Abstract
Objective
To determine the effect of enteral fish oil and safflower oil supplementation on the intestinal microbiome in premature infants with an enterostomy.
Study design
Premature infants with an enterostomy were randomized to receive early enteral supplementation with a high fat-polyunsaturated fatty acid (HF-PUFA) blend of fish oil and safflower oil versus standard nutritional therapy. We used 16S rRNA gene sequencing for longitudinal profiling of the microbiome from the time of study entry until bowel reanastomosis. We used weighted gene co-expression network analysis to identify microbial community modules that differed between study groups over time. We performed imputed metagenomic analysis to determine metabolic pathways associated with the microbial genes.
Results
Sixteen infants were randomized to receive enteral HF-PUFA supplementation and 16 infants received standard care. The intestinal microbiota of infants in the treatment group differed from those in the control group, with greater bacterial diversity and lower abundance of Streptococcus, Clostridium, and many pathogenic genera within the Enterobacteriaceae family. We identified four microbial community modules with significant differences between groups over time. Imputed metagenomic analysis of the microbial genes revealed metabolic pathways that differed between groups, including metabolism of amino acids, carbohydrates, fatty acids, and secondary bile acid synthesis.
Conclusion
Enteral HF-PUFA supplementation was associated with decreased abundance of pathogenic bacteria, greater bacterial diversity, and shifts in the potential metabolic functions of intestinal microbiota.
Trial registration
Keywords: nutrition, neonate, microbiota
Premature infants with necrotizing enterocolitis, spontaneous intestinal perforation, or intestinal atresia commonly require abdominal surgery and creation of a small bowel enterostomy.1 Following surgery, these infants are at high risk for a number of complications, including cholestasis, liver disease, sepsis, growth failure, and death.1–4 The etiology of these complications is multifactorial, but is believed to be in part related to prolonged reliance on parenteral soybean-based lipid formulations, which are rich in proinflammatory omega-6 fatty acids and phytosterols, and devoid of the omega-3 fatty acids eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA).5–8 Fish oil, which is rich in anti-inflammatory omega-3 fatty acids, has been associated with decreased cholestasis and liver injury in children with short bowel syndrome.9,10 A recent randomized controlled trial of early enteral supplementation with a high fat polyunsaturated fatty acid (HF-PUFA) blend of fish oil and safflower oil versus usual care in premature infants with an enterostomy found that infants treated with the HF-PUFA blend had lower conjugated bilirubin levels than infants in the control group.11 Furthermore, the infants who received HF-PUFA required fewer sepsis evaluations and had improved growth after bowel reanastomosis. The mechanisms underlying the beneficial effects of enteral HF-PUFA in this setting are unclear.
The intestinal microbiome has an essential role in intestinal function, including nutrient absorption, metabolism, maintenance of barrier integrity, and protection against infection.12,13 The microbiome is altered by diet, including fat intake.14,15 Infants who have undergone surgical enterostomy placement often have multiple predisposing factors that may perturb the intestinal microbiome, including premature birth, history of bowel injury or perforation, antibiotic exposure, prolonged withholding of enteral feeds, and intestinal surgery.4 The objective of our study was to determine the effects of enteral supplementation with a HF-PUFA blend containing fish oil and safflower oil, versus standard nutritional therapy, on the intestinal microbiome in premature infants with an enterostomy. We hypothesized that treatment with enteral HF-PUFA leads to functional and community-level changes in the microbiome, which may promote the development of functional microbial communities that contribute to the clinical benefits of HFPUFA that were observed in this cohort.
Methods
A randomized, controlled trial of enteral fish oil and safflower oil supplementation versus usual care was conducted in the Neonatal Intensive Care Unit of Brenner Children’s Hospital at Wake Forest Baptist Medical Center (ClinicalTrials.gov: NCT01306838). Details of the study design have been previously described by Yang et al.11 In brief, inclusion in the study required the presence of a jejunostomy or ileostomy, birth gestational age of less than 37 weeks, and postnatal age less than two months at the time of study enrollment. Infants were excluded if they had a colostomy, major congenital anomaly, or metabolic disease. Infants in the control group received usual nutritional care. Once tolerating minimal enteral feedings of 20 mL/kg/day, infants in the treatment group received enteral fat supplementation with fish oil (Major Fish Oil 500, Major Pharmaceuticals, Livonia, Michigan or Rugby Sea Omega 50, Rugby Laboratories, Inc, Corona, California) and safflower oil (Microlipid, Nestle Nutrition, Florham Park, New Jersey). The fish oil supplements contained EPA, DHA, and Vitamin E (tocopherol). Fish oil was initiated at a dose of 0.2 g every 12 hours for infants weighing <1000 g or 0.25 g every 12 hours for infants weighing >1000 g, and increased to a maximum dose of 0.5 g every 6 hours. Safflower oil, which is enriched in the omega-6 fatty acid linoleic acid, was started at 1 g/kg/d and increased by 0.5 g/kg/day to 2.5 g/kg/day, for a goal omega-6 to omega-3 fatty acid ratio of 3.75 to 5.1. Parenteral Intralipid (Baxter Healthcare, Deerfield, Illinois) was decreased by 0.5 g/kg/day as enteral fat supplementation advanced and discontinued when the dose reached <1 g/kg/day.
Enteral feedings were initiated upon recovery of bowel function after ostomy placement and gradually advanced. Infants received their mother’s milk when available or infant formula. As enteral nutrition advanced, parenteral nutrition and intravenous lipid were decreased to achieve a growth rate of 15 g/kg/d. Goal caloric intake was generally 120–130 kcal/kg/d. Stool ostomy output was collected weekly from the time of study enrollment to the time of bowel reanastomosis, up to a maximum of 10 weeks. Samples were stored at −80°C until further processing. Four subjects included in the original randomized trial were excluded from this analysis due to missing samples (two subjects in treatment group and two subjects in control group). The trial was approved by the Wake Forest University Health Science Institutional Review Board and written informed consent was obtained from parents. The microbiome analysis was a secondary study of the existing, de-identified fecal specimens and was deemed exempt research by the Duke Institutional Review Board.
DNA extraction, PCR amplification, and DNA sequencing
Total genomic DNA was extracted from fecal samples using a commercial bead-beating method (Zymo Research Soil Microbe DNA Kit, Irvine, California). PCR amplification of the V4 region of the 16S rRNA gene was performed using 12 nucleotide barcode-indexed primers and previously described standardized PCR conditions for the Earth Microbiome Project.16 PCR amplicons were pooled in equimolar concentrations and purified by gel extraction. Sequencing was performed on the Illumina HiSeq Platform using 150 nucleotide single end reads.
Sequences were split, quality-trimmed, demultiplexed, and chimera-reduced using QIIME tools.17 High-quality sequences sharing ≥99% nucleotide sequence identity were clustered into operational taxonomic units (OTUs) using USEARCH (version 6), aligned to the Greengenes database (version 13.8.99), and representative sequences were given a taxonomic assignment using BLAST against the SILVA bacterial database (Release 111).18,19 Sparse OTUs with <3 occurrences in at least 20% of samples were excluded from subsequent analyses as they were unlikely to significantly contribute to the overall composition and inferred metabolic capacity in any one sample. In the case of calculating alpha-diversity, however, only singleton OTUs were excluded. Counts were normalized using the cumulative sums scaling approach (percentile p=0.5, determined using the ‘cumNormStatFast’ function) in the metagenomeSeq package (version 1.12.0).20
Microbiome Composition and Diversity
Microbial composition and diversity were determined using tools within the phyloseq (version 1.14.0) and metagenomeSeq packages (version 1.12.0).20,21 Smoothing spline Analysis of Variance (SS-ANOVA) was employed from the metagenomeSeq and gss (version 2.1–5) packages in R to identify time intervals in which alpha-diversity and bacterial taxonomic groups differed between treatment groups, controlling for repeat sampling of individual infants as a random effect.20,22 The area between the observed distributions was measured for each time interval of difference. P values were calculated using a permutation-based method in which the groups were permutated 10,000 times and the observed areas were compared with the empiric null distributions. All analyses were performed using R statistical software. Statistical significance was considered P<0.05, with Benjamini-Hochberg correction for multiple testing.
Construction of Microbial Gene Co-Occurrence Modules
We applied weighted gene co-expression network analysis to group individual bacterial OTUs into microbial co-occurrence modules, using the WGCNA package (version 1.49).23 This network analysis method groups OTUs with highly correlated abundance patterns, likely reflecting interactions or interdependency, into modules, which we considered to represent microbial subcommunities. Samples were clustered using hclust with an “average” method on a Euclidian distance matrix. Clusters were assigned using “cutreestatic” to remove outlying samples. An unsigned weighted adjacency matrix was constructed using a soft-threshold power of 10, which was determined using the scale free topology criterion. Modules were selected using a minimum module size of 5 and a static merge cut height of 0.6. Each module was assigned a color label and summarized by the Module Eigengene (ME) value, which represents the first principal component of each module. SS-ANOVA was used to determine time intervals in which module abundance differed between treatment groups.
Imputed Metagenomic Functional Analysis
We performed imputed metagenomic analysis using Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) to investigate the effect of treatment on functional pathways.24 Sequences were assigned to OTUs using closed reference assignment with Greengenes in QIIME as per the original authors’ recommendations.19.24 The inferred metagenomes from microbial OTUs were sorted into Kyoto Encyclopedia of Genes and Genomes (KEGG) metabolic pathways. We then used SS-ANOVA to determine metabolic pathways that differed between treatment groups over time.
Results
Baseline infant characteristics were similar between the 16 infants in the control group and 16 infants in the treatment group (Table I). The most common indication for enterostomy was spontaneous intestinal perforation (56%), followed by necrotizing enterocolitis (38%) and intestinal atresia (6%). The median number of days of antibiotics over the study period was not significantly different between groups. Cumulative enteral fat intake was greater among infants in the treatment group. A further detailed description of the clinical characteristics and outcomes in the cohort has been previously reported.11
Table 1.
Infant Characteristics
| Control (n=16) |
Treatment (n=16) |
P | ||
|---|---|---|---|---|
| Birth weight (g), med (range) | 845 (530–1981) | 777 (601–2179) | 0.71 | |
| Birth gestational age (wks), med (range) |
26.5 (23–35) | 26.5 (22–34) | 0.49 | |
| Male sex, n (%) | 12 (75) | 9 (56) | 0.46 | |
| Surgical indication, n (%) | 0.85 | |||
| Necrotizing Enterocolitis | 7 (44) | 5 (31) | ||
| Spontaneous Intestinal Perforation | 8 (50) | 10 (63) | ||
| Intestinal Atresia | 1 (6) | 1 (6) | ||
| Ostomy site, n (%) | >0.99 | |||
| Ileostomy | 11 (69) | 11 (69) | ||
| Jejunostomy | 5 (31) | 5 (31) | ||
| Diet, n (%) | 0.33 | |||
| Human milk | 6 (38) | 6 (38) | ||
| Formula | 1 (6) | 4 (25) | ||
| Combination | 9 (56) | 6 (38) | ||
| Day of life at first feeding week sample collection, med (IQR) |
21 (14–30) | 19 (12.5–26) | 0.55 | |
| Cumulative enteral fat intake over study period* (g), med (IQR) |
110 (75–265) | 477 (256–610) | 0.002 | |
| Supplemental fat intake over study period** (g), med (IQR) |
0 (0–22) | 155 (120–169) | <0.001 | |
| Total antibiotic days over study period, med (IQR) |
7 (1.5–21.5) | 5.5 (1.5–10) | 0.35 | |
| Antibiotics received, n (%) | ||||
| Meropenem | 1 (6) | 3 (19) | 0.60 | |
| Ceftazidime | 1 (6) | 0 (0) | >0.99 | |
| Clindamycin | 0 (0) | 1 (6) | >0.99 | |
| Vancomycin | 12 (75) | 9 (56) | 0.46 | |
| Ampicillin | 2 (13) | 2 (13) | >0.99 | |
| Gentamicin | 11 (69) | 9 (56) | 0.72 | |
Includes dietary fat as well as supplemental fat. While fat content varies in human milk, calculations were based on 4 g fat/100 ml.
Includes fish oil and safflower oil intake. All supplemental fat in control group was safflower oil, which was given to infants with poor growth despite full enteral feedings with high calorie formula per standard of care.
Relative composition of the fecal microbiota over time in control and treatment groups
A total of 202 samples were available for microbial sequencing, with a median (IQR) of 6.5 (4–8) samples per subject. A total of 77,928 bacterial OTUs were identified including singletons, which was reduced to 436 total OTUs after removal of sparse OTUs. At the phylum level, Proteobacteria and Firmicutes accounted for the majority of bacterial OTUs in both infant groups throughout the study (Figure 1; available at www.jpeds.com). As expected for the preterm infant, Bacteroidetes were proportionately rare.25 Using SS-ANOVA, we modeled differences in phyla abundance between the control and treatment groups over time, controlling for repeat sampling from each infant. Proteobacteria, which includes pathogenic Gram-negative bacilli, had significantly lower levels in the treatment group (Weeks 2–8, Area=−74551.1, P=0.0072). Actinobacteria were significantly but transiently lower in the control group during study weeks 4 and 5 (Area=233.1, P=0.0419). Other phyla did not differ significantly between study groups.
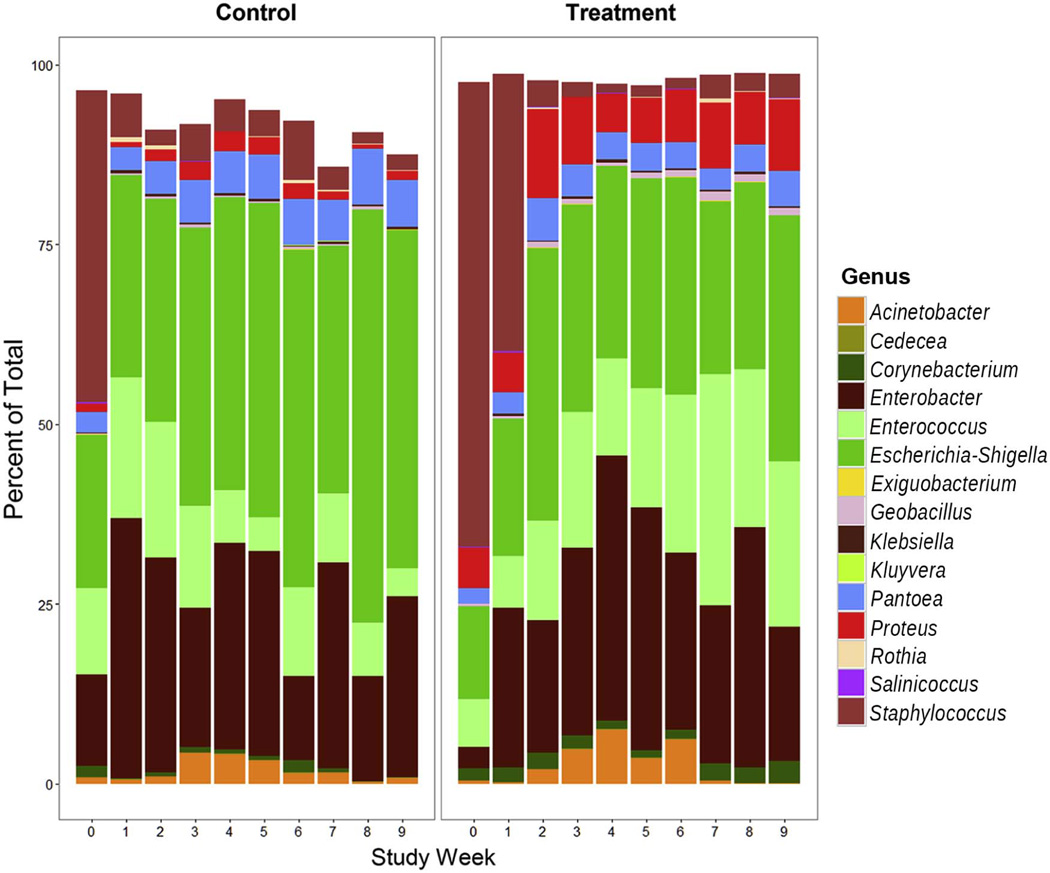
At the genus level, there were a number of changes in the microbiome over time (Figure 2). Many differences emerged between groups one to three weeks following initiation of HF-PUFA treatment, consistent with a therapeutic response (Table II; available at www.jpeds.com). Over time, genera with greater relative enrichment in the control group included Escherichia (Weeks 1–9, P<0.0001), Serratia (Weeks 2–9, P<0.0001), Pantoea (Weeks 2–9, P<0.0001), Clostridium (Weeks 2–9, P<0.0001), and Streptococcus (Weeks 3–9, P<0.0001). In the treatment group, there was enrichment of Corynebacterium (Weeks 1–6, P=0.0089). Proteus was also more abundant in the treatment group, but this difference was present even prior to initiation of therapy (Weeks 0–8, P<0.0001). Several differences were identified well over a month after treatment initiation, including greater enrichment of Enterococcus in the treatment group (Weeks 8–9, P<0.0001) and Haemophilus in the control group (Weeks 7–9, P=0.0125). Principal coordinates analysis (PCoA) was employed to examine longitudinal community-level dynamic changes and differences in treatment and control groups. The first principal component (PC1) diverged over time between groups with a significant difference over the full study period (weeks 0–9, P<0.0001) (Figure 3; available at www.jpeds.com).
Figure 2.
Changes in the relative proportion of the 15 most abundant bacteria genera over time in control and treatment groups.
Identification of microbial OTU co-occurrence modules
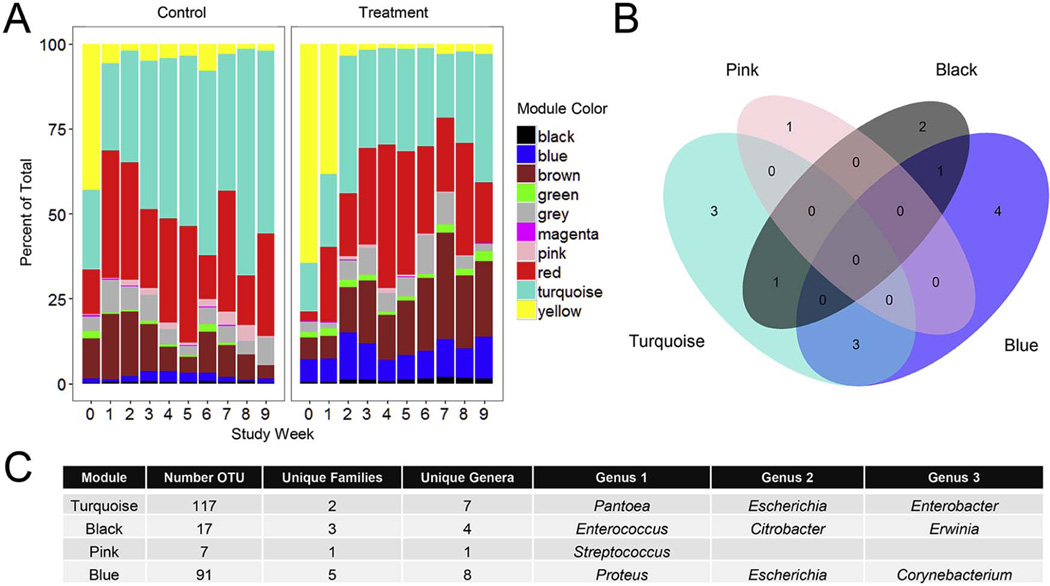
Although specific genera appeared to change in response to treatment, the identification of bacterial subgroups undergoing coordinated changes in the therapeutic response may more accurately reflect changes in physiology of the microbiota and explain medical outcomes.11 We used weighted gene co-expression network analysis to organize individual bacterial OTUs into community modules and then evaluated changes in these bacterial communities over time in response to treatment. We identified 10 community modules, with a median of 32.5 (range 6–117) unique bacterial OTUs per module (Figure 4; available at www.jpeds.com). We examined the relative proportion of the total OTUs accounted for by each module over time in treatment and control groups (Figure 5, A). Using SS-ANOVA, four modules had significant differences in abundance between the groups over some time interval. There was an expansion of the Turquoise module over time in the control group that was not observed in the treatment group (Weeks 2–9, Area=−12157.2, P<0.0001). The Pink module also had greater relative enrichment in the control group (Weeks 3–9, Area=−5161.9, P=0.0054). Modules with greater enrichment in the treatment group included Black (Weeks 1–9, Area=330.3, P=0.0054) and Blue (Weeks 0–8, Area=2068.2, P<0.0001).
Figure 5.
Weighted gene co-expression network analysis identified 10 unique bacterial community modules. A. The relative abundance of network modules in treatment and control group infants over time. B. Venn diagram showing overlap of genera within the four modules with significant differences between treatment and control groups. Many genera were represented in more than one module, and some were unique to a specific module. C. The composition of four modules with significant differences between treatment groups. The number of unique OTUs, unique families, and unique genera within each module are noted, followed by a list of three genera enriched in each module.
Many bacterial genera were represented in multiple modules, and others were unique to a specific module (Figure 5, B). For example, the Blue module contained all 75 OTUs within the Proteus genus, and also contained OTUs from Escherichia, Corynebacterium, and five other genera (Figure 5, C). The Turquoise module, which had greater abundance in the control group, was enriched in many pathogenic Gram-negative bacilli, including Pantoea, Escherichia, and Enterobacter. The Pink module, also enriched in the control group, was composed entirely of Streptococcus OTUs. The Black module, which was more abundant in the treatment group, was composed of Enterococcus, Citrobacter, Erwinia, and Geobacillus OTUs. The Yellow module, enriched in Staphylococcus, declined after the first study weeks in both groups.
The Turquoise module had the greatest magnitude of difference between groups. ME values in the Turquoise module diverged between the study groups over time (Figure 6; available at www.jpeds.com). The shifts in ME values were reflected in changes in the abundance of genera enriched in the Turquoise module, with Pantoea, Citrobacter, Escherichia, and Tatumella all increasing in abundance in the control group over time relative to the treatment group.
Alpha-diversity by treatment group
Changes in alpha-diversity and richness over time are presented in Figure 7 (available at www.jpeds.com). Many infants in both groups experienced a decline in diversity over the study period. Alpha-diversity was significantly higher among infants in the treatment group following initiation of therapy (Shannon Index: Weeks 3–9, P=0.007; Inverse Simpson Index: Weeks 1–9, P=0.0015). Richness as measured by the Chao1 Index did not differ significantly between groups.
Imputed metagenomic functional analysis
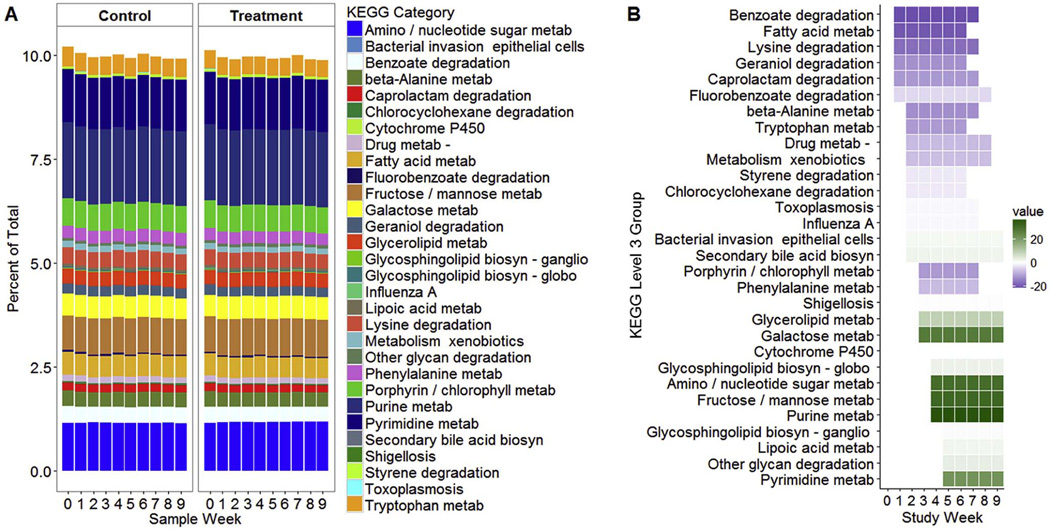
We used imputed metagenomic analysis to compare the predicted functions of the microbial genes in treatment and control groups. A number of pathways showed sustained differences between groups (Figure 8 and Table III; Table III available at www.jpeds.com). Lipid metabolic pathways that differed between groups included fatty acid metabolism (Weeks 1–6, P=0.0053), glycerolipid metabolism (Weeks 3–9, P=0.0052), arachidonic acid metabolism (Weeks 4–7, P=0.0045), steroid hormone biosynthesis (Weeks 2–5, P=0.0077), and secondary bile acid biosynthesis (Weeks 2–9, P<0.0001). In the control group, there was enrichment of butyrate metabolism (Weeks 2–5; P=0.0055), drug metabolism by cytochrome P450 (Weeks 2–8, P<0.0001), and metabolism of specific amino acids, including phenylalanine (Weeks 3–7, P=0.0065), lysine (Weeks 1–7, P=0.0025), and tryptophan (Weeks 2–6, P=0.0040). In the treatment group, there was enrichment of amino sugar and nucleotide sugar metabolism (Weeks 4–9, P=0.0060), purine metabolism (Weeks 4–9, P=0.0055), and pyrimidine metabolism (Weeks 5–9, P=0.0053).
Figure 8.
Metabolic pathways associated with microbial genes. A. The relative proportion of metabolic pathways with significant differences between treatment and control groups over time. B. KEGG metabolic pathways that differed between treatment and control groups. Shaded boxes represent the time interval with significant differences between groups by SS-ANOVA. Purple shading indicates pathways with greater abundance in the control group and green represents pathways with greater abundance in the treatment group. Darker shading indicates a greater magnitude of difference between groups.
Discussion
In this cohort of premature infants with small bowel enterostomies, enteral supplementation with a HF-PUFA blend containing fish oil and safflower oil was associated with lower conjugated bilirubin levels, fewer episodes of suspected sepsis, and improved growth after bowel reanastomosis.11 Here, we report the longitudinal changes in the intestinal microbiome of these infants over the course of treatment. We observed differences between treatment and control groups over time in both individual bacterial taxonomic groups as well as larger bacterial network modules, suggesting that HF-PUFA treatment has broad effects on intestinal microbial ecology that may contribute to the clinical benefits of HF-PUFA therapy observed in this cohort of infants.
Many of the bacteria that differed between treatment and control groups have important immunologic and metabolic functions in host health. Bacteria within the Enterobacteriaceae family are capable of provoking potent host inflammatory responses and are frequent invasive pathogens in premature infants and infants with short bowel syndrome.26–28 A relative expansion of many genera within the Enterobacteriaceae family was observed over time in the control group that was not seen in infants receiving enteral fish oil, including Escherichia, Pantoea, Serratia, and Citrobacter. In addition, we observed lower levels of Clostridium within the microbiota of treatment group infants. Bacteria within the Clostridium genus catalyze the conversion of primary bile acids to secondary bile acids.29,30 Bile acids have signaling and antimicrobial effects and their biotransformation by bacteria may alter host lipid metabolism, bile acid excretion, and intestinal barrier integrity.29–31 Clostridia also have a role in fermentation of dietary carbohydrates to butyrate, a short-chain fatty acid with generally beneficial effects on intestinal barrier integrity and immune regulation.32–35
We observed an overall decline in bacterial diversity among many infants in both groups over the study period. The reasons for this decline are unclear, but may have been related to antibiotic use or functional adaptive changes following surgery. HF-PUFA therapy was associated with more stable diversity, as infants in the treatment group maintained significantly higher diversity over time compared with infants in the control group. It is possible that the increased diversity in the treatment group was not solely due to direct effects on HF-PUFA on microbiota, but also indirect effects. For example, HF-PUFA treatment was associated with fewer suspected-sepsis events, and although the overall exposure to antibiotics was not different between the groups, differences in the timing of antibiotic exposure may have contributed to the higher bacterial diversity in the treatment group.
Along with the changes in bacterial composition, there were number of differences in predicted metabolic functions of the microbial genes in treatment and control group infants based on imputed metagenomic analysis. Among treatment group infants, there was relatively greater enrichment of genes involved in metabolism of nucleotides and specific carbohydrates, and in the control group there was enrichment of genes involved in metabolism of certain amino acids, xenobiotics, and fatty acids. This method of inferring metabolic functions from microbial genes has limitations as it cannot account for phenotypic variation between microbial communities based on expression level differences between and within species and the metabolic contributions of unmapped sequences. However, the approach suggests that the microbial changes seen following HF-PUFA therapy had significant effects on the overall functional capacity of the intestinal microbiome.
We suspect that the broad reduction in the relative burden of proinflammatory and pathogenic bacteria, changes in key bacteria with metabolic effects, and greater bacterial diversity in the treatment group all contributed to the clinical benefits observed in the treatment group. Further, these bacterial changes likely acted in concert with direct anti-inflammatory and molecular effects of fish oil on the host. Fish oil is enriched in the omega-3 long-chain polyunsaturated fatty acids (LC-PUFAs) EPA and DHA. Bioactive metabolites of these LC-PUFAs include resolvins and protectins that reduce inflammation and support brain and retinal development. Omega-3 LC-PUFAs in fish oil activate peroxisome proliferator-activated receptor alpha (PPARα) and alter expression of genes involved in fatty acid oxidation and glucose metabolism, reducing hepatic lipid accumulation and insulin resistance.36,37 Infants in the HF-PUFA treatment group also had reduced exposure to parenteral soybean-based lipids, which are lacking in omega-3 fatty acids but enriched in the omega-6 fatty acid linoleic acid. Linoleic acid is processed to arachidonic acid, which has terminal metabolites that promote inflammation. Combined, these prior observations and our measurements of the microbiome during enteral fish oil therapy raise the hypothesis that the more favorable balance of omega-6 to omega-3 fatty acids and decreased burden of proinflammatory bacteria in the treatment group infants led to reduced inflammation and bacterial translocation across the intestinal barrier.
There are many potential mechanisms by which the microbiome may have been altered by enteral HF-PUFA intake. The greater amount of enteral fat and greater omega-3 to omega-6 fatty acid ratio may have provided substrate or altered the local environment in a manner that fostered the growth of certain bacteria. We cannot say with certainty whether the microbial differences we observed were specific to fish oil or secondary to the earlier provision and increased dose of enteral fat in the treatment group, independent of the type of fat. The enteral fat supplements contain additional active components, including vitamin E and linoleic acid in safflower oil, that may have accounted for some of the differences in the microbiome between treatment and control groups. Analysis of intestinal tissue samples collected from the proximal ostomy at the time of bowel reanastomosis showed that infants in the treatment group had significantly higher tissue levels of DHA, EPA, and linoleic acid with a lower total omega-6 to omega-3 fatty acid ratio compared with infants in the control group (personal communication of unpublished data- Qing Yang).
Data from animal models indicate that fish oil in particular has effects on shaping the microbiome.38–40 Ghosh et al found that mice fed a diet supplemented with fish oil had reduced abundance of Enterobacteriaceae and Clostridia species compared with mice fed a diet rich in omega-6 fatty acids.38 Further, they found that fish oil supplementation reduced bacterial translocation and intestinal injury in a model of induced colitis. A more recent study utilized a fat-1 transgenic mouse model in which tissue omega-6 fatty acids are converted to omega-3 fatty acids, eliminating the confounding effects of diet.41 The transgenic mice had decreased markers of inflammation and metabolic endotoxemia compared with wild type mice fed the same diet. The gut microbiota appeared to mediate these differences, as the between-group differences could be reduced by co-housing or eliminated by provision of antibiotics. The microbiome of the wild type mice had significantly greater abundance of Proteobacteria, and the fat-1 mice had greater abundance of Bifidobacterium and Lactobacillus.
It is important to note that the changes we observed in the microbiome occurred in the context of the premature infant gut. Premature infants are still undergoing rapid intestinal growth and development, as well as maturation of the immune system. In addition, these infants were recovering from major intestinal injury and undergoing functional adaptation after loss of bowel length. It is possible that enteral fatty acids may have differential effects on the microbiome and host at different stages of development, and that some effects may take longer than the duration of this study. Our study had additional limitations. The sample size was small and investigators were not blinded to the group assignments. Given that the microbiome can vary significantly between individuals, a larger sample size and a multicenter study will be necessary to confirm the differences we observed between groups and further examine the influence of other confounding variables, such as other dietary and treatment factors, on the microbiome.
In conclusion, in this cohort of premature infants with enterostomies, enteral HF-PUFA treatment was associated with functional and community-level changes in the microbiome. Understanding these changes and their potential to modify growth, intestinal adaptation, and susceptibility to cholestasis and sepsis will increase our ability to minimize morbidities and improve outcomes in this high-risk population.
Supplementary Material
Figure 1, online. Operational taxonomic unit (OTU) counts within major bacterial phyla by study week and treatment group. An OTU is a cluster of sequences with a high degree of similarity (≥99% identity). Each point represents an OTU within the specified bacterial phyla. The overlying boxplots outline the interquartile range. Dotted rectangles indicate intervals with statistically different levels of a phylum between groups.
Figure 3, online. Principal coordinates analysis (PCoA) of the microbiome in control and treatment group infants over time. PCoA is a method to distill and visualize similarities and differences between microbiome sample sequencing data. The first principal component (PC1) accounts for as much of the variation in the data as possible. A. Box-plots of the first principle component (PC1) in control and treatment groups for each study week. B. PC1 values modeled for control and treatment groups, showing divergence over time. C. Difference in PC1 values in treatment and control group, showing that treatment and control groups become more dissimilar over time.
Figure 4, online. Hierarchical cluster dendrogram based on microbial co-occurrence networks in study infants.
Figure 6, online. Turquoise Microbial Module. A. Median module eigengene (ME) values over time in control and treatment groups. The ME represents the first principal component of the module. Error bars represent standard error of the mean. B. Abundance of five genera enriched in the turquoise module over time.
Figure 7, online. Alpha-diversity and richness over time. Measures of richness (Chao1) reflect the number of different types of bacteria (OTUs) present in a sample, whereas alpha-diversity indices (Shannon, Inverse Simpson) also take into account differences in the proportional abundance (ie. the evenness of distribution) of the different OTUs within the sample. Means and standard errors are shown. Dotted rectangles indicate intervals with statistically significant differences between treatment and control groups by SS-ANOVA.
Acknowledgments
Supported by the National Institutes of Health (T32 HD60558-5, K12 HD043494-14 [to N.Y.], and R01GM108494 [to P.S.]); Wake Forest University Health Science Interim/Venture and Center for Integrative Medicine Funds (to Q.Y.); Department of Pediatrics Developmental Fund (to Q.Y.); Gerber Foundation Fund (to Q.Y.); the March of Dimes (to P.S.); the Hartwell Foundation (to P.S.); and the Jean and George Brumley, Jr, Neonatal Perinatal Research Institute.
We thank Jessica McCann, MD, Michael Cotten, MD (serves on the Data Safety Monitoring Board for rEVO Biologics, Inc, and received personal fees), and Ronald Goldberg, MD
Abbreviations
- HF-PUFA
high fat-polyunsaturated fatty acid
- LC-PUFA
long chain-polyunsaturated fatty acid
- ME
module eigengene
- OTU
operational taxonomic unit
- SS-ANOVA
smoothing spline analysis of variance
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors declare no conflicts of interest.
References
- 1.Blakely ML, Tyson JE, Lally KP, McDonald S, Stoll BJ, Stevenson DK, et al. Laparotomy versus peritoneal drainage for necrotizing enterocolitis or isolated intestinal perforation in extremely low birth weight infants: outcomes through 18 months adjusted age. Pediatrics. 2006;117:e680–e687. doi: 10.1542/peds.2005-1273. [DOI] [PubMed] [Google Scholar]
- 2.Kelly DA. Liver complications of pediatric parenteral nutrition--epidemiology. Nutrition. 1998;14:153–157. doi: 10.1016/s0899-9007(97)00232-3. [DOI] [PubMed] [Google Scholar]
- 3.Wadhawan R, Oh W, Hintz SR, Blakely ML, Das A, Bell EF, et al. Neurodevelopmental outcomes of extremely low birth weight infants with spontaneous intestinal perforation or surgical necrotizing enterocolitis. J Perinatol. 2014;34:64–70. doi: 10.1038/jp.2013.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moss RL, Dimmitt RA, Barnhart DC, Sylvester KG, Brown RL, Powell DM, et al. Laparotomy versus peritoneal drainage for necrotizing enterocolitis and perforation. N Engl J Med. 2006;354:2225–2234. doi: 10.1056/NEJMoa054605. [DOI] [PubMed] [Google Scholar]
- 5.Gawecka A, Michalkiewicz J, Kornacka MK, Luckiewicz B, Kubiszewska I. Immunologic properties differ in preterm infants fed olive oil vs soy-based lipid emulsions during parenteral nutrition. JPEN J Parenter Enteral Nutr. 2008;32:448–453. doi: 10.1177/0148607108319802. [DOI] [PubMed] [Google Scholar]
- 6.Xu Z, Harvey KA, Pavlina T, Dutot G, Hise M, Zaloga GP, et al. Steroidal compounds in commercial parenteral lipid emulsions. Nutrients. 2012;4:904–921. doi: 10.3390/nu4080904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carter BA, Taylor OA, Prendergast DR, Zimmerman TL, Von Furstenberg R, Moore DD, et al. Stigmasterol, a soy lipid-derived phytosterol, is an antagonist of the bile acid nuclear receptor FXR. Pediatr Res. 2007;62:301–306. doi: 10.1203/PDR.0b013e3181256492. [DOI] [PubMed] [Google Scholar]
- 8.Clayton PT, Whitfield P, Iyer K. The role of phytosterols in the pathogenesis of liver complications of pediatric parenteral nutrition. Nutrition. 1998;14:158–164. doi: 10.1016/s0899-9007(97)00233-5. [DOI] [PubMed] [Google Scholar]
- 9.Diamond IR, Sterescu A, Pencharz PB, Kim JH, Wales PW. Changing the paradigm: omegaven for the treatment of liver failure in pediatric short bowel syndrome. J Pediatr Gastroenterol Nutr. 2009;48:209–215. doi: 10.1097/MPG.0b013e318182c8f6. [DOI] [PubMed] [Google Scholar]
- 10.Le HD, de Meijer VE, Robinson EM, Zurakowski D, Potemkin AK, Arsenault DA, et al. Parenteral fish-oil-based lipid emulsion improves fatty acid profiles and lipids in parenteral nutrition-dependent children. Am J Clin Nutr. 2011;94:749–758. doi: 10.3945/ajcn.110.008557. [DOI] [PubMed] [Google Scholar]
- 11.Yang Q, Ayers K, Welch CD, O'Shea TM. Randomized Controlled Trial of Early Enteral Fat Supplement and Fish Oil to Promote Intestinal Adaptation in Premature Infants with an Enterostomy. J Pediatr. 2014 doi: 10.1016/j.jpeds.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Human Microbiome Project C. Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–214. doi: 10.1038/nature11234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nat Rev Immunol. 2009;9:313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu T, Hougen H, Vollmer AC, Hiebert SM. Gut bacteria profiles of Mus musculus at the phylum and family levels are influenced by saturation of dietary fatty acids. Anaerobe. 2012;18:331–337. doi: 10.1016/j.anaerobe.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Turnbaugh PJ, Ridaura VK, Faith JJ, Rey FE, Knight R, Gordon JI. The effect of diet on the human gut microbiome: a metagenomic analysis in humanized gnotobiotic mice. Sci Transl Med. 2009;1:6ra14. doi: 10.1126/scitranslmed.3000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 2013;41:D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeSantis TZ, Hugenholtz P, Larsen N, Rojas M, Brodie EL, Keller K, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–5072. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paulson JN, Stine OC, Bravo HC, Pop M. Differential abundance analysis for microbial marker-gene surveys. Nat Methods. 2013;10:1200–1202. doi: 10.1038/nmeth.2658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.McMurdie PJ, Holmes S. phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS One. 2013;8:e61217. doi: 10.1371/journal.pone.0061217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gu C. Smoothing Spline ANOVA Models: R Package gss. J Stat Softw. 2014;58:1–25. [Google Scholar]
- 23.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langille MG, Zaneveld J, Caporaso JG, McDonald D, Knights D, Reyes JA, et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat Biotechnol. 2013;31:814–821. doi: 10.1038/nbt.2676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Drell T, Lutsar I, Stsepetova J, Parm U, Metsvaht T, Ilmoja ML, et al. The development of gut microbiota in critically ill extremely low birth weight infants assessed with 16S rRNA gene based sequencing. Gut Microbes. 2014;5:304–312. doi: 10.4161/gmic.28849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith A, Saiman L, Zhou J, Della-Latta P, Jia H, Graham PL., 3rd Concordance of Gastrointestinal Tract Colonization and Subsequent Bloodstream Infections With Gram-negative Bacilli in Very Low Birth Weight Infants in the Neonatal Intensive Care Unit. Pediatr Infect Dis J. 2010;29:831–835. doi: 10.1097/INF.0b013e3181e7884f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stoll BJ, Hansen N, Fanaroff AA, Wright LL, Carlo WA, Ehrenkranz RA, et al. Late-onset sepsis in very low birth weight neonates: the experience of the NICHD Neonatal Research Network. Pediatrics. 2002;110:285–291. doi: 10.1542/peds.110.2.285. [DOI] [PubMed] [Google Scholar]
- 28.Cole CR, Frem JC, Schmotzer B, Gewirtz AT, Meddings JB, Gold BD, et al. The rate of bloodstream infection is high in infants with short bowel syndrome: relationship with small bowel bacterial overgrowth, enteral feeding, and inflammatory and immune responses. J Pediatr. 2010;156:941–947. 7, e1. doi: 10.1016/j.jpeds.2009.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kakiyama G, Pandak WM, Gillevet PM, Hylemon PB, Heuman DM, Daita K, et al. Modulation of the fecal bile acid profile by gut microbiota in cirrhosis. J Hepatol. 2013;58:949–955. doi: 10.1016/j.jhep.2013.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swann JR, Want EJ, Geier FM, Spagou K, Wilson ID, Sidaway JE, et al. Systemic gut microbial modulation of bile acid metabolism in host tissue compartments. Proc Natl Acad Sci U S A. 2011;108:4523–4530. doi: 10.1073/pnas.1006734107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sayin SI, Wahlstrom A, Felin J, Jantti S, Marschall HU, Bamberg K, et al. Gut microbiota regulates bile acid metabolism by reducing the levels of tauro-beta-muricholic acid, a naturally occurring FXR antagonist. Cell Metab. 2013;17:225–235. doi: 10.1016/j.cmet.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 32.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, deRoos P, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, Takahashi D, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 34.Atarashi K, Tanoue T, Shima T, Imaoka A, Kuwahara T, Momose Y, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peng L, Li ZR, Green RS, Holzman IR, Lin J. Butyrate enhances the intestinal barrier by facilitating tight junction assembly via activation of AMP-activated protein kinase in Caco-2 cell monolayers. J Nutr. 2009;139:1619–1625. doi: 10.3945/jn.109.104638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jump DB. N-3 polyunsaturated fatty acid regulation of hepatic gene transcription. Curr Opin Lipidol. 2008;19:242–247. doi: 10.1097/MOL.0b013e3282ffaf6a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Svegliati-Baroni G, Candelaresi C, Saccomanno S, Ferretti G, Bachetti T, Marzioni M, et al. A model of insulin resistance and nonalcoholic steatohepatitis in rats: role of peroxisome proliferator-activated receptor-alpha and n-3 polyunsaturated fatty acid treatment on liver injury. Am J Pathol. 2006;169:846–860. doi: 10.2353/ajpath.2006.050953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ghosh S, DeCoffe D, Brown K, Rajendiran E, Estaki M, Dai C, et al. Fish oil attenuates omega-6 polyunsaturated fatty acid-induced dysbiosis and infectious colitis but impairs LPS dephosphorylation activity causing sepsis. PLoS One. 2013;8:e55468. doi: 10.1371/journal.pone.0055468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu HN, Zhu J, Pan WS, Shen SR, Shan WG, Das UN. Effects of fish oil with a high content of n-3 polyunsaturated fatty acids on mouse gut microbiota. Arch Med Res. 2014;45:195–202. doi: 10.1016/j.arcmed.2014.03.008. [DOI] [PubMed] [Google Scholar]
- 40.Patterson E, RM OD, Murphy EF, Wall R, O OS, Nilaweera K, et al. Impact of dietary fatty acids on metabolic activity and host intestinal microbiota composition in C57BL/6J mice. Br J Nutr. 2014;111:1905–1917. doi: 10.1017/S0007114514000117. [DOI] [PubMed] [Google Scholar]
- 41.Kaliannan K, Wang B, Li XY, Kim KJ, Kang JX. A host-microbiome interaction mediates the opposing effects of omega-6 and omega-3 fatty acids on metabolic endotoxemia. Sci Rep. 2015;5:11276. doi: 10.1038/srep11276. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure 1, online. Operational taxonomic unit (OTU) counts within major bacterial phyla by study week and treatment group. An OTU is a cluster of sequences with a high degree of similarity (≥99% identity). Each point represents an OTU within the specified bacterial phyla. The overlying boxplots outline the interquartile range. Dotted rectangles indicate intervals with statistically different levels of a phylum between groups.
Figure 3, online. Principal coordinates analysis (PCoA) of the microbiome in control and treatment group infants over time. PCoA is a method to distill and visualize similarities and differences between microbiome sample sequencing data. The first principal component (PC1) accounts for as much of the variation in the data as possible. A. Box-plots of the first principle component (PC1) in control and treatment groups for each study week. B. PC1 values modeled for control and treatment groups, showing divergence over time. C. Difference in PC1 values in treatment and control group, showing that treatment and control groups become more dissimilar over time.
Figure 4, online. Hierarchical cluster dendrogram based on microbial co-occurrence networks in study infants.
Figure 6, online. Turquoise Microbial Module. A. Median module eigengene (ME) values over time in control and treatment groups. The ME represents the first principal component of the module. Error bars represent standard error of the mean. B. Abundance of five genera enriched in the turquoise module over time.
Figure 7, online. Alpha-diversity and richness over time. Measures of richness (Chao1) reflect the number of different types of bacteria (OTUs) present in a sample, whereas alpha-diversity indices (Shannon, Inverse Simpson) also take into account differences in the proportional abundance (ie. the evenness of distribution) of the different OTUs within the sample. Means and standard errors are shown. Dotted rectangles indicate intervals with statistically significant differences between treatment and control groups by SS-ANOVA.