). As shown in Figure 5B, Arhgap42 mRNA levels were significantly upregulated in SMC cultures subjected to cyclic stretch using the FX-4000T Flexcell system, and this effect was also ROCK-dependent. Arhgap42 mRNA levels were also upregulated in isolated portal vein segments subjected to static stretch (Figure 5C). The fact that LacZ expression was upregulated by stretch in portal veins isolated from Arhgap42 LacZ gene-trap mice strongly indicated that this effect was mediated transcriptionally (Figure 5D).
Figure 5. ARHGAP42 expression is activated by RhoA signaling and cell stretch.
(A) Primary rat aortic SMCs were treated with 10 μM sphingosine 1-phosphate (S1P) with or without the ROCK inhibitor Y-27632. Arhgap42 expression was measured after 72 hours by semiquantitative PCR. Data represent mean ± SEM of 4 experiments; *P < 0.001 vs. control; **P < 0.001 vs. S1P-treated (ANOVA). n = 4. (B) Using the FX-4000T Flexcell system, primary rat aortic SMCs were subjected to 0 (Ctrl) or 20% equibiaxial elongation at 1 Hz (cyclic strain). Arhgap42 message was measured at 18 hours by quantitative PCR. Data represent mean ± SEM; n = 3 from 2 independent experiments; *P < 0.001 vs. no cyclic strain; **P < 0.05 vs. minus Y-27632 (ANOVA). (C) Rat portal veins placed in ex vivo culture were subjected to 0 or 600 mg of static stretching force. At 72 hours Arhgap42 message was measured by semiquantitative RT-PCR. Graph shows ImageJ-based quantification of 3 independent experiments. *P < 0.05 (Student’s t test). (D) Portal veins isolated from Arhgap42+/gt mice were cultured ex vivo and subjected to 0 or 300 mg of static stretching force for 5 days. After LacZ staining, tissues were processed for standard microscopy including H&E staining. Scale bar: 200 μm. Data are representative of 3 independent experiments.
Activation of ARHGAP42 expression attenuates the development of hypertension.
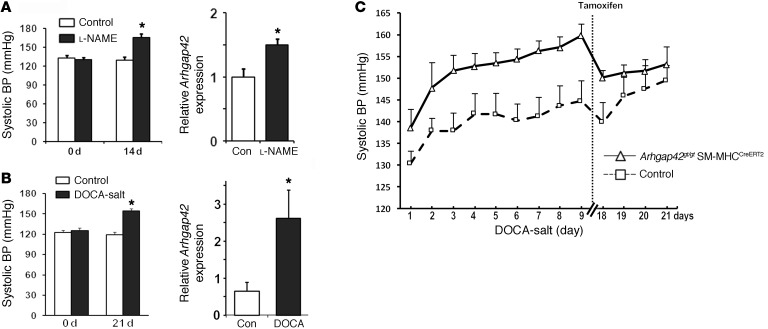
Since RhoA/MRTF/SRF-dependent upregulation of ARHGAP42 would serve as a transcriptionally mediated negative-feedback loop for RhoA signaling in SMCs, we postulated that this mechanism prevents excessive SMC constriction by hypertensive signals. In support of this idea, and in accordance with the results of our cell stretch experiments, arterial Arhgap42 mRNA levels were significantly increased in mice made hypertensive by l-NAME or deoxycorticosterone acetate (DOCA)–salt regimens (Figure 6, A and B). To test whether upregulation of Arhgap42 expression under these conditions counteracted the development of hypertension, we subjected WT and Arhgap42-deficient mice (Arhgap42gt/gt SM-MHCCreERT2) to a DOCA-salt regimen and monitored BP by telemetry. Although the initial DOCA-salt–mediated increase in BP was similar in both groups (Figure 6C), BP in Arhgap42gt/gt SM-MHCCreERT2 continued to increase over the next week at a rate of 1.5 mmHg/d compared with 0.9 mmHg/d in WT mice (P < 0.05). As previously observed, the BP pressure difference between these 2 groups was completely reversed by treatment of Arhgap42gt/gt SM-MHCCreERT2 with tamoxifen, which also restored Arhgap42 expression (Supplemental Figure 4).
Figure 6. Arhgap42 expression limits the development of hypertension.
(A) WT mice were treated with l-NAME (450 mg/l in drinking water). After 14 days, BP was measured by tail cuff method, and Arhgap42 message in isolated mesenteric arteries was measured by quantitative PCR. Data represent mean ± SEM; n = 4 per group; *P < 0.05 vs. untreated (Student’s t test). (B) WT mice were implanted with a 50-mg slow-release DOCA pellet and then fed 0.9% NaCl in drinking water. After 3 weeks, BP was measured by tail cuff method, and Arhgap42 message in aorta was measured by quantitative PCR. *P < 0.05 vs. untreated; n = 6 per group (Student’s t test). (C) Following radiotelemeter implantation and equilibration, 3 WT and 5 Arhgap42gt/gt SM-MHCCreERT2 mice were implanted with a 50-mg slow-release DOCA pellet and then fed 0.9% NaCl in drinking water for 3 weeks. Ten days after the start of the DOCA-salt regimen, both groups were treated with tamoxifen (100 mg/kg) by oral gavage for 3 consecutive days. Graph represents average mean arterial BP averaged over each 24-hour period. P < 0.05 (slopes test).
Discussion
We have previously shown that the Rho-specific GTPase-activating protein ARHGAP42 is highly and selectively expressed in SMCs and that global gene-trap–mediated reduction in ARHGAP42 levels resulted in hypertension (11). By characterizing the transcription mechanisms that control human ARHGAP42 expression, we have now identified a novel mechanism for the BP-associated locus within the ARHGAP42 first intron. Our data strongly indicate that rs604723 is the causative SNP at this locus and that the minor T allele variation increases ARHGAP42 expression by promoting SRF binding to an SMC-selective intronic regulatory element. Our demonstration that the minor ARHGAP42 allele is more highly expressed in HuAoSMCs, when coupled with similar data from human artery samples, strongly supports our hypothesis that this variation reduces BP by inhibiting RhoA-dependent constriction of resistance vessels.
To our knowledge, this is the first demonstration of a genetic variant that alters SRF binding and that directly connects SRF function to a GWAS locus. Although we have not yet identified the precise mechanisms that control ARHGAP42 transcription, our data are in excellent agreement with the known role of SRF in the regulation of SMC-specific gene expression (25). DHS2 transcriptional activity was responsive to myocardin overexpression, and knockdown of SRF clearly decreased the activity of the DHS2 regulatory element and of endogenous ARHGAP42 mRNA containing the minor T allele. Because most SMC marker genes are regulated by multiple SRF-binding CArG elements (31–33), it will be important to determine whether additional CArGs are critical for endogenous ARHGAP42 expression. Interestingly, we saw little effect of SRF knockdown on ARHGAP42 mRNA containing the major C allele. Although this may be explained by residual SRF expression in SRF knockdown cells, it may suggest that SRF is not absolutely required for expression of the major ARHGAP42 allele or that enhanced ARHGAP42 expression of the minor allele is more sensitive to SRF levels. The latter possibility would be consistent with the low binding affinity of this degenerate CArG element. The DHS2 fragment containing the major allele exhibits relatively high transcriptional activity suggesting that sequences outside of the SRF binding region are important, and we are currently attempting to identify the transcriptional mechanisms involved. It is also interesting to note that the DHS2 element had significant transcriptional activity on its own and when cloned upstream of a heterologous minimal promoter (data not shown), suggesting that its contributions to ARHGAP42 transcription are complex and that it does not function as an enhancer in the traditional sense.
We cannot rule out the possibility that other SNPs in the rs604723 LD block play a role in the control of ARHGAP42 expression and/or BP regulation. During the completion of the current studies, Kato et al. demonstrated that a minor allele variation 7.5 kb upstream from the ARHGAP42 TSS (rs2055450) was associated with decreased BP in European, East Asian, and South Asian populations as well as methylation of a DNA region within the ARHGAP42 first intron (6). According to the 1000 Genomes database and additional genotyping of our mixed-European-descent and African American patient populations (Supplemental Table 2), rs2055450 is in relatively high LD with rs604723, significantly extending the functional significance of the minor ARHGAP42 allele in the control of BP in diverse human populations. However, like the other SNPs within the ARHGAP42 BP locus, rs2055450 is not located in a region that exhibits sequence conservation or open/active chromatin signatures. Thus, when coupled with our extensive data on the role of the DHS2 and rs604723, it is likely that rs2055450 and the other variations in this LD block are “bystanders” in regard to the regulation of ARHGAP42 expression and BP. In support of this conclusion, the rs633185 SNP (the variation nearest to a potential regulatory element) had no effect on the transcriptional activity of the DHS1 fragment or protein binding to a 100-bp probe containing this sequence (Supplemental Figure 3).
Interestingly, the GTEx database indicates that minor ARHGAP42 allele LD block was significantly associated with increased expression of TMEM133, a hypothetical, uncharacterized RNA just downstream of ARHGAP42. However, as shown in Supplemental Figure 5, our RNA sequencing and targeted RT-PCR data indicate that TMEM133 is not transcribed as a separate gene, but is actually an extension of the ARHGAP42 3′-UTR. These data fully explain why variations that alter ARHGAP42 expression also affect TMEM133, and, by default, eliminate changes in TMEM133 expression as an explanation for the BP locus within ARHGAP42.
Our data add to a growing body of evidence that common noncoding variants alter cardiovascular risk by altering transcription factor binding and gene expression (34, 35), and support previous studies implicating RhoA signaling in the regulation of BP homeostasis in mice (14, 36, 37). Our human genetic data from well-characterized untreated patients confirmed that the minor ARHGAP42 allele was associated with decreased BP. Although the effect size may be relatively small, it has been observed in multiple diverse populations (4–6), indicating that ARHGAP42 genotype plays an important role. Although our data support the possibility that ARHGAP42 genotype contributes to the susceptibility of African Americans to the development of hypertension (1), the very low MAF in this population has also made it difficult to confirm this idea. Given the paucity of genetic mechanisms that explain this susceptibility, we are expanding our genotyping and BP analyses in this population. Our results also suggest that ARHGAP42 genotyping could also prove useful for individualizing antihypertensive therapies. For example, we predict that hypertensive patients homozygous for the major allele would respond better to antihypertensive agents that target SMC contractility (an endpoint directly controlled by RhoA) and that hypertensive patients homozygous for the minor allele may respond better to agents that target cardiac or kidney function. Finally, the regulation of ARHGAP42 expression and/or activity provides a novel therapeutic target for the development of antihypertensive therapies.
Methods
Cell culture.
Human aortic, coronary, and bronchial SMCs were purchased from Lonza and maintained in smooth muscle growth medium-2 (SmGM-2) supplemented with growth factors and 5% FBS. Primary aortic SMCs were isolated from 2-month-old Wistar rats as previously described (24). In brief, thoracic aortae were stripped of endothelial and adventitial layers by microdissection, and then SMCs were dispersed by treatment with trypsin and collagenase. Cells were maintained in DMEM supplemented with F12 and 10% FBS and used from passages 5–15. SMC preparations are routinely tested for smooth muscle differentiation marker gene expression, and only those that are deemed at least 85% pure by these measurements are used for further experimentation. Primary mouse ECs were immortalized by transformation with large T antigen (Gibco).
Generation and characterization of ArhGAP42gt/gt SM-MHCCreERT2 mice.
The Arhgap42 gene-trap mouse line was created as previously described (11). In brief, Arhgap42 gene-trap embryonic stem (ES) cells (SIGTR ES CE0477) were obtained from the Mutant Mouse Regional Resource Center (University of California, Davis). Chimeric mice were produced in house by blastocyst injection of Arhgap42+/gt ES cells using standard procedures, and an Arhgap42gt/gt line on a C57BL/6 background was generated by backcrossing for at least 8 generations. For ARHGAP42 rescue experiments, Arhgap42gt/gt mice were bred to a tamoxifen-inducible SM-MHCCreERT2 line provided by Stefan Offermanns (University of Heidelberg, Germany) on a pure C57BL/6 background. Cre activity in this model was controlled by treatment with tamoxifen (100 mg/kg) by i.p. injection for 5 consecutive days or oral gavage for 3 consecutive days. All experiments were performed in male mice 2–4 months old using age- and sex-matched littermate genetic controls. Genotyping was performed using DNA isolated from tail biopsies using either LacZ (5′-GCATCGAGCTGGGTAATAAGCGTTGGCAAT; 3′-GACACCAGACCAACTGGTAATGGTAGCGAC) or locus-specific primers (5′-TTCGTTGAGACAACTGCACACC; 3′-CCCTTCACACTTTGCTCTCTTAGC).
BP measurements.
Conscious BP measurements were made in mice aged 9–20 weeks by radiotelemetry using the PA-C10 telemeter (Data Sciences International) or tail cuff methods using the BP-100 probe (Iworx). For telemetry, after positioning of the telemetry catheter tip in the thoracic aorta (fed through the left carotid), the transmitter was inserted s.c. on the back/flank of mice. Mice were housed individually in a standard polypropylene cage placed on a radio receiver, maintained in a 12-hour light/dark cycle, and allowed 7 days of recovery/equilibration before BP measurements were begun. All BP measurements were recorded and stored using the Ponemah v6.10 system (Data Sciences International). Five-minute recordings were collected every 30 minutes, and data were analyzed using Excel software. Tail cuff measurements were performed as previously described (11). In brief, mice were fitted with the BP-100 probe and subjected to 20 BP measurements over 20 minutes every day (between 1400 and 1600 hours) for 7 consecutive days.
DNase hypersensitivity.
Approximately 80 million fresh HuAoSMCs were serum-starved for 24 hours and then washed twice with cold 1× PBS. Cells were pooled and collected in cold RSB buffer (10 mM Tris-Cl, pH 7.4, 10 mM NaCl, 3 mM MgCl2). Nuclei were isolated by lysing of cells in RSB buffer plus 0.1% NP-40 and centrifuging at 500 g for 10 minutes at 4°C. Supernatant was removed from the pellet. Pelleted nuclei were suspended in cold 1 DNase incubation buffer (NEB) and mixed. The nuclei suspension was treated with 1 U DNase I (Sigma-Aldrich; D5025) for 15 minutes at 37°C. After 15 minutes, 50 mM EDTA was added to stop the digestion reaction. DNase I–digested DNA was embedded in low-melt agarose plugs to minimize additional shearing, and then blunt-ended, extracted, and ligated to biotinylated linkers (Linker 1a: 5′-Bio-ACAGGTTCAGAGTTCTACAGTCCGAC-3′; Linker 1b: 5′-GTCGGACTGTAGAACTCTGAAC-Amm-3′). Biotinylated fragments were digested with MmeI and incubated with streptavidin-coated DynaI beads (Invitrogen) to purify digested DNA fragments. A second linker was ligated to the MmeI-digested DNA-linker-bead complex, and purified DNA fragments were PCR-amplified, sequenced by Illumina/Solexa (Linker 2a: 5′-P-TCGTATGCCGTCTTCTGCTTG-3′; Linker 2b: 5′-CAAGCAGAAGACGGCATACGANN-3′), and then aligned to the human genome using Bowtie.
For targeted, allele-specific DNase hypersensitivity assays, nuclei from heterozygous HuAoSMCs were treated with increasing amounts of DNase I from 0 to 1 μg. Reactions were subjected to PCR using primers specific to the major C or minor T alleles, and PCR band intensities were expressed relative to untreated samples set to 1.
Generation of luciferase constructs, DNA transfection, and reporter assays.
Regulatory regions of interest were PCR-amplified from human aortic smooth muscle genomic DNA and then cloned into the pGL3 basic vector (Promega). HuBrSMCs or ECs were seeded in 24-well culture plates the day prior to transfections at a density of approximately 2.5 × 104 cells per well. Cells were transfected with 50 ng DNA per well. In coexpression experiments, cells were transfected with 25 ng of luciferase reporter DNA and 25 ng of FLAG-tagged myocardin or empty expression construct per well. In all experiments, cells were transfected in triplicate and a promoterless pGL3 basic vector was transfected in parallel. After 48 hours of incubation at 37°C, luciferase assays were performed using the Steady-Glo Luciferase Kit (Promega) following the manufacturer’s instructions. Raw luciferase values were normalized to the activity of the promoterless pGL3 vector.
Electrophoretic mobility shift assays.
HuAoSMC nuclear lysates were prepared using the Nuclear Isolation kit from Thermo Fisher Scientific. Lysates were dialyzed in Dignam Buffer D (20 mM HEPES, pH 7.9, 20% [vol/vol] glycerol, 0.1 M KCl, 0.2 mM EDTA, 0.5 mM PMSF, and 0.5 mM DTT). FLAG-tagged SRF was translated in vitro using the TnT kit (Promega). Major and minor allele rs633185 and rs604723 gel shift probes were prepared by PCR amplifying a 100-bp fragment from the corresponding DHS1 or DHS2 luciferase reporter plasmids with the polymorphism at the center. Each binding reaction contained 10 μg lysate or 1 μl in vitro–translated SRF, 20,000 cpm of 32P-labeled oligonucleotide probe, and 0.20 μg dIdC in binding buffer (10 mmol/l Tris, pH 7.5, 50 mmol/l NaCl, 100 mmol/l KCl, 1 mmol/l DDT, 1 mmol/l EDTA, 5% glycerol).
ChIP experiments.
ChIP assays were performed according to X-ChIP protocol (Abcam) with slight modifications. In brief, HuAoSMCs or human coronary SMCs (HuCoSMCs) were fixed for 5 minutes in 0.7% formaldehyde. The cross-linking reaction was stopped by incubation of cells with 0.125 M glycine for 5 minutes. Cells were scraped in lysis buffer (5 mM PIPES, pH 8.0, 85 mM KCl, 0.5% NP-40) and then nuclei isolated by centrifugation at 2,300 g for 5 minutes. Nuclei lysis buffer (50 mM Tris-Cl, pH 8.1, 10 mM EDTA, 0.13% SDS) was added to pelleted nuclei. Chromatin was sheared into 500-bp fragments by sonication and immunoprecipitated with 1 μg of anti-SRF antibody (Santa Cruz Biotechnology; catalog sc-335) or normal rabbit IgG antibody (Cell Signaling Technology; catalog 2729) as a negative control for nonspecific binding.
SRF affinity precipitations.
Nuclear extracts from HuAoSMCs were prepared using NUPER nuclear extraction kit (Thermo Fisher Scientific) according to manufacturer protocol. For precipitations, 5′ biotinylated 20-bp double-stranded oligonucleotides (2.5 μM final concentration) were combined with 200 μg of nuclear extract in binding buffer (10 mM HEPES, pH 7.4, 8% glycerol, 1 mM MgCl2, 0.05% Triton X-100, 1.3 mM DTT, protease and phosphatase inhibitors) at room temperature with rotation. After 10 minutes, streptavidin agarose beads (CL-4B, Sigma-Aldrich, catalog 85881) were added, and reactions were incubated for another 30 minutes. After 2 washes (75 mM KCl, 5 mM HEPES, pH 7.4, 0.5 mM MgCl2, 4% glycerol, 0.05% Tween-20, 1 mM DTT), precipitates were eluted in 2× sample buffer and run in a 10% SDS-PAGE for Western blotting with an SRF antibody (Santa Cruz Biotechnology).
SRF knockdown.
The following siRNAs were obtained from Invitrogen: nontargeted control (NTC) siRNA (to GFP) 5′-GGUGCGCUCCUGGACGUAGCC-3′, SRF 5′-UAAUACUCAUGGCAAACAU[dT][dT]-3′ (sense) and 5′-AUGUUUGCCAUGAGUAUUA[dT][dT]-3′ (antisense). SRF siRNA was obtained as a sense/antisense mixture. HuBrSMCs were transfected with 50 nM SRF or NTC siRNA using Dharmafect siRNA transfection reagent (Dharmacon). For luciferase assays in SRF knockdown cells, siNTC or siSRF-treated HuBrSMCs were split 48 hours after knockdown and then seeded into 24-well culture plates. The following day, NTC and SRF knockdown cells were transfected with pGL3-promoterless, -DHS2C, or -DHS2T constructs. Luciferase experiments were performed 48 hours after transfection of pGL3 reporters. For allele-specific transcript measurements, HuAoSMCs were harvested 72 hours after knockdown.
Quantitative PCR.
RNA was isolated from cells or tissues using RNeasy Mini Kit (Qiagen). RNA was treated with DNase (Qiagen) to eliminate contaminating genomic DNA. RNA underwent first-strand cDNA synthesis using the iScript cDNA synthesis kit (Bio-Rad). Twenty nanograms cDNA was used in quantitative or semiquantitative PCR assays. Semiquantitative PCR primers used were: mouse Arhgap42 exons 1–4, 5′-CTGCCCACTCTGGAGTTCAGCG, 3′-GCTGCACCGATCTGTTCTTTTCG; mouse Arhgap42 exons 10–17, 5′-GAACCGATTTACACGTTACCCG, 3′-GGTTGGACCAAATATGACACCG; GAPDH, 5′-ATGGGTGTGAACCACGAGAA, 3′-GGCATGGACTGTGGTCATGA. Real-time PCR primers used were: ARHGAP42 exons 2–4, 5′-TTGGAGATGCAGAAACTGATGA, 3′-TTTGAATCAGTCTACGCCTTTCTTC; rat Arhgap42, 5′-TTCTGCATCTCCGATACAGTC, 3′-ATCAAAGAGCTGCTGAAGGATG; GAPDH, 5′-ATGGGTGTGAACCACGAGAA, 3′-GGCATGGACTGTGGTCATGA. Allele-specific PCR primers for rs604723 variation were 5′-TGTTGTTCCAAGGGTTCTT-3′ (T) and 5′-TGTTGTTCCAAGGGTTCTC-3′ (C).
CRISPR/Cas9–mediated gene editing.
Guide RNAs that flanked the 100-bp conserved DHS2 region (see below for sequences) were designed using sgRNA CRISPR design tool from the Zhang laboratory (38) and cloned into the sgRNA expression cassette of the pSpCas9(BB)-2A-Puro (PX459) plasmid (Addgene; catalog 62988). HuBrSMCs were transfected with either 15 μg of PX459 plasmid or 7.5 μg of PX459 plasmid containing each guide RNA. Forty-eight hours after transfection, cells were treated with puromycin (2 μg/ml) for 48 hours and then allowed to recover in normal growth media for an additional 24 hours. Genomic DNA and mRNA were isolated using the AllPrep DNA/RNA Kit (Qiagen). Deletion efficacy was tested by PCR using primers that flanked the deleted region. ARHGAP42 message was measured using semiquantitative PCR as described above. Guide RNAs used were as follows: gRNA1 sense, 5′-CACCGCTTGGAACAACATTAGACTG; gRNA1 antisense, 5′-AAACCAGTCTAATGTTGTTCC AAGC; gRNA2 sense, 5′-CACCGCAGCTGGACTGTGGGTCAGA; gRNA2 antisense, 5′-AAACTCTGACCCACAGTCCAGCTGC.
Cell and vessel stretch assays.
The Flexercell FX-4000 Tension apparatus (Flexcell International Corp.) uses regulated vacuum pressure to deform flexible-bottomed culture plates. Rat aortic SMCs were cultured on collagen I–coated BioFlex culture plates and were loaded into the FX-4000T cyclic tension device and subjected to 0 or 20% equibiaxial elongation at 1 Hz for 18 hours. Longitudinally cut rat portal vein strips were placed into ex vivo culture in DMEM/F12 with 2% dialyzed FCS and 10 nM insulin as previously described (30, 39). Strips were stretched for 3 days by attachment of a 600-mg stainless steel weight at 1 end of the vessel. Mouse portal vein rings were cultured as above and subjected to 300 mg of stretching force for 5 days. For LacZ staining, tissues were rinsed in PBS and fixed in 4% paraformaldehyde for 20 minutes at room temperature. After three 10-minute washes in PBS, tissues were incubated overnight at room temperature in X-gal staining solution (in PBS; 2 mM MgCl2, 5 mM ferrocyanide, 5 mM ferricyanide, 0.1% sodium deoxycholate, 0.2% NP-40, and 1 mg/ml X-gal). Tissues were processed for standard microscopy including H&E staining.
Hypertension models.
For DOCA-salt experiments, mice were implanted s.c. with a 50-mg 21-day slow-release DOCA pellet (Innovative Research of America) and fed 0.9% NaCl in drinking water for 3 weeks. Control mice were subjected to a sham operation and fed water. For l-NAME experiments, mice were treated with l-NAME in drinking water (450 mg/l) for 14 days. All groups were maintained on standard chow.
Human subjects.
In brief, participants were enrolled via a combination of passive (signs) and active (e-mail) recruitment. Potentially eligible participants included adults 30 years or older, with a recent office (clinic) BP measurement that was borderline high (120–149 mmHg systolic and/or 80–95 mmHg diastolic with neither value greater than 149/95 mmHg). Exclusion criteria included diabetes, pregnancy, dementia, any condition that would preclude wearing an ambulatory BP monitor, and persistent atrial fibrillation or other arrhythmia. Written informed consent took place during the first visit, during which time the patients were given ample opportunity to ask questions and to review the consent form in detail. In addition to carefully measured triplicate office BP measurements, participants underwent 24-hour ambulatory BP monitoring (ABPM) using the Oscar 2 oscillometric monitor (Suntech Medical). The monitors were programmed to measure BP at 30-minute intervals from 6 am to 10 pm and at 1-hour intervals from 10 pm to 6 am. The minimum number of readings we accepted as an adequate ABPM session was 14 for awake and 6 for sleep. ARHGAP42 genotype at rs604723 and rs2055450 was determined on patient blood samples as previously described (11).
Statistics.
All data represent at least 3 separate experiments presented as means ± SEM. Means were compared by 2-tailed Student’s t test or ANOVA (where indicated), and statistical significance was considered as a P value less than 0.05. All gels shown are representative of at least 3 individual experiments, and band intensities were quantified using ImageJ software (NIH).
Study approval.
The present studies in animals and/or humans were reviewed and approved by an appropriate IRB. All animal procedures were approved by the IACUC of the University of North Carolina at Chapel Hill. All animals were housed in facilities accredited by the Association for Assessment and Accreditation of Laboratory Care International. All clinical investigations were conducted according to Declaration of Helsinki principles and have been approved by the IRB at the University of North Carolina at Chapel Hill (IRB: UNC 10-0595). All human study procedures took place in a clinical research center, and all subjects provided informed consent prior to their participation in the study.
Author contributions
XB, KDM, and CPM designed research studies, conducted experiments, acquired data, analyzed data, and wrote the manuscript. AJV designed research studies, acquired blood pressure data, analyzed clinical data, and revised the manuscript. RAD and AOO obtained patient samples and analyzed patient samples. GAS, CRL, JCS, and CP established clinical cohorts, obtained patient samples, and analyzed patient samples. JMT designed research studies, analyzed data, and wrote the manuscript.
Supplementary Material
Acknowledgments
The authors thank Feng-Cheng Lin (University of North Carolina at Chapel Hill) for biostatistical support for these studies and Monte Willis (University of North Carolina at Chapel Hill) for aid in the use of the Flexercell tension apparatus. These studies were supported by NIH National Heart, Lung, and Blood Institute (NHLBI) R01 HL130367 (awarded to JMT, CPM, and AJV) and NIH-NHLBI R01 HL109607 (to CPM) as well as American Heart Fellowships to XB (14POST20380117), KDM (15PRE25340001), and RAD (15PRE25090045). Support was provided by an Institutional Development Grant awarded to JMT from the North Carolina Biotechnology Center (NCBC grant 2005-IDG-1008). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund (http://commonfund.nih.gov/GTEx/index) of the Office of the Director of the NIH. Additional funds were provided by the National Cancer Institute, National Human Genome Research Institute, NHLBI, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke. Donors were enrolled at Biospecimen Source Sites funded by NCI/SAIC-Frederick, Inc. (SAIC-F) subcontracts to the National Disease Research Interchange (10XS170), Roswell Park Cancer Institute (10XS171), and Science Care, Inc. (X10S172). The Laboratory, Data Analysis, and Coordinating Center (LDACC) was funded through a contract (HHSN268201000029C) to The Broad Institute, Inc. Biorepository operations were funded through an SAIC-F subcontract to the Van Andel Institute (10ST1035). Additional data repository and project management were provided by SAIC-F (HHSN261200800001E). Statistical Methods development grants were made to the University of Geneva (MH090941 and MH101814), the University of Chicago (MH090951, MH090937, MH101820, and MH101825), the University of North Carolina at Chapel Hill (MH090936 and MH101819), Harvard University (MH090948), Stanford University (MH101782), Washington University in St. Louis (MH101810), and the University of Pennsylvania (MH101822). The data used for the analyses described in this manuscript were obtained from the GTEx Portal (http://www.gtexportal.org) on 03/23/16.
Footnotes
Note regarding evaluation of this manuscript: Manuscripts authored by scientists associated with Duke University, The University of North Carolina at Chapel Hill, Duke-NUS, and the Sanford-Burnham Medical Research Institute are handled not by members of the editorial board but rather by the science editors, who consult with selected external editors and reviewers.
Conflict of interest: The authors have declared that no conflict of interest exists.
Reference information:J Clin Invest. 2017;127(2):670–680. https://doi.org/10.1172/JCI88899.
Contributor Information
Xue Bai, Email: baix@email.unc.edu.
George A. Stouffer, Email: rstouff@med.unc.edu.
Craig R. Lee, Email: clee1@email.unc.edu.
Akinyemi Oni-Orisan, Email: akind33@gmail.com.
Cam Patterson, Email: cpatters@nyp.org.
References
- 1.Writing Group Members. et al. Executive Summary: Heart Disease and Stroke Statistics — 2016 Update: A Report From the American Heart Association. Circulation. 2016;133(4):447–454. doi: 10.1161/CIR.0000000000000366. [DOI] [PubMed] [Google Scholar]
- 2.Padmanabhan S, Caulfield M, Dominiczak AF. Genetic and molecular aspects of hypertension. Circ Res. 2015;116(6):937–959. doi: 10.1161/CIRCRESAHA.116.303647. [DOI] [PubMed] [Google Scholar]
- 3.Franceschini N, Chasman DI, Cooper-DeHoff RM, Arnett DK. Genetics, ancestry, and hypertension: implications for targeted antihypertensive therapies. Curr Hypertens Rep. 2014;16(8):461. doi: 10.1007/s11906-014-0461-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wain LV, et al. Genome-wide association study identifies six new loci influencing pulse pressure and mean arterial pressure. Nat Genet. 2011;43(10):1005–1011. doi: 10.1038/ng.922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.International Consortium for Blood Pressure Genome-Wide Association Studies. et al. Genetic variants in novel pathways influence blood pressure and cardiovascular disease risk. Nature. 2011;478(7367):103–109. doi: 10.1038/nature10405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kato N, et al. Trans-ancestry genome-wide association study identifies 12 genetic loci influencing blood pressure and implicates a role for DNA methylation. Nat Genet. 2015;47(11):1282–1293. doi: 10.1038/ng.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Taylor JM, Hildebrand JD, Mack CP, Cox ME, Parsons JT. Characterization of graf, the GTPase-activating protein for rho associated with focal adhesion kinase. Phosphorylation and possible regulation by mitogen-activated protein kinase. J Biol Chem. 1998;273(14):8063–8070. doi: 10.1074/jbc.273.14.8063. [DOI] [PubMed] [Google Scholar]
- 8.Doherty JT, Lenhart KC, Cameron MV, Mack CP, Conlon FL, Taylor JM. Skeletal muscle differentiation and fusion are regulated by the BAR-containing Rho-GTPase-activating protein (Rho-GAP), GRAF1. J Biol Chem. 2011;286(29):25903–25921. doi: 10.1074/jbc.M111.243030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Taylor JM, Macklem MM, Parsons JT. Cytoskeletal changes induced by GRAF, the GTPase regulator associated with focal adhesion kinase, are mediated by Rho. J Cell Sci. 1999;112(pt 2):231–242. doi: 10.1242/jcs.112.2.231. [DOI] [PubMed] [Google Scholar]
- 10.Hildebrand JD, Taylor JM, Parsons JT. An SH3 domain-containing GTPase-activating protein for Rho and Cdc42 associates with focal adhesion kinase. Mol Cell Biol. 1996;16(6):3169–3178. doi: 10.1128/MCB.16.6.3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bai X, et al. The smooth muscle-selective RhoGAP GRAF3 is a critical regulator of vascular tone and hypertension. Nat Commun. 2013;4:2910. doi: 10.1038/ncomms3910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Somlyo AP, Somlyo AV. Signal transduction through the RhoA/Rho-kinase pathway in smooth muscle. J Muscle Res Cell Motil. 2004;25(8):613–615. doi: 10.1007/s10974-004-3146-1. [DOI] [PubMed] [Google Scholar]
- 13.GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348(6235):648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wirth A, et al. G12-G13-LARG-mediated signaling in vascular smooth muscle is required for salt-induced hypertension. Nat Med. 2008;14(1):64–68. doi: 10.1038/nm1666. [DOI] [PubMed] [Google Scholar]
- 15.Viera AJ, et al. Levels of office blood pressure and their operating characteristics for detecting masked hypertension based on ambulatory blood pressure monitoring. Am J Hypertens. 2015;28(1):42–49. doi: 10.1093/ajh/hpu099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Viera AJ, Hinderliter AL, Kshirsagar AV, Fine J, Dominik R. Reproducibility of masked hypertension in adults with untreated borderline office blood pressure: comparison of ambulatory and home monitoring. Am J Hypertens. 2010;23(11):1190–1197. doi: 10.1038/ajh.2010.158. [DOI] [PubMed] [Google Scholar]
- 17.Schisler JC, et al. Stable patterns of gene expression regulating carbohydrate metabolism determined by geographic ancestry. PLoS One. 2009;4(12):e8183. doi: 10.1371/journal.pone.0008183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Theken KN, et al. Evaluation of cytochrome P450-derived eicosanoids in humans with stable atherosclerotic cardiovascular disease. Atherosclerosis. 2012;222(2):530–536. doi: 10.1016/j.atherosclerosis.2012.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oni-Orisan A, et al. Cytochrome P450-derived epoxyeicosatrienoic acids and coronary artery disease in humans: a targeted metabolomics study. J Lipid Res. 2016;57(1):109–119. doi: 10.1194/jlr.M061697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halladay JR, et al. The Heart Healthy Lenoir project — an intervention to reduce disparities in hypertension control: study protocol. BMC Health Serv Res. 2013;13:441. doi: 10.1186/1472-6963-13-441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang M, Fang H, Zhou J, Herring BP. A novel role of Brg1 in the regulation of SRF/MRTFA-dependent smooth muscle-specific gene expression. J Biol Chem. 2007;282(35):25708–25716. doi: 10.1074/jbc.M701925200. [DOI] [PubMed] [Google Scholar]
- 22.Zhou J, et al. The SWI/SNF chromatin remodeling complex regulates myocardin-induced smooth muscle-specific gene expression. Arterioscler Thromb Vasc Biol. 2009;29(6):921–928. doi: 10.1161/ATVBAHA.109.187229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lockman K, Taylor JM, Mack CP. The histone demethylase, Jmjd1a, interacts with the myocardin factors to regulate SMC differentiation marker gene expression. Circ Res. 2007;101(12):e115–e123. doi: 10.1161/CIRCRESAHA.107.164178. [DOI] [PubMed] [Google Scholar]
- 24.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of SRF binding to CArG box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116(1):36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mack CP. Signaling mechanisms that regulate smooth muscle cell differentiation. Arterioscler Thromb Vasc Biol. 2011;31(7):1495–1505. doi: 10.1161/ATVBAHA.110.221135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinson JS, Medlin MD, Lockman K, Taylor JM, Mack CP. Smooth muscle cell-specific transcription is regulated by nuclear localization of the myocardin-related transcription factors. Am J Physiol Heart Circ Physiol. 2007;292(2):H1170–H1180. doi: 10.1152/ajpheart.00864.2006. [DOI] [PubMed] [Google Scholar]
- 27.Staus DP, Blaker AL, Taylor JM, Mack CP. Diaphanous 1 and 2 regulate smooth muscle cell differentiation by activating the myocardin-related transcription factors. Arterioscler Thromb Vasc Biol. 2007;27(3):478–486. doi: 10.1161/01.ATV.0000255559.77687.c1. [DOI] [PubMed] [Google Scholar]
- 28.Lagna G, Ku MM, Nguyen PH, Neuman NA, Davis BN, Hata A. Control of phenotypic plasticity of smooth muscle cells by bone morphogenetic protein signaling through the myocardin-related transcription factors. J Biol Chem. 2007;282(51):37244–37255. doi: 10.1074/jbc.M708137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lockman K, Hinson JS, Medlin MD, Morris D, Taylor JM, Mack CP. Sphingosine 1-phosphate stimulates smooth muscle cell differentiation and proliferation by activating separate serum response factor co-factors. J Biol Chem. 2004;279(41):42422–42430. doi: 10.1074/jbc.M405432200. [DOI] [PubMed] [Google Scholar]
- 30.Albinsson S, Nordström I, Hellstrand P. Stretch of the vascular wall induces smooth muscle differentiation by promoting actin polymerization. J Biol Chem. 2004;279(33):34849–34855. doi: 10.1074/jbc.M403370200. [DOI] [PubMed] [Google Scholar]
- 31.Mack CP, Owens GK. Regulation of smooth muscle α-actin expression in vivo is dependent on CArG elements within the 5′ and first intron promoter regions. Circ Res. 1999;84(7):852–861. doi: 10.1161/01.RES.84.7.852. [DOI] [PubMed] [Google Scholar]
- 32.Manabe I, Owens GK. CArG elements control smooth muscle subtype-specific expression of smooth muscle myosin in vivo. J Clin Invest. 2001;107(7):823–834. doi: 10.1172/JCI11385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li L, Liu Z, Mercer B, Overbeek P, Olson EN. Evidence for serum response factor-mediated regulatory networks governing SM22α transcription in smooth, skeletal, and cardiac muscle cells. Dev Biol. 1997;187(2):311–321. doi: 10.1006/dbio.1997.8621. [DOI] [PubMed] [Google Scholar]
- 34.Musunuru K, et al. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466(7307):714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Almontashiri NA, et al. 9p21.3 Coronary artery disease risk variants disrupt TEAD transcription factor-dependent transforming growth factor β regulation of p16 expression in human aortic smooth muscle cells. Circulation. 2015;132(21):1969–1978. doi: 10.1161/CIRCULATIONAHA.114.015023. [DOI] [PubMed] [Google Scholar]
- 36.Guilluy C, et al. The Rho exchange factor Arhgef1 mediates the effects of angiotensin II on vascular tone and blood pressure. Nat Med. 2010;16(2):183–190. doi: 10.1038/nm.2079. [DOI] [PubMed] [Google Scholar]
- 37.Pelham CJ, et al. Cullin-3 regulates vascular smooth muscle function and arterial blood pressure via PPARγ and RhoA/Rho-kinase. Cell Metab. 2012;16(4):462–472. doi: 10.1016/j.cmet.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8(11):2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeidan A, Nordström I, Dreja K, Malmqvist U, Hellstrand P. Stretch-dependent modulation of contractility and growth in smooth muscle of rat portal vein. Circ Res. 2000;87(3):228–234. doi: 10.1161/01.RES.87.3.228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.