Abstract
G protein-coupled receptors (GPCRs) detect a wide variety of physical and chemical signals and transmit that information across the cellular plasma membrane. Dimerization is a proposed modulator of GPCR signaling, but the structure and stability of class A GPCR dimerization have been difficult to establish. Here we investigated the dimerization affinity and binding interface of human cone opsins, which initiate and sustain daytime color vision. Using a time-resolved fluorescence approach, we found that human red cone opsin exhibits a strong propensity for dimerization, whereas the green and blue cone opsins do not. Through mutagenesis experiments, we identified a dimerization interface in the fifth transmembrane helix of human red cone opsin involving amino acids I230, A233, and M236. Insights into this dimerization interface of red cone opsin should aid ongoing investigations of the structure and function of GPCR quaternary interactions in cell signaling. Finally, we demonstrated that the same residues needed for dimerization are also partially responsible for the spectral tuning of red cone opsin. This last observation has the potential to open up new lines of inquiry regarding the functional role of dimerization for red cone opsin.
Graphical abstract
The spatial organization of cell surface receptors plays a major role in biological function. Organization can occur over a range of length scales, from the ligand-activated dimerization of growth factor receptors to micrometer-scale clusters of T-cell receptors during an immune response.1,2 G protein-coupled receptors (GPCRs) make up a large family of membrane proteins for which the general rules governing spatial organization have remained elusive. Long considered to act as monomers during activation, class A GPCRs have recently been investigated for their potential to form dimers or oligomers. However, answers to this line of inquiry have remained elusive because of the difficulty in identifying dimeric complexes in the heterogeneous environment of the plasma membrane,3–5 as well as linking dimerization to functional outcomes. Thus, more information is needed about the structure, stability, and function of GPCR dimerization.
Efforts have been made to characterize the dimerization interface for class A GPCRs. Many of these studies are based on crystal packing geometries6–10 and structural modeling,11–14 whereas others have included methods such as peptide disruption15–20 and Cys residue cross-linking.21–25 Only a limited number of studies have reported site-specific mutagenesis that disrupted hypothetical dimer contact regions in human GPCRs,26–31 all of which relied on FRET or BRET (Förster or bioluminescence resonance energy transfer, respectively) as the principle probe for dimerization. For instance, the 5-HT1A serotonin receptor was investigated with FRET, where it was shown that the wild-type (WT) receptor is dimeric and that mutations in transmembrane (TM) helices 4 and 5 (TM4 and TM5, respectively) disrupted dimerization.27 The M3 muscarinic receptor was probed with a BRET saturation assay after the introduction of successive point mutations into 70 lipid-facing amino acids.28 Many of these mutants decreased the intensity of the BRET signal, and TM5 was found to be particularly important. However, in many of these studies, the point mutants did not eliminate energy transfer completely, which was interpreted as evidence of higher-order oligomers and multiple dimerization interfaces.26–30 The difficulty with BRET and FRET studies is that they can be overly sensitive to weak and transient interactions and potentially overestimate the extent of dimerization. This interpretation was supported by data from several assays that are sensitive to dimer lifetimes.32–34 For example, FRAP microscopy of the β1 adrenergic receptor indicated that it forms only transient interactions.32 Single-molecule tracking of the M1 muscarinic receptor revealed that dimers are relatively short-lived.33 An in situ affinity assay showed that dopamine receptor D2 is not corecruited as a homo-oligomer, despite reports of high FRET that were originally posited as evidence of dimerization.34 Such conflicting observations have severely complicated the field of GPCR dimerization, leading to multiple reports for and against the significance of this process even for the same receptor.35–38
This lack of clarity from previous work on GPCR dimerization can be resolved by combining methods that are sensitive to receptor dimerization and distinguish transient collisions from stable dimeric complexes. Here we investigated the dimerization of visual cone opsins with time-resolved fluorescence assays, an approach that allowed us to simultaneously measure receptor concentration, mobility, and dimerization. We used two related techniques to quantify these properties: fluorescence correlation spectroscopy (FCS) and pulsed-interleaved excitation fluorescence cross-correlation spectroscopy (PIE-FCCS). Data were collected within small regions of a live cell plasma membrane, where the proteins are translationally mobile. Dimerization was measured in several ways. (i) It was measured by molecular brightness, defined as the average rate that photons are emitted from diffusing complexes. For receptors labeled with a fluorescent protein, dimeric species contain two fluorescent proteins (one fused to each receptor) and thus emit twice as many photons as monomeric receptors. (ii) Dimeric complexes also exhibit reduced mobility relative to that of monomeric receptors, revealed as a decrease in their effective diffusion coefficients. (iii) In two-color experiments, the lifetime of the donor fluorescent protein (eGFP) is sensitive to acceptor (mCherry)-labeled receptors in the proximity. Thus, fluorescence lifetimes were used to calculate FRET efficiencies, indicative of dimerization. (iv) PIE-FCCS can also be used to measure the population of receptors that undergo co-diffusion, a signature of stable dimerization. PIE-FCCS is exquisitely sensitive to pairs of receptors (one with eGFP and one with mCherry) stably diffusing together.
Here we investigated the dimerization of human visual cone opsins, which we term red, green, and blue (corresponding to genes OPN1LW, OPN1MW, and OPN1SW, respectively). Cone opsins endow humans with trichromatic vision and comprise the basis of daytime image formation. They share a high degree of sequence similarity with rhodopsin (~40%),39 the most abundant photoreceptor pigment responsible for night and peripheral vision. Rhodopsin has long served as a prototype for GPCR studies and was the first GPCR to be structurally resolved via X-ray crystallography.40 Evidence of rhodopsin dimerization includes results of atomic force microscopy (AFM) of isolated outer membrane disks,41 association assays in reconstituted liposomes,42 purification of dimeric rhodopsin imaged by transmission electron microscopy,43 and cryoelectron tomography of sectioned rod cells.44 Recently, we reported a live-cell PIE-FCCS study in which dimerization of retinal-free rod opsin was found to be density-dependent at concentrations 10–100 times lower than those in retinal membranes.45 This result suggests that though the affinity is relatively modest, it leads to a dominantly dimeric receptor at high densities. Although no point mutations have been reported that disrupt rhodopsin dimerization, recent work indicates that synthetic TM peptides mimicking TM4 and TM5, and TM1 and TM2, can disrupt dimerization.15 This finding is also consistent with crystal packing analysis and modeling of the AFM data indicating two dimerization interfaces for rhodopsin.46
Here we sought to elucidate dimerization in human cone opsins, which share ~40% sequence homology with rhodopsin.47 No quantitative biophysical study of human cone opsin dimerization has yet been reported. A recent study of murine S-opsin reported cotrafficking with R-opsin, suggesting that it dimerizes with rhodopsin in a mouse model.48 Another study noted blue cone opsin aggregation due to a Phe-rich region in this receptor,49 but the aggregated protein was located in the ER and trans-Golgi network instead of the plasma membrane. Using the approach outlined above, we found that red cone opsin was significantly dimerized, and that the average cross-correlation was higher than that of rhodopsin. Neither blue nor green cone opsin showed significant homodimerization in the plasma membrane. Using site-directed mutagenesis, we identified amino acid residues in TM5 that disrupted dimerization of red cone opsin but induced dimerization of green cone opsin. The same mutations also were responsible for the spectral shift between these two opsins, suggesting their possible functional role in cone opsin dimerization.
MATERIALS AND METHODS
DNA Constructs and Primers
Human blue, green, and red cone opsin cDNAs cloned into the pUC57 vector were synthesized and obtained from Genentech (San Francisco, CA). The last 14 amino acids in blue cone opsin and the last 12 amino acids in green and red cone opsins were replaced by a 1D4 tag, a TETSQVAPA amino acid sequence comprising the last nine amino acids of rod opsin. For the construction of cone opsin-eGFP and cone opsin-mCherry, cDNAs of human blue, green, and red cone opsins were amplified by PCR. EcoR1 and Sac2 restriction sites were then introduced at the 5′- and 3′- ends, with the following primers: for the cone opsin-eGFP construct, forward primer GTGGGGAATTCGCCATGAAGACCATCATCGCCCT and reverse primer TCTGGCCGCGGTGGCTGGAGCGACCTGA; for the cone opsin-mCherry construct, forward primer GTGGGGAATTCGCCATGAAGACCATCATCGCCCT and reverse primer TCTGGCCGCGGGGCTGGAGCGACCTGA. The resulting amplified DNA was then cloned into the peGFP-N3 and pmCherry-N1 original vectors (Clontech, Mountain View, CA).
Green cone opsin T230I, S233A, V236M (green-IAM) and red cone opsin I230T, A233S, M236V (red-TSV) were constructed with Phusion high-fidelity DNA polymerase (New England Biolabs, Ipswich, MA) using a standard protocol.50 These constructs were then used for PIE-FCCS experiments.
Additionally, a 1D4 tag was added at the C-terminus of eGFP in the cone opsin-eGFP constructs. These constructs then were used to test the expression levels of cone opsins following their transfection into HEK-293 cells, as well as for protein purification and ultraviolet–visible (UV–vis) spectroscopy experiments.
WT green, red, green-IAM, and red-TSV were also subcloned into a pcDNA3.1(+) vector following a standard protocol,50 and the resulting constructs were used for protein purification and UV–vis spectroscopy experiments. The composition of each construct was confirmed by DNA sequencing.
Cos-7 Cell Cultures and Data Collection
Cos-7 cells were cultured in Dulbecco’s modified Eagle’s medium (1× DMEM with GlutaMAX, Life Technologies, Carlsbad, CA) at a density of ~106 cells/mL and 37 °C. Both 10% fetal bovine serum (FBS, Life Technologies) and 1% penicillin/streptomycin (BioReagent, Sigma-Aldrich, St. Louis, MO) were added to the medium. Cells grown on 100 mm × 20 mm tissue culture plates (Falcon, Corning Inc., Corning, NY) at 37 °C were split when their confluency reached ~80–90%. Two to three days prior to being imaged, cells were removed from the 100 mm × 20 mm plates and transferred to 35 mm × 10 mm γ-irradiated glass bottom culture dishes (MatTek, Ashland, MA). Cells then were transfected 24 h prior to being imaged by using Opti-MEM I medium with reduced phenol red and Lipofectamine 2000 transfection reagent (Life Technologies) according to the supplied protocols. On the day of imaging, the medium was changed from DMEM to Opti-MEM I medium without phenol red (Life Technologies) at least 30 min prior to imaging.
Fluorescence Correlation Spectroscopy (FCS)
FCS was conducted with a custom-modified Eclipse Ti inverted microscope (Nikon Instruments, Tokyo, Japan) described in previous publications.45,51,52 The 488 nm laser was selected and filtered (LL01-488-12.5, Semrock, Rochester, NY) from a supercontinuum fiber laser with a pulse duration of 5 ps and a repetition rate of 10 MHz (SuperK NKT Photonics, Birkerød, Denmark). At the microscope, a dichroic beam splitter directed the 488 nm beam to the sample (zt488/561rpc, Chroma Technology Corp., Bellows Falls, VT). The fluorescence signal collected by the objective passed through a laser-blocking filter to remove scattered laser light (zet488/561m, Chroma Technology Corp.). The signal was then directed to a camera port at the side of the microscope where it was focused through a 50 μm confocal pinhole onto a bandpass-filtered SPAD detector (Micro Photon Devices, Bolzano, Italy). The time- correlated single-photon data were recorded with a four- channel routed time-correlated single-photon counting (TCSPC) device (Picoharp 300, PicoQuant, Berlin, Germany) at a timing resolution of 32 ps. TCSPC files were processed as described previously to produce the fluorescence correlation curves and fluorescence lifetime histograms.45,51
Pulsed-Interleaved Excitation Fluorescence Cross-Correlation Spectroscopy (PIE-FCCS)
PIE-FCCS was conducted on the same instrument described above, with a 561 nm laser excitation beam selected from the supercontinuum laser and delayed ~50 ns with respect to the 488 nm pulses (LL02-561-12.5, Semrock). The same dichroic and laser blocking filter was used for two-color excitation. The fluorescence signal emitting from eGFP and mCherry was split with a long-pass filter (FF560-FDi01-25×36, Semrock) after passing through a confocal pinhole. A second SPAD detector collected the long wavelength portion of the signal after it had passed through a bandpass filter (FF01-621/69-25, Semrock). The TCSPC files were used to construct FCS and FCCS data after time gating the fluorescence signal. Only photons arriving within 50 ns of the 488 nm pulse in the “green” detector and within 50 ns of the 561 nm pulse in the “red” detector were used to construct the time-dependent fluorescence signals [FG(t) and FR(t), respectively]. The correlation spectra were calculated, fit, and analyzed as previously described.45,51,52
Lifetime Fitting
To measure the fluorescence lifetimes of the various eGFP fusion constructs, we constructed lifetime histograms from the time-correlated single-photon counting data collected in the FCS and PIE-FCCS studies. Thus, we report only lifetime measurements for the same cells for which FCS or PIE-FCCS data are also provided. Photons were binned at 64 ps according to their arrival time relative to a synchronized pulse from the excitation laser. The lifetime was fit with a nonlinear least-squares routine in MatLab after deconvolution with an instrument response function collected from a scattering sample taken on the same microscope under an identical optical setup. Data were then fit to a single-component exponential decay function.
Expression of Cone Opsins in HEK-293 Cells; Pigment Reconstitution and Purification
HEK-293 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM) with 10% FBS (Hyclone, Logan, UT), 5 μg/mL plasmocin (InvivoGen, San Diego, CA), and 1 unit/mL penicillin and 1 μg/mL streptomycin (Life Technologies) at 37 °C under 5% CO2. Transient cell transfections were performed with polyethylenimine.53,54 Twenty-four h post-transfection, cone opsins were reconstituted with either 11-cis-retinal or 9-cis-retinal. Retinal was delivered to the cell culture from a DMSO stock solution to a final concentration of 10 or 20 μM for 11-cis-retinal or 9-cis-retinal, respectively, and then cells were incubated in the dark for 24 h at 37 °C with 5% CO2. Cells from 15 plates (10 cm) were harvested and centrifuged at 800g, and the pellet was suspended in 50 mM HEPES, 150 mM NaCl, 20 mM n-dodecyl β-D-maltopyranoside (DDM), and protease inhibitor cocktail (pH 7.0) before being incubated for 1 h at 4 °C on a rotating platform. The lysate was centrifuged at 100000g for 1 h at 4 °C, and cone opsins were purified from the supernatant with an anti-rhodopsin C-terminal 1D4 antibody immobilized on CNBr-activated agarose. Two hundred microliters of 2 mg of 1D4/mL of agarose beads was added to the supernatant and incubated for 1 h at 4 °C. The resin was then transferred to a column and washed with 10 mL of 50 mM HEPES, 150 mM NaCl, and 2 mM DDM (pH 7.0). Cone opsins were eluted with 150 mM HEPES, 150 mM NaCl, and 2 mM DDM (pH 7.0), supplemented with 0.6 mg/mL TETSQVAPA peptide.
UV–Vis Spectroscopy of Cone Opsin Pigments
UV–visible spectra were recorded from freshly purified cone pigment samples. The absorption spectrum between 260 and 700 nm was initially recorded in the dark to calculate the concentrations of purified cone opsins. To obtain difference spectra of purified cone pigments, a baseline was recorded first on the sample in the dark and then measured following exposure the sample to white light delivered from a Fiber-Light illuminator (150 W lamp) (Dolan-Jenner, Boxborough, MA) at a distance of 10 cm for 3 min.
Cross-Linking of Cone Opsins in Membranes
HEK-293 cells were transiently transfected with WT green, red, green-IAM, and red-TSV constructs cloned into the pcDNA3.1(+) vector. Forty-eight hours after transfection, cells from two 10 cm plates were harvested, washed with PBS, and suspended in 200 μL of 20 mM Bis-Tris-propane, 100 mM NaCl buffer (pH 7.5) containing a protease inhibitor cocktail. After gentle trituration with a syringe and needle, the membranous fraction was pelleted by centrifugation at 16000g for 10 min at 4 °C. The pellet was suspended in 200 μL of the Bis-Tris-propane buffer, and half of the resulting solution was used for cross-linking. Two millimolar disuccinimidyl glutarate (DSG) cross-linker (Thermo Scientific, Waltham, MA) was added to membranous samples, and the cross-linking reaction was allowed to proceed for 2 h on ice. The reaction was terminated with 1 M Tris-HCl (pH 8.0), added to a final concentration of 50 mM, and the samples were incubated for 15 min followed by membrane solubilization with 20 mM DDM. Cross-linked opsins (50 μg of total protein extract) were separated on a 10% SDS-PAGE gel and then transferred to polyvinyl difluoride (PVDF) membranes. Proteins were detected with an anti-rhodopsin C-terminal 1D4 antibody, an HRP-conjugated anti-immunoglobulin, and a chemiluminescence assay (Thermo Scientific). The amount of opsin dimer was quantified by densitometry using ImageJ.
RESULTS
Fluorescence Correlation Spectroscopy of Human Cone Opsins
We first created fluorescent protein (eGFP) fusions to the C-termini of three cone opsins, here termed red (OPN1LW), green (OPN1MW), and blue (OPN1SW). These fusions were then inserted into a vector plasmid for mammalian expression and transiently transfected into Cos-7 cells. Cos-7 cells, an immortalized cell line derived from African green monkey kidneys, were chosen mainly for their optimized geometry for optical imaging experiments. They express no endogenous visual opsins and are an ideal model system for investigating protein interactions in a native plasma membrane. We interrogated opsin-eGFP-expressing cells with FCS. In our instrument (Figure 1A), a focused laser is positioned near the edge of a live cell maintained at 37 °C. Single photons are recorded as they are emitted from fluorescent molecules diffusing in and out of a fixed region in space, defined by the excitation laser focus and confocal detection optics. The time-dependent intensity of the photon counts displays relatively large fluctuations that are indicative of single molecules diffusing in and out of the detection area (Figure 1B). These fluctuations have a characteristic size and time scale, which can be quantified by a time–time autocorrelation function (Figure 1C). Two key features of this correlation function are the decay time, τD, and the early time amplitude, G(0).
Figure 1.
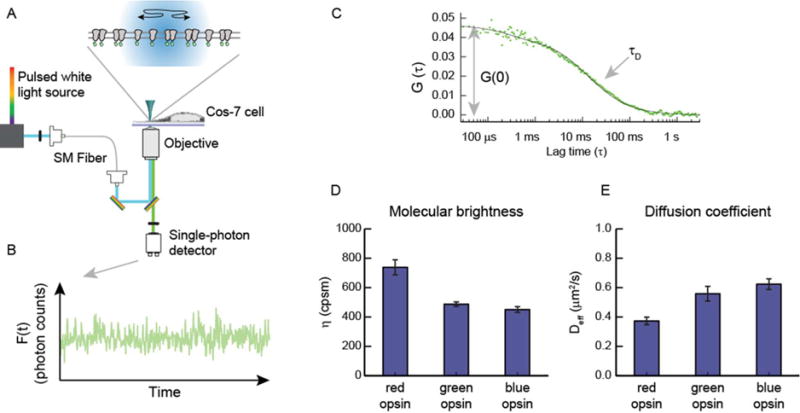
Schematic of FCS data collection and analysis. (A) eGFP-labeled GPCRs diffuse in and out of the confocal area (radius of 210 nm) illuminated by a 488 nm laser. Emission from the fluorescent proteins is collected by the objective and directed to a single photon detector. (B) Photon counts are binned into evenly spaced time segments to produce the time-varying fluorescence signal, F(t). (C) Fluctuations in the fluorescence intensity, δF(t) = F(t) − 〈F(t)〉, are used to calculate a time–time autocorrelation function, G(t) = 〈δF(t) × δF(t + τ)〉/F〈(t)〉2. The early time amplitude of G(τ) is used to obtain the average population of diffusing particles, whereas the decay time is proportional to the average dwell time of the particles in the confocal area. (D and E) Parameters extrapolated from the fitted data are used to quantify the average molecular brightness (η) and effective diffusion coefficient (Deff for each cell (error bars are standard errors of the means).
The amplitude of the correlation function, G(0), is proportional to the size of the intensity fluctuations and thus is inversely proportional to the average number of molecules in the laser focus, 〈N〉. We obtained the local concentration by dividing 〈N〉 by the diffraction-limited area (πω2), where ω is the beam radius. For the opsin-eGFP experiments, we observed cells with concentrations ranging from 80 to 2000 molecules/μm2. The concentration in this case refers to the number of diffusing species, not necessarily the number of opsin-eGFP proteins. To determine the dimerization state of the diffusing species, we took the average photon counts from a cell measurement and divided it by the average number of diffusing species, 〈N〉. The result is termed molecular brightness (symbolized by η) and has units of photon counts per second per molecule (cpsm). Molecular brightness is proportional to the number of protomers in a diffusing species; i.e., a dimer is ideally twice as bright as a monomer. Average molecular brightness values for green and blue cone opsins were 451 ± 19 and 461 ± 25 cpsm, respectively (Figure 1D), consistent with monomeric lipid-anchored eGFP measured on the same instrument under similar conditions (errors reported here and below are standard errors of the mean).45,51 For red cone opsin, the average molecular brightness was 738 ± 51 cpsm (Figure 1D), a value similar to that obtained from a dimer control protein consisting of a fluorescent protein fused to a leucine zipper dimerization motif and a lipid-anchored peptide.45,51 Our brightness data indicate that red cone opsin is dimeric in the plasma membrane. The molecular brightness is not quite double the monomer brightness, which could indicate that the protein is distributed into a mixture of monomer, dimer, and oligomer states. The observed brightness is also affected by fluorescent proteins in the dark state, which are reported to be >20% in single-particle counting studies of eGFP.55
To further characterize the dimerization state of the cone opsins, we analyzed their mobility using the FCS data. The decay time of the FCS correlation function, τd, is the average dwell time of a molecule in the detection area. The dwell time was converted to a diffusion coefficient through the equation Deff = ω2/4τD, where ω is the calibrated radius of the detection area, 210 nm in our instrument. There have been reports of transmembrane proteins displaying complex diffusion behavior in the cell plasma membrane.56,57 Nevertheless, our data fit well to the two-dimensional diffusion model, so we used this relationship and acknowledge that it is better considered as an effective diffusion coefficient, Deff. Diffusion coefficients are dependent on the size of the protein or protein complex, although the scaling laws for TM proteins are still under debate.58,59 Single-cell FCS measurements of red cone opsin displayed an average diffusion coefficient of 0.37 ± 0.03 μm2/s, whereas those of green and blue cone opsins were significantly larger, 0.56 ± 0.05 and 0.62 ± 0.04 μm2/s, respectively (Figure 1E). The lower diffusion coefficient of red cone opsin indicates a dimeric complex, consistent with conclusions drawn from the molecular brightness data.
PIE-FCCS Data Show That Human Red Cone Opsin, but Not Blue or Green Cone Opsin, Is Segregated into Dimeric Structures in the Plasma Membrane
Both the mobility and brightness data from FCS measurements are consistent with red cone opsin diffusing as a dimer, and green and blue cone opsins diffusing as monomers. We next turned to PIE-FCCS to further investigate the dimerization state of the visual cone opsins. PIE-FCCS is a variation of FCCS, which has been a powerful approach to measuring molecular associations in live cells.60 The unique advantage of PIE-FCCS is that it separates individual excitation events in time such that each photon collected can be assigned to an excitation source (i.e., 488 nm light or 561 nm light).61 In this way, photons emitted by eGFP that pass to the red detector (i.e., spectral cross-talk) are eliminated from the calculation of the cross-correlation function. Fluorescent proteins have broad emission spectra, and spectral cross-talk leads to an uncorrectable overestimation of the cross-correlation function.62 This effect is especially true for membrane proteins, which diffuse more slowly and in a more complicated matrix than water-soluble cytosolic proteins do.
To conduct these experiments, we cotransfected Cos-7 cells to express cone opsin-eGFP and cone opsin-mCherry and then measured them with a custom-built PIE-FCCS instrument (Figure 2A).45,51 In this instrument, 488 and 561 nm laser light was selected from a pulsed white light source and directed through fibers of different lengths before being focused at the same point within the sample. This configuration introduced a delay of ~50 ns such that individual photons arriving at the detectors could be assigned to an excitation source. Only photons arriving at the green detector within 50 ns of a 488 nm pulse and at the red detector within 50 ns of the 561 nm pulse were included in the time-dependent intensity traces (Figure 2B). Autocorrelation functions for the red and green channel were then calculated and fit to reveal the number of diffusing species labeled with an mCherry and an eGFP probe, respectively (Figure 2C, red and green dots). The autocorrelation functions for the eGFP- and mCherry-labeled proteins showed a fast (~500 μs) and a slow (~40 ms) time component, which we ascribed to photophysical dynamics and two-dimensional (2D) diffusion, respectively, as described in our previous publications.45,51 Variations in amplitude between the red and green autocorrelation functions reflect slight variations in expression level (more single-cell measurements are shown in Figures S1–S3). A cross-correlation function was also calculated (Figure 2C, blue dots). The amplitude of the cross-correlation function is proportional to the number of diffusing species containing at least one mCherry and one eGFP probe. The cross-correlation function rigorously identifies the degree to which proteins are associated into stable dimeric complexes. Sample FCCS data sets are shown in Figure S1 (red cone opsin), Figure S2 (green cone opsin), and Figure S3 (blue cone opsin). The receptor densities in single-cell measurements ranged from 65 to 2000 molecules/μm2.
Figure 2.
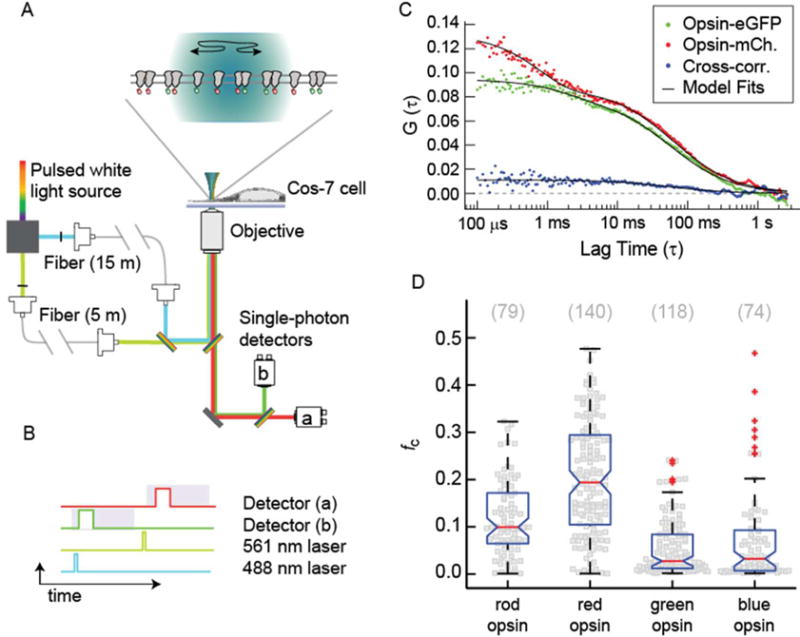
PIE-FCCS data collection and analysis. (A) Two-color excitation is used to illuminate the sample. Pulsed-interleaved excitation is achieved by increasing the optical path of the 488 nm light relative to the 561 nm light with single-mode optical fibers of different lengths. Fluorescence emission is spectrally separated and detected by two single-photon counting modules. (B) Diagram of single-photon events. Only photons collected in detector B after excitation by a 488 nm pulse were included in FG(t), and photons collected by detector A after excitation by a 561 nm pulse were included in FR(t). This strategy eliminates cross-talk from spectral leak-through and direct excitation. (C) Correlation functions and model fits [GG(τ), GR(τ), and GX(τ)] are shown for a representative single-cell measurement. The cross-correlation function is fit with a 2D Brownian model, whereas the autocorrelation functions are fit with an additional triplet term to account for the photophysics of the fluorescent protein. The amplitude of the CCF is related to the fraction of dimers through the fc parameter. (D) Single-cell fc values (gray squares) are shown for rod opsin and the three cone opsins. The total number of cells is shown in parentheses above the distributions. Boxplots are overlaid to indicate the median (red line) and percentiles (box, 25–75%; whiskers, 0–100%; red plus signs, outliers).
The relative cross-correlation, fC, was calculated for each cell by taking the amplitude of the cross-correlation function and dividing it by the time zero value of the autocorrelation function with the highest amplitude: fc = GX(0)/GR or G(0). For eGFP- and mCherry-labeled systems, we expect a median fc value of 0.167 for fully dimerized proteins.51 In Figure 2D, the fc distributions of rod opsin and the red, blue, and green cone opsins are shown as bee-swarm plots with a boxplot overlay for statistical comparison. The median fc value of rod opsin was comparable to the expected value for a dimeric protein as reported previously.45 The data for red cone opsin displayed a wide distribution of values, with a median fc of 0.19 (Figure 2D). This result was higher than that of rod opsin (P < 0.001), suggesting that red cone opsin forms dimers and possibly higher-order oligomers. The cross-correlation of green and blue cone opsins clustered near zero with median fc values of 0.028 and 0.032, respectively (Figure 2D), indicating that blue and green cone opsins are overwhelmingly monomeric in the plasma membrane.
In contrast to what was reported previously for rod opsin,45 red, blue, and green cone opsins did not show any significant trend between fc and receptor density. This can be visualized by plotting the dimer concentration as a function of monomer concentration (Figure S4). A correlated trend, as seen for rod opsin, suggests a modest driving force for dimerization. If the population is either dominantly monomeric or dominantly dimeric over the measured concentrations, there will not be a significant correlation. For green and blue cone opsins, a lack of correlation between monomer and dimer concentrations is consistent with a lack of significant dimerization. For red cone opsin, these data suggest that either the dimerization affinity is very high or red cone opsin forms higher-order oligomers.
Lifetime FRET Data of Cone Opsins
The time-correlated single-photon counting approach used for PIE-FCCS also allowed us to construct lifetime histograms for each cell measured in Figure 2. The eGFP fluorescence lifetime is sensitive to resonance energy transfer, i.e., FRET, and thus can be used as a probe for molecular associations. Our eGFP/mCherry system was not as efficient a FRET pair as, for example, cyan and yellow fluorescent proteins, but we did observe a significant decrease in the fluorescence lifetime of red cone opsin fused to eGFP in dual expression experiments. Using the acceptor-free lifetime, τ0, from the eGFP-only experiments described above, we calculated the FRET efficiency for each cell measurement (Figure S5A). For red cone opsin, the FRET efficiencies reached as high as 12%, with an average of 3.6 ± 0.9. The green and blue cone opsin FRET efficiencies never exceeded 3.5%, and their averages were near zero (−0.12 ± 0.01 and −0.06 ± 0.02%, respectively). These data provide further evidence that red opsin forms dimeric complexes, whereas blue and green cone opsins are mostly monomeric. The FRET efficiencies for each of the cone opsins did not show any significant trend with density over the range measured here (Figure S6).
A Triple-Point Swap Mutant Disrupts Dimerization of Red Cone Opsin and Increases the Level of Dimerization of Green Cone Opsin
Human red and green cone opsins differ by only 15 residues,63 as seen in the amino acid sequence alignment of human red cone opsin with human green cone opsin in Figure S7. On the basis of the data provided above, we found that among cone opsins, only red cone opsin forms dimers. Therefore, we determined which of the unique amino acids were located at the receptor surface and thus could be involved in receptor–receptor interactions. Three of those residues [230, 233, and 236; TSV in green and IAM in red cone opsin (Figure 3A)] were located on TM5, implicated in the formation of the dimer interface in other GPCRs.15 To test if these specific amino acids are responsible for red cone opsin dimerization, we constructed two triple-point swap mutants: a red I230T, A233S, M236V (red-TSV) to abolish receptor dimerization and a green T230I, S233A, V236M (green-IAM) to induce receptor dimerization.
Figure 3.
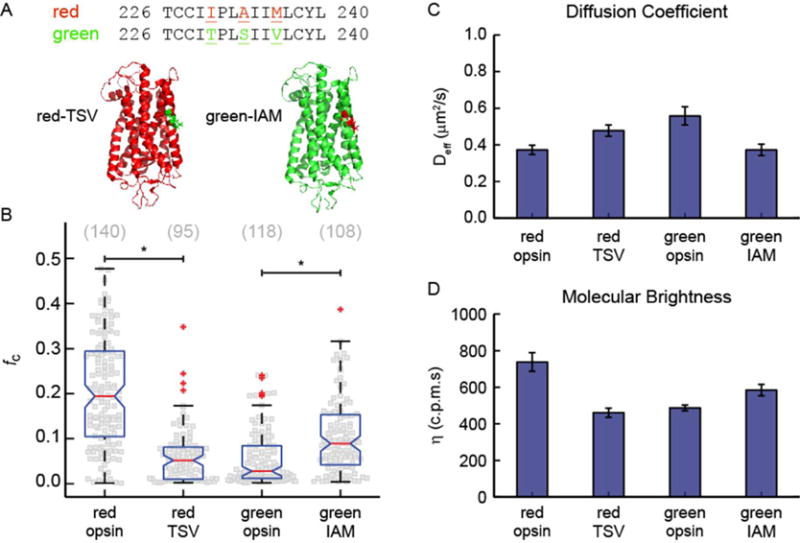
Triple-point red/green cone opsin mutants. (A) Three-dimensional homology model of red and green cone opsins highlighting the location of the three point mutations. (B) Summary of cross-correlation, fC, for WT cone opsins and triple-point mutants. A significant change in dimerization is caused by the triple-point mutation. The cross-correlation of red-TSV decreased compared to that of WT red cone opsin, whereas the cross-correlation of green-IAM increased relative to that of WT green cone opsin. This observation is consistent with the mutation disrupting dimerization in red-TSV and inducing dimerization in green-IAM. (C) Effective diffusion coefficients for eGFP-labeled opsins. These data indicate that the mobility of red-TSV increases relative to that of the WT, consistent with the loss of dimerization, whereas the mobility of green-IAM decreases relative to that of the WT, consistent with an increase in the level of dimerization. (D) Molecular brightness of eGFP-labeled opsins. The average molecular brightness of red-TSV decreased significantly compared to that of the WT, indicating a mostly monomeric population. The molecular brightness of green-IAM increased relative to that of the WT, indicating a significant increase in the level of dimeric species. For panels C and D, the error bars are the standard errors of the means.
We then repeated the FCS and PIE-FCCS experiments described above with the triple-point swap mutants, and the findings are summarized in Figure 3 (sample PIE-FCCS data are shown in Figures S8 and S9). The PIE-FCCS measurements of red-TSV displayed significantly reduced cross-correlation compared with those of WT red cone opsin (Figure 3B; P < 0.001). Moreover, the cross-correlation of green-IAM increased relative to that of the WT protein, indicating an increased level of dimerization (Figure 3B; P < 0.001). The median fc value of green-IAM was 0.10, which is greater than the median fc of WT green cone opsin (0.03), but less than the median fc for red cone opsin (0.19). The single-color FCS data for red-TSV and green-IAM are consistent with the conclusions drawn from the PIE-FCCS results. The diffusion coefficient of the red-TSV mutant increased, and its molecular brightness decreased, indicating a higher mobility and lower level of clustering relative to those of the WT protein (Figure 3C,D). Conversely, the diffusion coefficient of the green-IAM mutant decreased and the molecular brightness increased relative to those of the WT protein, denoting a lower mobility and an increased level of clustering (Figure 3C,D).
Blue cone opsin shares ~40% sequence homology with red and green cone opsins. According to our results obtained from FCS experiments, both green and blue cone opsins do not form dimers in the cell membrane. V211, S214, and C217 are three residues in blue cone opsin that are homologous to I230, A233, and M236 in red cone opsin and T230, S233, and V236 in green cone opsin (see the sequence alignment in Figure 3A). Interestingly, both green and blue cone opsins have an identical serine residue in the middle of this “triple-amino acid motif”. Moreover, both green and blue cone opsins contain a valine residue, although this is located on the opposite ends of this motif. V211 and C217 in blue cone opsin correspond to T230 and V236 in green cone opsin, respectively. All four residues are comparable in size and smaller than the isoleucine and methionine present in red cone opsin.
Our data are consistent with the triple-point mutation increasing the level of dimerization of green cone opsin, but not to the level observed for WT red cone opsin. This comparison suggests that there could be higher-order oligomerization in red cone opsin involving more than just TM5. To test this idea, we also conducted PIE-FCCS measurements of cells cotransfected with red-TSV and WT red cone opsins. If there is a TM5–TMX interface, we would expect there to be significant cross-correlation for the combination of a wild type with TM5 mutants. Instead, we observed a significant drop in the fc values, similar to the results of the red-TSV only experiments (Figure S10). Although this finding does not exclude possible combined interfaces with TM5 (i.e., TM4 and TM5 or TM5 and TM6), it does indicate that TM5 is part of a dimerization interface.
The ability of triple-point mutations to significantly modulate cone opsin dimerization affinity opens up new avenues for investigating the chemical interface between GPCRs in a dimeric complex. One possibility for future work is to probe the side chain dependence of the cross-correlation to resolve the local interactions that drive dimerization. As an initial experiment, we mutated the Ala residue to Val at position 230 in the green swap mutant to compare green-IAM with green-IVM. The level of dimerization of green-IVM was slightly lower than in green-IAM (Figure S11), suggesting that the steric bulk of the Val residue relative to Ala contributes to the dimerization affinity. More work will be needed to fully understand the chemical forces driving cone opsin dimerization.
Cross-Linking of Cone Opsins and the Effect of a Triple Mutation on Dimer Formation
PIE-FCCS experiments indicated that red cone opsin has a stronger propensity than green cone opsin to self-associate. This affinity was alleviated by introducing three mutations into the amino acids located at the potential dimer interface, which matched the sequence in green cone opsin. In contrast, changing the same amino acids in green cone opsin to those present in red cone opsin enhanced self-association. To confirm these results, we performed cross-linking of red, green, and mutated cone opsins expressed in HEK-293 cell membranes with a short (7.7 Å spacer arm) disuccinimidyl glutarate (DSG) cross-linker. This homobifunctional N-hydroxysuccinimide ester (NHS-ester) cross-linker reacts with primary amines on the N-termini of the ε-amine of Lys residues forming a stable covalent amide bond connecting the two protein molecules, but only if such molecules are in the proximity. Thus, only those receptors self-associating in the lipid bilayer (not freely diffusing monomers) are likely to be captured by the cross-linker to form stable dimers and/or oligomers. Before cross-linking, a dimer population of approximately 40–60% was found for the green and red WT and mutant cone opsin samples. This relatively high dimer content in all samples could result from nonspecific aggregation in the presence of SDS, commonly noted for many integral membrane proteins.64–66 These properties, however, could change after surface modification of the receptors by a bound cross-linker. Nevertheless, after incubation with a DSG cross-linker, an increased level of receptor DSG chemical dimers and a decrease in the level of monomers were observed for red cone opsin but not for green cone opsin (Figure 4A,B). The higher-order oligomers overlapped with a smear, possibly resulting from various extents of receptor N-glycosylation. The smear also could lead to an overestimation of their content, so we quantified only band intensities of monomers and dimers, finding 79 ± 9% of DSG dimers for red cone opsin but only ~21 ± 14% of DSG dimers for green cone opsin after cross-linking. Consistent with the fluorescence microscopy measurements described above, less dimer formation was found for red-TSV (48 ± 12% vs 79 ± 9% for WT) and enhanced dimerization was seen for green-IAM (43 ± 2% vs 21 ± 14% for WT) (Figure 4A,B, arrows).
Figure 4.

Effect of triple mutations on opsin cross-linking. (A) Effects of mutations on the formation of DSG-cross-linked opsin dimers. Membranes isolated from HEK-293 cells containing cone opsins were cross-linked with DSG (see dark lines above panels A and B). Fifty micrograms of the total protein extracts was loaded on each lane of the SDS–PAGE gel. (B) Quantification of DSG-cross-linked dimers. The amount of DSG-cross-linked opsin dimers was calculated from densitometric analyses of protein bands corresponding to the opsin monomer and dimer from three independent experiments. Results are presented as means ± standard deviations. Arrows indicate an increase in the extent of dimer formation in green-IAM opsin relative to green cone opsin and a decrease in the level of dimer formation in red-TSV opsin compared to red cone opsin.
Role of a Triple Mutant in Spectral Tuning
Because of the high degree of sequence similarity between red and green cone opsins, there has been significant interest in understanding the shift in their spectral sensitivities.39 The wavelength of maximal absorbance of red cone opsin is 564 nm, just 30 nm longer than the maximal absorbance wavelength of green cone opsin, 534 nm.39 Moreover, red and green cone opsin amino acid sequences are identical except for 15 residues. On the basis of functional mutagenesis work, the spectral distinction between red and green cone opsins distributed over precisely seven of those amino acid residues.67 The majority (~70%) of the spectral shift can be assigned to positions 277, 285, and 309. However, positions 116, 180, 230, and 233 are required to fully recapitulate the native absorbance profile.68 Of these seven amino acids, two (230 and 233) are part of the dimeric interface we identified above and also account for 4 nm of the spectral shift between red and green cone opsin.67,68
We performed absorbance measurements of the cone opsin constructs used in our study to verify their function and spectral sensitivity. We also sought to confirm that the triple-point mutant used in this work shows the expected spectral shift seen in previous studies. To purify the cone opsin-eGFP fusion proteins, a 1D4 tag was added at the C-terminus following the eGFP sequence. Expression levels of WT and mutant cone opsin-eGFP-1D4 constructs in transiently transfected HEK-293 cells were similar, and their membrane localization was unaffected by the tag (Figure S12). Transiently expressed cone opsins instead of native 11-cis-retinal were regenerated with the isochromophore 9-cis-retinal and purified by 1D4 immunoaffinity chromatography, after which their UV–vis absorption spectra were recorded (Figure S12C,D). Absorption maxima of retinal-regenerated cone pigments were difficult to discriminate because of the high absorbance of eGFP, with an absorption maximum at 488 nm. Therefore, to extract the absorption spectra of cone opsins, we recorded difference spectra. As shown in Figure S12, the absorption maximum of 9-cis-retinal-bound green opsin was ~493 nm, and it was ~525 nm for 9-cis-retinal-bound red opsin, in agreement with previous observations.69 A spectral shift of 6 nm toward shorter wavelengths was observed for red-TSV as compared to WT red cone opsin. However, no spectral shift was found for green-IAM compared to WT green cone opsin (Figure S12D). One explanation for this finding could be that an exchange of the native chromophore 11-cis-retinal with the 9-cis isochromophore avoided the spectral shift in the green-IAM mutant due to specific structural changes possibly occurring in isoopsins. Alternatively, the inability to detect a spectral shift could result from the overlap in the absorption maxima between 9-cis-green opsin (493 nm) and eGFP (488 nm).
To validate the red cone opsin observations and eliminate spectral interference from the eGFP tag, we recloned the green and red cone opsins and their respective mutants into a pcDNA3.1(+) vector without the eGFP fusion. Measured expression levels of WT and mutant cone opsins in transiently transfected HEK-293 cells were similar (Figure 5A). UV–vis absorption spectra were then recorded for purified cone pigments regenerated with the 11-cis-retinal chromophore (Figure 5B,C). Here, spectral shifts were found for both mutants: a 4 nm shift toward longer wavelengths for green-IAM opsin (from 530 to 534 nm) and a 5 nm shift toward shorter wavelengths for red-TSV opsin (from 560 to 555 nm) (Figure 5C). These results are consistent with those of an earlier study that linked residues I230 and A233 in exon 4 to the spectral sensitivity of red cone opsin.67 A direct connection between the two effects (dimerization and color tuning) remains to be established. The first step needed is to probe dimerization in the chromophore-bound state and then to determine the mechanism of how the residues mentioned above drive spectral sensitivity.
Figure 5.
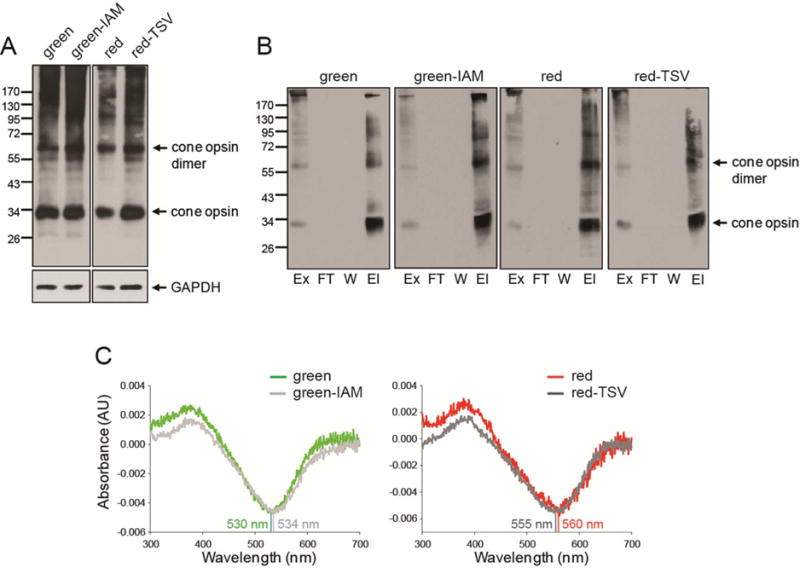
Biochemical characterization of green, red, and mutant cone opsins. (A) Immunoblots indicating expression levels of green cone opsin, green-IAM, red cone opsin, and red-TSV transiently expressed in HEK-293 cells. Fifty micrograms of total protein cell lysate obtained 48 h after transfection was used for detection with an anti-rhodopsin C-terminal 1D4 tag antibody. GAPDH was the protein loading control. (B) Immunoblots of green cone opsin, green-IAM, red cone opsin, and red-TSV regenerated with 11-cis-retinal and purified by immunoaffinity chromatography. Proteins were detected with an anti-rhodopsin C-terminal 1D4 tag antibody. Abbreviations: Ex, extract (total lysate); FT, flow-through; W, last wash; El, elution. (C) Difference absorption spectra of green cone opsin, green-IAM, red cone opsin, and red-TSV regenerated with 11-cis-retinal and purified by immunoaffinity chromatography. Spectra of green-IAM and red-TSV reveal shifts of 4–5 nm.
CONCLUSIONS
Evidence presented in this paper is consistent with human red cone opsin, but not green or blue cone opsin, forming a stable dimer in the live cell plasma membrane. This result was particularly surprising because of the high level of sequence similarity (>95%) between the red and green cone opsins. From the limited number of lipid-facing residues that differ between red and green cone opsins, we identified three amino acids in TM5 that are required for dimerization. By swapping these three amino acids from green cone opsin into red cone opsin, we observed a significant decrease in the level of dimerization. Conversely, the analogous green cone swap mutant showed a significant increase in the level of dimerization. All of our fluorescence methods, molecular brightness, mobility, FRET, and cross-correlation, yielded results consistent with this conclusion. Moreover, these microscopic observations were confirmed by membrane receptor cross-linking. This study is the first to show disruption of an opsin dimer with point mutations. It also is the first to identify a set of GPCRs for which receptor dimerization can be both eliminated and induced with point mutations.
Translating these results into the physiology of vision will require future investigation. For example, the lipid content of rod and cone membrane disks differs notably from that of the plasma membrane of Cos-7 cells,70 and the density range studied here is 2–3 orders of magnitude lower than the actual physiological densities.39 So, what physiological relevance could there be for red cone opsin dimerization? One possibility could be to stabilize the protein by reducing the effect of thermal fluctuations on receptor activation. For rhodopsin, the stable dimeric form is likely to prevent spontaneous activation and thus permits single-photon detection.71,72 Point mutations in TM5 of rhodopsin can increase dark noise and cause stationary night blindness before degenerative retinitis pigmentosa becomes evident.73,74 For green cone opsin, with an absorption spectrum similar to that of rhodopsin, dimerization could be less critical because it operates under high lighting conditions where low levels of spontaneous activation are less problematic. Red cone opsin also operates under high-light conditions, but its increased sensitivity to longer wavelengths makes it more susceptible to activation by thermal fluctuations. Red cone opsin does in fact have a significantly higher rate of thermal activation,75 and this likely needs to be suppressed for effective daytime vision. Consequently, stabilization in the dimeric conformation could be important for red cone opsin to prevent self-activation at body temperature.
Another potential physiological role for dimerization of red cone opsin is to modulate its spectral sensitivity. Our results show that the amino acids in TM5 responsible for dimerization also shift the maximal absorbance wavelength of the opsin–retinal complex. This finding suggests that dimerization could, through allosteric modulation of the protein structure, shift the transition energy of 11-cis-retinal-bound opsin. Alternatively, the dimeric forms of rhodopsin and red cone opsin could be essential for transport to the outer segment membranes or for development and stabilization of the photoreceptor cellular structure. One possibliy is that the dimeric form of cone opsin is a byproduct of the other evolutionary pressures that drove the spectral shift away from green cone opsin. Regardless of the answers to these questions, our results establish an essential link between the amino acid sequence of a functional GPCR and its propensity to form dimeric structures. They also provide a unique opportunity to investigate the chemical forces responsible for the lateral association of GPCRs and to further resolve the physiological role of dimerization.
Supplementary Material
Acknowledgments
We thank Dr. Leslie T. Webster, Jr., and members of the Palczewski laboratory for helpful comments on the manuscript and Dr. Wenyu Sun for the initial design of cone opsin DNA constructs. Dr. Jiane Zhang helped with the 11-cis-retinal regeneration experiments. We also thank Xiaojun Shi for providing feedback on the manuscript.
Funding
This work was supported by funding from National Institutes of Health Grants EY024451 (A.W.S.), EY025214 (B.J.), EY009339 (K.P.), EY024864 (K.P.), and EY021126 (K.P.) and by Research to Prevent Blindness (K.P.). K.P. is John H. Hord Professor of Pharmacology.
ABBREVIATIONS
- AFM
atomic force microscopy
- BRET
bioluminescence resonance energy transfer
- cpsm
counts per second per molecule
- CCF
cross-correlation function
- DDM
n-dodecyl β-D-maltoside
- DMSO
dimethyl sulfoxide
- DSG
disuccinimidyl glutarate
- DMEM
Dulbecco’s modified Eagle’s medium
- ER
endoplasmic reticulum
- eGFP
enhanced green fluorescence protein
- FCS
fluorescence correlation spectroscopy
- FCCS
fluorescence cross-correlation spectroscopy
- FRAP
fluorescence recovery after photobleaching
- FRET
Förster resonance energy transfer
- GPCRs
G protein-coupled receptors
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
- 5-HT
5-hydroxytryptamine
- NHS ester
N-hydroxysuccinimide ester
- PCR
polymerase chain reaction
- PVDF
polyvinyl difluoride
- PIE
pulsed-interleaved excitation
- PIE-FCCS
pulsed-interleaved excitation fluorescence cross-correlation spectroscopy
- RMSD
root-mean-square deviation
- SPAD
single-photon avalanche diode
- SDS–PAGE
sodium dodecyl sulfate– polyacrylamide gel electrophoresis
- TCSPC
time-correlated single-photon counting
- TM
transmembrane helix
- UV–vis
ultraviolet–visible
- WT
wild-type
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00877.
Figure S1–S12 (PDF)
ORCID
Adam W. Smith: 0000-0001-5216-9017
Author Contributions
B.J. and W.D.C. should be considered as joint first authors.
Notes
The authors declare no competing financial interest.
References
- 1.Endres NF, Barros T, Cantor AJ, Kuriyan J. Emerging concepts in the regulation of the EGF receptor and other receptor tyrosine kinases. Trends Biochem Sci. 2014;39:437–446. doi: 10.1016/j.tibs.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 2.Groves JT, Kuriyan J. Molecular mechanisms in signal transduction at the membrane. Nat Struct Mol Biol. 2010;17:659–665. doi: 10.1038/nsmb.1844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Khelashvili G, Dorff K, Shan J, Camacho-Artacho M, Skrabanek L, Vroling B, Bouvier M, Devi LA, George SR, Javitch JA, Lohse MJ, Milligan G, Neubig RR, Palczewski K, Parmentier M, Pin JP, Vriend G, Campagne F, Filizola M. GPCR-OKB: the G Protein Coupled Receptor Oligomer Knowledge Base. Bioinformatics. 2010;26:1804–1805. doi: 10.1093/bioinformatics/btq264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lambert NA. GPCR Dimers Fall Apart. Sci Signaling. 2010;3:e12. doi: 10.1126/scisignal.3115pe12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Milligan G. The Prevalence, Maintenance, and Relevance of G Protein—Coupled Receptor Oligomerization. Mol Pharmacol. 2013;84:158–169. doi: 10.1124/mol.113.084780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lodowski DT, Salom D, Le Trong I, Teller DC, Ballesteros JA, Palczewski K, Stenkamp RE. Crystal packing analysis of Rhodopsin crystals. J Struct Biol. 2007;158:455–462. doi: 10.1016/j.jsb.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu H, Wacker D, Mileni M, Katritch V, Han GW, Vardy E, Liu W, Thompson AA, Huang XP, Carroll FI, Mascarella SW, Westkaemper RB, Mosier PD, Roth BL, Cherezov V, Stevens RC. Structure of the human κ-opioid receptor in complex with JDTic. Nature. 2012;485:327–332. doi: 10.1038/nature10939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang J, Chen S, Zhang JJ, Huang XY. Crystal structure of oligomeric beta1-adrenergic G protein-coupled receptors in ligand-free basal state. Nat Struct Mol Biol. 2013;20:419–425. doi: 10.1038/nsmb.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu B, Chien EYT, Mol CD, Fenalti G, Liu W, Katritch V, Abagyan R, Brooun A, Wells P, Bi FC, Hamel DJ, Kuhn P, Handel TM, Cherezov V, Stevens RC. Structures of the CXCR4 Chemokine GPCR with Small-Molecule and Cyclic Peptide Antagonists. Science. 2010;330:1066–1071. doi: 10.1126/science.1194396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manglik A, Kruse AC, Kobilka TS, Thian FS, Mathiesen JM, Sunahara RK, Pardo L, Weis WI, Kobilka BK, Granier S. Crystal structure of the μ-opioid receptor bound to a morphinan antagonist. Nature. 2012;485:321–326. doi: 10.1038/nature10954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Periole X, Knepp AM, Sakmar TP, Marrink SJ, Huber T. Structural Determinants of the Supramolecular Organization of G Protein-Coupled Receptors in Bilayers. J Am Chem Soc. 2012;134:10959–10965. doi: 10.1021/ja303286e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simpson LM, Taddese B, Wall ID, Reynolds CA. Bioinformatics and molecular modelling approaches to GPCR oligomerization. Curr Opin Pharmacol. 2010;10:30–37. doi: 10.1016/j.coph.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 13.Nemoto W, Toh H. Prediction of interfaces for oligomerizations of G-protein coupled receptors. Proteins: Struct Funct Genet. 2005;58:644–660. doi: 10.1002/prot.20332. [DOI] [PubMed] [Google Scholar]
- 14.Filizola M, Weinstein H. Structural models for dimerization of G-protein coupled receptors: The opioid receptor homodimers. Biopolymers. 2002;66:317–325. doi: 10.1002/bip.10311. [DOI] [PubMed] [Google Scholar]
- 15.Jastrzebska B, Chen Y, Orban T, Jin H, Hofmann L, Palczewski K. Disruption of Rhodopsin Dimerization with Synthetic Peptides Targeting an Interaction Interface. J Biol Chem. 2015;290:25728. doi: 10.1074/jbc.M115.662684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Harikumar KG, Dong M, Cheng Z, Pinon DI, Lybrand TP, Miller LJ. Transmembrane Segment Peptides Can Disrupt Cholecystokinin Receptor Oligomerization without affecting Receptor Function. Biochemistry. 2006;45:14706–14716. doi: 10.1021/bi061107n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hayashi R, Osada S, Yoshiki M, Sugiyama D, Fujita I, Hamasaki Y, Kodama H. Superoxide production in human neutrophils is enhanced by treatment with transmembrane peptides derived from human formyl peptide receptor. J Biochem. 2006;139:981–988. doi: 10.1093/jb/mvj117. [DOI] [PubMed] [Google Scholar]
- 18.Wang J, He L, Combs CA, Roderiquez G, Norcross MA. Dimerization of CXCR4 in living malignant cells: control of cell migration by a synthetic peptide that reduces homologous CXCR4 interactions. Mol Cancer Ther. 2006;5:2474–2483. doi: 10.1158/1535-7163.MCT-05-0261. [DOI] [PubMed] [Google Scholar]
- 19.Baneres JL, Parello J. Structure-based Analysis of GPCR Function: Evidence for a Novel Pentameric Assembly between the Dimeric Leukotriene B4 Receptor BLT1 and the G-protein. J Mol Biol. 2003;329:815–829. doi: 10.1016/s0022-2836(03)00439-x. [DOI] [PubMed] [Google Scholar]
- 20.Hebert TE, Moffett S, Morello JP, Loisel TP, Bichet DG, Barret C, Bouvier M. A Peptide Derived from a β2-Adrenergic Receptor Transmembrane Domain Inhibits Both Receptor Dimerization and Activation. J Biol Chem. 1996;271:16384–16392. doi: 10.1074/jbc.271.27.16384. [DOI] [PubMed] [Google Scholar]
- 21.Xue L, Rovira X, Scholler P, Zhao H, Liu J, Pin JP, Rondard P. Major ligand-induced rearrangement of the heptahelical domain interface in a GPCR dimer. Nat Chem Biol. 2014;11:134–140. doi: 10.1038/nchembio.1711. [DOI] [PubMed] [Google Scholar]
- 22.Dong M, Pinon DI, Bordner AJ, Miller LJ. Elucidation of the active conformation of the amino terminus of receptor-bound secretin using intramolecular disulfide bond constraints. Bioorg Med Chem Lett. 2010;20:6040–6044. doi: 10.1016/j.bmcl.2010.08.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mancia F, Assur Z, Herman AG, Siegel R, Hendrickson WA. Ligand sensitivity in dimeric associations of the serotonin 5HT2c receptor. EMBO Rep. 2008;9:363–369. doi: 10.1038/embor.2008.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kota P, Reeves PJ, RajBhandary UL, Khorana HG. Opsin is present as dimers in COS1 cells: Identification of amino acids at the dimeric interface. Proc Natl Acad Sci U S A. 2006;103:3054–3059. doi: 10.1073/pnas.0510982103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo W, Shi L, Javitch JA. The Fourth Transmembrane Segment Forms the Interface of the Dopamine D2 Receptor Homodimer. J Biol Chem. 2003;278:4385–4388. doi: 10.1074/jbc.C200679200. [DOI] [PubMed] [Google Scholar]
- 26.Marsango S, Caltabiano G, Pou C, Varela Liste MJ, Milligan G. Analysis of Human Dopamine D3 Receptor Quaternary Structure. J Biol Chem. 2015;290:15146–15162. doi: 10.1074/jbc.M114.630681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gorinski N, Kowalsman N, Renner U, Wirth A, Reinartz MT, Seifert R, Zeug A, Ponimaskin E, Niv MY. Computational and Experimental Analysis of the Transmembrane Domain 4/5 Dimerization Interface of the Serotonin 5-HT1A Receptor. Mol Pharmacol. 2012;82:448–463. doi: 10.1124/mol.112.079137. [DOI] [PubMed] [Google Scholar]
- 28.McMillin SM, Heusel M, Liu T, Costanzi S, Wess J. Structural Basis of M3Muscarinic Receptor Dimer/Oligomer Formation. J Biol Chem. 2011;286:28584–28598. doi: 10.1074/jbc.M111.259788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Harikumar KG, Pinon DI, Miller LJ. Transmembrane Segment IV Contributes a Functionally Important Interface for Oligomerization of the Class II G Protein-coupled Secretin Receptor. J Biol Chem. 2007;282:30363–30372. doi: 10.1074/jbc.M702325200. [DOI] [PubMed] [Google Scholar]
- 30.Salahpour A, Angers S, Mercier JF, Lagacé M, Marullo S, Bouvier M. Homodimerization of the β2-Adrenergic Receptor as a Prerequisite for Cell Surface Targeting. J Biol Chem. 2004;279:33390–33397. doi: 10.1074/jbc.M403363200. [DOI] [PubMed] [Google Scholar]
- 31.Hernanz-Falcon P, Rodriguez-Frade JM, Serrano A, Juan D, del Sol A, Soriano SF, Roncal F, Gomez L, Valencia A, Martinez-A C, Mellado M. Identification of amino acid residues crucial for chemokine receptor dimerization. Nat Immunol. 2004;5:216–223. doi: 10.1038/ni1027. [DOI] [PubMed] [Google Scholar]
- 32.Dorsch S, Klotz KN, Engelhardt S, Lohse MJ, Bunemann M. Analysis of receptor oligomerization by FRAP microscopy. Nat Methods. 2009;6:225–230. doi: 10.1038/nmeth.1304. [DOI] [PubMed] [Google Scholar]
- 33.Hern JA, Baig AH, Mashanov GI, Birdsall B, Corrie JET, Lazareno S, Molloy JE, Birdsall NJM. Formation and dissociation of M1 muscarinic receptor dimers seen by total internal reflection fluorescence imaging of single molecules. Proc Natl Acad Sci U S A. 2010;107:2693–2698. doi: 10.1073/pnas.0907915107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fonseca JM, Lambert NA. Instability of a Class A G Protein-Coupled Receptor Oligomer Interface. Mol Pharmacol. 2009;75:1296–1299. doi: 10.1124/mol.108.053876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rashid AJ, So CH, Kong MMC, Furtak T, El-Ghundi M, Cheng R, O’Dowd BF, George SR. D1–D2 dopamine receptor heterooligomers with unique pharmacology are coupled to rapid activation of Gq/11 in the striatum. Proc Natl Acad Sci U S A. 2007;104:654–659. doi: 10.1073/pnas.0604049104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Frederick AL, Yano H, Trifilieff P, Vishwasrao HD, Biezonski D, Meszaros J, Urizar E, Sibley DR, Kellendonk C, Sonntag KC, Graham DL, Colbran RJ, Stanwood GD, Javitch JA. Evidence against dopamine D1/D2 receptor heteromers. Mol Psychiatry. 2015;20:1373–1385. doi: 10.1038/mp.2014.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lan TH, Liu Q, Li C, Wu G, Steyaert J, Lambert NA. BRET evidence that β2 adrenergic receptors do not oligomerize in cells. Sci Rep. 2015;5:10166. doi: 10.1038/srep10166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Angers S, Salahpour A, Joly E, Hilairet S, Chelsky D, Dennis M, Bouvier M. Detection of β2-adrenergic receptor dimerization in living cells using bioluminescence resonance energy transfer (BRET) Proc Natl Acad Sci U S A. 2000;97:3684–3689. doi: 10.1073/pnas.060590697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hofmann L, Palczewski K. Advances in understanding the molecular basis of the first steps in color vision. Prog Retinal Eye Res. 2015;49:46–66. doi: 10.1016/j.preteyeres.2015.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Palczewski K, Kumasaka T, Hori T, Behnke CA, Motoshima H, Fox BA, Trong IL, Teller DC, Okada T, Stenkamp RE, Yamamoto M, Miyano M. Crystal Structure of Rhodopsin: A G Protein-Coupled Receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 41.Fotiadis D, Liang Y, Filipek S, Saperstein DA, Engel A, Palczewski K. Atomic-force microscopy: Rhodopsin dimers in native disc membranes. Nature. 2003;421:127–128. doi: 10.1038/421127a. [DOI] [PubMed] [Google Scholar]
- 42.Mansoor SE, Palczewski K, Farrens DL. Rhodopsin self-associates in asolectin liposomes. Proc Natl Acad Sci U S A. 2006;103:3060–3065. doi: 10.1073/pnas.0511010103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jastrzebska B, Ringler P, Palczewski K, Engel A. The rhodopsin-transducin complex houses two distinct rhodopsin molecules. J Struct Biol. 2013;182:164–172. doi: 10.1016/j.jsb.2013.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gunkel M, Schoneberg J, Alkhaldi W, Irsen S, Noe F, Kaupp UB, Al-Amoudi A. Higher-Order Architecture of Rhodopsin in Intact Photoreceptors and Its Implication for Phototransduction Kinetics. Structure. 2015;23:628–638. doi: 10.1016/j.str.2015.01.015. [DOI] [PubMed] [Google Scholar]
- 45.Comar WD, Schubert SM, Jastrzebska B, Palczewski K, Smith AW. Time-Resolved Fluorescence Spectroscopy Measures Clustering and Mobility of a G Protein-Coupled Receptor Opsin in Live Cell Membranes. J Am Chem Soc. 2014;136:8342–8349. doi: 10.1021/ja501948w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fotiadis D, Jastrzebska B, Philippsen A, Muller DJ, Palczewski K, Engel A. Structure of the rhodopsin dimer: a working model for G-protein-coupled receptors. Curr Opin Struct Biol. 2006;16:252–259. doi: 10.1016/j.sbi.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 47.Stenkamp RE, Filipek S, Driessen CAGG, Teller DC, Palczewski K. Crystal structure of rhodopsin: a template for cone visual pigments and other G protein-coupled receptors. Biochim Biophys Acta, Biomembr. 2002;1565:168–182. doi: 10.1016/s0005-2736(02)00567-9. [DOI] [PubMed] [Google Scholar]
- 48.Zhang T, Cao LH, Kumar S, Enemchukwu NO, Zhang N, Lambert A, Zhao X, Jones A, Wang S, Dennis EM, Fnu A, Ham S, Rainier J, Yau KW, Fu Y. Dimerization of visual pigments in vivo. Proc Natl Acad Sci U S A. 2016;113:9093–9098. doi: 10.1073/pnas.1609018113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang T, Fu Y. A Phe-rich region in short- wavelength sensitive opsins is responsible for their aggregation in the absence of 11-cis-retinal. FEBS Lett. 2013;587:2430–2434. doi: 10.1016/j.febslet.2013.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Russell D. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 2001. [Google Scholar]
- 51.Marita M, Wang Y, Kaliszewski MJ, Skinner KC, Comar WD, Shi X, Dasari P, Zhang X, Smith AW. Class A Plexins Are Organized as Preformed Inactive Dimers on the Cell Surface. Biophys J. 2015;109:1937–1945. doi: 10.1016/j.bpj.2015.04.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith AW. Detection of rhodopsin dimerization in situ by PIE-FCCS, a time-resolved fluorescence spectroscopy. Methods Mol Biol. 2015;1271:205–219. doi: 10.1007/978-1-4939-2330-4_14. [DOI] [PubMed] [Google Scholar]
- 53.Boussif O, Lezoualc’h F, Zanta MA, Mergny MD, Scherman D, Demeneix B, Behr JP. A versatile vector for gene and oligonucleotide transfer into cells in culture and in vivo: polyethylenimine. Proc Natl Acad Sci U S A. 1995;92:7297–7301. doi: 10.1073/pnas.92.16.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen Y, Tang H. High-throughput screening assays to identify small molecules preventing photoreceptor degeneration caused by the rhodopsin P23H mutation. Methods Mol Biol. 2015;1271:369–390. doi: 10.1007/978-1-4939-2330-4_24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Di Rienzo C, Gratton E, Beltram F, Cardarelli F. Fast spatiotemporal correlation spectroscopy to determine protein lateral diffusion laws in live cell membranes. Proc Natl Acad Sci U S A. 2013;110:12307–12312. doi: 10.1073/pnas.1222097110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wawrezinieck L, Rigneault H, Marguet D, Lenne PF. Fluorescence Correlation Spectroscopy Diffusion Laws to Probe the Submicron Cell Membrane Organization. Biophys J. 2005;89:4029–4042. doi: 10.1529/biophysj.105.067959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Naji A, Levine AJ, Pincus PA. Corrections to the Saffman-Delbruck Mobility for Membrane Bound Proteins. Biophys J. 2007;93:L49–L51. doi: 10.1529/biophysj.107.119222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chung I, Akita R, Vandlen R, Toomre D, Schlessinger J, Mellman I. Spatial control of EGF receptor activation by reversible dimerization on living cells. Nature. 2010;464:783–787. doi: 10.1038/nature08827. [DOI] [PubMed] [Google Scholar]
- 60.Bacia K, Kim SA, Schwille P. Fluorescence cross-correlation spectroscopy in living cells. Nat Methods. 2006;3:83–89. doi: 10.1038/nmeth822. [DOI] [PubMed] [Google Scholar]
- 61.Muller BK, Zaychikov E, Brauchle C, Lamb DC. Pulsed Interleaved Excitation. Biophys J. 2005;89:3508–3522. doi: 10.1529/biophysj.105.064766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Endres NF, Das R, Smith AW, Arkhipov A, Kovacs E, Huang Y, Pelton JG, Shan Y, Shaw DE, Wemmer DE, Groves JT, Kuriyan J. Conformational Coupling across the Plasma Membrane in Activation of the EGF Receptor. Cell. 2013;152:543556. doi: 10.1016/j.cell.2012.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nathans J, Thomas D, Hogness DS. Molecular genetics of human color vision: the genes encoding blue, green, and red pigments. Science. 1986;232:193–202. doi: 10.1126/science.2937147. [DOI] [PubMed] [Google Scholar]
- 64.Weisz O, Swift A, Machamer C. Oligomerization of a membrane protein correlates with its retention in the Golgi complex. J Cell Biol. 1993;122:1185–1196. doi: 10.1083/jcb.122.6.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Soulie S, Denoroy L, Caer JPL, Hamasaki N, Groves JD, Maire Ml. Treatment with Crystalline Ultra-Pure Urea Reduces the Aggregation of Integral Membrane Proteins without Inhibiting N-Terminal Sequencing. J Biochem. 1998;124:417–420. doi: 10.1093/oxfordjournals.jbchem.a022128. [DOI] [PubMed] [Google Scholar]
- 66.Javitch JA. The Ants Go Marching Two by Two: Oligomeric Structure of G-Protein-Coupled Receptors. Mol Pharmacol. 2004;66:1077–1082. doi: 10.1124/mol.104.006320. [DOI] [PubMed] [Google Scholar]
- 67.Asenjo AB, Rim J, Oprian DD. Molecular determinants of human red/green color discrimination. Neuron. 1994;12:1131–1138. doi: 10.1016/0896-6273(94)90320-4. [DOI] [PubMed] [Google Scholar]
- 68.Neitz J, Neitz M. The genetics of normal and defective color vision. Vision Res. 2011;51:633–651. doi: 10.1016/j.visres.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Srinivasan S, Cordomí A, Ramon E, Garriga P. Beyond spectral tuning: human cone visual pigments adopt different transient conformations for chromophore regeneration. Cell Mol Life Sci. 2016;73:1253–1263. doi: 10.1007/s00018-015-2043-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jastrzebska B, Debinski A, Filipek S, Palczewski K. Role of membrane integrity on G protein-coupled receptors: Rhodopsin stability and function. Prog Lipid Res. 2011;50:267–277. doi: 10.1016/j.plipres.2011.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Baylor DA, Lamb TD, Yau KW. Responses of retinal rods to single photons. J Physiol. 1979;288:613–634. [PMC free article] [PubMed] [Google Scholar]
- 72.Rieke F, Baylor DA. Origin of Reproducibility in the Responses of Retinal Rods to Single Photons. Biophys J. 1998;75:1836–1857. doi: 10.1016/S0006-3495(98)77625-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang N, Kolesnikov AV, Jastrzebska B, Mustafi D, Sawada O, Maeda T, Genoud C, Engel A, Kefalov VJ, Palczewski K. Autosomal recessive retinitis pigmentosa E150K opsin mice exhibit photoreceptor disorganization. J Clin Invest. 2013;123:121–137. doi: 10.1172/JCI66176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Okada T, Ernst OP, Palczewski K, Hofmann KP. Activation of rhodopsin: new insights from structural and biochemical studies. Trends Biochem Sci. 2001;26:318–324. doi: 10.1016/s0968-0004(01)01799-6. [DOI] [PubMed] [Google Scholar]
- 75.Kefalov V, Fu Y, Marsh-Armstrong N, Yau KW. Role of visual pigment properties in rod and cone phototransduction. Nature. 2003;425:526–531. doi: 10.1038/nature01992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


