Abstract
Bakground:
Aspergillus fumigatus is an airborne opportunistic fungal pathogen that can cause fatal infections in immunocompromised patients. Although the current anti-fungal therapies are relatively efficient, some issues such as drug toxicity, drug interactions, and the emergence of drug-resistant fungi have promoted the intense research toward finding the novel drug targets.
Methods:
In search of new antifungal drug targets, we have used a bioinformatics approach to identify novel drug targets. We compared the whole proteome of this organism with yeast Saccharomyces cerevisiae to come up with 153 specific proteins. Further screening of these proteins revealed 50 potential molecular targets in A. fumigatus. Amongst them, RNA-binding protein (RBP) was selected for further examination. The aspergillus fumigatus RBP (AfuRBP), as a peptidylprolyl isomerase, was evaluated by homology modeling and bioinformatics tools. RBP-deficient mutant strains of A. fumigatus were generated and characterized. Furthermore, the susceptibility of these strains to known peptidylprolyl isomerase inhibitors was assessed.
Results:
AfuRBP-deficient mutants demonstrated a normal growth phenotype. MIC assay results using inhibitors of peptidylprolyl isomerase confirmed a higher sensitivity of these mutants compared to the wild type.
Conclusion:
Our bioinformatics approach revealed a number of fungal-specific proteins that may be considered as new targets for drug discovery purposes. Peptidylprolyl isomerase, as a possible drug target, was evaluated against two potential inhibitors, and the promising results were investigated mechanistically. Future studies would confirm the impact of such target on the antifungal discovery investigations
Keywords: Aspergillus fumigatus, RNA-binding protein, Peptidylprolyl isomerase, Juglone, RNA interference
INTRODUCTION
A spergillus fumigatus is currently the most important airborne fungal pathogen and the leading cause of invasive aspergillosis (~85% of cases)[1]. Despite the introduction of new effective antifungal drugs, the mortality rate of invasive aspergillosis still exceeds 40%[2]. Furthermore, the emergence of drug resistance adds more complications to the treatment process. Hence, new antifungal agents are urgently needed.
While the classical cell-based screening method has been successfully used in drug discovery process[3], it is an expensive, time-consuming and labor-intensive approach[4]. Recent advances in application of computation methods in genome, proteome, and metabolome mining coupled with complete genome sequences of pathogen organisms have opened an alternative path toward identifying new drug targets. For instance, several reports have confirmed the usefulness of comparative genome analysis in identification of pathogen-specific drug and vaccine candidates[5-7].
The genome of A. fumigatus contains ~9900 genes, but few antifungal targets have been developed yet[8]. In the present study, we compared the whole proteome of A. fumigatus with Saccharomyces cerevisiae, as a non-pathogen fungal organism, to identify Aspergillus-specific proteins. Amongst the proteins, those with close homologues to human were removed, and the final targets were selected. One novel target, the aspergillus fumigatus RNA-binding protein (AfuRBP), was selected and found to be a peptidylprolyl isomerase (PPI). For further functional studies, AfuRbp knock out and knock down mutant strains were generated. MIC assay with known inhibitors, phenylglyoxal and juglone, and a synthetic derivative of juglone confirmed the higher sensitivity of the mutants to these compounds when compared to the wild-type strain.
MATERIALS AND METHODS
Comparative analysis of A. fumigatus and S. cerevisiae proteome
The identification of novel drug targets against A. fumigatus, as a human pathogen, was based on alignment of its proteome with the proteome of S. cerevisiae. The proteomes of these two fungi were extracted from NCBI (https://www.ncbi.nlm.nih.gov/genome/browse/). Three different approaches were used to identify and confirm those sequences that are present in A. fumigatus but are absent or significantly diverge from S. cerevisiae. LAST (http://last.cbrc.jp)[9] has been used as a fast sequence alignment tool to identify unique sequences for A. fumigatus. LAST uses variable-length (spaced) seeds realized by a suffix array[10]. The expected value of 10 (-e84) was chosen to obtain higher sensitivity (-m1000000), and the other settings and parameters were used as default. In the next step, unique sequences from A. fumigatus were compared using suffix tree analysis by Mummer (version 3.0) (http://mummer.sourceforge. net)[11]. Minimum maximal match was seven to restrict output proteins, but other settings were chosen as default. Finally, the proteins that were unique to fungi were identified by BlastP (NCBI Blast v. 2.2.1)[12]. Modeling was carried out by I-TASSER server (Michigan University, USA)[13-15]. Docking of the compounds was performed by HEX 6.3 software when required.
Strains, plasmids, and culture conditions
A. fumigatus akuAKU80 and its pyrG- derivative (akuAKU80 pyrG-) with a highly efficient homologous recombination background were used for isolation of AfuRbp gene homologue and gene disruption or gene silencing experiments. E. coli Top10 (Invitrogen, USA) cells and pGEM–T Easy cloning system (Promega, USA) were used in all DNA recombinant procedures. Plasmid PGEM-GlaA comprising Aspergillus niger glucoamylase A (glaA) promoter and glaA termination signal was used for preparation of RNAi plasmid. All molecular methods including PCR and RT-PCR were performed based on established protocols[16]. In RT-PCR settings, 1 µg of total RNA was used in cDNA synthesis reactions and the A. fumigatus actin gene (AFUA_6G04740) was used as a loading control. Fungal strains were grown and kept on SAB agar or SAB agar medium supplemented with uridine and uracil. Modified Vogel’s medium[17] was used in isolation of fungal transformants.
Construction of AfuRbp deletion/silencing cassettes and strains
For construction of the AfuRbp deletion cassette, a sequential cloning strategy was applied as described before[18]. Briefly, 1.5 kb of 5’ and 1.2 kb of 3’ flanking regions of the gene were amplified separately, using RBP_KO1/RBP_KO2 (containing SphI/NcoI restriction sites) and RBP_KO3/RBP_KO4 (containing EcoRI/SalI restriction sites) primer sets, respectively (Table 1). These fragments were cloned next together in pGEM-Teasy vector. At the final step, the A. fumigatus pyrG gene, as a selection marker, with its own promoter and terminator containing EcoRI site at both ends was cloned into EcoRI site, between the 5’and 3’ flanking regions, resulted in pRBP-KO (Fig. 1A).
Table 1.
Primers used in this study
| Name | Sequence (5`-3`) | Enzymes |
|---|---|---|
| RBP_KO1 | GGGCCCGCATGCTCTCTTCAGCTCTGTGGTTG | SphI |
| RBP_KO2 | CCATGGCATACACTCGAGCACGGTCG | NcoI |
| RBP_KO3 | GAATTCAATGCGTGATGTTG | EcoRI |
| RBP_KO4 | GTCGACGTCAGTTGACAGAGCGAG | SalI |
| RBP_SENSE_F | GAGATCTCCATCCCATTGTTGTT | BglII |
| RBP_SENSE_R | TGATATCTGCTCCTGCTGTCGGCGT | EcoRV |
| RBP-F | AAGTTCATGGAAAAGTGGGC | ------- |
| RBP-R | AGTCAGTTGACAGAGCGAGG | ------- |
| RBP_rt1 | CCCCTGAGGTTGAGGAGCGG | ------- |
| RBP_rt2 | CGAGGTTCTGCACTTTGGCG | ------- |
| GFP_F | GATATCGTCCAGGAGCGCAC | EcoRV |
| GFP_R | AAGCTTCTCGATGCGGTT | HindIII |
| ACT-F | ATGTCACTGTGCAGATTGTC | ------- |
| ACT-R | CGTAGAGGGAGAGAACGGCC | ------- |
Fig. 1.
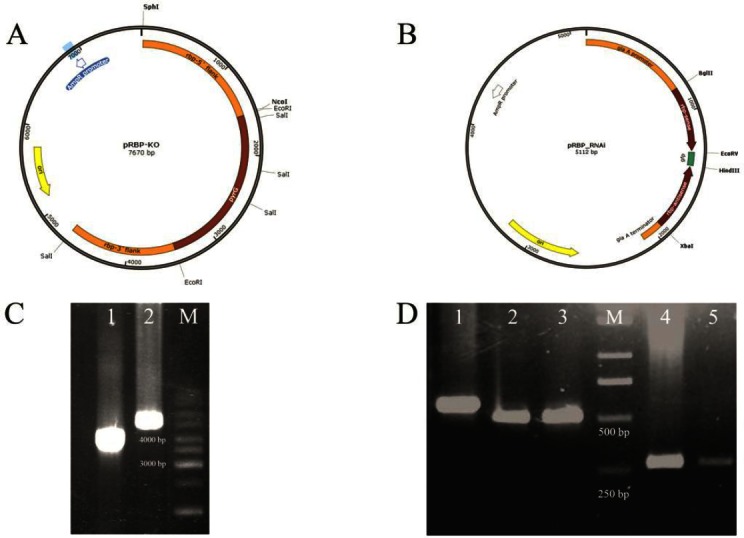
Gene constructs and molecular validations. Schematic representation of constructed plasmids used in gene deletion (pRBP-KO) (A) and RNAi (pRBP_RNAi) (B) experiments. (C) PCR analysis of wild-type (lane 1) and deletant strain (lane 2) genome using primers RBP-F1 and RBP-R1. The amplification of a 4.6-kb product confirmed the replacement of native AfuRbp with the disrupted fragment. (D) RT-PCR analysis of RNAi transformant grown in maltodextrin or glucose medium for 24 h. Lane 1, actin fragment amplified from the genome (560 bp); lane 2, actin fragment amplified from cDNA originated from glucose; lane 3, maltodextrin cultures (485 bp); lane 4, AfuRbp expression level in RNAi transformant grown in glucose medium; lane 5, maltodextrin medium. AfuRBP-specific RT-PCR primers, RBP_rt1/rt2, amplified a 300-bp product. M, DNA size marker
To generate the AfuRbp silencing cassette, an inducible RNAi transcriptional unit was designed based on a previously described protocol[19, 20]. This unit was driven by the glaA promoter and comprises an approximately 500 bp Rbp fragment (sense, nucleotides 342-841), followed by a 100-bp buffer EGFp fragment and the same ~500 bp Rbp fragment in the opposite direction (antisense). Sense and antisense fragments were amplified by PCR using primers RBP_SENSE_F and RBP_SENSE_R carrying appropriate restriction sites (Table 1) and cloned into PGEM-GlaA vector. A 100-bp EGFP fragment was then amplified by PCR using specific primers (Table 1) and cloned between the latter fragments to generate final silencing cassette (pRBP_RNAi) (Fig. 1B).
The prepared constructs were used to transform A. fumigatus KU80ΔpyrG strain as described before[21]. Positive transformants were selected on Vogel’s minimal medium lacking uracil/uridine supplements. To find the knock out strains, transformants were screened by PCR using primers RBP-F and RBP-R (Table 1). As the silencing construct, pRBP_RNAi did not contain any fungal selection marker, the pRG3- AMA1 vector containing the pyrG selection marker was used as the second plasmid in co-transformation reaction. RNAi transformants were also selected based on PCR using gfp- and AfuRbp-specific primers.
Susceptibility testing
MICs were determined based on CLSI Broth Microdilution Method (M38-A2 Document) with some modifications. Briefly, fungal spores (104/well) were inoculated in 96-well microtiter plates containing RPMI 1640 medium enriched by 2% glucose (or maltodextrin), and the susceptibility was assessed at a final compound concentration range of 0-200 µg/ml after 48 h. All MIC assays were performed in triplicates. Based on orthology and EC number (EC: 5.2.1.8) of AFUA_2G07650 (AfuRBP), two organic inhibitors of the PPI activity, i.e. phenylglyoxal and juglone (5-hydroxy-1,4-naphthoquinone), were found by searching BRENDA enzyme database (www.brenda-enzymes.info) and tested (Fig. 2). A synthetic derivative of juglone, 5-O-Acetoxy-1,4-naphthoquinone (juglone acetate, JA), was also prepared using a previously published method[13] and assessed in MIC tests. This derivative was made to make the compound more lipophilic.
Fig. 2.

List of enzymes (EC number) targeted by juglone or phenylglyoxal. The peptidylprolyl isomerase is the only overlap target for both juglone and phenylglyoxal as inhibitors.
Docking and homology modeling
For bioinformatics assessment of the chemical inhibitors used in MIC assays, the 3D structure of AfuRBP was predicted by I-TASSER. The predicted binding sites were subsequently used for docking of the inhibitors. HEX 6.3, ArgusLab (v.4.0.1, http://www.arguslab.com/arguslab.com/Publications.html), and WebLab Viewer Lite (v. 4.2) were used for docking of the inhibitors and manipulation of the protein.
RESULTS
Identification of target proteins based on in silico comparative analyses
Based on the LAST alignment of the A. fumigatus proteome (9630 proteins) against S. cerevisiae, 474 unique proteins were recognized in A. fumigatus as the homologous of these proteins were not present in S. cerevisiae. All of the 474 proteins were further examined in KEGG, NCBI, EXPASY, and EBI databases. It was found that the functions of 161 proteins have already been explained, and their accession numbers are available. The remaining 313 proteins were found to be hypothetical (data not shown). Finally, 50 out of 161 protein candidates were chosen as they were fungal-specific but not present in S. cerevisiae (Table 2). The RBP, as a potential drug target, was selected for further evaluation. This protein is encoded by an 1158-bp gene located on chromosome 2 and has been annotated as a PPI.
Table 2.
List of selected proteins specific to fungi other than S. cerevisiae
| No. | Accession no. | Description | Taxonomy | Orthology |
|---|---|---|---|---|
| 1 | AFUA_8G06570 | Acetylxylan esterase ((EC:3.-.-.-) | Dikarya | Acetylxylan esterase [EC:3.1.1.72] |
| 2 | AFUA_4G00860 | Cell surface protein | Pezizomycotina | |
| 3 | AFUA_1G13450 | Cell wall proline-rich protein | Pezizomycotina | |
| 4 | AFUA_6G14090 | CFEM domain protein | Leotiomyceta | |
| 5 | AFUA_4G12450 | Conserved lysine-rich protein | Pezizomycotina | |
| 6 | AFUA_4G13230 | Developmental regulatory protein WetA | Saccharomyceta† | |
| 7 | AFUA_8G00250 | Dimethylallyl tryptophan synthase (EC:2.5.1.-) | Leotiomyceta | N-dimethylallyltransferase [EC:2.5.1.-] |
| 8 | AFUA_7G01280 | DNA damage response protein RcaA | Metazoa | |
| 9 | AFUA_7G06740 | Endoglucanase (EC:3.2.1.4) | Metazoa | Endoglucanase [EC:3.2.1.4] |
| 10 | AFUA_6G11980 | Exo-beta-1,3-glucanase (EC:3.2.1.58) | Dikarya | glucan 1,3-beta-glucosidase [EC:3.2.1.58] |
| 11 | AFUA_6G14070 | Extracellular glycine/serine-rich protein | Neosartorya fischeri & Aspergillus fumigatus | |
| 12 | AFUA_5G01620 | Extracellular proline-rich protein | Eurotiomycetidae | |
| 13 | AFUA_8G07090 | Extracellular proline-serine-rich protein | Dikarya | |
| 14 | AFUA_7G02060 | Extracellular serine-rich protein | Leotiomyceta | |
| 15 | AFUA_7G02460 | Extracellular serine-rich protein | Trichocomaceae | |
| 16 | AFUA_6G00670 | Extracellular serine-rich protein | Dikarya | |
| 17 | AFUA_7G00320 | Extracelular proline-glycine-rich protein | Leotiomyceta | |
| 18 | AFUA_7G05650 | Glutamine-serine-rich protein MS8 | Leotiomyceta | |
| 19 | AFUA_7G04870 | Glutamine-serine-proline-rich protein | Saccharomyceta† | |
| 20 | AFUA_6G10580 | GPI anchored CFEM domain protein | Dikarya | |
| 21 | AFUA_8G04860 | GPI anchored glycoprotein | Leotiomyceta | |
| 22 | AFUA_6G14010 | GPI anchored protein | Leotiomyceta | |
| 23 | AFUA_4G09600 | GPI anchored protein | Metazoa | |
| 24 | AFUA_7G00970 | GPI anchored serine-threonine-rich protein | Leotiomyceta | |
| 25 | AFUA_4G08295 | Histone h1.3. | Leotiomyceta | |
| 26 | AFUA_8G07060 | Hydrophobin | Leotiomyceta | |
| 27 | AFUA_8G01790 | Integral membrane protein | Leotiomyceta | |
| 28 | AFUA_6G04280 | Integral membrane protein | Leotiomyceta | |
| 29 | AFUA_5G00620 | Integral membrane protein | Leotiomyceta | |
| 30 | AFUA_1G11380 | Integral membrane protein | Leotiomyceta | |
| 31 | AFUA_8G01190 | Isoamyl alcohol oxidase | Leotiomyceta | |
| 32 | AFUA_5G03980 | LysM domain protein | Leotiomyceta | |
| 33 | AFUA_6G03050 | Oleate delta-12 desaturase (EC:1.14.99.-) | Dikarya | omega-6 fatty acid desaturase (delta-12 desaturase) |
| 34 | AFUA_8G02230 | Serine-leucine-rich repeat protein | Aspergillus fumigatus | |
| 35 | AFUA_1G13830 | Threonine-rich protein | Eurotiomycetidae | |
| 36 | AFUA_1G02270 | ARS-binding protein Abp2 | Ascomycota | |
| 37 | AFUA_5G05990 | AT DNA-binding protein | Leotiomyceta | |
| 38 | AFUA_3G08110 | Cell wall protein | Ascomycota | |
| 39 | AFUA_5G09580 | Conidial hydrophobin Hyp1/RodA | Leotiomyceta | |
| 40 | AFUA_5G09010 | Conserved glutamic acid-rich protein | Pezizomycotina | |
| 41 | AFUA_3G12790 | Conserved glutamic acid-rich protein | Eurotiomycetidae | |
| 42 | AFUA_1G05290 | Endo-1,3(4)-beta-glucanase (EC:3.2.1.6); K01180 endo-1,3(4)-beta-glucanase [EC:3.2.1.6] | Fungi | endo-1,3(4)-beta-glucanase [EC:3.2.1.6] |
| 43 | AFUA_3G13110 | Extracellular serine-threonine-rich protein | Leotiomyceta | |
| 44 | AFUA_5G14210 | Glucose repressible protein Grg1 | Dikarya | |
| 45 | AFUA_1G11220 | GPI anchored protein | Trichocomaceae | |
| 46 | AFUA_1G03630 | GPI anchored serine-threonine-rich protein (EC:3.1.1.5) lysophospholipase | Fungi | |
| 47 | AFUA_5G13725 | Integral membrane protein | Pezizomycotina | |
| 48 | AFUA_3G08670 | Integral membrane protein | Leotiomyceta | |
| 49 | AFUA_3G00830 | Pfs domain protein | Leotiomyceta | |
| 50 | AFUA_2G07650 | RNA-binding protein (EC:5.2.1.8); K01802 Peptidylprolyl isomerase [EC:5.2.1.8] | Trichocomaceae | peptidylprolyl isomerase [EC:5.2.1.8] |
Orthology is specified where data exist.
excluding S. cerevisiae
Disruption of AfuRbp in A. fumigatus
The SphI/SalI pRBP-KO fragment was used for transformation of A. fumigatus (akuAKU80 pyrG-) protoplasts. Through the PCR screening of transformants, a deletion strain was isolated and examined further. In this transformant, RBP-F1 and RBP-R1 primers amplified a fragment of ~4.6 kb (KO) instead of a 3.8-kb wild-type fragment, confirming the replacement of native gene with disrupted one (Fig. 1C). RT-PCR analysis of the deletant revealed that AfuRbp does not express in this mutants (data not shown). The growth phenotype of this mutant in various media containing different carbon and nitrogen sources was similar to the parental strain, indicating that AfuRbp is not essential in A. fumigatus. In PCR screening of RNAi transformants, one positive integrant was selected. This transformant was grown in glaA-inducing medium containing 1% maltodextrin, as the sole carbon source. Semi-quantitative RT-PCR analysis using primers RPB-rt1 and RBP-rt2 showed the down-regulation of AfuRbp expression in the presence of maltodextrin (Fig. 1D). Phenotypic analysis of this transformant in inducing medium re-confirmed the non-essential role of AfuRbp in fungal growth.
Effect of juglone and the synthetic compound on the mutant strains
To test whether the deletion or down-regulation of AfuRbp has any effect on drug susceptibility of the fungus, MIC levels were determined for phenylglyoxal, juglone, and JA. Table 3 demonstrates MIC values for these compounds. The results demonstrated a higher sensitivity of all strains to juglone compared to phenylglyoxal and JA. While the phenylglyoxal compound did not appear as a strong antifungal inhibitor, the mutant strains showed more sensitivity to this chemical. MIC assays for RNAi transformant was performed in maltodextrin medium. The higher sensitivity of RNAi transformant in maltodextrin medium compared to the glucose medium indicated that the down-regulation of AfuRbp can affect the strain susceptibility to PPI inhibitors.
Table 3.
In vitro activity of compounds against wild-type and AfuRbp mutants of A. fumigatus by MIC test (µg/ml)
| Inhibitor | Wild strain | KO mutant | RNAi mutant | |||
|---|---|---|---|---|---|---|
| RPMI/glucose | RPMI/maltodextrin | RPMI/glucose | RPMI/maltodextrin | RPMI/glucose | RPMI/maltodextrin | |
| Juglone | 3.12 | 3.12 | 1.56 | 1.56 | 3.12 | 1.56 |
| Phenylglyoxal | 200 | 200 | 100 | 100 | 200 | 100 |
| Juglone acetate | 6.25 | 6.25 | 3.12 | 3.12 | 6.25 | 3.12 |
| Amphotericin B | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 | 1.56 |
Outcome of modeling and docking for AfuRBP as a model
As we could not find any template protein having a minimum 35% similarity to our target protein, a threading algorithm for RBP protein modeling was used by I-TASSER server. Five models were generated, and according to the I-TASSER validation criteria, model number one had the least calculation errors and fitted best within the X-ray/NMR 3D population criteria (Fig. 3). In this study, five top models proposed by I-TASSER were used as receptors of the docking experiments, and phenylglyoxal, juglone, and JA were used as ligands. Docking outcome showed that the interaction of model number 4 with JA produced a proper energy (-186.3), which indicates the highest absolute value among the docking results. Other docking outcomes that were sorted by energy are shown in Table 4.
Fig. 3.
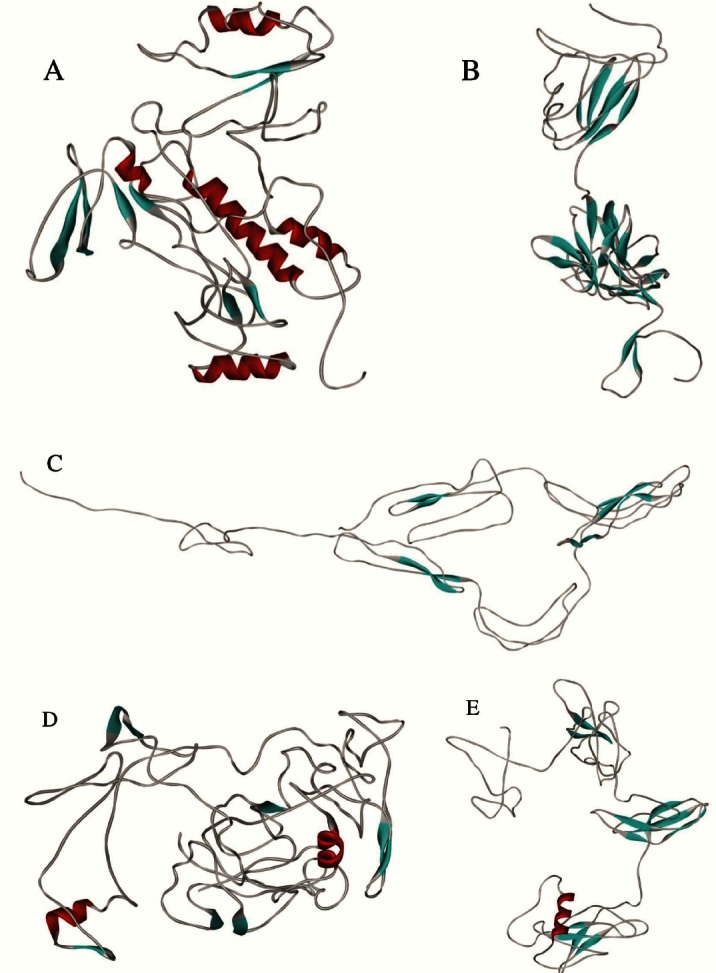
The five predicted three-dimensional structures of AfuRBP protein obtained by homology modeling with I-TASSER. Model numbers: 1 (A), 2 (B), 3 (C), 4 (D), 5 (E).
Table 4.
The results of Docking experiments
| Dock name | Produced energy of interaction (kJ/mol) | Position (supercoil) | Position (alphaa helix) | Position (beta sheet) |
|---|---|---|---|---|
| Model4 with JA | -186.3 | 8-9-10-11-243-244-275-317 | 245-246-247 | 276-277 |
| Model3 with JA | -184.7 | 347-348-349-350-366-367-368-369-370-371 | None | 317-318-319-320-321 |
| Model1 with JA | -174.5 | 220-221-222-223-224-225-226-227-228-229-230-231-232-233-234-235-236 | 237-238-239-240 | None |
| Model5 with JA | -174.4 | 109-110-111-112-113-114-115-131-132-133-134-135-136-181-182-183 | None | None |
| Model3 with juglone | -166.4 | 324-325-350-356-357-358-363-364 | None | None |
| Model2 with JA | -162.5 | 80-81-82-83-84-85-126-127-128 | None | 79-86-87 |
| Model4 with juglone | -161.7 | 74-75-156-157-158-351-352 | None | None |
| Model3 with phenylglyoxal | -153.12 | 354-355-356 | None | None |
| Model1 with juglone | -152 | 26-27-28-29-30-31-213-214-215-216 | None | None |
| Model4 with phenylglyoxal | -146.5 | 383-384-385 | None | None |
| Model1 with phenylglyoxal | -146.1 | 243-244-245-246-247 | 279-280 | None |
| Model2 with phenylglyoxal | -143.5 | 143-144-145-157-158 | 229-230-231 | 219-220-221 |
| Model5 with phenylglyoxal | -117.3 | 167-168-169-170-171-172-173-174-175 | None | None |
| Model5 with juglone | -31.9 | 82-83-84-85-86-87 | None | 126-127 |
| Model2 with juglone | 0 | None | None | None |
The synthetic compound (JA), juglone and phenylglyoxal, were used as ligands and the five predicted models (1, 2, 3, 4, and 5) were used as receptors. Position of docked ligand and binding sites for docking results with their energy values are indicated in the Table. Docking was performed by HEX 6.3 software.
DISCUSSION
Because of the common eukaryotic origin of fungi and mammals, the development of new antifungal drugs has been challenging. This may be the reason for the introduction of only five classes of antifungal agents, including fluorinated pyrimidine analogs, polyenes, allylamines, azoles, and echinocandins[22]. Here, we have tried to find some novel drug targets against A. fumigatus using bioinformatics. Our alignment has demonstrated a group of proteins/genes (Table 2) that are fungal-specific and may be essential to A. fumigatus. Our highlights differ from other studies such as Abadio and colleagues[23] in comparative alignment as we searched for those novel targets that only exist in A. fumigatus. In this study, we found a RNA-binding protein (AfuRBP) that has been annotated as a member of PPI enzyme family. The PPI family contains three members, including cyclophilins, FKBPs, and Parvulin[24]. Cyclophilins are described by an eight-standard β-barrel that forms a hydrophobic pocket. FKBPs, instead, contain an amphipatic, five-stranded β-sheet that enfolds around a single, short α-helix[25, 26]. Members of Parvulin family contain a PPI domain consisting of a half β-barrel and its four antiparallel strands surrounded by four α-helices. The results of I-TASSER modeling and docking demonstrated that the AfuRBP is structurally closer to Parvulin family members of PPI enzyme family. To elucidate the function of this protein, AfuRbp-deficient strains were generated using gene deletion and RNAi strategies. Normal phenotype of these strains indicated a non-essential role for this protein in growth physiology of Aspergillus fumigatus. Some studies have demonstrated that the inactivation of a drug target gene can result in increased sensitivity of the organism to the drug or other compounds that inhibit the same pathway[27, 28]. In this sense, the sensitivity of AfuRbp-disrupted strains to PPI inhibitors was assessed.
Juglone showed the highest inhibitory effect on the mutants and wild strain. In addition, we used JA as an inhibitor to investigate the change in the structure of juglone through modification of its hydroxyl group in order to increase the hydrophobicity. The decision to produce JA was partly due to the predicted activity of this derivative through the docking scores, which had a higher absolute energy of interaction than juglone. Overall, MIC test of JA demonstrated different results for the mutant and wild strains. Although docking showed a high potential for JA, the MIC test indicated slight decrease in activity, which might be due to lipophilic binding of JA to other cell components.
Considering the specific inhibitory effect of juglone on Parvulin[29] and the higher sensitivity of AfuRbp deficient mutants to this chemical compared to the parental strain, it can be concluded that juglone may act as a specific inhibitor of the AfuRBP. More detailed studies on recombinant AfuRBP may verify this claim.
In conclusion, in this study, the bioinformatics and biological assessments of AfuRBP were carried out, and the presented data can be considered as a start for pin pointing a target towards developing a new drug against A. fumigatus.
ACKNOWLEDGEMENTS
The first author wishes to thank Reza Ghamarian for his precious help. This work was partially supported by Pasteur Institute of Iran through Grant No. 504.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Nicolle LE, Rotstein C, Bourgault AM, St-Germain G, Garber G. Registry CIDSIF Or the Canadian Infectious Diseases Society Invasive Fungal Registry. Invasive fungal infections in Canada from 1992 to 1994. The canadian journal of infectious diseases. 1998;9(6):347–352. doi: 10.1155/1998/473219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nivoix Y, Velten M, Letscher-Bru V, Moghaddam A, Natarajan-Amé S, Fohrer C, Lioure B, Bilger K, Lutun P, Marcellin L, Launoy A, Freys G, Bergerat JP, Herbrecht R. Factors associated with overall and attributable mortality in invasive Aspergillosis. Clinical infectious diseases. 2008;47(9):1176–1184. doi: 10.1086/592255. [DOI] [PubMed] [Google Scholar]
- 3.Oliver JD, Sibley GE, Beckmann N, Dobb KS, Slater MJ, McEntee L, du Pre S, Livermore J, Bromley MJ, Wiederhold NP, Hope WW, Kennedy AJ, Law D, Birch M. F901318 represents a novel class of antifungal drug that inhibits dihydroorotate dehydrogenase. Proceedings of the national academy of sciences of the United States of America. 2016 Oct 25; doi: 10.1073/pnas.1608304113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lavecchia A, Di Giovanni C. Virtual screening strategies in drug discovery: a critical review. Current medicinal chemistry. 2013;20(23):2839–2860. doi: 10.2174/09298673113209990001. [DOI] [PubMed] [Google Scholar]
- 5.Barh D, Kumar A. In silico identification of candidate drug and vaccine targets from various pathways in Neisseria gonorrhoeae. In silico biology. 2009;9(4):225–231. [PubMed] [Google Scholar]
- 6.Carlton JM, Angiuoli SV, Suh BB, Kooij TW, Pertea M, Silva JC, Ermolaeva MD, Allen JE, Selengut JD, Koo HL, Peterson JD, Pop M, Kosack DS, Shumway MF, Bidwell SL, Shallom SJ, van Aken SE, Riedmuller SB, Feldblyum TV, Cho JK, Quackenbush J, Sedegah M, Shoaibi A, Cummings LM, Florens L, Yates JR, Raine JD, Sinden RE, Harris MA, Cunningham DA, Preiser PR, Bergman LW, Vaidya AB, van Lin LH, Janse CJ, Waters AP, Smith HO, White OR, Salzberg SL, Venter JC, Fraser CM, Hoffman SL, Gardner MJ, Carucci DJ. Genome sequence and comparative analysis of the model rodent malaria parasite Plasmodium yoelii yoelii. Nature. 2002;419(6906):512–519. doi: 10.1038/nature01099. [DOI] [PubMed] [Google Scholar]
- 7.Butt AM, Nasrullah I, Tahir S, Tong Y. Comparative genomics analysis of Mycobacterium ulcerans for the identification of putative essential genes and therapeutic candidates. PloS one. 2012;7(8):e43080. doi: 10.1371/journal.pone.0043080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nierman WC, Pain A, Anderson MJ, Wortman JR, Kim HS, Arroyo J, Berriman M, Abe K, Archer DB, Bermejo C, Bennett J, Bowyer P, Chen D, Collins M, Coulsen R, Davies R, Dyer PS, Farman M, Fedorova N, Fedorova N, Feldblyum TV, Fischer R, Fosker N, Fraser A, García JL, García MJ, Goble A, Goldman GH, Gomi K, Griffith-Jones S, Gwilliam R, Haas B, Haas H, Harris D, Horiuchi H, Huang J, Humphray S, Jiménez J, Keller N, Khouri H, Kitamoto K, Kobayashi T, Konzack S, Kulkarni R, Kumagai T, Lafon A, Latgé JP, Li W, Lord A, Lu C, Majoros WH, May GS, Miller BL, Mohamoud Y, Molina M, Monod M, Mouyna I, Mulligan S, Murphy L, O'Neil S, Paulsen I, Peñalva MA, Pertea M, Price C, Pritchard BL, Quail MA, Rabbinowitsch E, Rawlins N, Rajandream MA, Reichard U, Renauld H, Robson GD, Rodriguez de Córdoba S, Rodríguez-Peña JM, Ronning CM, Rutter S, Salzberg SL, Sanchez M, Sánchez-Ferrero JC, Saunders D, Seeger K, Squares R, Squares S, Takeuchi M, Tekaia F, Turner G, Vazquez de Aldana CR, Weidman J, White O, Woodward J, Yu JH, Fraser C, Galagan JE, Asai K, Machida M, Hall N, Barrell B, Denning DW. Genomic sequence of the pathogenic and allergenic filamentous fungus Aspergillus fumigatus. Nature. 2005;438(7071):1151–1156. doi: 10.1038/nature04332. [DOI] [PubMed] [Google Scholar]
- 9.Kiełbasa SM, Wan R, Sato K, Horton P, Frith MC. Adaptive seeds tame genomic sequence comparison. Genome research. 2011;21(3):487–493. doi: 10.1101/gr.113985.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Abouelhoda MI, Stefan Kurtz, Enno Ohlebusch. Replacing suffix trees with enhanced suffix arrays. Journal of discrete algorithms. 2004;2(1):53–86. [Google Scholar]
- 11.Kurtz S, Phillippy A, Delcher AL, Smoot M, Shumway M, Antonescu C, Salzberg SL. Versatile and open software for comparing large genomes. Genome biol. 2004;5(2):R12. doi: 10.1186/gb-2004-5-2-r12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic acids research. 2008;36(suppl 2):W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nature protocols. 2010;5(4):725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roy A, Dong Xu, Jonathan Poisson, Yang Zhang. A protocol for computer-based protein structure and function prediction. Journal of visualized experiments. 2011;57:e3259. doi: 10.3791/3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang Y. I-TASSER server for protein 3D structure prediction. BMC bioinformatics. 2008;9(1):40. doi: 10.1186/1471-2105-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Green MR, Sambrook J. Molecular cloning a laboratory manual. forth ed. cold spring harbor laboratory press; 2014. [Google Scholar]
- 17.Vogel H. A convenient growth medium for Neurospora (medium N) Microbial genetics bulletin. 1956;13:42–43. [Google Scholar]
- 18.Khalaj V, Azizi M, Enayati S, Khorasanizadeh D, Ardakani EM. NCE102 homologue in Aspergillus fumigatus is required for normal sporulation, not hyphal growth or pathogenesis. FEMS microbiology letters. 2012;329(2):138–145. doi: 10.1111/j.1574-6968.2012.02513.x. [DOI] [PubMed] [Google Scholar]
- 19.Khalaj V, Eslami H, Azizi M, Rovira-Graells N, Bromley M. Efficient downregulation of alb1 gene using an AMA1-based episomal expression of RNAi construct in Aspergillus fumigatus. FEMS microbiology letters. 2007;270(2):250–254. doi: 10.1111/j.1574-6968.2007.00680.x. [DOI] [PubMed] [Google Scholar]
- 20.Bromley M, Gordon C, Rovira-Graells N, Oliver J. The Aspergillus fumigatus cellobiohydrolase B (cbhB) promoter is tightly regulated and can be exploited for controlled protein expression and RNAi. FEMS microbiology letters. 2006;264(2):246–254. doi: 10.1111/j.1574-6968.2006.00462.x. [DOI] [PubMed] [Google Scholar]
- 21.van Hartingsveldt W, Mattern IE, van Zeijl CM, Pouwels PH, van den Hondel CA. Development of a homologous transformation system for Aspergillus niger based on the pyrG gene. Molecular genetics and genomics. 1987;206(1):71–75. doi: 10.1007/BF00326538. [DOI] [PubMed] [Google Scholar]
- 22.Cannon RD, Lamping E, Holmes AR, Niimi K, Baret PV, Keniya MV, Tanabe K, Niimi M, Goffeau A, Monk BC. Efflux-mediated antifungal drug resistance. Clinical microbiology reviews. 2009;22(2):291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abadio AK, Kioshima ES, Teixeira MM, Martins NF, Maigret B, Felipe MS. Comparative genomics allowed the identification of drug targets against human fungal pathogens. BMC genomics. 2011;12:75. doi: 10.1186/1471-2164-12-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaw PE. Peptidyl-prolyl isomerases: a new twist to transcription. EMBO reports. 2002;3(6):521–526. doi: 10.1093/embo-reports/kvf118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Michnick SW, Rosen MK, Wandless TJ, Karplus M, Schreiber SL. Solution structure of FKBP, a rotamase enzyme and receptor for FK506 and rapamycin. Science. 1991;252(5007):836–839. doi: 10.1126/science.1709301. [DOI] [PubMed] [Google Scholar]
- 26.Van Duyne GD, Standaert RF, Karplus PA, Schreiber SL, Clardy J. Atomic structure of FKBP-FK506, an immunophilin-immunosuppressant complex. Science. 1991;252(5007):839–842. doi: 10.1126/science.1709302. [DOI] [PubMed] [Google Scholar]
- 27.Giaever G, Shoemaker DD, Jones TW, Liang H, Winzeler EA, Astromoff A, Davis RW. Genomic profiling of drug sensitivities via induced haploinsufficiency. Nature genetics. 1999;21(3):278–283. doi: 10.1038/6791. [DOI] [PubMed] [Google Scholar]
- 28.Alam MK, El-Ganiny AM, Afroz S, Sanders DA, Liu J, Kaminskyj SG. Aspergillus nidulans galactofuranose biosynthesis affects antifungal drug sensitivity. Fungal genetics and biology. 2012;49(12):1033–1043. doi: 10.1016/j.fgb.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 29.Hennig L, Christner C, Kipping M, Schelbert B, Rücknagel KP, Grabley S, Küllertz G, Fischer G. Selective inactivation of parvulin-like peptidyl-prolyl cis/trans isomerases by juglone. Biochemistry. 1998;37(17):5953–5960. doi: 10.1021/bi973162p. [DOI] [PubMed] [Google Scholar]


