Abstract
Lung cancer is the leading cause of cancer mortality. Great advances in non-small cell lung cancer therapy have been seen in the last decade, beginning with the success in treating lung cancer harboring EGFR mutations and ALK-gene rearrangements. The potential of these biomarker-driven therapies has propelled research in biomarker targeted approaches to the forefront of lung cancer research. The successful development of immunotherapeutic agents targeting PD-L1 and PD-1 with an associated non-genomic biomarker has opened a new front in the effort for targeted approaches. Although early-phase lung cancer studies have hinted at the potential to use biomarkers to select patients for allocation to treatment in the conduct of clinical trials, data from late-phase studies have tempered expectations. The data leave unclear the wisdom of routinely restricting enrollment on lung cancer clinical trials to biomarker restricted populations, particularly non-genomic biomarkers.
Keywords: biomarker, non-small cell lung cancer, clinical trials, targeted therapy, mutations
I. INTRODUCTION
Non-small cell lung cancer (NSCLC) is the most common type of lung cancer, constituting roughly 85% of all lung cancer diagnoses.1 Lung cancer is the leading cause of cancer death in the United States, accounting for approximately one quarter of all cancer deaths.2 The five-year survival rate for patients diagnosed with lung cancer is 17.4%.3 This high mortality rate can be attributed to advancedstage at diagnosis and a lack of effective treatment options. While platinum-based chemotherapy has shown benefit in late-stage patients, life expectancy is extended by only a few months on average, with significant toxicity.4 These poor outcomes, in addition to success in biomarker-driven therapies,5–7 have led to a push for biomarker targeted therapies in lung cancer and customization of treatments based on molecular abnormalities.
About a decade ago, there was tremendous enthusiasm that our refined understanding of molecular abnormalities would lead to a renaissance in the way lung cancer is treated.6 Genetic profiles of various cell lines with known treatment-specific sensitivities were analyzed by gene expression microarray, and matched to genetic expression observed in individuals’ malignant tissue.8,9 Information from these analyses formed the scientific basis of multiple lung cancer clinical trials in which treatment allocation was based on the results of microarray analysis. However, on further investigation, errors were identified in the data and results could not be reproduced.10 Ultimately, this discovery led to much of this research being retracted and the discontinuation of multiple clinical trials.11 Despite this setback, in a disease with limited options, the appetite for clinical trials in which biomarkers guided allocation to treatment arm persisted.
In the intervening period, the field of lung cancer has seen significant change, and is often considered a model for the use of biomarkers to guide therapy.6 However, while single gene abnormalities in tumors have significantly altered the therapeutic approach for many patients, biomarker-based trials have not been uniformly successful in guiding patients to appropriate therapies. The successes and failures of recent trials have highlighted the need for careful assessment of the premise that patients should be allocated to treatment arms on clinical trials based on biomarker expression.
II. TARGETING SINGLE GENE MUTATIONS HAS PROVEN EFFECTIVE FOR SELECTION OF AGENTS
Targeted therapies have induced dramatic responses in patients whose tumors harbor specific genetic abnormalities, namely, epidermal growth factor receptor (EGFR) mutations and anaplastic lymphoma kinase (ALK)-gene rearrangements. EGFR mutations are seen in over 10% of all NSCLC patients in some series, and are found more frequently in East Asian populations. Patients with these mutations experience disproportionate benefit with tyrosine kinase inhibitors (TKIs) such as gefitinib, erlotinib, and afatinib.5,6,12 These agents provide longer progression-free survival (PFS) and increased response rate (RR) when compared to conventional chemotherapy in this population. As a result, patients whose tumors harbor an EGFR mutation generally receive a TKI as initial treatment. Rearrangements involving the ALK gene occur in about 5% of NSCLC patients,5–7 and when compared to standard of care chemotherapy, ALK gene-rearranged patients treated with crizotinib have longer PFS (10.9 versus 7.0 months in frontline,13 7.7 versus 3.0 months in second line7) and higher RR (74% versus 45% in frontline; 65% versus 20% in second line).
Additional examples of targetable molecular abnormalities include ROS1 gene rearrangements and BRAF mutations. ROS-1 gene rearrangements, first discovered in glioblastoma, are seen in approximately 1–3% of NSCLC patients.14,15 Despite no U.S. Food and Drug Administration (FDA) approved treatment, crizotinib has shown promise in vitro and in early clinical studies for the treatment of ROS-1 gene rearrangements.14–16 These successes have led to crizotinib receiving the breakthrough therapy designation from the FDA for patients with ROS-1 rearrangements.17 BRAF mutations, though more commonly seen in melanoma, occur in about 4% of NSCLC patients.18 The BRAF inhibitor dabrafenib and the combination of dabrafenib plus trametinib have shown promising results.19,20
III. RECENT SUCCESS IN TRIALS INVOLVING IMMUNOTHERAPEUTIC AGENTS
Trials of anti-PD-1 (programmed cell death) and anti-PD-L1 (programmed cell death ligand) inhibitory antibodies have evaluated immunohistochemistry (IHC) to test for PD-L1 expression in tumors.21,22
Immune checkpoint inhibitors have recently been shown to improve clinical outcomes.23–25 These treatments target an inhibitory interaction involving T cells, allowing the T cells to recognize and destroy tumor cells. KEYNOTE-001 is a recent study that assessed the efficacy of pembrolizumab in patients. Along with assessing efficacy, the trial was designed to establish and validate a threshold level of PD-L1 expression that could be associated with a higher likelihood of clinical benefit. Four hundred ninety-five patients enrolled in this study, all of whom were treated with pembrolizumab. Response rate, PFS, and OS were all significantly greater in patients who had expression of PD-L1 in at least half of their cells.23 Although some cohorts enrolled patients with known PD-L1 expression, others enrolled without respect to PD-L1 expression or specifically patients without PD-L1 expression on their tumors.23
The POPLAR and CheckMate 017 and 057 studies compared inhibitors of the PD-1/PD-L1 checkpoint to docetaxel in a population that was not restricted based on any biomarker. The phase II POPLAR study enrolled previously treated, NSCLC patients who were stratified by PD-L1 expression using immunohistochemistry assays on tumor infiltrating immune cells (ICs) and tumor cells (TCs), histology, and previous therapy.2 Incrementally improved efficacy over docetaxel was seen with increasing PD-L1 expression.4 Similarly, in the phase III CheckMate 057 study, the PD-1 inhibitor nivolumab showed a greater overall survival (OS) and overall response rate (ORR) compared to docetaxel.25 Higher levels of PD-L1 expression were associated with greater benefit from nivolumab. Patients with PD-L1 expression in at least 10% of their tumor cells had a hazard ratio (HR) for OS of 0.4, as compared to 1.0 in those below that cutoff.25 Although nivolumab was superior to docetaxel at all levels of PD-L1 expression in the CheckMate 017 trial, there was again a suggestion of greater benefit in those above the 10% cut point (HR 0.5 versus 0.7) than below.26
Rather than restricting enrollment based on biomarker status, these studies enrolled a broad population, and used the resultant data to characterize the biomarker. Although KEYNOTE-001 allocated patients to some cohorts based on PD-L1 status, the distribution of PD-L1 expression of study participants was similar to the general population.
IV. OUTSIDE OF IMMUNE CHECKPOINT INHIBITORS, SELECTION BY IHC HAS NOT LED TO IMPROVED OUTCOMES IN BIOMARKER-DRIVEN THERAPIES
Excision repair cross-complementation group 1 (ERCC1) is felt to repair lesions formed by cisplatin-DNA adducts, and was proposed to interfere with the efficacy of cisplatin-based therapy. Of the four ERCC1 isoforms (201, 202, 203, 204), only ERCC1-202 serves this repair function. In 2006, Olaussen et al.27 retrospectively tested hundreds of tissue samples from patients enrolled in the International Adjuvant Lung Cancer Trial using IHC to test for ERCC1 expression. An association between ERCC1-negative tumors and extended survival with cisplatin-based adjuvant chemotherapy was identified.28 However, five years after the initial publication, when tissue was retested with the same 8F1 antibody, 77% of samples, as opposed to the original 44%, had positive results.19 Additionally, the investigators could not differentiate the functional ERCC1-202 isoform from the other isoforms. In a subsequent clinical trial in which patients were randomized to chemotherapy treatments driven by ERCC1 and ribonucleotide reductase M1 (RRM1) expression versus a control arm of carboplatin and gemcitabine (Fig. 1), no benefit was found from biomarker-driven therapy in either PFS (6.9 versus 6.1 months) or OS (11.3 versus 11 months).29
FIG. 1.
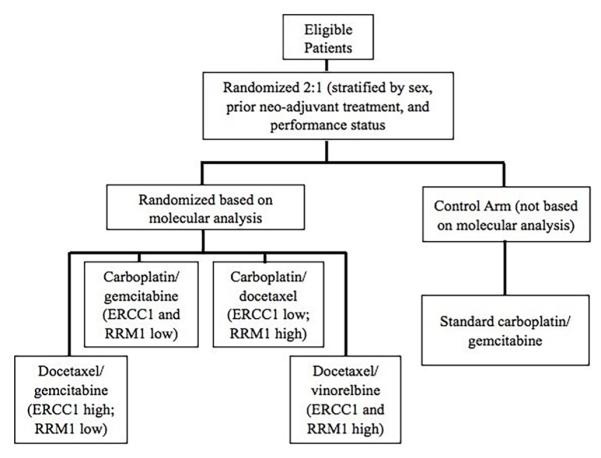
Study diagram for the randomized phase III multicenter trial of RRM1 & ERCC1 directed customized chemotherapy versus standard of care for first line treatment of patients with advanced non-small-cell lung cancer
IHC analysis was also used to evaluate whether high EGFR expression was predictive of improved outcome for NSCLC patients receiving cetuximab, an antibody targeting EGFR.30 In the First-Line ErbituX (FLEX) study, high EGFR expression was defined as an H score of 200 or above, while low expression was any score below 200. An association between increased overall survival and high EGFR expression was found in patients receiving cetuximab and chemotherapy.30 However, these results could not be replicated in subsequent studies with other chemotherapy backbones, in which a high H score was not found to be predictive of a favorable outcome with chemotherapy and cetuximab.31
Many trials of agents targeting MET protooncogene (MET), a proto-oncogene that codes for the hepatocyte growth factor receptor, used IHC to analyze MET overexpression as a potential predictor of outcome.32 A phase II study of onartuzumab in combination with erlotinib showed a numerically unfavorable HR for the entire population. However, patients with a MET IHC score of +2 or +3 showed significantly improved PFS and OS compared to patients receiving erlotinib alone.32,33 Despite the success in the phase II trial, several NSCLC studies were stopped, including a phase III study of erlotinib with or without onartuzumab in patients with this high degree of MET staining due to lack of efficacy.34
V. STUDIES SELECTING TREATMENT BASED ON MULTIPLE BIOMARKERS HAVE BEEN COMPLETED
Well-run multiple therapy studies have been conducted in which treatment selection was based on multiple biomarkers. The BATTLE (Biomarker-Integrated Approaches of Targeted Therapy for Lung Cancer Elimination) I and II studies highlight these efforts, the first of which has been completed and reported. From November 2006 to October 2011, 341 patients enrolled in BATTLE I, the first completed prospective, biopsy-mandated, biomarker-based, adaptively randomized clinical study in patients with pretreated, advanced NSCLC.35,36 Treatment arms consisted of erlotinib, vandetanib [vascular endothelial growth factor (VEGFR), EGFR, and RET (rearranged during transfection) proto-oncogene inhibitor], erlotinib plus bexarotene (retinoid × receptor inhibitor), and sorafenib (RAF and VEGFR inhibitor). The primary endpoint of the study was overall eight-week disease control rate (DCR) with a secondary endpoint of PFS.
Patients were enrolled under an umbrella protocol, and after a mandated core biopsy and associated molecular testing, the first 97 patients were randomly assigned to one of the four treatment arms in an equal ratio (excluding patients with prior erlotinib treatment who were excluded from erlotinib-containing arms). The subsequent 158 patients were adaptively randomized to study arms according to a Bayesian adaptive algorithm, informed by associations between the biomarker groups and the DCR for the first 97 patients. This adaptive randomization was continually updated to allow for more appropriate biomarker prediction. The biomarker profiles used to adaptively randomize patients to the most effective study drug consisted of EGFR mutation/copy number, KRAS/BRAF mutation, VEGF/VEGFR-2 expression, RXRs/Cyclin D1 (CCND1) expression, and CCND1 copy number. In 244 patients eligible for analysis, overall eight-week DCR was 46% and the median PFS was 1.9 months (95% CI, 1.8–2.4).
BATTLE successfully employed a novel research design demonstrating (i) the feasibility of performing biopsies and real-time biomarker analyses in previously treated lung cancer patients, (ii) identifying interactions between the treatments and markers, and (iii) confirming the prespecified hypotheses of treatment efficacy in the presence of individual markers related to the treatments’ mechanism of action.35 It incorporated four different treatment arms with the cooperation of four different pharmaceutical companies within a single study, leading to a significant decrease in screen fails.36
Unfortunately, several of the prespecified markers were ultimately found to have little, if any, predictive value in optimizing treatment selection.36 The authors concluded that, “biomarker groups were less predictive than were individual biomarkers, which diluted the impact of strong predictors in determining treatment probabilities.”36 The greatest example of this was erlotinib in the EGFR arm. Despite extensive data correlating erlotinib response with EGFR mutations, erlotinib did not meet the predicted efficacy for the EGFR group. In fact, the highest DCR for erlotinib (40%) was in the VEGF/VEGFR 2 biomarker group. This example underscores the fact that even in adaptive trial designs like BATTLE, assumptions are made, and failures of these assumptions can lead to the same pitfalls seen with more traditional study designs.
VI. ONGOING CLINICAL TRIALS WILL EXPAND ON THE BATTLE-1 APPROACH
The BATTLE-2 trial contains similar aims to BAT-TLE-I. Patients with an ALK gene rearrangement or an EGFR mutation are excluded unless they previously failed treatment with an inhibitor targeting the mutation. The primary endpoint is again DCR at eight weeks and the secondary endpoint is PFS. Patients with at least one prior chemotherapy treatment are allocated to erlotinib alone, erlotinib with the Akt inhibitor MK-2206, the MEK inhibitor selumetinib with MK-2206, or sorafenib alone based on their molecular profile. Patients in the first cohort are adaptively randomized into one of the four treatment arms based on eight-week DCR and KRAS mutation status. Patients in the second cohort are assigned by a refined adaptive randomization based on the most predictive biomarkers tested on patients in the first cohort. Patients that progress are allowed to reenroll into a different randomized arm.
However, at this time, erlotinib is not as promising an agent in EGFR wild-type patients as when the study was designed. The Tarceva Italian Lung Optimization trial (TAILOR) trial was a phase III trial randomizing to erlotinib or docetaxel as second line treatment for NSCLC patients without an EGFR mutation. Docetaxel led to improved outcomes compared to erlotinib.37 Less data are available regarding the other arms, and data are eagerly awaited on results of this trial.
VII. LARGER MULTICENTER MULTIARM STUDIES ARE PLANNED WITH TREATMENT ALLOCATION BASED ON BIOMARKERS
The phase II/III “master protocol” is directed at squamous cell disease, a notoriously difficult group of patients to treat.8 EGFR and ALK mutations are rare in lung SQCC;38 however, a broad pathologic assessment has elucidated potential therapeutic targets.38 PIK3CA copy number increases or mutations have been identified in approximately one third of patients.39 CDKN2A, a tumor suppressor protein, is inactivated in up to 72% of cases.40,41 Other studies have discovered correlations between HGF/c-Met overexpression and progression, as well as an association between FGFR amplification and mutations with tumor growth and survival.42,43 Based on these genetic markers, the original master protocol was devised to include five different substudies.44 Eligible patients are either assigned to one of four biomarker-specific substudies or to the “nonmatch” therapy substudy. Within each substudy, patients are then randomized to a biomarker-driven targeted therapy or standard of care (Soc) therapy.45
The master protocol provides a process by which a large number of patients with advanced-stage lung SQCC are screened for enrollment.44 The primary objective of the protocol is to evaluate PFS between targeted therapy (TT) versus SoC docetaxel with other objectives to evaluate toxicities associated with TT versus SoC. Each arm opens and closes independent of other arms.44 Substudies can also be added to the protocol as continual discovery of drug-biomaker pairs occur.45 During the phase II portion, positive results at “rolling” interim analysis will determine if a protocol arm enters a phase III portion. At the phase III stage, the primary objective will be OS for TT versus SoC within each biomarker-defined subgroup with other objectives including PFS, toxicities, and RR with TT versus SoC. It is estimated that 500–1000 patients will be screened for enrollment per year.45
Before a patient receives treatment, a common biomarker profile is generated in a CLIA certified laboratory, and results are expected to be obtained within approximately two weeks. If the patient is positive for certain specified mutational targets (PI3K, CDK4/6, FGFR, HGF), the patient will go on the corresponding substudy, and be randomized to either the targeted therapy drug or chemotherapy.45 In the original protocol, if the patient was a nonmatch for those mutations, the patient was placed in the nonmatch substudy and randomized to receive either MEDI4736 (an anti-PD- L1) or chemotherapy.45
Size and scope are major assets in the search for suitable treatments for lung SQCC. The high potential rate (70%) of patients selected for a biomarker-driven trial, the large number of patients screened, and the five trials promise a decreased screen failure rate. Another advantage involves the structure of this umbrella trial, which enables multiple experimental agents to be tested simultaneously. If there appears to be a significant treatment response in a phase II trial, phase III trials will be implemented quickly, which accelerates drug and biomarker developments.
Yet, there are also suppositions regarding the function of biomarkers in various portions of the trial, such as how specific biomarkers can predict response to treatment. The reliance of lung SQCC on these individual mutations has yet to be fully understood. Additionally, anti-PD-L1 treatment has exhibited robust response rates that may diminish the perceived impact of the results of other drug treatments.27
In response to the recent approval of the immunotherapy, nivolumab, for patients diagnosed with SQCC, the master protocol study team has implemented major revisions.46 Now, the docetaxel arm of the nonmatch substudy has been closed, and the trial is now open to patients on their second or greater lines of therapy.46
VIII. CONCLUSIONS
The progress seen in both EGFR mutant and ALK-gene rearranged NSCLC has provided a great deal of excitement regarding biomarker-driven research in NSCLC. The advances in using immune check-point inhibitors have led to further enthusiasm. However, despite increasing scientific knowledge, many high-profile studies in which enrollment has been based on one non-genomic biomarker or several biomarkers have not had an impact on disease management. Though the research community continues to design exciting and ambitious trials in which treatment allocation is based on biomarkers, the limited success of these trials suggests that we must continually evaluate the approach to assure that it is generating the best options for patients and the most robust research results possible.
ACKNOWLEDGMENT
This work was supported by NIH Grant No. 1K23CA149079 awarded to Dr. Edward Garon.
ABBREVIATIONS
- ALK
anaplastic lymphoma kinase
- CCND1
cyclin D1
- DCR
death control rate
- EGFR
epidermal growth factor receptor
- ERCC1
excision repair cross-complementation group 1
- IC
immune cell
- MET
MET proto-oncogene
- NSCLC
non-small cell lung cancer
- PFS
progression-free survival
- PD-1
programmed cell death
- PD-L1
programmed cell death ligand
- ROS-1
ROS proto-oncogene 1
- RR
response rate
- RRM1
ribonucleotide reductase M1
- SQCC
squamous cell carcinoma
- TC
tumor cell
- TKI
tyrosine-kinase inhibitor
- TT
targeted therapy
REFERENCES
- 1.Roy S, Herbst J, Heymach V. Lung cancer. N Engl J Med. 2008;359:1367–80. doi: 10.1056/NEJMra0802714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R, Miller KD, Jemal A. Cancer statistics, 2015. CA: Cancer J Clin. 2015;65(1):5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 3.Howlader N, Noone AM, Krapcho M, Garshell J, Miller D, Altekruse SF, Kosary CL, Yu M, Ruhl J, Tatalovich Z, Mariotto A, Lewis DR, Chen HS, Feuer EJ, Cronin KA. SEER cancer statistics review, 1975–2012. National Cancer Institute; Maryland: 2015. [Google Scholar]
- 4.Schiller JH, Harrington D, Belani CP, Langer C, Sandler A, Krook J, Zhu J, Johnson DH. Comparison of four chemotherapy regimens for advanced non-small cell lung cancer. N Engl J Med. 2002;346(2):92–8. doi: 10.1056/NEJMoa011954. [DOI] [PubMed] [Google Scholar]
- 5.Mok TS, Wu Y, Thongprasert S, Yang C, Chu D, Saijo N, Sunpaweravong P, Han B, Margono B, Ichinose Y, Nishiwaki Y, Ohe Y, Yang JJ, Chewaskulyong B, Jiang H, Duffield EL, Watkins CL, Armour AA, Fukuoka M. Gefitinib or carboplatin paclitaxel in pulmonary adenocarcinoma. N Engl J Med. 2009;361(10):947–57. doi: 10.1056/NEJMoa0810699. [DOI] [PubMed] [Google Scholar]
- 6.Hensing T, Chawla A, Batra R, Salgia R. A personalized treatment for lung cancer: Molecular pathways, targeted therapies, and genomic characterization. Syst Anal Hum Multigene Disorders. 2014;799:85–117. doi: 10.1007/978-1-4614-8778-4_5. [DOI] [PubMed] [Google Scholar]
- 7.Shaw AT, Kim DW, Nakagawa K, Seto T, Crinó L, Ahn M, De Pas T, Besse B, Solomon B, Blackhall F, Wu YL, Thomas M, O’Byrne K, Moro-Sibilot D, Camidge R, Mok T, Hirsh V, Riely GJ, Lyer S, Tassell V, Polli A, Wilner DK, Jänne Crizotinib versus chemotherapy in advancedALK positive lung cancer. N Engl J Med. 2013;368(25):2385–94. doi: 10.1056/NEJMoa1214886. [DOI] [PubMed] [Google Scholar]
- 8.Ledford H. ‘Master protocol’ aims to revamp cancer trials. Nature. 2013;498(7453):146–7. doi: 10.1038/498146a. [DOI] [PubMed] [Google Scholar]
- 9.Potti A, Mukherjee S, Petersen R, Dressman HK, Bild A, Koontz J, Kratzke R, Watson MA, Kelley M, Ginsburg GS, West M, Harpole DH, Nevins JR. A genomic strategy to refineprognosis in early-stage non–small-cell lung cancer. N Engl J Med. 2006;355(6):570–80. doi: 10.1056/NEJMoa060467. [DOI] [PubMed] [Google Scholar]
- 10.Potti A, Dressman HK, Bild A, Riedel RF, Chan G, Sayer R, Cragun J, Cottrill H, Kelley MJ, Petersen R, Harpole D, Marks J, Berchuck A, Ginsburg GS, Febbo P, Lancaster J, Nevins JR. Genomic signatures to guide the use of chemotherapeutics. Nat Med. 2006;12(11):1294–300. doi: 10.1038/nm1491. [DOI] [PubMed] [Google Scholar]
- 11.Hutson S. Data handling errors spur debate over clinical trial. Nat Med. 2010;16(6):618. doi: 10.1038/nm0610-618a. [DOI] [PubMed] [Google Scholar]
- 12.Sequist LV, Yang JC, Yamamoto N, O’Byrne K, Hirsh V, Mok T, Geater SL, Orlov S, Tsai CM, Boyer M, Su WC, Bennouna J, Kato T, Gorbunova V, Lee KH, Shah R, Massey D, Zazulina V, Shahidi M, Schuler M. Phase III study of afatinib or cisplatinplus pemetrexed in patients with metastatic lung adenocarcinoma with EGFR mutations. J Clin Oncol. 2013;31(27):3327–34. doi: 10.1200/JCO.2012.44.2806. [DOI] [PubMed] [Google Scholar]
- 13.Mok T, Kim D, Wu Y, Solomon B, Nakagawa K, Mekhail T. First-line crizotinib versus pemetrexed–cisplatin or pemetrexed–carboplatin in patients (pts) with advanced ALK-positive non-squamous non-small cell lung cancer (NSCLC): Results of a phase III study (PROFILE 1014) J Clin Oncol. 2014;32(5Suppl) [Google Scholar]
- 14.Oxnard GR, Binder A, Janne PA. New targetable oncogenes in non-small-cell lung cancer. J Clin Oncol. 2013 Mar 10;31(8):1097–104. doi: 10.1200/JCO.2012.42.9829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bergethon K, Shaw AT, Ou SH, Katayama R, Lovly CM, McDonald NT, Massion PP, Siwak-Tapp C, Gonzalez A, Fang R, Mark EJ, Batten JM, Chen H, Wilner KD, Kwak EL, Clark JW, Carbone DP, Ji H, Engelman JA, Mino-Kenudson M, Pao W, Iafrate AJ. ROS1 rearrange-ments define a unique molecular class of lung cancers. J Clin Oncol. 2012;30(8):863–70. doi: 10.1200/JCO.2011.35.6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ou S, Bang Y, Camidge DR, Riely G, Salgia R, Shapiro G, Solomon BJ, Engelman JA, Kwak EL, Clark JW, Tye L, Wilner KD, Stephenson P, Varella-Garcia M, Bergethon K, Iafrate AJ, Shaw AT. Efficacy and safety of crizotinib in patientswith advanced ROS1-rearranged non-small cell lung cancer (NSCLC) J Clin Oncol. 2013;31(Suppl) abstr.8032. [Google Scholar]
- 17.Pfizer Receives U.S FDA Breakthrough Therapy Designation for XALKORI (crizotinib) For The Treatment Of Patients With ROS1-Positive Non-Small Cell Lung Cancer. Pfizer. 2015 Apr; [accessed 2015 August 5]. http://www.pfizer.com/news/press-release/press-release-detail/pfizer_receives_u_s_fda_breakthrough_therapy_designation_for_xalkori_crizotinib_for_the_treatment_of_patients_with_ros1_positive_non_small_cell_lung_cancer.
- 18.Cardarella S, Ogino A, Nishino M, Butaney M, Shen J, Lydon C, Yeap BY, Sholl LM, Johnson BE, Jänne PA. Clinical, pathologic, and biologic features associated with BRAF mutations in non-small cell lung cancer. Clin Cancer Res. 2013;19(16):4532–40. doi: 10.1158/1078-0432.CCR-13-0657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Planchard D, Mazieres J, Riely G, Rudin CM, Barlesi F, Quoix E, Souquet PJ, Socinski MA, Switzky J, Ma B, Goodman VL, Carson SW, Curtis CM, Streit MRW, Johnson BE. Interim results of phase II studyBRF113928 of dabrafenib in BRAF V600E mutation–positive non-small cell lung cancer (NSCLC)patients. J Clin Oncol. 2013;31(Suppl.):8009. [Google Scholar]
- 20.Planchard D, Groen HJM, Kim TM, Rigas J, Souquet PJ, Baik C, Barlesi F, Mazieres J, Quoix E, Curtis CM, Mookerjee B, Pandite L, Tucker C, D’Amelio A, Johnson BE. Interim results of a phase II study of the BRAF inhibitor (BRAFi) dabrafenib (D) in combination with the MEK inhibitor trametinib (T) in patients (pts) with BRAF V600E mutated (mut) metastatic non-small cell lung cancer (NSCLC) J Clin Oncol. 2015;33(suppl) abstr. 8006. [Google Scholar]
- 21.Topalian SL, Hodi FS, Brahmer JR, Gettinger SN, Smith DC, McDermott DF, Powderly JD, Carvajal RD, Sosman JA, Atkins MB, Leming PD, Spigel DR, Antonia SJ, Horn L, Drake CG, Pardoll DM, Chen L, Sharfman WH, Anders RA, Taube JM, McMiller TL, Xu H, Korman AJ, Jure-Kunkel M, Agrawal S, McDonald D, Kollia GD, Gupta A, Wigginton JM, Sznol M. Safety, activity, and immune correlates of anti–PD-1 antibody in cancer. N Engl J Med. 2012;366(26):2443–54. doi: 10.1056/NEJMoa1200690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Soria J, Cruz C, Bahleda R, Delord J, Horn L, Herbst R, Spigel D, Mokatrin A, Fine G, Gettinger S. Clinical activity, safety and biomarkers of PD-L1 blockade in non-small cell lung cancer (NSCLC): additional analyses from a clinical study of theengineered antibody MPDL3280A (anti-PDL1) Eur J Cancer. 2013;49(2 suppl):S3798. [Google Scholar]
- 23.Garon EB, Rizvi NA, Hui R, Leighl N, Balmanoukian AS, Eder JP, Patnaik A, Aggarwal C, Gubens M, Horn L, Carcereny E, Ahn MJ, Felip E, Lee JS, Hellmann MD, Hamid O, Goldman JW, Soria JC, Dolled-Filhart M, Rutledge RZ, Zhang J, Lunceford JK, Rangwala R, Lubiniecki GM, Roach C, Emancipator K, Gandhi L. Pembrolizumab for the treatment of non–small-cell lung cancer. N Engl J Med. 2015;372(21):2018–28. doi: 10.1056/NEJMoa1501824. [DOI] [PubMed] [Google Scholar]
- 24.Spira AI, Park K, Mazieres J, Vansteenkiste JF, Rittmeyer A, Ballinger M, Waterkamp D, Kowanetz M, Mokatrin A, Fehrenbacher L. Efficacy, safety and predictive biomarker results from a randomized phase II study comparing MPDL3280A vs docetaxel in 2L/3L NSCLC (POPLAR) J Clin Oncol. 2015;33(suppl) abstr. 8010. [Google Scholar]
- 25.Paz-Ares L, Horn L, Borghaei H, Spigel DR, Steins M, Ready N, Chow LQM, Voke EE, Felip E, Holgado E, Barlesi F, Kohlhaeufl M, Rodriguez O, Burgio MA, Fayette J, Gettinger SN, Harbison C, Dorange C, Finckenstein FG, Brahmer JR. Phase III, randomized trial (CheckMate 057) of nivolumab (NIVO) versus docetaxel (DOC) in advanced non-squamous cell (non-SQ) non-small cell lung cancer (NSCLC) J Clin Oncol. 2015;33(suppl) abstr. LBA109. [Google Scholar]
- 26.Spigel DR, Reckamp KL, Rizvi NA, Poddubskaya E, West HJ, Eberhardt WEE, Baas P, Antonia SJ, Pluzanski A, Vokes EE, Holgado E, Waterhouse DM, Ready N, Gainor JF, Aren OR, Horn L, Paz-Ares L, Baudelet C, Lestini BJ, Brahmer JR. A phase III study (CheckMate 017) of nivolumab (NIVO; anti-programmed death-1 [PD-1]) vs docetaxel (DOC) in previously treated advanced or metastatic squamous (SQ) cell non-small cell lung cancer (NSCLC) J Clin Oncol. 2015;33(suppl) abstr. 8009. [Google Scholar]
- 27.Olaussen KA, Dunant A, Fouret P, Brambilla E, André F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper MM, Stahel R, Sabatier L, Pignon JP, Tursz T, Le Chevalier T, Soria JC. DNA repair by ERCC1 in non–small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med. 2006;355(10):983–91. doi: 10.1056/NEJMoa060570. [DOI] [PubMed] [Google Scholar]
- 28.Friboulet L, Olaussen KA, Pignon J, Shepherd FA, Tsao M, Graziano S, Kratzke R, Douillard JY, Seymour L, Pirker R, Filipits M, Andre F, Solary E, Ponsonnailles F, Robin A, Stoclin A, Dorvault N, Commo F, Adam J, Vanhecke E, Saulnier P, Thomale J, Le Chevalier T, Dunant A, Rousseau V, Le Teuff G, Brambilla E, Soria JC. ERCC1 isoform expression and DNA repair in non–small-cell lung cancer. N Engl J Med. 2013;368(12):1101–10. doi: 10.1056/NEJMoa1214271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bepler G, Williams C, Schell MJ, Chen W, Zheng Z, Simon G, Gadgeel S, Zhao X, Schreiber F, Brahmer J, Chiappori A, Tanvetyanon T, Pinder-Schenck M, Gray J, Haura E, Antonia S, Fischer JR. Randomized international phase III trial of ERCC1 and RRM1 expression-based chemotherapy versus gemcitabine/carboplatin in advancednon-small-cell lung cancer. J Clin Oncol. 2013;31(19):2404–12. doi: 10.1200/JCO.2012.46.9783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pirker R, Pereira JR, von Pawel J, Krzakowski M, Ramlau R, Park K, de Marinis F, Eberhardt WE, Paz-Ares L, Störkel S, Schumacher KM, von Heydebreck A, Celik I, O’Byrne KJ. EGFR expression as a predictor ofsurvival for first-line chemotherapy plus cetuximab in patients with advanced non-small-cell lung cancer: Analysis of data from the phase 3 FLEX study. Lancet Oncol. 2012;13(1):33–42. doi: 10.1016/S1470-2045(11)70318-7. [DOI] [PubMed] [Google Scholar]
- 31.Kim ES, Neubauer M, Cohn A, Schwartzberg L, Garbo L, Caton J, Robert F, Reynolds C, Katz T, Chittoor S, Simms L, Saxman S. Docetaxel or pemetrexed with orwithout cetuximab in recurrent or progressive non-small-cell lung cancer after platinum-based therapy: Aphase 3, open-label, randomised trial. Lancet Oncol. 2013;14(13):1326–36. doi: 10.1016/S1470-2045(13)70473-X. [DOI] [PubMed] [Google Scholar]
- 32.Gelsomino F, Facchinetti F, Haspinger E, Garassino M, Trusolino L, De Braud F, Tiseo M. Targeting the MET gene for the treatment of non-small-cell lung cancer. Crit Rev Oncol. 2014;89(2):284–99. doi: 10.1016/j.critrevonc.2013.11.006. [DOI] [PubMed] [Google Scholar]
- 33.Spigel DR, Edelman MJ, Mok T, O’Byrne K, Paz-Ares L, Yu W, Rittweger K, Thurm H. Treatment rationale study design for the MetLung trial: A randomized, double-blind phase III study of onartuzumab (MetMAb) in combination with erlotinib versus erlotinib alone in patients who have received standard chemotherapy for stage IIIB orIV met-positive non-small-cell lung cancer. Clin Lung Cancer. 2012;13(6):500–4. doi: 10.1016/j.cllc.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 34.Spigel D, Edelman M, O’Byrne K, Paz-Ares L, Shames D, Yu W, Paton VE, Mok T. Onartuzumab plus erlotinib versuserlotinib in previously treated stage IIIb or IV NSCLC: results from the pivotal phase III randomized,multicenter, placebo-controlled METLung (OAM4971g) global trial. J Clin Oncol. 2014;32(5 Suppl):367–485. [Google Scholar]
- 35.Kim ES, Herbst RS, Wistuba II, Lee JJ, Blumenschein GR, Jr, Tsao A, Stewart DJ, Hicks ME, Erasmus J, Jr, Gupta S, Alden CM, Liu S, Tang X, Khuri FR, Tran HT, Johnson BE, Heymach JV, Mao L, Fossella F, Kies MS, Papadimitrakopoulou V, Davis SE, Lippman SM, Hong WK. The BATTLE trial: personalizing therapy for lung cancer. Cancer Discov. 2011;1(1):44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin EH, Anderson KM, Gause CK. The BATTLE trial: A bold step toward improving the efficiency of biomarker-based drug development. Cancer Discov. 2011;1(1):17–20. doi: 10.1158/2159-8274.CD-11-0036. [DOI] [PubMed] [Google Scholar]
- 37.Garassino MC, Martelli O, Broggini M, Farina G, Veronese S, Rulli E, Bianchi F, Bettini A, Longo F, Luca M, Tomirotti M, Marabese M, Ganzinelli M, Lauricella C, Labianca R, Floriani I, Giaccone G, Torri V, Scanni A, Marsoni S. Erlotinib versus docetaxel assecond-line treatment of patients with advanced non-small-cell lung cancer and wild-type EGFR tumours (TAI-LOR): a randomised controlled trial. Lancet Oncol. 2013;14(10):981–8. doi: 10.1016/S1470-2045(13)70310-3. [DOI] [PubMed] [Google Scholar]
- 38.Rekhtman N, Paik PK, Arcila ME, Tafe LJ, Oxnard GR, Moreira AL, Travis WD, Zakowski MF, Kris MG, Ladanyi M. Clarifying the spectrum of driver oncogene mutations in biomarker-verified squamous carcinoma of lung: lack of EGFR/KRAS and presence of PIK3CA/AKT1 mutations. Clin Cancer Res. 2012;18(4):1167–76. doi: 10.1158/1078-0432.CCR-11-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yamamoto H, Shigematsu H, Nomura M, Lockwood WW, Sato M, Okumura N, Suzuki M, Wistuba II, Fong KM, Lee H, Toyooka S, Date H, Lam WL, Minna JD, Gazdar AF. PIK3CA mutations and copy number gains in human lung cancers. Cancer Res. 2008;68(17):6913–21. doi: 10.1158/0008-5472.CAN-07-5084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Choi Y, Anders L. Signaling through cyclin D-dependent kinases. Oncogene. 2014;33(15):1890–903. doi: 10.1038/onc.2013.137. [DOI] [PubMed] [Google Scholar]
- 41.Cancer Genome Atlas Research Network Comprehensive genomic characterization of squamous cell lung cancers. Nature. 2012;489(7417):519–25. doi: 10.1038/nature11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gumustekin M, Kargi A, Bulut G, Gozukizil A, Ulukus C, Oztop I, Atabey N. HGF/c-met overexpressions, but not met mutation, correlates with progression of non-small cell lung cancer. Pathol Oncol Res. 2012;18(2):209–18. doi: 10.1007/s12253-011-9430-7. [DOI] [PubMed] [Google Scholar]
- 43.Weiss J, Sos ML, Seidel D, Peifer M, Zander T, Heuckmann JM, Ullrich RT, Menon R, Maier S, Soltermann A, Moch H, Wagener P, Fischer F, Heynck S, Koker M, Schöttle J, Leenders F, Gabler F, Dabow I, Querings S, Heukamp LC, Balke-Want H, Ansén, Rauh D, Baessmann I, Altmüller J, Wainer Z, Conron M, Wright G, Russell P, Solomon B, Brambilla E, Brambilla C, Lorimier P, Sollberg S, Brustugun OT, Engel-Riedel W, Ludwig C, Petersen I, Sänger J, Clement J, Grown H, Timens W, Sietsma H, Thunnissen E, Smit E, Heide-man D, Cappuzzo F, Ligorio C, Damiani S, Hallek M, Beroukhim R, Pao W, Klebl B, Baumann M, Buettner R, Ernestus K, Stoelben E, Wolf J, Nürnberg P, Perner S, Thomas RK. Frequent and focal FGFR1amplification associates with therapeutically tractable FGFR1 dependency in squamous cell lung cancer. Sci Transl Med. 2010;2(62):62ra93. doi: 10.1126/scitranslmed.3001451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Papadimitrakopoulou V, Gandara D, Hirsch F, Sigal E, Redman M, Allen J, Mack P, Wistuba I, Herbst R. Phase ii/iii biomarker driven master protocol for second line therapy of squamous cell lung cancer (scca) J Thoracic Oncol. 2013;8(2 suppl):S368. [Google Scholar]
- 45.Herbst RS, Gandara DR, Hirsch FR, Redman MW, LeB-lanc M, Mack PC, Schwartz LH, Vokes E, Ramalingam SS, Bradley JD, Sparks D, Zhou Y, Miwa C, Miler MA, Yelensky R, Li Y, Allen JD, Sigal EV, Wholley D, Sigman CC, Blumenthal GM, Malik S, Kelloff GJ, Abrams JS, Blanke CD, Papadimitrakopoulou VA. Lung master protocol (lung-MAP)—a biomaker-driven protocol for accelerating development of therapies for squamous cell lung cancer: SWOG S1400. Clin Cancer Res. 2015;21(7):1514–1524. doi: 10.1158/1078-0432.CCR-13-3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Southwest Oncology Group Summary of S1400 Revision 2 Changes (Protocol Version Dated 4/22/15) [accessed 2015 August 5]. http://www.lungmap.org/sites/default/files/S1400%20Lung-MAP%20--%20Summary%20of%20Revision%202%20Changes%206-8-15.pdf.


