Abstract
Population-based carrier screening is limited to well-studied or high-impact genetic conditions for which the benefits may outweigh the associated harms and costs. As the cost of genome sequencing declines and availability increases, the balance of risks and benefits may change for a much larger number of genetic conditions, including medically actionable additional findings. We designed an RCT to evaluate genomic clinical sequencing for women and partners considering a pregnancy. All results are placed into the medical record for use by healthcare providers. Through quantitative and qualitative measures, including baseline and post result disclosure surveys, post result disclosure interviews, 1–2 year follow-up interviews, and team journaling, we are obtaining data about the clinical and personal utility of genomic carrier screening in this population. Key outcomes include the number of reportable carrier and additional findings, and the comparative cost, utilization, and psychosocial impacts of usual care vs. genomic carrier screening. As the study progresses, we will compare the costs of genome sequencing and usual care as well as the cost of screening, pattern of use of genetic or mental health counseling services, number of outpatient visits, and total healthcare costs. This project includes novel investigation into human reactions and responses from would-be parents who are learning information that could both affect a future pregnancy and their own health.
Keywords: Genetic testing, Genome sequencing, Preconception, Genetics, Clinical trial
1. Background
Current professional recommendations for population-based carrier screening in the United States limit this approach to a small number of well-studied higher impact genetic conditions for which the potential benefits outweigh the potential harms and costs associated with the screening. However, as the cost of genome sequencing declines and availability increases, it is unclear whether the balance of benefits and harms will change for additional conditions.
Carrier screening using genome sequencing (which we refer to as “genomic carrier screening”) rather than panel screening could identify more mutations and identify more couples at risk of bearing children affected with genetic conditions. Thus, a benefit of genomic carrier screening is the detection of more couples at risk of conceiving affected children, which could provide greater and more informed choice in reproductive decision-making. Additionally, screening allows for earlier diagnosis and treatment of an affected child. However, there is also potential for harms due to screening, including the additional healthcare costs associated with genetic counseling, costs and anxiety related to partner genetic testing in low prevalence situations, subsequent service utilization associated with knowledge of this information, and psychosocial harms from learning about carrier risks.
We describe how we developed an experimental randomized controlled trial (RCT) at Kaiser Permanente Northwest (KPNW), designed to investigate the clinical implementation of genomic carrier screening to aid in reproductive decision-making in preconception adults. This National Human Genome Research Institute-funded study is part of the Clinical Sequencing Exploratory Research (CSER) consortium to address the clinical implications of genome sequencing. Every woman in the study receives limited carrier screening (usually for cystic fibrosis) completed by a healthcare provider and then, as part of the study activities, is randomly allocated to receive either genome sequencing or no additional testing. Through this study, we will learn the level of interest that healthy couples have in receiving information from genomic carrier screening. We will obtain information about how they prefer to receive the carrier information, what they do with the results (e.g., how they react, who they share them with), and how the results influence their plans for a future pregnancy.
The prima’ry outcome is the mean number of variants reported for carrier status. Key secondary outcomes include the comparative cost, utilization, and psychosocial impacts of usual care vs. genomic carrier screening. In an effort to obtain patient’s perspectives about the downsides of broad genomic carrier screening, we also evaluate why women and their partners decline genomic carrier screening. As a part of the study, we place the order for genomic carrier screening and the results into the participant’s electronic medical record so that the information will be available for future patient care. In this article, we describe the methods to address these goals.
2. Clinical context
The only condition that is recommended by the American College of Obstetricians and Gynecologists to be offered for carrier screening in all couples in the United States who are considering pregnancy is cystic fibrosis (CF) [1]. CF has a relatively high carrier prevalence of about 3% in non-Hispanic Caucasians and 1–2% for Asian Americans, African Americans, and Hispanic Caucasians [2]. Carrier screening has also been recommended to be offered for specific contexts with high prevalence, such as Tay Sachs disease for individuals with Ashkenazi Jewish, French Canadian, or Cajun ancestry [3]. Additionally, carrier screening is typically offered to family members of an identified carrier or affected individual or members of ethnic or racial groups known to have a higher prevalence of a condition [4–6].
According to Online Mendelian Inheritance in Man (OMIM), there are currently listings for 2933 X-linked and autosomal recessive genetic conditions [7], most of which are not covered by current professional guidelines. Although commercially available panels incorporate tens or hundreds of conditions, the panels are not widely used and testing is often limited to well-known variants in the genes examined [8–10]. Because the conditions included in OMIM are caused by variants in many different genes, genomic carrier screening greatly expands the breadth of traditional clinical carrier screening to identify carriers of conditions with rare or novel variants.
While many research and clinical programs are beginning to use whole exome or whole genome sequencing, almost all do so for patients with a distinct clinical phenotype, such as cancer or a developmental disability [11–15]. In that context, carrier status is an additional finding that often is not immediately relevant and is overshadowed by the clinical context that led to the genetic testing in the first place. Although these programs are uncovering important information about how sick patients and their families respond to genome sequencing, very little is known about the acceptability and interest for genome sequencing in a healthy population.
3. Overview of experimental design
Our RCT is unique in that we select participants based on planning for a future pregnancy. Thus, our population can provide a perspective that focuses on the near-term relevance of and acceptance for carrier screening results. KPNW’s comprehensive medical care, which is delivered in a managed care setting, will help us maximize the potential relevance of the genomic data to the patient and to investigate the impact of genomic carrier screening by evaluating downstream healthcare utilization and costs. Carrier screening may be broadly relevant to all individuals when they or their close relatives may plan a future pregnancy, and carrier results can be reported back any time whole genome or exome sequencing is performed as a secondary finding. By assessing the acceptability of broad preconception carrier screening in a healthy population, we will obtain information about the average number of pathogenic and likely pathogenic carrier variants as well as the frequency of medically actionable findings.
The purpose of this study is to inform future implementation efforts and understand how patients and clinicians react to, understand, and use information from genomic carrier screening compared with usual care (which includes screening for a small number of targeted conditions). Our study design allows for exploration of the advantages and disadvantages of expanded testing using genomic carrier screening compared with tests for individual or a small number of conditions. This study was reviewed and approved by the Kaiser Permanente Northwest Institutional Review Board, and all participants completed informed consent.
3.1. Inclusion and exclusion criteria
Women eligible for the study have a clinical carrier screening test (usually CF) ordered and completed as part of a preconception planning visit or during pregnancy and, if they were pregnant at the time of the test, are now post-partum. Additionally, women must be current health system members, not currently pregnant, and planning a future pregnancy. Women are excluded if their genetic test was for diagnostic purposes. Male partners are eligible if their enrolled female partner is found to be a carrier of an autosomal recessive genetic condition. Having a male partner is not an eligibility criterion, as we are also interested in measuring changes in reproductive plans for women seeking pregnancy through a sperm donor.
3.2. Enrollment
Women who have had a carrier test completed as part of a preconception visit or who had the test during pregnancy and are at least six months post-partum are identified daily from the electronic medical record. An initial chart review to confirm the eligibility of a woman to participate in the trial is performed, and she is mailed a brochure with information about the study. She is then called, screened for eligibility, and, if interested in joining the study, mailed the consent form to review. About one week later, she is again called to schedule an in-person consent visit with a genetic counselor. At this visit, the potential participant reviews information about participation in the study with the genetic counselor and has her questions answered. If she decides to join the study and signs the consent form, then she completes a baseline survey and is randomly allocated into the genome sequencing (GS) arm or the usual care (UC) arm. Women in the GS arm have their blood drawn at the consent visit. Women in the UC arm receive no additional testing via the study; their only additional follow-up is an online survey six months later (Fig. 1).
Fig. 1.
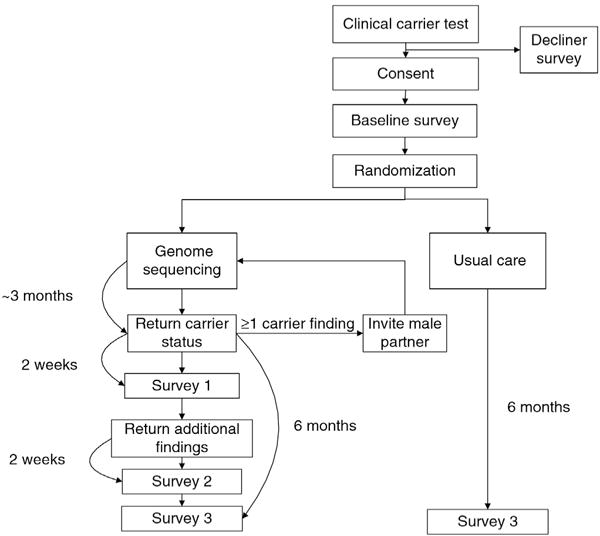
Flow chart of experimental randomized control trial for genomic carrier screening in healthy patients seeking preconception genetic testing.
3.3. Sample size
We determined the minimum number of participants required to detect an effect on the primary endpoint (i.e., number of results disclosed). On the basis of an initial population mean (i.e., number of results disclosed) of 0.02 and a clinically significant interval of 0.1 carrier results returned, we calculated the required sample size for a range of values for the power and standard deviation (0.1 to 0.5, since we do not know how sensitive the GS screen will be) and a 5:8 allocation ratio of intervention to control. With a target sample size of 325 participants (125 GS arm, 200 UC arm), we have at least 95% power to detect a clinically significant change in the number of results disclosed. Assuming a 50% carrier rate, we also expect to enroll 63 male partners.
3.4. Statistical analysis for primary endpoint
We will use an independent sample t-test to compare the mean number of validated findings for carrier status for participants in each arm of the study. Rank-based methods (Mann-Whitney U) will be used for sensitivity analysis if the distribution is highly skewed or there are a large number of observations with no findings. In addition, we will determine whether participants in the GS arm are more likely to have one or more findings compared to people in the control arm using logistic regression.
4. Intervention
Individuals in the GS arm receive results from a blood sample sequenced at Illumina (San Diego, CA) on the HiSeq platform. Genomes are sequenced to an average fold coverage of at least 30X (average coverage of 37X) with a minimum of 87% of bases having at least 20X coverage. Genomic alignment (Burrows-Wheeler Aligner) [16] and variant calls (GATK) [17] are performed at the University of Washington. Variants are annotated by SeattleSeqAnnotation138 [18,19] and augmented by the most recent ClinVar and HGMD releases. All variants undergo tertiary review at Oregon Health & Science University (OHSU) with a combination of automated and human curation to identify variants in genes. Sanger sequencing is performed at OHSU’s CLIA laboratory to confirm any variants before they are returned to a participant.
We disclose carrier results for about 750 gene/condition pairs. The list of gene/condition pairs included those genes for which national professional organizations recommended screening in at least one population, genes present on carrier tests from commercial laboratories which offered expanded carrier testing in 2013, genes associated with conditions that are commonly found on newborn screening panels, and genes associated with serious childhood recessive diseases. Members of the study advisory committee also nominated genes for inclusion based on professional expertise. The list is dynamic but currently includes 728 autosomal recessive, X-linked, and mitochondrial conditions. All gene/condition pairs have been categorized as lifespan limiting, serious, mild, unpredictable, or adult onset [20]. Conditions in the category of lifespan limiting are required to be disclosed as part of participation in this study. However, a participant can decide whether to receive results from any of the other categories; for a participant who opts out of a specific category at the consent visit, the variants in genes in that category are not analyzed or validated. Only variants that are pathogenic or likely pathogenic according to American College of Medical Genetics and Genomics criteria [21] are disclosed to participants.
If there is at least one carrier result to disclose to a woman, she receives an in-person genetic counseling session. If there are no results to return, a letter is mailed to inform her. If a participant is found to be a carrier of an autosomal recessive condition, her male partner is then eligible to participate and is invited into the study to undergo genetic carrier screening. Like the women, male partners select the categories of results they wish to receive and are required to receive the results of lifespan limiting conditions. In addition, the men must receive results for which their partner was a carrier.
In addition to carrier results, participants can choose to receive additional findings. The additional findings list includes medically actionable and highly penetrant Mendelian conditions that affect adults. The list currently includes 121 genes, including most of the 56 genes recommended by the American College of Medical Genetics and Genomics for return [22]. Changes to the secondary findings list since the Gallego paper was published include the addition of PRRT2, PRKG2, COL5A1, COL5A2, APOB, PCSK9, and SLC3A1 and the deletion of KCNJ2, SMARCB1, TTN due to changes in evidence. Criteria for inclusion for disclosure as an additional finding has been published [23]. Additional findings are returned about one month after carrier results are disclosed. Similar to the disclosure of carrier status, all additional findings are disclosed in person during a genetic counseling session; if there are no results a letter is mailed to inform the participant. We intentionally separated the result visits (carrier status and additional findings) in order to not potentially overwhelm the participant with too much information at one visit.
All participants in the GS arm complete surveys at enrollment in the study, two weeks after carrier results are returned, two weeks after additional findings are returned, and six months after carrier results are returned (Fig. 1). All testing is paid for by the study and participants are compensated $10 for each survey completed, with an additional $20 if all surveys are completed.
After both carrier information and additional findings have been returned to a participant, OHSU provides a final report that is placed in the participant’s electronic medical record. The report includes information about a gene, condition, specific variant, frequency of the condition, and pathogenicity of the variant. All supporting information for the pathogenicity classification is provided and cited, including a list of all genes that were reviewed for variants. Reports for participants who had no pathogenic or likely pathogenic variants are also included in the participant’s medical record.
Finally, women who are eligible to participate in our RCT but decline to do so are asked to complete a brief survey so that we can understand why they chose not to participate in the study. This survey includes questions on basic demographic characteristics, current number of children, whether a family member has an inherited recessive or X-linked condition, and reasons for declining. We also ask these patients whether they know anyone with a child with an inherited recessive or X-linked condition and how they perceive the impact it has on that family.
5. Outcomes
5.1. Patient-reported outcomes (PROs)
We chose several instruments to measure potential psychosocial impacts on patients receiving genetic results according to the approach of Patrick and Erickson (1993) [24]. These items include 1) symptoms (anxiety, depression), 2) perceptions (conflict with decision making, self-rated health status, satisfaction with genetic counseling, and amount of information received in genetic counseling), and 3) resilience (positive health attitudes). We chose these measures because they are relevant to receiving genetic testing, brief, and adaptable for administration via the web.
By definition, a PRO is any report of the status of a patient’s health condition that comes directly from the patient and without interpretation by a clinician or anyone else [25]. We previously reviewed the literature to identify standardized outcome measures for patients receiving carrier status and secondary findings, select PRO measures that allow for consistency across CSER projects, and address issues specific to our unique healthy population. We use multiple instruments to quantitatively measure a range of health and psychosocial impacts that genome sequencing or CF carrier testing could have on participants while minimizing the respondent’s burden by keeping the time to complete the surveys to under 20 min (Table 1).
Table 1.
Patient survey measures.
| Domain | Measure | Survey Timepointa |
|---|---|---|
| Health-related quality of life | BRFSS general health status BREF [26–28] |
BL, FU BL, FU |
| Anxiety | STAI 6 [29] | BL, FU |
| Depression | PHQ-8 [30] | BL, FU |
| Satisfaction – decision regret | Decision conflict scale [31] | BL, CS, AF, FU |
| Satisfaction – decision | Satisfaction with decision instrument [32] | BL, CS, AF, FU |
| Satisfaction – genetic counseling | Genetic counseling satisfaction scale [33] | CS, AF |
| Information seeking | Health information orientation scale [34] | BL |
| Decision style preference | Control preferences scale [35] | BL |
| Understanding | Genetic knowledge – original contribution specific to carrier testing Subjective numeracy scale [36] |
BL, FU BL, FU BL, FU |
| Religiosity or spirituality | Brief multidimensional measure of religiousness/spirituality [37] | BL |
| Attitudes toward genetic testing | Perceived vulnerability Perceived benefits/utility [38] Original contributions |
BL, FU BL, FU BL, FU |
| Uncertainty | Original contributions | CS, AF |
| Decisional impact | Original contributions | FU |
| Information sharing | Original contributions | BL, CS, AF, FU |
| Reaction to results | Original contributions | CS, AF |
| Expected reaction to results | Original contributions | BL |
| Decision | Categories of conditions participants chose to receive results on | BL |
BL = baseline, CS = 2 weeks after carrier results were returned, AF = 2 weeks after additional findings results were returned, FU = 6-month follow-up from baseline (UC) or carrier results (GS).
PROs are obtained through surveys and interviews regarding the effect genetic carrier screening has on participants’ quality of life, satisfaction with care, timeliness of reporting, and how genomic information was used. PROs are measured at baseline and six months later for all study participants. Participants in the GS arm also complete surveys two weeks after being provided with the results of carrier screening and another two weeks after provision of additional findings. All surveys are conducted online via a locally created online survey tool. We conducted focus groups to gather preliminary reactions to the study recruitment materials, consent forms, and overall design [39].
All variables from the PRO survey are electronically coded. Data are cleaned to check for unrealistic or outlying values and to assess for missing data. For PROs collected at only one timepoint and for the GS arm, we will examine descriptive statistics for scale scores and originally contributed items. For PROs collected at only one timepoint and across both arms, we will compare the arms by their responses (χ2, Mann-Whitney U, and t-tests for univariate analyses of discrete, ordinal, and continuous variables, respectively). For PROs that are collected from both arms at both baseline and follow-up, we will use generalized estimating equations [40,41] to test whether there is a differential change across time (i.e., time × arm effect). We will use the appropriate form of generalized estimating equation depending on the nature of the variable (e.g., identity link and Gaussian distribution for normally distributed continuous PROs). A significant coefficient for the time × arm product term would indicate that one arm changed more than the other from baseline to follow-up. We will graph any significant interactions to interpret the nature of the differential change. We will also examine whether there were differences in PROs between those with one or more positive results to those with negative results. Male partner data will be included in all analyses whenever appropriate. In analyses utilizing both male partners and females, analyses will control for the dependency within partner dyads.
5.2. Results disclosure: qualitative data
Case conferences and visits to KPNW sites for results disclosure with a genetic counselor are audio-recorded; a subset is observed by study staff with expertise in qualitative data collection. All observations are described using ethnographic fieldnotes or transcription. When result disclosure visits are observed, qualitative study staff conduct interviews with both participants and genetic counselors soon after the visit. The interviews of participants and genetic counselors explore similar topics about the experience of communicating/receiving the results (Table 2). Visit conversations between participants and genetic counselors are documented through detailed notes. On occasion, when the conversations are extensive, these interviews or segments of interviews may be transcribed verbatim.
Table 2.
Post genetic carrier screening qualitative interview questions.
| Patient debrief | Genetic counselor debrief |
|---|---|
| What did you learn? | What do you think you conveyed? |
| What were your expectations, concerns, or worries coming in to visit? | What were your concerns or worries about the visit? |
| What was surprising? | What was surprising? |
| What was hard to understand, or did you have questions about the results? | What was hard to convey? |
| What was the most helpful or useful information? | What did you perceive to be the most helpful or useful information? |
| What will your future actions or decisions be? | What will the patients’ future actions or decisions be? |
| Given what you know now, would you have done anything differently? | Would you have done anything differently if this visit was part of clinical practice (and not research)? |
| Is there anything you would prefer not to know? | Did any ethical issues arise from this case? |
| Do you have suggestions for improving the process? | Do you have suggestions for improving the process? |
Additionally, genetic counselors and other study staff were trained by the lead study ethnographer to write ethnographic field notes. They are prompted every other week to complete “journal entries” so that we can document their reflections about the RCT, interactions with participants, discussions and deliberations among staff, and suggestions for improvement. These notes are compiled and analyzed as part of our larger qualitative dataset.
We re-interview a subset of the couples one to two years after results disclosure. Female participants and their partners are interviewed separately with a semi-structured, in-depth interview guide that addresses the recall, sense, concerns, and utility of the results; genetic knowledge; additional findings; value of genome sequencing for carrier testing; and process improvement. All interviews are audio-recorded and transcribed.
Data gleaned from study staff’s journal entries are coded and summarized according to themes as they arise. Data relating to patients (or patient dyads, when appropriate) are summarized into a structured summary to facilitate case-based analysis.
5.3. Economic outcomes
We will compare the costs of genome sequencing and usual care and explore whether the healthcare services are used differently. Little is known about the cost of providing genome sequencing and particularly about the amount of time and other resources needed by providers to evaluate test results and prepare to communicate with patients about the results. Specifically, we will compare the cost of screening, the pattern of use of genetic counseling services and of mental health counseling services, total number of outpatient visits received, and total healthcare costs. We will create utilization profiles for each patient by using electronic medical records and other healthcare system administrative data. These data are generated at each patient contact with the healthcare system.
We expect participants in the GS arm will use more genetic counseling services compared to participants in the UC arm, because compared to UC, genome sequencing will provide more information and may identify unknown risk factors for study subjects. We will evaluate whether participants in the GS arm use more genetic services and mental health counseling compared with participants in the UC arm. We will also assess whether participants in the GS arm are more likely to access other types of outpatient care, such as primary care visits or other medical specialty visits related to their genome sequencing, compared with participants in the UC arm. Finally, participants in the GS arm may have greater use of medical care because they want to access services related to additional findings beyond carrier results.
The distribution of each utilization outcome, such as total outpatient visits, will be examined prior to analyses. These data are often characterized by a large number of zeros and skewed distribution, so we expect to use a two-part model [42]. The first stage in the model is typically a logistic regression to assess whether there is a greater probability of any use of the service category in one arm compared to the other. In the second stage, we will use ordinary least squares for costs and negative binomial models to examine whether the arms differ on the total number of services used in a service category. We will examine whether the arms differ by any use of each category of service use and by the number of visits in each category.
6. Discussion
The NextGen study examines patient interest and utility of genomic carrier screening in a healthy preconception population. This study is different than most others because genome or exome sequencing is often used with an obvious clinical phenotype (e.g., cancer, developmental disabilities, cardiac conditions) and not in a healthy population [11–15]. The results will allow us to explore how couples use genetic information as they prepare to have children. Also, the additional findings will allow us to see how physicians and patients use health information that is directly relevant to the patient’s own health.
The primary outcome of our trial is the number of results disclosed per participant. Several studies have reported on the average number of results disclosed per participant [23,43–46], but those have important differences, including being limited to medically actionable results, much smaller lists of genes that are reviewed for pathogenic variants, or limiting the population to people with specific disease phenotypes. Our study will help determine the average number of variants for recessive conditions in healthy individuals of reproductive age who have a near-term interest in having children; subsequently, we will learn how this information is received, interpreted, and acted upon through immediate post-result interviews, PRO survey questions, open-ended survey responses, downstream interviews of couples, and impact on healthcare utilization.
Key secondary outcomes of our trial include the comparative cost, utilization, and psychosocial impacts of genomic carrier sequencing and usual care. Because KPNW is an integrated healthcare system, we have the advantage of being able to look at downstream use of a wide variety of types of care related to both the direct effects of genomic carrier screening (e.g., genetic counseling visits) and the more tangential use of services (e.g., mental health services, primary care visits, specialty care visits).
Because of the rapidly changing field of genomics, we anticipate needing to make changes in the study over time. In particular, the genes with relevant findings to disclose for carrier status will change as new evidence for disease association is published. In addition, the ability of the electronic medical record software to handle genetic and genomic data is continuously evolving as new modules are created and implemented, so we may adapt our approaches to ordering and integrating genetic test results as technology advances. Finally, our reports are currently tailored to each participant; if the field of genetic testing moves toward a more standardized report format, further changes to the individual reports may be needed.
Our study is taking part in parallel with other CSER consortium projects, so we anticipate leveraging data from measures and outcomes common to KPNW and CSER, which will allow for comparison of outcomes across a broad range of populations and settings. Our collaboration with the CSER consortium will also allow us to begin creating best practices regarding genome and exome sequencing in clinical practice.
Our study has some limitations. First, the KPNW adult population is mainly white and has a higher socio-economic status than the overall US population. This mirrors research participants in general, which makes it challenging to know how genomic carrier sequencing would be accepted in a more diverse population. Second, the carrier frequency is low for most autosomal recessive conditions, so it is unlikely we will find more than a handful of couples that are carriers of the same condition. Third, interpretation of variants is challenging due to the incomplete knowledge base for variant classification, and interpretation is even more challenging in the absence of a clinical disease phenotype. We anticipate that variant classification will change over the course of the study. Part of our work will be to see how change of variant classification affects participants.
In summary, we designed an RCT to evaluate genomic clinical sequencing for women and their partners considering a future pregnancy in a real-world clinical setting. We developed and examined tools and procedures that could aid in standardized dissemination of clinically relevant genetic information to patients. These tools could be adopted into widespread clinical care which in turn could help to harness the potential benefits of genomic medicine. This project includes novel investigation into human reactions and responses from would-be parents learning information that could affect a future pregnancy. We are conducting this essential groundwork to help determine how future implementation of expanded use of genomic information can become a routine aspect of individual patients’ reproductive and medical decisions.
Acknowledgments
This work was supported by the National Human Genome Research Institute [grant number UM1HG007292; co-PIs: Wilfond, Goddard]; and the Clinical Sequencing Exploratory Research (CSER) consortium [grant numbers U01HG006507 (PI: Jarvik), U01HG007307 (Coordinating center)]. The sponsor had no role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the article for publication.
Abbreviations
- KPNW
Kaiser Permanente Northwest
- CSER
Clinical Sequencing Exploratory Research
- CF
cystic fibrosis
- OMIM
Online Mendelian Inheritance in Man
- GS
genome sequencing
- OHSU
Oregon Health & Science University
- PRO
Patient-reported outcome
Footnotes
Conflict of interest
None of the authors has any conflicts of interests to disclose.
References
- 1.American College of Medical Genetics and American College of Obstetricians and Gynecologists. Preconception and Prenatal Carrier Screening for Cystic Fibrosis: Clinical and Laboratory Guidelines. 2001 [Google Scholar]
- 2.American College of Obstetricians and Gynecologists. Update on carrier screening for cystic fibrosis, ACOG. 2011;486:1028–1031. doi: 10.1097/AOG.0b013e31821922c2. [DOI] [PubMed] [Google Scholar]
- 3.ACOG Committee on genetics. ACOG committee opinion. 318. Screening for Tay-Sachs Disease; Oct, 2005. 2005. [DOI] [PubMed] [Google Scholar]
- 4.The American Congress of Obstetricians and Gynecologists. Spinal Muscular Atrophy. 2009 [Google Scholar]
- 5.The American Congress of Obstetricians and Gynecologists. Carrier Screening for Fragile X Syndrome. 2010 [Google Scholar]
- 6.The American Congress of Obstetricians and Gynecologists. Preconception and Prenatal Carrier Screening for Genetic Diseases in Individuals of Eastern European Jewish Descent. 2009 doi: 10.1097/AOG.0b013e3181bd12f4. [DOI] [PubMed] [Google Scholar]
- 7.McKusick-Nathans Institute of Genetic Medicine Johns Hopkins University. Online Mendelian Inheritance in Man. OMIM; Baltimore, MD: 2016. [Google Scholar]
- 8.Arup Laboratories. Prenatal and Expanded Carrier Screening Panels. 2015 [Google Scholar]
- 9.Lazarin GA, Haque IS. Expanded carrier screening: a review of early implementation and literature. Semin Perinatol. 2016;40:29–34. doi: 10.1053/j.semperi.2015.11.005. [DOI] [PubMed] [Google Scholar]
- 10.Progenity, Pan-Ethnic Carrier Screening Panel. Ann Arbor, MI: Progenity.com; 2015. [Google Scholar]
- 11.Johnston JJ, Rubinstein WS, Facio FM, Ng D, Singh LN, Teer JK, et al. Secondary variants in individuals undergoing exome sequencing: screening of 572 individuals identifies high-penetrance mutations in cancer-susceptibility genes. Am J Hum Genet. 2012;91:97–108. doi: 10.1016/j.ajhg.2012.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang Y, Muzny DM, Xia F, Niu Z, Person R, Ding Y, et al. Molecular findings among patients referred for clinical whole-exome sequencing. JAMA. 2014;312:1870–1879. doi: 10.1001/jama.2014.14601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan CM, Moore EG, Leos C, Rini C. Patient hopes for diagnostic genomic sequencing: roles of uncertainty and social status. Eur J Hum Genet. 2015 doi: 10.1038/ejhg.2015.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McLaughlin HM, Ceyhan-Birsoy O, Christensen KD, Kohane IS, Krier J, Lane WJ, et al. A systematic approach to the reporting of medically relevant findings from whole genome sequencing. BMC Med Genet. 2014;15:134. doi: 10.1186/s12881-014-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gray SW, Park ER, Najita J, Martins Y, Traeger L, Bair E, et al. Oncologists’ and cancer patients’ views on whole-exome sequencing and incidental findings: results from the CanSeq study. Genet Med. 2016;18:1011–1019. doi: 10.1038/gim.2015.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.SeattleSeq Annotation. Input Variation List File for Annotation. 2016 [Google Scholar]
- 19.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Himes P, Kauffman TL, Muessig KR, Amendola L, Berg JS, Dorschner MO, et al. Genome sequencing and carrier testing: decisions on categorization and whether to disclose results of carrier testing. Genet Med. 2016 doi: 10.1038/gim.2016.198. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and guidelines for the interpretation of sequence variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17:405–424. doi: 10.1038/gim.2015.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gallego CJ, Bennette CS, Heagerty P, Comstock B, Horike-Pyne M, Hisama F, et al. Comparative effectiveness of next generation genomic sequencing for disease diagnosis: design of a randomized controlled trial in patients with colorectal cancer/polyposis syndromes. Control Clin Trials. 2014;39:1–8. doi: 10.1016/j.cct.2014.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dorschner MO, Amendola LM, Turner EH, Robertson PD, Shirts BH, Gallego CJ, et al. Actionable, pathogenic incidental findings in 1,000 participants’ exomes. Am J Hum Genet. 2013;93:631–640. doi: 10.1016/j.ajhg.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Patrick DL, Erickson P. Health status and health policy: quality of life in health care evaluation and resource allocation. Oxford, England: 1993. [Google Scholar]
- 25.Patient-Centered Outcomes Research Institute. Patient-centered Outcomes Research Working Definition. 2012 [Google Scholar]
- 26.World Health Organization. Development of the World Health Organization WHOQOL-BREF Quality of Life Assessment, 28, The WHOQOL Group. Psychological Medicine. 1998:551–558. doi: 10.1017/s0033291798006667. [DOI] [PubMed] [Google Scholar]
- 27.Skevington SM, Lotfy M, O’Connell KA. The World Health Organization’s WHOQOL-BREF quality of life assessment: psychometric properties and results of the international field trial. A report from the WHOQOL group. Qual Life Res Int J Qual Life Asp Treat Care Rehab. 2004;13:299–310. doi: 10.1023/B:QURE.0000018486.91360.00. [DOI] [PubMed] [Google Scholar]
- 28.Berlim MT, Pavanello DP, Caldieraro MA, Fleck MP. Reliability and validity of the WHOQOL BREF in a sample of Brazilian outpatients with major depression. Qual Life Res Int J Qual Life Asp Treat Care Rehab. 2005;14:561–564. doi: 10.1007/s11136-004-4694-y. [DOI] [PubMed] [Google Scholar]
- 29.Marteau TM, Bekker H. The development of a six-item short-form of the state scale of the Spielberger State-Trait Anxiety Inventory (STAI) Br J Clin Psychol/Br Psychol Soc. 1992;31(Pt 3):301–306. doi: 10.1111/j.2044-8260.1992.tb00997.x. [DOI] [PubMed] [Google Scholar]
- 30.Instruction Manual: Instructions for Patient Health Questionnaire (PHQ) and GAD-7 Measures. https://phqscreeners.pfizer.edrupalgardens.com/sites/g/files/g10016261/f/201412/instructions.pdf. 2015 Accessed on July 15, 2015.
- 31.Brehaut JC, O’Connor AM, Wood TJ, Hack TF, Siminoff L, Gordon E, et al. Validation of a decision regret scale. Med Decis Mak. 2003;23:281–292. doi: 10.1177/0272989X03256005. [DOI] [PubMed] [Google Scholar]
- 32.Holmes-Rovner M, Kroll J, Schmitt N, Rovner DR, Breer ML, Rothert ML, et al. Patient satisfaction with health care decisions: the satisfaction with decision scale. Med Decis Mak. 1996;16:58–64. doi: 10.1177/0272989X9601600114. [DOI] [PubMed] [Google Scholar]
- 33.Tercyak KP, Johnson SB, Roberts SF, Cruz AC. Psychological response to prenatal genetic counseling and amniocentesis. Patient Educ Couns. 2001;43:73–84. doi: 10.1016/s0738-3991(00)00146-4. [DOI] [PubMed] [Google Scholar]
- 34.DuBenske LL, Burke Beckjord E, Hawkins RP, Gustafson DH. Psychometric evaluation of the Health Information Orientation Scale: a brief measure for assessing health information engagement and apprehension. J Health Psychol. 2009;14:721–730. doi: 10.1177/1359105309338892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Degner LF, Sloan JA, Venkatesh P. The control preferences scale. Can J Nurs Res. 1997;29:21–43. [PubMed] [Google Scholar]
- 36.Fagerlin A, Zikmund-Fisher BJ, Ubel PA, Jankovic A, Derry HA, Smith DM. Measuring numeracy without a math test: development of the subjective numeracy scale. Med Decis Mak. 2007;27:672–680. doi: 10.1177/0272989X07304449. [DOI] [PubMed] [Google Scholar]
- 37.Fetzer Institute/National Institute on Aging Working Group. Multidimensional Measurement of Religiousness/Spirituality for Use in Health Research. John E Fetzer Institute; Kalamazoo, MI: 2003. [Google Scholar]
- 38.Fang CY, Dunkel-Schetter C, Tatsugawa ZH, Fox MA, Bass HN, Crandall BF, et al. Attitudes Toward Genetic Carrier Screening for Cystic Fibrosis among Pregnant Women: the Role of Health Beliefs and Avoidant Coping StyleWomen’s health (Hillsdale, NJ) 1997;3:31–51. [PubMed] [Google Scholar]
- 39.Schneider JL, Goddard KA, Davis J, Wilfond B, Kauffman TL, Reiss JA, et al. “Is it worth knowing?” Focus group participants’ perceived utility of genomic preconception carrier screening. J Genet Couns. 2016;25:135–145. doi: 10.1007/s10897-015-9851-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hardin J, Hilbe J. Generalized Estimating Equations Wiley Encyclopedia of Clinical Trials 1–8. Chapman and Hall/CRC; London: 2003. [Google Scholar]
- 41.Liang KY, Zeger SL. Longitudinal data analysis using generalized linear models. Biometrika. 1986;73:13–22. [Google Scholar]
- 42.Mullahy J. Econometric modeling of health care costs and expenditures: a survey of analytical issues and related policy considerations. Med Care. 2009;47:S104–S108. doi: 10.1097/MLR.0b013e31819c9593. [DOI] [PubMed] [Google Scholar]
- 43.Lazarin GA, Haque IS, Nazareth S, Iori K, Patterson AS, Jacobson JL, et al. An empirical estimate of carrier frequencies for 400+ causal Mendelian variants: results from an ethnically diverse clinical sample of 23,453 individuals. Genet Med. 2013;15:178–186. doi: 10.1038/gim.2012.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jang MA, Lee SH, Kim N, Ki CS. Frequency and spectrum of actionable pathogenic secondary findings in 196 Korean exomes. Genet Med. 2015;17:1007–1011. doi: 10.1038/gim.2015.26. [DOI] [PubMed] [Google Scholar]
- 45.Amendola LM, Dorschner MO, Robertson PD, Salama JS, Hart R, Shirts BH, et al. Actionable exomic incidental findings in 6503 participants: challenges of variant classification. Genome Res. 2015;25:305–315. doi: 10.1101/gr.183483.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gambin T, Jhangiani SN, Below JE, Campbell IM, Wiszniewski W, Muzny DM, et al. Secondary findings and carrier test frequencies in a large multiethnic sample. Genome Med. 2015;7:54. doi: 10.1186/s13073-015-0171-1. [DOI] [PMC free article] [PubMed] [Google Scholar]


