Abstract
Objective
A major barrier to genetic studies of OA is the need to obtain large numbers of individuals with standardized radiographic evaluations for OA. To address this gap, we performed a genome-wide association study (GWAS) of radiographically-defined tibiofemoral knee OA in 3,898 cases and 3,168 controls from four well-characterized North American cohorts, and replication analysis of published OA loci.
Methods
We performed meta-analysis using a two-stage design. Stage 1 (discovery) consisted of a GWAS meta-analysis of radiographic knee OA carried out in the Osteoarthritis Initiative and the Johnston County Osteoarthritis Project. Knee OA was defined as definitive osteophytes and possible joint space narrowing or total joint replacement in one or both knees. Stage 2 (validation) was performed in the Multicenter Osteoarthritis Study and Genetics of Osteoarthritis Study. We genotyped lead meta-analysis variants (P-value<1×10−4) from Stage 1 and tested the association between these variants and knee OA. We then combined results from all cohorts in a meta-analysis.
Results
Lead variants from Stage 1, representing 49 unique loci, were analyzed in Stage 2; none met genome-wide significance in the combined analysis of Stage 1 and 2. We validated one locus with nominal significance (P-value<0.05), which was also our top finding in the combined meta-analysis: rs4867568 (LSP1P3, OR[95% CI]=0.84[0.79–0.91], P-value=3.02×10−6). We observed nominally significant associations (P-value<0.05) with two published OA loci: rs143383 (GDF5, OR[95% CI]=1.12[1.04–1.21], P-value=2.13×10−3) and rs1558902 (FTO, OR[95% CI]=1.10[1.02–1.18], P-value=0.01).
Conclusion
These findings provide suggestive evidence for a novel knee OA locus and confirm previously published associations in GDF5 and FTO.
It has long been recognized that there is a strong genetic component to osteoarthritis (OA) as evidenced by the clustering of hand osteoarthritis within families (1). More recent twin studies have estimated that the heritability of hip and knee OA is 60% for hip OA (2) and 39% for knee OA (3). These estimates suggest that genetic factors may play a large role in the development of OA, although this may differ by joint site (4). Large-scale genome-wide association studies (GWAS) of hand, hip, and knee osteoarthritis have been conducted in European Caucasians, providing a dozen genome-wide significant loci that include ALDH1A2 for hand OA (5), DOT1L, NCOA3, ASTN2, FILIP1/SENP6, KLHDC5/PTHLH, and CHST11 for hip OA (6–9), and GDF5, chromosome 7q22, and MCF2L for knee OA (10–13). Variants in two other genes, GLT8D1 and GNL3, have been associated at genome-wide levels of significance with total joint replacement (9).
Despite numerous efforts to identify genetic factors associated with OA, robust replication of findings has been difficult. This is likely due to the highly heterogeneous nature and phenotype specificity of OA, as well as potentially different environmental effects on weight-bearing and non-weight-bearing joint sites (14). The genetic architecture of OA likely involves many loci, each having small effect sizes. Nevertheless, identifying even small effect size loci may provide insights into aspects of etiology and pathogenesis of OA that in some cases may suggest targets for prevention and treatment. A major barrier to large-scale genetic studies of OA has been the difficulty in obtaining large numbers of subjects who have undergone rigorous phenotyping using standardized evaluation. To address this gap, we performed a two-stage genome-wide association study (GWAS) of radiographically-defined tibiofemoral knee OA in 3,898 cases and 3,168 controls from four well-characterized North American OA cohorts. We also evaluated evidence for the association of knee OA with previously published OA loci.
PATIENTS AND METHODS
We conducted a GWAS of radiographic tibiofemoral knee OA in Caucasian subjects using a two-stage design. Stage 1 (discovery) consisted of a GWAS meta-analysis carried out in two independent populations: the Osteoarthritis Initiative (OAI) and the Johnston County Osteoarthritis Project (JoCo). In Stage 2 (validation), lead SNPs from the most significantly associated loci identified from the meta-analysis were genotyped and tested for association in two independent cohorts: the Multicenter Osteoarthritis Study (MOST) and The Genetics of Osteoarthritis (GO) study. Finally, we conducted a full meta-analysis of the lead SNPs by combining study results from all four cohorts. Knee OA was evaluated with fixed-flexion posteroanterior (PA) radiographs. Definitive knee OA was defined as having definite osteophytes and possible joint space narrowing (Kellgren-Lawrence [KL] grade ≥ 2) or total joint replacement in one or both knees. We defined controls as having in both knees no or doubtful evidence for OA (KL grade = 0 or 1) at all available time points. The same definition for cases and controls were used in Stage 1 and Stage 2 analyses.
Stage 1 cohorts included 2,672 cases (2,014 from OAI and 658 from JoCo) and 1,776 controls (953 from OAI and 823 from JoCo). Discovery loci that met suggestive evidence (P-value < 1×10−4) in Stage 1 were brought forward for de novo genotyping in Stage 2 cohorts. Stage 2 cohorts included 1,226 cases (709 from MOST and 517 from GO) and 1,392 controls (405 from MOST and 987 from GO). In total, there were 3,898 cases and 3,168 controls. We considered associations to be genome-wide significant if they reached 5 × 10−8 in the combined meta-analysis of Stage 1 and Stage 2 results.
Discovery cohorts
The Osteoarthritis Initiative (OAI) is a prospective longitudinal study designed to identify risk factors for the incidence and progression of symptomatic tibiofemoral knee OA. A total of 4,796 men and women of any race/ethnicity aged 45 – 79 years were enrolled into pre-defined progression or incidence subcohorts (15). Briefly, the progression subcohort included individuals who had symptomatic radiographic knee OA while the incidence subcohort included individuals who were considered to be at increased risk for developing symptomatic radiographic knee OA based on weight, knee symptoms, history of knee injuries/surgeries, family history of knee replacement and hand OA. Participants were recruited at four different clinical sites: 1) Brown University (Providence, RI); 2) The Ohio State University (Columbus, OH); 3) University of Maryland and The Johns Hopkins University (Baltimore, MD); and 4) University of Pittsburgh (Pittsburgh, PA). Details of the study protocol, including recruitment procedures and eligibility criteria are available on the OAI web site (http://oai.epi-ucsf.org/datarelease/docs/StudyDesignProtocol.pdf).
A total of 4,492 subjects aged 45–79 years received bilateral PA weight-bearing fixed-flexion knee radiograph at baseline between 2004 and 2006 and were invited back to assess incidence or progression of OA annually for up to 96 months. We restricted analyses to centrally read annual assessments up to 48 months. Central image assessment data releases were version 0.5 for baseline, 1.5 for 12-month, 3.4 for 24-month, 5.4 for 36-month, and 6.2 for 48-month visits. For this study, radiographs obtained at baseline were used to define OA cases; baseline and follow-up radiographs were used to define controls.
The Johnston County Osteoarthritis Project (JoCo) is an ongoing, community-based study of the occurrence of knee and hip OA in African American and Caucasian residents, aged 45 years and above, in a rural county in North Carolina. A detailed description of participant recruitment has been reported (16). Briefly, participants were recruited by probability sampling, with oversampling of African Americans. A total of 3,068 individuals were recruited at baseline. Similar to the OAI, cases in JoCo were defined at baseline (1991–1998), and controls were required to be OA-free at baseline and up to two subsequent follow-up exams (1999–2004 and 2006–2010). The current analysis includes 1,481 Caucasian participants with genotype data and radiographic information obtained from weight-bearing anteroposterior (AP) extended radiographs at baseline and fixed-flexion PA radiographs at follow-up. There is substantial agreement by KL grade between AP extended and PA fixed-flexion radiographs (17).
Validation cohorts
The Multicenter Osteoarthritis Study (MOST) is a longitudinal, prospective, observational study of knee OA in older individuals from the general population who either have OA or are at increased risk for developing knee OA based on weight, knee symptoms, or history of knee injuries/surgeries (18). Additional information regarding recruitment and study protocols are available on the MOST website (http://most.ucsf.edu/default.asp). A total of 3,026 participants were enrolled. Baseline examinations began in 2003 and follow-up visits were attempted at 15, 30, 60, and 84 months after the initial visit to collect clinical measurements and radiological data. Similar to the OAI and JoCo, cases were defined at baseline and controls were required to be OA-free at baseline and subsequent follow-up visits up to 60 months.
The Genetics of Osteoarthritis (GO) Study is a case-control based genetic association study of OA. The goal of this study was to identify genetic variations associated with OA, with careful attention to rigorous phenotyping of controls in the same manner as the cases (19). The GO study recruited approximately 1,000 OA participants with hand osteoarthritis with or without OA in knees, hips, and lumbosacral spine in the Caucasian population and 1,000 unaffected controls with similar age, gender and ethnicity.
Genotyping
The OAI was genotyped on the Illumina Omni-Quad 2.5M array at the Translational Genomics Research Institute (Phoenix, AZ) and the JoCo study was genotyped on the Illumina Infinium 1M-Duo bead array at Expression Analysis (Morrisville, NC). Genotypes for both studies were called using the Illumina BeadStudio software. The total number of genotyped SNPs was 2,440,283 in OAI and 1,199,187 in JoCo.
Samples that had call rates across all SNPs of <95% were removed (185 samples in OAI and 13 samples in JoCo). We additionally excluded from analysis potentially problematic samples based on 1) apparent mismatches between self-reported and genetically determined gender or 2) detection of second degree or higher relationships with other samples (56 samples in OAI and 40 samples in JoCo). An additional 34 OAI samples were excluded in whom we detected large chromosomal abnormalities using Log R Ratio (LRR) and B Allele Frequency (BAF), as described by others (20–22). Genotypes for both cohorts were imputed to the 1000 genomes CEU reference panel (June 2011 release) using the Minimac software program (http://genome.sph.umich.edu/wiki/Minimac), resulting in a total of 8,248,570 and 8,349,255 imputed SNPs in the OAI and JoCo, respectively, available for analyses after removal of SNPs with low minor allele frequencies (<1%) and poor imputation quality scores (<0.3).
Genotyping for Stage 2 was performed at the Translational Genomics Research Institute (Phoenix, AZ) on a customized Illumina array, which was designed to capture 49 top loci (P-value ≤ 1×10−4) identified in Stage 1 and previously reported OA variants from large-scale genome-wide association studies (9, 23). We removed SNPs with call rates <99% and samples with genotyping rates <97%. Furthermore, SNPs with extreme deviation from Hardy-Weinberg equilibrium (P-value < 1×10−7) and minor allele frequencies <1% were removed.
Statistical analysis
Association analysis for the OAI and JoCo were conducted using PLINK and ProbABEL, respectively, with adjustment for baseline age, sex, study site, and principal components (PCs). PCs estimated from the genetic data were included to account for unobserved population sub-structure and were derived from the genome-wide SNPs, following LD pruning and removal of SNPs with minor allele frequencies <5%. Only SNPs that had minor allele frequencies >1% and imputation quality score >0.3 were included in Stage 1 analyses. Inverse variance fixed-effects meta-analysis was carried out using METAL (24), which weights the contribution of both studies by the observed standard error. Heterogeneity between studies was assessed using Cochran’s Q statistic. We performed a meta-analysis of genome-wide association results from the OAI and JoCo and brought forward the most strongly associated loci (P-value < 10−4) for de novo genotyping on a customized array in two independent cohorts, MOST and GO. These top loci were pruned so that only the most significant SNP in a pair of SNPs in high linkage disequilibrium (LD), r2 > 0.8, was selected. If the selected SNP could not be genotyped, then the second most significant SNP was selected or a proxy in high LD, r2 > 0.8, was chosen as a tag SNP. We also performed association analysis of previously reported OA loci including three knee OA loci (10–13) and eight loci from the arcOGEN (Arthritis Research Council Osteoarthritis Genetics) study (9). After validation analyses of top loci and previously reported loci in Stage 2, we then combined results from Stage 1 and Stage 2 in a meta-analysis to determine whether any loci from Stage 1 could be elevated to genome-wide significance with the addition of data from Stage 2. We also performed secondary analyses additionally adjusting for body mass index (BMI) and history of knee injury or surgery.
We estimated that our two-stage design totaling 3,898 cases and 3,168 controls provided 80% power to detect odds ratios ranging from 1.24 – 1.39 for OA-associated SNPs at an alpha = 5×10−8 (i.e., conventional thresholds for genome-wide statistical significance) across a range of allele frequencies, and odds ratios ranging from 1.13 – 1.24 for OA-associated SNPs at an alpha = 1×10−5.
RESULTS
Our sample included 2,672 cases and 1,776 controls in Stage 1 (discovery), and 1,226 cases and 1,392 controls in Stage 2 (validation); all were self-reported Caucasians. In total, there were 3,898 cases and 3,168 controls in the full meta-analysis across Stages 1 and 2. Clinical characteristics of Stage 1 (OAI and JoCo) and Stage 2 (MOST and GO) samples are provided in Table 1. OA cases had a mean age of 63–64 years in OAI, JoCo, and MOST, and 72 years in GO. Cases were more likely than controls to be women in both the OAI (56% vs. 54%) and MOST (60% vs. 54%). The proportion of women was similar between cases and controls in JoCo (61%) and GO (70%). Across all studies, cases had a higher body mass index than controls (OAI: 29.0 vs. 27.0 kg/m2, JoCo: 30.2 vs. 28.0 kg/m2, MOST: 31.4 vs. 28.4 kg/m2, GO: 28.9 vs. 26.9 kg/m2). Also, cases in all studies had a higher proportion of individuals who had a history of knee injury (OAI: 50.8% vs. 38.5%, JoCo: 27.2% vs 15.9%, MOST: 53.5% vs. 36.7%, GO: 14.3% vs. 6.0%) or knee surgery (OAI: 32.2% vs. 12.2%, JoCo: 15.4% vs 2.6%, MOST: 35.0% vs. 11.1%, GO: 20.9% vs. 4.6%).
Table 1.
Baseline characteristics of OAI, JoCo, MOST, and GO study participants
| Stage 1: Discovery | Stage 2: Validation | |||||||
|---|---|---|---|---|---|---|---|---|
| OAI | JoCo | MOST | GO | |||||
| Cases (n = 2,014) |
Controls (n = 953) |
Cases (n =658) |
Controls (n = 823) |
Cases (n=709) |
Controls (n=405) |
Cases (n=517) |
Controls (n=987) |
|
| Mean age (SD) in years at baseline |
63.1 (9.0) | 59.5 (9.1) | 63.1 (10.4) | 58.3 (9.1) | 64.3 (7.7) | 60.8 (7.2) | 72.5 (8.3) | 68.0 (7.8) |
| % female | 55.7 | 54.0 | 61.1 | 61.2 | 59.9 | 54.1 | 69.6 | 70.5 |
| Mean BMI (SD) in kg/m2 |
29.0 (4.6) | 27.0 (4.3) | 30. 2 (6.0) | 28.0 (5.3) | 31.4 (5.9) | 28.4 (4.3) | 28.9 (5.6) | 26.9 (4.7) |
| Mean weight (SD) in kg |
82.9 (16.1) | 77.3 (15.9) | 83.9 (19.0) | 77.8 (17.5) | 89.5 (18.7) | 82.9 (15.7) | 79.0 (16.6) | 73.4 (15.4) |
| % Current smokers |
4.8 | 4.0 | 12.3 | 22.1 | 11.2 | 11.1 | NA | NA |
| % History of prior knee surgery |
32.2 | 12.2 | 15.4 | 2.6 | 35.0 | 11.1 | 20.9 | 4.6 |
| % History of prior knee injury |
50.8 | 38.5 | 27.2 | 15.9 | 53.5 | 36.7 | 14.3 | 6.0 |
| % Frequent knee pain** |
59.0 | 47.3 | 54.4 | 33.8 | 65.0 | 34.6 | NA | NA |
Knee pain, aching, or stiffness in or around the knee on most days of the week during the past month
Stage 1 (discovery)
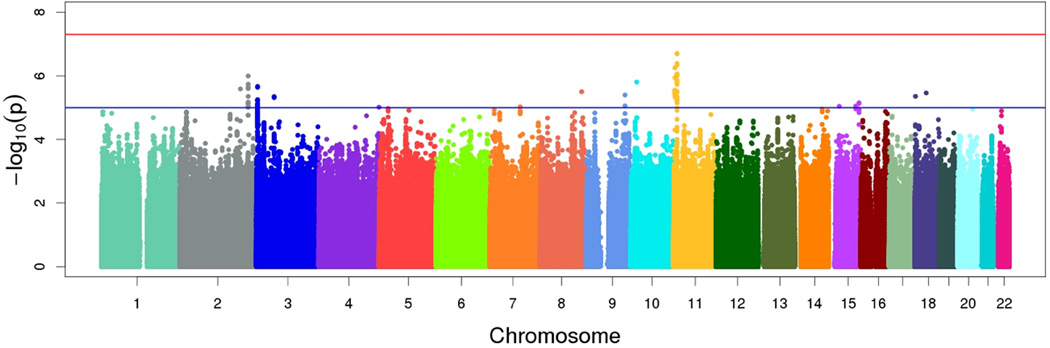
Results of Stage 1 genome-wide association meta-analysis of knee OA in OAI and JoCo are summarized in the Manhattan plot shown in Figure 1. No SNP achieved genome-wide significance (i.e., 5×10−8). The most significant finding (rs274508, OR [95% CI]=0.77 [0.70–0.85], P-value=2.00×10−7) was located within an intergenic region on chromosome 11 between ZBED5 (zinc finger BED domain-containing protein 5) and GALNT18 (polypeptide N-Acetylgalactosaminyltransferase 18). The minor allele frequency at this SNP was 0.32.
Figure 1.
Manhattan plot for Stage 1 (discovery) GWAS. The red horizontal line depicts genome-wide significance (P-value < 5×10−8). The blue horizontal line depicts suggestive significance (P-value < 1×10−5).
Following genome-wide association analyses in the discovery set, we identified 823 SNPs associated with knee OA at a P-value threshold ≤ 1×10−4. We used linkage disequilibrium pruning (removing SNPs with r2 < 0.80 to an already captured SNP) to reduce this number to 49 uncorrelated SNPs. We carried forward these 49 unique loci for de novo genotyping in MOST and GO.
Stage 2 (validation)
Of the 49 SNPs tested, five were associated at nominal levels of statistical significance (P-value < 0.05) in Stage 2, including rs4867568 near LSP1P3 (OR [95% CI]=0.88 [0.78–0.99]), rs1026407 in COL27A1 (OR [95% CI]=1.57 [1.09–2.27]), rs1026407 in UBE2E1 (OR [95% CI]=1.16 [1.01–1.32]), rs1628543 near TBK1/RASSF3 (OR [95% CI]=1.13[1.00–1.27]), and rs6892607 near FAM173B (OR [95% CI]=0.80 [0.68–0.94]) (Table 2). However, only one of these SNPs, rs4867568, was validated in the same direction of effect as in Stage 1, yielding a combined Stage 1 and Stage 2 meta-analysis P-value of 3.02×10−6 (OR [95% CI]=0.84 [0.79–0.91]). None of the nominally validated SNPs (nor any of the other SNPs from Stage 1) achieved genome-wide levels of significance in the combined Stage 1 and 2 meta-analysis.
Table 2.
Combined Stage 1 and Stage 2 meta-analysis, adjusted for age, sex, study site, and PCs
| Stage 1: Discovery (OAI and JoCo) |
Stage 2: Validation (MOST and GO) |
Stage 1+2: Meta-analysis (OAI, JoCo, MOST, and GO) |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Marker | Chr | Nearest Gene(s) | A1/ A2 |
A1 freq |
OR (95% CI) |
P-value | OR (95% CI) |
P-value | OR (95% CI) |
P-value |
| rs4867568 | 5 | LSP1P3 | t/c | 0.50 | 0.82 (0.75–0.90) |
1.97E-05 | 0.88 (0.78–0.99) |
0.04 | 0.84 (0.79–0.91) |
3.02E-06 |
| rs1352413 | 7 | DYNC1I1 | a/g | 0.07 | 1.51 (1.26–1.81) |
9.40E-06 | 1.22 (0.97–1.54) |
0.09 | 1.39 (1.21–1.61) |
5.58E-06 |
| rs274508 | 11 | ZBED5/ GALNT18 |
a/c | 0.68 | 0.77 (0.70–0.85) |
2.00E-07 | 0.96 (0.84–1.09) |
0.53 | 0.84 (0.77–0.90) |
6.04E-06 |
| rs7079380 | 10 | FRMD4A | c/g | 0.19 | 0.78 (0.70–0.88) |
3.00E-05 | 0.88 (0.76–1.03) |
0.10 | 0.82 (0.75–0.90) |
1.59E-05 |
| rs11258527 | 10 | FRMD4A | a/g | 0.11 | 0.72 (0.61–0.84) |
5.23E-05 | 0.85 (0.70–1.03) |
0.09 | 0.77 (0.68–0.87) |
2.78E-05 |
| rs28480016 | 16 | WWOX | t/c | 0.88 | 1.38 (1.19–1.59) |
1.32E-05 | 1.08 (0.89–1.30) |
0.45 | 1.26 (1.12–1.41) |
9.12E-05 |
| rs7636780 | 3 | CRBN/LRRN1 | t/c | 0.76 | 1.29 (1.16–1.43) |
2.30E-06 | 1.01 (0.87–1.16) |
0.93 | 1.18 (1.08–1.28) |
1.23E-04 |
| rs3829930 | 10 | FRMD4A | t/c | 0.27 | 0.81 (0.73–0.89) |
2.74E-05 | 0.95 (0.84–1.09) |
0.49 | 0.86 (0.79–0.93) |
1.72E-04 |
| rs974515 | 3 | CRBN/LRRN1 | t/c | 0.28 | 0.79 (0.72–0.88) |
5.61E-06 | 0.99 (0.86–1.13) |
0.85 | 0.86 (0.79–0.93) |
1.80E-04 |
| rs6920712 | 6 | ZBTB24/AK9 | t/c | 0.26 | 1.24 (1.12–1.37) |
5.68E-05 | 1.06 (0.93–1.21) |
0.40 | 1.17 (1.08–1.27) |
2.18E-04 |
| rs60278383 | 2 | LINC00607 | c/g | 0.10 | 1.48 (1.27–1.74) |
1.02E-06 | 0.99 (0.83–1.19) |
0.94 | 1.25 (1.11–1.41) |
2.61E-04 |
| rs73210326 | 3 | XXYLT1 | t/c | 0.77 | 0.79 (0.71–0.88) |
4.00E-05 | 0.96 (0.83–1.11) |
0.54 | 0.85 (0.78–0.93) |
2.90E-04 |
| rs7111344 | 11 | TRIM21/OR52K2 | a/g | 0.07 | 0.65 (0.55–0.77) |
5.63E-07 | 1.07 (0.86–1.34) |
0.53 | 0.78 (0.68–0.89) |
3.30E-04 |
| rs35672585 | 2 | STAT4 | a/g | 0.19 | 1.37 (1.20–1.56) |
2.58E-06 | 1.00 (0.85–1.16) |
0.95 | 1.20 (1.08–1.33) |
3.80E-04 |
| rs709339 | 3 | ERC2 | a/t | 0.64 | 1.25 (1.14–1.38) |
4.85E-06 | 0.99 (0.87–1.12) |
0.87 | 1.15 (1.06–1.24) |
4.27E-04 |
| rs17651945 | 15 | LOC101929701 | a/g | 0.95 | 1.60 (1.30–1.97) |
1.10E-05 | 0.99 (0.75–1.32) |
0.96 | 1.35 (1.14–1.60) |
4.32E-04 |
| rs3788387 | 22 | SEZ6L | t/g | 0.56 | 0.82 (0.74–0.89) |
1.27E-05 | 1.00 (0.89–1.12) |
0.98 | 0.88 (0.82–0.95) |
5.56E-04 |
| rs2781102 | 9 | BRINP1/ MIR147A |
a/g | 0.53 | 1.24 (1.13–1.36) |
4.01E-06 | 0.98 (0.88–1.10) |
0.77 | 1.13 (1.06–1.22) |
5.83E-04 |
| rs62447871 | 7 | ARL4A/ETV1 | a/t | 0.50 | 0.80 (0.72–0.88) |
1.55E-05 | 0.99 (0.88–1.11) |
0.85 | 0.88 (0.81–0.95) |
7.16E-04 |
| rs73226453 | 8 | ENTPD4/ SLC25A37 |
c/g | 0.96 | 1.65 (1.30–2.10) |
4.09E-05 | 1.00 (0.72–1.40) |
0.99 | 1.39 (1.15–1.69) |
8.30E-04 |
| rs1396415 | 3 | CRBN/LRRN1 | a/g | 0.20 | 0.78 (0.70–0.88) |
1.97E-05 | 1.02 (0.88–1.19) |
0.79 | 0.86 (0.79–0.94) |
1.03E-03 |
| rs4984434 | 15 | NR2F2 | a/g | 0.75 | 0.78 (0.70–0.87) |
1.43E-05 | 1.01 (0.89–1.16) |
0.83 | 0.87 (0.80–0.95) |
1.28E-03 |
| chr14:87364089 | 14 | LOC101928767/ LOC283585 |
a/g | 0.01 | 0.37 (0.24–0.58) |
1.11E-05 | 0.99 (0.64–1.54) |
0.96 | 0.61 (0.44–0.83) |
1.61E-03 |
| rs6963954 | 7 | NOD1 | a/g | 0.93 | 0.67 (0.55–0.80) |
1.87E-05 | 1.02 (0.82–1.27) |
0.85 | 0.80 (0.69–0.92) |
1.70E-03 |
| rs10189819 | 2 | STAT4 | t/c | 0.80 | 0.77 (0.68–0.87) |
2.62E-05 | 1.01 (0.87–1.17) |
0.88 | 0.86 (0.78–0.95) |
1.71E-03 |
| rs186095 | 5 | MCTP1 | a/c | 0.92 | 0.68 (0.57–0.81) |
1.22E-05 | 1.04 (0.84–1.28) |
0.71 | 0.81 (0.71–0.92) |
1.75E-03 |
| rs17089295 | 8 | ENTPD4/ SLC25A37 |
a/g | 0.04 | 0.61 (0.48–0.78) |
6.27E-05 | 1.05 (0.76–1.47) |
0.76 | 0.74 (0.61–0.90) |
2.13E-03 |
| rs6005018 | 22 | SEZ6L | t/c | 0.63 | 0.82 (0.75–0.90) |
4.43E-05 | 1.02 (0.90–1.15) |
0.79 | 0.89 (0.83–0.96) |
2.29E-03 |
| rs4908382 | 1 | SMPDL3B | a/g | 0.38 | 0.82 (0.75–0.90) |
4.84E-05 | 1.02 (0.90–1.16) |
0.72 | 0.89 (0.82–0.96) |
2.54E-03 |
| rs17436401 | 16 | RBFOX1 | t/c | 0.66 | 0.82 (0.74–0.90) |
4.18E-05 | 1.02 (0.90–1.15) |
0.74 | 0.89 (0.83–0.96) |
2.61E-03 |
| rs16857702 | 2 | DIRC3 | t/g | 0.03 | 0.57 (0.43–0.75) |
7.15E-05 | 1.10 (0.76–1.60) |
0.60 | 0.72 (0.58–0.90) |
4.12E-03 |
| chr16:79894219 | 16 | LOC101928248/ LOC102724084 |
a/g | 0.02 | 0.43 (0.29–0.64) |
2.88E-05 | 1.17 (0.72–1.88) |
0.53 | 0.64 (0.48–0.87) |
4.62E-03 |
| rs6664358 | 1 | AJAP1/MIR4417 | t/c | 0.55 | 0.82 (0.75–0.90) |
1.66E-05 | 1.07 (0.95–1.20) |
0.29 | 0.90 (0.84–0.97) |
5.53E-03 |
| rs73105763 | 3 | FOXP1 | a/c | 0.08 | 1.48 (1.23–1.79) |
3.18E-05 | 0.94 (0.75–1.16) |
0.54 | 1.22 (1.06–1.40) |
5.88E-03 |
| rs7075619 | 10 | ADRA2A/GPAM | t/g | 0.25 | 0.81 (0.73–0.90) |
8.22E-05 | 1.04 (0.91–1.19) |
0.55 | 0.89 (0.82–0.97) |
6.16E-03 |
| rs11096655 | 2 | PUM2 | a/g | 0.31 | 0.81 (0.74–0.90) |
2.58E-05 | 1.06 (0.94–1.20) |
0.33 | 0.90 (0.83–0.97) |
6.53E-03 |
| rs2900162 | 9 | BRINP1/ MIR147A |
t/c | 0.51 | 0.83 (0.75–0.90) |
3.10E-05 | 1.06 (0.94–1.19) |
0.34 | 0.91 (0.84–0.97) |
6.54E-03 |
| rs72862801 | 18 | METTL4 | t/c | 0.15 | 1.35 (1.17–1.57) |
6.97E-05 | 0.96 (0.82–1.13) |
0.65 | 1.16 (1.04–1.29) |
8.58E-03 |
| rs2184221 | 9 | BRINP1/ MIR147A |
a/g | 0.48 | 0.82 (0.75–0.90) |
1.53E-05 | 1.08 (0.96–1.21) |
0.21 | 0.91 (0.85–0.98) |
8.73E-03 |
| rs11844656 | 14 | RPS6KA5 | c/g | 0.48 | 0.83 (0.76–0.91) |
7.40E-05 | 1.05 (0.94–1.18) |
0.37 | 0.91 (0.85–0.98) |
0.01 |
| rs62355519 | 5 | PRLR | t/c | 0.08 | 0.73 (0.62–0.85) |
7.85E-05 | 1.16 (0.93–1.44) |
0.19 | 0.85 (0.75–0.97) |
0.02 |
| rs2040287 | 18 | MIR4318/ LINC00669 |
t/c | 0.03 | 0.50 (0.38–0.67) |
3.47E-06 | 1.28 (0.93–1.77) |
0.13 | 0.77 (0.62–0.95) |
0.02 |
| rs4756129 | 11 | ABTB2 | a/g | 0.66 | 0.79 (0.70–0.89) |
6.33E-05 | 1.06 (0.94–1.20) |
0.34 | 0.91 (0.83–0.99) |
0.02 |
| rs3104589 | 11 | CDON/RPUSD4 | t/c | 0.11 | 1.40 (1.18–1.65) |
7.26E-05 | 0.86 (0.71–1.06) |
0.15 | 1.15 (1.01–1.31) |
0.03 |
| rs79516393 | 11 | ABTB2 | a/g | 0.13 | 1.50 (1.23–1.84) |
8.32E-05 | 0.93 (0.78–1.11) |
0.40 | 1.14 (1.00–1.31) |
0.05 |
| rs3736250 | 9 | COL27A1 | a/g | 0.03 | 0.62 (0.48–0.79) |
9.10E-05 | 1.57 (1.09–2.27) |
0.02 | 0.82 (0.67–1.00) |
0.05 |
| rs1026407 | 3 | UBE2E1 | c/g | 0.74 | 0.81 (0.73–0.89) |
4.62E-05 | 1.16 (1.01–1.32) |
0.03 | 0.92 (0.85–1.00) |
0.06 |
| rs1628543 | 12 | TBK1/RASSF3 | a/g | 0.56 | 0.83 (0.76–0.91) |
8.94E-05 | 1.13 (1.00–1.27) |
0.05 | 0.93 (0.87–1.00) |
0.06 |
| rs6892607 | 5 | FAM173B | t/g | 0.16 | 1.30 (1.14–1.47) |
6.60E-05 | 0.80 (0.68–0.94) |
0.01 | 1.08 (0.97–1.19) |
0.15 |
In secondary analyses, we included additional adjustments for BMI and history of knee injury or surgery, which resulted in no to slight attenuation of top associations (P-value < 1×10−5) in the combined Stage 1 and 2 meta-analysis (Table 3). There were four loci that reached suggestive significance (P-value < 1×10−5) only after adjusting for BMI (rs7079380 and rs11258527 in FRMD4A) and history of knee injury/surgery (rs6963954 in NOD1 and rs974515 near CRBN/LRRN1) (Table 3).
Table 3.
Associations with knee OA adjusted for BMI and knee injury/surgery in the combined Stage 1 and 2 meta-analysis*
| Adjusted for age, sex, study site, and PCs |
Adjusted for age, sex, study site, PCs, and BMI |
Adjusted for age, sex, study site, PCs, and knee trauma |
||||||
|---|---|---|---|---|---|---|---|---|
| Marker | Chr | Nearest Gene(s) | OR (95% CI) |
P-value | OR (95% CI) |
P-value | OR (95% CI) |
P-value |
| rs4867568 | 5 | LINC01021/LSP1P3 | 0.84 (0.79–0.91) | 3.02E-06 | 0.84 (0.78–0.90) | 2.91E-06 | 0.86 (0.80–0.92) | 2.52E-05 |
| rs1352413 | 7 | DYNC1I1 | 1.39 (1.21–1.61) | 5.58E-06 | 1.37 (1.19–1.59) | 2.53E-05 | 1.31 (1.13–1.52) | 2.85E-04 |
| rs274508 | 11 | ZBED5/GALNT18 | 0.84 (0.77–0.90) | 6.04E-06 | 0.84 (0.77–0.91) | 1.84E-05 | 0.83 (0.77–0.90) | 5.39E-06 |
| rs7079380 | 10 | FRMD4A | 0.82 (0.75–0.90) | 1.59E-05 | 0.80 (0.73–0.88) | 5.79E-06 | 0.82 (0.75–0.90) | 3.74E-05 |
| rs11258527 | 10 | FRMD4A | 0.77 (0.68–0.87) | 2.78E-05 | 0.75 (0.66–0.85) | 7.22E-06 | 0.76 (0.67–0.86) | 1.60E-05 |
| rs6963954 | 7 | NOD1 | 0.80 (0.69–0.92) | 1.70E-03 | 0.79 (0.68–0.92) | 1.83E-03 | 0.71 (0.62–0.83) | 5.12E-06 |
| rs974515 | 3 | CRBN/LRRN1 | 0.86 (0.79–0.93) | 1.80E-04 | 0.85 (0.78–0.93) | 1.42E-04 | 0.83 (0.76–0.90) | 7.81E-06 |
Only SNPs that achieved a P-value < 1×10−5 in any one of these three adjusted analyses are presented
Replication of previously reported OA loci
We also tested associations between previously reported OA SNPs that were genome-wide significant or replicated across several studies (9–13), which were mostly conducted in European cohorts. We detected nominally significant associations (P-value < 0.05) with three of the 11 previously reported OA SNPs: rs143383 near GDF5 (OR [95% CI]=1.12 [1.04–1.21], P-value=2.13×10−3); rs835487 near CHST11 (OR [95% CI]=0.93 [0.85–0.99], P-value=0.03); and rs8044769 near FTO (OR [95% CI]=1.10 [1.03–1.19], P-value=6.13×10−3). Associations observed for GDF5 and FTO, but not CHST11, were directionally consistent with previously reported effects (Table 4). Notably, the odds ratios at all loci estimated in our study were smaller than the published results.
Table 4.
Associations between radiographic knee OA and previously reported OA loci, combined Stage 1 and 2 meta-analysis
| Previously reported associations from the literature |
Adjusted for age, sex, study site, and PCs |
Adjusted for age, sex, study site, PCs, and knee trauma |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Marker | Chr | Nearest Gene(s) |
A1/ A2 |
OR (95% CI) |
Trait | OR (95% CI) |
P-value | OR (95% CI) |
P-value |
| rs143383* | 20 | UQCC1/ GDF5 |
t/c | 1.16 (1.11–1.22) |
Knee | 1.12 (1.04–1.21) |
2.13E-03 | 1.15 (1.05–1.25) |
1.58E-03 |
| rs11842874 | 13 | MCF2L | a/g | 1.17 (1.11–1.23) |
Knee | 0.91 (0.79–1.04) |
0.16 | 0.91 (0.79–1.05) |
0.21 |
| rs4730250 | 7 | 7q22 (DUS4L) |
g/a | 1.18 (1.11–1.23) |
Knee | 1.02 (0.93–1.12) |
0.70 | 1.02 (0.92–1.12) |
0.75 |
| rs6976/ rs11177** |
3 | GLT8D1/ GNL3 |
t/c, a/g |
1.12 (1.08–1.16) |
TJR | 1.06 (0.98–1.14) |
0.14 | 1.03 (0.95–1.11) |
0.49 |
| rs4836732 | 9 | ASTN2 | c/t | 1.20 (1.14–1.27) |
THR-female | 0.96 (0.90–1.04) |
0.34 | 0.96 (0.89–1.03) |
0.25 |
| rs9350591† | 6 | FILIP1/ SENP6 |
t/c | 1.18 (1.12–1.25) |
Hip | -- | -- | -- | -- |
| rs10492367 | 12 | KLHDC5/ PTHLH |
t/g | 1.14 (1.09–1.20) |
Hip | 1.05 (0.96–1.16) |
0.30 | 1.08 (0.98–1.19) |
0.11 |
| rs835487 | 12 | CHST11 | g/a | 1.13 (1.09–1.18) |
THR | 0.93 (0.85–0.99) |
0.03 | 0.93 (0.86–1.00) |
0.06 |
| rs12107036 | 3 | TP63 | g/a | 1.21 (1.13–1.29) |
TKR-female | 1.01 (0.94–1.09) |
0.78 | 1.04 (0.96–1.12) |
0.34 |
| rs8044769 | 16 | FTO | c/t | 1.11 (1.07–1.15) |
Knee/hip- female |
1.10 (1.03–1.19) |
6.13E-03 | 1.11 (1.03–1.19) |
5.27E-03 |
| rs10948172 | 6 | SUPT3H/ CDC5L |
g/a | 1.14 (1.09–1.20) |
Male | 0.98 (0.91–1.06) |
0.69 | 0.96 (0.88–1.04) |
0.29 |
rs143383 was not genotyped in the validation cohorts, but is in LD with rs6087704 (r2=0.96), which was genotyped; meta-analysis association results are presented for rs6087704
rs11177 and rs6976 represent the same signal (r2=1)
rs9350591 was genotyped, but did not pass quality control
DISCUSSION
We performed the largest and most comprehensive GWAS of radiographic tibiofemoral knee OA yet to be carried out in North American Caucasians, incorporating four independent cohorts and a two-stage design. Our study provides suggestive evidence for a novel knee OA locus on chromosome 5p13 near LSP1P3 (lymphocyte-specific protein 1 pseudogene 3), which has not been identified by any other GWAS to date. In contrast to previously published large-scale OA GWAS, which largely included cases that underwent total joint replacement, our study focused particularly on tibiofemoral knee OA cases defined radiographically by definite osteophytes and possible joint space narrowing; less than 5% had total joint replacements. We also provide modest replication for two previously reported OA loci, including GDF5 and FTO. Even with a large sample size of 3,898 cases, we did not identify any loci at genome-wide levels of significance and replicated only two loci at nominal levels of significance. These findings highlight the polygenic nature of knee OA and the need for even larger studies to achieve sufficient power to detect small effect sizes.
Similar to other complex diseases, the genetic architecture of OA is characterized by many common genetic variants (minor allele frequencies greater than 5%) with small effect sizes. So far, only a dozen genome-wide significant loci have been identified for knee and hip OA in European Caucasians, ranging in effect sizes from 1.12 to 1.28 (25). The largest GWAS of OA to date was conducted by the arcOGEN (Arthritis Research Council Osteoarthritis Genetics) study in the United Kingdom, which included 7,410 cases and about 11,000 population controls (9). The arcOGEN study identified five novel loci at genome-wide significance (GLT8D1/GNL3, ASTN2, FILIP1/SENP6, KLHDC5/PTHLH, and CHST11) and three novel loci at near genome-wide significance (TP63, FTO, and SUPT3H/CDC5L). Of these eight loci, we replicated only one, the FTO locus, despite having enough statistical power to detect odds ratios greater than 1.10 at nominal significance. One possible explanation for this is that some of the OA-associated loci reported in arcOGEN were specific to hip OA and some were only significant in sub-analyses restricted to men or women. We also used a slightly different, less severe, radiographic tibiofemoral knee OA phenotype. Fewer than 5% of our cases had total knee replacements while the majority of arcOGEN cases (~80%) had total hip and/or knee replacements.
One of the major challenges facing genetic studies of knee OA is the high degree of phenotype heterogeneity that exists due to differences in phenotype definition. Recent efforts have focused on standardizing phenotype definitions across genetic studies of OA so as to reduce phenotype heterogeneity and facilitate replication (14). Both symptomatic and radiographic definitions are used in genetic studies of OA and may yield different associations. Furthermore, even within the commonly used KL grading system for radiographic OA, KL grades may be interpreted differently across cohorts. For example, in the TREAT-OA (Translational Research in Europe Applied Technologies for Osteoarthritis) consortium, there were at least three different interpretations of KL grade 2, including “one definite osteophyte”, “definite osteophytes”, and “definite osteophytes with possible joint space narrowing” (14). In our study of North American OA cohorts, radiographic knee OA was defined as definite osteophytes with possible joint space narrowing at the tibiofemoral joint, the classic interpretation of KL grade 2 knee OA (26). This definition was standardized across all cohorts to limit phenotype heterogeneity and improve power to detect genetic associations.
Another source of phenotype heterogeneity may be confounding by risk factors that contribute to OA liability. A significant proportion of cases and controls in this analysis had a history of previous knee injury, one of the primary risk factors for knee OA aside from advanced age, overweight, obesity, and female gender (27). There are several models by which knee trauma may affect genetic associations. One model may consider knee trauma as an independent risk factor for OA that does not operate through genetics and is itself sufficient for putting one on the path to OA. Under this model the most appropriate analysis would be not to adjust for history of knee trauma, as this assumes the impact of OA susceptibility loci would be identical in those with and without knee injury, but rather to exclude those with knee injury. A second model is that OA risk alleles influence OA risk independently of prior knee trauma. Under this model, adjustment for prior knee injury should not alter the effects of OA risk loci on OA risk. Yet a third model is that knee trauma provides a permissive milieu in which OA risk alleles are more likely to be expressed. Under this model, a follow-up analysis that adjusts for prior knee injury on OA risk should attenuate the effects of these loci. Associations with our top meta-analysis finding and replicated findings in GDF5 and FTO remained largely unchanged after adjusting for knee trauma, suggesting that these loci operate independently of prior knee trauma. While history of knee injury or surgery may not necessarily lead to post-traumatic OA, our findings are consistent with other reports that the genetic contribution to post-traumatic and non-traumatic knee OA may be similar (28).
The strength of our GWAS is in fact the careful phenotyping of all subjects using standardized radiography methods across all four meta-analysis cohorts and the availability of longitudinal data to radiographically confirm the absence of knee OA in controls at all follow-up exams over an average of five years. However, the time between the initiating event and onset of knee OA is often much longer. Controls were younger than cases and it may be only a matter of time before they develop knee OA. It is therefore possible that individuals classified as controls may eventually become cases at a later time point than we were able to capture from existing longitudinal data. This is especially relevant to OAI and MOST studies that ascertained participants who either have or are at high risk for symptomatic OA. Controls from OAI and MOST may be more likely to be misclassified as disease-free than controls from JoCo and GO, resulting in less power to detect significant associations. Including older, disease-free controls without predisposing risk factors would be ideal and help reduce misclassification bias.
While one of the strengths of our analysis was the standardized radiographic assessment of knee OA across all four studies, the major limitation was the small sample size. Even with nearly 4,000 cases, we were powered to detect odds ratios of only 1.24 to 1.39 at genome-wide levels of significance. Odds ratios for prior loci associated with knee OA range from 1.12 to 1.28. Since genome-wide genotyping was available for OAI and JoCo studies only, we were limited to a two-stage GWAS that has less power than a single-stage GWAS with the same number of samples. Also, Stage 2 was smaller in sample size than Stage 1 and would have less power to detect significant associations, further hampering replication. Additional large replication samples will be needed to increase sample size and power to elevate small-effect loci from suggestive to genome-wide significance.
In summary, we conducted the largest GWAS study of tibiofemoral knee OA in North American Caucasian OA cohorts to date based on standardized radiographic phenotypes. Our study validated two previously reported OA-associated loci in GDF5 and FTO, the latter likely exerting its effects through body mass index (29). The small effect sizes identified in this study are in line with the highly polygenic nature of knee OA, and even larger scale GWAS meta-analysis of knee OA will be needed to provide genome-wide statistical evidence.
Acknowledgments
We thank Uzma Atif, PhD, formerly of GlaxoSmithKline; William Maixner, DDS PhD and Shad Smith, PhD, Algynomics, Inc. for their contributions.
Funding sources: This study was supported by the American Recovery and Reinvestment Act (ARRA) through grant number RC2-AR-058950 from NIAMS/NIH. The OAI is public-private partnership comprised of five contracts (N01-AR-2-2258; N01-AR-2-2259; N01-AR-2-2260; N01-AR-2-2261; N01-AR-2-2262) funded by the NIH. The JoCo is supported in part by S043, S1734, & S3486 from the CDC/Association of Schools of Public Health; 5-P60-AR30701 & 5-P60-AR49465-03 from NIAMS/NIH; genotyping was supported by Algynomics, Inc. Additional support was obtained from NIH grant P30-DK072488. The MOST study was supported by U010AG18820, U01AG18832, U01AG18947, U01AG19069, and R01AG028359. The GO study was funded by GlaxoSmithKline. Study sponsors were not involved in the study design, analysis, or interpretation of the data for this analysis. MSY was supported by NIA/NIH T32AG00262, Arthritis Foundation Doctoral Dissertation Award (6081), NIA/NIH T32AG023480, and Friends of Hebrew SeniorLife.
Footnotes
Competing interests: None of the authors has any relevant conflicts of interest to report.
CONTRIBUTIONS
All authors were involved in the conception, design, analysis, and interpretation of the data. MSY and BDM drafted the article. LMY-A, YL, CEL, DJD, JBR, JT, DTF, CEM, CKK, MCN, MCH, JMJ, and RDJ critically revised the article for important intellectual content. All authors provided final approval of the article.
REFERENCES
- 1.Stecher RM. Heberden's nodes; the clinical characteristic of osteo-arthritis of the fingers. Ann Rheum Dis. 1948;7(1):1–8. [PMC free article] [PubMed] [Google Scholar]
- 2.MacGregor AJ, Antoniades L, Matson M, Andrew T, Spector TD. The genetic contribution to radiographic hip osteoarthritis in women: results of a classic twin study. Arthritis Rheum. 2000;43(11):2410–2416. doi: 10.1002/1529-0131(200011)43:11<2410::AID-ANR6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 3.Spector TD, Cicuttini F, Baker J, Loughlin J, Hart D. Genetic influences on osteoarthritis in women: a twin study. BMJ. 1996;312(7036):940–943. doi: 10.1136/bmj.312.7036.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.MacGregor AJ, Li Q, Spector TD, Williams FM. The genetic influence on radiographic osteoarthritis is site specific at the hand, hip and knee. Rheumatology (Oxford) 2009;48(3):277–280. doi: 10.1093/rheumatology/ken475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Styrkarsdottir U, Thorleifsson G, Helgadottir HT, Bomer N, Metrustry S, Bierma-Zeinstra S, et al. Severe osteoarthritis of the hand associates with common variants within the ALDH1A2 gene and with rare variants at 1p31. Nat Genet. 2014;46(5):498–502. doi: 10.1038/ng.2957. [DOI] [PubMed] [Google Scholar]
- 6.Castano Betancourt MC, Cailotto F, Kerkhof HJ, Cornelis FM, Doherty SA, Hart DJ, et al. Genome-wide association and functional studies identify the DOT1L gene to be involved in cartilage thickness and hip osteoarthritis. Proc Natl Acad Sci U S A. 2012;109(21):8218–8223. doi: 10.1073/pnas.1119899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Evangelou E, Valdes AM, Castano-Betancourt MC, Doherty M, Doherty S, Esko T, et al. The DOT1L rs12982744 polymorphism is associated with osteoarthritis of the hip with genome-wide statistical significance in males. Ann Rheum Dis. 2013;72(7):1264–1265. doi: 10.1136/annrheumdis-2012-203182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Evangelou E, Kerkhof HJ, Styrkarsdottir U, Ntzani EE, Bos SD, Esko T, et al. A meta-analysis of genome-wide association studies identifies novel variants associated with osteoarthritis of the hip. Ann Rheum Dis. 2014;73(12):2130–2136. doi: 10.1136/annrheumdis-2012-203114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.arcOGEN Consortium. Zeggini E, Panoutsopoulou K, Southam L, Rayner NW, Day-Williams AG, et al. Identification of new susceptibility loci for osteoarthritis (arcOGEN): a genome-wide association study. Lancet. 2012;380(9844):815–823. doi: 10.1016/S0140-6736(12)60681-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Valdes AM, Evangelou E, Kerkhof HJ, Tamm A, Doherty SA, Kisand K, et al. The GDF5 rs143383 polymorphism is associated with osteoarthritis of the knee with genome-wide statistical significance. Ann Rheum Dis. 2011;70(5):873–875. doi: 10.1136/ard.2010.134155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kerkhof HJ, Lories RJ, Meulenbelt I, Jonsdottir I, Valdes AM, Arp P, et al. A genome-wide association study identifies an osteoarthritis susceptibility locus on chromosome 7q22. Arthritis Rheum. 2010;62(2):499–510. doi: 10.1002/art.27184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Evangelou E, Valdes AM, Kerkhof HJ, Styrkarsdottir U, Zhu Y, Meulenbelt I, et al. Meta-analysis of genome-wide association studies confirms a susceptibility locus for knee osteoarthritis on chromosome 7q22. Ann Rheum Dis. 2011;70(2):349–355. doi: 10.1136/ard.2010.132787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Day-Williams AG, Southam L, Panoutsopoulou K, Rayner NW, Esko T, Estrada K, et al. A variant in MCF2L is associated with osteoarthritis. Am J Hum Genet. 2011;89(3):446–450. doi: 10.1016/j.ajhg.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerkhof HJ, Meulenbelt I, Akune T, Arden NK, Aromaa A, Bierma-Zeinstra SM, et al. Recommendations for standardization and phenotype definitions in genetic studies of osteoarthritis: the TREAT-OA consortium. Osteoarthritis Cartilage. 2011;19(3):254–264. doi: 10.1016/j.joca.2010.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lester G. The Osteoarthritis Initiative: A NIH Public-Private Partnership. HSS J. 2012;8(1):62–63. doi: 10.1007/s11420-011-9235-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jordan JM, Helmick CG, Renner JB, Luta G, Dragomir AD, Woodard J, et al. Prevalence of knee symptoms and radiographic and symptomatic knee osteoarthritis in African Americans and Caucasians: the Johnston County Osteoarthritis Project. J Rheumatol. 2007;34(1):172–180. [PubMed] [Google Scholar]
- 17.Nelson AE, Renner JB, Shi XA, Shreffler JH, Schwartz TA, Jordan JM. Cross-sectional comparison of extended anteroposterior and posteroanterior fixed flexion positioning to assess radiographic osteoarthritis at the knee: the Johnston County Osteoarthritis Project. Arthritis Care Res (Hoboken) 2010;62(9):1342–1345. doi: 10.1002/acr.20210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Segal NA, Nevitt MC, Gross KD, Hietpas J, Glass NA, Lewis CE, et al. The Multicenter Osteoarthritis Study: opportunities for rehabilitation research. PM R. 2013;5(8):647–654. doi: 10.1016/j.pmrj.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jordan JM, Atif U, Li H, Renner JB, Liu Y, Hill D, et al. Gender differences in radiographic OA phenotypes in a genetic association study: The Genetics of Osteoarthritis (GO) Osteoarthritis Cartilage. 2007;15(Supp C):C136. [Google Scholar]
- 20.Conlin LK, Thiel BD, Bonnemann CG, Medne L, Ernst LM, Zackai EH, et al. Mechanisms of mosaicism, chimerism and uniparental disomy identified by single nucleotide polymorphism array analysis. Hum Mol Genet. 2010;19(7):1263–1275. doi: 10.1093/hmg/ddq003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peiffer DA, Le JM, Steemers FJ, Chang W, Jenniges T, Garcia F, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16(9):1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, et al. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34(6):591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Speliotes EK, Willer CJ, Berndt SI, Monda KL, Thorleifsson G, Jackson AU, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42(11):937–948. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26(17):2190–2191. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zengini E, Finan C, Wilkinson JM. The Genetic Epidemiological Landscape of Hip and Knee Osteoarthritis: Where Are We Now and Where Are We Going? J Rheumatol. 2016;43(2):260–266. doi: 10.3899/jrheum.150710. [DOI] [PubMed] [Google Scholar]
- 26.Kellgren JH, Lawrence JS. Radiological assessment of osteo-arthrosis. Ann Rheum Dis. 1957;16(4):494–502. doi: 10.1136/ard.16.4.494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Silverwood V, Blagojevic-Bucknall M, Jinks C, Jordan JL, Protheroe J, Jordan KP. Current evidence on risk factors for knee osteoarthritis in older adults: a systematic review and meta-analysis. Osteoarthritis Cartilage. 2015;23(4):507–515. doi: 10.1016/j.joca.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 28.Valdes AM, Doherty SA, Muir KR, Wheeler M, Maciewicz RA, Zhang W, et al. The genetic contribution to severe post-traumatic osteoarthritis. Ann Rheum Dis. 2013;72(10):1687–1690. doi: 10.1136/annrheumdis-2012-202562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Panoutsopoulou K, Metrustry S, Doherty SA, Laslett LL, Maciewicz RA, Hart DJ, et al. The effect of FTO variation on increased osteoarthritis risk is mediated through body mass index: a mendelian randomisation study. Ann Rheum Dis. 2014;73(12):2082–2086. doi: 10.1136/annrheumdis-2013-203772. [DOI] [PMC free article] [PubMed] [Google Scholar]