Abstract
The role of mesenchymal stem cells (MSC) in osteosarcoma (OS), the most common primary tumor of bone, has not been extensively elucidated. We have recently shown that OS is characterized by interstitial acidosis, a microenvironmental condition that is similar to a wound setting, in which mesenchymal reactive cells are activated to release mitogenic and chemotactic factors. We therefore intended to test the hypothesis that, in OS, acid-activated MSC influence tumor cell behavior. Conditioned media or co-culture with normal MSC previously incubated with short-term acidosis (pH 6.8 for 10 hours, H+-MSC) enhanced OS clonogenicity and invasion. This effect was mediated by NF-κB pathway activation. In fact, deep-sequencing analysis, confirmed by Real-Time PCR and ELISA, demonstrated that H+-MSC differentially induced a tissue remodeling phenotype with increased expression of RelA, RelB, and NF-κB1, and downstream, of CSF2/GM-CSF, CSF3/G-CSF, and BMP2 colony-promoting factors, and of chemokines (CCL5, CXCL5, and CXCL1), and cytokines (IL6 and IL8), with an increased expression of CXCR4. An increased expression of IL6 and IL8 were found only in normal stromal cells, but not in OS cells, and this was confirmed in tumor-associated stromal cells isolated from OS tissue. Finally, H+-MSC conditioned medium differentially promoted OS stemness (sarcosphere number, stem-associated gene expression), and chemoresistance also via IL6 secretion. Our data support the hypothesis that the acidic OS microenvironment is a key factor for MSC activation, in turn promoting the secretion of paracrine factors that influence tumor behavior, a mechanism that holds the potential for future therapeutic interventions aimed to target OS.
Keywords: tumor microenvironment, mesenchymal stroma, inflammation, cancer stemness
Introduction
Tumors are complex structures in which cancer cells are intermingled with different reactive cell types, the so-called tumor stroma1 interactions between cancer cells and tumor stroma and that involve soluble mediators, intercellular contacts, and changes in physical, chemical, and biochemical parameters, such as pH, oxygen levels, and substrates such as glucose and amino acids. These interactions are drawing an increasing interest as it is now recognized that they play a key role in tumor growth, progression, invasiveness, immune escape, and drug resistance.2, 3 The importance of tumor stroma has also been recently suggested in osteosarcoma (OS)4–6 ; the most common primary malignant bone tumor. OS treatment includes surgical removal of the primary plus multiagent chemotherapy to prevent or inhibit the development of metastases. Resistance to cytotoxic agents occurs frequently and negatively affects the outcome of OS,7 and targeting molecular pathways has not produced significant improvements. Indeed, the prognosis for OS has not significantly changed in 40 years.8 despite numerous promising therapeutic avenues in vitro. The discrepancy between successful preclinical results and actual clinical outcomes may be explained by the difference between the optimal in vitro conditions (homotypic cultures of cancer cells under standard environmental settings), and the complexity of tumors, including the interactions among different cell types under different environmental conditions that, in tumor microenvironment (TME) of OS, give rise to a complex, still largely unpredictable behavior.
In sarcomas, endothelial and immune cells are commonly considered the major components of the tumor supporting stroma.9 However, as for other solid cancers, stromal cells of mesenchymal origin, like tumor-associated fibroblasts (TAF) and MSC, are also recruited at the site of OS growth.10 In a mesenchymal cancer like OS, mesenchymal reactive stromal cells have been largely neglected and poorly investigated. When isolated from tissue samples, they are not easily distinguished from tumor cells, both sharing the same immunophenotypic features. However, OS arises and develops within the bone marrow, that is very rich in MSC, and it has been observed that OS are able to actively recruit circulating MSC.11–13
The role of reactive stroma on tumor progression is debatable.11 Two populations of stromal mesenchymal cells appear to co-exist in the TME of OS: a naïve MSC component from normal tissues (naïve-MSC) and, in addition, tumor tissue-derived MSC (tumor tissue educated MSC, T-MSC). Naive-MSC have been described as both promoting and inhibiting tumor progression, while T-MSC is thought to promote tumor progression as a result of their reprogramming by the tumor.11 The existence of naïve MSC in OS is supported by the high number of circulating MSC.12 When injected in OS xenografts, MSC migrate toward the tumor site,13 as in general they migrate toward sites of inflammation.14 Indeed, OS, like other malignancies, acts as a” wound that does not heal”.15, 16 At the tumor site, MSCs recognize exogenous and endogenous inflammatory signals, activate, and acquire a myofibroblast-like phenotype. In the TME, extracellular acidosis is a strong exogenous signal emanating from the growing tumors, that can induce reprogramming of naïve MSC into T-MSC. Indeed, we have previously demonstrated that MSC cooperate with OS cells by fueling their metabolism,6 and that sarcoma cells strongly acidify TME.17, 18 Cancer-associated extracellular acidosis is a general feature of cancer that mainly derives from the metabolic switch to persistent aerobic glycolysis even under adequate oxygen conditions, the so-called “Warburg effect”.19 Acidosis plays a critical role in the progression of many cancers as it is able to foster chemo/radioresistance, neo-angiogenesis, invasion, and stemness.20
To date, the exact mechanisms of the interaction between OS and mesenchymal stroma remain unclear, especially under an acidic TME. In this study, we demonstrate that OS cells reprogram naïve MSC to T-MSC through the acidification of the extracellular environment, thereby prompting T-MSC to release a plethora of growth factors, cytokines, and chemokines that eventually support OS proliferation, migration, and stemness.
Materials and Methods
Reagents
Dulbecco's Modified Eagle Medium (DMEM); penicillin, streptomycin (Thermo Fisher Scientific); fetal bovine serum (Euroclone); Iscove's Modified Dulbecco's Medium (IMDM) (Life Technologies); TruSeq RNA Sample Prep Kit v2 (Illumina); NucleoSpin RNA II (Macherey-Nagel); MuLV Reverse Transcriptase (Applied Biosystems); Flowcytomix Multiple Analyte Detection Kit (eBioscience); Human Inflammatory Cytokines Multi-Analyte ELISArray™ Kit (Qiagen); Human IL6 DuoSet and the Human CXCL8/IL8 Quantikine ELISA Kits (R&D Systems Inc); MMP11 ELISA kit (Emelca Bioscience); ELISA-based TransAM NFkB Family kit (Active Motif); transwell for migration assay (Costar, Corning); EGF, bFGF, IL6 and IL8 (PeproTech); anti-human Ki67 monoclonal antibody (MAb), diaminobenzidine (DAB), (Dako); anti-human vimentin (clone V9, Santa Cruz); anti-human alpha smooth muscle actin, α-SMA (clone 1A4, R&D System), anti-human Ki67 (clone MIB-1, Dako); anti-human LAMP2 (#HPA029100) and anti-human NF-κB1 (#HPA027305, Sigma-Aldrich); anti-human NF-κB p65 RelA (#8242, Cell Signalling); Tocilizumab (Roche); Matrigel (BD Biosciences); all other reagents were obtained from Sigma-Aldrich.
Immunohistochemistry
Canine OS paraffin embedded samples were obtained from the Department of Animal Pathology of the University of Turin at the time of the surgical treatment (amputation) before adjuvant chemotherapy. Representative 5 µm thick sections were mounted on a glass slide covered with 2% silane solution in acetone. After dewaxing in xylene and rehydration in ethanol for antigen retrieval, the slides were heated in a microwave oven in 0.02 M citrate buffer, pH 6.0. After cooling, the sections were incubated in 3% perhydrol solution to block the endogenous peroxidase reaction. Non-specific binding was blocked by incubation in 5% bovine serum albumin. The following primary antibodies were used: MAb anti-human vimentin, anti-human alpha smooth muscle actin, α-SMA, anti-human Ki67, anti-human LAMP2, anti-human NF-κB1, and NF-κB p65 RelA. Sections were washed, incubated with a biotinylated secondary antibody, covered with DAB and counterstained with Mayer's hematoxylin. Negative controls were also performed by omitting the primary antibody.
Cell cultures
Human skin fibroblasts (FB, 2 lots) were provided by the Japanese Collection of Research Bioresources (Osaka, JP) and cultured in DMEM plus 50 units/mL penicillin, 50 mg/mL streptomycin, and 10% fetal bovine serum (FBS). Human MSC from bone marrow (BM-MSC) and adipose tissue (AT-MSC) were purchased from Lonza (Euroclone) and from the American Type Cell Culture Collection (ATCC) respectively, and cultured in Minimum Essential Medium Eagle Alpha Modified (Alpha-MEM) plus 100 U/mL penicillin, 100 mg/mL streptomycin, 10% FBS and buffered at pH 7.4 (complete) or 0.1% FCS (low-serum). MSC were always used before the 5–6 passage in culture. The human OS cell lines MG-63, Saos-2, and HOS were purchased from ATCC and cultured in IMDM plus 100 units/mL penicillin, 100 mg/mL streptomycin, and 10% FBS (complete). Cells were maintained at 37°C, 5% CO2 in humidified atmosphere. Culture medium at specific pH was obtained by adjusting the concentration of sodium bicarbonate according to the Henderson-Hasselbach equation, as previously described.17 When not differently specified, acidic medium was buffered at pH 6.8, and physiological medium was buffered at pH 7.4. At the endpoint of each experiment, the final pH of the supernatant was always measured to ascertain the maintenance of the pH value along the incubation time.
To collect the conditioned medium (CM) of MSC pretreated with short-term acidosis (CM MSCpH 6.8), MSC were seeded (3×105 cells) in complete medium incubated, after adhesion, for 20 hrs in low-serum medium, washed with PBS, and incubated with low-serum medium at pH 6.8 or at pH 7.4 for 10 hrs. At the end of the acidic stress, the medium was replaced with low-serum medium at pH 7.4 for additional 48 hrs for all the conditions. This pH neutral supernatant, that we named as H+-MSC CM, was finally collected, centrifuged, and stored at −80°C until use.
T-MSC were isolated from an OS sample after signed informed consent regarding the use for research studies was obtained, after the institutional ethical committee approval (n. 20204 of 31.07.09), and according to the method described by previous authors.21 The patient was enrolled in the study on 2009, and only cells at 4th, 5th passages from the first seeding were used. Briefly, tissue specimens were minced and mechanically dissociated. The suspension was filtered using a 100-µm cell strainer, and then cells were seeded in standard MSC medium (complete Alpha-MEM) and cultured at 37°C, 5% CO2.
Q-RT-PCR
Total RNA was extracted by using the NucleoSpin RNA II and reverse transcribed with MuLV Reverse Transcriptase. Quantitative Reverse Transcription Polymerization Chain Reaction (Q-RT-PCR) was performed by amplifying 1 µg of cDNA using the Light Cycler instrument and the Universal Probe Library system (Roche Applied Science). Probes and primers were selected using a web-based assay design software (ProbeFinder, http://www.roche-applied-science.com). Results were normalized to 18S rRNA or β-actin, according to the 2−ΔΔCT method, as previously described.22 All primer sequences and probes are listed in Table 1. Each assay was repeated at least three times.
Table 1.
Probes and primers
| Gene | Full name | Accession Number |
Primers | Probe |
|---|---|---|---|---|
| 18S rRNA |
18S ribosomal RNA | X03205.1 | F = gcaattattccccatgaacg R = gggacttaatcaacgcaagc |
48 |
| ACTB | Actin, beta | NM_001101.2 | F = ccaccgcgagaagatga R = ccagaggcgtacagggatag |
64 |
| 18S rRNA |
18S ribosomal RNA | X03205.1 | F = gcaattattccccatgaacg R = gggacttaatcaacgcaagc |
48 |
| ACTB | Actin, beta | NM_001101.2 | F = ccaccgcgagaagatga R = ccagaggcgtacagggatag |
64 |
| NFKB1 | Nuclear factor kappa B subunit 1 |
NM_003998.3 | F = cctggaaccacgcctcta R = ggctcatatggtttcccattta |
49 |
| RelA | RELA proto-oncogene, NF-kB subunit |
NM_021975.3 | F = actgtgtgacaaggtgcagaa R = cacttgtcggtgcacatca |
64 |
| RelB | RELB proto-oncogene, NF-kB subunit |
NM_006509.3 | F = gattgtcgagcccgtgac R = ccacgccgtagctgtcat |
4 |
| CSF2 | Colony stimulating factor 2 |
NM_000758.2 | F = tctcagaaatgtttgacctcca R = gcccttgagcttggtgag |
1 |
| CSF3 | Colony stimulating factor 3 |
M17706.1 | F = gagcaagtgaggaagatccag R = cagcttgtaggtggcacaca |
1 |
| BMP2 | Bone morphogenetic protein 2 |
NM_001200.2 | F = cggactgcggtctcctaa R = ggaagcagcaacgctagaag |
49 |
| CCL5 | Chemokine (C-C motif) ligand 5 |
NM_002985.2 | F = tgcccacatcaaggagtattt R = ctttcgggtgacaaagacg |
59 |
| CXCL5 | Chemokine (C-X-C motif) ligand 5 |
NM_002994.3 | F = ggtccttcgagctccttgt R = gcagctctctcaacacagca |
71 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 |
NM_001511.2 | F = tcctgcatcccccatagtta R = cttcaggaacagccaccagt |
52 |
| CXCR4 | Chemokine (C-X-C motif) receptor 4 |
NM_001008540 .1 |
F = ggtggtctatgttggcgtct R = actgacgttggcaaagatga |
18 |
| MMP11 | Matrix metallopeptidase 11 |
NM_005940.3 | F = tgctgacatcatgatcgactt R = agtggacatccccttctcg |
60 |
| IL6 | Interleukin 6 | NM_000600.3 | F = gatgagtacaaaagtcctgatcca R = ctgcagccactggttctgt |
40 |
| IL8/CXC L8 |
C-X-C motif chemokine ligand 8 |
NM_000584.3 | F = gagcactccataaggcacaaa R = atggttccttccggtggt |
72 |
| OCT4/P OU5F1 |
POU class 5 homeobox 1 |
NM_002701.4 | F = cttcgcaagccctcatttc R = gagaaggcgaaatccgaag |
60 |
| NANOG | Nanog homeobox | NM_024865.2 | F = atgcctcacacggagactgt R = agggctgtcctgaataagca |
69 |
Protein analysis
3 MSC and 2 FB lots were seeded at 3×104 cells/well into 24-well plates. After adhesion, cells were incubated with low-serum medium at different pH. After 24 hrs, culture supernatants were collected, centrifuged and assayed for the analysis of inflammatory cytokines. For FB and MG-63 OS cells we used the Human Th1/Th2 cytokine 11plex Flowcytomix Multiple Analyte Detection Kit, and protein concentration was normalized to the number of cells counted by using erythrosin B dye vital staining. For MSC, we used the Human Inflammatory Cytokines Multi-Analyte ELISArray™ Kit, and protein concentration was normalized to the total protein content quantified by the breadford assay. We also quantified IL6 with Human IL6 DuoSet, IL8 with Human CXCL8/IL8 Quantikine ELISA Kits, and MMP11 with human MMP11 ELISA kit. Data were normalized to the total protein content. The nuclear NF-κB-related proteins NF-κB1, RelA, and RelB were quantified with ELISA-based TransAM NF-κB Family kit. Briefly, nuclear protein extracts (8 µg/well) were added to a 96-well plate with immobilized oligonucleotide containing the NF-κB consensus site. The primary antibodies were then added followed by horseradish peroxidase (HRP)-conjugated antibody. Colorimetric reactions for all the ELISA assays were quantified by the Tecan Infinite F200pro microplate reader.
RNA extraction and RNA-seq analysis
Cells were incubated for 24 hrs in complete medium at low pH (6.5) or at physiological pH (7.4). At the end of the incubation, total RNA was extracted using guanidinium thiocyanate-phenol-chloroform and quantified with Bioanalyzer (Agilent) following the manufacturer's instructions. RIN (RNA Integrity Number) and A260/A280 ratios of total RNA were all 10, and over 1.8, respectively. The library of template molecules for high throughput DNA sequencing was converted from the total RNA using TruSeq RNA Sample Prep Kit v2 following the manufacturer's protocol. The library was quantified with Bioanalyzer (Agilent) following the manufacturer's instruction. Library (3 pM) was subjected to cBot clonal amplification system for cluster generation on a Single Read Flow Cell v4 with a cluster generation instrument (Illumina). Sequencing was performed on a Genome Analyzer GAIIx for 76 cycles using Cycle Sequencing v4 regents (Illumina). Image analysis and base calling were performed using Off-Line Basecaller Software 1.6 (Illumina). Reads were aligned using ELAND v2 of CASAVA Software 1.7 with the sequence data sets. Human genome build 19 (hg19) were downloaded from genome browser (http://genome.ucsc.edu/), University of California, Santa Cruz, as the analytic reference. Transcript coverage for every gene locus was calculated from the total number passing filter reads that mapped to exons, and quantified with Quantile normalization algorithm, by Avadis NGS software (version1.5, Strand Scientific Intelligence Inc). The filtering was performed using default parameters. All new data has been deposited in DDBJ/EMBL/GenBank under DRA004087 and DRA004091.
Clonogenic assay
MSC (3 lots, 760 cells/well) or Saos-2 and HOS OS cells (280 cells/well both) were seeded at low density and incubated at 37 °C in a humidified 5% CO2 atmosphere. After 24 hrs, medium was changed with complete medium at different pH values, for MSC, or with 25% CM MSCpH 6.8 or with 25% CM MSCpH 7.4 plus 75% IMDM supplemented with 2% FBS, for Saos-2 or HOS cells. Colonies were fixed with methanol, stained with crystal violet, and counted after 14 days for MSC and 6 days for OS cells. Three experiments were independently performed, with two technical replicates.
Migration assay
Migration assay on MSC (BM-MSC, 1 lot) and HOS was performed using transwells with 8 µm pores. Before the seeding of cells on the membrane of the transwells, cells were pre-treated with CM MSCpH7.4 or CM MSCpH6.8 for 24 hrs. In the lower compartment, 800 µL of CM MSCpH7.4 or CM MSCpH6.8 were placed. Cells were resuspended in 200 µL of medium containing 0.1% bovine serum albumin and seeded into the transwells (1.5×104 cells for MSC and 3×104 cells for HOS). Cells were then incubated in 5% CO2 at 37°C, and allowed to migrate (20 hrs for MSC and 6 hrs for HOS). At the endpoint, migrated cells on the lower side were fixed in methanol, stained with crystal violet solution and counted from nine random fields (20× lens for HOS cells and 10× lens for MSC) in each well. All experiments were performed in triplicates.
Invasion assay
Invasion assay was performed using transwells with 8 µm pores that were pre-coated with 1:3 diluted Matrigel. Before the seeding on matrigel, HOS cells were pre-treated with CM MSCpH7.4 or CM MSCpH6.8 for 24 hrs. In the lower compartment, 800 µL of CM MSCpH7.4 or CM MSCpH6.8 were placed. 1.6×105 cells were diluted in 200 µL of medium containing 0.1% bovine serum albumin and seeded onto matrigel pre-coated transwells. Cells were then incubated in 5% CO2 at 37°C, and allowed to migrate for 36 hrs. At the endpoint, migrated cells were fixed in methanol, stained with crystal violet solution and counted. Three technical replicates were performed.
Serum analysis
Five patients with newly diagnosed high-grade OS were seen at our institution, signed informed consent regarding the use of their biological materials for research studies, and entered the study. Blood samples were collected after the institutional ethical committee approval (n. 0000184, 07/01/2015). Serum was also collected 12 months after diagnosis, after treatment completion. Serum levels of IL6 and IL8 were measured by ELISA, according to the manufacturer instructions.
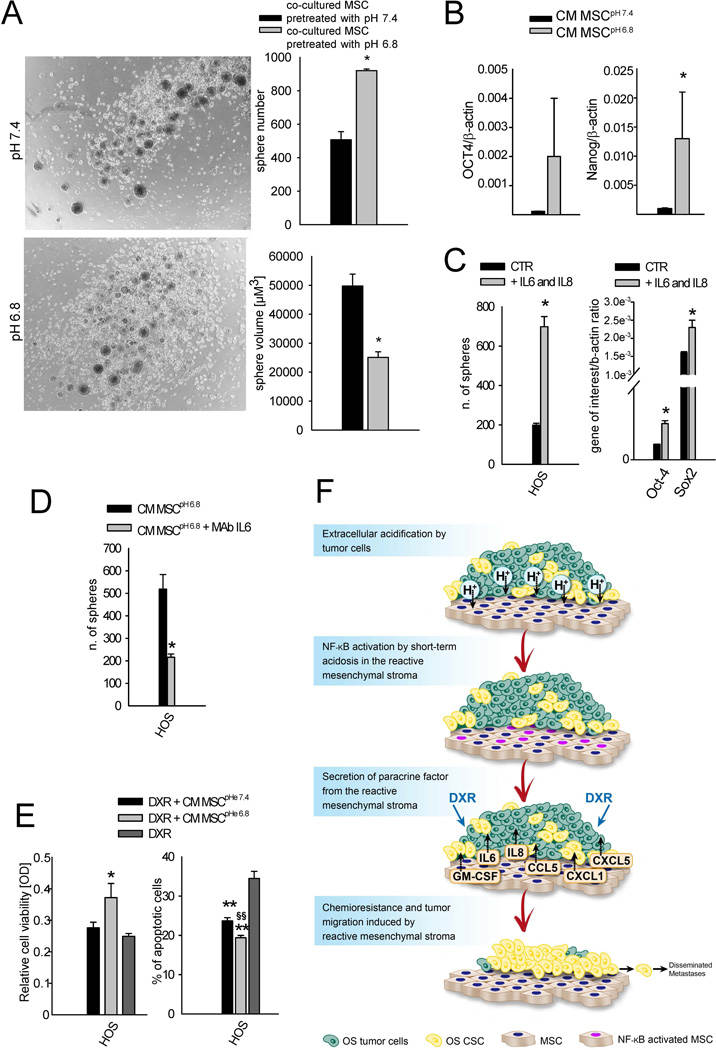
Sarcosphere-forming efficiency
To evaluate the effect of MSC secretome after the exposure to acidosis on OS spherogenicity, we performed a co-culture of MSC and HOS cells using transwell inserts with a 0.4 µm porous membrane. MSC were seeded (1.3×104 cells/transwell) in complete medium. After adhesion, cells were incubated for 20 hrs in low-serum medium, and then washed with PBS and treated with low-serum medium at pH 6.8 or pH 7.4 for 10 hrs. The medium was then replaced with low-serum medium at pH 7.4. At this time, HOS cells were seeded at 3×104 cells in DMEM:F12 medium at pH 7.4 with progesterone (20 nM), putresceine (10 mg/mL), sodium selenite (30 nM), apo-transferrin (100 µg/mL), and insulin (25 µg/mL) in low attachment 12-well plates, and the transwell inserts were placed over these wells. After 6 days, the total number of HOS spheres was counted and the radius of each spheroid was measured using NIS-Elements Microscope Imaging Software (Nikon). At the same endpoint, HOS spheres were characterized for the expression of octamer-binding transcription factor 4 (OCT-4) and Nanog stem cell-related markers by Q-RT-PCR, as previously described. To evaluate if sphere formation induced by the MSC secretome relays on IL-6 specific secretion, Tocilizumab neutralizing anti-IL-6 monoclonal antibody was added to both MSC and HOS cells [100 µg/mL] every 24 hrs. Untreated cells were considered as negative control. For each assays three replicates were performed. Furthermore, in another assay, HOS were seeded (3×104 cells) in DMEM:F12 medium at pH 7.4 with progesterone (20 nM), putresceine (10 mg/mL), sodium selenite (30 nM), apo-transferrin (100 µg/mL), insulin (25 µg/mL), recombinant human IL6 (50 ng/mL) and recombinant human IL8 (100 ng/mL), in low attachment 12-well plates. IL6 and IL8 were added every 24 hrs. Untreated cells were considered as negative control. After 6 days, the total number of HOS spheres was counted and the radius of each spheroid was measured as previously described. At the same endpoint, HOS spheres were characterized for the expression of OCT-4 and Sox2 stem cell-related markers by Q-RT-PCR, as previously described. For each assays three replicates were performed.
Cell viability
Cell viability after doxorubicin (DXR) treatment was determined using the acid phosphatase indirect assay. HOS cells were seeded in 96-well plates (2×103 cells/well). After 24 hrs, the medium was changed with 50% complete IMDM plus 50% CM MSCpH 6.8 or CM MSCpH 7.4 (negative control), added with DXR (50 ng/mL). After 72 hrs, cells were washed in PBS and incubated at 37°C with 100 µL of buffer containing 0.1 M sodium acetate (pH 5.0), 0.1% Triton X-100, and 5 mM p-nitrophenil phosphate. The reaction was stopped by adding 10 µL of 1 N NaOH, and absorbance read using a microplate reader (Tecan Infinite F200pro).
Apoptosis assay
HOS cells (5×103 cells) were seeded on glass coverslips in complete IMDM. After cell adhesion, medium was changed with 50% complete IMDM plus 50% CM MSCpH 6.8, CM MSCpH 7.4 (negative control), or 50% complete IMDM plus 50% Alpha-MEM supplemented with 0.1% FBS. After 24 hrs, medium was replaced with the same conditions, added with DXR (10 ng/mL). After 48 hrs, cells were fixed with 3.7% paraformaldehyde for 20 min, permeabilized with PBS containing 0.1% TritonX-100 for 10 min, and stained with 2.25 µg/mL of Hoechst 33258 at RT for 10 min. Cells were observed under fluorescence microscopy and cells with apoptotic nuclear bodies were counted in at least five different fields by using 20× objective. Results were expressed as percentage of apoptotic cells over the fields. Three technical replicates were performed.
Statistical analysis
Statistical analysis was performed with the StatView™ 5.0.1 software (SAS Institute Inc.). Mann-Whitney U test was used as unpaired comparison for two independent variables. Data were expressed as mean ± standard error (SE). Only P values < 0.05 were considered for statistical significance.
Results and Discussion
Reactive stroma in OS
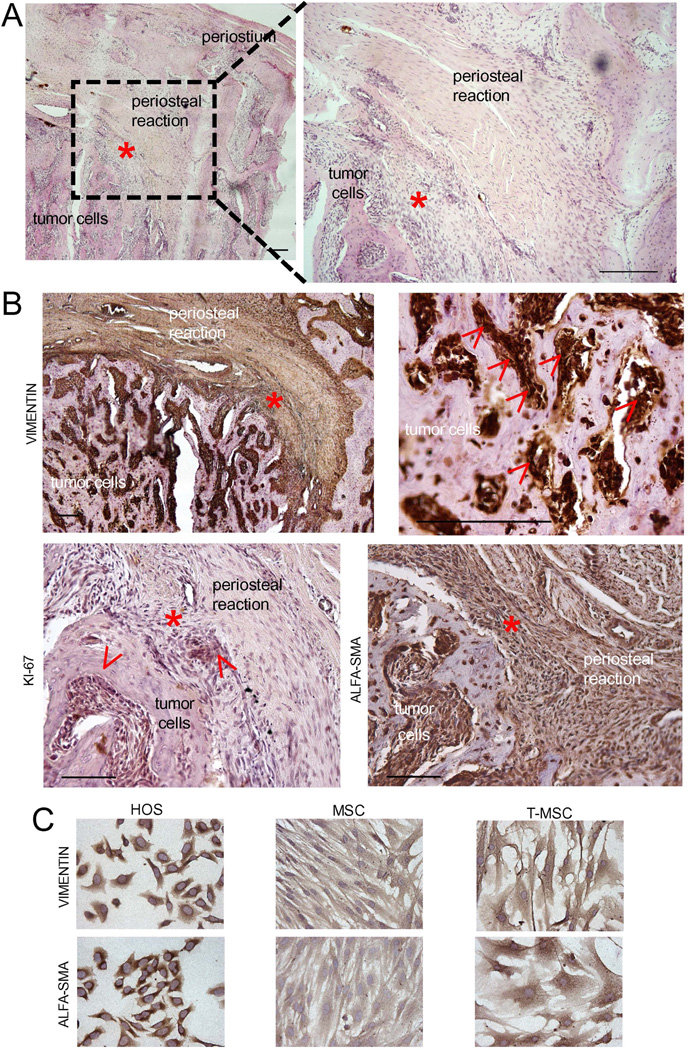
In OS TME, surrounding and infiltrating non-transformed host stroma is composed of immune cells, endothelial cells, and cells of mesenchymal origin, including MSC.23 The reactive mesenchymal stroma is recognized to exert powerful effects on tumor biology in several solid cancers and can be easily recognized by the staining of specific markers, like α-SMA. In OS, the differential identification MSCs is almost impossible, as they share the same cellular origin as cancer cells, and therefore also the same antigens. Furthermore, analysis of human specimens of OS is largely prevented by the extensive necrosis that develops following preoperative chemotherapy. We therefore considered spontaneous canine OS, whose standard treatment does not include drug administration before surgery. Comparative pathology is a useful approach to study human cancer, especially in OS, given the striking analogies between human and dog disease.24 Canine OS exhibits clinical and molecular features that closely resemble the corresponding human disease. Indeed, both clinical and molecular evidence suggest that human and canine OS share many key features, including anatomic location, presence of microscopic metastatic disease at diagnosis, development of chemotherapy-resistant metastases, altered expression/activation of several proteins (e.g. Met, PTEN, STAT3), and p53 inactivation, among others.25, 26 We identified a favorable anatomical site to observe the tumor-stroma interaction, a tumor area in which reactive mesenchymal cells are clearly distinguishable from OS cells even on morphologic criteria, the area of periosteal reaction (Fig. 1A). Here, proliferating tumor cells stimulate a periosteal reaction that contains a consistent number of reactive cells of mesenchymal origin.27 In the examined specimen of canine OS, both tumor cells and reactive mesenchymal stroma were strongly positive for vimentin and α-SMA expression (Fig. 1B), whereas Ki-67-positive cycling cells were predominantly prevalent in the tumor (Fig. 1B). Interestingly, the intensity of vimentin immunostaining was remarkably higher in the tumor than in the reactive stroma (Fig. 1B). We also isolated T-MSC from a tissue sample of human OS. T-MSC can be isolated from sarcoma tissues and, although not transformed, they may contain minor cytogenetic mutations.21 In our case, T-MSC were not immortalized since after 14–15 passages they did not further propagate. Human OS cells, T-MSC, and MSC were all positive for vimentin and α-SMA expression (Fig. 1C).
Figure 1. Morphology and immunostaining of canine OS tissue sample.
A red asterisk (*) indicates the same area in all the histological sections and includes a borderline zone between the tumor area and the periosteal stromal reaction. (A) Histological sample of canine OS stained by standard hematoxylin and eosin (H & E). Scale bar 500 µm; (B) Immunohistochemistry for vimentine, α-SMA, and Ki67 of the same sections shown in panel A. For vimentin, both high and low magnifications are shown. Arrows indicate positive staining. Scale bar 200 µm; (C) Cell cultures of normal human MSC, of T-MSC derived from a human OS tissue samples, and OS cells (HOS) stained for the same markers shown in panel B. Scale bar 200 µm.
Short-term acidosis activates downstream signaling of the NFkB pathway in MSC but not in OS cells
Short-term acidosis induced by cancer cells has significant effects on the surrounding stroma. Tissue acidification (pH decrease of at least 0.5–1.0 pH unit) is commonly associated with inflammation,28 and acts as a driving force for stem cells during the regenerative process.29 Acidification per se can activate the NF-κB inflammatory pathway30 that is commonly activated in TME, including in cancer-associated fibroblasts.31 Thus, we hypothesized that OS cells induce the T-MSC phenotype in the mesenchymal stroma by secreting protons that, in turn, activate the NK-κB-mediated pro-inflammatory response.
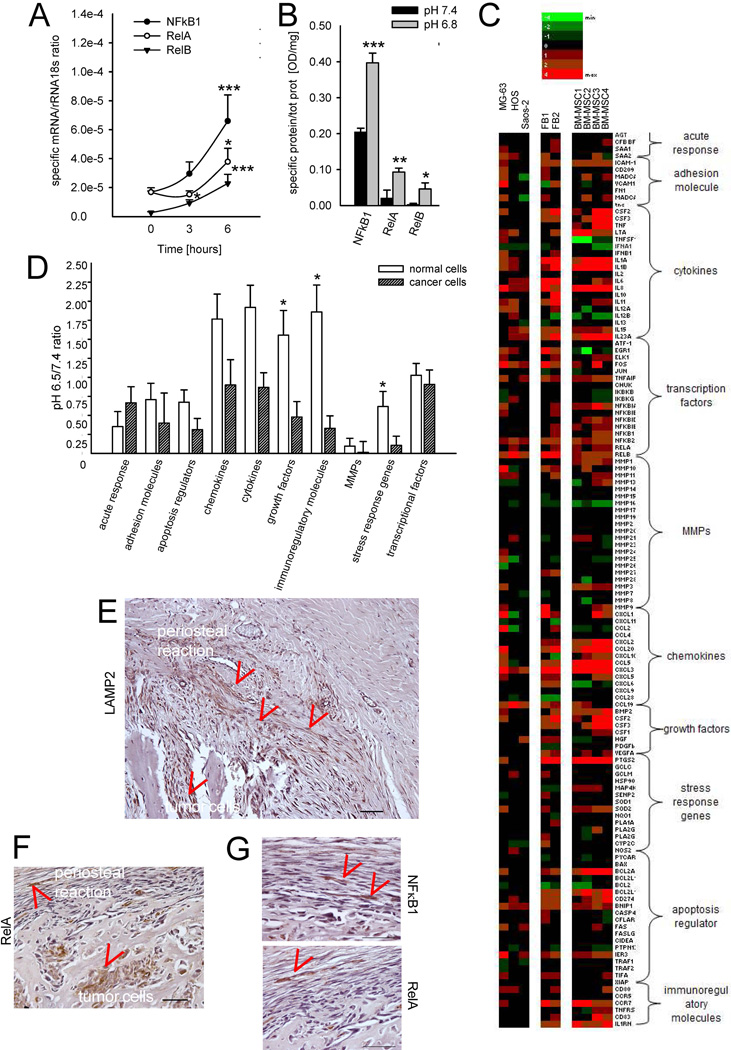
As in vitro models of MSC that can reach and infiltrate OS, we used fibroblasts, FB, and MSC. We considered both BM-MSC and AT-MSC, as naïve MSC in OS TME might derive from either origin. Incubation with low pH was sufficient to significantly and promptly induce RelB (for mRNA P=0.01 at 3 hrs vs. T0, P=0.0009 at 6 hrs vs. T0; for protein P=0.0377 vs. pH 6.8 after 24 hrs), NF-κB1 (for mRNA P<0.0001 at 6 hrs vs. T0; for protein P<0.0001 vs. pH 6.8 after 24 hrs), and RelA (for mRNA P=0.0094 at 6 hrs vs. T0; for protein P=0.0094 vs. pH 6.8 after 24 hrs) in MSC (Fig. 2A and B). According to the literature, in our experimental conditions, NF-κB activation might be promoted by ROS production that is enhanced under acidosis.32 We also performed a deep sequencing analysis on both tumor and normal cells to study the effects of acidosis on transcription of all downstream genes that are directly and indirectly regulated by NF-κB transcription factors (Fig. 2C).32 In normal cells, we observed a selective and significant increase for the gene categories of: growth factors, stress response genes, and immunoregulatory molecules, whereas tumor cells were less unaffected by acidosis (Fig. 2D). We then looked at the localization of intratumoral acidosis in canine tissue sections of osteosarcoma in the area of the periosteal reaction that was already seen in Fig. 1. We used LAMP2 staining at the plasma membrane as a marker of the acidic microenvironment,33 which was evident both in the cells of the tumor area, and in the area of stromal reaction close to the tumor (red signs, Fig. 2E). Similarly, RelA and NF-κB1 staining localized in the nuclei of normal cells in the reactive tissue of the same area (red signs, Fig. 2F and G).
Figure 2. Short-term acidosis induces NF-κB activation in MSC.
(A) mRNA expression of NF-κB transcription factors in MSC (n=3, 2 from bone marrow and 1 from adipose tissue) exposed to acidic medium at different times. Mean ± SE (n=12), ***P<0.001, and *P<0.05; (B) Quantification of the signal for NF-κB transcription factors translocated into the nucleus of MSC (n=3) after 24 hrs of culture with medium at different pH. Mean ± SE (n=12); ***P<0.001, **P<0.01, and *P<0.05; (C) Heat map representation of the fold increase, by deep-sequencing analysis, of the mRNA levels of NF-κB-related genes of OS cells (MG63, HOS, Saos-2) and normal stromal cells (2 lots of skin fibroblasts, FB, and 4 lots of BM-MSC) after short-term acidosis (pH 6.5) compared to physiological medium (pH 7.4). Colors on the heat map indicate the log2 ratios of expression (representing normalized read counts). Red, upregulation; green, downregulation. (D) Clustering in gene categories of the results obtained by the deep-sequencing analysis of the transcriptional levels of NF-κB-related genes. Mean ± SE; *P<0.05. (E) LAMP2 staining of histological sample of canine OS and an indirect marker of acidic area in the tumor microenvironment; the same sections was also stained for (F) NF-κB p65 (RelA, B1 scale bar 100 µm) also shown at higher magnification (scale bar 50 µm) (G) and NF-κB (G). Arrows indicate positive staining.
When genes were considered individually, the trend of increase in cultured MSC was significant also for several cytokines and chemokines, including CSF3, IL1A, IL23A, CXCL2, and CCL5, in addition to the metalloproteinase MMP2 and the immunomodulatory molecules CCR7 and IL1RN (Supplementary Fig. 1). It has already been shown that, once they have homed to the tumor site, MSC are a source of cytokines and chemokines,34, 35 and have an immunomodulatory action, including a pro-inflammatory profile.36 However, this is the first time that the role of tumoral acidosis is demonstrated to play a major role in this process.
Acidic TME promotes the secretion of high levels of IL6 and IL8 by the mesenchymal stroma
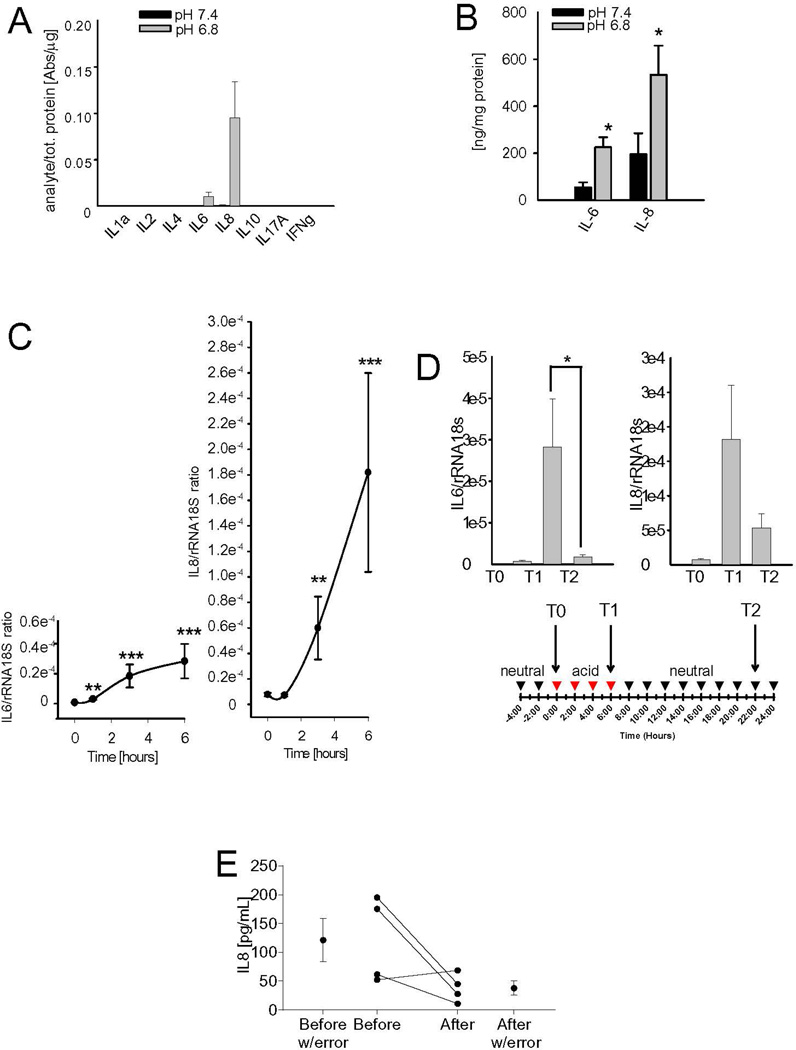
Results obtained from global screening approach on the downstream genes related to the NF-κB pathway were further validated by Q-RT-PCR and/or protein analyses. In human MSC, at the protein level, IL6 and IL8 were the only cytokines that were clearly induced by short-term acidosis (Fig. 3A). These increases were also significant when analyzed by ELISA (Fig. 3B). IL6 had a ~3 fold induction (P=0.0177), and IL8 had a ~2.5 fold induction (P=0.0116) by acid pH. At the mRNA level, acid-promoted induction was also time-dependent, and was significant already at 1 or 3 hrs, for IL6 or IL8, respectively (Fig. 3C). Notably, in FB we observed a similar trend for both IL6 and IL8, although IL6 was more induced by low pH than IL8, whereas the other cytokines were unaffected (Supplementary Fig. 2). The induction of mRNA expression for IL6 and IL8 was very rapid and time-dependent (Fig. 3C, P-values for IL6 at 1 hr were P=0.0012, at 3 hrs P=0.0002, at 6 hrs P<0.0001, and for IL8 at 3 hrs P=0.0018, and at 6 hrs P<0.0001). These effects were reversible, as replacement of the acid medium with the neutral medium restored the original level of expression after 16 hrs (Fig. 3D, for IL6 P=0.0495). Interestingly, the increased expression after short-term acidosis (pH 6.3 for 16 hrs) has already been demonstrated in human airway smooth muscle cells, which are normal cells of mesenchymal origin.37 High levels of circulating IL6 and IL8 are adverse prognostic factors in OS patients,38, 39 and, accordingly, when we evaluated IL8 levels in a small series of patients, we observed a consistent but not significant decrease of IL8 following the disappearance of the disease in a small cohort of patients (Fig. 3E). To date, IL6 and IL8 have been assumed to be secreted and released into the blood by tumor cells, although very few reports have actually demonstrated IL6 and IL8 secretion by isolated OS cells. In contrast, IL6 and IL8 have been frequently detected in the cancer-associated stroma.40 Also in OS we found a very low expression of these cytokines in cancer cells, both at neutral and acidic pH, by deep sequence analysis (Fig. 2C), protein analysis (Supplementary Fig. 2), and Q-RT-PCR (Supplementary Fig. 3A). Similarly, when not exposed to acidosis, MSC expressed very low levels of both IL6 and IL8 mRNA (Supplementary Fig. 3A). However, T-MSC isolated from OS tissue samples and analyzed at early passages (before the 6th passage), showed a higher expression of IL6 in respect to normal MSC (Supplementary Fig. 3B, P=0.0203), even at neutral pH.
Figure 3. Short-term acidosis promotes the release of the inflammatory cytokines IL6 and IL8 by the tumor-associated mesenchymal stroma.
(A) Human Inflammatory Cytokines Multi-Analyte ELISArray™ performed on MSC cultured in acidic or physiological medium for 24 hours (n=3); (B) IL6 and IL8 protein expression in the same samples after 24 hrs from the start of the incubation with acidic medium by using specific ELISA. Mean ± SE (n=12), *P<0.05; (C) IL6 and IL8 mRNA analysis of MSC at different time points after the incubation with acidic medium. Mean ± SE (n=12), ***P<0.001, **P<0.01, and *P<0.05 vs T0; (D) The induction of IL6 and IL8 mRNA expression by the acidic medium is reversible: mRNA levels of IL6 and IL8 were analyzed at T0, when the medium was change with acidic medium, at T1 at the end of the incubation period, and at T2, after additional 16 hours from the interruption of the acidic stimulus. Mean ± SE of each different lot of MSC (n=12), CSF2/GM-CSF, CSF3/G-CSF, BMP2 mRNA analysis of MSC after 6 hours of culture in acidic medium. Mean ± SE (n=12); (E) IL8 serum level trend in patients at diagnosis and at 12 months since diagnosis (after treatment).
Altogether, these data indicate that the previously reported prognostic significance of IL8 and IL6 serum levels in OS patients may be due to release from mesenchymal stroma in response to tumor acidification, rather than to release from tumor cells per se. Thus, we hypothesize that the mesenchymal reaction to acidosis may play a critical role in tumor aggressiveness and the clinical outcome. Therefore, we investigated the effect of H+-MSC secretome on OS aggressiveness, in terms of migration and stemness.
Short-term acidosis turns MSC into T-MSC that secrete pro-clonogenic and pro-migratory factors
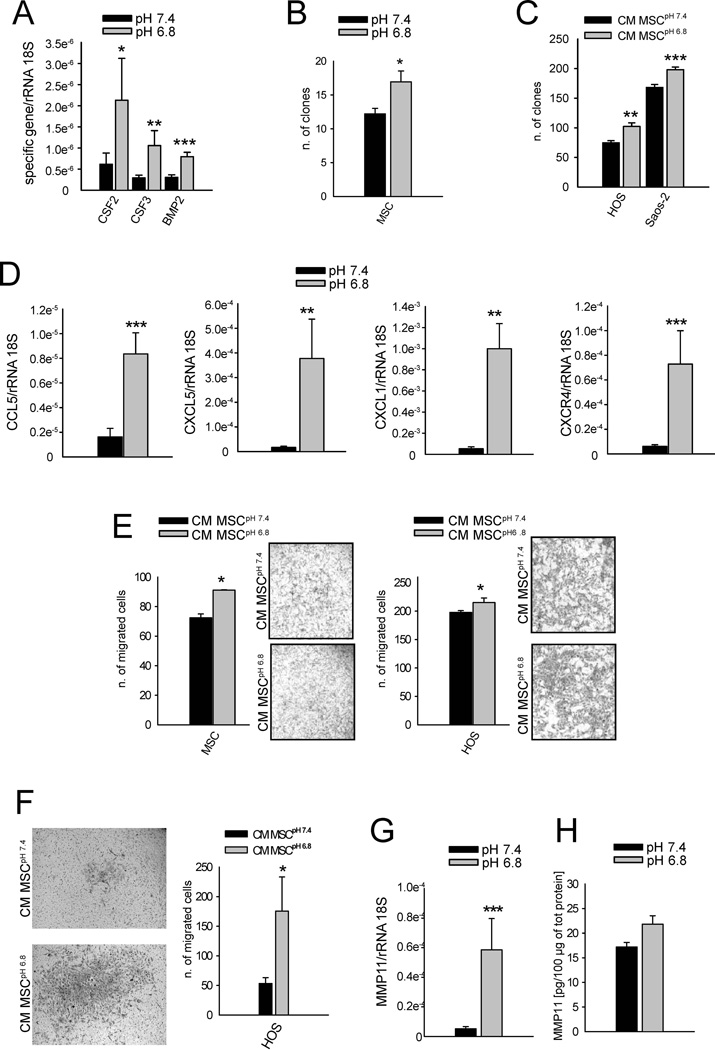
Among the NF-κB-related genes included in the growth factor category of deep-sequencing analysis, we found a significant acid-mediated increase of CSF2/GM-CSF (P=0.0209), CSF3/G-CSF (P=0.0047), and BMP2 (P=0.0007) in MSC after 6 hrs (Fig. 4A). GM-CSF and G-CSF are directly related to the clonogenicity of both normal and transformed cells,41 and BMP2 to the clonogenicity of mesenchymal progenitors.42 Indeed, when cultured in acidic medium, the clonogenic potential of MSC was significantly augmented (Fig. 4B, P=0.0148), as were OS cells when treated with CM MSCpH 6.8 (Fig. 4C, P=0.0035 for HOS, and P=0.0007 for Saos-2). Our results on mesenchymal cells are similar to the behavior of hematopoietic progenitors that are significantly more clonogenic at low pH (7.1) in comparison with high pH (7.6).43 GM-CSF secretion by T-MSC might also impact the antitumor immune response, since it induces the migration of monocytes, macrophages, and neutrophils to the tumor.44 Similarly, the mRNA expression of the chemokines CCL5 (P=0.0003), CXCL5 (P=0.0065), and CXCL1 (P=0.0014) was induced when MSC were cultured at low pH, together with an increased expression of the CXCR4 (P=0.0002), the putative receptor for the stromal-derived-factor-1 (SDF1) ligand (Fig. 4D). The release of these chemokines might support not only the recruitment of inflammatory cells, but also further stimulate tumor growth and progression. Indeed, CXCL1 and CXCL5 variably affect tumor growth, vascularity, and apoptosis,45 whereas CCL5 promotes metastasis formation in several cancers,46 including OS.13 Finally, the increased expression of CXCR4 receptor in MSC might recruit additional MSC at the tumor site. According to recent data, CXCR4 is needed for MSC homing to tumor sites, for their differentiation into myofibroblasts, and/or for their survival,47 and CXCR4 mRNA in OS samples negatively correlates with patient survival.48 Here, we demonstrated an improvement of migratory potential of both MSC and OS cells (Fig. 4E), and of the invasive potential of HOS cells (Fig. 4F) when exposed to the secretome of MSC that were previously stressed by short-term acidosis (P=0.0495 for both). Thus, the stimulus induced by acid in the TME might provide a niche containing MSC that can recreate a provisional ECM, upon which additional chemoattracted FB and MSC may seed and proliferate. Finally, among the matrix metalloproteinases that by deep sequencing analysis were shown to be upregulated by acidosis, we confirmed the increased expression of MMP11 at the mRNA (Fig. 4G P=0.0003 vs pH 6.8). There were also increases at the protein level, yet the differences were not significant (Fig. 4H). In contrast to other metalloproteinases, MMP11 has been implicated in the survival and expansion of the pro-tumorigenic stroma rather than to ECM degradation.49 Thus, MMP-11 expression by acidosis may represent an autocrine survival mechanism for MSC in the hostile TME of OS.
Figure 4. Short-term acidosis induces a MSC-derived secretome that boosts the migration and invasive potential and the colony formation of OS cells.
(A) CSF2/GM-CSF, CSF3/G-CSF, BMP2 mRNA analysis of MSC after 6 hours of culture in acidic medium. Mean ± SE (n=12); ***P<0.001, **P<0.01, and * P<0.05 vs pH 7.4; (B) Clonal efficiency of MSC in acidic medium. Mean ± SE (n=18); *P<0.05 vs pH 7.4; (C) Clonal efficiency of OS cells when cultured in CM of MSC that were pre-treated (CM MSCpH 6.8) or not (CM MSCpH 7.4) with acidic medium. Mean ± SE (n=12), ***P<0.001, **P<0.01 vs CM MSCpH 7.4. (D) CCL5, CXCL5, CXCL1, and CXCR4 mRNA analysis of MSC after 6 hours of culture in acidic medium. Mean ± SE (n=12), ***P<0.001, **P<0.01 vs pH 7.4; (E) Migration of MSC and HOS cells and (F) invasion ability of HOS cells exposed to CM of MSC pre-treated (CM MSCpH 6.8) or not (CM MSCpH 7.4) with acidic medium that here were used as chemotactic stimulus. Mean ± SE (n=3). In the migration assay the right panels are representative images of the porous membrane with migrated cells stained with crystal violet (for MSC 10× lens, for HOS 20× lens), whereas in the invasion assay the same type of images are on the left panel (20× lens). *P<0.05 vs CM MSCpH 7.4 (G) MMP11 mRNA analysis of MSC after 6 hrs of culture in acidic medium. Mean ± SE (n=12); ***P<0.001 vs pH 7.4. (H) MMP11 protein expression in supernatant of MSC cultured or not in acidic medium for 24 hrs. Mean ± SE (n=12).
Overall, these data suggest that in OS the secretome of mesenchymal stroma reactive to tumor-associated acidosis causes the attraction of additional reactive cells and the migration of tumor cells from the primary site, possibly contributing to the enhancement of circulating tumor cells and ultimately leading to metastasis.
MSC pro-inflammatory response to short-term acidosis favors OS stemness
A recent study on a breast cancer model has shown that injected MSC distribute throughout the tumor-associated stroma mostly as single cells, and closely associate with cancer stem cells (CSC), possibly implying a supporting function of MSC for CSC maintenance.50 Analogously, we have demonstrated that when co-cultured with OS cells, MSC further expand the CSC subpopulation of OS through IL6 secretion (paper submitted). We thus sought to explore the impact of IL6 and IL8 secretion by MSC in inducing OS stemness in the context of the acidic TME of OS. Indeed, OS cells express both IL6 and IL8 receptors,51 as confirmed by deep sequencing analysis (IL6R gene ID ENSG00000160712: 11.7 for MG-63 and HOS, and 11.2 for Saos-2; IL6ST gene ID ENSG00000134352: 13.7 for MG-63, 12.8 for HOS, and 12.9 for Saos-2), and thus might react to the cytokines released by the acid-induced mesenchymal stroma. To evaluate the effect of the secretome of H+-MSC, we used the spherogenic assay and the expression of stem-related genes by Q-RT-PCR to measure the stemness potential of OS cells.22 HOS cells co-cultured with H+-MSC formed a significantly smaller (P=0.0495, Fig. 5A) but higher number of spheres (P=0.0495, Fig. 5A). Furthermore, when incubated with CM from MSCpH 6.8, these cells expressed higher levels of OCT4 and Nanog stem cell-related markers compared to those incubated with CM from MSCpH 7.4 (P=0.0495 for Nanog, Fig. 5B). The increased stemness of HOS cells is possibly mediated by IL650 and/or by the previously mentioned increase of BMP2 expression (Fig. 4A). To further validate this hypothesis we incubated HOS cells under spherogenic conditions with the addition of only IL6 and IL8 factors in the medium, or with or without anti-IL6 specific antibody added to the acid-induced MSC secretome: the exposure to IL6 and IL8 as the unique stimulating factors or, the blockage of IL6 with specific antibody enhanced and inhibited sphere formation, respectively (P=0.0495, Fig. 5C and D), and the treatment with IL6 and IL8 also enhanced the expression of the stem-related markers Oct4 and Sox2 (P=0.0495, Fig. 5C), respectively. The increased number of formed spheres under these conditions paralleled with a decrease of sphere volume, implying that an augment of sphere number was due to an increased clonogenicity potential rather than to an increase of cell proliferation (P = 0.0495, Supplementary Fig. 4). The expansion of the CSC fraction in tumor-associated stroma has also been shown in gastric and pancreatic carcinomas.52, 53, 52 In ovarian cancer, it was mediated by the expression of BMP2, BMP4, and BMP6.543 Since stemness is naturally associated with chemoresistance, we sought to verify the effect of MSC stressed by short-term acidosis on OS cell sensitivity to DXR. Indeed, recent studies focused on the screening of several drug libraries have considered TME as an unexplored target for drug discovery.54 As an example, exploitation of 23 different stromal cells in inducing innate resistance of 45 cancer cell lines with several anticancer drugs has successfully demonstrated a strong correlation between stroma-derived HGF factor and innate resistance to RAF inhibitor treatment.55 Here we further emphasized the concept of mimicking the real TME (including an acidic pH) under in vitro conditions. The use of MSC for drug screening should be coupled with additional variables that are present in the TME and that can activate chemoresistance-promoting activity of MSC, including local acidosis. In our experimental conditions, CM from MSCpH 6.8 significantly reduced DXR toxicity in HOS cells compared to CM MSCpH 7.4 as measured by a viability assay (P=0.0375, Fig. 5E, left panel), and an apoptosis assay (P=0.0039 vs DXR treatment alone for both CMpH 6.8 and CMpH 7.4, and P=0.0065 vs CM pH 7.4, Fig. 5F, right panel). Taken together, these results emphasize the importance of the stromal environment to chemotherapy resistance of OS and suggest that the targeting of MSC might be an interesting, unexplored strategy to affect the OS CSC population or niche, and that MSC might be even considered as an effective tumor- or, more specifically, CSC-targeted vehicle for anti-cancer drugs.55
Figure 5. The secretome of acid-stressed MSC promote a stem-like phenotype in HOS cells.
(A). Sphere-forming efficiency assay in HOS cells co-cultured with MSC that were previously stressed or not with acidic medium. Left panel, representative images of the obtained spheres (4× lens). Mean ± SE (n=9), *P<0.05; (B) mRNA expression of the stem-related genes OCT4 and Nanog in HOS co-cultured for 6 days with MSC that were pretreated (CM MSCpH 6.8) or not (CM MSCpH 7.4) with acidic medium. Mean ± SE (n=4), *P<0.05; (C) Sphere-forming efficiency (left panel) and stem-related gene expression (right panel) of HOS cells treated with IL6 and IL8. Mean ± SE (n=3), *P<0.05; (D) Sphere-forming efficiency of HOS co-cultured for 6 days with MSC that were pretreated (CM MSCpH 6.8) with acidic medium with or w/o Tocilizumab (anti-IL6 antibody). Mean ± SE (n=3), *P<0.05; (E) Indirect viability assay (left panel) and apoptosis assay (right panel) to measure the effect of CM obtained from MSC treated with acidic medium (MSC CMpH 6.8) on HOS resistance to doxorubicin (DXR). Mean ± SE (n=8 for viability test and n=6 for apoptosis test), *P<0.05 and **P<0.01 vs DXR, §§P<0.01 vs CM MSCpH 7.4; (F) Due to the high glycolytic metabolism, OS cells strongly acidify the TME. The local increased concentration of protons is sensed by MSC in the TME as a strong exogenous stress. The acid-stressed mesenchymal stroma might be therefore reprogrammed into T-MSC by the activation of the NF-κB inflammatory pathway that triggers the transcription and the secretion of several cytokines and chemomikes, among which IL6, IL8, CCL5, GM-CSF, CXCL1, and CXCL5. Through a paracrine circuit, the secretome derived from the reactive mesenchymal stroma, in turn, might promote the stemness and migration of OS cells.
Conclusion
For the first time, this study focused on the response of cancer-associated mesenchymal reactive cells to acidic TME and its effect on OS progression. Our data suggest a novel concept for OS-stroma interaction, and we propose a model with OS cells producing extracellular acidosis that, in turn, might reprogram normal mesenchymal cells to reactive, inflammatory cancer-promoting cells that might support tumor stemness and migration (Fig. 5D). We believe that dissecting the complexity of MSC interactions with cancer cells will have terrific benefits for the understanding and treatment of life-threatening cancers, including OS.
Supplementary Material
Novelty and impacts.
This study explores the role of acidic OS microenvironment in promoting tumor aggressiveness via activation of reactive MSC, and, for the first time, demonstrates that, under such conditions, these elements secrete a plethora of clonogenic, chemotactic, and pro-migratory factors via NF-κB pathway, in turn fostering tumor migration, stemness, and chemoresistance. Understanding tumor-stroma interactions is crucial for the identification of novel therapeutic interventions.
Acknowledgments
The work was supported by the Italian Association for Cancer Research (n. 11426 to N.B.), and by the financial support for Scientific Research “5 per mille 2012” (to N.B.), and by National Institutes of Health/National Cancer Institute (NIH/NCI) grants (n R01CA077575 and R01CA187532 to R.J.G.). We thank Dr. Emanuela Morello and Dr. Marina Martano, the University of Turin, for providing the tumor tissue sample from spontaneous canine OS, and Maria Pia Cumani for the graphical assistance.
Abbreviations
- OS
Osteosarcoma
- TME
tumor microenvironment
- CSC
cancer stem cells
- MSC
mesenchymal stem cells
- BM-MSC
bone marrow MSC
- AD-MSC
adipose tissue MSC
- T-MSC
tumor tissue educated MSC
- TAF
tumor-associated fibroblasts
- FB
fibroblasts
- IL6
interleukin 6
- CM
conditioned medium
- OCT-4
octamer-binding transcription factor 4
- DXR
doxorubicin
- AP
acid phosphatase
- SDF1
stromal-derived-factor-1
- CXCR4
C-X-C chemokine receptor type 4
References
- 1.Bissell MJ, Radisky D. Putting tumours in context. Nature reviews Cancer. 2001;1:46–54. doi: 10.1038/35094059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marx J. Cancer biology. All in the stroma: cancer's Cosa Nostra. Science. 2008;320:38–41. doi: 10.1126/science.320.5872.38. [DOI] [PubMed] [Google Scholar]
- 3.Bonuccelli G, Whitaker-Menezes D, Castello-Cros R, Pavlides S, Pestell RG, Fatatis A, Witkiewicz AK, Vander Heiden MG, Migneco G, Chiavarina B, Frank PG, Capozza F, et al. The reverse Warburg effect: glycolysis inhibitors prevent the tumor promoting effects of caveolin-1 deficient cancer associated fibroblasts. Cell Cycle. 2010;9:1960–1971. doi: 10.4161/cc.9.10.11601. [DOI] [PubMed] [Google Scholar]
- 4.Bian ZY, Fan QM, Li G, Xu WT, Tang TT. Human mesenchymal stem cells promote growth of osteosarcoma: involvement of interleukin-6 in the interaction between human mesenchymal stem cells and Saos-2. Cancer science. 2010;101:2554–2560. doi: 10.1111/j.1349-7006.2010.01731.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tsukamoto S, Honoki K, Fujii H, Tohma Y, Kido A, Mori T, Tsujiuchi T, Tanaka Y. Mesenchymal stem cells promote tumor engraftment and metastatic colonization in rat osteosarcoma model. International journal of oncology. 2012;40:163–169. doi: 10.3892/ijo.2011.1220. [DOI] [PubMed] [Google Scholar]
- 6.Bonuccelli G, Avnet S, Grisendi G, Salerno M, Granchi D, Dominici M, Kusuzaki K, Baldini N. Role of mesenchymal stem cells in osteosarcoma and metabolic reprogramming of tumor cells. Oncotarget. 2014;5:7575–7588. doi: 10.18632/oncotarget.2243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Baldini N, Scotlandi K, Barbanti-Brodano G, Manara MC, Maurici D, Bacci G, Bertoni F, Picci P, Sottili S, Campanacci M, et al. Expression of P-glycoprotein in high-grade osteosarcomas in relation to clinical outcome. The New England journal of medicine. 1995;333:1380–1385. doi: 10.1056/NEJM199511233332103. [DOI] [PubMed] [Google Scholar]
- 8.Hogendoorn PC, Athanasou N, Bielack S, De Alava E, Dei Tos AP, Ferrari S, Gelderblom H, Grimer R, Hall KS, Hassan B, Jurgens H, Paulussen M, et al. Bone sarcomas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO. 2010;21(Suppl 5):v204–v213. doi: 10.1093/annonc/mdq223. [DOI] [PubMed] [Google Scholar]
- 9.Ehnman M, Larsson O. Microenvironmental Targets in Sarcoma. Frontiers in oncology. 2015;5:248. doi: 10.3389/fonc.2015.00248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.D'Souza N, Burns JS, Grisendi G, Candini O, Veronesi E, Piccinno S, Horwitz EM, Paolucci P, Conte P, Dominici M. MSC and Tumors: Homing, Differentiation, and Secretion Influence Therapeutic Potential. Advances in biochemical engineering/biotechnology. 2013;130:209–266. doi: 10.1007/10_2012_150. [DOI] [PubMed] [Google Scholar]
- 11.Sun Z, Wang S, Zhao RC. The roles of mesenchymal stem cells in tumor inflammatory microenvironment. Journal of hematology & oncology. 2014;7:14. doi: 10.1186/1756-8722-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bian ZY, Li G, Gan YK, Hao YQ, Xu WT, Tang TT. Increased number of mesenchymal stem cell-like cells in peripheral blood of patients with bone sarcomas. Archives of medical research. 2009;40:163–168. doi: 10.1016/j.arcmed.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 13.Xu WT, Bian ZY, Fan QM, Li G, Tang TT. Human mesenchymal stem cells (hMSCs) target osteosarcoma and promote its growth and pulmonary metastasis. Cancer letters. 2009;281:32–41. doi: 10.1016/j.canlet.2009.02.022. [DOI] [PubMed] [Google Scholar]
- 14.McAnulty RJ. Fibroblasts and myofibroblasts: their source, function and role in disease. The international journal of biochemistry & cell biology. 2007;39:666–671. doi: 10.1016/j.biocel.2006.11.005. [DOI] [PubMed] [Google Scholar]
- 15.Whiteside TL. The tumor microenvironment and its role in promoting tumor growth. Oncogene. 2008;27:5904–5912. doi: 10.1038/onc.2008.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dvorak HF. Tumors: wounds that do not heal. Similarities between tumor stroma generation and wound healing. The New England journal of medicine. 1986;315:1650–1659. doi: 10.1056/NEJM198612253152606. [DOI] [PubMed] [Google Scholar]
- 17.Avnet S, Di Pompo G, Lemma S, Salerno M, Perut F, Bonuccelli G, Granchi D, Zini N, Baldini N. V-ATPase is a candidate therapeutic target for Ewing sarcoma. Biochimica et biophysica acta. 2013;1832:1105–1116. doi: 10.1016/j.bbadis.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Perut F, Avnet S, Fotia C, Baglio SR, Salerno M, Hosogi S, Kusuzaki K, Baldini N. V-ATPase as an effective therapeutic target for sarcomas. Experimental cell research. 2014;320:21–32. [PubMed] [Google Scholar]
- 19.Warburg O. On the origin of cancer cells. Science. 1956;123:309–314. doi: 10.1126/science.123.3191.309. [DOI] [PubMed] [Google Scholar]
- 20.Sedlakova O, Svastova E, Takacova M, Kopacek J, Pastorek J, Pastorekova S. Carbonic anhydrase IX, a hypoxia-induced catalytic component of the pH regulating machinery in tumors. Frontiers in physiology. 2014;4:400. doi: 10.3389/fphys.2013.00400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brune JC, Tormin A, Johansson MC, Rissler P, Brosjo O, Lofvenberg R, von Steyern FV, Mertens F, Rydholm A, Scheding S. Mesenchymal stromal cells from primary osteosarcoma are non-malignant and strikingly similar to their bone marrow counterparts. International journal of cancer. 2011;129:319–330. doi: 10.1002/ijc.25697. [DOI] [PubMed] [Google Scholar]
- 22.Salerno M, Avnet S, Bonuccelli G, Eramo A, De Maria R, Gambarotti M, Gamberi G, Baldini N. Sphere-forming cell subsets with cancer stem cell properties in human musculoskeletal sarcomas. International journal of oncology. 2013;43:95–102. doi: 10.3892/ijo.2013.1927. [DOI] [PubMed] [Google Scholar]
- 23.Joyce JA, Pollard JW. Microenvironmental regulation of metastasis. Nature reviews Cancer. 2009;9:239–252. doi: 10.1038/nrc2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chun R, Kurzman ID, Couto CG, Klausner J, Henry C, MacEwen EG. Cisplatin and doxorubicin combination chemotherapy for the treatment of canine osteosarcoma: a pilot study. Journal of veterinary internal medicine / American College of Veterinary Internal Medicine. 2000;14:495–498. doi: 10.1892/0891-6640(2000)014<0495:cadccf>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 25.Fenger JM, London CA, Kisseberth WC. Canine osteosarcoma: a naturally occurring disease to inform pediatric oncology. ILAR J. 2014;55:69–85. doi: 10.1093/ilar/ilu009. [DOI] [PubMed] [Google Scholar]
- 26.Withrow SJ, Powers BE, Straw RC, Wilkins RM. Comparative aspects of osteosarcoma. Dog versus man. Clin Orthop Relat Res. 1991:159–168. [PubMed] [Google Scholar]
- 27.Ragsdale BD, Madewell JE, Sweet DE. Radiologic and pathologic analysis of solitary bone lesions. Part II: periosteal reactions. Radiologic clinics of North America. 1981;19:749–783. [PubMed] [Google Scholar]
- 28.Punnia-Moorthy A. Evaluation of pH changes in inflammation of the subcutaneous air pouch lining in the rat, induced by carrageenan, dextran and Staphylococcus aureus. Journal of oral pathology. 1987;16:36–44. doi: 10.1111/j.1600-0714.1987.tb00674.x. [DOI] [PubMed] [Google Scholar]
- 29.Chakkalakal DA, Mashoof AA, Novak J, Strates BS, McGuire MH. Mineralization and pH relationships in healing skeletal defects grafted with demineralized bone matrix. Journal of biomedical materials research. 1994;28:1439–1443. doi: 10.1002/jbm.820281209. [DOI] [PubMed] [Google Scholar]
- 30.Bischoff DS, Zhu JH, Makhijani NS, Yamaguchi DT. Acidic pH stimulates the production of the angiogenic CXC chemokine, CXCL8 (interleukin-8), in human adult mesenchymal stem cells via the extracellular signal-regulated kinase, p38 mitogen-activated protein kinase, and NF-kappaB pathways. Journal of cellular biochemistry. 2008;104:1378–1392. doi: 10.1002/jcb.21714. [DOI] [PubMed] [Google Scholar]
- 31.Karin M, Greten FR. NF-kappaB: linking inflammation and immunity to cancer development and progression. Nature reviews Immunology. 2005;5:749–759. doi: 10.1038/nri1703. [DOI] [PubMed] [Google Scholar]
- 32.Gupta SC, Singh R, Pochampally R, Watabe K, Mo YY. Acidosis promotes invasiveness of breast cancer cells through ROS-AKT-NF-kappaB pathway. Oncotarget. 2014;5:12070–12082. doi: 10.18632/oncotarget.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Damaghi M, Tafreshi NK, Lloyd MC, Sprung R, Estrella V, Wojtkowiak JW, Morse DL, Koomen JM, Bui MM, Gatenby RA, Gillies RJ. Chronic acidosis in the tumour microenvironment selects for overexpression of LAMP2 in the plasma membrane. Nat Commun. 2015;6:8752. doi: 10.1038/ncomms9752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mueller L, Goumas FA, Affeldt M, Sandtner S, Gehling UM, Brilloff S, Walter J, Karnatz N, Lamszus K, Rogiers X, Broering DC. Stromal fibroblasts in colorectal liver metastases originate from resident fibroblasts and generate an inflammatory microenvironment. The American journal of pathology. 2007;171:1608–1618. doi: 10.2353/ajpath.2007.060661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gallagher PG, Bao Y, Prorock A, Zigrino P, Nischt R, Politi V, Mauch C, Dragulev B, Fox JW. Gene expression profiling reveals cross-talk between melanoma and fibroblasts: implications for host-tumor interactions in metastasis. Cancer research. 2005;65:4134–4146. doi: 10.1158/0008-5472.CAN-04-0415. [DOI] [PubMed] [Google Scholar]
- 36.Uccelli A, de Rosbo NK. The immunomodulatory function of mesenchymal stem cells: mode of action and pathways. Annals of the New York Academy of Sciences. 2015;1351:114–126. doi: 10.1111/nyas.12815. [DOI] [PubMed] [Google Scholar]
- 37.Ichimonji I, Tomura H, Mogi C, Sato K, Aoki H, Hisada T, Dobashi K, Ishizuka T, Mori M, Okajima F. Extracellular acidification stimulates IL-6 production and Ca(2+) mobilization through proton-sensing OGR1 receptors in human airway smooth muscle cells. American journal of physiology Lung cellular and molecular physiology. 2010;299:L567–L577. doi: 10.1152/ajplung.00415.2009. [DOI] [PubMed] [Google Scholar]
- 38.Rutkowski P, Kaminska J, Kowalska M, Ruka W, Steffen J. Cytokine and cytokine receptor serum levels in adult bone sarcoma patients: correlations with local tumor extent and prognosis. Journal of surgical oncology. 2003;84:151–159. doi: 10.1002/jso.10305. [DOI] [PubMed] [Google Scholar]
- 39.Xiao H, Chen L, Luo G, Son H, Prectoni JH, Zheng W. Effect of the cytokine levels in serum on osteosarcoma. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine. 2014;35:1023–1028. doi: 10.1007/s13277-013-1136-x. [DOI] [PubMed] [Google Scholar]
- 40.Erez N, Glanz S, Raz Y, Avivi C, Barshack I. Cancer associated fibroblasts express pro-inflammatory factors in human breast and ovarian tumors. Biochemical and biophysical research communications. 2013;437:397–402. doi: 10.1016/j.bbrc.2013.06.089. [DOI] [PubMed] [Google Scholar]
- 41.Joraschkewitz M, Depenbrock H, Freund M, Erdmann G, Meyer HJ, De Riese W, Neukam D, Hanauske U, Krumwieh M, Poliwoda H, et al. Effects of cytokines on in vitro colony formation of primary human tumour specimens. Eur J Cancer. 1990;26:1070–1074. doi: 10.1016/0277-5379(90)90054-w. [DOI] [PubMed] [Google Scholar]
- 42.Rui YF, Lui PP, Lee YW, Chan KM. Higher BMP receptor expression and BMP-2-induced osteogenic differentiation in tendon-derived stem cells compared with bone-marrow-derived mesenchymal stem cells. International orthopaedics. 2012;36:1099–1107. doi: 10.1007/s00264-011-1417-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McAdams TA, Miller WM, Papoutsakis ET. Variations in culture pH affect the cloning efficiency and differentiation of progenitor cells in ex vivo haemopoiesis. British journal of haematology. 1997;97:889–895. doi: 10.1046/j.1365-2141.1997.1372951.x. [DOI] [PubMed] [Google Scholar]
- 44.Rudisch A, Dewhurst MR, Horga LG, Kramer N, Harrer N, Dong M, van der Kuip H, Wernitznig A, Bernthaler A, Dolznig H, Sommergruber W. High EMT Signature Score of Invasive Non-Small Cell Lung Cancer (NSCLC) Cells Correlates with NFkappaB Driven Colony-Stimulating Factor 2 (CSF2/GM-CSF) Secretion by Neighboring Stromal Fibroblasts. PloS one. 2015;10:e0124283. doi: 10.1371/journal.pone.0124283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coussens LM, Werb Z. Inflammation and cancer. Nature. 2002;420:860–867. doi: 10.1038/nature01322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Karnoub AE, Dash AB, Vo AP, Sullivan A, Brooks MW, Bell GW, Richardson AL, Polyak K, Tubo R, Weinberg RA. Mesenchymal stem cells within tumour stroma promote breast cancer metastasis. Nature. 2007;449:557–563. doi: 10.1038/nature06188. [DOI] [PubMed] [Google Scholar]
- 47.Quante M, Tu SP, Tomita H, Gonda T, Wang SS, Takashi S, Baik GH, Shibata W, Diprete B, Betz KS, Friedman R, Varro A, et al. Bone marrow-derived myofibroblasts contribute to the mesenchymal stem cell niche and promote tumor growth. Cancer cell. 2011;19:257–272. doi: 10.1016/j.ccr.2011.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lu Y, Guan GF, Chen J, Hu B, Sun C, Ma Q, Wen YH, Qiu XC, Zhou Y. Aberrant CXCR4 and beta-catenin expression in osteosarcoma correlates with patient survival. Oncology letters. 2015;10:2123–2129. doi: 10.3892/ol.2015.3535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motrescu ER, Rio MC. Cancer cells, adipocytes and matrix metalloproteinase 11: a vicious tumor progression cycle. Biological chemistry. 2008;389:1037–1041. doi: 10.1515/BC.2008.110. [DOI] [PubMed] [Google Scholar]
- 50.Liu S, Ginestier C, Ou SJ, Clouthier SG, Patel SH, Monville F, Korkaya H, Heath A, Dutcher J, Kleer CG, Jung Y, Dontu G, et al. Breast cancer stem cells are regulated by mesenchymal stem cells through cytokine networks. Cancer research. 2011;71:614–624. doi: 10.1158/0008-5472.CAN-10-0538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Han XG, Du L, Qiao H, Tu B, Wang YG, Qin A, Dai KR, Fan QM, Tang TT. CXCR1 knockdown improves the sensitivity of osteosarcoma to cisplatin. Cancer letters. 2015;369:405–415. doi: 10.1016/j.canlet.2015.09.002. [DOI] [PubMed] [Google Scholar]
- 52.Nishimura K, Semba S, Aoyagi K, Sasaki H, Yokozaki H. Mesenchymal stem cells provide an advantageous tumor microenvironment for the restoration of cancer stem cells. Pathobiology : journal of immunopathology, molecular and cellular biology. 2012;79:290–306. doi: 10.1159/000337296. [DOI] [PubMed] [Google Scholar]
- 53.Kabashima-Niibe A, Higuchi H, Takaishi H, Masugi Y, Matsuzaki Y, Mabuchi Y, Funakoshi S, Adachi M, Hamamoto Y, Kawachi S, Aiura K, Kitagawa Y, et al. Mesenchymal stem cells regulate epithelial-mesenchymal transition and tumor progression of pancreatic cancer cells. Cancer science. 2013;104:157–164. doi: 10.1111/cas.12059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McLean K, Gong Y, Choi Y, Deng N, Yang K, Bai S, Cabrera L, Keller E, McCauley L, Cho KR, Buckanovich RJ. Human ovarian carcinoma-associated mesenchymal stem cells regulate cancer stem cells and tumorigenesis via altered BMP production. The Journal of clinical investigation. 2011;121:3206–3219. doi: 10.1172/JCI45273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Grisendi G, Spano C, Rossignoli F, D'Souza N, Golinelli G, Fiori A, Horwitz EM, Guarneri V, Piacentini F, Paolucci P, Dominici M. Tumor Stroma Manipulation By MSC. Current drug targets. 2016 doi: 10.2174/1389450117666160307143226. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.