Abstract
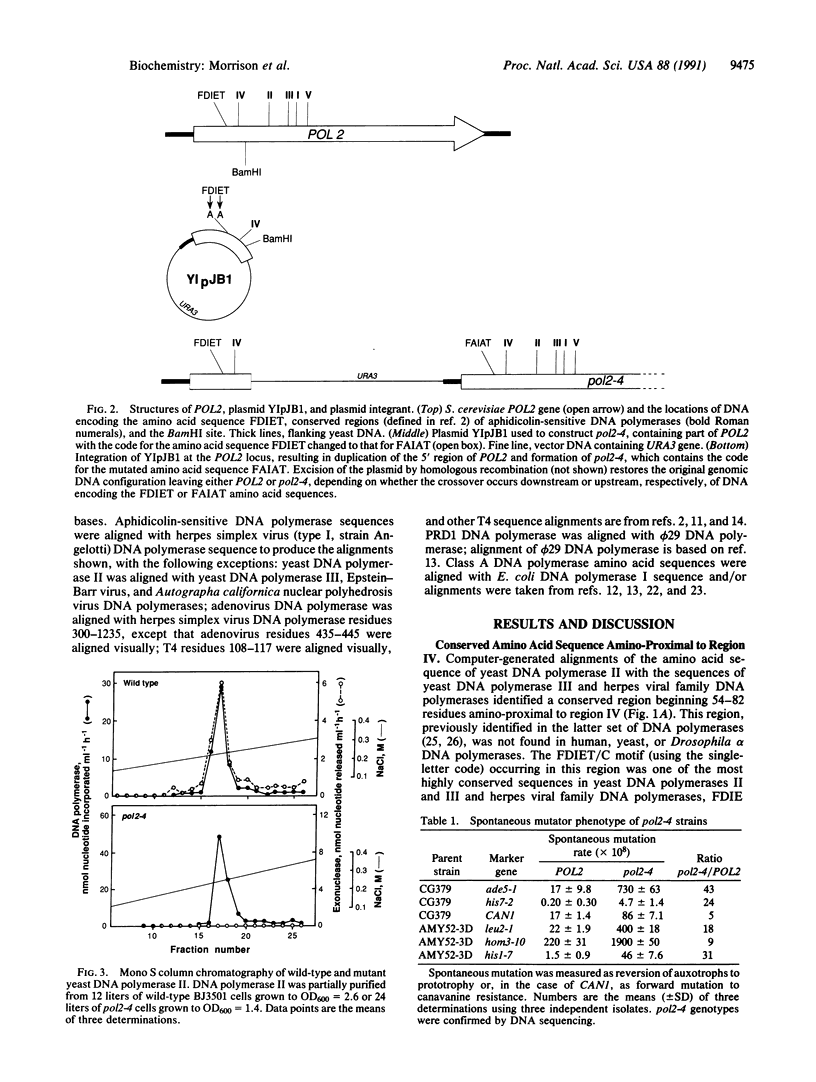
We have identified an amino-proximal sequence motif, Phe-Asp-Ile-Glu-Thr, in Saccharomyces cerevisiae DNA polymerase II that is almost identical to a sequence comprising part of the 3'----5' exonuclease active site of Escherichia coli DNA polymerase I. Similar motifs were identified by amino acid sequence alignment in related, aphidicolin-sensitive DNA polymerases possessing 3'----5' proofreading exonuclease activity. Substitution of Ala for the Asp and Glu residues in the motif reduced the exonuclease activity of partially purified DNA polymerase II at least 100-fold while preserving the polymerase activity. Yeast strains expressing the exonuclease-deficient DNA polymerase II had on average about a 22-fold increase in spontaneous mutation rate, consistent with a presumed proofreading role in vivo. In multiple amino acid sequence alignments of this and two other conserved motifs described previously, five residues of the 3'----5' exonuclease active site of E. coli DNA polymerase I appeared to be invariant in aphidicolin-sensitive DNA polymerases known to possess 3'----5' proofreading exonuclease activity. None of these residues, however, appeared to be identifiable in the catalytic subunits of human, yeast, or Drosophila alpha DNA polymerases.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernad A., Blanco L., Lázaro J. M., Martín G., Salas M. A conserved 3'----5' exonuclease active site in prokaryotic and eukaryotic DNA polymerases. Cell. 1989 Oct 6;59(1):219–228. doi: 10.1016/0092-8674(89)90883-0. [DOI] [PubMed] [Google Scholar]
- Bernad A., Lázaro J. M., Salas M., Blanco L. The highly conserved amino acid sequence motif Tyr-Gly-Asp-Thr-Asp-Ser in alpha-like DNA polymerases is required by phage phi 29 DNA polymerase for protein-primed initiation and polymerization. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4610–4614. doi: 10.1073/pnas.87.12.4610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke J. D., LaCroute F., Fink G. R. A positive selection for mutants lacking orotidine-5'-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197(2):345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Boulet A., Simon M., Faye G., Bauer G. A., Burgers P. M. Structure and function of the Saccharomyces cerevisiae CDC2 gene encoding the large subunit of DNA polymerase III. EMBO J. 1989 Jun;8(6):1849–1854. doi: 10.1002/j.1460-2075.1989.tb03580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brooke R. G., Singhal R., Hinkle D. C., Dumas L. B. Purification and characterization of the 180- and 86-kilodalton subunits of the Saccharomyces cerevisiae DNA primase-DNA polymerase protein complex. The 180-kilodalton subunit has both DNA polymerase and 3'----5'-exonuclease activities. J Biol Chem. 1991 Feb 15;266(5):3005–3015. [PubMed] [Google Scholar]
- Budd M. E., Sitney K. C., Campbell J. L. Purification of DNA polymerase II, a distinct DNA polymerase, from Saccharomyces cerevisiae. J Biol Chem. 1989 Apr 15;264(11):6557–6565. [PubMed] [Google Scholar]
- Chang L. M. DNA polymerases from bakers' yeast. J Biol Chem. 1977 Mar 25;252(6):1873–1880. [PubMed] [Google Scholar]
- Delarue M., Poch O., Tordo N., Moras D., Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990 May;3(6):461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Freemont P. S., Sanderson M. R., Beese L., Friedman J. M., Joyce C. M., Steitz T. A. Genetic and crystallographic studies of the 3',5'-exonucleolytic site of DNA polymerase I. Science. 1988 Apr 8;240(4849):199–201. doi: 10.1126/science.2832946. [DOI] [PubMed] [Google Scholar]
- Derbyshire V., Grindley N. D., Joyce C. M. The 3'-5' exonuclease of DNA polymerase I of Escherichia coli: contribution of each amino acid at the active site to the reaction. EMBO J. 1991 Jan;10(1):17–24. doi: 10.1002/j.1460-2075.1991.tb07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorsky D. I., Crumpacker C. S. Site-specific mutagenesis of a highly conserved region of the herpes simplex virus type 1 DNA polymerase gene. J Virol. 1990 Mar;64(3):1394–1397. doi: 10.1128/jvi.64.3.1394-1397.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs J. S., Chiou H. C., Bastow K. F., Cheng Y. C., Coen D. M. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci U S A. 1988 Sep;85(18):6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribskov M., Burgess R. R. Sigma factors from E. coli, B. subtilis, phage SP01, and phage T4 are homologous proteins. Nucleic Acids Res. 1986 Aug 26;14(16):6745–6763. doi: 10.1093/nar/14.16.6745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall J. D., Wang Y. S., Pierpont J., Berlin M. S., Rundlett S. E., Woodward S. Aphidicolin resistance in herpes simplex virus type I reveals features of the DNA polymerase dNTP binding site. Nucleic Acids Res. 1989 Nov 25;17(22):9231–9244. doi: 10.1093/nar/17.22.9231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamatake R. K., Hasegawa H., Clark A. B., Bebenek K., Kunkel T. A., Sugino A. Purification and characterization of DNA polymerase II from the yeast Saccharomyces cerevisiae. Identification of the catalytic core and a possible holoenzyme form of the enzyme. J Biol Chem. 1990 Mar 5;265(7):4072–4083. [PubMed] [Google Scholar]
- Ito J., Braithwaite D. K. Yeast mitochondrial DNA polymerase is related to the family A DNA polymerases. Nucleic Acids Res. 1990 Nov 25;18(22):6716–6716. doi: 10.1093/nar/18.22.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki H., Ishino Y., Toh H., Nakata A., Shinagawa H. Escherichia coli DNA polymerase II is homologous to alpha-like DNA polymerases. Mol Gen Genet. 1991 Apr;226(1-2):24–33. doi: 10.1007/BF00273583. [DOI] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Hsieh J. C., Ito J. Bacteriophage PRD1 DNA polymerase: evolution of DNA polymerases. Proc Natl Acad Sci U S A. 1987 Dec;84(23):8287–8291. doi: 10.1073/pnas.84.23.8287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung G. H., Leavitt M. C., Schultz M., Ito J. Site-specific mutagenesis of PRD1 DNA polymerase: mutations in highly conserved regions of the family B DNA polymerase. Biochem Biophys Res Commun. 1990 Aug 16;170(3):1294–1300. doi: 10.1016/0006-291x(90)90534-t. [DOI] [PubMed] [Google Scholar]
- Kouzarides T., Bankier A. T., Satchwell S. C., Weston K., Tomlinson P., Barrell B. G. Sequence and transcription analysis of the human cytomegalovirus DNA polymerase gene. J Virol. 1987 Jan;61(1):125–133. doi: 10.1128/jvi.61.1.125-133.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Leavitt M. C., Ito J. T5 DNA polymerase: structural--functional relationships to other DNA polymerases. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4465–4469. doi: 10.1073/pnas.86.12.4465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcy A. I., Hwang C. B., Ruffner K. L., Coen D. M. Engineered herpes simplex virus DNA polymerase point mutants: the most highly conserved region shared among alpha-like DNA polymerases is involved in substrate recognition. J Virol. 1990 Dec;64(12):5883–5890. doi: 10.1128/jvi.64.12.5883-5890.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A., Araki H., Clark A. B., Hamatake R. K., Sugino A. A third essential DNA polymerase in S. cerevisiae. Cell. 1990 Sep 21;62(6):1143–1151. doi: 10.1016/0092-8674(90)90391-q. [DOI] [PubMed] [Google Scholar]
- Patel S. S., Wong I., Johnson K. A. Pre-steady-state kinetic analysis of processive DNA replication including complete characterization of an exonuclease-deficient mutant. Biochemistry. 1991 Jan 15;30(2):511–525. doi: 10.1021/bi00216a029. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz L. J., Stocki S., Nonay R. L., Dimayuga E., Goodrich L. D., Konigsberg W. H., Spicer E. K. DNA polymerization in the absence of exonucleolytic proofreading: in vivo and in vitro studies. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2417–2421. doi: 10.1073/pnas.88.6.2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Sanjanwala B., Ganesan A. T. DNA polymerase III gene of Bacillus subtilis. Proc Natl Acad Sci U S A. 1989 Jun;86(12):4421–4424. doi: 10.1073/pnas.86.12.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicer E. K., Rush J., Fung C., Reha-Krantz L. J., Karam J. D., Konigsberg W. H. Primary structure of T4 DNA polymerase. Evolutionary relatedness to eucaryotic and other procaryotic DNA polymerases. J Biol Chem. 1988 Jun 5;263(16):7478–7486. [PubMed] [Google Scholar]
- Syväoja J., Suomensaari S., Nishida C., Goldsmith J. S., Chui G. S., Jain S., Linn S. DNA polymerases alpha, delta, and epsilon: three distinct enzymes from HeLa cells. Proc Natl Acad Sci U S A. 1990 Sep;87(17):6664–6668. doi: 10.1073/pnas.87.17.6664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Von Borstel R. C. Measuring spontaneous mutation rates in yeast. Methods Cell Biol. 1978;20:1–24. doi: 10.1016/s0091-679x(08)62005-1. [DOI] [PubMed] [Google Scholar]
- Wang T. S., Wong S. W., Korn D. Human DNA polymerase alpha: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989 Jan;3(1):14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- Wintersberger U., Wintersberger E. Studies on deoxyribonucleic acid polymerases from yeast. 1. Parial purification and properties of two DNA polymerases from mitochondria-free cell extracts. Eur J Biochem. 1970 Mar 1;13(1):11–19. doi: 10.1111/j.1432-1033.1970.tb00893.x. [DOI] [PubMed] [Google Scholar]






