Abstract
In the era of applied meta-omics and personalized medicine, the oral microbiome is a valuable asset. From biomarker discovery to being a powerful source of therapeutic targets, and to presenting an opportunity for developing non-invasive approaches to health care, it has become clear that oral microbes may hold the answer for understanding disease, even beyond the oral cavity. Although our understanding of oral microbiome diversity has come a long way in the past 50 years, there are still many areas that need to be fine-tuned for better risk assessment and diagnosis, especially in early developmental stages of human life. Here, we discuss the factors that impact development of the oral microbiome, and explore oral markers of disease, with a focus on the early oral cavity. Our ultimate goal is to put different experimental and methodological views into perspective for better assessment of early oral and systemic disease at an early age, and discuss how oral microbiomes- at the community level, could provide improved assessment in individuals, and populations at risk.
Introduction
Leeuwenhoek’s microscope observations in his own dental plaque, over 3 centuries ago, suggested an unprecedented diversity of microorganisms (or “animalcules”) in the human oral cavity. Indeed, W.D Miller, in his seminal account of the microorganisms of the human mouth ‘Die Mikroorganismen der Mundhohle’ (1892)[1], suggests that a great number of bacterial morphotypes obtained in the mouth could make their classification impossible. Since then, microbiologists and microbial ecologists have come a long way - from targeting specific members of the “culturable” fraction of the oral cavity [2], to using cloning [3, 4], fingerprinting [5] and high throughput sequencing technologies, to reveal that this microecosystem may harbor over 800 to 1,000 different oral bacterial taxa (as sharing >98.5% 16S rRNA sequence identity)[3], with varying abundance and diversity patterns across age and health status. Consequently, this vast microbial diversity, which includes bacteria, archaea, viruses, phage and various microeukaryotes, makes it a challenge for clinicians and microbial ecologists to look for valid markers of oral health and disease, not to mention the hurdles associated with the significant variation found between and within (different oral cavity sites) subjects [6, 7]. Diversity surveys on healthy oral microbiomes do suggest a core group of phylotypes [8, 9], different from those found under diseased conditions[5, 10]. Further, an extensive body of literature has focused on reviewing and surveying the oral microbiome in health and disease [3, 11–14]. However, finding consistent bacterial markers, across studies and cohorts, is still a daunting task. Moreover, translating these findings into effective treatment of oral, and even systemic health is a significant challenge.
In this review we attempt to dissect relationships between oral bacteria and health by exploring the factors that shape the human oral microbiome, with a focus on the early oral cavity. Thus, we review studies on the oral microbiome of children, and the ecological and developmental aspects of this microecosystem that relate to oral and systemic health. To accomplish these goals, we 1) examine the factors impacting development and maturation of the oral microbiome since birth; 2) explore biomarkers influencing oral health and disease in children and 3) inspect how oral microbial markers affect health beyond the child’s oral cavity into adulthood. Finally, 4) we discuss the technical and analytical areas where the microbial ecology and clinical field should focus to translate biomarker discovery into preventive therapeutics in oral and systemic health in an individual's lifetime.
1. Development and maturation of the oral microbiome following birth
How we acquire our microbiome during pregnancy and after birth has been the subject of numerous research studies [15], the majority focusing on microbes populating the lower gastrointestinal (GI) tract, with far more limited information on the microorganisms populating of the oral cavity following birth. Interestingly, emerging evidence has pointed to a connection between the placental environment and the oral microbiome. For instance, 16S rRNA gene sequencing analysis has revealed the presence of several prominent oral commensals such as Streptococcus, Fusobacterium, Neisseria, Prevotella and Porphyromonas in the human and murine placenta[16–18]. Indeed, close similarities were found between the presumed placental microbiome and that in the mother oral cavity, compared to any other body site[18]. The hypotheses behind these findings propose that during pregnancy, oral commensals reach the amniotic fluid through blood (low grade bacteremia), a condition that may be exacerbated during periodontal disease and oral infection in mothers, with potential deleterious consequences for term delivery [19–22]. Moreover, an association has been found between mode of delivery and specific oral microbiome patterns in three to six month old infants, specifically as far as the abundances of Streptococcus, Fusobacterium and Slackia [23].
Nonetheless, the hypotheses of a placental microecosystem of hematogenic origin, and establishment of the newborn oral cavity influenced by mother-derived oral bacteria are controversial. The critiques mainly rely on a lack of evidence that bacterial DNA found in amniotic fluid is that of live bacteria, as one would expect that high throughput sequencing technologies would detect any bacterial DNA present regardless of cell viability, so the signal obtained may be merely cell debris. Additionally, there is the possibility of contamination of placenta with maternal blood [24]. Despite these observations, it is a fact that the both the placental and oral microecosystem are particularly unique in diversity compared to any other body site, including the vaginal microbiome [7], and that the microbial signal obtained in the amniotic fluid includes bacteria not usually found in the urogenital tract. Thus, the connection between maternal oral microbes and the seeding of the oral microbiome in newborns is potentially likely, but needs to be further tested using mechanistic, rather than association-type, models.
Assembly of microbes in the oral cavity. After initial seeding, assemblage of the human microbiome seems to be significantly influenced by environmental exposure. In this scenario, specific body habitats create a set of physical, chemical, and biological conditions that “filter” body microecosystems, to allow colonization by a very particular set of environmentally-derived bacteria [25]. In the case of the oral cavity, it is likely that a specific set of environmental stimuli and immune filtering, after initial seeding, shapes the oral environment into adulthood, potentially impacting oral health. In this regard, a combination of different mother microbiome sources including gut, vaginal, skin, as well as breast milk and early foods may shape the early oral microbiome [26, 27]. For instance, just as it happens with the gut microbiome in infants, the early oral cavity is also influenced by mode of delivery [15, 23, 28]. Specifically, vaginally delivered infants tend to show increased abundances of taxa such as Prevotella, Bacteroides and TM7, while C-section-delivered babies show increased colonization by Propionibacterium Staphylococcus, Slackia and Veillonella. The way these signals impact children oral health later in life, as it has been suggested with gut microbial communities, requires further investigation.
Right after birth, the newborn oral cavity seems to be rapidly dominated by Streptococcus (S. mutans, S. epidermidis and S. salivarius) and Fusobacterium [28–30]. Apparently, the bloom of Streptococcus spp. (particularly S. salivarius) is associated with the first oligosaccharide stimuli in the infant oral cavity [31]. Possibly, the metabolic products by Streptococcus on dietary oligosaccharides in breast milk or formula (fermentation) pave the way for other oral commensals to thrive. Furthermore, different species of pioneer Streptococcus spp. isolated from both breast- and formula-fed neonates have the metabolic capability to cleave immunoglobulin A1 (IgA1) [32], which would suggest Streptococci could bloom in an IgA1-rich environment, likely triggered by breast milk[33]. However, it is unclear whether potential differential abundances in diverse Streptococci species arise in response to formula or breast milk stimuli, or if these differences could impact oral health.
From the first months of life (3-4 months) into the first years and adulthood, the oral microbiome becomes significantly more diverse and undergoes very particular succession mechanisms [34, 35]. For instance, at the genera and species levels, E. coli, Staphylococcus and Pseudomonas, as well as lactic acid producing bacteria such as Lactobacillus crispatus, Lactobacillus gasseri and Streptococcus, all of which are associated with the gut, skin or breast milk [15, 36–38] are prevalent in the first months, even before tooth eruption. Conversely, Fusobacteria, Tenericutes, Synergistetes, TM7 and SR1 start dominating the oral microecosystem towards the first years of life, specifically, Veillonella, Fusobacterium, Neisseria, Prevotella, Rothia, Treponema and Streptococcus mutans, linked to more mature oral microbiomes and with potentially cariogenic microbiomes seem to emerge as infants transition into more mature stages, and likely more exposure to the external environment [34, 35, 39–42].
Nonetheless, increasing attention has been given to possible links between host genetic factors and the composition of the human microbiome [43–45], with some studies of this kind specifically targeting the oral cavity. For example, studies on healthy monozygotic and dizygotic infant twins using high-throughput sequencing and fingerprinting methods have shown that host genetic background (as opposed to the shared environment) have no significant effect in shaping salivary or plaque bacterial communities [46, 47]. These findings are in contrast to other twin cohort studies, in which checkerboard assays point to taxa in saliva and plaque that are potentially heritable and that may have role in the pathology of caries (e.g. Mutans streptococci) [48, 49]. Likewise, positive associations between some Major Histocompatibility Complex (HMC) class II alleles and salivary levels of culturable S. mutans and lactobacilli have been found in caries bearing adult women and young adults [50, 51]. Consequently, the extent to which the shared environment and the host genetic landscape influence colonization of the early oral cavity is still unclear and should be further investigated based on standardized deep sequencing techniques.
2. The oral microenvironment and biomarkers of oral disease in children
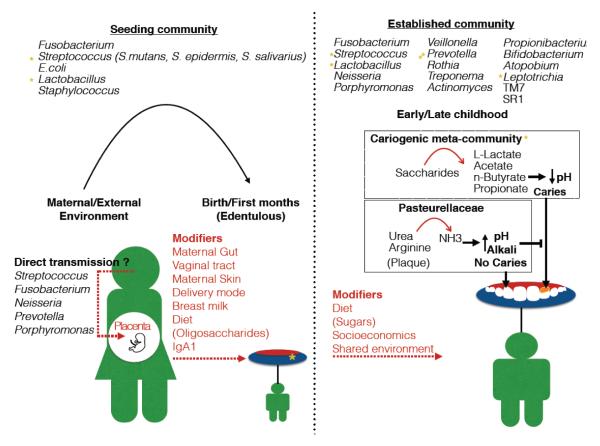
Thus, most evidence available shows that an early oral environment, strongly shaped by maternal sources rapidly transitions into a more complex, mature microecosystem influenced by the external environment. A critical question then is to identify, at an early age, the factors that trigger oral disease later in life. For instance, healthy children (from 3 months old) already harbor potentially cariogenic bacteria (S. mutans), which increase in abundance with age [35, 52]. Additionally, among 3-month old children, Increased presence of Lactobacilli, a taxon that tends to dominate carious lesions [41, 53] may be predictive of caries at 3 years [34]. These observations suggest that a combination of extrinsic and intrinsic factors determine oral disease phenotypes later in childhood. Figure 1 offers a schematic view of oral microbiome development at an early age, and subsequent maturation, including influence of oral disease markers.
Figure 1.
Oral microbiome development, maturation, and emergence of oral disease markers. The early oral microbiome is mainly shaped by maternal stimuli including a possible signal from the maternal oral cavity (via placenta), as well as her gut, skin and vaginal microbiomes. During the first months, diet further modifies the oral microecosystem, specifically as far as oligosaccharides present in formula and breast milk and Immunoglobulin A1 from the later. These maternal and dietary stimuli give rise to the seeding oral community of infants, which include potentially cariogenic bacteria (e.g. S. mutans). Later in childhood, diet and environment determine the mature oral microbiome, which increases significantly in diversity including colonization with potentially cariogenic taxa. This stage is also critical for the emergence of oral disease, with sugar consumption and fermentation by potentially cariogenic taxa (along with an acidic pH) being the main determinants. However, the establishment of oral caries may be reversed by ureases encoded by certain commensals in plaque (e.g. Pasteurellaceae).
Childhood oral diseases
Caries, the most common chronic childhood disease, is of dietary/bacterial origin, and it occurs as a result of cariogenic diets (sugar-related) and the metabolism of specific bacteria on dietary sugars in susceptible hosts [54, 55]. Thus, it is reasonable to expect that the first dietary stimuli have a strong influence on the etiology of caries at later developmental stages. For instance, meta-analyses have shown evidence that bottle fed children exhibit more early childhood caries (p<0.05), with additional supporting evidence to show that breastfeeding may prevent caries in early childhood[56]. Indeed, a suppressive effect on cariogenic S. mutans has been observed by Lactobacilli isolated only from the oral cavity of breast fed children but not from formula fed children [57], implying potential benefits of breast milk in the oral ecosystem. However, other studies have suggested that a possible positive effect of breastfeeding on the incidence of caries in children may be masked by other confounding socio-economical factors such as poverty, ethnicity and maternal prenatal smoking[58].
All in all, it seems that frequent sucrose consumption at early age (from 1.5 to 3 years old) is one of the leading causes influencing increased colonization with S. mutans, the main causing and predictive factor of oral caries [59–63]. During this process, particular community dynamics involving sucrose metabolism, pH homeostasis and biofilm formation, impact oral disease development [55, 64]. For instance, tooth decay and enamel demineralization seem to be promoted by a community shift in biofilm populations towards acidogenic (acid producing) and aciduric (acid tolerant) cariogenic bacteria, likely triggered by low pH after sucrose fermentation [65]. Additionally, sucrose seems to be a substrate for the production of extracellular and intracellular polysaccharides, two components that determine biofilm formation and structure [66, 67]. Indeed, children with nursing caries (12-48 month old with primary dentition), who are also under increased sucrose exposure, not only have higher incidences of S. mutans but also an altered biofilm structure characterized low Ca, Pi and F and higher levels of insoluble polysaccharides compared to caries free children [68, 69].
Thus, a constant influx of fermentable sugars into the oral cavity results in increased carbohydrate fermentation (lactic acid production), followed by longer periods of oral exposure to low pH (Stephan curve [70]) and the selection of bacteria able to thrive on these conditions (aciduric taxa), such as S. mutans, Lactobacilli and Bifidobacteria [71, 72]. These taxa constitute some of the main biomarkers in caries lesions biofilms. For instance, S. mutans can trigger pH conditions as low as 3.0 after sugar stimuli through fermentation, a scenario that has been linked to enamel demineralization [73, 74]. In contrast, other species within the Streptococcus genus, such as S. salivarius and S. mitis are associated with an increase of pH through alkali generating pathways, and therefore are linked to a protective effect against caries[64, 65].
However, in epidemiological experiments or animal models of oral disease, it is not immediately clear whether the bloom in the aforementioned cariogenic taxa occurs in response to increased sugar availability, or if low pH after sugar metabolism creates an acidogenic environment that displaces non-aciduric, health-associated taxa [75]. In this regard, controlled culture systems of defined mixed inoculum have shown that constant glucose influx, while maintaining neutral pH, keeps the bacterial community in balance, and the proportions of potentially cariogenic taxa at low levels (e.g. S. mutans and L. rhamnosus) [76]. In contrast, allowing the pH to drop, proportional to glucose fermentation, results in significant blooms of caries-associated (aciduric) taxa, at the expense of health-associated bacteria [77]. Thus, low pH due to sugar catabolism, rather than substrate availability alone, is a determining factor in promoting the rise of potentially cariogenic taxa in the oral cavity [75].
These observations denote the high complexity of caries as far as the physicochemical triggers and taxa involved, and suggest the adoption of an ecology-wise approach to understand oral disease [71, 72]. For example, despite the traditional central role of S. mutans in childhood caries, oral bacteria belonging to other genera such as Granulicatella, Actinomyces, Actinobaculum, Scardovia, Atopobium, Aggregatibacter, Slackia, Bifidobacteria and Prevotella have also been associated with early childhood caries and severe early childhood caries (S-ECC, an extremely destructive form of early-childhood caries involving multiple teeth) [10, 34, 78, 79]. Interestingly, however, the role of Lactobacilli on childhood caries seems ambiguous. On the one hand there is the potential suppressive effect of Lactobacilli isolated from the oral cavity of breast-milk fed 3-month old infants on cariogenic Streptococci [57], but also the predicted high incidence of caries at three years of age in three month old infants with higher proportions of Lactobacilli [34]. Furthermore, Lactobacillus is dominant in deep lesions on permanent dentition of children with S-ECC, suggesting an even more prominent potential role than S. mutans [80].
Other conflicting results have shown similar bacterial genera in caries-positive (n=10; 19.1±3.5 months) and caries-free (n=9; 19.3±3.2 months) children previous to eruption of secondary primary molars, with a few major genera making up more than 60% of the total microbiota in both groups [81]. Furthermore, although the group of children positive for caries showed higher prevalence of Streptococcus, Neisseria and Veillonella, compared to higher levels of Leptotrichia, Actinomyces, Prevotella and Porphyromonas in the caries-free children, these differences were not statistically significant [81]. Additional inconsistent results suggest windows of infectivity, strong interpersonal differences, and many different bacteria as being responsible for disease onset [82]. These apparent incongruences may correspond to known differences in microbial diversity coverage provided by culture-based and molecular methods, and by diverse molecular methods (e.g., cloning, fingerprinting and high-throughput sequencing). Thus, it is necessary to improve and standardize microbial community profiling in early oral disease by refining diversity at deeper taxonomic levels (e.g., species and strains). Additionally, adopting a system-level approach to profile microbemicrobe and microbe-metabolite interactions in the early oral ecosystem should improve our understanding of microbial function in disease, beyond taxonomic characterizations [65, 83].
As far as meta-community dynamics, we are starting to realize that it is a dysbiotic oral ecosystem (microbial imbalance), rather than the detection of a few bugs, what characterizes and triggers oral disease. For instance, early studies using culture based and fingerprinting approaches (Denaturing Gradient Gel Electrophoresis (DGGE)) demonstrated a higher level of microbial diversity in the dental plaque of healthy children (2-8 years old) over those with severe dental caries[84]. These results contrast with the microarray detection of higher bacterial diversity in saliva of caries-affected 6 to 8 year olds with mixed dentition compared to healthy controls[85]. Likewise, although S. mutans has been the central subject of microbe connections to caries, cloning approaches to analyze the microbiome of children (primary and permanent teeth) and young adults have suggested that other taxa such as Atopobium, Propionibacterium and Lactobacillus are more abundant in S. mutans - caries-bearing subjects, while other Lactobacillus spp., Bifidobacterium and non-mutans streptococci were predominant on affected subjects not harboring S. mutans [82]. Altogether, these observations highlight the non-specific source, polymicrobial nature, and complex metabolic and community dynamics of oral disease [71, 86].
In this regard, analyses of metabolic homeostasis and community ecology (454-pyrosequencing) in the early oral environment (3-6 year olds) have revealed that urease activity and alkali production is significantly associated with the bacterial composition of dental plaque [87]. It seems that urease-alkali activity by certain commensals (e.g. Pasteurellaceae family) positively impacts the oral acid-base homeostasis to inhibit aciduric and caries-associated taxa (e.g. Leptotrichia and S. mutans) in plaque under low sugar consumption[88]. However, the opposite scenario characterizes saliva, in which increased urease activity is associated with high caries incidence and levels of S. mutans [89]. This scenario poses a very complex framework to understand community and metabolic interactions in the early oral cavity and to implement valid biomarkers of caries risk in children in relation to alkali balance. A useful approach would be to characterize microbial communities using shotgun metagenomic sequencing in addition to enzymatic activity profiling. However, up to date, no studies exist on the metagenome of the early oral cavity.
As far as whole-metabolome analyses in the early oral cavity, studies have shown that the saliva of caries-bearing children with primary, mixed, and permanent dentition exhibit different metabolomic profiles (Nuclear magnetic resonance) and increased levels of lactate, n-butyrate, and acetate compared to healthy matches[90]. Furthermore, the salivary metabolome of caries-affected children after a 3 month treatment with composite resin exhibits significant changes that correlate with a reduction in propionate, acetate, n-butyrate, and saccharides along with decreased levels of cultured S. mutans and Lactobacillus sp. [91]. These observations correlate with the proposed links between bacterial metabolism of sugars (glycolysis), fermentation, and an increased abundance of acidogenic taxa (e.g. Propionibacterium and Lactobacillus) in early oral disease [71, 92, 93]. Other saliva metabolome analyses (Gas Chromatography Mass Spectrometry [GCMS]) on children with tooth decay highlighted a prominent role of metabolites involved in the arginine and proline metabolic pathway, and connected arginine to alkali production and hence, acid-base balance [64, 94].
Indeed, a metaproteomic analysis of oral biofilm in caries-bearing and healthy adults identified proteins involved in the L-lactate dehydrogenase and arginine deiminase systems. These systems aid in pH buffering and show the highest expression in the supragingival dental plaque of healthy individuals[64, 95]. Interestingly, in these studies, overexpression of the L-lactate dehydrogenase and arginine deiminase systems correlated negatively with high abundance of sugar transporters (ATP-binding cassettes and phosphotransferase systems) and other sugar degrading proteins from Actinomyces, Corynebacterium, Rothia and Streptococcus. Thus, the combined power of meta-omics and high throughput approaches to community ecology could provide a clearer, system-like picture of early oral disease and potential marker discovery in early diagnostics. These approaches are also key to determine how early oral disease provides clues to understand systemic health from childhood into adulthood.
Finally, meta-OMIC integration analysis and system-level approaches should also take into account biogeographical aspects of early oral disease. For example, in adults, there is a distinct microbial composition between different healthy tooth surfaces and different tooth types in a single individual [96, 97] and between carious lesions in dentine and enamel [98]. In children, healthy permanent and deciduous teeth also harbor different supragingival microbial profiles [99]. Furthermore, analyses of tooth decay in children have shown that caries in a given tooth not only impacts the microbial ecology and health onsite but also on nearest teeth [78, 80, 100]. Likewise, studies on temporal and spatial microbiome variation in preschool children show that both plaque and saliva acquire distinct microbiome configurations with age and transitions to cariogenic stages [101]. These observations demonstrate that targeting single sites in the early oral cavity to assess oral disease may only offer a partial view of disease biomarkers.
3. Bacterial markers of early oral health and disease and implications for systemic health
The associations between periodontal and systemic disease have been the subject of numerous studies including oral marker links to cardiovascular, respiratory, immune, metabolic, osteopathic and obstetric complications [102–106]. The proposed mechanisms of these oral-systemic links include the spread of infection from the oral cavity in the form of bacteremia or circulating bacterial toxins, triggering increased circulating pro-inflammatory cytokines and a weakened immune system, as well as cross-reactivity (molecular mimicry) between bacterial and self-antigens [104, 107]. Although associations between oral disease markers and systemic health in children are scarce, some studies have provided important insights.
For instance children with juvenile idiopathic arthritis (13 years old in average), who typically resemble adults with rheumatoid arthritis, have increased antibody response to Porphyromonas gingivalis, one of the main taxon involved in periodontal disease [108, 109], and higher incidence of symptoms related to periodontal disease [110]. Likewise, children with celiac disease (1.4 years old in average) under gluten free diets show a less diverse salivary microbiome compared to healthy controls, and increased abundance of oral caries-related taxa (e.g. Rothia, Porphyromonas, Gemellaceae, Prevotella, Streptococcus and Lachnospiraceae) [111]. The abundances of these taxa also correlated with higher levels of organic volatile compounds in the saliva and fecal samples of affected children detected through GC-MS [112]. A similar scenario takes place in the tongue microbiome of children with Crohn's disease, who exhibit reduced diversity and decreased abundance of Fusobacteria and Firmicutes compared with healthy controls [113]. However, it is unclear whether these apparent dysbiosis leads to more susceptibility to oral disease (e.g. caries) in Crohn’s patients. Therefore, these findings, which mirror what takes place in the intestinal mucosa during Crohn's, have more important implications for developing diagnostic tools of systemic disease based on oral biomarkers.
As such, the assessment of oral risk factors in children could provide important information on current and future systemic health status, however, further understanding of directionality in these mechanisms, and caution in diagnostics are necessary. For instance, it is likely that a generally weakened immune system compromises oral health, as it is the case of HIV-infected children [114]. Likewise, bacteremia can be the result of periodontal trauma in children (teeth extraction) [115], with potential detrimental consequences for cardiovascular disease (platelet aggregation induced by oral commensals) and endocarditis in adulthood [102, 116]. Thus, although multiple avenues are open to use oral disease biomarkers in the diagnosis of chronic disorders such as cancer and liver disease [117–119], several technical, methodological, experimental and therapeutic challenges remain.
4.Challenges: Techniques, analyses, biomarker discovery and translational preventive therapeutics in oral and systemic health
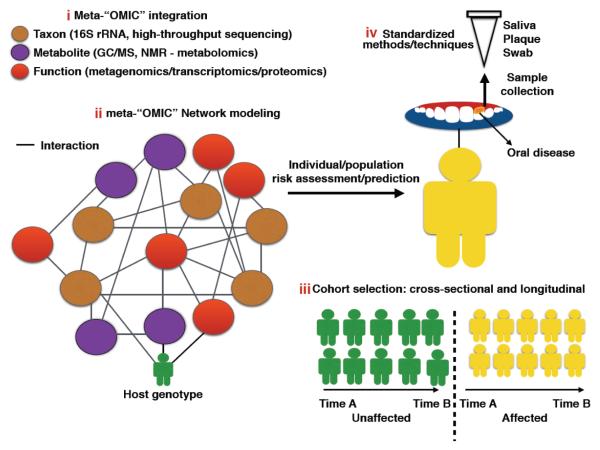
Even though the application of molecular-based techniques in the last 20 years has allowed us to have a better understanding of the complex oral microecosystem, we are just beginning to unravel the role that multiple intrinsic and extrinsic layers play in oral disease. This awareness requires knowledge of a multi-system approach that integrates different techniques to characterize the host-microbe entity [120], and analyses that extend into an individual’s or population’s environment (socioeconomics) - a challenge that is most relevant in childhood health [83]. We propose that such framework should build on four main stakes to guarantee more effective risk assessment, bring precision at the individual level and minimize population-level health disparities [121] (Figure 2).
Figure 2.
Biomarker discovery and translational preventive therapeutics in oral and systemic health. Effective risk assessment of oral disease and diagnostics precision at the individual level and specific populations at risk may be accomplished by implementing a framework that builds on four main stakes: i) High resolution tools to study the oral microbiome at the compositional and functional levels (“omic” techniques that simultaneously unravel community composition, potential and encoded functions); ii) Effective meta-omic integration modeling to extrapolate/translate the data to ecosystem (host) phenotype (Computational and statistical models to integrate various “omic” datasets and algorithms that allow risk prediction based on meta-omic integration networks); iii) Studies that not only include cross-sectional but also longitudinal data in larger cohorts (Valid biomarkers of oral disease in real time (pre and post disease) and at a global scale); and iv) Standardization of methods (Sample storage methods, standardization of phylogenetic markers and sequencing techniques, building of comprehensive oral microbe databases).
i) High resolution tools to study the oral microbiome at the compositional and functional levels
As characterizations of oral microbiomes move from strictly culture-based methods to using the Human Oral Microbiome Identification Microarray (HOMIM)[122] and, lately, whole community profiling using high throughput 16S rRNA sequencing and shotgun metagenomics, the concepts of the “ecological plaque hypothesis”[71] and a super complex oral ecosystem have become more relevant. This complexity, however, has also pointed out that understanding microecosystem functioning requires a multi level view that includes taxonomic assessment (composition), potential (metagenome -transcriptome), and encoded functions (proteome and metabolome) [123, 124]. This view is essential to shift current disease models from focusing on pathobiomes rather than on pathogens [125]. Thus, a complete understanding of early oral disease may not be accomplished by applying one of these techniques, but by combining two or more, an approach that is seldom implemented in early oral community ecology assessment (but see [126]).
ii) Effective meta-omic integration to extrapolate/translate the data to ecosystem (host) phenotype
Likewise, the use of one or more “omic” techniques requires effective models to integrate these datasets and predict host phenotype and potential disease risks. This view takes into account that system components (modules, layers) cannot be considered from isolated perspectives[127]. In this regard, Meta-OMIC modeling and network-view approaches are just beginning to emerge to characterize oral microbiome dynamics and predict health and disease states [64, 126, 128]. However, there is a clear necessity for more applications of such models into risk assessment in the early oral cavity. Moreover, given the difficulty of implementing mechanistic views of disease onset in the early oral cavity, meta-OMIC network modeling becomes an attractive tool to understand ecosystem functioning and potential causal factors [129]. In this regard, efforts in assessing disease biomarkers in the early oral cavity are already exploiting machine learning models for disease risk prediction [101]. However, these efforts have been so far limited to microbial community composition analyses.
iii) Studies that not only include cross-sectional but also longitudinal data in larger cohorts
Most of the molecular and meta-OMIC based reports on the oral microbiome in health and disease have traditionally relied on small cohorts. This scenario may improve as sequencing and other high-throughput techniques decrease in cost, which will result in improved resolution power. Likewise, efficient risk assessment for individuals and specific vulnerable populations may requires longitudinal approaches [83], which is key to identify valid biomarkers of disease in real time (pre and post disease) [130, 131]. In this regard, the use of larger cohorts, in combination with longitudinal sampling, has revealed the corresponding weight shared environment and genetics have on the early oral microbiome[46]. Furthermore, despite the realization that there may be little geographic structure in global diversity patterns in the human salivary microbiome [4], global surveys of diverse human populations at a larger scale may provide important clues to better understand the influence of cultural and genetic factors in impacting health and disease [132].
iv) Need for standardization of technical methods: An issue that spans all areas of microbiome research in general
The necessity to find reproducible results, and the ability to compare different datasets are huge challenges that the microbiome field faces today [133]. These concerns have increased efforts to evaluate better sample storage methods [134–137], to select appropriate phylogenetic markers and techniques and effectively assess microbial community composition[138] and build comprehensive databases for the vast number of uncharacterized and uncultured oral organisms (e.g. The Human Oral Microbiome Database (HOMD) [139] and the Core Oral Microbiome Database (CORE) [140]). As all microbiome fields (and researchers) make significant progress towards standardization of methods, the chances for better risk assessment and prediction of oral disease in children will improve significantly.
Conclusion: The future of microbiome studies for oral health and childhood diseases
As the challenges mentioned above are surpassed, microbiome research on early oral health and disease should move beyond profiling oral community patterns in cross-sectional and longitudinal cohorts. For instance, as we standardize techniques to find valid oral disease biomarkers, efforts should be focused on using biomarker information to apply effective therapeutics in specific individuals and populations at most risk. Specially, the concept of precision medicine has been proposed to minimize health disparities in the most vulnerable populations, taking into account subject-specific variability in genes and environment [83]. Along these lines, we are already making efforts to identify oral microbial markers at increased taxonomic depth and diversity coverage, a particularly relevant issue in oral disease. For instance, the improved Human Oral Microbiome using next generation sequencing (HOMINGS), from the Forsyth institute (http://forsyth.org) [122], harbors a well-curated collection of sequences from oral bacteria at the species level, allowing researchers to accurately expand marker identification in the highly diverse oral ecosystem. Likewise, more rigorous studies on infant twin cohorts and using genome-wide analyses, host gene expression levels and deep sequencing in the oral cavity could provide clues on the extent to which oral disease in children is explained by heritable or environmentally-acquired taxa. Finally, researchers involved in oral microbiome research should visualize the value of oral samples beyond the oral cavity - a key concept in the era of applied meta-omics and personalized medicine.
References
- 1.Miller WD. Die Mikroorganismen der Mundhöhle. DMW-Deutsche Medizinische Wochenschrift. 1892;18:1016–1018. [Google Scholar]
- 2.Kligler IJ. A Biochemical Study and Differentiation of Oral Bacteria: With Special Reference to Dental Caries. Columbia University; 1915. [Google Scholar]
- 3.Dewhirst FE, Chen T, Izard J, et al. The human oral microbiome. J Bacteriol. 2010;192:5002–5017. doi: 10.1128/JB.00542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nasidze I, Li J, Quinque D, et al. Global diversity in the human salivary microbiome. Genome Res. 2009;19:636–643. doi: 10.1101/gr.084616.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ling Z, Kong J, Jia P, et al. Analysis of oral microbiota in children with dental caries by PCR-DGGE and barcoded pyrosequencing. Microb Ecol. 2010;60:677–690. doi: 10.1007/s00248-010-9712-8. [DOI] [PubMed] [Google Scholar]
- 6.Bik EM, Long CD, Armitage GC, et al. Bacterial diversity in the oral cavity of 10 healthy individuals. ISME J. 2010;4:962–974. doi: 10.1038/ismej.2010.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Human Microbiome Project Consortium Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zaura E, Keijser BJF, Huse SM, Crielaard W. Defining the healthy “core microbiome” of oral microbial communities. BMC Microbiol. 2009;9:1–12. doi: 10.1186/1471-2180-9-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keijser BJF, Zaura E, Huse SM, et al. Pyrosequencing analysis of the oral microflora of healthy adults. J Dent Res. 2008;87:1016–1020. doi: 10.1177/154405910808701104. [DOI] [PubMed] [Google Scholar]
- 10.Becker MR, Paster BJ, Leys EJ, et al. Molecular analysis of bacterial species associated with childhood caries. J Clin Microbiol. 2002;40:1001–1009. doi: 10.1128/JCM.40.3.1001-1009.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zarco MF, Vess TJ, Ginsburg GS. The oral microbiome in health and disease and the potential impact on personalized dental medicine. Oral Dis. 2012;18:109–120. doi: 10.1111/j.1601-0825.2011.01851.x. [DOI] [PubMed] [Google Scholar]
- 12.Wade WG. The oral microbiome in health and disease. Pharmacol Res. 2013;69:137–143. doi: 10.1016/j.phrs.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 13.Costalonga M, Herzberg MC. The oral microbiome and the immunobiology of periodontal disease and caries. Immunol Lett. 2014;162:22–38. doi: 10.1016/j.imlet.2014.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Paster BJ, Olsen I, Aas JA, Dewhirst FE. The breadth of bacterial diversity in the human periodontal pocket and other oral sites. Periodontol 2000. 2006;42:80–87. doi: 10.1111/j.1600-0757.2006.00174.x. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez-Bello MG, Costello EK, Contreras M, et al. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc Natl Acad Sci U S A. 2010;107:11971–11975. doi: 10.1073/pnas.1002601107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bearfield C, Davenport ES, Sivapathasundaram V, Allaker RP. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG. 2002;109:527–533. doi: 10.1111/j.1471-0528.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- 17.Fardini Y, Chung P, Dumm R, et al. Transmission of diverse oral bacteria to murine placenta: evidence for the oral microbiome as a potential source of intrauterine infection. Infect Immun. 2010;78:1789–1796. doi: 10.1128/IAI.01395-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aagaard K, Ma J, Antony KM, et al. The placenta harbors a unique microbiome. Sci Transl Med. 2014;6:237ra65. doi: 10.1126/scitranslmed.3008599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldenberg RL, Hauth JC, Andrews WW. Intrauterine Infection and Preterm Delivery. N Engl J Med. 2000;342:1500–1507. doi: 10.1056/NEJM200005183422007. [DOI] [PubMed] [Google Scholar]
- 20.Han YW, Redline RW, Li M, et al. Fusobacterium nucleatum induces premature and term stillbirths in pregnant mice: implication of oral bacteria in preterm birth. Infect Immun. 2004;72:2272–2279. doi: 10.1128/IAI.72.4.2272-2279.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han YW, Ikegami A, Bissada NF, et al. Transmission of an uncultivated Bergeyella strain from the oral cavity to amniotic fluid in a case of preterm birth. J Clin Microbiol. 2006;44:1475–1483. doi: 10.1128/JCM.44.4.1475-1483.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Han YW, Shen T, Chung P, et al. Uncultivated bacteria as etiologic agents of intra-amniotic inflammation leading to preterm birth. J Clin Microbiol. 2009;47:38–47. doi: 10.1128/JCM.01206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lif Holgerson P, Harnevik L, Hernell O, et al. Mode of birth delivery affects oral microbiota in infants. J Dent Res. 2011;90:1183–1188. doi: 10.1177/0022034511418973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kliman HJ. Comment on “The placenta harbors a unique microbiome”. Sci Transl Med. 2014;6:254le4–254le4. doi: 10.1126/scitranslmed.3009864. [DOI] [PubMed] [Google Scholar]
- 25.Costello EK, Stagaman K, Dethlefsen L, et al. The application of ecological theory toward an understanding of the human microbiome. Science. 2012;336:1255–1262. doi: 10.1126/science.1224203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Isolauri E. Development of healthy gut microbiota early in life. J Paediatr Child Health. 2012;48(Suppl 3):1–6. doi: 10.1111/j.1440-1754.2012.02489.x. [DOI] [PubMed] [Google Scholar]
- 27.Munyaka PM, Khafipour E, Ghia J-E. External influence of early childhood establishment of gut microbiota and subsequent health implications. Front Pediatr. 2014;2:109. doi: 10.3389/fped.2014.00109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hegde S, Munshi AK. Influence of the maternal vaginal microbiota on the oral microbiota of the newborn. J Clin Pediatr Dent. 1998;22:317–321. [PubMed] [Google Scholar]
- 29.Merglova V, Polenik P. Early colonization of the oral cavity in 6- and 12-month-old infants by cariogenic and periodontal pathogens: a case-control study. Folia Microbiol. 2016 doi: 10.1007/s12223-016-0453-z. [DOI] [PubMed] [Google Scholar]
- 30.Wan AK, Seow WK, Purdie DM, et al. Oral colonization of Streptococcus mutans in six-month-old predentate infants. J Dent Res. 2001;80:2060–2065. doi: 10.1177/00220345010800120701. [DOI] [PubMed] [Google Scholar]
- 31.Carlsson J, Grahnén H, Jonsson G, Wikner S. Early Establishment of Streptococcus salivarius in the Mouths of Infants. J Dent Res. 1970;49:415–418. doi: 10.1177/00220345700490023601. [DOI] [PubMed] [Google Scholar]
- 32.Cole MF, Evans M, Fitzsimmons S, et al. Pioneer oral streptococci produce immunoglobulin A1 protease. Infect Immun. 1994;62:2165–2168. doi: 10.1128/iai.62.6.2165-2168.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hanson LA, Söderström T. Human milk: Defense against infection. Prog Clin Biol Res. 1981;61:147–159. [PubMed] [Google Scholar]
- 34.Lif Holgerson P, Öhman C, Rönnlund A, Johansson I. Maturation of Oral Microbiota in Children with or without Dental Caries. PLoS One. 2015;10:e0128534. doi: 10.1371/journal.pone.0128534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cephas KD, Kim J, Mathai RA, et al. Comparative analysis of salivary bacterial microbiome diversity in edentulous infants and their mothers or primary care givers using pyrosequencing. PLoS One. 2011;6:e23503. doi: 10.1371/journal.pone.0023503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward TL, Hosid S, Ioshikhes I, Altosaar I. Human milk metagenome: a functional capacity analysis. BMC Microbiol. 2013;13:116. doi: 10.1186/1471-2180-13-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martín R, Langa S, Reviriego C, et al. Human milk is a source of lactic acid bacteria for the infant gut. J Pediatr. 2003;143:754–758. doi: 10.1016/j.jpeds.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 38.Jost T, Lacroix C, Braegger C, Chassard C. Assessment of bacterial diversity in breast milk using culture-dependent and culture-independent approaches. Br J Nutr. 2013;110:1253–1262. doi: 10.1017/S0007114513000597. [DOI] [PubMed] [Google Scholar]
- 39.Yang F, Zeng X, Ning K, et al. Saliva microbiomes distinguish caries-active from healthy human populations. ISME J. 2012;6:1–10. doi: 10.1038/ismej.2011.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Struzycka I. The oral microbiome in dental caries. Pol J Microbiol. 2014;63:127–135. [PubMed] [Google Scholar]
- 41.Rôças IN, Alves FRF, Rachid CTCC, et al. Microbiome of Deep Dentinal Caries Lesions in Teeth with Symptomatic Irreversible Pulpitis. PLoS One. 2016;11:e0154653. doi: 10.1371/journal.pone.0154653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Crielaard W, Zaura E, Schuller AA, et al. Exploring the oral microbiota of children at various developmental stages of their dentition in the relation to their oral health. BMC Med Genomics. 2011;4:22. doi: 10.1186/1755-8794-4-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goodrich JK, Waters JL, Poole AC, et al. Human genetics shape the gut microbiome. Cell. 2014;159:789–799. doi: 10.1016/j.cell.2014.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodrich JK, Davenport ER, Beaumont M, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host Microbe. 2016;19:731–743. doi: 10.1016/j.chom.2016.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Blekhman R, Goodrich JK, Huang K, et al. Host genetic variation impacts microbiome composition across human body sites. Genome Biol. 2015;16:191. doi: 10.1186/s13059-015-0759-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stahringer SS, Clemente JC, Corley RP, et al. Nurture trumps nature in a longitudinal survey of salivary bacterial communities in twins from early adolescence to early adulthood. Genome Res. 2012;22:2146–2152. doi: 10.1101/gr.140608.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Papapostolou A, Kroffke B, Tatakis DN, et al. Contribution of host genotype to the composition of health-associated supragingival and subgingival microbiomes. J Clin Periodontol. 2011;38:517–524. doi: 10.1111/j.1600-051X.2011.01718.x. [DOI] [PubMed] [Google Scholar]
- 48.Corby PM, Bretz WA, Hart TC, et al. Heritability of oral microbial species in caries-active and caries-free twins. Twin Res Hum Genet. 2007;10:821–828. doi: 10.1375/twin.10.6.821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corby PMA, Bretz WA, Hart TC, et al. Mutans streptococci in preschool twins. Arch Oral Biol. 2005;50:347–351. doi: 10.1016/j.archoralbio.2004.08.010. [DOI] [PubMed] [Google Scholar]
- 50.Acton RT, Dasanayake AP, Harrison RA, et al. Associations of MHC genes with levels of caries-inducing organisms and caries severity in African-American women. Hum Immunol. 1999;60:984–989. doi: 10.1016/s0198-8859(99)00088-9. [DOI] [PubMed] [Google Scholar]
- 51.Ozawa Y, Chiba J, Sakamoto S. HLA class II alleles and salivary numbers of mutans streptococci and lactobacilli among young adults in Japan. Oral Microbiol Immunol. 2001;16:353–357. doi: 10.1034/j.1399-302x.2001.160606.x. [DOI] [PubMed] [Google Scholar]
- 52.Gizani S, Papaioannou W, Haffajee AD, et al. Distribution of selected cariogenic bacteria in five different intra-oral habitats in young children. Int J Paediatr Dent. 2009;19:193–200. doi: 10.1111/j.1365-263X.2008.00956.x. [DOI] [PubMed] [Google Scholar]
- 53.Kianoush N, Nguyen K-AT, Browne GV, et al. pH gradient and distribution of streptococci, lactobacilli, prevotellae, and fusobacteria in carious dentine. Clin Oral Investig. 2014;18:659–669. doi: 10.1007/s00784-013-1009-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Takahashi N, Nyvad B. The role of bacteria in the caries process: ecological perspectives. J Dent Res. 2011;90:294–303. doi: 10.1177/0022034510379602. [DOI] [PubMed] [Google Scholar]
- 55.Burne RA, Marquis RE. Alkali production by oral bacteria and protection against dental caries. FEMS Microbiol Lett. 2000;193:1–6. doi: 10.1111/j.1574-6968.2000.tb09393.x. [DOI] [PubMed] [Google Scholar]
- 56.Avila WM, Pordeus IA, Paiva SM, Martins CC. Breast and Bottle Feeding as Risk Factors for Dental Caries: A Systematic Review and Meta-Analysis. PLoS One. 2015;10:e0142922. doi: 10.1371/journal.pone.0142922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Holgerson PL, Vestman NR, Claesson R, et al. Oral microbial profile discriminates breast-fed from formula-fed infants. J Pediatr Gastroenterol Nutr. 2013;56:127–136. doi: 10.1097/MPG.0b013e31826f2bc6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iida H, Auinger P, Billings RJ, Weitzman M. Association between infant breastfeeding and early childhood caries in the United States. Pediatrics. 2007;120:e944–52. doi: 10.1542/peds.2006-0124. [DOI] [PubMed] [Google Scholar]
- 59.Alaluusua S, Mättö J, Grönroos L, et al. Oral colonization by more than one clonal type of mutans streptococcus in children with nursing-bottle dental caries. Arch Oral Biol. 1996;41:167–173. doi: 10.1016/0003-9969(95)00111-5. [DOI] [PubMed] [Google Scholar]
- 60.Burt BA, Eklund SA, Morgan KJ, et al. The effects of sugars intake and frequency of ingestion on dental caries increment in a three-year longitudinal study. J Dent Res. 1988;67:1422–1429. doi: 10.1177/00220345880670111201. [DOI] [PubMed] [Google Scholar]
- 61.Naidu R, Nunn J, Kelly A. Socio-behavioural factors and early childhood caries: a cross-sectional study of preschool children in central Trinidad. BMC Oral Health. 2013;13:30. doi: 10.1186/1472-6831-13-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kanasi E, Dewhirst FE, Chalmers NI, et al. Clonal analysis of the microbiota of severe early childhood caries. Caries Res. 2010;44:485–497. doi: 10.1159/000320158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Harris R, Nicoll AD, Adair PM, Pine CM. Risk factors for dental caries in young children: a systematic review of the literature. Community Dent Health. 2004;21:71–85. [PubMed] [Google Scholar]
- 64.Edlund A, Yang Y, Yooseph S, et al. Meta-omics uncover temporal regulation of pathways across oral microbiome genera during in vitro sugar metabolism. ISME J. 2015;9:2605–2619. doi: 10.1038/ismej.2015.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.McLean JS. Advancements toward a systems level understanding of the human oral microbiome. Front Cell Infect Microbiol. 2014;4:98. doi: 10.3389/fcimb.2014.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Leme AFP, Koo H, Bellato CM, et al. The Role of Sucrose in Cariogenic Dental Biofilm Formation—New Insight. J Dent Res. 2006;85:878–887. doi: 10.1177/154405910608501002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Krzyściak W, Jurczak A, Kościelniak D, et al. The virulence of Streptococcus mutans and the ability to form biofilms. Eur J Clin Microbiol Infect Dis. 2014;33:499–515. doi: 10.1007/s10096-013-1993-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nobre dos Santos M, Melo dos Santos L, Francisco SB, Cury JA. Relationship among dental plaque composition, daily sugar exposure and caries in the primary dentition. Caries Res. 2002;36:347–352. doi: 10.1159/000065959. [DOI] [PubMed] [Google Scholar]
- 69.Mattos-Graner RO, Smith DJ, King WF, Mayer MP. Water-insoluble glucan synthesis by mutans streptococcal strains correlates with caries incidence in 12- to 30-month-old children. J Dent Res. 2000;79:1371–1377. doi: 10.1177/00220345000790060401. [DOI] [PubMed] [Google Scholar]
- 70.Stephan RM, Miller BF. A Quantitative Method for Evaluating Physical and Chemical Agents which Modify Production of Acids in Bacterial Plaques on Human Teeth. J Dent Res. 1943;22:45–51. [Google Scholar]
- 71.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Adv Dent Res. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 72.Marsh PD. Dental Biofilms in Health and Disease. In: Goldberg M, editor. Understanding Dental Caries. Springer International Publishing; 2016. pp. 41–52. [Google Scholar]
- 73.Matsui R, Cvitkovitch D. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 2010;5:403–417. doi: 10.2217/fmb.09.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Forssten SD, Björklund M, Ouwehand AC. Streptococcus mutans, caries and simulation models. Nutrients. 2010;2:290–298. doi: 10.3390/nu2030290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Marsh PD. Are dental diseases examples of ecological catastrophes? Microbiology. 2003;149:279–294. doi: 10.1099/mic.0.26082-0. [DOI] [PubMed] [Google Scholar]
- 76.Bradshaw DJ, McKee AS, Marsh PD. Effects of carbohydrate pulses and pH on population shifts within oral microbial communities in vitro. J Dent Res. 1989;68:1298–1302. doi: 10.1177/00220345890680090101. [DOI] [PubMed] [Google Scholar]
- 77.Bradshaw D, Marsh PD. Analysis of pH–Driven Disruption of Oral Microbial Communities in vitro. Caries Res. 1998;32:456–462. doi: 10.1159/000016487. [DOI] [PubMed] [Google Scholar]
- 78.Jiang W, Zhang J, Chen H. Pyrosequencing analysis of oral microbiota in children with severe early childhood dental caries. Curr Microbiol. 2013;67:537–542. doi: 10.1007/s00284-013-0393-7. [DOI] [PubMed] [Google Scholar]
- 79.Tanner ACR, Kent RL, Jr, Holgerson PL, et al. Microbiota of severe early childhood caries before and after therapy. J Dent Res. 2011;90:1298–1305. doi: 10.1177/0022034511421201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gross EL, Leys EJ, Gasparovich SR, et al. Bacterial 16S sequence analysis of severe caries in young permanent teeth. J Clin Microbiol. 2010;48:4121–4128. doi: 10.1128/JCM.01232-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Xu H, Hao W, Zhou Q, et al. Plaque bacterial microbiome diversity in children younger than 30 months with or without caries prior to eruption of second primary molars. PLoS One. 2014;9:e89269. doi: 10.1371/journal.pone.0089269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Aas JA, Griffen AL, Dardis SR, et al. Bacteria of dental caries in primary and permanent teeth in children and young adults. J Clin Microbiol. 2008;46:1407–1417. doi: 10.1128/JCM.01410-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Divaris K. Predicting Dental Caries Outcomes in Children: A “Risky” Concept. J Dent Res. 2015 doi: 10.1177/0022034515620779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Y, Ge Y, Saxena D, Caufield PW. Genetic profiling of the oral microbiota associated with severe early-childhood caries. J Clin Microbiol. 2007;45:81–87. doi: 10.1128/JCM.01622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Luo AH, Yang DQ, Xin BC, et al. Microbial profiles in saliva from children with and without caries in mixed dentition. Oral Dis. 2012;18:595–601. doi: 10.1111/j.1601-0825.2012.01915.x. [DOI] [PubMed] [Google Scholar]
- 86.Theilade E. The non-specific theory in microbial etiology of inflammatory periodontal diseases. J Clin Periodontol. 1986;13:905–911. doi: 10.1111/j.1600-051x.1986.tb01425.x. [DOI] [PubMed] [Google Scholar]
- 87.Morou-Bermudez E, Rodriguez S, Bello AS, Dominguez-Bello MG. Urease and Dental Plaque Microbial Profiles in Children. PLoS One. 2015;10:e0139315. doi: 10.1371/journal.pone.0139315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Morou-Bermudez E, Elias-Boneta A, Billings RJ, et al. Urease activity in dental plaque and saliva of children during a three-year study period and its relationship with other caries risk factors. Arch Oral Biol. 2011;56:1282–1289. doi: 10.1016/j.archoralbio.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Morou-Bermudez E, Elias-Boneta A, Billings RJ, et al. Urease activity as a risk factor for caries development in children during a three-year study period: a survival analysis approach. Arch Oral Biol. 2011;56:1560–1568. doi: 10.1016/j.archoralbio.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Fidalgo TKS, Freitas-Fernandes LB, Angeli R, et al. Salivary metabolite signatures of children with and without dental caries lesions. Metabolomics. 2012;9:657–666. [Google Scholar]
- 91.Fidalgo TKS, Freitas-Fernandes LB, Almeida FCL, et al. Longitudinal evaluation of salivary profile from children with dental caries before and after treatment. Metabolomics. 2014;11:583–593. [Google Scholar]
- 92.McLean JS, Fansler SJ, Majors PD, et al. Identifying low pH active and lactate-utilizing taxa within oral microbiome communities from healthy children using stable isotope probing techniques. PLoS One. 2012;7:e32219. doi: 10.1371/journal.pone.0032219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Takahashi N, Washio J, Mayanagi G. Metabolomic approach to oral biofilm characterization—A future direction of biofilm research. J Oral Biosci. 2012;54:138–143. [Google Scholar]
- 94.Foxman B, Srinivasan U, Wen A, et al. Exploring the effect of dentition, dental decay and familiality on oral health using metabolomics. Infect Genet Evol. 2014;22:201–207. doi: 10.1016/j.meegid.2013.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Belda-Ferre P, Williamson J, Simón-Soro Á , et al. The human oral metaproteome reveals potential biomarkers for caries disease. Proteomics. 2015;15:3497–3507. doi: 10.1002/pmic.201400600. [DOI] [PubMed] [Google Scholar]
- 96.Simón-Soro A, Tomás I, Cabrera-Rubio R, et al. Microbial geography of the oral cavity. J Dent Res. 2013;92:616–621. doi: 10.1177/0022034513488119. [DOI] [PubMed] [Google Scholar]
- 97.Sato Y, Yamagishi J, Yamashita R, et al. Inter-Individual Differences in the Oral Bacteriome Are Greater than Intra-Day Fluctuations in Individuals. PLoS One. 2015;10:e0131607. doi: 10.1371/journal.pone.0131607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Simón-Soro A, Guillen-Navarro M, Mira A. Metatranscriptomics reveals overall active bacterial composition in caries lesions. J Oral Microbiol. 2014;6:25443. doi: 10.3402/jom.v6.25443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Shi W, Qin M, Chen F, Xia B. Supragingival Microbial Profiles of Permanent and Deciduous Teeth in Children with Mixed Dentition. PLoS One. 2016;11:e0146938. doi: 10.1371/journal.pone.0146938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Afroughi S, Faghihzadeh S, Khaledi MJ, Motlagh MG. Dental caries analysis in 3–5 years old children: A spatial modelling. Arch Oral Biol. 2010/5;55:374–378. doi: 10.1016/j.archoralbio.2010.03.008. [DOI] [PubMed] [Google Scholar]
- 101.Teng F, Yang F, Huang S, et al. Prediction of Early Childhood Caries via Spatial-Temporal Variations of Oral Microbiota. Cell Host Microbe. 2015;18:296–306. doi: 10.1016/j.chom.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 102.Li X, Kolltveit KM, Tronstad L, Olsen I. Systemic diseases caused by oral infection. Clin Microbiol Rev. 2000;13:547–558. doi: 10.1128/cmr.13.4.547-558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Beck JD, Slade G, Offenbacher S. Oral disease, cardiovascular disease and systemic inflammation. Periodontol 2000. 2000;23:110–120. doi: 10.1034/j.1600-0757.2000.2230111.x. [DOI] [PubMed] [Google Scholar]
- 104.Seymour GJ, Ford PJ, Cullinan MP, et al. Relationship between periodontal infections and systemic disease. Clin Microbiol Infect. 2007;13(Suppl 4):3–10. doi: 10.1111/j.1469-0691.2007.01798.x. [DOI] [PubMed] [Google Scholar]
- 105.Iacopino AM, Cutler CW. Pathophysiological relationships between periodontitis and systemic disease: recent concepts involving serum lipids. J Periodontol. 2000;71:1375–1384. doi: 10.1902/jop.2000.71.8.1375. [DOI] [PubMed] [Google Scholar]
- 106.Shen H, Ye F, Xie L, et al. Metagenomic sequencing of bile from gallstone patients to identify different microbial community patterns and novel biliary bacteria. Sci Rep. 2015;5:17450. doi: 10.1038/srep17450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Slots J, Rams TE. New views on periodontal microbiota in special patient categories. J Clin Periodontol. 1991;18:411–420. doi: 10.1111/j.1600-051x.1991.tb02309.x. [DOI] [PubMed] [Google Scholar]
- 108.Tanaka S, Murakami Y, Seto K, et al. The detection of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans in the supragingival plaque of children with and without caries. Pediatr Dent. 2003;25:143–148. [PubMed] [Google Scholar]
- 109.Merchant AT. Periodontitis and dental caries occur together. J Evid Based Dent Pract. 2012;12:18–19. doi: 10.1016/S1532-3382(12)70005-2. [DOI] [PubMed] [Google Scholar]
- 110.Lange L, Thiele GM, McCracken C, et al. Symptoms of periodontitis and antibody responses to Porphyromonas gingivalis in juvenile idiopathic arthritis. Pediatr Rheumatol Online J. 2016;14:8. doi: 10.1186/s12969-016-0068-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Francavilla R, Ercolini D, Piccolo M, et al. Salivary microbiota and metabolome associated with celiac disease. Appl Environ Microbiol. 2014;80:3416–3425. doi: 10.1128/AEM.00362-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Di Cagno R, De Angelis M, De Pasquale I, et al. Duodenal and faecal microbiota of celiac children: molecular, phenotype and metabolome characterization. BMC Microbiol. 2011;11:219. doi: 10.1186/1471-2180-11-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Docktor MJ, Paster BJ, Abramowicz S, et al. Alterations in diversity of the oral microbiome in pediatric inflammatory bowel disease. Inflamm Bowel Dis. 2012;18:935–942. doi: 10.1002/ibd.21874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Hicks MJ, Carter AB, Rossmann SN, et al. Detection of fungal organisms in saliva from HIV-infected children: a preliminary cytologic analysis. Pediatr Dent. 1998;20:162–168. [PubMed] [Google Scholar]
- 115.Roberts GJ, Holzel HS, Sury MR, et al. Dental bacteremia in children. Pediatr Cardiol. 1997;18:24–27. doi: 10.1007/s002469900103. [DOI] [PubMed] [Google Scholar]
- 116.Joshipura K, Ritchie C, Douglass C. Strength of evidence linking oral conditions and systemic disease. Compend Contin Educ Dent Suppl. 2000:12–23. quiz 65. [PubMed] [Google Scholar]
- 117.Fábián TK, Fejérdy P, Csermely P. Salivary Genomics, Transcriptomics and Proteomics: The Emerging Concept of the Oral Ecosystem and their Use in the Early Diagnosis of Cancer and other Diseases. Curr Genomics. 2008;9:11–21. doi: 10.2174/138920208783884900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gao S, Li S, Ma Z, et al. Presence of Porphyromonas gingivalis in esophagus and its association with the clinicopathological characteristics and survival in patients with esophageal cancer. Infect Agent Cancer. 2016;11:3. doi: 10.1186/s13027-016-0049-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Nakajima M, Arimatsu K, Kato T, et al. Oral Administration of P. gingivalis Induces Dysbiosis of Gut Microbiota and Impaired Barrier Function Leading to Dissemination of Enterobacteria to the Liver. PLoS One. 2015;10:e0134234. doi: 10.1371/journal.pone.0134234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Ibrahim R, Pasic M, Yousef GM. Omics for personalized medicine: defining the current we swim in. Expert Rev Mol Diagn. 2016:1–4. doi: 10.1586/14737159.2016.1164601. [DOI] [PubMed] [Google Scholar]
- 121.Lee JY, Divaris K. The ethical imperative of addressing oral health disparities: a unifying framework. J Dent Res. 2014;93:224–230. doi: 10.1177/0022034513511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Mougeot J-LC, Stevens CB, Cotton SL, et al. Concordance of HOMIM and HOMI N GS technologies in the microbiome analysis of clinical samples. J Oral Microbiol. 2016 doi: 10.3402/jom.v8.30379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Duran-Pinedo AE, Frias-Lopez J. Beyond microbial community composition: functional activities of the oral microbiome in health and disease. Microbes Infect. 2015;17:505–516. doi: 10.1016/j.micinf.2015.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Wong DTW. Salivaomics. J Am Dent Assoc. 2012;143:19S–24S. doi: 10.14219/jada.archive.2012.0339. [DOI] [PubMed] [Google Scholar]
- 125.Vayssier-Taussat M, Albina E, Citti C, et al. Shifting the paradigm from pathogens to pathobiome: new concepts in the light of meta-omics. Front Cell Infect Microbiol. 2014;4:29. doi: 10.3389/fcimb.2014.00029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hart TC, Corby PM, Hauskrecht M, et al. Identification of microbial and proteomic biomarkers in early childhood caries. Int J Dent. 2011;2011:196721. doi: 10.1155/2011/196721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Gehlenborg N, O’Donoghue SI, Baliga NS, et al. Visualization of omics data for systems biology. Nat Methods. 2010;7:S56–S68. doi: 10.1038/nmeth.1436. [DOI] [PubMed] [Google Scholar]
- 128.Dimitrov DV, Hoeng J. Systems approaches to computational modeling of the oral microbiome. Front Physiol. 2013;4:172. doi: 10.3389/fphys.2013.00172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bartel J, Krumsiek J, Schramm K, et al. The Human Blood Metabolome-Transcriptome Interface. PLoS Genet. 2015;11:e1005274. doi: 10.1371/journal.pgen.1005274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ferreira Zandoná A, Ando M, Gomez GF, et al. Longitudinal analyses of early lesions by fluorescence: an observational study. J Dent Res. 2013;92:84S–9S. doi: 10.1177/0022034513490167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Härkäne T, Larmas MA, Virtanen JI, Arjas E. Applying modern survival analysis methods to longitudinal dental caries studies. J Dent Res. 2002;81:144–148. [PubMed] [Google Scholar]
- 132.Nasidze I, Li J, Schroeder R, et al. High diversity of the saliva microbiome in Batwa Pygmies. PLoS One. 2011;6:e23352. doi: 10.1371/journal.pone.0023352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Nunes AP, Oliveira IO, Santos BR, et al. Quality of DNA extracted from saliva samples collected with the Oragene\texttrademark DNA self-collection kit. BMC Med Res Methodol. 2012;12:1–6. doi: 10.1186/1471-2288-12-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Rogers NL, Cole SA, Lan H-C, et al. New saliva DNA collection method compared to buccal cell collection techniques for epidemiological studies. Am J Hum Biol. 2007;19:319–326. doi: 10.1002/ajhb.20586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ng DPK, Koh D, Choo SGL, et al. Effect of storage conditions on the extraction of PCR-quality genomic DNA from saliva. Clin Chim Acta. 2004;343:191–194. doi: 10.1016/j.cccn.2004.01.013. [DOI] [PubMed] [Google Scholar]
- 137.Quinque D, Kittler R, Kayser M, et al. Evaluation of saliva as a source of human DNA for population and association studies. Anal Biochem. 2006;353:272–277. doi: 10.1016/j.ab.2006.03.021. [DOI] [PubMed] [Google Scholar]
- 138.Ahn J, Yang L, Paster BJ, et al. Oral microbiome profiles: 16S rRNA pyrosequencing and microarray assay comparison. PLoS One. 2011;6:e22788. doi: 10.1371/journal.pone.0022788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Chen T, Yu W-H, Izard J, et al. The Human Oral Microbiome Database: a web accessible resource for investigating oral microbe taxonomic and genomic information. Database. 2010;2010:baq013. doi: 10.1093/database/baq013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Griffen AL, Beall CJ, Firestone ND, et al. CORE: a phylogenetically-curated 16S rDNA database of the core oral microbiome. PLoS One. 2011;6:e19051. doi: 10.1371/journal.pone.0019051. [DOI] [PMC free article] [PubMed] [Google Scholar]