Much work over the last three decades has established that chemical modifications of DNA and histones are intimately linked with various facets of DNA metabolism, most notably transcription. In addition to chromatin components, RNA has long been known to be chemically modified [1]. The N6-methyladenosine (m6A) RNA modification has recently emerged as a widely prevalent RNA mark [2]. First discovered in the 1970s, m6A is found in many eukaryotes, as well as in some viruses, and characterizes many RNA species, including mRNAs, tRNAs, rRNAs, snRNAs, and lncRNAs [1, 2]. Although broadly distributed, assigning a functional role to m6A has remained elusive. To this end, a recent study by Deepak Patil, Samie Jaffrey and colleagues has intriguingly found that the mammalian RNA with the most mapped m6A nucleotides is the X-inactive specific transcript (Xist) lncRNA [3]. Xist RNA is essential for X-chromosome inactivation, which equalizes X-linked gene expression between male and female mammals by silencing most genes on one of the two X-chromosomes in females [4].
m6A enrichment within Xist RNA offered the authors an opportunity to dissect the functional importance of m6A. Xist RNA ‘coats’ the X-chromosome from which it is expressed and enables silencing of genes on that X-chromosome [5, 6]. In recent years, advances in technology have revealed the diverse mechanisms by which Xist RNA silences transcription of X-linked genes. Xist RNA recruits silencing protein complexes, modifies the 3D structure of the X-chromosome, and repositions the X-chromosome to the nuclear periphery [7]. Although undoubtedly illuminating, these studies left open the question of precisely how Xist RNA executes its many functions.
To elucidate m6A function, Patil et al. expanded upon the discovery, from the labs of Mitch Guttman and Jeannie Lee, that Xist RNA binds the RNA-Binding Motif protein 15 (RBM15). The Guttman and Lee labs separately pioneered affinity purification protocols to capture proteins that bind RNAs and identified RBM15 as a high-confidence Xist-interacting protein [8, 9]. RBM15 is a member of the SPEN family of proteins that recognize RNA molecules based on the structure of the RNA [10]. Patil et al. found that knock-down of RBM15 and its homolog RBM15B reduced the silencing of genes on the Xist RNA-decorated X-chromosome. The authors identified the exact binding sites within Xist RNA through the iCLIP (individual-nucleotide resolution UV crosslinking and immunoprecipitation) technique. Intriguingly, RBM15/15B iCLIP clusters circumscribed the m6A sites in Xist RNA.
Patil et al. next attempted to connect RBM15 to m6A catalysis by interrogating other proteins associated with the m6A RNA modification. They honed in on WTAP (Wilm’s Tumor Associated Protein), which binds the A-repeat region within the 5′end of Xist RNA[11]. This region is known to be enriched for the m6A modification [12]. In addition to Xist RNA, WTAP was previously found to interact with RBM15/15B and, importantly, with METTL3, an m6A catalyst [13–16]. Indeed, the authors showed that WTAP can recruit METTL3 to catalyze m6A within Xist RNA. siRNA-based knock-down of WTAP or METTL3 reduced m6A deposition in Xist RNA as well as X-linked gene silencing by Xist RNA.
These results in turn raised the question of how the m6A modification can facilitate gene silencing. The m6A moiety was previously found to be ‘read’ by the YTH family of proteins [17]. Amongst the YTH family, the DC1 protein became of interest due to its localization predominantly in the nucleus. Patil and colleagues therefore tested DC1 binding within Xist RNA and found that it was enriched on m6A-marked residues. siRNA knock-down of DC1 showed that it is necessary for X-linked gene silencing upon Xist RNA coating. To determine if DC1 recruitment is sufficient for X-linked gene silencing, the authors tethered DC1 to Xist RNA through the viral BoxB-λN RNA-protein binding system [18]. This experiment rescued Xist RNA-induced gene silencing, even upon siRNA reduction of METTL3. In these experiments, measuring the degree of m6A reduction upon METTL3 knock-down would have further bolstered the role of METTL3-catalyzed m6A in X-inactivation.
Although sufficient to mediate the m6A-independent Xist RNA gene silencing, DC1 does not have intrinsic silencing function. The authors postulated that DC1 must act via secondary effectors. DC1 is known to bind with SHARP, LBR, HNRNPU, and HNRNPK proteins and to interact with components of the Polycomb repressive complexes 1 and 2 – all of which either directly or indirectly associate with Xist RNA and effect gene silencing [8, 9, 12, 19]. Additional work will be required to connect DC1 directly to these factors.
In sum, the study by Patil et al. demonstrates the functional importance of the m6A through investigations of the modification in Xist RNA. In Xist RNA, m6A likely acts as a scaffold for the assembly of silencing protein complexes. These findings will undoubtedly spur the discovery of similar and additional roles for m6A in other RNA species.
Figure 1.
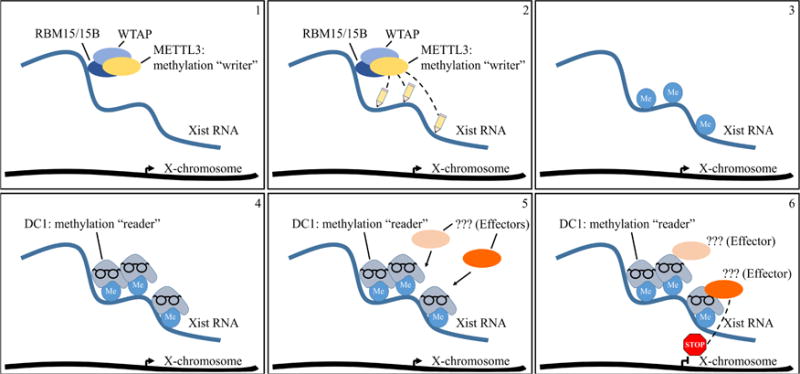
Role of m6A RNA methylation in X-chromosome inactivation.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Liu N, Pan T. N6-methyladenosine-encoded epitranscriptomics. Nat Struct Mol Biol. 2016;23:98–102. doi: 10.1038/nsmb.3162. [DOI] [PubMed] [Google Scholar]
- 2.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Patil DP, et al. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:369–373. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maclary E, et al. Long nonoding RNAs in the X-inactivation center. Chromosome Res. 2013;21:601–614. doi: 10.1007/s10577-013-9396-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clemson CM, et al. XIST RNA paints the inactive X chromosome at interphase: evidence for a novel RNA involved in nuclear/chromosome structure. J Cell Biol. 1996;132:259–275. doi: 10.1083/jcb.132.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brown CJ, et al. The human XIST gene: analysis of a 17 kb inactive X-specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell. 1992;71:527–542. doi: 10.1016/0092-8674(92)90520-m. [DOI] [PubMed] [Google Scholar]
- 7.Engreitz JM, et al. Long non-coding RNAs: spatial amplifiers that control nuclear structure and gene expression. Nat Rev Mol Cell Biol. 2016;17:756–770. doi: 10.1038/nrm.2016.126. [DOI] [PubMed] [Google Scholar]
- 8.McHugh CA, et al. The Xist lncRNA interacts directly with SHARP to silence transcription through HDAC3. Nature. 2015 doi: 10.1038/nature14443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Minajigi A, et al. Chromosomes. A comprehensive Xist interactome reveals cohesin repulsion and an RNA-directed chromosome conformation. Science. 2015;349 doi: 10.1126/science.aab2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ariyoshi M, Schwabe JW. A conserved structural motif reveals the essential transcriptional repression function of Spen proteins and their role in developmental signaling. Genes Dev. 2003;17:1909–1920. doi: 10.1101/gad.266203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Moindrot B, et al. A Pooled shRNA Screen Identifies Rbm15, Spen, and Wtap as Factors Required for Xist RNA-Mediated Silencing. Cell reports. 2015;12:562–572. doi: 10.1016/j.celrep.2015.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chu C, et al. Systematic discovery of xist RNA binding proteins. Cell. 2015;161:404–416. doi: 10.1016/j.cell.2015.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horiuchi K, et al. Identification of Wilms’ tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J Biol Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhong S, et al. MTA is an Arabidopsis messenger RNA adenosine methylase and interacts with a homolog of a sex-specific splicing factor. The Plant cell. 2008;20:1278–1288. doi: 10.1105/tpc.108.058883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Agarwala SD, et al. RNA methylation by the MIS complex regulates a cell fate decision in yeast. PLoS Genet. 2012;8:e1002732. doi: 10.1371/journal.pgen.1002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ping XL, et al. Mammalian WTAP is a regulatory subunit of the RNA N6-methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C, et al. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat Chem Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 18.Baron-Benhamou J, et al. Using the lambdaN peptide to tether proteins to RNAs. Methods Mol Biol. 2004;257:135–154. doi: 10.1385/1-59259-750-5:135. [DOI] [PubMed] [Google Scholar]
- 19.Hasegawa Y, et al. The matrix protein hnRNP U is required for chromosomal localization of Xist RNA. Dev Cell. 2010;19:469–476. doi: 10.1016/j.devcel.2010.08.006. [DOI] [PubMed] [Google Scholar]


