Abstract
Mutations/deletions in the SHANK3 gene are associated with autism spectrum disorders and intellectual disability. Here, we present electrophysiological and behavioral consequences in novel heterozygous and homozygous mice with a transcriptional stop cassette inserted upstream of the PDZ domain-coding exons in Shank3 (Shank3E13). Insertion of a transcriptional stop cassette prior to exon 13 leads to loss of the two higher molecular weight isoforms of Shank3. Behaviorally, both Shank3E13 heterozygous (HET) and homozygous knockout (KO) mice display increased repetitive grooming, deficits in social interaction tasks, and decreased rearing. Shank3E13 KO mice also display deficits in spatial memory in the Morris water maze task. Baseline hippocampal synaptic transmission and short-term plasticity are preserved in Shank3E13 HET and KO mice, while both HET and KO mice exhibit impaired hippocampal long-term plasticity. Additionally, Shank3E13 HET and KO mice display impaired striatal glutamatergic synaptic transmission. These results demonstrate for the first time in this novel Shank3 mutant that both homozygous and heterozygous mutation of Shank3 lead to behavioral abnormalities with face validity for autism along with widespread synaptic dysfunction.
Keywords: autism spectrum disorder, Shank3, Phelan-McDermid syndrome, mouse model, social interaction, grooming
Introduction
Shank3 is a multidomain protein in the postsynaptic density (PSD) of excitatory synapses [Boeckers et al., 2002; Naisbitt et al., 1999]. It contains functional and diverse binding motifs such as an ankyrin repeat, SH3, prolinerich, PDZ, and SAM domains. Through its PDZ domain, Shank3 interacts with GKAP/PSD-95/glutamate receptor complexes [Ehlers, 1999; Naisbitt et al., 1999], and through its Homer binding domain, it binds to the Homer-group1 metabotropic glutamate receptor (mGluR1) complex [Tu et al., 1999]. Through these interactions Shank3 aids in assembly of signaling complexes at the synapse to govern spine development, protein targeting, and signal transduction [Arons et al., 2012; Duffney et al., 2013; Ehlers, 1999, 2002; Park et al., 2003; Roussignol et al., 2005; Verpelli et al., 2011].
Genetic alteration of the SHANK3 gene is associated with neuropsychiatric disorders [Bonaglia et al., 2001; Durand et al., 2007; Gong, Lippa, Zhu, Lin, & Rosso, 2009; Gauthier et al., 2010; Grabrucker et al., 2014; Misceo et al., 2011; Moessner et al., 2007; Vucurovic et al., 2012]. Heterozygous deletion of SHANK3 is causative in the 22q13 deletion syndrome (Phelan-McDermid syndrome, PMS), characterized by hypotonia, social withdrawal, delayed psychomotor development, repetitive behaviors, and mild dysmorphic features [Kolevzon et al., 2011; Sarasua et al., 2011; Soorya et al., 2013]. Approximately 75% of PMS patients meet autism diagnostic criteria [Soorya et al., 2013]. Mutations/deletions of SHANK3 are also associated with autism spectrum disorders (ASD) and schizophrenia [Bozdagi et al., 2010; Gauthier et al., 2010; Grabrucker et al., 2014; Guilmatre et al., 2014; Herbert, 2011].
Tremendous phenotypic heterogeneity exists in patients with alterations in SHANK3, which can pose challenges to diagnosis and treatment. One potential contributor is the extensive array of SHANK3 isoforms. ASD-associated SHANK3 alterations can disrupt isoform expression or domain structure and function in the various isoforms. Typically, studies involving Shank3 protein expression focus on three major isoforms (α β γ) [Han et al., 2013; Peca et al., 2011; Wang et al., 2011]. Additional studies, however, reveal that the Shank3 gene has multiple promoters and is alternatively spliced, suggesting that the number of Shank3 isoforms can be extensive [Kouser et al., 2013; Speed et al., 2015; Wang et al., 2011; Wang, Xu, Bey, Lee, & Jiang, 2014].
To further evaluate Shank3 isoform-specific phenotypes we generated a Shank3 mutant mouse targeting disruption of the PDZ domain with a transcriptional stop cassette prior to exon 13, leading to loss of the two higher molecular weight isoforms. These Shank3E13 heterozygous (HET) and homozygous (KO) mutants display phenotypes with face validity for autism such as increased repetitive grooming and deficits in social interaction. Shank3E13 KO mice also display spatial memory deficits while HET and KO mice display impaired hippocampal LTP and striatal glutamatergic synaptic transmission.
Materials and Methods
Shank3E13 Mutant Mice
The construct inserted a neo-stop cassette into a unique Bgl II site in intron 12 of the Shank3 gene [Dragatsis & Zeitlin, 2001; Guy, Selfridge, Cobb, & Bird, 2007]. The neo-Stop cassette was flanked by loxP sites. Targeting vector was pBluescript II SK (±) (Agilent). The final construct had a 5′ homology (1042 bp) and a 3′ homology (5823 bp) arm with a 3020 bp DNA fragment containing floxed neo-stop cassette inserted. A DT (diphtheria toxin) cassette was included for negative selection. The linearized construct (by Sal1) was electroporated into embryonic stem (ES) cells (129s6SvEvTac) and clones were selected for G418 resistance. The correctly targeted ES clones were identified by PCR using three primers (Forward: screen-13E-s-1 CATGGTAAGTGACTCCAG TTTTTGCTCAGATAC, Reverse (1): screen-13E-s-2-1 GAATTCGATGACCTCGAGGATCTATAACTTCG, Reverse (2): screen-13E-s-3 CAACTTAAAGTCGAG GTTGACAGA GGTTACCAC); WT5921 bp band, KI (knock-in)51267 bp band. Correct targeting was confirmed by sequencing PCR products, and genomic DNA was analyzed by Southern blot. Positive ES clones were injected into blastocysts (C57BL6J) generating chimeras at the UT Southwestern Transgenic Facility. Chimeras were bred with C57BL/6J to confirm germline transmission identified by PCR with three primers: Forward (1): loxp1-sequence CCATGCGTCAAACTTGATGGATGCCTAG, Forward (2): 24M–up-loxp2–501 CTTACTCCACACAGGCATAGAGTGTCTG, Reverse: 13E-Gtyping-antisense-1 GAGCACTAACTGCTCTTCTGAAGGTC; WT DNA = 332 bp band, KI DNA = 576 bp band. The KI mice (Shank3E13) were backcrossed with C57BL/6J for at least four generations.
Other Shank3 Mutant Mice
Previously we generated and characterized three other Shank3 mutant mice e4–9 [Jaramillo et al., 2016] and G/G [Speed et al., 2015], ΔC/ΔC [Kouser et al., 2013]. The ΔC/ΔC Shank3 mice were a collaborative gift from Dr. Paul Worley. These mice were all used in this study to compare exon presence using Southern blot analysis in Figure 1.
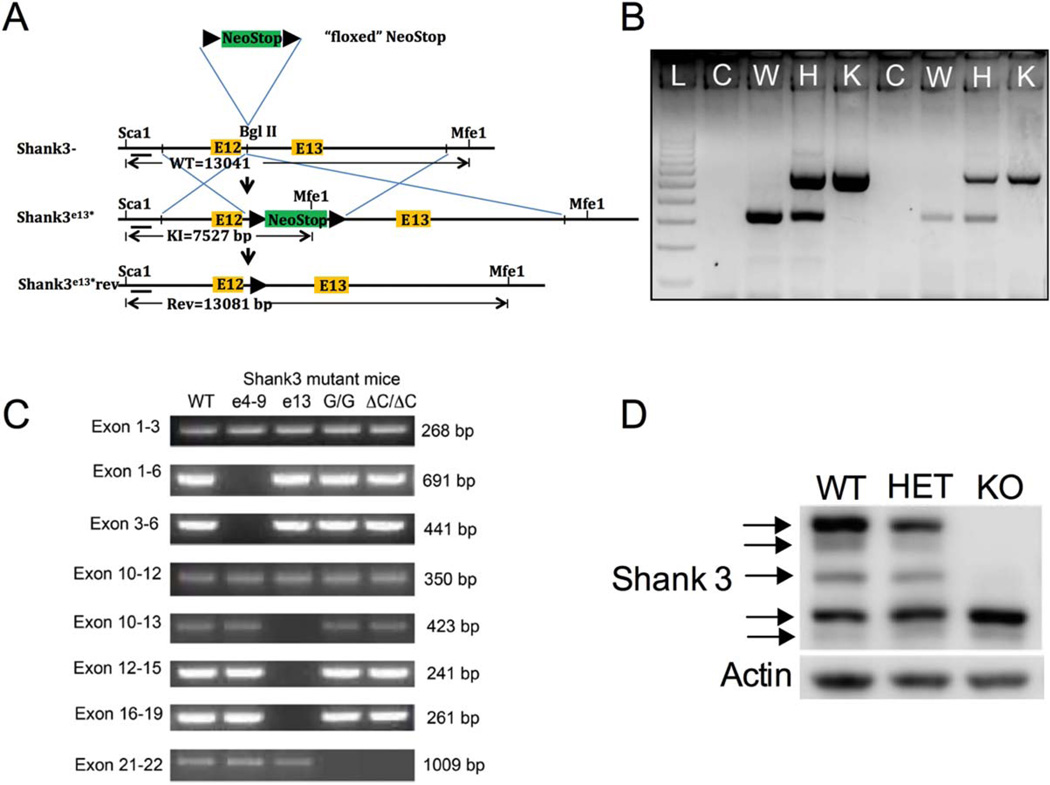
Figure 1.
Generation of Shank3E13 mice. (A) The schema showing the location of the Neo-Stop cassette within intron 12 (Bgl II) near WT exon 13 of Shank3, wild-type Shank3 allele (Shank3-wt), product of homologous recombination (Shank3E13*) and hypothetical genetic reversal (Shank3E13*rev) by cre recombinase. The position of a Shank3 5′ flanking probe along with the size of diagnostic restriction fragments identifying the wild type, Shank3E13* and Shank3E13*rev are indicated. The restriction sites are: Sca1 and Mfe1. The black arrowheads represent loxP sites. (B) Genotyping analysis of WT (W), HET (H), and KO (K). WT mice produced a single 332 bp band while the KO produced a 576 bp band. HET produced bands of both sizes. A sample with no DNA was loaded as a control (C). “L” designates the 100bp ladder. (C) RT-PCR of cDNA from brain tissue of 4-week old (WT): wildtype, (e4–9):Shank3e4–9 (Jaramillo et al., 2015a), (e13):Shank3e13, (G/G):Shank3G/G (Speed et al., 2015), ΔC/ΔC: Shank3ΔC/ΔC (Kouser et al., 2013) mice. The Shank3 gene contains five functional domains; Ankyrin binding domain from exons 4 to 9, SH3 domain from exons 11 to 12, PDZ domain from exons 14 to 16, Proline rich domain from exon 21, and SAM domain from exon 22. Shank3e13 mice show a loss of bands from exons 13 to 19 which indicates a lack of PDZ domain-encoding exons. Shank3e4–9 shows a loss of bands from exons 4 to 9. Shank3G/G and Shank3ΔC/ΔC show a loss of band containing exons 21–22. (D) Whole brain cell lysates from WT, HET, and KO Shank3E13 mice probed with C-terminal Shank3 antibody.
Western Blot
Synaptosome and whole cell lysate and preparation
Synaptosomes and whole cell lysates were prepared as previously described [Kouser et al., 2013].
Western blot and antibodies
For Western blotting, samples were diluted in 2 × Laemmli sample buffer containing 2-mercaptoethanol (Bio-Rad, Hercules, CA, USA) and boiled for 10 min. Five to twenty microgram of protein were loaded and blotted with antibodies specific for synaptic proteins: anti-GluR1 (Abcam, Cambridge, England), anti-GluR2 (Invitrogen, Carlsbad, CA, USA), anti-GluR3 (Cell Signaling, Danvers, MA, USA), anti-NR1 (Sigma, St. Louis, MO, USA), anti-NR2A (Sigma, St. Louis, MO, USA), anti-NR2B (NeuroMAB, Davis, CA, USA), anti-Shank3 C-terminal (gift from P Worley; raised against residues 1379–1740 and 1379–1675), anti-mGluR5 (Millipore, Billerica, MA, USA), anti-PSD-95 (Calbiochem, Billerica, MA, USA), anti-Homer 1 b/c (Santa Cruz, Santa Cruz, CA, USA). Anti-β-Actin (Sigma, St. Louis, MO, USA) was used as loading control. Blots were incubated with HRP-conjugated anti-rabbit, anti-mouse secondary antibodies (GE Healthcare, Dallas, TX, USA). For some samples, we used clarity western ECL (Bio-Rad, Hercules, CA, USA), and exposed film was developed by an Image Works film processor. We also used fluorescent secondary IRDye® anti-Rabbit (product #: 926–32212) or anti-mouse (product #: 926–68071) at 1:10,000 (LI-COR) for some. Fluorescence was visualized with Odyssey fc® . Signals were quantified using Image Studio (LiCor, Lincoln, Nebraska, USA), normalized to β–actin, and analyzed using Microsoft Excel (Redmond, WA) and Graphpad Prism (La Jolla, CA).
Behavioral overview
All mice were age- and sex-matched, littermate progeny of heterozygous Shank3E13 mutants. Behaviors were performed by experimenters blind to genotypes. The behavioral cohort consisted of 55 mice; n = 20 wildtype (WT, ten female and ten male), n = 15 heterozygous (HET, seven male and eight female), n = 20 homozygous (HOMO, ten female and ten male), that were 5.4– 9 weeks of age to start. Thus, we tested seven male littermate trios (WT/het/homo), eight female littermate trios (WT/het/homo), three male littermate pairs (WT/homo), and two female littermate pairs (WT/homo). Behaviors were tested in the following order: elevated plus maze, dark/light, open field, locomotor, rearing, grooming, 3-box social interaction, caged conspecific (social interaction with a caged adult), rotarod, juvenile social interaction, genotype/sex-matched adult social interaction, marble burying, Morris water maze, prepulse inhibition of startle, and startle threshold. Results are presented in different order than tested to ease the flow of presentation. Analysis used StatPlus software (Version 2015, AnalystSoft, Alexandria, VA, USA) using either two-way ANOVAs or three-way repeated measures ANOVA with genotype and sex as the main variables and appropriate repeated measure. Post-hoc planned comparisons were applied for significant effects and interactions. For detailed statistical results see Table 1.
Table 1.
Statistical Analysis Details
| Social Interaction Test | ||||
|---|---|---|---|---|
| N | Test | Data | Variables | Statistical Test (significant data in bold) |
| N = 20 WT, 20 HET, 15 KO |
3-Box Social Interaction |
Trial 1: Base- Line Fig. 2A |
Sex, Genotype, & Chamber | 3-way rmANOVA; Main effect of Sex: F(1,129) = 0.00118, P = 0.972; Main effect of Genotype: F(2,129) = 0.00112, P = 0.998; Main effect of Chamber: F(1,129) = 7.37, P < 0.00093; Main effect of Sex × Genotype Interaction: F(2,129) = 0.00137, P = 0.998; Main effect of Genotype × Chamber: F(4,129) = 1.59, P = 0.180; Main effect of Sex × Chamber: F(2,129) 5 0.028, P = 0.9719; Main effect of Sex × Genotype × Chamber: F(4,129) = 0.213, P = 0.9304 |
| Posthoc Scheffe contrast of means (Chamber) | ||||
| Back vs. Front | P = 0.447 | |||
| Back vs. Middle | P < 0.00112 | |||
| Front vs. Middle | P < 0.046 | |||
| Trial 2: Social Preference Fig. 2B |
Sex, Genotype, & Interaction Time |
3-way rmANOVA; Main effect of Sex: F(1,129) = 0.557, P = 0.457; Main effect of Genotype: F(2,129) = 1.149, P = 0.286; Main effect of Chamber: F(1,129) = 13.74, P < 0.00036; Main effect of Sex × Genotype Interaction: F(2,129) = 0.289, P = 0.591; Main effect of Genotype × Chamber: F(4,129) =0.0026, P = 0.959; Main effect of Sex × Chamber: F(2,129) = 1.003, P = 0.319; Main effect of Sex × Genotype × Chamber: F(4,129) = 0.894, P = 0.346 |
||
| Interaction zone (Social Time vs. Inanimate Time) within each Genotype |
Planned comparisons: WT: P < 0.010, HET: P = 0.575, KO: P < 0.0091 |
|||
| Trial 3: Social Novelty Fig. 2C |
Sex, Genotype, & Interaction Target |
3-way rmANOVA; Main effect of Sex: F(1,129) = 0.802, P = 0.3727; Main effect of Genotype: F(2,129) = 1.18, P = 0.311; Main effect of Target: F(1,129) = 0.801, P < 0.00009; Main effect of Sex × Genotype Interaction: F(2,129) = 1.344, P = 0.266; Main effect of Genotype × Target: F(4,129) = 1.94, P = 0.149; Main effect of Sex × Target: F(2,129) = 0.163, P = 0.686; Main effect of Sex × Genotype × Target: F(4,129) = 0.555, P = 0.575 |
||
| Interaction zone (Novel vs. Familiar) within each Genotype |
Planned comparisons: WT: P < 0.016, HET: P = 0.768, KO: P = 0.352 |
|||
| N = 20 WT, 20 HET, 15 KO |
Caged Conspecific |
Interaction w/caged mouse Fig 2D |
Sex & Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 1.48, P = 0.228; Main effect of Genotype: F(2,54) = 2.03, P = 0.14; Sex × Genotype Interaction: F(2,54) = 2.69, P = 0.077 |
| Planned Comparison | ||||
| WT vs. HET | P = 0.686 | |||
| WT vs. KO | P < 0.0066 | |||
| HET vs. KO | P < 0.042 | |||
| N = 10 WT, 10 HET, 7 KO (All Pairs) |
Genotype-Sex match |
Duration of total interac- tion 2E |
Sex & Genotype | 2-way ANOVA; Main effect of Sex: F(1,55) = 1.23, P = 0.272; Main effect of Genotype: F(2,55) = 8.073, P < 0.0009; Sex × Genotype Interaction: F(2,55) = 0.325, P = 0.723 |
| Posthoc Scheffe contrast of means | ||||
| WT vs. HET | P = 0.99 | |||
| WT vs. KO | P < 0.003 | |||
| HET vs. KO | P < 0.005 | |||
| Number of Interaction bouts 2F |
Sex & Genotype | 2-way ANOVA; Main effect of Sex: F(1,55) = 12.54, P < 0.0008; Main effect of Genotype: F(2,55) = 25.69, P < 2.1–08; Sex × Genotype Interaction: F(2,55) = 3.08, P = 0.054 |
||
| Posthoc Scheffe contrast of means (genotype) |
||||
| WT vs. HET | P = 0.062 | |||
| WT vs. KO | P < 2.09–08 | |||
| HET vs. KO Posthoc Scheffe contrast of means (sex) |
P < 0.0004 | |||
| WT Male vs. Female | P < 0.00079 | |||
| HET Male vs. Female | P = 0.919 | |||
|
KO Male vs. Female Posthoc Scheffe contrast of means (female) |
P < 0.017 | |||
| WT vs. HET | P < 0.0048 | |||
| WT vs. KO | P < 0.00001 | |||
| HET vs. KO | P = 0.205 | |||
| WT/HET/KO Mean/SEM Posthoc Scheffe contrast of means (Male) |
27/1.01, 19/2.01, 14.8/1.46 | |||
| WT vs. HET | P = 0.999 | |||
| WT vs. KO | P < 0.00028 | |||
| HET vs. KO | P < 0.00013 | |||
| WT/HET/KO Mean/SEM | 19.2/1.33, 19.25/1.56, 9.4/2.08 | |||
| N = 20 WT, 20 HET, 15 KO |
Social Interaction w/Juvenile |
Social Interac- tion Fig. 2G |
Sex, Genotype, & Trial | 3-way rmANOVA; Main effect of Sex: F(1,88) =0.056, P = 0.812; Main effect of Genotype: F(2,88) = 0.198, P = 0.820; Main effect of Trial: F(1,88) = 3.587, P = 0.061; Main effect of Sex × Genotype Interaction: F(2,88) = 1.37, P = 0.259; Main effect of Genotype × Trial: F(2,88) = 3.74, P < 0.027; Main effect of Sex × Trial: F(1,88) = 0.115, P = 0.735; Main effect of Sex × Genotype × Trial: F(2,88) = 1.175, P = 0.313 |
| Trial (Initial Test Interaction time vs. Recognition Test Interaction time) within each Genotype |
Planned comparisons: WT: P < 0.000523, HET: P = 0.901, KO: 0.974. |
|||
| Repetitive Behavior Test | ||||
| N | Test | Data Analyzed | Variables | Statistic Test |
| N = 20 WT, 20 HET, 15 KO |
Grooming | Time Spent Grooming (sec) Fig. 3A |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,53) =21.50, P < 0.04; Main effect of Genotype: F(2,53) = 88.40, P < 0.011; Main effect of Sex × Genotype Interaction: F(2,53) = 0.295, P = 0.E+0 |
| Planned Comparison | ||||
| WT vs. KO | P < 1.21E-08 | |||
| WT vs. HET | P < 0.0008 | |||
| HET vs. KO Posthoc Scheffe Contrast of means (sex) |
P = 0.136 | |||
| WT Male vs Female | P = 0.398 | |||
| HET Male vs Female | P < 0.0454 | |||
| KO Male vs Female | P = 0.162 | |||
| Posthoc Scheffe Contrast of means (sex) Female |
||||
| WT vs HET | P < 0.0264 | |||
| WT vs KO | P < 0.00017 | |||
| HET vs KO | P < 0.2976 | |||
| WT/HET/KO Mean/SEM Posthoc Scheffe Contrast of means (sex) Male |
71.27/10.8, 140.5/21.24, 180.55/20.34 | |||
| WT vs HET | P < 0.00077 | |||
| WT vs KO | P < 0.00001 | |||
| HET vs KO | P < 0.7714 | |||
| WT/HET/KO Mean/SEM | 91.1/14.7, 195.99/21.63, 214.59/17.03 | |||
| Anxiety and Motor Behavior Test | ||||
| N | Test | Data Analyzed | Variables | Statistical Test |
| N = 20 WT, 20 HET, 15 KO |
Open field | Time in center Fig. 3B |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 0.189, P = 0.66; Main effect of Genotype: F(2,54) = 1.35, P = 0.266; Sex × Genotype Interaction: F(2,54) = 0.198, P = 0.82 |
| Total Distance Traveled Fig. 3C |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 0.3.76, P = 0.19; Main effect of Genotype: F(2,54) = 8.41, P = 0.10; Main effect of Sex × Genotype Interac- tion: F(2,54) = 0.20, P = 0.09 |
||
| Latency to enter light chamber Fig. 3D |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 2.40, P = 0.127; Main effect of Genotype: F(2,54) = 0.109, P = 0.896; Sex × Genotype Interaction: F(2,54) = 0.56, P = 0.56 |
||
| N = 20 WT, 20 HET, 15 KO |
Dark/Light | Time in Light & Dark Chambers Fig. 3E |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 4.84, P < 0.032; Main effect of Genotype: F(2,54) = 1.549, P50.222; Sex × Genotype Interaction: F(1,52) = 1.70, P = 0.192 |
| Time in light chamber |
Posthoc Scheffe contrast of means (sex) |
|||
| WT Male vs. Female | P = 0.168 | |||
| HET Male vs. Female | P = 0.903 | |||
| KO Male vs. Female | P < 0.015 | |||
| KO Mean/SEM | Male:21.35/12.81 Femlae: 110.37/46.8 |
|||
| Posthoc Scheffe contrast of means (sex): |
||||
|
Female: WT vs. Het |
P = 0.174 | |||
| WT vs. KO | P = 0.864 | |||
| Het vs. KO | P = 0.070 | |||
|
Male: WT vs. Het |
P = 0.910 | |||
| WT vs. KO | P = 0.844 | |||
| Het vs. KO | P = 0.996 | |||
| Time in dark chamber |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 4.84, P < 0.03; Main effect of Genotype: F(2,54) = 1.54, P = 0.222; Sex × Genotype Interaction: F(2,54) = 0.1.70, P = 0.192 |
||
| Posthoc Scheffe contrast of means (group): |
||||
|
Female: WT vs. Het |
P = 0.174 | |||
| WT vs. KO | P = 0.864 | |||
| Het vs. KO | P = 0.070 | |||
|
Male: WT vs. Het |
P = 0.910 | |||
| WT vs. KO | P = 0.844 | |||
| Het vs. KO | P = 0.996 Posthoc Scheffe contrast of means (sex): |
|||
| WT Male vs. Female | P = 0.168 | |||
| HET Male vs. Female | P = 0.903 | |||
| KO Male vs. Female | P < 0.015 | |||
| KO Mean/SEM | Male:578.65/12.81 Femlae: 489.62/46.82 | |||
| N = 20 WT, 20 HET, 15 KO |
Elevated Plus Maze | Duration in Closed arms (%) Fig. 3F |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 1.48, P = 0.228; Main effect of Genotype: F(2,54) = 2.03, P = 0.14; Sex × Genotype Interaction: F(2,54) = 2.69, P = 0.077 |
| Duration in Open arms (%) Fig. 3F |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 0.0042, P = 0.948; Main effect of Genotype: F(2,54) = 3.15, P = 0.051; Sex × Genotype Interaction: F(2,54) = 1.43, P = 0.24 |
||
| Planned comparison | ||||
| WT vs. Het | P < 0.026 | |||
| Het vs. KO | P = 0.708 | |||
| WT vs. KO | P = 0.063 | |||
| N = 20 WT, 20 HET, 15 KO |
Rearing | Beam Breaks Fig. 3G |
Sex, Genotype, & Bin | 3-way rmANOVA; Main effect of Sex: F(1,659) = 48.88, P < 7.412E-12; Main effect of Genotype: F(2, 659) = 30.59, P < 2.29E-13; Main effect of Bin: F(11,659) = 25.09, P < 0.000001; Main effect of Sex × Genotype Interaction: F(2,659) = 4.72, P < 0.00925; Main effect of Sex × Bin Interaction: F(11,659) = 0.674, P = 0.763; Main effect of Genotype × Bin Interaction: F(22,659) = 2.039, P < 0.003; Main effect of Sex × Genotype × Bin Interac- tion: F(22,659) = 0.431, P = 0.989 |
| Posthoc Scheffe contrast of means (genotype) |
||||
| WT vs. HET | P < 1.814E-5 | |||
| WT vs. KO | P < 4.21E-13 | |||
| HET vs. KO | P = 0.052 | |||
| Posthoc Scheffe contrast of means (Sex: Male) |
||||
| WT vs. HET | P = 0.846 | |||
| WT vs. KO | P = 0.999 | |||
| HET vs. KO | P = 0.802 | |||
| Posthoc Scheffe contrast of means (Sex: Female) |
||||
| WT vs. HET | P < 0.047 | |||
| WT vs. KO | P < 0.016 | |||
| HET vs. KO | P = 0.957 | |||
| Posthoc Scheffe contrast of means (sex): |
||||
| WT Male vs. Female | P < 0.00064 | |||
| HET Male vs. Female | P = 0.921 | |||
| KO Male vs. Female | P = 0.604 | |||
| Total Beam Breaks Fig. 3H |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 7.28, P < 0.0095; Main effect of Genotype: F(2,54) = 4.556, P < 0.0153; Sex × Genotype Interaction: F(2,54) = 0.703, P = 0.500 |
||
| Posthoc Scheffe contrast of means (genotype) |
||||
| WT vs. HET | P = 0.201 | |||
| WT vs. KO | P < 0.016 | |||
| HET vs. KO Posthoc Scheffe contrast of means (Sex: Male) |
P = 0.646 | |||
| WT vs. HET | P = 0.887 | |||
| WT vs. KO | P = 0.475 | |||
| HET vs. KO Posthoc Scheffe contrast of means (Sex: Female) |
P = 0.830 | |||
| WT vs. HET | P = 0.105 | |||
| WT vs. KO | P < 0.023 | |||
| HET vs. KO Posthoc Scheffe contrast of means (sex): |
P = 0.852 | |||
| WT Male vs. Female | P < 0.0099 | |||
| HET Male vs. Female | P = 0.433 | |||
|
KO Male vs. Female Posthoc Scheffe contrast of means (Sex: Male) |
P = 0.344 | |||
| WT vs. HET | P = 0.105 | |||
| WT vs. KO | P < 0.023 | |||
| HET vs. KO Posthoc Scheffe contrast of means (Sex: Female) |
P = 0.852 | |||
| WT vs. HET | P = 0.887 | |||
| WT vs. KO | P = 0.475 | |||
| HET vs. KO | P = 0.830 | |||
| N = 20 WT, 20 HET, 15 KO |
Marbles burying | Marbles buried Fig. 3I |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,54) = 0.772, P = 0.384; Main effect of Genotype: F(2,54) = 21.60, P < 0.00001; Sex × Genotype Interac- tion: F(2,54) = 0.333, P = 0.718 |
| Posthoc Scheffe contrast of means (genotype): |
||||
| WT vs. Het | P = 0.301 | |||
| WT vs. KO | P < 3.57E-07 | |||
| Het vs. KO | P < 0.00037 | |||
| N = 20 WT, 20 HET, 15 KO |
Locomotor | Beam Breaks Fig. 3J |
Sex and Genotype | 3-way rmANOVA; Main effect of Sex: F(1,54) = 8.66, P < 0.0033; Main effect of Genotype: F(2,54) = 12.81, P < 3.1E- 06; Main effect of Bin: F(23,1008) = 65.69, P < 0.000001; Main effect of Sex × Genotype Interac- tion: F(2,54) = 10.30, P < 0.00004; Main effect of Sex × Bin Interaction: F(23,1008) = 1.17, P = 0.261; Main effect of Genotype × Bin Interaction: F(46,1008)50.959, P = 0.550; Main effect of Sex × Genotype × Bin Interac- tion: F(46,1008) = 0.49, P = 0.99 |
| Posthoc Scheffe contrast of means (genotype): |
||||
| WT vs. Het | P = 0.301 | |||
| WT vs. KO | P < 3.57E-07 | |||
| KO Het vs. KO Posthoc Scheffe contrast of means (Sex: Male) |
P < 0.00037 | |||
| WT vs. HET | P = 0.432 | |||
| WT vs. KO | P = 0.726 | |||
| HET vs. KO Posthoc Scheffe contrast of means (Sex: Female) |
P = 0.849 | |||
| WT vs. HET | P = 0.989 | |||
| WT vs. KO | P = 0.914 | |||
| HET vs. KO Posthoc Scheffe contrast of means (sex): |
P = 0.967 | |||
| WT Male vs. Female | P = 0.122 | |||
| HET Male vs. Female | P = 0.939 | |||
| KO Male vs. Female | P = 0.697 | |||
| Total Beam Breaks Fig. 3K |
Sex and Genotype | 2-way ANOVA; Main effect of Sex: F(1,47) = 1.02, P = 0.318; Main effect of Genotype: F(2,47) = 1.50, P = 0.232; Sex × Genotype Interaction: F(2,47) = 1.213, P = 0.307 |
||
| N = 20 WT, 20 HET, 15 KO |
Rotarod | Latency to Fall Fig. 3L |
Sex, Genotype, and Trial | 3-way rmANOVA; Main effect of Sex: F(1,52) = 19.46, P < 0.00001; Main effect of Genotype: F(2,52) = 14.47, P < 8.57E-07; Main effect of Trial: F(7,408) = 9.986, P < 0.1.70E-11; Main effect of Sex × Genotype Interaction: F(2,52) 518.99, P < 1.33E-08; Main effect of Sex × Trial Interaction: F(7,408) = 5.05, P < 0.00002; Main effect of Genotype × Trial Interaction: F(14,408) = 3.511, P < 0.00002, Main effect of Sex × Genotype × Trial Inter- action: F(14,408) = 3.197, P < 0.00008 |
| Posthoc Scheffe contrast of means (genotype) |
||||
| WT vs. HET | P = 0.608 | |||
| WT vs. KO | P < 0.00016 | |||
| HET vs. KO Posthoc Scheffe contrast of means (Sex: Female) |
P < 0.00001 | |||
| WT vs. HET | P = 0.385 | |||
| WT vs. KO | P < 0.003 | |||
| HET vs. KO Posthoc Scheffe contrast of means (Sex: Male) |
P = 0.180 | |||
| WT vs. HET | P < 1.79E-07 | |||
| WT vs. KO | P = 0.903 | |||
| HET vs. KO Posthoc Scheffe contrast of means (sex): |
P < 8.31E-07 | |||
| WT Male vs. Female | P < 0.0022 | |||
| HET Male vs. Female | P < 0.00007 | |||
| KO Male vs. Female | P = 0.397 | |||
| N = 20 WT, 20 HET, 15 KO |
Startle | Avg Startle Ampli- tude Fig. 1M |
Sex, Genotype, and dB | 3-way rmANOVA; Main effect of Sex: F(1,259) = 7.98, P < 0.0051; Main effect of Genotype: F(2,259) = 14.47, P < 0.0388; Main effect of dB: F(8, 259) = 75.15, P < 0.00001; Main effect of Sex × Genotype Interaction: F(2,259) = 0.855, P = 0.426; Main effect of Sex × dB Interaction: F(4,259) = 1.61, P = 0.172; Main effect of Genotype × dB Interaction: F(8,259) = 1.019, P = 0.429, Main effect of Sex × Genotype × Trial Interaction: F(8,259) = 0.158, P = 0.995 |
| N = 20 WT, 20 HET, 15 KO |
Prepulse Inhibition | % Inhibition Fig. 3N | Sex, Genotype, and dB | 3-way rmANOVA; Main effect of Sex: F(1,143) = 0.254, P = 0.614; Main effect of Genotype: F(2,143) = 0.102, P = 0.902; Main effect of dB: F(2, 143) = 48.48, P < 2.22E-16; Main effect of Sex × Genotype Interaction: F(2,143) = 0.831, P = 0.437; Main effect of Sex × dB Interaction: F(4,143) = 0.038, P = 0.961; Main effect of Genotype × dB Interaction: F(4,143) = 0.667, P = 0.616, Main effect of Sex × Genotype × Trial Interaction: F(4,143) = 0.245, P = 0.912 |
| N = 20 WT, 20 HET, 15 KO |
Morris Water Maze | Latency to Plat- form Fig. 4A |
Sex, Genotype, and Day | 3-way rmANOVA; Main effect of Sex: F(1,363)51.812, P50.179; Main effect of Genotype: F(2,363) = 27.07, P < 1.350E-11; Main effect of Day: F(6,363) =7.659, P < 1.053E–7; Main effect of Sex × Genotype Interaction: F(2,363) = 1.239, P = 0.290; Main effect of Sex × Day Interaction: F(6,363) = 1.05, P = 0.387; Main effect of Genotype × Day Interaction: F(12,363) = 2.11, P < 0.042; Main effect of Sex × Genotype × Day Interac- tion: F(12,363) = 0.818, P = 0.631 |
| Posthoc Scheffe contrast of means (Genotype) |
||||
| WT vs. HET | P = 0.191 | |||
| WT vs. KO | P < 3.701–11 | |||
| HET vs. KO | P < 1.547–5 | |||
| Total Distanced Traveled Fig. 4B |
Sex, Genotype, and Day | 3-way rmANOVA; Main effect of Sex: F(1,363) = 8.375, P < 0.004; Main effect of Genotype: F(2,363) = 18.78, P < 1.914e-8; Main effect of Day: F(6,363) = 19.45, P < 0.00001; Main effect of Sex × Genotype Interaction: F(2,363) = 4.87, P < 0.008; Main effect of Sex × Day Interaction: F(6,363) = 0.984, P = 0.435; Main effect of Genotype × Day Interaction: F(12,363) = 3.036, P < 0.0004; Main effect of Sex × Genotype × Day Interac- tion: F(12,363) = 0.808, P = 0.649 |
||
| Posthoc Scheffe contrast of means (Genotype) |
||||
| WT vs. HET | P = 0.095 | |||
| WT vs. KO | P < 2.16e-8 | |||
| HET vs. KO | P < 0.003 | |||
| Posthoc Scheffe contrast of means (Sex: Male) |
||||
| WT vs. HET | P = 0.789 | |||
| WT vs. KO | P = 0.464 | |||
| HET vs. KO | P = 0.908 | |||
| Posthoc Scheffe contrast of means (Sex: Female) |
||||
| WT vs. HET | P = 0.635 | |||
| WT vs. KO | P = 0.124 | |||
| HET vs. KO | P = 0.628 | |||
| Posthoc Scheffe contrast of means (sex): |
||||
| WT Male vs. Female | P = 0.732 | |||
| Percent Thigmo- taxis Fig. 4C |
HET Male vs. Female | P = 0.577 | ||
| KO Male vs. Female | P = 0.217 | |||
| Probe Trial Fig. 4D | Sex, Genotype, and Day | 3-way rmANOVA; Main effect of Sex: F(1,363) = 4.05, P < 0.044; Main effect of Genotype: F(2,363) = 44.96, P < 0.00001; Main effect of Day: F(6,363) = 11.07, P < 3.08e–7; Main effect of Sex × Genotype Interaction: F(2,363) = 1.133, P = 0.323; Main effect of Sex = Day Interaction: F(6,363) = 0.378, P = 0.892; Main effect of Genotype × Day Interaction: F(12,363) × 2.088, P < 0.017; Main effect of Sex × Genotype × Day Interac- tion: F(12,363) = 0.394, P = 0.965 |
||
| Posthoc Scheffe contrast of means (Sex: Male) |
||||
| WT vs. HET | P = 0.175 | |||
| WT vs. KO | P < 1.11e016 | |||
| HET vs. KO | P < 0.00001 | |||
| Sex, Genotype, and Quadrant |
3-way rmANOVA; Main effect of Sex: F(1,207) = 2.78e-5, P = 0.995; Main effect of Genotype: F(2,207) = 3.05e- 5, P = 0.99; Main effect of Quadrant: F(3,207) = 19.28, P < 6.43e-11; Main effect of Sex × Genotype Interaction: F(2,207) = 5.01e-6, P = 0.999; Main effect of Sex × Quadrant Interaction: F(3,207) = 1.469, P = 0.224; Main effect of Genotype × Quadrant Interac- tion: F(6,207) = 2.623, P < 0.01; Main effect of Sex × Genotype × Quadrant Interaction: F(6, 207) =0.523, P = 0.789 |
|||
| Planned comparison (WT) | ||||
| Target vs. Near | P < 2.01e-9 | |||
| Target vs. Far | P < 0.0001 | |||
| Target vs. Opposite (HET) | P < 2.02e–7 | |||
| Target vs. Near | P < 0.0001 | |||
| Target vs. Far | P < 0.0074 | |||
| Target vs. Opposite (KO) | P < 0.001 | |||
| Target vs. Near | P = 0.105 | |||
| Target vs. Far | P = 0.618 | |||
| Target vs. Opposite | P = 0.476 | |||
Behavioral tests
Elevated plus maze
was conducted as described previously [Etherton, Blaiss, Powell, & Sudhof, 2009]. Briefly, mice were placed in the center of the maze (each arm was 30 cm long and 5 cm wide with 25 cm high walls on the closed arms) and allowed to freely explore for 5 min. All mice were tested under dim white light at ~7 lux. Noldus Ethovision version 3.1 (Leesburg, VA, USA) was used to track and record mouse behavior.
Locomotor and Rearing activity
was measured as described previously [Etherton et al., 2009; Powell et al., 2004; Tabuchi et al., 2007]. Mice were placed in novel cages (a clean cage with the same dimensions as their home cage; L × W × H = 27.3 × 16.5 × 12.7 cm3) with minimal bedding and allowed to freely explore for 2hr under red lighting. Horizontal locomotor activity (i.e. the number of photobeam breaks) was measured by computer software (San Diego Instruments; San Diego, CA, USA), and data were analyzed in 5-min bins.
Rearing activity
was measured in the same manner as locomotor activity, however recording only occurred over a 1-hr time period. Additionally, the photobeams were raised to 3 cm higher to detect rearing of the mouse but not locomotor activity.
Dark/Light test
was conducted as described previously [Blundell et al., 2009; Powell et al., 2004]. Mice were placed in the dark chamber (each chamber was 25 × 26 cm with 2066 lux on the light side and ~1 lux on the dark side) and allowed to habituate for 2 min. After habituation, mice were allowed to freely explore both chambers for 10 min.
Rotarod
was conducted as described previously [Powell et al., 2004]. Briefly, mice were placed on a stationary rotarod (IITC Life Science) that was then activated and accelerated from 0 to 45 revolutions/min over 5 min. The latency for the mice to fall off the rod was measured. If a mouse held onto the rotating rod for one complete revolution it was scored as a fall. Each mouse received four trials per day for 2 days. Within a day, each trial had an intertrial interval of ~45 min.
Open Field
was conducted as described previously [Etherton et al., 2009]. Briefly, mice were placed along the edge of an open arena (44×44×44 cm, ~7 lux) and allowed to freely explore for 10 min. Time spent in the center of the arena (15 × 15 cm) as well as locomotor activity were measured. Mice were monitored using CleverSys TopScan Software (Reston VA).
Social interaction with a novel juvenile target mouse
was performed essentially as described [Blundell et al., 2009; Kwon et al., 2006; Tabuchi et al., 2007]. Briefly, following a 15-min habituation under red light, the experimental and target mice were placed in a novel cage for 2 min and allowed to directly interact with a novel C57BL6J juvenile mouse (3 weeks of age; Jackson labs). Interaction was scored by observing the duration and number of times the test mouse initiated contact with or sniffed the juvenile mouse. Contact was considered as any part of the body touching the other mouse. Three days later the same experimental and novel juvenile mice were paired again in a novel cage for 2 min and scored in the same manner.
Social interaction with a caged adult
was performed as described [Blundell et al., 2010a]. Briefly, the test was performed in a 48 × 48 cm2 white plastic arena under red light using 3.5′′ × 30′′ × 2′′ (w × h × l) clear rectangular cage containing a novel adult mouse. The lower half of the rectangular cage has small openings to allow for olfactory and minimal tactile interaction. Initially mice were allowed to explore the arena for 5 min with an empty rectangular cage. Then, mice were allowed to approach a novel adult mouse housed in the rectangular cage for another 5 min.
Social interaction with genotype- and sex-matched pairs
was performed by pairing mice with a sex- and genotype-matched partner within the experimental cohort. Matched pairs were derived from separate cages and were never previously housed together. Mouse pairs were placed at separate ends in an open field arena (44 × 44 × 44 cm) and allowed to interact for 5 min under dim lighting (~7 lux).
Three-chamber social approach
social preference and social novelty was tested using a three-chambered box as described previously [Blundell et al., 2009] and based to a large extent on the original descriptions [Moy et al., 2004; Nadler et al., 2004]. This test consisted of three 10-min trials. During the first trial, the mouse was allowed to explore the 3 chamber box in which each end-chamber contained an empty cage (upside down pencil holder). In the second trial, the 3-chamber box contained a novel stimulus mouse under a cage in one of the end-chambers and an empty cage in the opposite end-chamber. The test mouse was free to choose between an inanimate cage and a caged, social target. For the third trial, the test mouse was free to choose between a caged novel social target (novel mouse) versus the same caged mouse in trial 2 (familiar social target). Locations of empty cages and social targets were counterbalanced, and mice were placed back into the home cage for very brief intervals between trials. Social interactions/approaches were objectively monitored and scored using video-tracking and automated CleverSys Social Scan Software (Reston VA).
Grooming
as performed by placing mice into an empty cage and monitoring and video recording for 10 min. Time spent grooming the face, head, body, or tail, are all considered grooming.
Morris water maze
was conducted as previously described [Powell et al., 2004]. Briefly, in a 120 cm diameter pool a 10-cm diameter circular platform was submerged ~1 cm below the surface of the water (22 ± 1 °C) made opaque with white, nontoxic tempra paint. After finding the platform or being guided by the experimenter to the platform if the 60-sec trial elapsed, mice remained on the platform for 15 sec before being removed and returned to their home cage. Training was conducted over 7 consecutive days, followed by a probe trail on day 8 (60-sec swim with no platform). To test basic visual function, we measured the latency to reach the platform with a visible cue atop the platform in the water maze on day 14.
Prepulse inhibition and startle
Both prepulse inhibition and startle response were conducted as previously described [Blundell, Kaeser, Sudhof, & Powell, 2010b]. Briefly, mice were placed inside a cylinder mounted atop a piezoelectric accelerometer that detected and transduced animal movements inside the startle chambers (San Diego Instruments, San Diego, CA, USA). Acoustic stimuli varying in decibels (dB) were delivered by high-frequency speakers mounted 33 cm above the cylinder. To test startle response mice were exposed to stimuli ranging from 0 to 120 db and the amplitude of the startle responses was obtained in arbitrary units. For prepulse inhibition (PPI), mice were subjected to five trial types in a 22-min session: pulse alone (40 msec, 120 dB, white noise pulse), three different prepulse/pulse trials (20 msec prepulse of 4, 8, or 16 dB above background noise level of 70 dB preceded the 120 dB pulse by 100 msec onset-onset interval), and no stimulus. All trials were presented pseudorandomly with an average of 15 sec (7–23 sec) between the 62 trials. Prior to testing, mice were acclimated to the cylinders for 5 min then tested in four blocks of test trials. The first and last blocks consisted of six pulse-alone trials. Blocks 2 and 3 each contained six pulse-alone trials, five of each level of prepulse/pulse trials, and five no-stimulus trials.
Marble burying
As described previously [Blundell et al., 2010a], individual mice were placed in a novel home cage with 5 cm of bedding. Twenty marbles were evenly placed on top of the bedding throughout the home cage, and mice were free to explore the cage for 30 min. After 30 min the number of marbles buried was recorded. A marble was defined as buried when <25% of the marble was visible. This test was conducted in a well-lit room (~80 lux).
Physiology
Acute slice preparation
Four to six week old male mice were anaesthetized with 8% chloral hydrate (400 mg/kg) and transcardially perfused with artificial cerebral spinal fluid (ACSF) containing (in mM): 120 NaCl, 3.5 KCl, 1.25 NaH2PO4, 1 MgSO4, 26 NaHCO3, 10 dextrose, and 2 CaCl2 adjusted to pH 7.4 and saturated with 95% O2/5% CO2. Brains were rapidly removed and placed in ice-cold sucrose ACSF containing (in mM): 75 sucrose, 87 NaCl, 3 KCl, 1.25 NaH2PO4, 7 MgSO4, 26 NaHCO3, 20 dextrose, and 0.5 CaCl2 adjusted to pH 7.4 and saturated with 95% O2/5% CO2. Coronal slices containing dorsal hippocampus or thalamocortical slices containing dorsal striatum (300–400 µm) were made using VT1000s Vibratome (Leica Biosystems, Buffalo Grove, IL, USA). Slices recovered at 35°C for 30–45 min in ACSF and slowly cooled to room temperature before recording.
Extracellular “field” recordings
were performed as previously described [Kouser et al., 2013; Speed et al., 2015]
Whole-cell patch clamp recordings
Excitatory postsynaptic currents were recorded from cell bodies identified at 80 × on an AxioExaminer D1 Differential Interference Contrast (DIC) microscope (Zeiss, Thornwood, New York, USA). CA1 neurons were identified by triangular appearance and large apical dendrite. Striatal medium spiny neurons (MSNs) were identified by their oval shape, multiple dendrites, and hyperpolarized resting membrane potential (approximately −85mV) measured after break-in. Stimulating electrode was placed just below the corpus callosum (Fig. 10A) ~150–200µm from the recorded MSN as described [Jaramillo et al., 2016]. Borosilicate glass electrodes (4–6MX) were filled with internal solution containing (in mM): 110 CsMethanesulfonate, 15 CsCl, 8 NaCl, 10 TEA-Cl, 2 EGTA, 10 HEPES, 3 QX 314, 2 ATP, 0.3 GTP. Observed junction potential was ~10 mV and was compensated. Cells with > 25% change in access resistance (15–25 MΩ) or holding current were not included. Stimulation was controlled via a Model 2200 stimulus isolator (A-M Systems, Sequim, WA, USA) though a monopolar tungsten microelectrode (FHC, Bowdoin, ME) with a 0.1 msec biphasic pulse. 100 µM picrotoxin was in bath for all experiments. Sample size (n) indicates number of cells with no more than five cells per mouse and five or more mice per genotype.
Data collection and analysis
All recordings were in shallow submersion chambers perfused with oxygenated ACSF at 32 ± 0.5 °C with a flow rate of 3–5 mL/min. Data were sampled at 10 kHz using Digidata 1440 digitizers with Clampex 10.3 and filtered at 1–5 kHz with either Model 1800 amplifiers (A-M Systems, Sequim, WA, USA) for field recordings or Mutliclamp 700B amplifiers (Molecular Devices, Sunnyvale, CA, USA). Raw traces were analyzed using Clampfit 10.3 (Molecular Devices, Sunnyvale, CA, USA). Statistical analysis and graphing were performed in GraphPad 6 (GraphPad Software, San Diego, CA, USA). For statistical analysis, we used genotype and sex as main factors for ANOVA analysis. In most cases, there was not a significant main effect of sex. In those cases, where there was a main effect of sex, most did not demonstrate an interaction between sex and genotype. Thus, in our figures we have combined males and females into single graphs. For those tasks where we identified an interaction between sex and genotype, we have provided average values and standard error of the mean in our statistical table as well as separate figures of the male and female data for those most interested in sex differences. We never “combined” our male and female data in our statistical analyses, rather maintaining sex as an independent variable as is appropriate for ANOVA statistics.
Compounds and drugs
Octahydro-12-(hydroxymethyl)-2-imino-5,9:7,10a-dimethano-10aH[1,3]dioxocino[6,5-d] py rimidine-4,7,10,11,12-pentolTetrodotoxin (TTX, Tetrodotoxin), picrotoxin, N-(2,6-Dimethylphenylcarbamoylmethyl)triethylammonium chloride (QX 314), (R,S)-3,5-Di hydroxyphenylglycine (DHPG), CsMethanesulfonate, and CsCl were obtained from Sigma-Aldrich (Minneapolis, MN). All other reagents were obtained from Fisher Scientific (Waltham, MA).
Results
Generation of Shank3E13 Mice
We disrupted the Shank3 gene in mice by insertion of a transcriptional neo-stop cassette between exon 12 (E12) and exon 13 (E13) (Fig. 1A). To ensure proper incorporation of the neo-stop cassette, we used PCR primers spanning the loxP sites (Fig. 1B). To verify correct targeting, we performed RT-PCR on brain cDNA of 4-week old Shank3e13 mice. Loss expression of mRNA containing exons 13–19 in Shank3e13 mice demonstrated correct targeting (Fig. 1C). We also show correct targeting in the Shank3e4–9 [Jaramillo et al., 2015], an autism-associated insertion mutation in exon 21 in G/G mice [Speed et al., 2015], and Shank3 exon 21 deletion mice (ΔC/ΔC) [Kouser et al., 2013] (Fig. 1C). Western blots of whole brain lysate using C-terminal Shank3 antibody (gift from Paul Worley) showed loss of the higher molecular weight bands in the KO mice (Fig. 1D, KO). The same antibody on whole brain lysate from a complete Shank3 KO (gift from Yong-hui Jiang) showed loss of the same bands (not shown) indicating that these bands are representative of Shank3. Remaining bands in the “KO” lane in Figure 1D were also absent from the complete Shank3 knockout, demonstrating these bands represent remaining isoforms of Shank3.
Shank3E13 HET and KO Mice Display Social Interaction Deficits
Because social differences are a core feature of autism and prominent features of PMS, we tested Shank3E13 mutant mice in a number of social tasks. In the 3-chamber test of sociability, Shank3E13 HET and KO mice displayed abnormalities. None of the groups displayed an initial bias for either side chamber (Fig. 2A). When one chamber contained a novel caged adult mouse and the other contained an empty cage, only WT and KO mice displayed a social preference. Shank3E13 HET mice did not display significant sociability (Social vs. Inanimate—WT: P<0.005; HET: P=0.532, KO: P<0.003; Fig. 2B). When one chamber contained a familiar caged adult mouse and the other contained a novel caged adult mouse, only WT mice showed social novelty preference. Shank3E13 HET and KO showed no social novelty preference (Novel vs. Familiar WT: P<0.016, HET: P=0.532, KO: P=0.352; Fig. 2C).
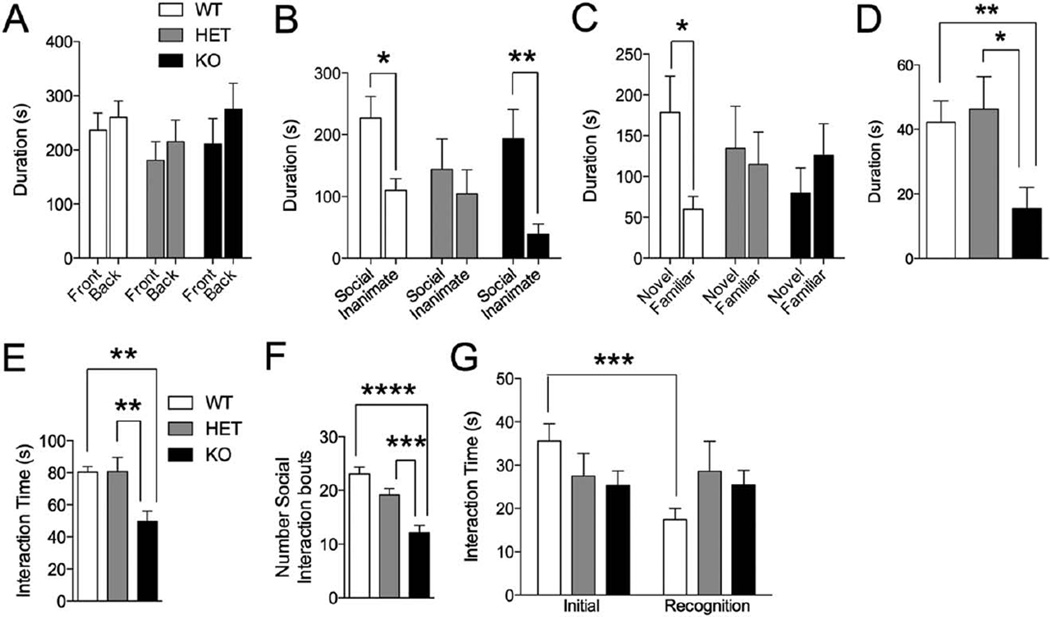
Figure 2.
Social test in Shank3E13 mutant mice. (A) In the 3-chamber test of sociability, none of the three mouse groups displayed an aberrant chamber preference in the initial trial. In second trial of this task, Shank3E13 HET mice did not display a preference for the social target over the inanimate object (B). In the third trial of this task, neither Shank3E13 HET nor KO mice displayed a preference for social novelty (C). Shank3E13 KO mice also displayed a significant decrease in approach of a novel, caged social target in the caged conspecific test (D). In the genotype and sex-matched social interaction test, Shank3E13 KO mice displayed a significant reduction in social interaction time (E), and a decrease in number of social bouts (F). Both HET and KO mice display deficits in social recognition in the social interaction with juvenile test of social memory (G). (*P<0.05; **P<0.01, ***P< 0.001, ****P< 0.0001; n = 20 WT, 20 HET, 15 KO).
We also examined approach behavior of Shank3E13 mutants in an open field arena in the presence of a novel, caged adult target mouse. Shank3E13 KO mice displayed decreased approach of the caged adult mouse when compared to WT and HET (Social: WT vs. KO: P<0.007; HET vs. KO: P<0.011; Fig. 2D).
We also assessed reciprocal social interaction by pairing mice of the same genotype/sex for an initial encounter. Shank3E13 KO pairs displayed decreased social interaction compared to WT/WT and HET/HET pairs (total interaction: WT vs. KO: P<0.0059; HET vs. KO: P<0.0037; 2E). Furthermore, Shank3E13 KO mice displayed decreased number of social interaction bouts (WT vs. KO: P<2.09E-8; HET vs. KO: P<0.0004; Fig. 2F).
We next tested Shank3E13 mutants in a social recognition memory task in which novel juvenile mice (3 wk old) were introduced to experimental mice, and then 3 days later the same juvenile mice were reintroduced to the same experimental mouse. WT mice displayed a significant decrease in interaction time when the juvenile mouse was reintroduced, suggesting that WT mice recognized the juvenile as familiar. HET and KO Shank3E13 mice displayed a lack of social recognition memory (Initial vs. recognition time—WT: P<0.0005, HET: P = 0.901, KO: P=0.974; Fig. 2G).
Shank3E13 HET and KO Mice Display Increased Repetitive Grooming
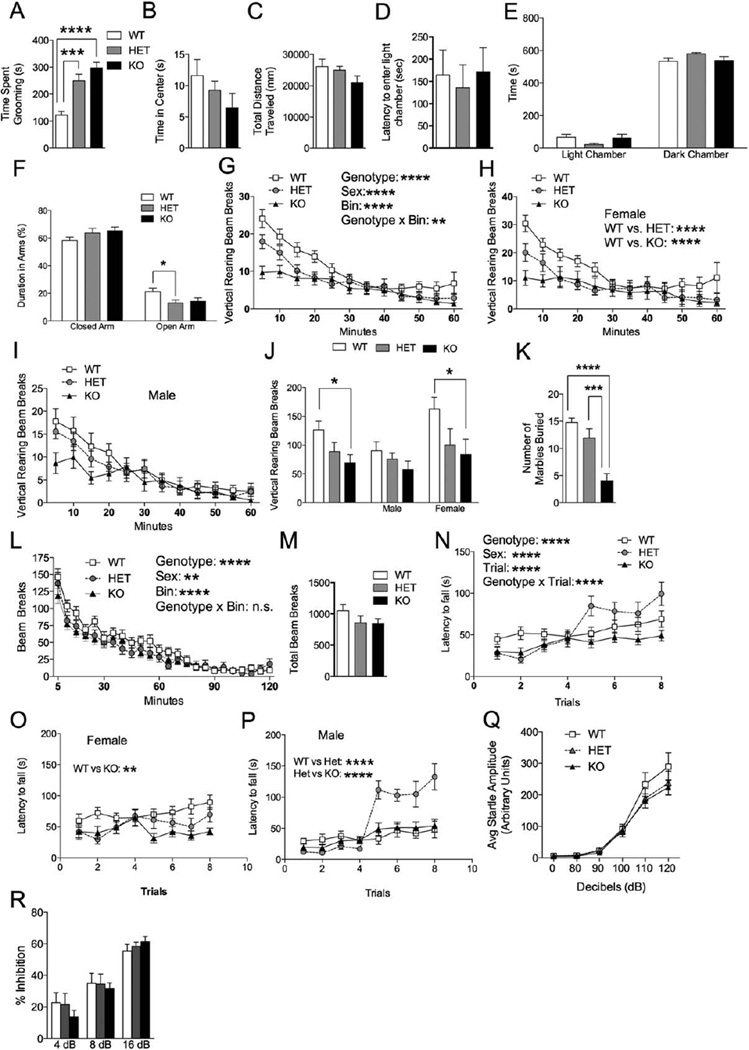
Because restricted and repetitive behaviors are a core feature of autism and PMS, we monitored repetitive grooming behavior in Shank3E13 mutants over a 10-min period. Both HET and KO mice displayed increased time spent grooming compared to WT (WT vs. HET: P <0.0008; WT vs. KO: P<1.21E-8; Fig. 3A).
Figure 3.
Grooming, anxiety and motor behavior in Shank3E13 mutant mice. (A) Shank3E13 HET and KO mice displayed a significant increase in time spent grooming compared to WT littermate mice. In the open field test all genotypes spent similar time in the center (B). Additionally they all traveled similar distances (C). In the dark/light test all three genotypes had similar latencies to enter into the light chamber (D). All three genotypes spent a similar amount of time in light and dark chambers (E). In the elevated plus maze all three genotypes spent a similar percentage of time in the closed arms, however, there was a significant decrease in time spent in the open arm in Shank3E13 HET mice when compared to WT mice (F). Shank3E13 HET and KO mice also displayed less rearing over a 1hr time period (G). Female Shank3E13 HET and KO mice displayed less rearing (H) while male mutant mice only displayed a trend toward less rearing (I). Total vertical rearing beam breaks was decreased in KO mice when analyzing the cohort. Further analysis revealed a decrease in female KO mice (J). In the marble-burying test Shank3E13 KO mice buried significantly less marbles than the WT and HET mice (K). In the locomotor test all three genotypes displayed similar levels of habituation over a 2-hr time period (L). There was no difference in the total number of beam breaks during the 2-hr locomotor test (M). In the rotarod test KO mice displayed decreased latency to fall compared to WT and HET mice (N). Female KO mice showed a significant decrease in latency to fall compared to WT (O). Male KO mice showed a significant decrease in latency to fall compared to HET and KO mice (P). All mouse groups displayed a similar startle response to auditory stimuli ranging from 0 to 120 db (Q). All three groups displayed similar % inhibition in the prepulse inhibition test (R). (*P<0.05, **P<0.01, ***P<0.001, n=20 WT, 20 HET, 15 KO).
Anxiety-Like Behavior in Shank3E13 Mutants
The social and grooming differences in Shank3E13 mice are not explained by a more generalized anxiety-like state. In the open field (OF), all three groups spent similar time in the central, anxiogenic region (Fig. 3B). Additionally, there was no difference in total distance traveled (Fig. 3C). In the dark/light test, all groups displayed a similar latency to enter the anxiety-provoking, light chamber (Fig. 3D). All spent similar time in the light and dark chambers (Fig. 3E), although Shank3E13 HETs displayed a trend toward reduced time in the light chamber (Het vs. WT: P = 0.057; Fig. 3E). Similarly, WT and KO mice displayed similar time in open and closed arms of the elevated plus maze (Fig. 3F). HET mice, however, displayed decreased time in the open arms compared to WT (WT VS HET: P>0.026; WT vs. KO: P = 0.093; Fig. 3F).
Rearing activity in mice is thought to be a form of exploratory activity possibly linked to anxiety. Both Shank3E13 HET and KO mice displayed reduced rearing over successive 5-min bins during a 1-hr testing period compared to WT (WT vs. HET: P<0.00002; WT vs. KO: P < 4.21E-13; Fig. 3G). Further analysis of the cohort showed a significant sex difference with HET and KO mice displaying a significant decrease in rearing compared to female WT mice (WT vs. HET: P<0.04, WT vs. KO: P < 0.01; Fig. 3H). Male mutant mice displayed a trend of decrease rearing however it was not significant (Fig. 3I). Correspondingly, total rearing activity was significantly decreased in the Shank3E13 KO but not HET mice (WT vs. KO: P<0.016; WT vs. HET: P=0.201; Female: WT vs. KO: P < 0.023; WT vs. HET: P=0.105; Fig. 3J).
Lastly, we tested mice using a marble-burying task, results of which are ascribed to behavioral domains including neophobia, anxiety, or repetitive behavior. Shank3E13 KO mice displayed a decrease in number of marbles buried compared to WT and HET mice (WT vs. KO: P<3.5E–7; HET vs. KO: P<0.00037; Fig. 3K).
Motor Skills and Auditory Responses in Shank3E13 Mutants
Because incoordination and gait abnormalities are present in some children with autism [Shetreat-Klein, Shinnar, & Rapin, 2014] and many children with PMS [Soorya et al., 2013], we tested locomotor activity and coordination in Shank3E13 mutants. All groups displayed similar habituation in activity over a 2-hr period (3-way ANOVA: genotype × bin interaction P=0.550, Fig. 3L). Additionally all displayed similar total beam breaks (Fig. 3M).
To assess for coordination and motor learning deficits, we used the accelerating rotarod. Shank3E13 KOs displayed decreased latency to fall from the rotarod over a two-day, 8-trial period compared to WT and HETs (WT vs. KO: P<0.00016; HET vs. KO: P<0.00001; Fig. 3N). Further analysis revealed a significant difference in sex in the rotarod test. Female KO mice displayed decreased latency to fall compared to WT mice (WT vs KO: P<0.003; Fig. 3O). Male HET mice showed a significant increase in time spent on the rotarod compared to WT mice, particularly on the second day of testing (WT vs. HET: P<1.79e–7; Fig. 3P).
We next assessed auditory startle responses and sensorimotor gating. All three groups displayed the same levels of acoustic startle response to a range of auditory stimuli from 0 to 120 dB (Fig. 3Q) and completely normal PPI (Fig. 3R).
Shank3E13 KO Mice Display Deficits in Spatial Learning
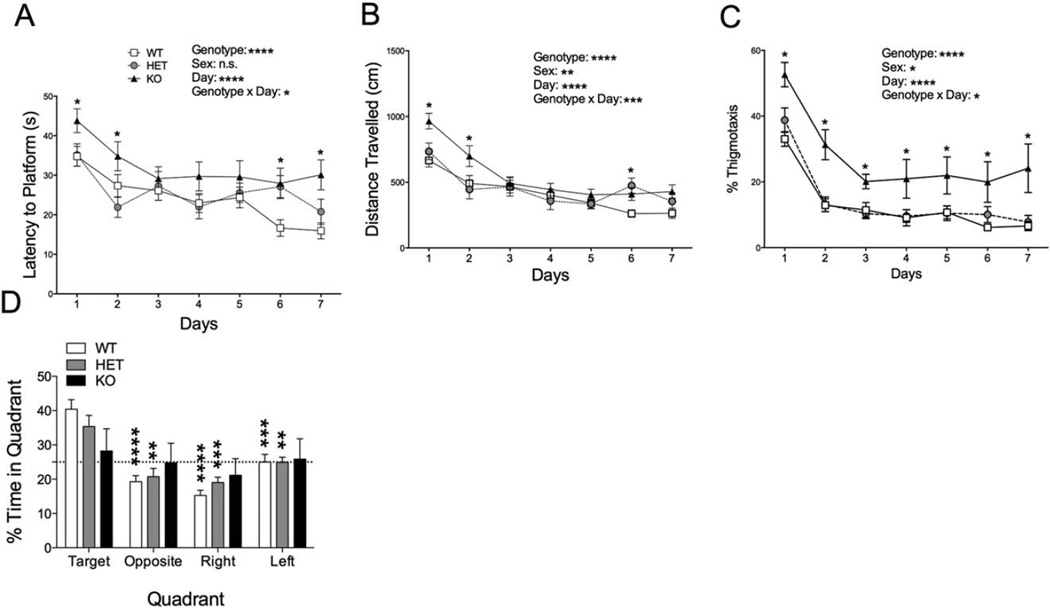
A significant percentage of patients with autism have comorbid intellectual disability [Leyfer et al., 2006; Srivastava & Schwartz, 2014], the prevalence of which is even higher in PMS [Phelan & Rogers, 1993]. Thus, we tested spatial learning and memory ability in Shank3E13 mutants using the Morris water maze. Shank3E13 KO mice displayed a significantly increased latency to reach the hidden platform compared to WT and HET (WT vs. KO: P<6.52E-09; HET vs. KO: P<0.0004; Fig. 4A). Using distance traveled prior to reaching the target platform, a measure that eliminates swim speed as a confound, KO mice also traveled longer distances during training compared to WT and HET mice (WT vs. KO: P<1.1E-16, HET vs. KO: P<1.8E-9; Fig. 4B). Consistent with decreased transition to a spatial learning strategy, KO mice spent more time in the periphery or thigmotaxis zone (WT vs. KO: P<2.1E-8, HET vs. KO: P<0.003; Fig. 4C). During the probe test for spatial memory 24 hr following the final training session, Shank3E13 KOs displayed no significant preference for the target quadrant, while both WT and HET mice showed clear spatial preferences (WT vs. KO: P<0.005; Fig. 4D).
Figure 4.
Spatial learning behavior in Shank3E13 mutant mice. Shank3E13 KO mice displayed increased latency to reach the hidden platform (A, asterisks in graph near symbols refer to WT vs. KO). KO mice also displayed an increase in distance traveled before reaching the hidden platform (B, asterisks in graph near symbols refer to WT vs. KO) and % of time in thigmotaxis (C, asterisks in graph near symbols refer to WT vs. KO) during the training phase of the Morris water maze test. In the probe trial KO mice did not show a preference for the target quadrant compared to the other quadrants (D, asterisks in graph near bars denote comparison to Target quadrant within each group). (*P <0.05; **P<0.01, ***P <0.001, ****P<0.0001; n = 20 WT, 20 HET, 15 KO).
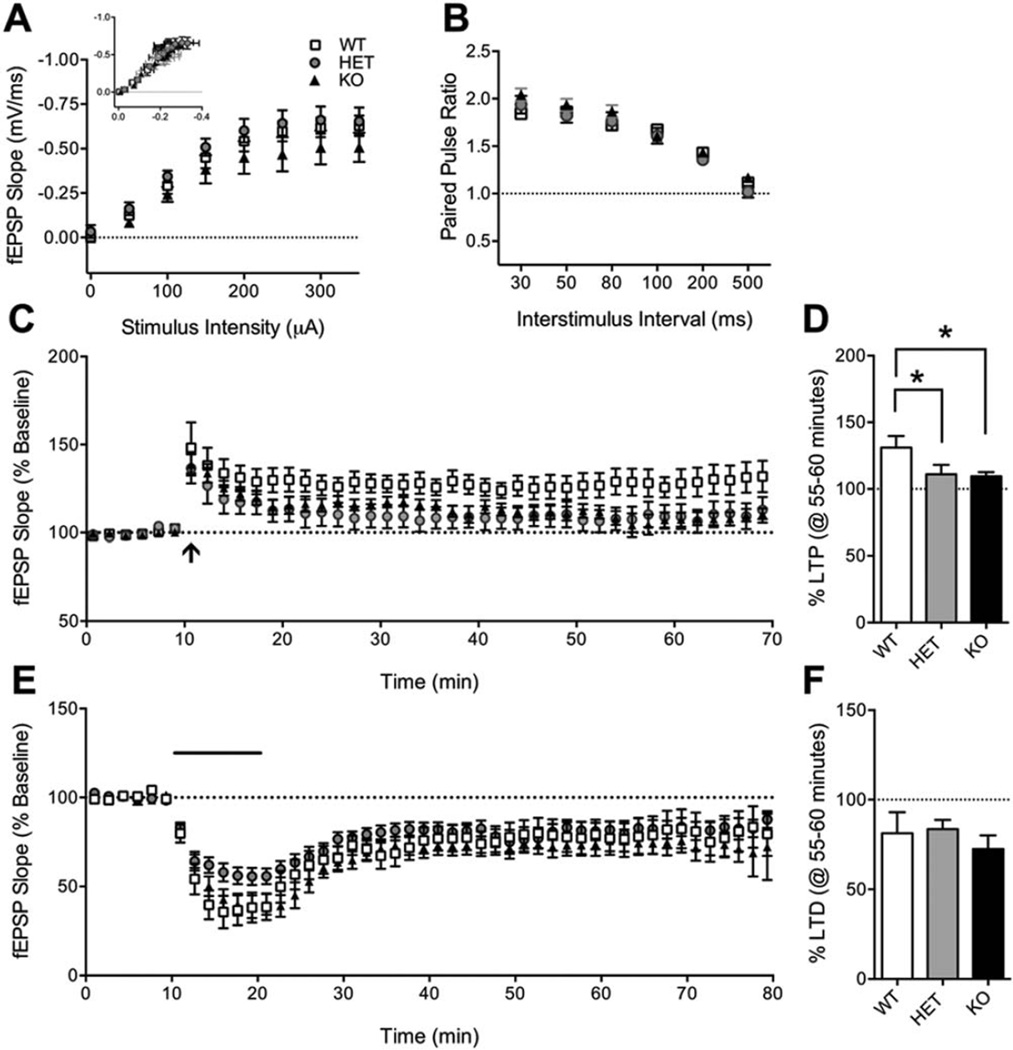
Intact Basal Synaptic Transmission and Short-Term Plasticity at Hippocampal Synapses in Shank3E13 HET and KO Mice
Because spatial learning is known to be dependent on hippocampal function and plasticity [Morris, Garrud, Rawlins, & O’Keefe, 1982; Moser & Moser, 1998; Moser, Krobert, Moser, & Morris, 1998] we examined these in Shank3E13 mutants. We first measured basal synaptic transmission using input/output (I/O) curves to measure responsiveness of CA1 pyramidal neurons to Schaffer collateral stimulation at 0.05 Hz (Fig. 5A). We found no difference between WT and Shank3E13 HET or KO mice for field excitatory postsynaptic potential (fEPSP) slope vs. stimulus intensity (Two-Way RM ANOVA. Genotype: P=0.2407; Stimulus Intensity: P<0.0001; Genotype×Stimulus Intensity: P=0.8857) or fEPSP slope vs. fiber volley amplitude (Two-Way RM ANOVA. Genotype: P=0.6258; Stimulus Intensity: P<0.0001; Genotype×Stimulus Intensity: P=0.9673).
Figure 5.
Hippocampal synaptic function in Shank3E13 mice. (A) Input/output (I/O) curves show no effect of genotype on fEPSP slope at stimulus intensities 0–350 µA. Inset: fEPSP slope (mV/ms) plotted against fiber volley amplitude (mV). WT: 9 slices/6 mice, HET: 12 slices/7 mice, KO: 12 slices/7 mice. (B) Paired-pulse ratio (PPR, slope 2/slope 1) is not affected by the Shank3E13 mutation at interstimulus intervals 30–500 ms. WT: 8 slices/4 mice, HET: 14 slices/8 mice, KO: 17 slices/9 mice. (C) Long-term potentiation (LTP) induced by a single 1 sec, 100Hz train (arrow) is decreased in Shank3E13 HET and KO mice. Each data point is shown as the average of five consecutive traces for clarity. (D) Mean normalized fEPSP at 55–60 min following conditioning stimulus showed decreased LTP in Shank3E13 HET and KO mice. WT: 11 slices/7 mice, Het: 9 slices/8 mice, KO: 12 slices/9 mice. (E) mGluR5-dependent long-term depression induced by 10-min bath application of Group I mGluR agonist DHPG (100 µM, solid bar) is not affected in Shank3E13 mutant mice. (F) Mean normalized fEPSP at 55–60 min following the start of DHPG washout showing normal mGluR-LTD in Shank3E13 mice. WT: 7 slices/5 mice, HET: 13 slices/8 mice, KO: 9 slices/6 mice. *P<0.05.
We also examined short-term plasticity as an indicator of presynaptic function. Paired-pulse ratio (PPR) was measured at 0.05Hz with interstimulus intervals (ISI) 30–500 msec in pseudorandom order (Fig. 5B). Stimulus intensity was set at 50% of the maximum fEPSP and adjusted to prevent epileptiform activity at shorter interstimulus intervals. The average of five trials at each ISI is reported per slice. However, no difference in PPR was found at any ISI (Two-Way RM ANOVA. Genotype: P = 0.4139; ISI: P<0.0001; Genotype X ISI: P=0.3638).
Shank3E13 HET and KO Mice Exhibit Impaired Hippocampal Long-Term Potentiation
Next we investigated NMDA receptor-dependent long-term potentiation (LTP) and Group I mGluR-dependent long-term depression (mGluR-LTD). Stimulus intensity was adjusted to generate 50–75% of the maximum fEPSP for LTP, and a stable baseline was recorded for 20 min prior to the 1-sec, 100 Hz conditioning stimulus (Fig. 5C). Potentiation was measured as the average fEPSP slope at 55–60 min post-tetanus normalized to the average fEPSP of the baseline (Fig. 5D). We observed significantly decreased LTP in Shank3E13 HET and KO mice (One-Way ANOVA, P=0.0438), indicating that NMDA receptor-mediated LTP is impaired in these mice and may underlie hippocampus-dependent learning and memory deficits.
Increases [Huber, Gallagher, Warren, & Bear, 2002] and decreases [Bateup, Takasaki, Saulnier, Denefrio, & Sabatini, 2011] in LTD induced by application of Group 1 mGluR agonists have been identified in genetic models of autism. Furthermore, Group I mGluRs, as well as AMPARs and NMDARs, are being targeted for their therapeutic potential in ASD [Connor, Bariselli, & Bellone, 2014]. To determine whether Group I mGluR function is altered in Shank3E13 mutants 100µM DHPG was bath applied for 10min and LTD was determined as the average fEPSP at 55–60 min following the start of DHPG washout, normalized to the average baseline fEPSP (Fig. 5E). LTD was similar across all three genotypes (Fig. 5F), indicating no effect on Group I mGluR-mediated long-term plasticity (One-Way ANOVA, P=0.5382).
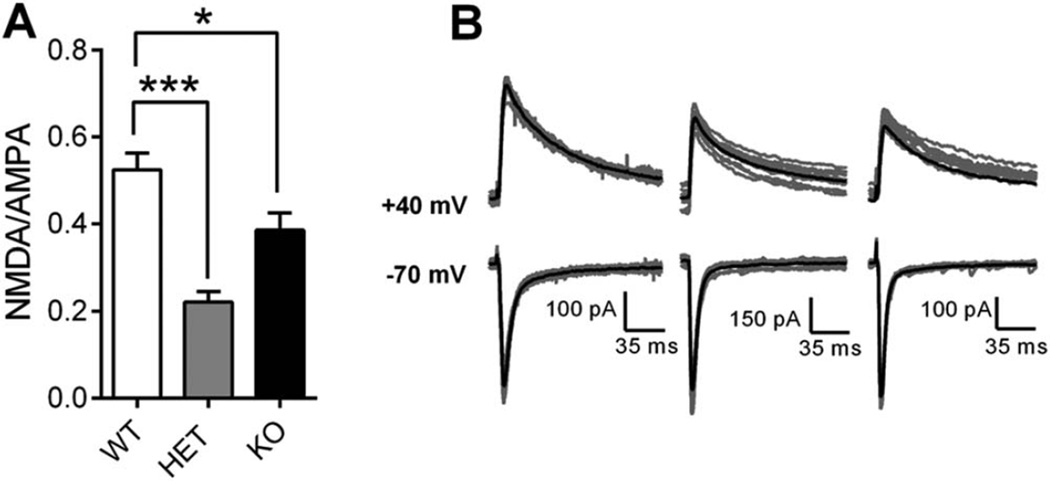
NMDAR/AMPAR Ratio is Decreased in Dorsal Striatum of Shank3E13 Mice
Shank3E13 HET and KO mice demonstrate increased repetitive grooming behavior, an obsessive-compulsive disorder-like phenotype attributed, in part, to deficits in striatal synaptic transmission in SAPAP3 mutant mice [Wan, Feng, & Calakos, 2011; Wan et al., 2014; Welch et al., 2007]. To determine whether abnormal striatal synaptic transmission correlates with repetitive grooming in Shank3E13 HET and KO mice, we used intrastriatal stimulation to evoke EPSCs from MSNs of dorsal striatum. Stimulus intensity (230–250 µA) was set to generate 200–500 pA peak amplitude of AMPAR-mediated currents at −70 mV holding potential. After achieving a stable baseline (5–10 min), 20 consecutive traces were recorded at 0.1 Hz, and peak amplitude measured 10–15 msec after stimulus onset was taken as the AMPAR-mediated component of the EPSC. Holding potential was gradually increased to +40 mV and the NMDAR-mediated component of the EPSC was measured at 40 msec following stimulus onset. The resulting NMDA/AMPA ratio was lower in MSNs from Shank3E13 HET and KO mice compared to WT [One-Way ANOVA, P<0.0001; Bonferroni’s multiple comparisons. P<0.001 (WT vs. HET), P<0.05 (WT vs. KO)] as shown in mean NMDA/AMPA ratio (Fig. 6A) and in representative traces (Fig. 6B). Thus baseline synaptic function is altered in striatum due to either decreased NMDAR-mediated responses or increased AMPAR-mediated responses.
Figure 6.
Striatal synaptic transmission is altered in Shank3E13 mutant mice. (A) Mean NMDA/AMPA ratio is decreased in Shank3E13 HET and KO mice. (B) Ten consecutive raw traces (gray) with overlaid average traces (black) at −70 mV (bottom) and +40 mV (top) from WT (left), HET (middle), and KO (right) mice. WT: 28 cells/9 mice, HET: 18 cells/4 mice, KO: 25 cells/5 mice (*P < 0.05, **P< 0.01, ***P < 0.001).
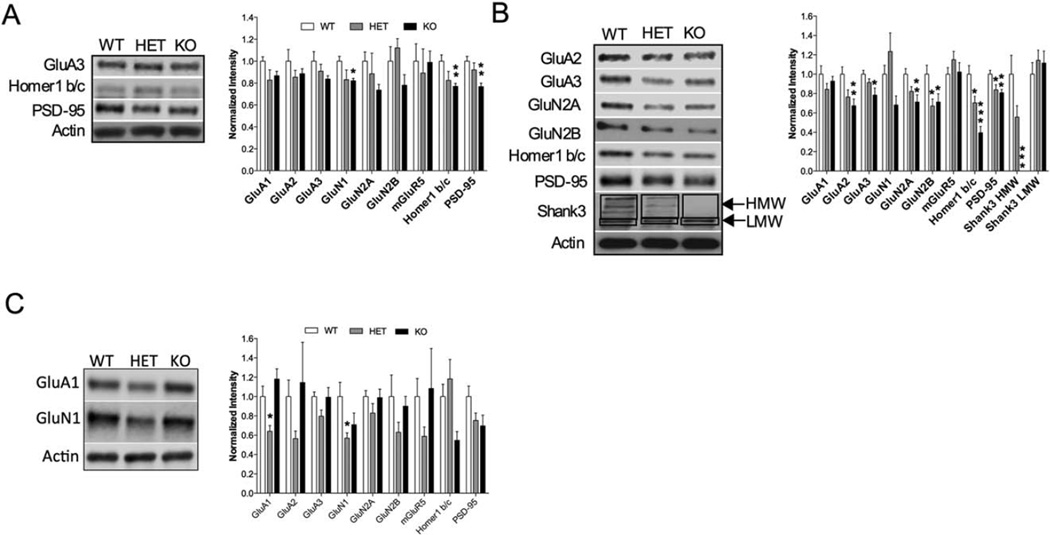
Altered Protein Expression in Striatal Tissue Lysates from Shank3E13 Mutants
To further delineate the impaired baseline striatal synaptic function in the Shank3E13 mutant mice, we investigated striatal protein expression levels focusing on synaptic proteins that interact directly or indirectly with Shank3. Whole tissue lysates from striatal tissue showed a significant reduction in GluN1 (WT vs. KO P<0.01), Homer 1b/c (WT vs. KO P<0.006) and PSD-95 (WT vs. KO P<0.001) in Shank3E13 KO mice compared to WT mice (Fig. 7A). Shank3E13 HET mice also show a trend toward a decrease in GluN1 (WT vs. HET P=0.078; Fig. 7A). To determine whether synaptosomal protein expression was altered in Shank3E13 mutant mice, we used striatal synaptosome preparations and observed a significant reduction in GluA2 (WT vs. KO P<0.006), GluA3 (WT vs. KO P<0.016), GluN2A (WT vs. KO P<0.005), GluN2B (WT vs. KO P<0.046), Homer 1 b/c (WT vs. KO P<0.0001), and PSD-95 (WT vs. KO P<0.0033) in Shank3E13 KO mice compared to WT mice (Fig. 7B). Shank3E13 HET mice also show a decrease in GluN2B (WT vs. HET P<0.038), Homer 1 b/c (WT vs. HET P<0.0351), and PSD-95 (WT vs. HET P<0.0288) when compared to WT. To determine whether synaptosomal protein expression was also altered in Shank3E13 mutant mice, we used hippocampal synaptosome preparations and observed a significant reduction in GluA1 (WT vs. HET P<0.03) and GluN1 (WT vs. HET P<0.03) in Shank3E13 HET mice compared to WT mice (Fig. 7C). Thus, our biochemical studies indicate that synaptic protein composition is altered in the striatum of both Shank3E13 HET and KO and the hippocampus of Shank3E13 HET mice.
Figure 7.
Biochemical analysis of striatal and hippocampal synaptic proteins in Shank3E13 mutant mice. (A) Quantification of whole cell lysates derived from striatal tissue shows significant decreases in GluN1, Homer 1 b/c, and PSD-95. Sample blots are shown on the left with grouped data on the right. (B) Quantification of synaptosome preparations from striatal tissue shows significant decrease in GluA2, GluA3, GluN2A, GluN2B, Homer 1b/c, PSD-95, and Shank3 HMW bands (*P<0.05, **P<0.01, ***P<0.001; for whole cell lysates n=8 WT, 3 HET, 8 KO; for Synaptosome lysates n=16 WT, 11 HET, 16 KO for all samples except blots for Shank3 protein, n=8 WT, 8 HET, 8 KO). (C) Quantification of synaptosome preparations from hippocampal tissue shows significant decrease in GluA1 and GluN1 in heterozygous mice. (*P<0.05 for synaptosome lysates n = 8WT, 8HET, 8KO).
Discussion
We target deletion of Shank3’s PDZ domain by inserting a premature stop codon in Shank3 prior to exon 13. An additional feature of this model is that the neo-Stop cassette is flanked by loxP sites, allowing future studies to feature regional and temporal control over the mutation. We show that this genetic disruption results in loss of the two highest molecular weight isoforms of Shank3 along with behaviors with face validity for ASD, deficits in hippocampal plasticity, altered striatal glutamatergic synaptic transmission, and altered synaptic glutamate receptor subunit expression.
We should note that while generating our Shank3E13 mutant mouse, another group successfully generated and published a similar mouse and characterized the homozygous knockout [Peca et al., 2011]. Thus, our study confirms and expands on the deficits described in the homozygous state [Peca et al., 2011]. The present study, however, represents the first characterization of the more clinically relevant heterozygous mutant model in which multiple important phenotypes were discovered.
Both Shank3E13 HET and KO mice displayed increase repetitive grooming and social deficits. Peca et al. found increased grooming behaviors to varying degrees in 35% of their homozygous mice colony that led to skin lesions [Peca et al., 2011]. [Schmeisser et al., 2012] also observed skin lesions in a similar Shank3 mutant mouse model in which the PDZ domain was disrupted. Curiously, we found that marble burying was not increased, but rather decreased in both heterozygous and homozygous mutants. Such decreased marble burying has been observed by us in other Shank3 mutant models [Kouser et al., 2013; Speed et al., 2015]. We observed increased grooming but no consistent skin lesions in our mutants. We also did not observe an anxiety-like phenotype in our mutant mice. Further validating the lack of anxiety in our mutant mice was the decrease in rearing observed in both HET and KO mice. Decreased rearing was also observed in the homozygous mice in Peca et al. [2011]. Additionally, we observed differences in motor learning and spatial memory in Shank3E13 KO mice that differ from similar studies in Peca et al. [2011]. Our deficits in Shank3E13 KO motor learning were observed on the second day of rotarod testing, whereas [Peca et al., 2011] did not conduct a second day of testing. It may be of interest that initial coordination on day one of the rotarod was decreased in both heterozygotes and homozygotes, while heterozygotes seems to have improved motor learning retention on day 2 of testing. Additionally Peca et al. did not use females in their behavioral testing [Peca et al., 2011], and the females appeared to drive rotarod differences in our model. Differences in protocol and cohort makeup were also present in the Morris water maze in which we observe spatial memory deficits in Shank3E13 KOs not seen in Peca et al. [2011]. Differences in the two studies may have also manifested as a result of the two different breeding and housing strategies. Our Shank3E13 mice were generated from 12 heterozygous breeding pairs generating littermate triplets (WT/HET/KO) or littermate pairs (WT/HET; WT/KO). Peca et al. generated their behavioral cohort from homozygous breeder pairs (WT×WT, KO×KO) [Peca et al., 2011]. Also, [Peca et al., 2011] housed mice by genotype, whereas we housed mice as littermate triplets or pairs to ensure that environmental variables are equally experienced. Subtle genetic background differences or differences in stress levels and husbandry conditions could also contribute to observed differences.
Regardless of the differences, we both demonstrated altered behaviors with ASD face validity in our models. Thus, where homozygous mutants are concerned, our two studies largely replicate across similar tests and should be viewed as complimentary.
Perhaps more important is our characterization of the clinically relevant heterozygous Shank3E13 mutants. Shank3E13 HET mice demonstrate a robust increase in repetitive grooming and deficits in social tasks including social preference and social recognition of a juvenile mouse. A lack of social preference in the HET but not the KO further supports the importance of characterizing the heterozygous mouse model and suggests variation in gene expression can contribute to heterogeneity in phenotypes.
In addition, we observed lack of sociability in the heterozygotes, but not the homozygotes. This is a puzzling observation but may be explained by differential compensation in the heterozygotes vs. homozygotes by other Shank isoforms. Direct interaction between two freely moving mice provides a more reciprocal interaction and was abnormal in homozygous Shank3E13 mutants. While this and other mutant mouse studies involving Shank3 truncations seem to mimic some of the behavioral phenotypes observed in ASD and PMS (incoordination, learning and memory deficits, repetitive behaviors, and social deficits), the phenotypes among mouse models and even patients with Shank3 can be highly variable. More clinical data are needed on genotype/phenotype relationships in patients with complete deletion of Shank3 vs. mutations in order to further determine how different Shank3 mutations affect behavior in patients [Harony-Nicolas, De Rubeis, Kolevzon, & Buxbaum, 2015].
In eight of our behavioral tests, we identified a main effect of sex that is consistent with overall differences in behavior between males and females in some paradigms. Only in rearing, locomotor activity, and rotarod was there an actual interaction between genotype and sex. This is consistent with roughly equal male/female ratios in patients with Phelan-McDermid Syndrome.
Our study also demonstrates deficits in CA1 hippocampal LTP in Shank3E13 HET and KO mice. Deficits in LTP have been fairly consistently observed across multiple Shank3 mouse models [Bozdagi et al., 2010; Jaramillo et al., 2015; Kouser et al., 2013; Yang et al., 2012].
Additionally, Shank3E13 HET and KO mice demonstrate deficits in striatal NMDAR/AMPAR-ratio in dorsal striatum. We also observed reduction in glutamatergic receptors including GluA2, GluA3, GluN2A, and GluN2B in western blots from Shank3E13 KO mice synaptosomes, a finding similar to that presented in Peca et al. [2011]. Furthermore, Shank3E13 HET mice displayed a reduction in synaptosomal GluN2B.
Overall, this study identifies the first robust, ASD-relevant behavioral abnormalities in the heterozygous model of a Shank3 PDZ domain mutant. We find clear differences in striatal synaptic function in heterozygous mice for the first time. In terms of the homozygous mutants, we feel this study compliments and extends Peca et al. [2011] and other publications involving mutation/deletion of Shank3. Thus, our study provides behavioral, biochemical, and physiological alterations in a clinically relevant Shank3 mutant model. Furthermore, this model was created with the ability to genetically reverse the mutation, providing a means to examine both regional and temporal genetic rescue of the identified phenotypes in future studies. We note that a key finding in a similar Shank3 mutant model has recently been published demonstrating that some behavioral and other phenotypes can be reversed by adult-onset temporal reversal of Shank3 deletion/mutation [Mei et al., 2016]. Replication of such groundbreaking findings across laboratories and in multiple Shank3 mutant mouse models will be key to move the field forward toward novel therapeutics.
Acknowledgments
We thank Dr. Paul Worley for Shank3 antibody. Also to Dr. Yong-hui Jiang for providing whole brains from complete Shank3 knockouts. Supported by NIH (R01HD069560, R21HD065290, & R01HD069560-S1 to C.M.P.), Autism Speaks (C.M.P.), Autism Speaks and Autism Science Foundation Postdoctoral Fellowships (HES), The Hartwell Foundation (C.M.P.), Ed and Sue Rose Distinguished Professorship in Neurology (C.M.P.), and gifts from Drs. Clay Heighten and Debra Caudy and BRAINS for Autism (C.M.P.). C.M.P. has accepted travel funds and honoraria to speak once at each of the following companies: Psychogenics, Inc.; Astra-Zeneca; Roche; Pfizer; and Dainippon Sumitomo Pharma Co. Dr. Powell also has investigator-initiated grant funding through a contract for clinical research with Novartis.
Footnotes
Author contributions: T.C.J., H.E.S., J.M.R., and C.M.P. designed research; T.C.J., H.E.S., Z.X., J.M.R., C.F.O., S.L., and I.F. performed research; T.C.J., H.E.S., and J.M.R. analyzed data; T.C.J., H.E.S., J.M.R., and C.M.P wrote the paper.
References
- Arons MH, Thynne CJ, Grabrucker AM, Li D, Schoen M, Cheyne JE, et al. Autism-associated mutations in ProSAP2/Shank3 impair synaptic transmission and neurexin-neuroligin-mediated transsynaptic signaling. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2012;32:14966–14978. doi: 10.1523/JNEUROSCI.2215-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1 in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. Journal of Neuroscience. 2011;31:8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Blaiss CA, Etherton MR, Espinosa F, Tabuchi K, Walz C, et al. Neuroligin-1 deletion results in impaired spatial memory and increased repetitive behavior. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2010a;30:2115–2129. doi: 10.1523/JNEUROSCI.4517-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Kaeser PS, Sudhof TC, Powell CM. RIM1alpha and interacting proteins involved in presynaptic plasticity mediate prepulse inhibition and additional behaviors linked to schizophrenia. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2010b;30:5326–5333. doi: 10.1523/JNEUROSCI.0328-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blundell J, Tabuchi K, Bolliger MF, Blaiss CA, Brose N, Liu X, et al. Increased anxiety-like behavior in mice lacking the inhibitory synapse cell adhesion molecule neuroligin 2. Genes, Brain, and Behavior. 2009;8:114–126. doi: 10.1111/j.1601-183X.2008.00455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeckers TM, Bockmann J, Kreutz MR, Gundelfinger ED. ProSAP/Shank proteins: A family of higher order organizing molecules of the postsynaptic density with an emerging role in human neurological disease. Journal of Neurochemistry. 2002;81:903–910. doi: 10.1046/j.1471-4159.2002.00931.x. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Borgatti R, Felisari G, Gagliardi C, Selicorni A, Zuffardi O. Disruption of the ProSAP2 gene in a t(12;22)(q24.1;q13.3) is associated with the 22q13.3 deletion syndrome. The American Journal of Human Genetics. 2001;69:261–268. doi: 10.1086/321293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozdagi O, Sakurai T, Papapetrou D, Wang X, Dickstein DL, Takahashi N, et al. Haploinsufficiency of the autism-associated Shank3 gene leads to deficits in synaptic function, social interaction, and social communication. Molecular Autism. 2010;1:15. doi: 10.1186/2040-2392-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragatsis I, Zeitlin S. A method for the generation of conditional gene repair mutations in mice. Nucleic Acids Research. 2001;29:E10. doi: 10.1093/nar/29.3.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffney LJ, Wei J, Cheng J, Liu W, Smith KR, Kittler JT, Yan Z. Shank3 deficiency induces NMDA receptor hypofunction via an actin-dependent mechanism. The Journal of Neuroscience: The Official Journal of the Society for Neuroscience. 2013;33:15767–15778. doi: 10.1523/JNEUROSCI.1175-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, et al. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nature Genetics. 2007;39:25–27. doi: 10.1038/ng1933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlers MD. Synapse structure: Glutamate receptors connected by the shanks. Current Biology. 1999;9:R848–R850. doi: 10.1016/s0960-9822(00)80043-3. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Molecular morphogens for dendritic spines. Trends Neuroscience. 2002;25:64–67. doi: 10.1016/s0166-2236(02)02061-1. [DOI] [PubMed] [Google Scholar]
- Etherton MR, Blaiss CA, Powell CM, Sudhof TC. Mouse neurexin-1alpha deletion causes correlated electrophysiological and behavioral changes consistent with cognitive impairments. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:17998–18003. doi: 10.1073/pnas.0910297106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier J, et al. De novo mutations in the gene encoding the synaptic scaffolding protein SHANK3 in patients ascertained for schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:7863–7868. doi: 10.1073/pnas.0906232107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Y, Lippa CF, Zhu J, Lin Q, Rosso AL. Disruption of glutamate receptors at Shank-postsynaptic platform in Alzheimer’s disease. Brain Research. 2009;1292:191–198. doi: 10.1016/j.brainres.2009.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabrucker S, Proepper C, Mangus K, Eckert M, Chhabra R, Schmeisser MJ, et al. The PSD protein Pro-SAP2/Shank3 displays synapto-nuclear shuttling which is deregulated in a schizophrenia-associated mutation. Experimental Neurology. 2014;253:126–137. doi: 10.1016/j.expneurol.2013.12.015. [DOI] [PubMed] [Google Scholar]
- Guilmatre A, Huguet G, Delorme R, Bourgeron T. The emerging role of SHANK genes in neuropsychiatric disorders. Developmental Neurobiology. 2014;74:113–122. doi: 10.1002/dneu.22128. [DOI] [PubMed] [Google Scholar]
- Guy J, Gan J, Selfridge J, Cobb S, Bird A. Reversal of neurological defects in a mouse model of Rett syndrome. Science. 2007;315:1143–1147. doi: 10.1126/science.1138389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Holder JL, Jr, Schaaf CP, Lu H, Chen H, Kang H, et al. SHANK3 overexpression causes manic-like behaviour with unique pharmacogenetic properties. Nature. 2013;503:72–77. doi: 10.1038/nature12630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harony-Nicolas H, De Rubeis S, Kolevzon A, Buxbaum JD. Phelan McDermid syndrome: From genetic discoveries to animal models and treatment. Journal of Child Neurology. 2015;30:1861–1870. doi: 10.1177/0883073815600872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herbert MR. SHANK3, the synapse, and autism. The New England Journal of Medicine. 2011;365:173–175. doi: 10.1056/NEJMcibr1104261. [DOI] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proceeding of National Academic Science United States of Amaerica. 2002;99:7746–7750. doi: 10.1073/pnas.122205699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Lui S, Powell C. Altered striatal synaptic function and abnormal behaviour in Shank3 exon4–9 deletion mouse model of autism. Autism Research. 2015 doi: 10.1002/aur.1529. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaramillo TC, Speed HE, Xuan Z, Reimers JM, Liu S, Powell CM. Altered striatal synaptic function and abnormal behaviour in Shank3 exon4–9 deletion mouse model of autism. Autism Research. 2016;9:350–375. doi: 10.1002/aur.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolevzon A, Cai G, Soorya L, Takahashi N, Grodberg D, Kajiwara Y, et al. Analysis of a purported SHANK3 mutation in a boy with autism: Clinical impact of rare variant research in neurodevelopmental disabilities. Brain Research. 2011;1380:98–105. doi: 10.1016/j.brainres.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Kouser M, Speed HE, Dewey CM, Reimers JM, Widman AJ, Gupta N, et al. Loss of predominant shank3 isoforms results in hippocampus-dependent impairments in behavior and synaptic transmission. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2013;33:18448–18468. doi: 10.1523/JNEUROSCI.3017-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon CH, Luikart BW, Powell CM, Zhou J, Matheny SA, Zhang W, et al. Pten regulates neuronal arborization and social interaction in mice. Neuron. 2006;50:377–388. doi: 10.1016/j.neuron.2006.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leyfer OT, Folstein SE, Bacalman S, Davis NO, Dinh E, Morgan J, et al. Comorbid psychiatric disorders in children with autism: Interview development and rates of disorders. Journal of Autism and Developmental Disorders. 2006;36:849–861. doi: 10.1007/s10803-006-0123-0. [DOI] [PubMed] [Google Scholar]
- Mei Y, Monteiro P, Zhou Y, Kim JA, Gao X, Fu Z, Feng G. Adult restoration of Shank3 expression rescues selective autistic-like phenotypes. Nature. 2016;530:481–484. doi: 10.1038/nature16971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misceo D, Rodningen OK, Baroy T, Sorte H, Mellembakken JR, Stromme P, et al. A translocation between Xq21.33 and 22q13.33 causes an intragenic SHANK3 deletion in a woman with Phelan-McDermid syndrome and hypergonadotropic hypogonadism. American Journal of Medical Genetics A. 2011;155A:403–408. doi: 10.1002/ajmg.a.33798. [DOI] [PubMed] [Google Scholar]
- Moessner R, Marshall CR, Sutcliffe JS, Skaug J, Pinto D, Vincent J, et al. Contribution of SHANK3 mutations to autism spectrum disorder. American Journal of Human Genetics. 2007;81:1289–1297. doi: 10.1086/522590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris RG, Garrud P, Rawlins JN, Keefe J. Place navigation impaired in rats with hippocampal lesions. Nature. 1982;297:681–683. doi: 10.1038/297681a0. [DOI] [PubMed] [Google Scholar]
- Moser EI, Krobert KA, Moser MB, Morris RG. Impaired spatial learning after saturation of long-term potentiation. Science. 1998;281:2038–2042. doi: 10.1126/science.281.5385.2038. [DOI] [PubMed] [Google Scholar]
- Moser MB, Moser EI. Functional differentiation in the hippocampus. Hippocampus. 1998;8:608–619. doi: 10.1002/(SICI)1098-1063(1998)8:6<608::AID-HIPO3>3.0.CO;2-7. [DOI] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ, Perez A, Barbaro RP, Johns JM, Magnuson TR, et al. Sociability and preference for social novelty in five inbred strains: An approach to assess autistic-like behavior in mice. Genes Brain and Behavior. 2004;3:287–302. doi: 10.1111/j.1601-1848.2004.00076.x. [DOI] [PubMed] [Google Scholar]
- Nadler JJ, Moy SS, Dold G, Trang D, Simmons N, Perez A, et al. Automated apparatus for quantitation of social approach behaviors in mice. Genes Brain and Behavior. 2004;3:303–314. doi: 10.1111/j.1601-183X.2004.00071.x. [DOI] [PubMed] [Google Scholar]
- Naisbitt S, Kim E, Tu JC, Xiao B, Sala C, Valtschanoff J, et al. Shank, a novel family of postsynaptic density proteins that binds to the NMDA receptor/PSD-95/GKAP complex and cortactin. Neuron. 1999;23:569–582. doi: 10.1016/s0896-6273(00)80809-0. [DOI] [PubMed] [Google Scholar]
- Connor EC, Bariselli S, Bellone C. Synaptic basis of social dysfunction: A focus on postsynaptic proteins linking group-I mGluRs with AMPARs and NMDARs. The European Journal of Neuroscience. 2014;39:1114–1129. doi: 10.1111/ejn.12510. [DOI] [PubMed] [Google Scholar]
- Park E, Na M, Choi J, Kim S, Lee JR, Yoon J, et al. The Shank family of postsynaptic density proteins interacts with and promotes synaptic accumulation of the beta PIX guanine nucleotide exchange factor for Rac1 and Cdc42. Journal of Biological Chemistry. 2003;278:19220–19229. doi: 10.1074/jbc.M301052200. [DOI] [PubMed] [Google Scholar]
- Peca J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, et al. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472:437–442. doi: 10.1038/nature09965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phelan K, Rogers RC. In: Phelan-McDermid syndrome. Pagon RA, Adam MP, Ardinger HH, Bird TD, Dolan CR, Fong CT, Smith RJH, Stephens K, editors. Seattle (WA): GeneReviews(R); 1993. [Google Scholar]
- Powell CM, Schoch S, Monteggia L, Barrot M, Matos MF, Feldmann N, et al. The presynaptic active zone protein RIM1alpha is critical for normal learning and memory. Neuron. 2004;42:143–153. doi: 10.1016/s0896-6273(04)00146-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, et al. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. Journal of Neuroscience. 2005;25:3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarasua SM, Dwivedi A, Boccuto L, Rollins JD, Chen CF, Rogers RC, et al. Association between deletion size and important phenotypes expands the genomic region of interest in Phelan-McDermid syndrome (22q13 deletion syndrome) Journal of Medical Genetics. 2011;48:761–766. doi: 10.1136/jmedgenet-2011-100225. [DOI] [PubMed] [Google Scholar]
- Schmeisser MJ, Ey E, Wegener S, Bockmann J, Stempel AV, Kuebler A, et al. Autistic-like behaviours and hyperactivity in mice lacking ProSAP1/Shank2. Nature. 2012;486:256–260. doi: 10.1038/nature11015. [DOI] [PubMed] [Google Scholar]
- Shetreat-Klein M, Shinnar S, Rapin I. Abnormalities of joint mobility and gait in children with autism spectrum disorders. Brain & #Development. 2014;36:91–96. doi: 10.1016/j.braindev.2012.02.005. [DOI] [PubMed] [Google Scholar]
- Soorya L, Kolevzon A, Zweifach J, Lim T, Dobry Y, Schwartz L, et al. Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Molecular Autism. 2013;4:18. doi: 10.1186/2040-2392-4-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speed HE, Kouser M, Xuan Z, Reimers JM, Ochoa CF, Gupta N, et al. Autism-associated insertion mutation (InsG) of Shank3 exon 21 causes impaired synaptic transmission and behavioral deficits. Journal of Neuroscience. 2015;35:9648–9665. doi: 10.1523/JNEUROSCI.3125-14.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava AK, Schwartz CE. Intellectual disability and autism spectrum disorders: causal genes and molecular mechanisms. Neuroscience and Biobehavioral Reviews. 2014;46(Pt 2):161–174. doi: 10.1016/j.neubiorev.2014.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Sudhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318:71–76. doi: 10.1126/science.1146221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu JC, Xiao B, Naisbitt S, Yuan JP, Petralia RS, Brakeman P, et al. Coupling of mGluR/Homer and PSD-95 complexes by the Shank family of postsynaptic density proteins. Neuron. 1999;23:583–592. doi: 10.1016/s0896-6273(00)80810-7. [DOI] [PubMed] [Google Scholar]
- Verpelli C, Dvoretskova E, Vicidomini C, Rossi F, Chiappalone M, Schoen M, et al. Importance of Shank3 protein in regulating metabotropic glutamate receptor 5 (mGluR5) expression and signaling at synapses. Journal of Biological Chemistry. 2011;286:34839–34850. doi: 10.1074/jbc.M111.258384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vucurovic K, Landais E, Delahaigue C, Eutrope J, Schneider A, Leroy C, et al. Bipolar affective disorder and early dementia onset in a male patient with SHANK3 deletion. European Journal of Medical Genetics. 2012;55:625–629. doi: 10.1016/j.ejmg.2012.07.009. [DOI] [PubMed] [Google Scholar]
- Wan Y, Ade KK, Caffall Z, Ilcim Ozlu M, Eroglu C, Feng G, Calakos N. Circuit-selective striatal synaptic dysfunction in the Sapap3 knockout mouse model of obsessive-compulsive disorder. Biological Psychiatry. 2014;75:623–630. doi: 10.1016/j.biopsych.2013.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan Y, Feng G, Calakos N. Sapap3 deletion causes mGluR5-dependent silencing of AMPAR synapses. The Journal of Neuroscience : The Official Journal of the Society for Neuroscience. 2011;31:16685–16691. doi: 10.1523/JNEUROSCI.2533-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, et al. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Human Molecular Genetics. 2011;20:3093–3108. doi: 10.1093/hmg/ddr212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Xu Q, Bey AL, Lee Y, Jiang YH. Transcriptional and functional complexity of Shank3 provides a molecular framework to understand the phenotypic heterogeneity of SHANK3 causing autism and Shank3 mutant mice. Molecular Autism. 2014;5:30. doi: 10.1186/2040-2392-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welch JM, Lu J, Rodriguiz RM, Trotta NC, Peca J, Ding JD, et al. Cortico-striatal synaptic defects and OCD-like behaviours in Sapap3-mutant mice. Nature. 2007;448:894–900. doi: 10.1038/nature06104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang M, Bozdagi O, Scattoni ML, Wohr M, Roullet FI, Katz AM, et al. Reduced excitatory neurotransmission and mild autism-relevant phenotypes in adolescent Shank3 null mutant mice. The Journal of Neuroscience : the Official Journal of the Society for Neuroscience. 2012;32:6525–6541. doi: 10.1523/JNEUROSCI.6107-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]