Abstract
Perrault syndrome (PS) is a genetically heterogeneous disorder characterized by primary ovarian insufficiency (POI) in females and sensorineural hearing loss in males and females. In many PS subjects, causative variants have not been found in the five reported PS genes. The objective of this study was to identify the genetic cause of PS in an extended consanguineous family with six deaf individuals. Whole exome sequencing (WES) was completed on four affected members of a large family, and variants and co-segregation was confirmed by Sanger sequencing. All hearing impaired individuals, including the proband, are homozygous for a pathogenic variant of CLDN14, but this only explains the deafness. The PS proband is also homozygous for a frameshift variant (c.1453_1454delGA, p.Glu485Lysfs*5) in exon 7 of SGO2 encoding shugoshin 2, which is the likely cause of her concurrent ovarian insufficiency. In mouse, Sgol2a encoding shugoshin-like 2a is necessary during meiosis in both sexes to maintain the integrity of the cohesin complex that tethers sister chromatids. Human SGO2 has not previously been implicated in any disorder, but in this case of POI and perhaps others, it is a candidate for unexplained infertility.
Keywords: coincidental syndrome; cohesin; CLDN14; ovarian insufficiency; Perrault syndrome; SGO2, Shugoshin-2; Sgol2a
Graphical Abstract
INTRODUCTION
Perrault syndrome (PS) is a genetically heterogeneous disorder characterized clinically by sensorineural hearing loss (SNHL) in males and females and primary ovarian insufficiency (POI) (1). Individuals with PS may also manifest neurological features including peripheral neuropathy, ataxia, mild learning disability (2). Moreover, growth hormone deficiency and Marfanoid features have also been reported as part of the phenotypic spectrum of PS (3, 4). Four of the five genes (CLPP, MIM 614129; HARS2, MIM 614926; LARS2, MIM 615300; C10orf2, MIM 616138) encode proteins that function in mitochondrial proteostasis and when mutated cause Perrault syndrome. The first reported mutated gene associated with PS (HSD17B4, MIM 233400) encodes a peroxisomal protein (5). There are also several individuals with PS who have unresolved genetic etiologies that warrant a search for additional PS genes (6).
In this study we have identified additional genetic complexity in a PS proband of a large Pakistani family segregating deafness. The highly consanguineous pedigree includes three affected males and three affected females, only one of which has POI. We considered the possibility either that there is a single PS gene segregating with incomplete penetrance for POI or one recessive mutant gene is responsible for the hearing loss and another is independently responsible for the POI. Using whole exome sequence (WES) data we show that all of the hearing loss segregating in family PKDF063, including the proband, is attributable to a well-characterized pathogenic homozygous missense variant of CLDN14 (7–9). Homozygosity for a two nucleotide deletion in the protein coding exon 7 of SGO2, a gene hitherto not involved in any human disorder, independently is the likely cause of POI in the hearing impaired female proband.
PATIENTS AND METHODS
Family PKDF063 includes 153 individuals of which 39 individuals were consented for participation in our study. Hearing loss segregating in members of family PKDF063 was noted during their first year after birth by their mothers. Bilateral severe to profound deafness was confirmed by an audiologist. Although not an objective evaluation, parents said that their deaf children, including the PS proband, have normal intelligence and motor development.
Genomic DNA was extracted from peripheral blood leukocytes. One affected individual and the proband were initially screened by di-deoxy sequencing using BigDye (Applied Biosystems, Foster City, CA, USA) for pathogenic variants in CLPP, one of the five reported PS genes. We also screened for variants of GJB2 (DFNB1A, MIM 220290) and HGF (DFNB39, MIM 608265) since in Pakistan mutant alleles of these two genes are common causes of deafness. WES was performed with genomic DNA from five individuals (four affected; one unaffected) of family PKDF063 (Fig. 1) using a Nextera Rapid Capture Exome kit and a HiSeq1500 instrument (Illumina San Diego, CA, USA). Computational analyses used a GATK (Genome Analysis Toolkit) pipeline followed by variant calls that were annotated with Annovar v2014_07_14. Short-listed variants were verified by di-deoxy sequencing using an ABI3730XL genetic analyzer (Applied Biosystems).
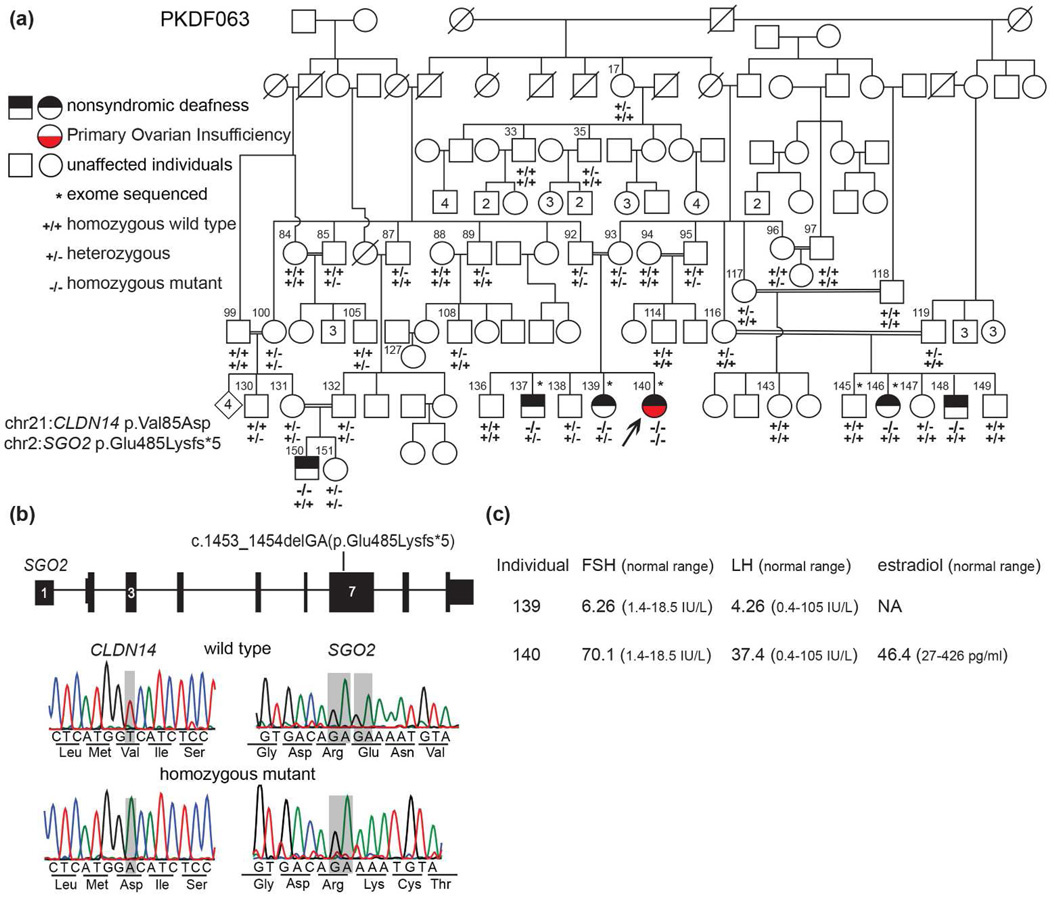
Fig. 1. Pedigree, clinical data and molecular genetics of family PKDF063.
(a) The proband with Perrault syndrome is indicated by an arrow. She has normal breast development (Tanner stage 5), secondary sexual hair and no evidence of cognitive impairment, sensory or motor peripheral neuropathy or Marfanoid features. Genotypes of CLDN14 and SGO2 for each of the ascertained 39 individuals are provided beneath their symbols. Individual 96 developed secondary amenorrhea of unknown etiology after the birth of her first daughter. Her serum levels were LH of 5.80 IU/L, FSH of 6.56 IU/L and estradiol of 770 pg/ml, indicative of hypogonadotropic hypogonadism suggesting a pituitary etiology acquired later in life. Squares and circles represent male and female individuals, respectively and numbers inside squares and circles are additional siblings. (b) Gene structure of SGO2 along with the location of a frameshift deletion in exon 7. Horizontal lines represent introns whereas thin and thick bars represent untranslated regions and exons, respectively. DNA chromatogram showing the missense variant of CLDN14 and a novel frameshift deletion of SGO2. The sites of the two variants are denoted by semitransparent gray rectangles. (c) Hormonal profiles of individuals 139 (22 yo) and 140 (23 yo) and the normal reference ranges. A raised FSH level is a key biochemical indicator of primary ovarian insufficiency while LH can remain within the normal range or be raised. Chr; Chromosome.
RESULTS
Clinical data
The proband (Fig. 1a, individual 140) is twenty-four years old and has a clinical diagnosis of PS (Fig. 1c) characterized by SNHL and POI. She was born to a consanguineous union and has two normal hearing and two hearing impaired siblings. Two additional branches of this pedigree include three additional deaf subjects (Fig. 1a, individuals 146, 148 and 150). The deaf proband presented at 24 years of age with a history of oligomenorrhoea. She had a delayed menarche at 17 years of age and reported irregular menstrual cycles during the succeeding few years followed by oligomenorrhoea and now menstruates once every 5–7 months. The endocrine profile of the proband revealed a low progesterone level (<0.2ng/ml; reference range of 0.2–1.4 ng/ml in the follicular stage, 4 to 25 ng/ml luteal phase, 0.1–1 post-menopausal), an estradiol level of 46.4pg/ml, follicle stimulating hormone (FSH) of 70.1mIU/ml that is in the reference range for post-menopausal women and a luteinizing hormone (LH) level of 37.4mIU/ml consistent with hypergonadotropic hypogonadism. The proband’s pelvic ultrasound shows normal appearing ovaries and a small uterus with thin endometrial lining measuring 5.7 cm × 2.3 cm × 3.0 cm (reference size 7.6 cm length × 4.5 cm width × 3 cm thick and an endometrial thickness of 2 mm to 16 mm depending on the stage of the menstrual cycle). The proband’s deaf sister (individual 139, 22 yo) has normal reproductive cycles, a hormone profile that falls within the normal reference range for her reproductive age, used a hearing aid since she was a three-year old and communicates verbally and in sign language.
Massively parallel and Sanger sequencing
Variants in two genes survived the initial filtering criteria (Table 1). The c.1664C>T variant in SON (rs13049658; p.(Thr555Met; NM_138927)) has an allele frequency of 14% (21 of 150 chromosomes) in Pakistani controls indicating that it is a benign polymorphism. However, the c.254T>A variant in CLDN14 (rs74315437; p.(Val85Asp)) is a known pathogenic allele (7, 8, 11) and co-segregates with hearing loss in family PKDF063. The six individuals with bilateral severe to profound hearing loss are homozygous for c.254T>A, whereas 33 hearing individuals in this family are either heterozygous for c.254T>A or homozygous for the reference allele (Fig. 1a). Homozygous or compound heterozygous pathogenic variants of human CLDN14 cause nonsyndromic deafness DFNB29 (MIM605608), and deaf females homozygous for c.254T>A are fertile (11). Moreover, claudin 14 null mice are fertile (9).
Table 1.
Filtering criteria to evaluate variant pathogenicity in WES data from family PKDF063
| Step | Variant filtering criteria |
|---|---|
| 1 | Depthof ≥ 20 unique reads with no strand bias and a minimum quality score > 20 for GATK to call the variant |
| 2 | Variant is homozygous or in compound heterozygosity in the four deaf PKDF063 individuals |
| 3 | An allele frequency < 0.005 in the ExAC database |
| 4 | Absence of variant in 94 ethnically matched control individuals |
| 5 | Missense variant predicted to be pathogenic by Polyphen-2, SIFT, Mutation Taster, FATHMM and Mutationassessor followed by manual inspection of conservation of wild type residue among vertebrates |
Next we pursued an explanation for the POI in the deaf proband (individual 140) diagnosed with PS by reanalyzing her WES data. There was no pathogenic variant in the five reported PS genes. However, using the aforementioned filtering criteria we found a frameshift variant that deserved further consideration. This variant (Fig. 1a) is a deletion in exon 7 of SGO2 at chr2: 201,436,522_523delGA (GRCh37/hg19), c.1453_1454delGA (NM_152524.5), p.Glu485Lysfs*5 (NP_689737.4) encoding shugoshin 2 (MIM 612425; previously symbol was SGOL2 encoding shugoshin-like 2). We then analyzed WES data for pathogenic variants of SGO2 in five individuals with a diagnosis of PS, but lacking a molecular genetic etiology. In addition, Sanger sequencing of SGO2 was performed in five individuals with POI of unknown etiology ascertained in the UK (6). In these ten individuals, no pathogenic variants in SGO2 were identified.
The proband in family PKD063 is homozygous for SGO2 p.(Glu485Lysfs*5). Her parents are carriers as are at least 14 others in this family (Fig. 1a). Variant c.1453_1454delGA is not present in 188 ethnically matched control chromosomes and is absent in the ExAC database of 121,412 chromosomes. Moreover, there are no homozygous loss of function variants of SGO2 reported in ExAC indicating that biallelic loss of function variants of SGO2 are likely to result in a clinically apparent phenotype (12).
DISCUSSION
We describe the first example of an individual with a clinical diagnosis of PS that can be ascribed to a combination of homozygous variants of two unlinked genes, CLDN14 and SGO2 encoding shugoshin 2. Cldn14 encodes a tight junction protein that is expressed abundantly in hair cells of the inner ear (https://shield.hms.harvard.edu/viewgene.html?gene=Cldn14) and is essential for the integrity of the cation-restrictive paracellular barrier of the reticular lamina in the organ of Corti (9). In the inner ear of postnatal Cldn14-null mice, the neurosensory outer and inner hair cells, which are responsible for sound transduction, rapidly degenerate resulting in profound deafness (9).
In mouse, Sgol1 encodes Shugoshin-like 1, which is necessary during mitosis for chromosome segregation. The absence of meiosis-specific shugoshin-like 2a in mouse encoded by Sgol2a (orthologue of human SGO2) causes infertility in males and females resulting from the premature loss of centromeric cohesion and an inability to bi-orient at the equatorial plate (13, 14). Consequently, gametes are produced with an abnormal number of chromosomes. In elderly human oocytes, a reduced level of shugoshin 2 was suggested to have a role in premature separation of dyads resulting in aneuploidy (15). Recently, frameshift variants of STAG3, also necessary for sister chromatid cohesion and synapsis, were associated with infertility in two consanguineous families (16, 17). Additionally, Stag3 null female and male mice are sterile, an observation that further strengthens the argument for the association of pathogenic variants of genes encoding the other members of the cohesin ring complex in POI and infertility (16).
Hearing loss and infertility are both genetically heterogeneous human disorders (18). In retrospect, given the large number of genes necessary for normal hearing and independently for fertility (19), it is not surprising that a person diagnosed clinically with Perrault syndrome would have biallelic pathogenic variants of two different genes, potentially assorting and acting independently. The proband of family PKDF063 (Fig. 1) is not masquerading as a PS subject. PS is her diagnosis and it is not an example of digenic inheritance where variants of two genes acting in concert explain a phenotype (20). The term “blended” has been suggested for the independent contributions to phenotype of two or more genes (21), but “blended” calls to mind a discredited nineteenth century notion of “blending inheritance”. In 1979, John M. Opitz suggested “coincidental syndrome” to describe the co-occurrence of more than one etiological distinct disorder in a patient. In the case described here, the proband manifests two genetically distinct disorders, deafness and POI, that co-occur by chance and yet are the clinical hallmarks of PS (22).
In summary, we observed that homozygosity for a rare protein-truncating DNA variant of SGO2 as the likely cause of POI and in combination with a reported pathogenic recessive variant of CLDN14 associated with SNHL coincidentally causes PS. Coincidence may explain undetermined genetic underpinning in other subjects with definitive audiological and hormonal findings typical of PS and the presumption of a single etiology. Finally, among several genes necessary for fertility (23), our data indicate that unexplained non-syndromic infertility or subfertility may also result from pathogenic variants of SGO2.
Acknowledgments
The authors thank the subjects in this study for their participation, Andrew J. Griffith and Leslie G. Biesecker for their critiques of our manuscript and Drs. Mehwish Pervaiz and Shazia Khalid Khan, Department of Gynecology, Jinnah Hospital Lahore, Pakistan for clinical evaluation of patients. This research was supported (in part) by the Intramural Research Program of the NIDCD/NIH to TBF (DC000048-19) and RJM (DC000086-01), the Higher Education Commission of Pakistan to SR, and an Action on Hearing Loss, UK grant to WGN. Written informed consents were obtained from all participants in this study. Institutional Review Board approval was obtained from the Combined Neurosciences Blue Panel at the National Institutes of Health (OH-93-N-016), from National Centre of Excellence in Molecular Biology at the University of the Punjab and from the National Health Service Research Ethics Committee (05/Q1404/49) and the University of Manchester. TBF takes responsibility for the integrity and accuracy of the data analyses. Research concept and study design: TBF, SR, RF, AUR. Analyses and interpretation of the data: RF, AUR, PLF, RJM, LD, SZ, AAK, DT, MZA, GB, SNK, WGN. RF, AUR, SR, TBF and WGN wrote the manuscript and all authors reviewed the manuscript. This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Footnotes
Conflict of interest
The authors declare no conflict of interests.
REFERENCES
- 1.Jenkinson EM, Clayton-Smith J, Mehta S, et al. Perrault syndrome: further evidence for genetic heterogeneity. J Neurol. 2012;259:974–976. doi: 10.1007/s00415-011-6285-5. [DOI] [PubMed] [Google Scholar]
- 2.Fiumara A, Sorge G, Toscano A, et al. Perrault syndrome: evidence for progressive nervous system involvement. Am J Med Genet A. 2004;128A:246–249. doi: 10.1002/ajmg.a.20616. [DOI] [PubMed] [Google Scholar]
- 3.Agrawala RK, Choudhury AK, Mohanty BK, et al. Perrault syndrome with growth hormone deficiency: a rare autosomal recessive disorder. J Pediatr Endocrinol Metab. 2015;28:1005–1007. doi: 10.1515/jpem-2014-0292. [DOI] [PubMed] [Google Scholar]
- 4.Jacob JJ, Paul TV, Mathews SS, et al. Perrault syndrome with Marfanoid habitus in two siblings. J Pediatr Adolesc Gynecol. 2007;20:305–308. doi: 10.1016/j.jpag.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 5.Konkolova J, Petrovic R, Chandoga J, et al. Peroxisomal D-bifunctional protein deficiency: First case reports from Slovakia. Gene. 2015;568:61–68. doi: 10.1016/j.gene.2015.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Demain LA, Urquhart JE, O'Sullivan J, et al. Expanding the Genotypic Spectrum of Perrault syndrome. Clin Genet. 2016 doi: 10.1111/cge.12776. [DOI] [PubMed] [Google Scholar]
- 7.Wilcox ER, Burton QL, Naz S, et al. Mutations in the gene encoding tight junction claudin-14 cause autosomal recessive deafness DFNB29. Cell. 2001;104:165–172. doi: 10.1016/s0092-8674(01)00200-8. [DOI] [PubMed] [Google Scholar]
- 8.Lee K, Ansar M, Andrade PB, et al. Novel CLDN14 mutations in Pakistani families with autosomal recessive non-syndromic hearing loss. Am J Med Genet A. 2012;158a:315–321. doi: 10.1002/ajmg.a.34407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Yosef T, Belyantseva IA, Saunders TL, et al. Claudin 14 knockout mice, a model for autosomal recessive deafness DFNB29, are deaf due to cochlear hair cell degeneration. Hum Mol Genet. 2003;12:2049–2061. doi: 10.1093/hmg/ddg210. [DOI] [PubMed] [Google Scholar]
- 10.Griffith AJ, Friedman TB. Hereditary Hearing Loss. In: Wackym PA, Snow JB Jr, editors. Ballenger’s Otorhinolaryngology Head and Neck Surgery. 18th. Chapter 26. People’s Medical Publishing House – USA; 2016. pp. 329–345. [Google Scholar]
- 11.Bashir ZE, Latief N, Belyantseva IA, et al. Phenotypic variability of CLDN14 mutations causing DFNB29 hearing loss in the Pakistani population. J Hum Genet. 2013;58:102–108. doi: 10.1038/jhg.2012.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Narasimhan VM, Hunt KA, Mason D, et al. Health and population effects of rare gene knockouts in adult humans with related parents. Science. 2016;352(6284):474–477. doi: 10.1126/science.aac8624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salic A, Waters JC, Mitchison TJ. Vertebrate shugoshin links sister centromere cohesion and kinetochore microtubule stability in mitosis. Cell. 2004;118:567–578. doi: 10.1016/j.cell.2004.08.016. [DOI] [PubMed] [Google Scholar]
- 14.Llano E, Gomez R, Gutierrez-Caballero C, et al. Shugoshin-2 is essential for the completion of meiosis but not for mitotic cell division in mice. Genes Dev. 2008;22:2400–2413. doi: 10.1101/gad.475308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yun Y, Lane SI, Jones KT. Premature dyad separation in meiosis II is the major segregation error with maternal age in mouse oocytes. Development. 2014;141:199–208. doi: 10.1242/dev.100206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Le Quesne Stabej P, Williams HJ, James C, et al. STAG3 truncating variant as the cause of primary ovarian insufficiency. Eur J Hum Genet. 2016;24:135–138. doi: 10.1038/ejhg.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caburet S, Arboleda VA, Llano E, et al. Mutant cohesin in premature ovarian failure. The New England Journal of Medicine. 2014;370:943–949. doi: 10.1056/NEJMoa1309635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Newman WG, Friedman TB, Conway GS. Perrault Syndrome. In: Pagon RA, Adam MP, Ardinger HH, et al., editors. GeneReviews(R) Seattle (WA): 2014. [Google Scholar]
- 19.Venkatesh T, Suresh PS, Tsutsumi R. New insights into the genetic basis of infertility. Appl Clin Genet. 2014;7:235–243. doi: 10.2147/TACG.S40809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schaffer AA. Digenic inheritance in medical genetics. J Med Genet. 2013;50:641–652. doi: 10.1136/jmedgenet-2013-101713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yang Y, Muzny DM, Reid JG, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369:1502–1511. doi: 10.1056/NEJMoa1306555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Opitz JM, Herrmann J, Pettersen JC, et al. Terminological, diagnostic, nosological, and anatomical developmental aspects of developmental defects in man. Adv Hum Genet. 1979;9:71–164. doi: 10.1007/978-1-4615-8276-2_2. [DOI] [PubMed] [Google Scholar]
- 23.Kosova G, Scott NM, Niederberger C, et al. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genet. 2012;90:950–961. doi: 10.1016/j.ajhg.2012.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]