Abstract
Background:
When Staphylococcus aureus is grown in the presence of high concentration of external glucose, this sugar is phosphorylated by glucokinase (glkA) to form glucose-6-phosphate. This product subsequently enters into anabolic phase, which favors biofilm formation. The presence of ROK (repressor protein, open reading frame, sugar kinase) motif, phosphate-1 and -2 sites, and tyrosine kinase sites in glkA of S. aureus indicates that phosphorylation must regulate the glkA activity. The aim of the present study was to identify the effect of phosphorylation on the function of S. aureus glkA and biofilm formation.
Methods:
Pure glkA and protein-tyrosine kinase (BYK) of S. aureus ATCC 12600 were obtained by fractionating the cytosolic fractions of glkA1 and BYK-1 expressing recombinant clones through nickel metal chelate column. The pure glkA was used as a substrate for BYK, and the phosphorylation of glkA was confirmed by treating with reagent A and resolving in SDS-PAGE, as well as staining with reagent A. The kinetic parameters of glkA and phosphorylated glkA were determined spectrophotometrically, and in silico tools were used for validation. S. aureus was grown in brain heart infusion broth, which was supplemented with glucose, and then biofilm units were calculated.
Results:
Fourfold elevated glkA activity was observed upon the phosphorylation by BYK. Protein-protein docking analysis revealed that glkA structure docked close to the adenosine triphosphate-binding site of BYK structure corroborating the kinetic results. Further, S. aureus grown in the presence of elevated glucose concentration exhibited an increase in the rate of biofilm formation.
Conclusion:
The elevated function of glkA is an essential requirement for increased biofilm units in S. aureus, a key pathogenic factor that helps its survival and the progress of infection.
Keywords: Glucokinase, Protein-tyrosine kinase, in silico analysis
INTRODUCTION
Staphylococcus aureus, a leading Gram-positive pathogen capable of infecting any anatomical locales, lives as a biofilm in the nasopharyngeal tract and causes a plethora of infections[1-3]. Biofilm formation is facilitated by secretory products such as hemolysins and toxins, as well as by the redox status in the S. aureus. The redox status is determined by the tricarboxylic acid (TCA) cycle, and this pathogen possesses a complete Krebs cycle[4,5]. As indicated in earlier studies, isocitrate dehydrogenase activity is decreased with the increase of biofilm units in an anaerobic condition[6-8]. The lactate dehydrogenase activity can be observed with the increments of glucose concentration and the upregulation of lactate, which is known to inhibit TCA cycle inhibitor and to create high reductive conditions in organism[6-8].
In S. aureus, in low concentration of external glucose, the glucose is metabolized through phosphotransferase system, while in high concentration, it is metabolized through the activity of glucokinase (glkA) and glucose permease enzymes[9]. The glkA catalyses the formation of glucose-6-phosphate, which plays a pivotal role in the biosynthesis of the cell wall, capsular polysaccharide, exopolysaccharide, and polysaccharide intracellular adhesion molecules[5,10,11]. All the mentioned molecules are key players in the formation of biofilms[7].
In the current study, we observed elevated biofilm units when this pathogen was grown in the presence of increasing glucose concentrations. In our previous study, we cloned glkA gene from S. aureus. The glkA gene sequence of S. aureus revealed the presence of adenosine triphosphate (ATP), phosphate 1- and 2-binding sites, and protein-tyrosine kinase (BYK) sites[12], which are known as the characteristic features of archaea glkAs. The glkA protein of the repressor protein, open reading frame, and sugar kinase (ROK) family has a regulatory function that is absent in other non-ROK glks.
In different bacteria, glkA plays an essential role in the rate-limiting reactions regulating the carbon flux into the pentose phosphate pathway, which is mostly involved in the anabolic biosynthesis[6,13,14]. In Gram-positive bacteria, the active glkA exposes Y residues distinct from normal conditions[15], which makes this enzyme highly vulnerable for phosphorylation by tyrosine kinases. Therefore, the existence of ROK motif and protein tyrosine phosphorylation sites in glkA may indicate that glkA is regulated by BYK.
BYK of S. aureus is a unique enzyme involving in the biosynthesis of capsular polysaccharide. This enzyme or CapB protein functions in association with its cognate transmembrane CapA. CapB has also the phosphorylation property and acts as an octamer of 230 kDa, which gets dissociated upon auto-phosphorylation[16,17]. BYK is actively involved in the pathogenesis and facilitates the internalization of S. aureus by epithelial cells in association with fibronectin-binding protein[10,11,18]. This BYK gene (CapB), which is 0.7 kb in size, has two conspicuous motifs referred to as Walkers A and B in the ATP-binding region, which is absent in its eukaryotic counterparts. These enzymes are phosphorylated at the tyrosine residues on the proteins, as well as on its own tyrosine residue[16,17]. These tyrosine phosphorylation and dephosphorylation are involved in the exo-polysaccharide biosynthesis, biofilm formation, and community development[19,20].
In order to understand the BYK kinetics in S. aureus, we developed a unique method using peptide BYKs as a substrate treated with reagent A, which specifically interacts with phosphorylated proteins to develop deep blue color. The absorbance of blue color can be measured spectrophotometrically at 820 nm[8]. The phosphorylated proteins can be also resolved in SDS-PAGE and identified by staining with reagent A. As a matter of fact, the phosphorrylated protein has slower mobility in acrylamide gel compared to non-phosphorylated counterparts. Moreover, the reagent A particularly reacts with phosphorylated molecules and as a result, the bands appeared on the SDS-PAGE gel are only phosphorylated proteins[8]. In the present study, BYK gene from S. aureus ATCC 12600 was cloned, sequenced, expressed and characterized. Furthermore, the effect of BYK-mediated phosphorylation on glkA activity and the enzyme kinetics of phosphorylated glkA were determined. The formation of S. aureus biofilm in BHI (brain heart infusion) broth and in BHI broth supplemented with glucose was also examined. Further, to substantiate these results, in silico tools were used.
MATERIALS AND METHODS
Cultural characterization of S. aureus ATCC 12600
S. aureus ATCC 12600 was propagated on modified Baird-Parker agar media at 37°C overnight. After an overnight incubation, a single black shiny colony with distinct zone was inoculated in BHI broth and grown at 37°C for 15 h. Subsequently, the chromosomal DNA, cytosolic and membrane fractions were extracted[21].
Amplification and sequencing of BYK gene from S. aureus ATCC 12600
The amplification of BYK gene was carried out usingthe primers used for BYK amplification were forward primer BYK-1: 5’-CATGACGAATACACG-3’ and reverse primer BYK-2: 5’-TCATGATTCATCA GT-3’. Polymerase chain reaction (PCR) was performed in Master Gradient thermocycler (Eppendorf) in a 50-µl volume. The reaction mixture contained 0.5 µg chromosomal DNA, 100 µM dNTPs mix, 100 pM forward and reverse primers, 15 mM Tris-HCl (pH 8.8), 2.5 mM MgCl2, and 1 U HotStart Taq DNA polymerase. PCR was set under the following conditions: an initial denaturation at 94°C for 10 minutes, followed by 40 cycles of 94°C for 60 seconds, 38.5°C for 30 seconds, 72°C for 90 seconds, and a final extension at 72°C for 7 minutes. The purification of PCR products was achieved using Nucleo-pore® Quick PCR purification kit (Genetix Biotech Asia Pvt. Ltd., India). The sequence of PCR products deciphered using dye terminating method at commercial sequencing facility, MWG BioTech Ltd., Bengaluru, India. The obtained sequences were analyzed and deposited at NCBI GenBank (accession No. GU353130).
Multiple sequence alignments of BYK sequence was performed using ClustalX v. 1.83 software. For this purpose, the BYK sequences from gi.284520866 (S. aureus ATCC 12600), gi.157154711 (Escherichia coli), gi.255767013 (Bacillus subtilis), gi.161612313 (Salmonella enteric), gi.218708088 (Vibrio splendidus), gi.206575712 (Klebsiella pneumonia), and gi.229587578 (Pseudomonas fluorescens) were retrieved from NCBI database and then aligned.
Cloning of S. aureus ATCC 12600 BYK gene into pQE-30 vector
The amplified PCR product of 0.7 kb corresponding to BYK gene was extracted from the agarose gel and treated with Klenow fragment (New England Bio labs, USA) following the manufacturer’s protocol. The blunt-ended PCR product was cloned in the SmaI site of pQE-30 plasmid (QIAGEN Inc., USA) and transformed into E. coli DH5α, and the obtained clone named as BYK-1[7,21].
Over-expression of BYK and glkA genes from BYK-1 and glkA1 clones
The BYK-1 clone was grown in Luria-Bertani broth containing 50 µg/ml ampicillin (OD540nm=0.6) at 37°C for 5 h. At this point, the expression of BYK was initiated by adding 0.75 mM isopropyl β-D-1-thiogalactopyranoside and incubated for 4 h. Then the culture was centrifuged, and the pellet was resuspended in 8 ml sonication buffer (0.1 M Tris-HCl, pH 7.5, 0.05M EDTA, and 0.25% sucrose) and sonicated to release the cytosolic fraction. The recombinant BYK (rBYK) was then purified from the cytosolic fraction of BYK-1 clone by passing through nickel metal chelate agarose column. Next, purified protein was dialysed against 0.1 M Tris-HCl (pH 7.5) and analyzed on 10% SDS-PAGE[7,21].
The recombinant glkA, which was prepared from glkA1 clone generating from earlier study[12] The related cytosolic fraction was purified by affinity chromatography through nickel metal chelate agarose column and the pure enzyme was used to determine the glkA kinetics and the regulation studies by BYK. The glkA assay was performed in 2 ml reaction mixture containing 60 mM Tris-HCl (pH 7.5), 0.5 mM MgCl2, 0.2 M ATP, 0.9 mM nicotinamide adenine dinucleotide phosphate, 10 units glucose-6-phosphate dehydro-genase, 12 mM glucose (substrate), and 10 ml enzyme (pure glkA obtained from glkA1 clone) and then incubated at 37°C for 30 min. The absorbance was measured at 340 nm against blank (without enzyme). Kinetic parameters, Km and Vmax, for glkA were determined using HaneseWoolf plot ([S] vs. [S]/V). Hills coefficient was also calculated to accentuate the specificity of glkA towards glucose[12].
BYK enzyme assay
The BYK activity was detected in both S. aureus ATCC 12600 and in rBYK. Enzyme assay was performed at 30°C using a novel non-radiolabeled synthetic peptide acted as a substrate on a Cyber lab spectrophotometer (USA). BYK assay mixture containing 0.1 M Tris-HCl (pH 7.5), 10 mM MgCl2, 100 mM ATP, 22.3 μM (30 µg/μl) BYKs peptide (BYKs=HGLDNYRGYSLG), and 0.5 µg/µl enzyme fraction (pure His tag BYK and membrane fraction) was incubated at 30°C for 10 minutes. The phosphorylated peptide was purified by passing through Sephadex G-25 column (1 cm×15 cm), and the fractions were eluted with 0.1 M Tris-HCl (pH 7.5) and 150 mM NaCl. The enzyme fraction was appeared in the void volume, while the phosphorylated peptide was obtained in the first elution volume. Also, bound phosphorous was estimated by adding freshly prepared reagent A (3.4 mM ammonium molybdate, 0.5 mM sulphuric acid, 0.5 M SDS, and 0.6 M L-ascorbic acid) and incubated at 30°C for 15 minutes. The absorbance was measured at 820 nm against blank (0.1 M Tris-HCl [pH 7.5] and 150 mM each of NaCl and reagent A)[8,22,23].
The BYK activity was defined as the amount of phosphate added to 1 µg peptide per ml at 30°C for 60 s. Standard KH2PO4 was used to develop the calibration curve for the estimation of the inorganic phosphate and free phosphate by adding reagent A[8]. The phosphorylation of peptide was demonstrated by fractionating the eluted peptide on 15% SDS-PAGE and subsequentstaining of the gel with reagent A. The appearance of blue colored bands in the gel indicated the presence of phosphorylated peptide. Similarly, the same reaction mixture without BYKs estimated the auto-phosphorylation property of BYK resolving the phosphorylated BYK on 15% SDS-PAGE gel. The enzyme activity was measured in 1 ml as the amount of phosphate added per microgram enzyme at 30°C in one minute. The effect of substrate concentration on enzyme activity was determined by taking different substrate concentrations (20, 40, 60, 80, 100, and 120) of synthetic peptide keeping the ATP concentration at a constant level. The kinetic parameters, Km and Vmax, were calculated from [S] vs. [S/V] (Hanes-Woolf) plot for both BYKs and auto-phosphorylation activity of BYK.
In vitro phosphorylation of glkA by BYK and its effect on the glkA kinetics
The BYK enzyme assay was carried out using 10 μg/ml pure glkA as substrate and 30 μg/ml pure BYK. The phosphorylated glkA purified from the column was used to carry out glkA enzyme assay[12]. Further, the phosphorylated enzyme was fractionated in 10% SDS-PAGE.
Biofilm assay
The biofilm assay was carried out for S. aureus ATCC 12600 grown in BHI broth and BHI broth enriched with 0.15% glucose[7]. The biofilm formation assay was performed in 96-well flat-bottomed polystyrene plates. S. aureus overnight cultures were diluted to 1:100 in BHI broth, and 200 µl diluted culture was added to each well and incubated at 37°C for 24 h. The supernatant from each well was carefully removed, and the biofilms formed was washed with phosphate buffered saline, pH 7.4. The wells were air dried and stained with 0.4% crystal violet and the distaining was completed with sterile distilled water. The absorbance at 570 nm was recorded in a microplate reader and referred as Abiofilm. Simultaneously, S. aureus was grown in BHI broth in static conditions at 37°C overnight, and the absorbance was recorded at 570 nm and referred as Agrowth. In addition, biofilm units were calculated as Abiofilm/Agrowth[7]. Bradford’s method was also used to determine the protein concentrations in all steps[24].
In silico analysis of BYK
The annotated BYK protein sequence was analyzed by using open-source software, including BLAST (v. 2.1.1), BioEdit (v. 7.2), Mega 4.1, ClustalW and ClustalX, and Modeller 9v8. The BYKs structure was built using Gallus gallus proteolyzed lysozyme protein data bank (PDB) (ID: 3ZVQ) as a template, which showed more than 90% homology. The built structure was validated by PROCHECK[25].
Protein-protein docking
The molecular docking was carried out using Hex v6.3 software between BYK, BYKs, and glkA structures to predict the possible modes of interactions[26]. To initiate the protein-protein docking, BYK structure was considered as the receptor and also synthetic peptides (BYKs) and glkA as ligand. Both were loaded into HEX working environment, and docking search was started by rotating the receptor and ligand about their centroids at their intermolecular distances. Both ligands were assigned to Euler rotation angles and a six-dimensional search was carried out over full rotational ranges. An initial steric scan at n=18 was followed by a final search at n=25 using the steric contribution to the docking energy. Each orientation was evaluated using a steric and an electrostatic correlation to order N for the significant increment of total docking times. High-resolution final search correlation was performed using smaller distance increments for the fast low-resolution steric scan phase for the rapid and finer coverage of search space at the final phase. This process was controlled by the distance range of 40, scan step of 0.8, and two sub-steps using docking controls. The angular increments of 7.5 degrees and a twist angle of 5.5 degrees were used in the rotational search in each of the ligand and receptor rotational angles. Newton-like energy minimization was applied, and single molecular mechanics energy was calculated for each docking conformation using Lennard-Jones and hydrogen bond potentials, adapted from the OPLS force-field limits, along with a clear charge-charge electrostatic input. Global rotational search and translational space scanning were carried out using spherical Polar Fourier transformations, which can rank the output based on surface complementarities and electrostatic potentials.
RESULTS
S. aureus biofilm formation
S. aureus grown in different concentrations of glucose showed a gradual increase in biofilm units, which was the highest at 0.15% glucose (0.059±0.04 and 0.161±0.05 biofilm units in BHI broth containing 0.15% glucose). Further, the increase of the glucose concentration reduced the bacterial growth, indicating that the formation of glucose-6-phosphate by glkA must be a regulated function in this pathogen.
Sequence analysis, expression and characterization of glkA from glkA1 clone
The glkA amino acid sequence (GenBank: JN645812) was scanned in PROSITE and the results indicated the presence of BYK sites, “KGIYDS” and “IKTEYHN”, phosphate-1 and phosphate-2 motifs, and sub-domains connect-1 and connect-2, which attach β- and γ-phosphates of ATP (Fig. 1). This may highlight the possible regulation of glkA through phosphorylation. The glkA of S. aureus ATCC 12600 was over-expressed in E. coli and the purified glkA was characterized (Fig. 2 and Table 1) and used in the phosphorylation experiment.
Fig. 1.
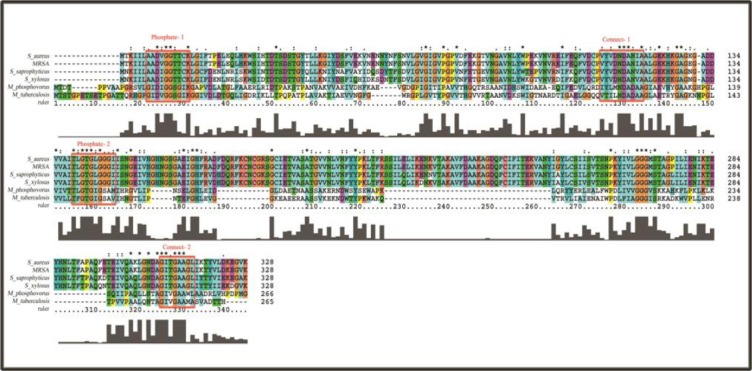
The multiple sequence alignments showing the presence of phosphate-1 and phosphate-2 motifs and the sub-domains connect-1 and connect-2 in the glkA sequence of Staphylococcus aureus, as well as Poly(P) ATP-dependent glkA of Microlunatus phosphovorus and M. tuberculosis. Boxes show that the phosphate-1 and phosphate-2 motifs and the sub-domains connect-1 and connect-2 in the glkA sequence are conserved in all these organisms.
Fig. 2.
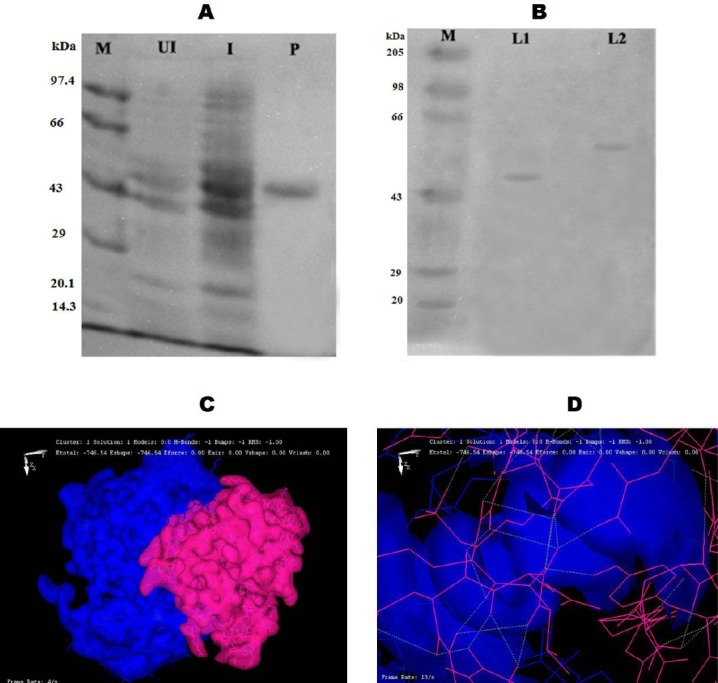
Structural and functional analysis of phosphorylated glkA. A) The expression and purification of rglkA from glkA1 clone using SDS-PAGE (10%). Lane M, protein molecular weight markers (Merck Bioscience, India Pvt. Ltd.); Lane UI, uninduced cytosolic fraction of glkA1 clone.; lane I, isopropyl-D-1-thiogalactopyranoside-induced cytosolic fraction of glkA1 clone; Lane P, nickel metal agarose column purified glkA. B) SDS-PAGE (10%) gel showing phosphorylated rglkA identified by reagent A, followed by Coomassie Brilliant Blue R250 staining. Lane M, protein molecular weight markers (Merck Bioscience, India Pvt. Ltd.); lane L1, normal rglkA; lane L2, phosphorylated rglkA. C) An image showing the formation of hydrogen bond between the glkA protein (blue) and BYK protein (pink). D) An image showing the interaction of ATP-binding site of glkA protein (blue) with BYK protein (pink).
Table 1.
Enzyme kinetics of recombinant glucokinases (glkA)
| Source of glkA | Enzyme activity (mMNADH/ml/min) | Vmax (mMNADH/mg/min) | KM (mM) | Hill coefficient S0.5 (mM) |
|---|---|---|---|---|
| rglkA | 1.05± 0.05 | 3.5±0.2 | 5.10±0.06 | 1.66±0.032 |
| Phosphorylated rglkA | 3.89±0.50 | 11.65±1.5 | 5.18±0.50 | 1.62±0.024 |
Values are the mean±SD from three determinations
Cloning, expression, and characterization of BYK and glkA genes of S. aureus ATCC 12600
The BYK gene (0.7 kb) was isolated from S. aureus ATCC 12600 chromosomal DNA using PCR and sequenced (GenBank accession number: GU353130). The sequence analysis demonstrated a complete homology with CapB gene sequence reported in all S. aureus strains, indicating that only one BYK is present in S. aureus[16,17,27]. The BYK gene was cloned in the SmaI site of pQE-30 vector, and the clone was named as BYK-1. The purified rBYK enzyme exhibited a single band with a molecular weight of 27 kDa corresponding to the monomeric form of BYK (Fig. 3). The annotated protein sequence of BYK showed the presence of Walker A and B motifs; Walker A “APGAGKST” was located between 50th and 57th amino acid sequences of BYK, and Walker B “FVIIDTP” was found between 153rd and 158th. These conserved motifs are specific for all BYKs[16]. The multiple sequence alignment results indicated that S. aureus BYK exhibits considerable variation in the sequence with other Gram-positive and Gram-negative bacteria. Also, phylogenetic analysis showed the substantial distance from other bacteria (Fig. 4A and 4B)[16,17,27]. To define the enzyme kinetics of BYK, synthetic peptide BYKs were synthesized and used in phosphorylation experiment. The results (Table 2 and Fig. 5) showed complete phosphorylation of BYKs by the enzyme, which could be identified with reagent A. In addition, the phosphorylated BYKs and BYK could be fractionated in SDS-PAGE, and on staining with reagent A, blue colored bands appeared, indicating the phosphorylation of substrate and similarly, the auto-phosphorylation of the enzyme (Fig. 6).
Fig. 3.
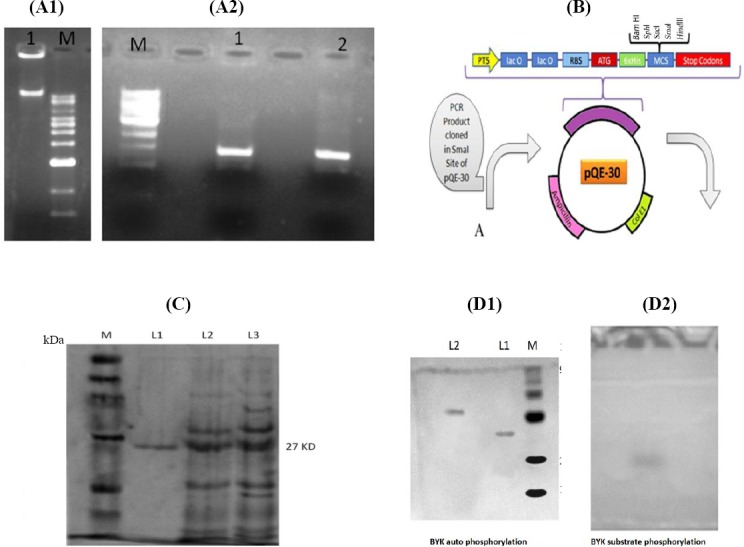
Cloning, expression, and characterization of BYK. A1) 1% agarose gel showing the presence of chromosomal DNA from S. aureus ATCC 12600. Lane 1, chromosomal DNA of S. aureus ATCC 12600; Lane M, molecular size marker. A2) PCR amplification of BYK gene of S. aureus ATCC 12600. Lane M, molecular size marker (33,500-500 bp); Lanes 1 and 2, PCR-amplified products. B) The schematic representation of pQE-30 plasmid vector. A represents PCR product cloned in the SmaI site of pQE vector. C) Expression and purification of rBYK from BYK-1 clone using SDS-PAGE (10%). Lane M, protein molecular weight markers (Merck Bioscience Private Ltd., India); Lane L1, nickel metal agarose column purified BYK; Lane L2, cytosolic fraction of uninduced BYK-1 clone; Lane L3, cytosolic fraction of isopropyl-D-1-thiogalactopyranoside-induced BYK-1 clone. D1) SDS-PAGE gel showing phosphorylated rBYK identified by reagent A, followed by Coomassie Brilliant Blue R250 staining. Lane M, protein molecular weight marker; Lane L1, normal rBYK; Lane 2, phosphorylated rBYK. D2) Pure phosphorylated rBYK and substrate BYKs separated in SDS-PAGE. All markers were obtained from Bangalore Genei Private Ltd. (India) otherwise stated.
Fig. 4.
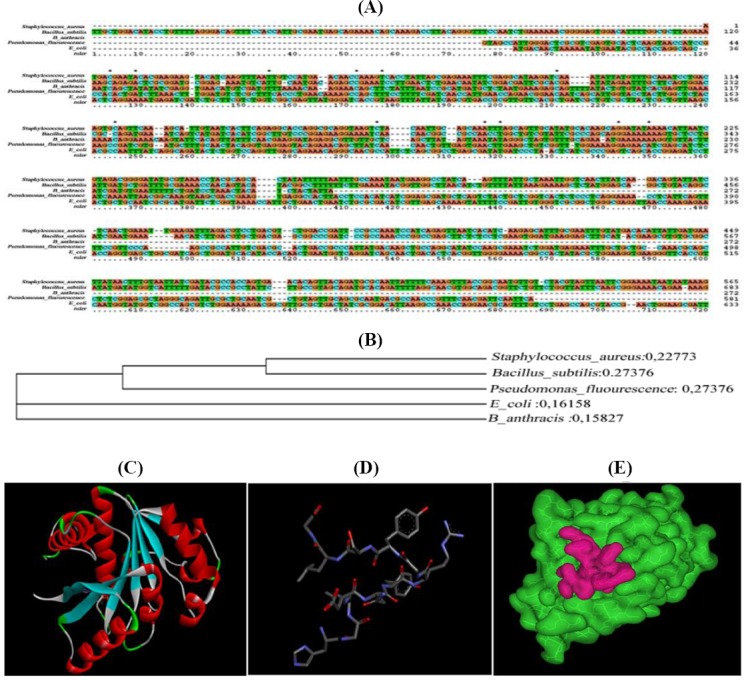
Evolutionary relationship of BYK and docking analysis of substrate BYKs. A) Multiple sequence alignments of 5 representative bacterial BYK. S. aureus ATCC 12600 (from the present study), Escherichia coli, Bacillus subtilis, Pseudomonas fluorescens, and Bacillus anthrasis BYK reported in the database were retrieved, and sequence alignment file was generated in protein information resource format for Query and template sequences using ClustalX tool. The homologous regions are shown in brown color, and variations are depicted in other colors. B) Phylogenetic analysis of S. aureus BYK with other bacterial BYKs; C) Three-dimentional structure of BYK catalytic domain; D) Three-dimentional structure of synthetic peptide; E) An iage showing the formation of hydrogen bond between the BYK protein (green) and synthetic peptide (red).
Table 2.
Enzyme kinetics of pure recombinant BYK (rBYK)
| Source of BYK | Enzyme activity (mM/ml/min) | KM (mM) | Vmax (mM/mg/min) |
|---|---|---|---|
| rBYK | 0.020±0.0100 | 0.750±0.10 | 0.42±0.10 |
| rBYK+membrane fraction | 0.064±0.009 | 0.206±0.07 | 1.006±0.43 |
| rBYK+membrane fraction+ substrate BYKs | 0.703±0.080 | 0.333±0.08 | 9.53±1.45 |
Values are the mean±SD from three determinations. 1, basal rBYK kinetics; 2, auto-phosphorylation kinetics; 3, substrate-level phosphorylation kinetics.
Fig. 5.
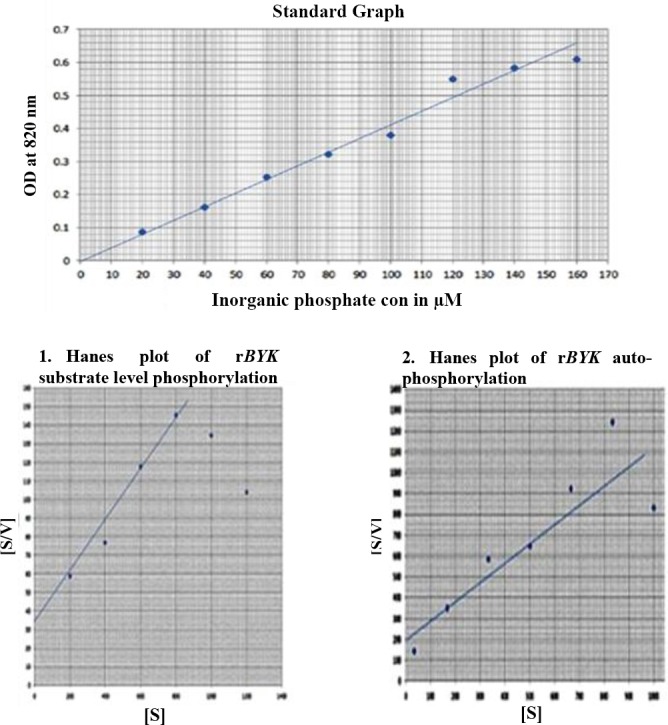
Kinetic parameters of rBYK.
Fig. 6.
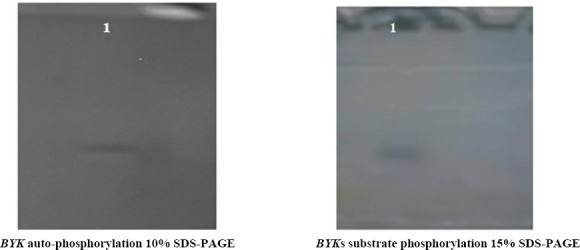
Auto-phosphorylation and substrate-level phosphor-rylation of BYK. 1, lane 1.
The BYKs structure was built by Modeller 9v8 using the template Gallus gallus proteolyzed lysozyme PDB (ID: 3ZVQ). The predicted structure of BYKs was validated through the PROCHECK, and the resultant Ramachandran plot indicated that 87.5% of the residues were structurally in favored region, and 12.5% were in additional allowed regions. This result suggested that the predicted model was valid with good stereochemical quality (Fig. 4C). The synthetic peptide BYKs structure used as a substrate and was docked with the BYK structure (PDB ID: 3BFV) (Fig. 4D). The results showed that BYKs were interacting with BYK structure very close to the ATP-binding site (Fig. 4E).
A list of 500 docked complexes was produced that are spatially having similar docking orientations. These conformations were grouped and ordered by energy. Among all the conformations, the first one with the lowest complex energy was chosen for the analysis. There was an initial distance of 28.9Å among the centroids of the two molecules when loaded into the working environment. After docking process, the first docked conformation demonstrated a distance of 13.6Å. From all the docked conformations, the interacting residues were found to be S9, Q30, T57, K82, T84, Q188, K192, R212, and Y222 from BYK and N5, Y6, R7, Y9, and G12 from BYKs, which interact together by forming hydrogen bonds with the best minimum complex energy of -240.44 kcal/mol (Fig. 4E). The interaction of these residues with BYKs substrate, which is close to the ATP-binding site, induced catalysis and phosphorylation of the substrate. This validates the use of synthetic peptide BYKs as a substrate, which corroborates with enzyme kinetic results (Fig. 4E and Table 2). The CapB protein expressed in cytosolic fraction in rBYK-1 clone showed maximum activity in the presence of membrane fraction. The enzyme kinetics of BYK indicated that similar to all phosphokinases, the auto-phosphorylation is followed by substrate-level phosphorylation[16,17,27] (Table 2 and Fig. 6).
The effect of BYK-mediated phosphorylation on glkA
The phosphorylating property of rBYK was demonstrated using pure glkA as substrate (Fig. 2A). The phosphorylated glkA was purified by passing through Sephadex G-25 column and reacted with reagent A both in the solution and in resolved SDS-PAGE gel. In SDS-PAGE, the phosphorylated glkA exhibited reduced mobility compared to non-phosphorylated glkA (Fig. 2B), which is similar to that shown by many phosphorylated proteins[8,23] The glkA structure docked very close to the ATP-binding site of BYK structure supporting the phosphorylation of glkA by BYK (Fig. 2C and 2D). This phosphate bound enzyme exhibited fourfold increased enzyme activity compared to the normal glkA with no change in KM (Table 1).
DISCUSSION
In S. aureus, glucose is catabolized properly under aerobic conditions for energy generation with elevated TCA cycle activity[28,29]. However, under high external glucose concentrations, the glkA and the glucose permease system are optimally functional[9], and thus the formed glucose-6-phosphate is pushed into pentose phosphate pathway. This process can result in building up lactate and nicotinamide adenine dinucleotide phosphate, as well as decreasing the TCA cycle activity, which may create an anabolic redox status, favoring more biosynthesis rather than energy generation[5-8]. Also, under the stress conditions in S. aureus, the elevated levels of glycolysis and reduced TCA cycle activities were observed[7,8]. All of these conditions support the increased rate of biofilm formation and the expression of virulence factors[5-8,13,14,19,20,30]. Studies have shown that phosphorylation in S. aureus controls TCA cycle activity[8,16-20], which has a direct influence on biofilm formation. Furthermore, the elevated levels of external glucose increases biofilm formation[7]. Interestingly, in our study, the elevated biofilm units were observed when S. aureus ATCC 12600 was grown in BHI broth supplemented with 0.15% glucose. It is clear that phosphorylation of glucose in this bacterium is catalyzed by glkA[5], and the protein sequence of glkA showed the presence of ROK motif, phosphate-1 and -2, connect-1 and -2, and tyrosine phosphorylation sites[12]. Such features are commonly present in poly-phosphate/ATP-dependent glkAs[31-34]. The presence of ROK site in the glkA explains that the activity of the enzyme is highly regulated[13]; moreover, this enzyme shows a very high affinity towards glucose compared with any other counterpart[25]. To validate these findings, BYK gene (CapB) was cloned, expressed, and the resulting protein was purified. The purified BYK protein exhibited the substrate-level phosphorylation, a well-known feature of BYK. The phosphorylated proteins were detected by reagent A, which binds specifically to the bound phosphate, confirming the BYK auto-phosphorylation and substrate (BYKs)-level phosphorylation properties.
Similarly, upon the phosphorylation of glkA by BYK, the phosphorylated glkA exhibited retarded mobility in SDS-PAGE compared to normal glkA and reacted with reagent A to give distinct blue colored bands, indicating the presence of phosphate moiety bound to the glkA (Fig. 2). This phosphorylated glkA showed fourfold elevated activity compared to unphosphorylated one (Table 1, Figs. 2 and 3)[12,16,17,27]. Y phosphorylation of glkA has activated the enzyme without changing the Km of the enzyme, which means that phosphorylation has increased the glkA activity (Table 1). To substantiate these findings, glkA structure docked very close to the ATP-binding site of BYK structure (Fig. 2C and 2D). Comparable functional property was observed in C321A mutated Bacillus subtilis glkAs, which showed enhanced enzyme activity with more exposure of Y residue[15]. These observations aptly concur with our results. Such elevated glucose-6-phosphate formation is essential for S. aureus to grow in increased external glucose condition and to push the anabolic reductive condition, which favors rapid biofilm formation[13,14,20], corroborating with our findings of elevated biofilm units observed in the presence of glucose (Fig. 7).
Fig. 7.
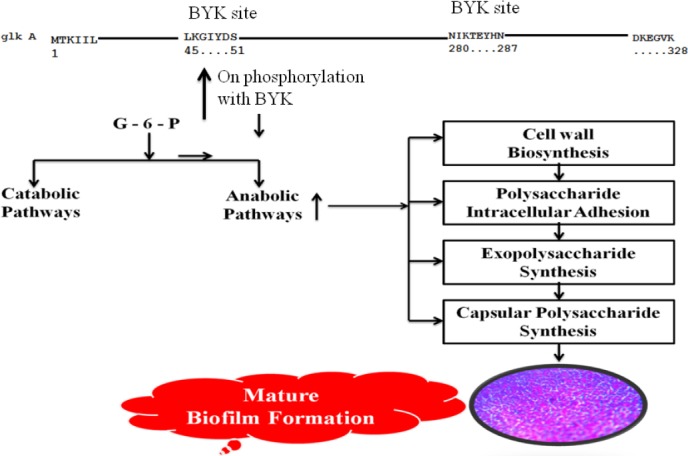
The effect of glkA phosphorylation on S. aureus, which leads to the increased biosynthesis of exopolysaccharide, capsular polysaccharide, cell wall, and polysaccharide intracellular adhesion, resulting in elevated rate of Biofilm formation.
In conclusion, phosphorylation of glkA has increased its activity by fourfold, which correlated with elevated biofilm formation in the presence of glucose. This attribute of glkA was further validated as glkA structure docked close to the ATP-binding site of BYK structure. Also, in our observations, the Km of glkA was unchanged upon phosphorylation.
ACKNOWLEDGMENTS
We sincerely acknowledge Sri Venkateswara Institute of Medical Sciences and University (Tirupati, Andhra Pradesh, India) for providing facilities to carry out this work.
Footnotes
CONFLICT OF INTEREST. None declared.
REFERENCES
- 1.Lowy FD. Staphylococcus aureus infections. The New England Journal of Medicine. 1998;339(8):520–532. doi: 10.1056/NEJM199808203390806. [DOI] [PubMed] [Google Scholar]
- 2.Götz F. Staphylococcus and biofilms. Molecular microbiology. 2002;43(6):1367–1378. doi: 10.1046/j.1365-2958.2002.02827.x. [DOI] [PubMed] [Google Scholar]
- 3.Knobloch JK, Horstkotte MA, Rohde H, Mack D. Evaluation of different detection methods of biofilm formation in Staphylococcus aureus. Medical microbiology and immunology. 2002;191(2):101–106. doi: 10.1007/s00430-002-0124-3. [DOI] [PubMed] [Google Scholar]
- 4.Caiazza NC, O’Toole GA. Alpha-toxin is required for biofilm formation by Staphylococcus aureus. Journal of bacteriology. 2003;185(10):3214–3217. doi: 10.1128/JB.185.10.3214-3217.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadykov MR, Mattes TA, Luong TT, Zhu Y, Day SR, Sifri CD, Lee CY, Somerville GA. Tricarboxylic acid cycle-dependent synthesis of Staphylococcus aureus Type 5 and 8 capsular polysaccharides. Journal of bacteriology. 2010;192(5):1459–1462. doi: 10.1128/JB.01377-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pagels M, Fuchs S, Pané-Farré J, Kohler C, Menschner L, Hecker M, McNamarra PJ, Bauer MC, von Wachenfeldt C, Liebeke M, Lalk M, Sander G, von Eiff C, Proctor RA, Engelmann S. Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Molecular microbiology. 2010;76(5):1142–1161. doi: 10.1111/j.1365-2958.2010.07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yeswanth S, Nanda Kumar Y, Venkateswara Prasad U, Swarupa V, Koteswara rao V, Venkata Gurunadha, Krishna Sarma P. Cloning and characterization of L-lactate dehydrogenase gene of Staphylococcus aureus. Anaerobe. 2013;24:43–48. doi: 10.1016/j.anaerobe.2013.09.003. [DOI] [PubMed] [Google Scholar]
- 8.Prasad UV, Vasu D, Yeswanth S, Swarupa V, Sunitha MM, Chaudhary A, Sarma PV. Phosphorylation controls the functioning of Staphylococcus aureus Isocitrate dehydrogenase - favours biofilm formation. Journal of enzyme inhibition and medicinal chemistry. 2015;30(4):655–661. doi: 10.3109/14756366.2014.959945. [DOI] [PubMed] [Google Scholar]
- 9.Götz F, Bannerman T, Schleifer KH. The genera Staphylococcus and Macrococcus. Prokaryotes. 2006;4:5–75. [Google Scholar]
- 10.McKenney D, Hübner J, Muller E, Wang Y, Goldmann DA, Pier GB. The ica locus of Staphylococcus epidermidis encodes production of the capsular polysaccharide/adhesin. Infection and immunity. 1998;66(10):4711–4720. doi: 10.1128/iai.66.10.4711-4720.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyafusa T, Caaveiro JM, Tanaka Y, Tsumoto K. Dynamic elements govern the catalytic activity of CapE, a capsular polysaccharide-synthesizing enzyme from Staphylococcus aureus. FEBS letters. 2013;587(23):3824–3830. doi: 10.1016/j.febslet.2013.10.009. [DOI] [PubMed] [Google Scholar]
- 12.Lakshmi HP, Yeswanth S, Prasad UV, Vasu D, Swarupa V, Kumar PS, Narasu ML, Krishna Sarma PV. Cloning, expression and characterization of glucokinase gene involved in the glucose-6-phosphate formation in Staphylococcus aureus. Bioinformation. 2013;9(4):169–173. doi: 10.6026/97320630009169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park SY, Kim HK, Yoo SK, Oh TK, Lee JK. Characterization of glk, a gene coding for glucose kinase of Corynebacterium glutamicum. FEMS Microbiology letters. 2000;188(2):209–215. doi: 10.1111/j.1574-6968.2000.tb09195.x. [DOI] [PubMed] [Google Scholar]
- 14.Fuchs S, Pané-Farré J, Kohler C, Hecker M, Engelmann S. Anaerobic gene expression in Staphylococcus aureus. Journal of bacteriology. 2007;189(11):4275–4289. doi: 10.1128/JB.00081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesak LR, Felix M, Mesak FM, Dahl MK. Bacillus subtilis GlcK activity requires cysteines within a motif that discriminates microbial glucokinases into two lineages. BMC microbiology. 2004;4:6. doi: 10.1186/1471-2180-4-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soulat D, Jault JM, Duclos B, Geourjon C, Cozzone AJ, Grangeasse C. Staphylococcus aureus operates protein-tyrosine phosphorylation through a specific mechanism. The Journal of biological chemistry. 2006;281(20):14048–14056. doi: 10.1074/jbc.M513600200. [DOI] [PubMed] [Google Scholar]
- 17.Olivares-Illana V, Meyer P, Bechet E, Gueguen-Chaignon V, Soulat D, Lazereg-Riquier S, Mijakovic I, Deutscher J, Cozzone AJ, Laprévote O, Morera S, Grangeasse C, Nessler S. Structural basis for the regulation mechanism of the tyrosine kinase CapB from Staphylococcus aureus. PLOS biology. 2008;6(6):e143. doi: 10.1371/journal.pbio.0060143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dziewanowska K, Patti JM, Deobald CF, Bayles KW, Trumble WR, Bohach GA. Fibronectin binding protein and host cell tyrosine kinase are required for internalization of Staphylococcus aureus by epithelial cells. Infection and Immunity. 1999;67(9):4673–4678. doi: 10.1128/iai.67.9.4673-4678.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whitmore SE, Lamont R. Tyrosine phosphorylation and bacterial virulence. International journal of oral science. 2012;4(1):1–6. doi: 10.1038/ijos.2012.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Standish AJ, Morona R. The role of bacterial protein tyrosine phosphatases in the regulation of the biosynthesis of secreted polysaccharides. Antioxidants and redox signaling. 2014;20(14):2274–2289. doi: 10.1089/ars.2013.5726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hari Prasad O, Nanda Kumar Y, Reddy OV, Chaudhary A, Sarma PV. Cloning, expression, purification and characterization of UMP kinase from Staphylococcus aureus. The protein journal. 2012;31(4):345–352. doi: 10.1007/s10930-012-9410-0. [DOI] [PubMed] [Google Scholar]
- 22.Clore JN, Stillman J, Sugerman H. Glucose-6-phosphatase flux in vitro is increased in type 2 diabetes. Diabetes. 2000;49(6):969–974. doi: 10.2337/diabetes.49.6.969. [DOI] [PubMed] [Google Scholar]
- 23.Vasu D, Sunitha MM, Srikanth L, Swarupa V, Prasad UV, Sireesha K, Yeswanth S, Kumar PS, Venkatesh K, Chaudhary A, Sarma PVGK. In Staphylococcus aureus the regulation of pyruvate kinase activity by serine/threonine protein kinase favors biofilm formation. 3 Biotech. 2015;5(4):505–512. doi: 10.1007/s13205-014-0248-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical biochemistry. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 25.Kumar PS, Kumar YN, Prasad UV, Yeswanth S, Swarupa V, Vasu D, Venkatesh K, Srikanth L, Rao VK, Sarma PV. Comparative structural and functional analysis of Staphylococcus aureus glucokinase with other bacterial glucokinases. Indian journal of pharmaceutical sciences. 2014;76(5):430–436. [PMC free article] [PubMed] [Google Scholar]
- 26.Macindoe G, Mavridis L, Venkatraman V, Devignes MD, Ritchie DW. HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic acids research. 2010;38:W445–449. doi: 10.1093/nar/gkq311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ilan O, Bloch Y, Frankel G, Ullrich H, Geider K, Rosenshine I. Protein tyrosine kinases in bacterial pathogens are associated with virulence and production of exopolysaccharide. The EMBO journal. 1999;18(12):3241–3248. doi: 10.1093/emboj/18.12.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ferreira MT, Manso AS, Gaspar P, Pinho MG, Neves AR. Effect of Oxygen on Glucose Metabolism: Utilization of Lactate in Staphylococcus aureus as revealed by in vivo NMR studies. PLoS One. 2013;8(3):e58277. doi: 10.1371/journal.pone.0058277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledala N, Zhang B, Seravalli J, Powers R, Somerville GA. Influence of iron and aeration on Staphylococcus aureus growth, metabolism, and transcription. Journal of bacteriology. 2014;196(12):2178–2189. doi: 10.1128/JB.01475-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Islam N, Kim Y, Ross JM, Marten MR. Proteomic analysis of Staphylococcus aureus biofilm cells grown under physiologically relevant fluid shear stress conditions. Proteome science. 2014;12:21. doi: 10.1186/1477-5956-12-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hsieh PC, Shenoy BC, Samols D, Phillips NF. Cloning, expression, and characterization of polyphosphate glucokinase from Mycobacterium tuberculosis. The journal of biological chemistry. 1996;271(9):4909–4915. doi: 10.1074/jbc.271.9.4909. [DOI] [PubMed] [Google Scholar]
- 32.Tanaka S, Lee SO, Hamaoka K, Kato J, Takiguchi N, Nakamura K, Ohtake H, Kuroda A. Strictly polyphosphate-dependent glucokinase in a polyphosphate-accumulating bacterium, Microlunatus phosphovorus. Journal of bacteriology. 2003;185(18):5654–5656. doi: 10.1128/JB.185.18.5654-5656.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hansen T, Schönheit P. ATP-dependent glucokinase from the hyperthermophilic bacterium Thermotoga maritima represents an extremely thermophilic ROK glucokinase with high substrate specificity. FEMS microbiology letters. 2003;226(2):405–411. doi: 10.1016/S0378-1097(03)00642-6. [DOI] [PubMed] [Google Scholar]
- 34.Mukai T, Kawai S, Mori S, Mikami B, Murata K. Crystal structure of bacterial inorganic polyphosphate/ATP-glucomannokinase. Insights into kinase evolution. The journal of biological chemistry. 2004;279(48):50591–50600. doi: 10.1074/jbc.M408126200. [DOI] [PubMed] [Google Scholar]


