Abstract
Epigenetic silencing of fragile X mental retardation 1 (FMR1) causes fragile X syndrome (FXS), a common inherited form of intellectual disability and autism. FXS correlates with abnormal synapse and dendritic spine development, but the molecular link between the absence of the FMR1 product FMRP, an RNA binding protein, and the neuropathology is unclear. We found that the messenger RNA encoding bone morphogenetic protein type II receptor (BMPR2) is a target of FMRP. Depletion of FMRP increased BMPR2 abundance, especially that of the full-length isoform that bound and activated LIM domain kinase 1 (LIMK1), a component of the noncanonical BMP signal transduction pathway that stimulates actin reorganization to promote neurite outgrowth and synapse formation. Heterozygosity for BMPR2 rescued the morphological abnormalities in neurons both in Drosophila and in mouse models of FXS, as did the postnatal pharmacological inhibition of LIMK1 activity. Compared with postmortem prefrontal cortex tissue from healthy subjects, the amount of full-length BMPR2 and of a marker of LIMK1 activity was increased in this brain region from FXS patients. These findings suggest that increased BMPR2 signal transduction is linked to FXS and that the BMPR2-LIMK1 pathway is a putative therapeutic target in patients with FXS and possibly other forms of autism.
INTRODUCTION
Fragile X syndrome (FXS) is the most common heritable form of intellectual disability and autism spectrum disorder (ASD) (1–3). FXS is caused by an expansion of trinucleotide (CGG) repeats in the fragile X mental retardation 1 (FMR1) gene that leads to transcriptional silencing and loss of the FMR1 protein (FMRP) (3–7). FMRP is an RNA binding protein that contains three RNA binding domains: two K homology (KH) domains (KH1 and KH2) and one arginine-glycine-glycine–rich domain (8–11). FMRP binds mRNAs directly, modulating transport and stability and negatively regulating translation (12–15). A missense mutation in the KH2 domain (I304N) that abolishes RNA binding activity has been identified in a patient with FXS (16, 17). This suggests that the pathogenesis of FXS might be caused by the absence of the RNA binding function of FMRP. Both postmortem brain tissues from FXS patients (18, 19) and from young adult mice lacking FMR1 (20, 21) exhibit widespread defects in synaptic plasticity and development, including an increased number of long and thin dendritic spines instead of the mature and strong mushroom-shaped spines. Transcripts of 40 genes have been identified as FMRP targets (22–25). Most encode proteins that are implicated in synaptic growth, plasticity, cell adhesion, or cytoskeletal structure and remodeling (15, 26). However, it is not known whether an increase in the abundance of these targets is responsible for the FXS phenotype or whether there are additional FMRP targets that contribute to the disease.
Bone morphogenetic proteins (BMPs) are members of the transforming growth factor–β (TGFp) superfamily of secreted proteins (27, 28). Their receptors consist of type I and type II subunits, which are both serine/threonine kinases (28). BMPR2 is a type II subunit that, in addition to the kinase domain (KD) in its C-terminal domain (CTD) that is typical of type II subunits, has a unique, evolutionarily conserved ~500–amino acid region. BMP binding leads to activation of the type I receptor and activation of the canonical pathway through phosphorylation of SMAD1, SMAD5, and SMAD8 (28); it also leads to a “noncanonical” signal that is mediated by proteins, such as LIM (Lin-11 Isl-1 Mec-3) domain kinase 1 (LIMK1), that interacts with the CTD (29). Once activated by BMPR2, LIMK1 phosphorylates and inhibits cofilin to promote neurite outgrowth and dendritogenesis (29). In cortical neurons, activation of the non-canonical pathway also involves the p21-activated kinase 1 (PAK1), which facilitates the recruitment of the type I BMP receptor to BMPR2 and the subsequent activation of LIMK1 (30).
The BMPR2-CTD-LIMK1 signaling pathway is conserved between human BMPR2 and the Drosophila BMPR2 ortholog wishful thinking (Wit). Although the CTD sequence homology is only ~30%, both CTDs interact with LIMK1, promote cofilin phosphorylation, and induce actin remodeling (29–36). At the Drosophila neuromuscular junction (NMJ), Glass bottom boat, a Drosophila BMP ortholog that is released from the postsynaptic muscle, binds Wit and regulates neuromuscular synapse formation, stability, and function (31, 32, 37, 38). Mutant wit larvae exhibit a reduced number of mature boutons, and this phenotype can be rescued by Wit but not by Wit that lacks the CTD (32), supporting an essential role of the CTD-LIMK1 interaction and noncanonical signaling in synaptic stability and growth at the NMJ. BMPR2 mRNA is not among the previously known targets of FMRP. However, because published genome-wide analysis data imply that BMPR2 mRNA could be an FMRP binding partner and because BMP signaling is involved in synaptic formation in Drosophila, we investigated the role of BMPR2 and noncanonical BMP signaling in FXS.
RESULTS
Two isoforms of BMPR2 are differentially expressed
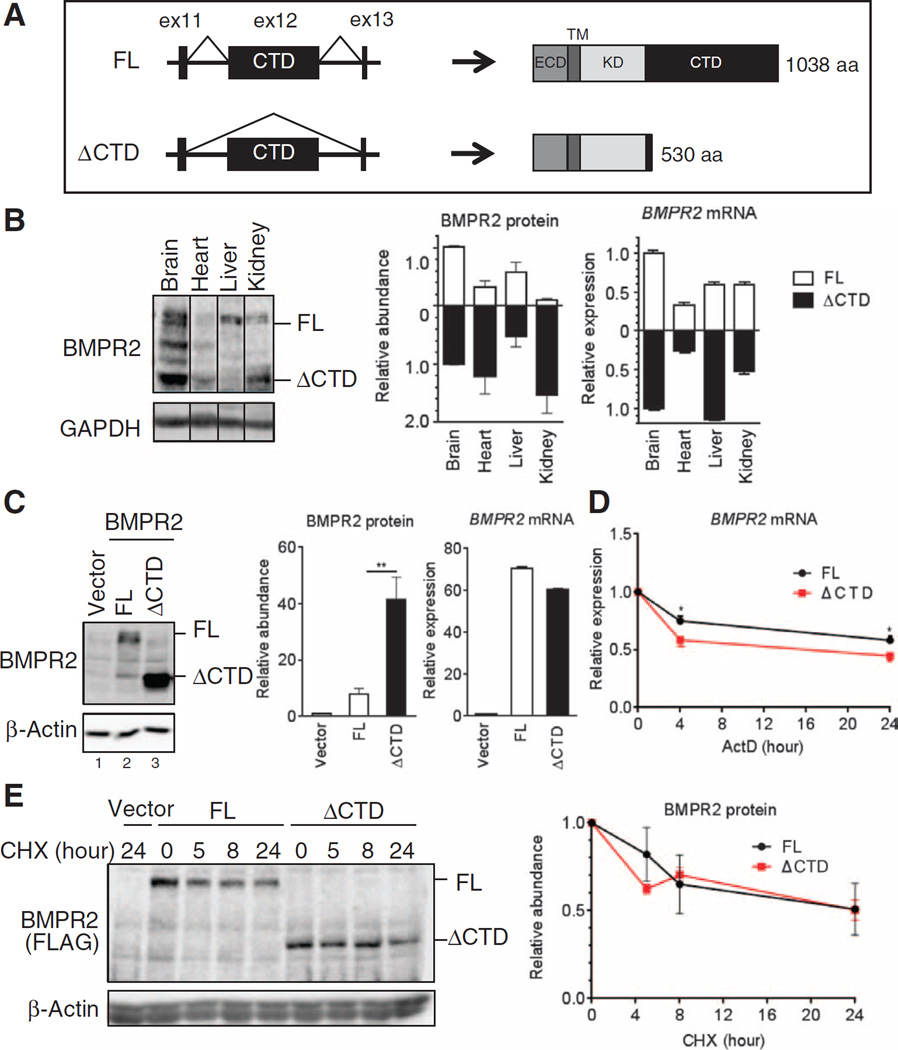
In mammals, alternative splicing of BMPR2 produces two mRNA forms: one “full length” (FL) that contains 13 exons and encodes a 1038–amino acid protein, and a shorter one (ΔCTD) that encodes a 530–amino acid protein. ΔCTD lacks exon 12 (ex12), which encodes 80% of the entire CTD (Fig. 1A). Immunoblot analysis of various tissues from adult mice reveals that the two isoforms are differentially expressed (Fig. 1B). However, although the ratio between BMPR2 protein isoforms varied from tissue to tissue, it did not correlate with the amount of the corresponding mRNA isoform (for example, ΔCTD in liver and heart; Fig. 1B). To investigate the cause of this discrepancy, we expressed the two BMPR2 mRNA isoforms from cytomegalovirus promoter complementary DNA (cDNA) constructs in COS-7 cells. Despite similar mRNA amounts, the ΔCTD protein was ~7 times more abundant than the FL protein in immunoblots probed with the BMPR2 antibody (Fig. 1C, lanes 2 and 3), or with a FLAG antibody and FLAG-tagged BMPR2 protein constructs (fig. S1, lanes 2 and 3). The different amounts of the two protein isoforms could not be explained by the stability of the correspondent transcripts because the ΔCTD mRNA was slightly less stable than the FL mRNA in an experiment in which de novo mRNA synthesis was blocked by actinomycin D, an inhibitor of RNA polymerase II (Fig. 1D). Furthermore, the stability of FL and ΔCTD proteins was similar over a period of 24 hours when new protein synthesis was inhibited by cycloheximide (Fig. 1E). These results strongly suggest that the BMPR2 isoforms are regulated posttranscriptionally.
Fig. 1. Different abundance of BMPR2 isoforms.
(A) Schematic of the exon/intron structure (left) and the protein structure (right) of two isoforms of BMPR2: the FL and the CTD-truncated isoform (ΔCTD). ECD, extracellular domain; TM, transmembrane domain; aa, amino acid. (B) Immunoblot analysis and quantitative reverse transcription polymerase chain reaction (qRT-PCR) for FL and ΔCTD BMPR2 in brain, heart, liver, and kidney isolated from 12-week-old wild-type (WT) mice. Data are means ± SD of five experiments. (C) Immunoblot analysis and qRT-PCR in lysates from COS-7 cells transfected with vectors encoding FL or ΔCTD BMPR2 normalized to β-actin and GAPDH, respectively. Data are means ± SD of five experiments. **P < 0.05 by Student’s t test. GAPDH, glyceraldehyde-3-phosphate dehydrogenase. (D) qRT-PCR analysis for BMPR2 expression in COS-7 cells transfected with vectors encoding MBP-BMPR2 [in which the BMPR2 ECD was replaced by maltose-binding protein (MBP)], FL or MBP-ΔCTD (ΔCTD) BMPR2, and treatment with actinomycin D (ActD) (5 µg/ml) 24 hours afterward for the period of time indicated. Expression was normalized to GAPDH. Data are means ± SD of five experiments. *P < 0.05 by Student’s t test. (E) Immunoblot analysis for BMPR2 in COS-7 cells transfected with optimized amount of MBP-BMPR2-FLAG (FL) or MBP-ΔCTD-FLAG (ΔCTD) expression vectors (to adjust for similar abundance of expressed protein) and treated with cycloheximide (CHX) (10 µg/ml) 48 hours afterward for the period of time indicated. Protein was normalized to β-actin. Data are means ± SD of four experiments.
BMPR2 translation is inhibited by the mRNA sequence encoding the CTD
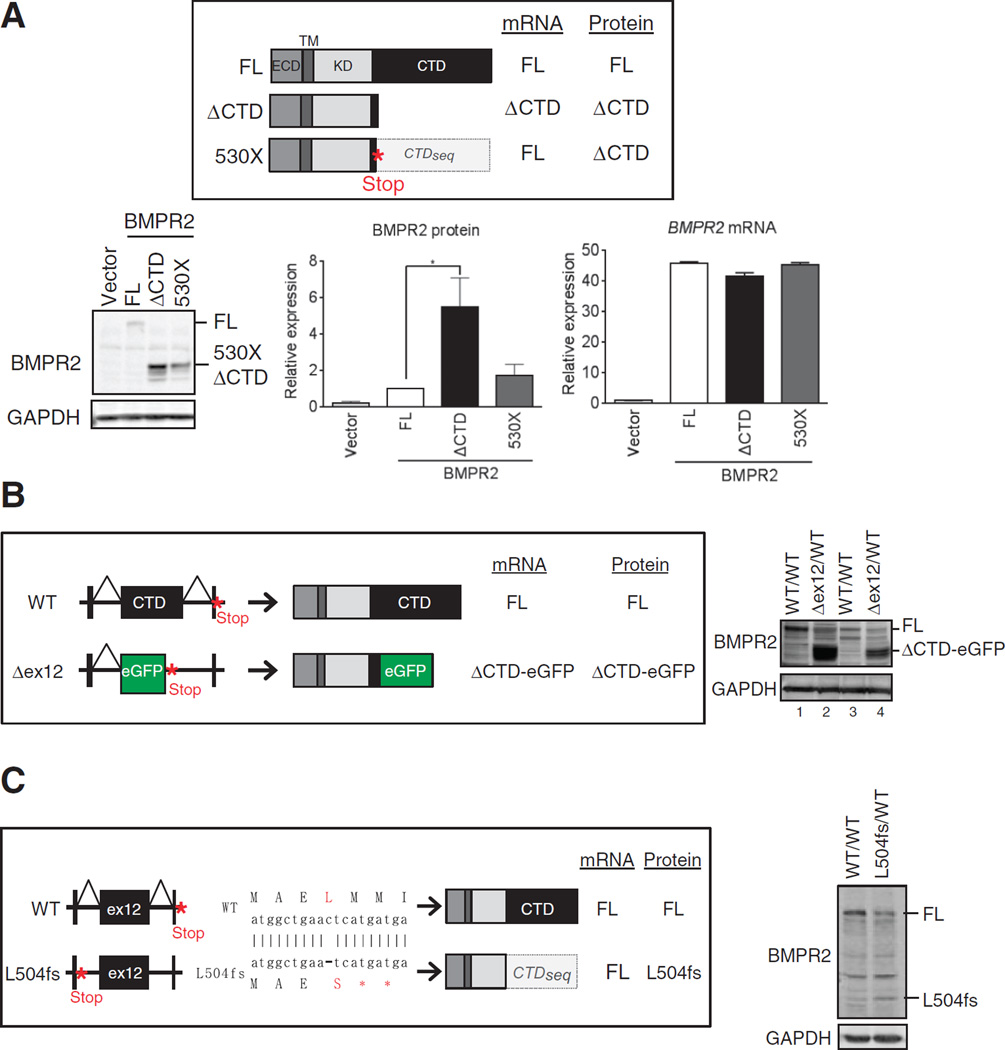
To test whether this posttranscriptional regulation is caused by protein size or by the presence of the CTD coding sequence (hereinafter CTDseq), we introduced a nonsense mutation at amino acid 530 in the FL construct (530X; Fig. 2A). The 530X transcript matches the FL transcript in length, but the 530X-encoded protein has the size of the ΔCTD deletion mutant (Fig. 2A). The difference between the transcripts encoding ΔCTD and 530X is the presence of CTDseq (Fig. 2A). Upon transfection in COS-7 cells, we observed that the ΔCTD protein (Fig. 2A) was about three times more abundant than the FL protein (Fig. 2A), which is consistent with the expression data above (Fig. 1C). On the contrary, the expression of 530X and FL (Fig. 2A) was similar. Similar results were obtained by in vitro transcription and translation, suggesting that a cis-acting sequence (or sequences) within the CTDseq affects translational efficiency.
Fig. 2. Translational regulation of BMPR2 through the mRNA sequence encoding the CTD.
(A) Top: Schematic of the expected mRNA and protein structures of C-terminal FLAG-tagged FL, ΔCTD, and 530X BMPR2. CTDseq is the mRNA sequence encoding CTD. Bottom: Immunoblotting and qRT-PCR of FLAG or BMPR2, respectively, in transfected COS-7 cells. BMRP2 mRNA was normalized to GAPDH. Data are means ± SD of three experiments. *P < 0.05 by analysis of variance (ANOVA) with post hoc Tukey’s test. (B) Top: schematic of WT and BMPR2 ex12 deletion mutant (Δex12) allele. In the Δex12/WT mice, the ex12 of one allele of the BMPR2 gene was replaced with eGFP. Right: Immunoblot for BMPR2 abundance in two independent primary vascular smooth muscle cell cultures generated from WT (WT/WT) and heterozygous (Δex12/WT) mice. (C) Top: Schematic of WT and BMPR2 L504fs allele. A frameshift mutation was introduced in at Leu504 (ex11) of BMPR2 gene using TALEN-mediated genome editing technology (L504fs) in rat vascular smooth muscle PAC1 cells. Consequently, L504fs allele produces mutant BMPR2 lacking 504 to 1038 amino acids; however, the transcripts contain CTDseq. Right: Immunoblots for FL BMPR2 and L504fs in WT and heterozygous cells. Blots in (B) and (C) are representative of two experiments.
To test the effect of the CTDseq on transcripts originating from the endogenous genomic locus, we used two in vivo systems expressing a ΔCTD protein, one in which the CTDseq is deleted and one in which a frameshift mutation truncates the BMPR2 protein but preserves the CTDseq in the mRNA. We established two independent lines of primary pulmonary artery smooth muscle cells (PASMCs) from a heterozygous knock-in mouse in which ex12 of the BMPR2 gene has been replaced by a cassette encoding the enhanced green fluorescent protein (eGFP) with a stop codon (39). In these cells (Δex12), one BMPR2 allele generates transcripts that lack ex12 and encode a ΔCTD (amino acids 1 to 529)–eGFP fusion protein (ΔCTD-eGFP); the other allele is intact (Fig. 2B, top). Independent cell lines were established from two wild-type (Fig. 2B, Fig. 1 lanes and 3) and two Δex12 mice (Fig. 2B, lanes 2 and 4). In both wild-type PASMCs lines (Fig. 2B, lanes 1 and 3), only the FL protein is detected by immunoblot, indicating that FL is the dominant form of BMPR2 protein in PASMCs (Fig. 2B, right). In the two Δex12 cells lines (Fig. 2B, lanes 2 and 4), the amount of FL protein is reduced to ~50% (Fig. 2B, lanes 2 and 4) compared to wild-type cells (Fig. 2B, lanes 1 and 3), consistent with the gene dosage of wild-type BMPR2, but the ΔCTD-eGFP protein accumulates ~7 times more than the FL protein (Fig. 2B, right). This result demonstrates a more efficient translation of the Δex12 transcripts compared to wild-type transcripts, presumably because of the lack of CTDseq (Fig. 2B). We used our second in vivo approach to test this hypothesis. Using TALEN (transcription activator-like effector nucleases)–mediated genome editing in rat PAC1 pulmonary arterial smooth muscle cells, we engineered a frameshift mutation at Leu504 in ex11 of BMPR2 (L504fs; Fig. 2C), which results in a premature termination at Met505. L504fs transcripts encode the ΔCTD protein but still contain the CTDseq (Fig. 2C). Therefore, if the CTDseq mediates translation inhibition, the L504fs protein should be expressed less efficiently than the ΔCTD-eGFP protein in Δex12 cells, compared to the respective wild-type controls. In L504fs/wild-type heterozygous cells, L504fs and wild-type proteins were expressed at about a 1:1 ratio (Fig. 2C), in marked contrast to the Δex12 cells (Fig. 2B). Thus, these genome-driven expression results confirm a role of the CTDseq in the BMPR2 mRNA as a negative translational regulator.
FMRP associates with BMPR2 mRNA through CTDseq and controls its translation
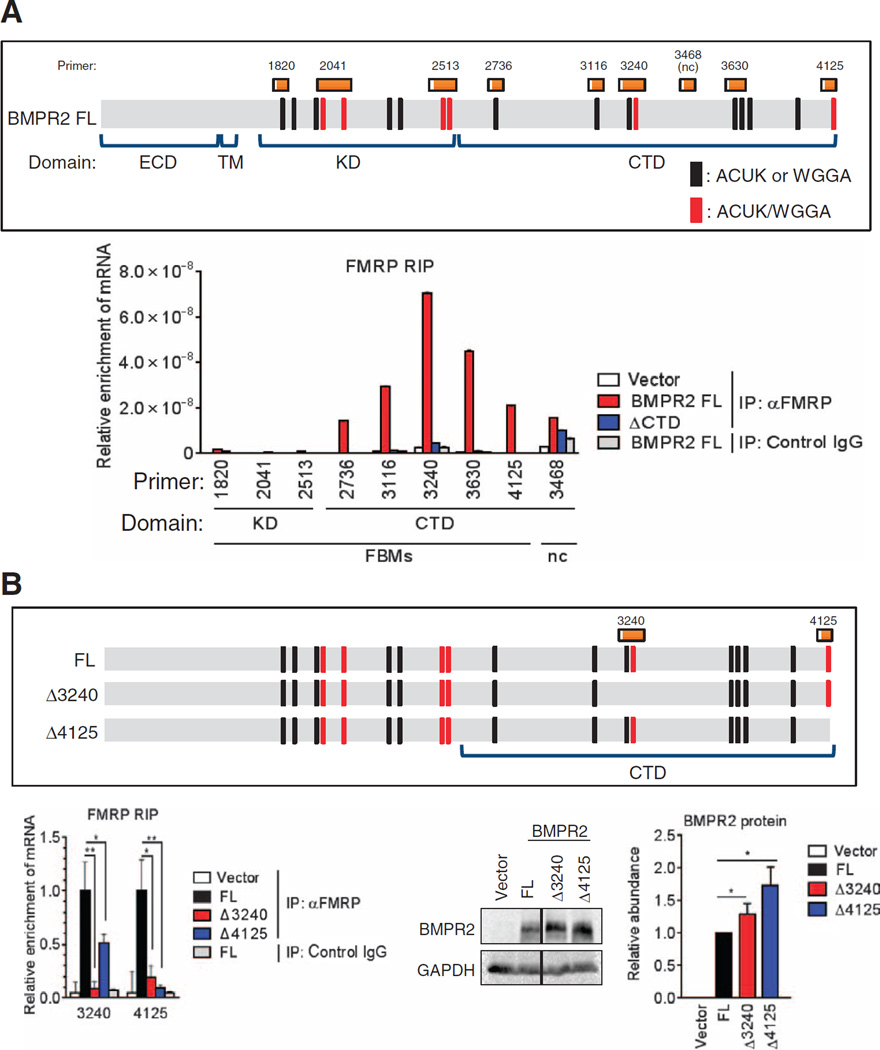
FMRP binds a small subset of mRNAs and inhibits their translation (10, 14, 22, 40, 41). On the basis of the results of two independent high-throughput RNA immunoprecipitation (RIP) analyses (11, 22) in which BMPR2 appeared in the list of potentially FMRP-bound mRNAs, we hypothesized that FMRP might be involved in the translational inhibition of FL BMPR2. To first test whether FMRP associates with the CTDseq through putative FMRP binding motifs (FBMs; ACUK, WGGA, or a combination of both) (22), we transfected human embryonic kidney (HEK) 293 cells with the FL or ΔCTD expression constructs and performed an RIP assay. The FMRP-bound mRNAs were sheared to an average length of ~200 nucleotides to compare the enrichment of different regions of the FL or ΔCTD mRNAs (Fig. 3A). We investigated eight putative FBMs (22) that span the entire length of the BMPR2 transcript (Fig. 3A and Table 1). Primer set #3468, which amplifies a region of the CTDseq that does not contain FBMs and was not enriched in a high-throughput RIP study (22), was added as negative control (Fig. 3A). The result indicated that all the five tested FBMs located in the CTDseq (#2736, #3116, #3240, #3630, and #4125; Fig. 3A) were highly enriched, demonstrating that FMRP binds to the CTDseq (Fig. 3A). No enrichment of RNAs was observed for the negative control primer #3468 (Fig. 3A), suggesting that an FBM is required for FMRP binding within the CTDseq. The three FBMs found in the KD were not enriched. Thus, it appears that the presence of short FBMs is not sufficient to bind FMRP to the 5′ region of the BMPR2 transcript and suggests that additional determinants are required for FMRP recruitment. This result is consistent with our finding that the CTDseq in FL transcripts [as well as in 530X (Fig. 2A) and L504fs (Fig. 2C)] inhibits translation, whereas translation of ΔCTD and Δex12 (Fig. 2B) transcripts was not inhibited because they lack the CTDseq. To examine whether the binding of FMRP to FBMs localized in the CTDseq is required for translational control, we generated deletion mutants, Δ3240 and Δ4125, in which sets of ACUK/WGGA combination FBMs, which are reported to be potent FBMs (22), were deleted (Fig. 3B) and subjected them to FMRP-RIP, as well as protein and mRNA expression analyses (Fig. 3B and fig. S2). Each mutant produces a three–amino acid deletion but preserves the translation reading frame in FL BMPR2. Either mutant decreased the association with FMRP by 75 to 90% (Fig. 3B), suggesting that FMRP binds synergistically to RNA and both clusters of FBMs are required for FMRP binding. Concurrently, the amount of both Δ3240 and Δ4125 mutant proteins was increased by 1.3- and 1.8-fold, respectively, in comparison with wild type (Fig. 3B). Thus, we conclude that the binding of FMRP to FBMs in the CTDseq results in translational inhibition of FL transcripts.
Fig. 3. FMRP binds BMPR2 CTDseq and negatively regulates translation.
(A) RIP assay for the relative enrichment of FL BMPR2 mRNA regions [schematic (top) marked with FMRP binding regions (black or red) and corresponding primers (orange; see table S2)] in FMRP immunoprecipitates from HEK293 cells transfected with FLAG-tagged FL or ΔCTD BMPR2 constructs. Data are representative of two experiments. Pulldown with nonspecific immunoglobulin G (IgG) served as the control. nc, negative control primer; IP, immunoprecipitation. (B) RIP assay as in (A) and immunoblotting in HEK293 cells transfected with FL or mutant (Δ3240 or Δ4125) BMPR2. Data are means ± SD of three experiments. *P < 0.05; **P < 0.01 by ANOVA with post hoc Tukey’s test.
Table 1. List of potential FMRP binding sites identified in the BMPR2 transcripts by RIP sequence.
On the basis of a previous research (16), potential FMRP binding sites were searched in the BMPR2 transcripts. Its core sequence (WGGA or ACUK) is underlined. Location of the RIP primers and the enrichment of RNA fragments are shown. NA, not applicable.
| Domain | Start | End | FBM sequence | FBM type |
RIP primer |
Enrichment | T-to-C conversion/ read count |
Enrichment |
|---|---|---|---|---|---|---|---|---|
| ECD | 203329583 | 203329597 | NA | NA | 0.83 | |||
| ECD | 203329616 | 203329667 | NA | NA | 0.71 | |||
| ECD | 203329669 | 203329687 | NA | NA | 0.22 | |||
| ECD | 203332327 | 203332341 | NA | NA | 0.83 | |||
| TM | 203378479 | 203378507 | NA | NA | 0.69 | |||
| KD | 203383687 | 203383725 | ACAACAUUGCCCGCUUUAUAG UUGGAGAUGAGAGAGUCA |
WGGA | 1820 | No | 0.95 | No |
| KD | 203383744 | 203383768 | AAUAUUUGCUUGUGAUGGAGUACUA | WGGA | 0.76 | |||
| KD | 203384831 | 203384856 | AGUCUCCACACAAGUGACUGGGUAAG | ACUK | 0.36 | |||
| KD | 203384869 | 203384893 | UGCUCAUUCUGUUACUAGAGGACUG | Mixed | 2041 | No | 0.33 | No |
| KD | 203395532 | 203395544 | NA | NA | 0.25 | |||
| KD | 203395592 | 203395623 | UUAUUAGUGACUUUGGACUGU CCAUGAGGCUGA |
Mixed | 0.62 | |||
| KD | 203397352 | 203397369 | UGUGAACUUGAGGGACUG | ACUK | 0.5 | |||
| KD | 203397390 | 203397420 | UAGACAUGUAUGCUCUUGGACUA AUCUAUUG |
WGGA | 0.88 | |||
| KD | 203417463 | 203417487 | AUCGAAGACUGUUGGGACCAGGAUG | Mixed | 2513 | No | 0.83 | No |
| KD | 203417515 | 203417538 | CUGAGGAAAGGAUGGCUGAACUUA | Mixed | 2513 | No | 0.71 | No |
| KD | 203417570 | 203417588 | NA | NA | 0.6 | |||
| CTD | 203420039 | 203420070 | UACAUUGAAGACUCUAUCCAU CAUACUGACAG |
ACUK | 2736 | Yes | 0.65 | Yes |
| CTD | 203420082 | 203420103 | NA | NA | 0.71 | |||
| CTD | 203420438 | 203420465 | AAUCUCAUGGAGCACUCUCUUAAACAGU | WGGA | 3116 | Yes | 0.8 | Yes |
| CTD | 203420507 | 203420530 | CUUUACCCACUCAUAAAACUUGCA | ACUK | 3240 | Yes | 1 | Yes |
| CTD | 203420532 | 203420570 | UAGAAGCAACUGGACAGCAGG ACUUCACACAGACUGCAA |
Mixed | 3240 | Yes | 0.56 | Yes |
| CTD | 203420588 | 203420610 | NA | NA | 0.78 | |||
| CTD | 203420887 | 203420913 | NA | NA | 0.38 | |||
| CTD | 203420939 | 203420961 | UUUAUUGGUGAGGACACCCGGCU | WGGA | 3630 | Yes | 0.63 | Yes |
| CTD | 203420995 | 203421016 | UUUACUGAGACGAGAGCAACAA | ACUK | 3630 | Yes | 0.67 | Yes |
| CTD | 203421033 | 203421052 | GUGUUCUGGAUCGUCUUGUG | WGGA | 0.82 | |||
| CTD | 203421200 | 203421222 | CCUAAUUCUCUGGAUCUUUCAGC | WGGA | 0.73 | |||
| CTD | 203424647 | 203424668 | AUAUAGGAAUGAACUGUCUGUG | Mixed | 4125 | Yes | 0.63 | Yes |
Modulation of FMRP alters FL-BMPR2 protein abundance
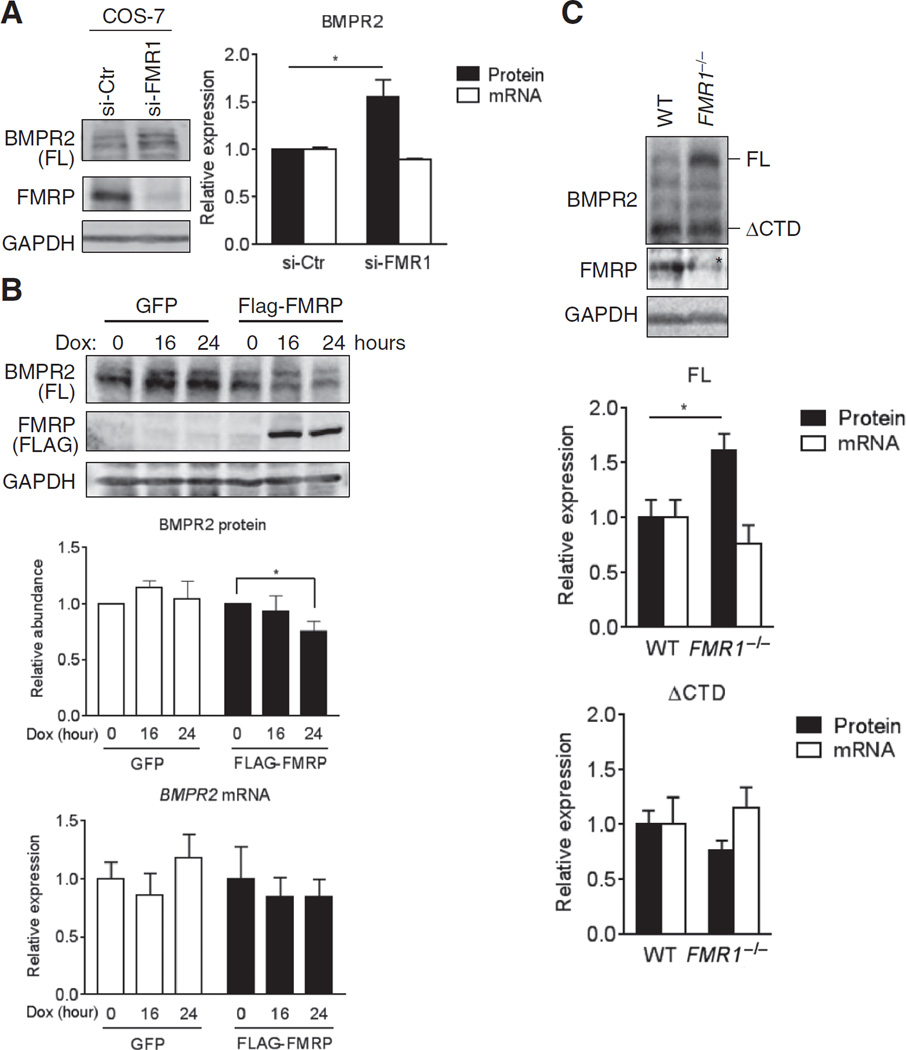
Next, we examined whether loss or gain of FMRP dictates the BMPR2 protein amount and the signal downstream of BMPR2. FMRP was depleted in COS-7 cells (Fig. 4A) by small inhibitory RNAs (siRNAs) (si-FMR1). Whereas si-FMR1 did not affect FL mRNA abundance, FL protein was increased ~1.5-fold over controls (Fig. 4A). Conversely, expressing exogenous FMR1 under doxycycline control (Tet-On system) in a stable HEK293 cell line reduced FL protein by 30%, compared to control cells expressing GFP. The expression of FL mRNA was unchanged (Fig. 4B). These results confirmed that the presence of FL protein inversely correlates with the abundance of FMRP and provided the rationale to examine the effect of FMRP loss on BMPR2 in FMR1-null mice. Mouse embryonic fibroblasts (MEFs) derived from FMR1-null mice exhibited ~2-fold increase in BMPR2 (FL) protein compared to MEFs derived from wild-type litter-mates (fig. S3A). Upon BMP treatment, the phosphorylation of SMAD1/5, a hallmark of the “canonical” pathway downstream of BMPR2, was also increased (fig. S3A), whereas TGFβ signaling was not affected, suggesting that the effect of the loss of FMRP is BMP-specific (fig. S3B). Next, we collected brain samples from FMR1-null or control (wild-type) mice at postnatal day 7 (P7) and subjected them to immunoblot and qRT-PCR analysis of BMPR2 abundance and expression, respectively. At protein but not at mRNA level, a greater amount of FL isoform was detected in the brain of FMR1-null mice in comparison with wild-type mice (Fig. 4C). Unlike FL, the amount of ΔCTD isoform was similar in both FMR1-null and wild-type mice (Fig. 4C). These results confirm an increase of FL isoform in the FMRP-depleted mouse brain and, thus, suggest that the increase in the CTD-mediated signal might contribute to the development of the neuronal phenotypes associated with FXS.
Fig. 4. FMR1 is a negative regulator of FL BMPR2 protein expression.
(A) Im-munoblotting and qRT-PCR for BMPR2 in COS-7 cells transfected with siRNAs against FMR1 (si-FMR1) or a control sequence (si-Ctr). Data are means ± SD of three experiments. *P < 0.01 by Student’s t test. (B) Immunoblotting and qRT-PCR for BMPR2 in stable HEK293 cell lines with doxycycline (Dox)–inducible expression of FLAG-tagged FMRP or GFP (control). Protein and mRNA abundance was normalized to GAPDH. Data are means ± SEM of three experiments. *P < 0.05 by ANOVA with post hoc Tukey’s test. (C) Im-munoblotting and qRT-PCR for BMPR2 in brain lysates from P7 FMR1−/− or WT mice. Data are means ± SEM of seven individual mice. *P < 0.05 by Student’s t test. Asterisk (*) indicates a nonspecific band in the FMRP immunoblot panel. In (A) to (C), protein and mRNA abundance was normalized to GAPDH.
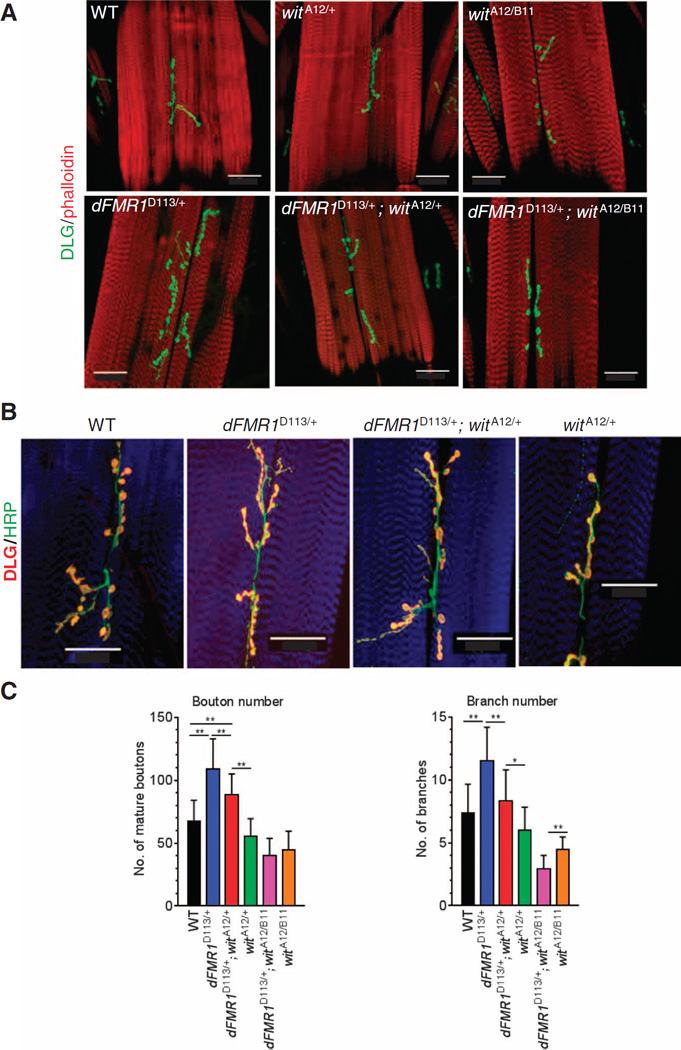
Abnormalities at the NMJ in a loss-of-expression FMR1 Drosophila mutant are rescued by reducing Wit
The Drosophila NMJ is a powerful model system for uncovering and characterizing genetic and molecular mechanisms that regulate synaptic formation (36, 42). A loss-of-expression mutation of dFMR1, the Drosophila ortholog of FMR1, causes overgrowth of synaptic boutons in the larval NMJ (43). In contrast, loss-of-expression mutants of the Drosophila ortholog of BMPR2 Wit exhibit reduced numbers of synaptic boutons (31, 37, 38). Because both Wit and dFMR1 are expressed in presynaptic motor neurons, we speculated that translational regulation of Wit by dFMR1 might be important for synaptic growth and stability at the NMJ. More precisely, if the overgrowth of boutons in dFMR1 mutants was due to increases of Wit and BMP signaling in presynaptic motor neurons, this phenotype could be rescued by reducing Wit abundance. To test this hypothesis, we crossed heterozygous (witA12/+) or homozygous (witA12/B11) wit mutants with heterozygous dFMR1 mutants (dFMR1D113/+) and analyzed the number of mature synaptic boutons and branching synapses by immunofluorescence staining of the postsynaptic marker discs large (DLG; Fig. 5A, green) and phalloidin (Fig. 5A, red) on muscle 6/7 in the abdominal segment 3 (A3) of third instar larvae. We also costained samples with DLG (Fig. 5B, red) and the presynaptic marker horseradish peroxidase (HRP) (Fig. 5B, green) to distinguish mature synaptic boutons, which are costained with both HRP and DLG, from “synaptic footprints,” which are stained only with DLG (32). As previously reported (43), loss of expression of one copy of dFMR1 (dFMR1D113/+) resulted in ~75% increase in the number of mature boutons and ~60% increase in branching compared to wild-type animals (Fig. 5C). When one allele of wit was mutated in the heterozygous dFMR1 mutant background (dFMR1D113/+; witA12/+), the bouton number decreased by ~20% (Fig. 5C). Mutation of both wit alleles in the dFMR1 mutant background (dFMR1D113/+ ; witA12/B11) reduced both boutons and branching numbers to indistinguishable quantity from those observed in wit mutants (witA12/+ or witA12/B11) with wild-type background (Fig. 5C). Thus, the augmented synaptic morphology in the dFMR1 mutant is, at least in part, due to increased Wit abundance and signaling in presynaptic neurons, and both can be decreased by reducing wit gene dosage. Considering that they are orthologs, the epistatic relationship between dFMR1 and wit in the regulation of synaptic development at the NMJ is consistent with the translational regulation of BMPR2 by FMRP.
Fig. 5. Epistasis between dFMR1 and wit during the formation of NMJ in Drosophila.
(A) Confocal images of muscle 6/7 in segment A3 from larvae of the indicated genotype. Boutons and muscle were stained with DLG antibody (green) and Alexa Fluor 568–conjugated phalloidin (red), respectively. Scale bars, 50 µm. (B) As in (A), except presynaptic and postsynaptic boutons and muscle were stained with HRP (green) and DLG (red) antibodies and Alexa Fluor 647–conjugated phalloidin (blue), respectively. Scale bars, 50 µm. (C) Bouton (left) and branch (right) numbers in muscle 6/7 in segment A3. Data are means ± SD of the following numbers of larvae: n =11 for WT, n = 27 for dFMR1D113M/+, n = 41 for witA12/+; dFMR1D113M/+, n = 10 for witA12/+, n = 10 for witA12/B11; dFMR1D113M/+, and n = 10 for witA12/B11. *P < 0.05; **P < 0.01 by ANOVA with post hoc Tukey’s test.
CTD-LIMK1 –dependent signal promotes augmented dendritogenesis in FMR1-null neurons
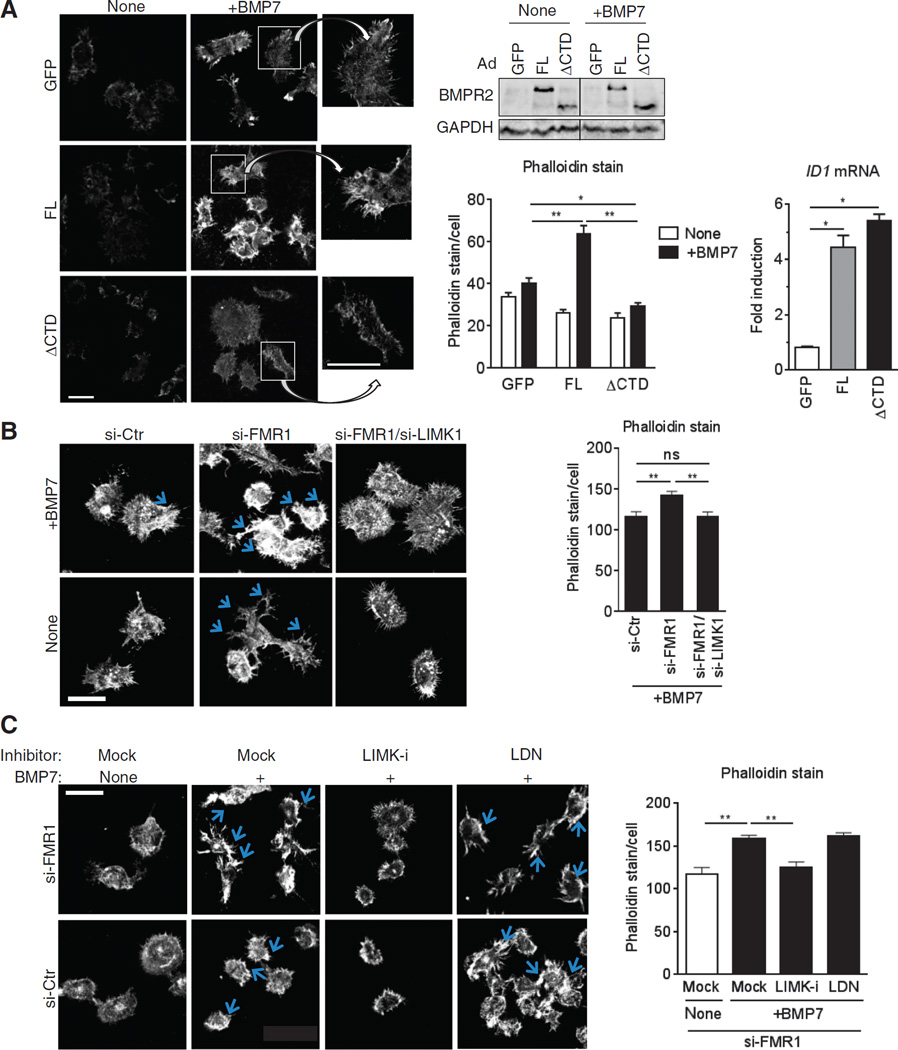
In the Drosophila NMJ, presynaptic expression of a Wit mutant lacking the CTD fails to rescue synaptic stability in wit-null mutants (32). This failed rescue has been attributed to the failure of the Wit ΔCTD mutant to activate dLIMK1 (the Drosophila ortholog of LIMK1), which is essential for promoting the stability of neuromuscular synapses (32). To compare the effect of FL and ΔCTD in BMP-mediated actin remodeling and filopodia formation, we expressed each isoform in the neural crest–derived neuroblastoma cell line N1E-115 (N1E) and subjected them to BMP7 (also known as Osteogenic protein-1) stimulation followed by phalloidin staining (Fig. 6A). BMP7 was used in this experiment because it is expressed in the developing nervous system and is a potent inducer of dendritic morphogenesis in N1E cells (29) and rat sympathetic neurons (44). Phalloidin staining in control cells showed that BMP7 induced the formation of filopodia, which are fundamental for neurite outgrowth (Fig. 6A). In comparison, FL-expressing cells showed an increased intensity of both basal and BMP7-induced phalloidin staining and filopodia formation (Fig. 6A, top). We observed proper plasma membrane localization (45) and similar amounts of ΔCTD and FL proteins (Fig. 6A, bottom left), which correlated to similar induction of the ID1 gene (Fig. 6A, bottom right), a readout of the SMAD-dependent signaling. However, ΔCTD did not mimic the FL effect but, rather, inhibited BMP7-mediated filopodia formation (Fig. 6A), indicating that ΔCTD may act as a dominant negative against endogenous BMP7-induced noncanonical signaling activation. These results suggest that FL, but not ΔCTD, promotes actin remodeling presumably through CTD-mediated activation of LIMK1 and phosphorylation of cofilin, which is fundamental for dendritogenesis and synaptogenesis.
Fig. 6. BMPR2 modulates actin remodeling through activation of the LIMK1-cofilin pathway.
(A) Alexa Fluor 568–conjugated phalloidin staining in N1E cells infected with adenovirus (Ad) expressing GFP (control), FL, or ΔCTD, and treated with BMP7 (10 ng/ml) for 24 hours. Scale bars, 40 µm. Insets are magnified ×2. Data are means ± SEM of ≥33 cells per condition. *P < 0.05, **P < 0.01 by ANOVA with post hoc Tukey’s test. A fraction of N1E cells was subjected to immunoblotting for BMPR2 (bottom left) and qRT-PCR for ID1 expression (bottom right), each was normalized to GAPDH. Data are means ± SD of three experiments. *P < 0.001 by ANOVA with post hoc Tukey’s test. (B and C) Phalloidin staining in N1E cells transfected with targeted siRNA(s) or control (si-Ctr) and treated 48 hours later with BMP7 (10 ng/ml) (B) for 24 hours or LIMKi-3 (LIMK-i; 3 µM) (C) or BMP type I receptor kinase inhibitor LDN (100 nM) for 12 hours and then BMP7 (10 ng/ml) for 24 hours. Scale bars, 50 µm. Blue arrows indicate filopodia. Phalloidin stain was blindly assessed and quantitated by ImageJ. Data are means ± SEM of ≥26 (B) or 29 (C) cells per condition. **P < 0.01 by ANOVA with post hoc Tukey’s test. ns, not significant.
To test whether filopodia formation is augmented upon down-regulation of FMR1, we transfected N1E cells with si-FMR1 and stained with phalloidin after BMP7 stimulation. Filopodia formation was increased in si-FMR1 cells with and without BMP7 stimulation (Fig. 6B) but reverted to the morphology of control cells when LIMK1 was simultaneously silenced by siRNA (Fig. 6B). These results suggest that the increase of the FL-LIMK1 signal leading to altered actin dynamics may be an underlying cause of dendritic spine abnormalities in cortical and hippocampal neurons in animal model of FXS and patients (46–49). To test whether perturbation of LIMK1 activity could modulate actin dynamics in FMRP-depleted cells, we exposed si-FMR1–treated N1E cells to LIMKi-3, a non-cytotoxic small-molecule inhibitor of LIMK1 (50), and then treated them with BMP7 and scored for filopodia formation (Fig. 6C). As a control, cells were treated with LDN-193189 (LDN), a potent inhibitor of the BMP type I receptor–mediated, SMAD-dependent signaling pathway (51, 52). LIMK-i inhibited BMP7-mediated filopodia formation (Fig. 6C), similarly to si-LIMK1 (Fig. 6B). Phosphorylation of cofilin, but not SMAD1/5, was reduced by LIMK-i, indicating an effective and specific inhibition of a BMPR2 non-canonical pathway mediated by LIMK1 activity by LIMK-i treatment (fig. S4A, top). On the contrary, LDN effectively inhibited both SMAD1/5 phosphorylation and transcriptional activation of the SMAD target gene Id3 (fig. S4, A and B) but did not inhibit filopodia formation (Fig. 6C). Thus, the LIMK1-cofilin pathway, but not the SMAD pathway, plays an essential role in promoting BMP7-BMPR2–mediated filopodia formation.
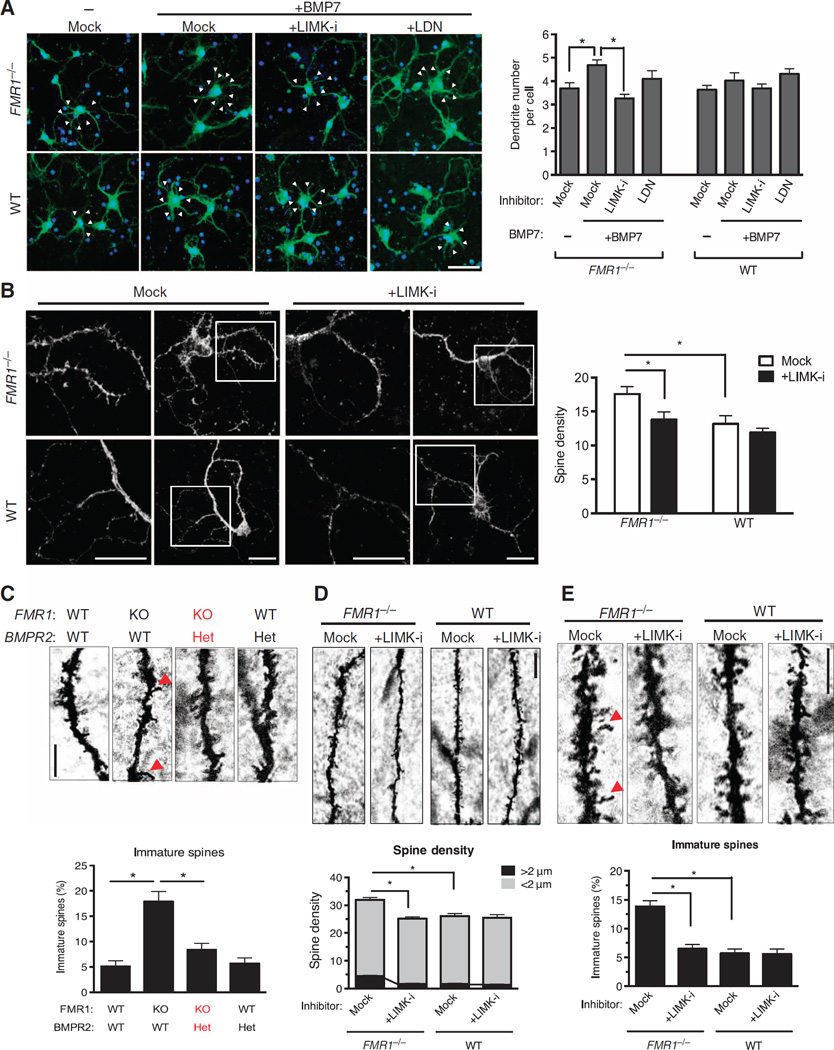
Reduction of the BMPR2-LIMK1 pathway rescues abnormalities of neuronal development in FMR1-null mice
To examine whether the aberrant dendritogenesis in FMR1-null neurons (53, 54) can be rescued by inhibition of LIMK1, we isolated cortical neurons from FMR1-null or wild-type mice at P0, followed by BMP7 and LIMK-i or LDN treatment for 24 hours (Fig. 7A). BMP7 treatment significantly increased the dendrite number in FMR1-null neurons, whereas it had only a modest effect on wild-type neurons (Fig. 7A). Treatment with LIMK-i, but not LDN, abolished the BMP7-dependent increase of dendritogenesis in FMR1-null neurons (Fig. 7A). The density of spines on the dendrites of cortical and hippocampal neurons is abnormally high in FMR1-null mice (46–49) and FXS patients (55). Staining with phalloidin revealed a ~40% increase in spine density in FMR1-null cortical neurons compared to that in wild-type neurons after 16 days in culture (Fig. 7B), resembling the morphology described in vivo (46–49). LIMK-i treatment reversed the abnormal dendritic spine morphology of FMR1-null neurons to the wild-type phenotype (Fig. 7B), suggesting that LIMK-i treatment could ameliorate the abnormally high turnover of dendritic protrusions observed in FMR1-null neuron (54). These results suggest that FMRP-deficient neurons develop irregular dendrites and spines at least in part as a result of increased FL BMPR2–LIMK1 signaling activity.
Fig. 7. Inhibition of BMPR2-LIMK1 pathway rescues abnormal neuronal development in the mouse model of FXS.
(A) Confocal microscopy detecting the number of dendrites (arrowheads) in neurons isolated from the cortex of FMR1−/− or littermate WT P0 mice, cultured for 3 days, and then transfected with a GFP expression plasmid and treated 24 hours later with LIMK-i (3µM) or LDN (100 nM) for 12 hours, followed by BMP7 (10 ng/ml) for 24 hours. Cells were stained and imaged at day 6 in vitro. Data are means + SEM from ≥30 GFP-positive neurons. *P < 0.01 by ANOVA with post hoc Tukey’s test. Scale bar, 30 µm. (B) Alexa Fluor 568– conjugated phalloidin staining (left) and confocal microscopy detecting spine density (right) in neurons isolated from the cortex of FMR1−/− or littermate WT P0 mice, cultured for 14 days, and then treated with LIMK-i (3µM) for 12 hours, followed by BMP7 (10 ng/ml) for 24 hours. Insets are magnified ×2. The minimum was counted. Data are means + SEM from ≥25 neurons. *P < 0.05 by ANOVA with post hoc Tukey’s test. Scale bar, 30 µm. (C) Golgi staining (left) in brain tissue isolated at P7 from FMR1−/−, FMR1−/−; BMPR2+/−, BMPR2+/−, or WT littermate mice. Dendritic spines in DG neuron were classified as either short (<2 µm) or long immature (>2 µm, red arrowheads), and the percentage of long immature spines was calculated (right). Data are means + SEM from more than seven DG neurons from three mice. *P < 0.01 by ANOVA with post hoc Tukey’s test. Scale bar, 5 µm. KO, FMR1 homozygous-null mice; Het, BMPR2 heterozygous mice. (D and E) Golgi staining (left) in brain tissue isolated at P7 from FMR1−/− or littermate WT mice treated with LIMK-i by intracerebroventricular injection at P1 and P4. Spine density was calculated as the total number of spines in a 30-µm stretch (D). The percentage of long immature spines (>2 µm) indicated by red arrowheads was calculated (E). Data are means and SEM from more than seven DG neurons from three mice. *P < 0.01 by ANOVA with post hoc Tukey’s test. Scale bars, 10 µm (D) and 5 µm (E).
Thus, we postulated that reducing the abundance of FL BMPR2 or the activation of LIMK1 in vivo might prevent some aspects of the FMR1-null phenotype. We first tested whether a reduction of the BMPR2 gene dosage could rescue the neuronal morphology in FMR1-null mice. We crossed FMR1 homozygous-null and BMPR2 heterozygous mice to generate FMR1−/−; BMPR2+/− mice. BMPR2 homozygous-null mice die during early embryogenesis and could not be tested. As previously reported (46–49), granule neurons in the dentate gyrus (DG) of FMR1-null mice exhibit an increased dendritic spine density (Fig. 7C, left) with a higher percentage of long thin (>2 µm) immature spines compared to control (wild-type) mice (Fig. 7C, right). The fraction of long immature spines of DG neurons observed in the FMR1−/−; BMPR2+/− mice was similar to that seen in wild-type litter-mates or BMPR2 heterozygous mice (Fig. 7C). These results demonstrate that genetic perturbations of the BMPR2 signaling pathway are sufficient to modify the abnormal dendrite morphology in an FXS mouse model and in the fly, implying that the pathway is evolutionally conserved.
Next, we examined whether postnatal administration of the LIMK1 inhibitor could rescue the abnormal morphology of hippocampal neurons in the FMR1-null mouse. LIMK-i or vehicle (dimethyl sulfoxide) was injected into the cranium of P1 and P4 mice, followed at P7 by Golgi’s staining of the central nervous system (CNS) and biochemical analysis. Brain samples from FMR1-null mice treated with LIMK-i showed markedly reduced abundance of phosphorylated cofilin (fig. S5), indicating an effective inhibition of the catalytic activity of LIMK1. As previously reported (46–49), compared to those in wild-type mice, granule neurons in the DG of FMR1-null mice exhibited increased dendritic spine density (Fig. 7D) and an increased proportion of long, thin (>2 µm), and immature spines (Fig. 7E). In FMR1-null mice treated with LIMK-i, both spine density (Fig. 7D) and the proportion of immature spines (Fig. 7E) were reversed to those observed in mock-treated wild-type mice (46–49). These results demonstrate that genetic and pharmacological perturbations of the BMPR2-LIMK1-cofilin pathway are sufficient to modify the abnormal dendrite morphology of an FXS mouse model.
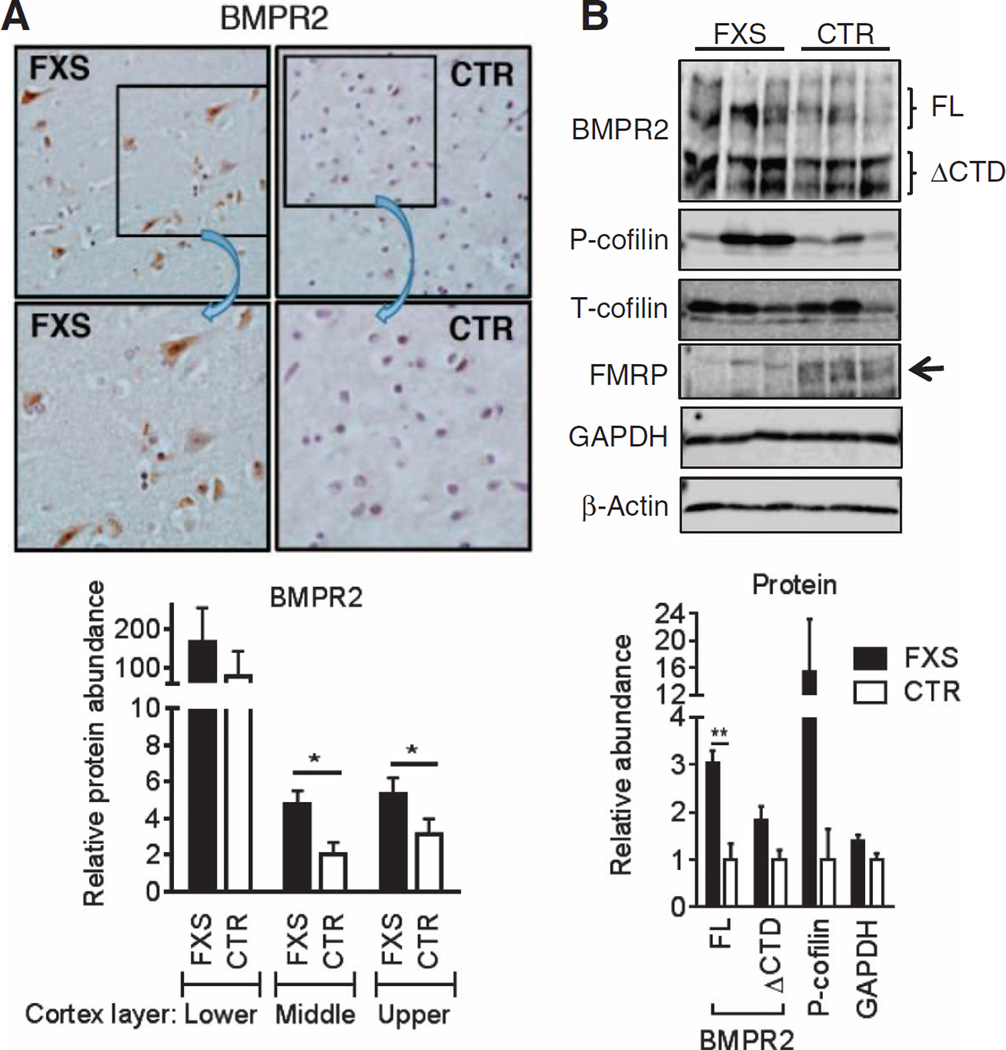
Induction of the BMPR2-LIMK1-cofilin pathway is seen in brain tissue from FXS patients
Finally, to test the hypothesis that the activation of the BMPR2-LIMK1-cofilin pathway found in the mouse FXS model also occurs in human FXS patients, we performed immunohistochemical (IHC) analysis to compare the amount of BMPR2 in the prefrontal cortexes of FXS patients and of gender- and age-matched control individuals. We observed ~2-fold more FL BMPR2 in FXS compared to control cortexes in the upper to middle layers (Fig. 8A). Consistent with the result of IHC, an immunoblot analysis confirmed that loss of FMRP in FXS patients’ cortexes correlated with a ~3-fold increase of FL BMPR2 protein (Fig. 8B) and 16-fold increase of phosphorylated cofilin in FXS cortexes compared to control cortexes (Fig. 8B). These results demonstrate that loss of FMRP leads to an increase of FL BMPR2, which in turn augments the phosphorylation of cofilin, presumably through activation of LIMK1, in the brain tissue of FXS patients. Thus, they support our model of a critical contribution of the BMPR2-LIMK1 signaling axis to the pathology of human FXS and offer a novel opportunity to a therapy for FXS and possibly other ASD conditions.
Fig. 8. Induction of the BMPR2-LIMK1-cofilin pathway is seen in brain tissue from FXS patients.
(A) Representative images of BMPR2 staining on the middle layer prefrontal cortex from FXS patients and control subjects (CTR) (top). Insets are magnified ×2. Average intensity of BMPR2 signals of lower, middle, or upper layer of prefrontal cortex of three FXS and three controls was quantitated (bottom). Data are means ± SD of three samples. *P < 0.01 by Student’s t test. (B) Immunoblotting in lysates from frontal cortexes from three FXS patients and three control subjects. Data are means ± SEM of three samples. **P < 0.01 by Student’s t test. P-cofilin, phosphorylated cofilin; T-cofilin, total cofilin.
DISCUSSION
The BMP signaling pathway controls morphogenesis of nearly every tissue and organ by coordinating basic properties of the cell, such as differentiation, proliferation, motility, morphology, and death, either during development and in the adult (27, 28). Here, we demonstrate that the BMPR2 mRNA is a target of translational regulation by FMRP and provide evidence supporting a link between augmented BMP signaling and neurological disorder in humans. The epistatic relationship between FMR1 and BMPR2 and the physiological significance of the FMRP-mediated down-regulation of BMPR2 during neuronal development have been conserved during evolution from Drosophila to mammals. In particular, the noncanonical signaling pathway downstream of BMPR2, which includes LIMK1, appears to play an essential role in the development of the neuropathology of patients with FXS and in the mouse model of FXS. Tempering this pathway, either by reducing the BMPR2 gene dosage or applying a small-molecule inhibitor of LIMK1, rescues the neuromorphological abnormality in the FXS mouse model, indicating that the BMPR2-LIMK1 axis is an intervention site for development of an FXS and ASD therapy. A previous study, which finds that BMPR2 deletion in the mouse hippocampus and forebrain produce reduced anxiety-related behaviors and increased exploration, supports a role of BMPR2 signaling in modulating cognitive and behavioral phenotypes (56).
CTDseq contains nine FBMs (three ACUK, four WGGA, and two mixed; Table 1). Although the mRNA sequence encoding the KD (KDseq) also contains FBMs (two ACUK, three WGGA, and four mixed; Table 1), association of FMRP with KDseq was not detectable (Fig. 3A), strongly suggesting the potential involvement of other RNA sequence or structural elements adjacent to FBMs for recruitment and stable association of FMRP with target mRNAs. Deletion analysis of individual FBMs suggests that FMRP binds multiple sites and represses translation without affecting mRNA stability. Furthermore, deletion of one FBM (Δ4125) or two FBMs (Δ3240) produces a smaller reduction of BMPR2 protein compared to the deletion of all nine FBMs (ΔCTD), suggesting that the recruitment of FMRP to multiple FBMs in CTDseq results in more potent repression of BMPR2 translation. Several modes of translational inhibition by FMRP have been reported (57). In some cases, FMRP binds the coding sequence, stalls polysomes, and represses active translation, whereas on some other targets, FMRP binds the 5′ or 3′ untranslated region and inhibits translation initiation (12–15). More recently, FMRP has been shown to bind directly to the L5 protein on the 80S ribosome and inhibit translation (58). We speculate that FMRP stalls polysomes or directly inhibits the activity of the 80S ribosome associating with the BMPR2 mRNA, as previously proposed (26). Ribosome profiling analysis in the presence or absence of FMRP will give a snapshot of the translation status of the BMPR2 mRNA and help understand the mode of translational inhibition by FMRP.
It has been reported that higher amounts of FMR1 mRNA and FMRP correlate with a more aggressive form of breast cancer (59). This correlation was attributed to FMRP binding to E-cadherin and vimentin mRNAs and regulating epithelial-to-mesenchymal transition, a hallmark of cancer invasion and metastasis,(57, 59–62). Conversely, loss of BMP signaling in mammary carcinomas can accelerate metastasis, indicating that BMP plays a tumor suppressor role (63). Therefore, it is conceivable that increased FMR1 transcripts might permit more invasive breast cancer by reducing the tumor suppressive activity of the BMP signal. Another disease potentially associated with the FMRP-BMPR2 axis is pulmonary artery hypertension (PAH). More than 80% of patients with heritable PAH and ~20% with sporadic PAH carry a loss-of-expression or loss-of-function mutation in the BMPR2 gene (64), but the penetrance of PAH among BMPR2 carriers is less than 20% (64). Therefore, we speculate that expression of genetic variants of FMR1 or dysregulation of FMRP might determine the development of PAH among a small subset of BMPR2 carriers.
The kinase PAK1, which phosphorylates and activates LIMK1, also appears to play a role in FXS (65). It has been reported that PAK1 binds the KH2 domain of FMRP and phosphorylates it, allowing it to be active (66). Furthermore, expression of a dominant-negative PAK1 mutant in the forebrain (67) or pharmacological inhibition of PAK1 rescues irregular dendritic spine morphology and behavioral abnormalities in FMR1-null mice (68), supporting the inhibition of PAK1 as a potential therapy for FXS and ASDs. However, there is yet no evidence of abnormal activity and/or amount of PAK1 in human patients with FXS or other neurological diseases. PAK1 is ubiquitously expressed, activated by various extracellular stimuli, and upstream of various essential biological processes, including cell proliferation, motility, and apoptosis, in addition to the control of the actin cytoskeleton (69). Thus, compared to LIMK1, PAK1 might be a less ideal therapeutic target for FXS because of the expected secondary effects of its inhibition. In contrast, LIMK1, which is expressed mainly in the CNS (70), appears to phosphorylate only cofilin, which directly controls actin polymerization (71). LIMKi-3, which we used in this study, was originally identified as an anticancer drug that inhibits LIMK1 and LIMK2 activity (median inhibitory concentration equals 7 and 8 nM for LIMK1 and LIMK2, respectively) and blocks tumor cell invasion (72). Under the condition used in mouse primary neuron culture, we detected no cytotoxicity; however, development of LIMK1-specific inhibitor might be more desirable for FXS therapy because of the ubiquitous presence of LIMK2.
MATERIALS AND METHODS
Animals
B6.129P2-FMR1tm1Cgr/J, FVB.129P2-Pde6b+ Ty rc-ch FMR1tm1Cgr/J, FVB.129P2-Pde6b+ Ty r c-ch/AntJ (The Jackson Laboratory), B6.129S4(Cg)-BMPR2tm1.1Enl/Mmnc (MMRC), and Tg(ACTB-cre)2Mrt mice were maintained according to the University of California, San Francisco (UCSF) Laboratory Animal Resource Center guidelines. To obtain FMR1 knockout littermates, we used wild-type and knockout (FMR1−/−) male mice derived from FMR1 heterozygous female and hemizygous male mating. To obtain FMR1−/−; BMPR2+/− mutant mice and littermates, we first mated B6.129S4(Cg)-Bmpr2tm1.1Enl/Mmnc mice with Tg(ACTB-cre)2Mrt mice, followed by mating with FMR1 knockout mice, and finally FMR1+/−; BMPR2+/− female and FMR1−/y; BMPR2+/− male mice were mated. Genotyping was performed following the Jackson Laboratory protocol.
Drosophila lines and genetic crosses
Flies were raised at 25°C. Control wild-type flies used in this study were the w1118 strain. Mutant alleles witA12, witB11, and dFMR1D113M were obtained from the Bloomington Stock Center. These lines were crossed to generate witA12/+ [witA12/+], dFMR1D113M/+ [dFMR1D113M/+], witA12/+; dFMR1D113M/+ [witA12;dFMR1D113M/+], witA12/B11 [witA12/witB11], and witA12/B11; dFMR1D113M/+ [witA12; dFMR1D113M/witB11].
Cell culture and transfection
COS-7, HEK293, N1E, and PAC1 cells were purchased from American Type Culture Correction and maintained in Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS). HEK293 cell lines stably expressing FLAG-HA-FMR1 isoform 1 and eGFP were reported previously (22). Primary MEFs were prepared from embryonic day 14.5 FMR1−/− and control wild-type mouse embryos and cultured in DMEM containing 10% FBS. PASMCs from heterozygous Dex12 and control wild-type mice were obtained from K. D. Bloch and cultured as reported previously (39). Postnatal (P0) cortical neurons were prepared from littermate FMR1−/− and wild-type mice and cultured in Neurobasal Medium (Life Technologies). Cells were cultured at 37°C in the presence of 5% CO2. Plasmid and siRNA transfections were performed using FuGENE 6 (Promega) or Lipofectamine 2000 (Life Technologies) and Lipofectamine RNAiMAX (Life Technologies) reagents, respectively.
TALEN-mediated targeted gene mutagenesis in PAC1 cell line
TALEN arms were designed using TAL Effector Nucleotide Targeter 2.0 (Cornell University). PAC1 cells were transfected with two targeting TALEN effectors [TAL_17Q (NN HD NI HD NI NN NG NN NG NN HD NG NN NI NN NI NN NI) and TAL_18R (NG HD NG HD NG HD NI NG NI NG HD NI] and with pcDNA3.1-eGFP in a ratio of 5:5:1. Forty-eight hours after transfection, eGFP-positive cells were sorted by flow cytometry, and single cells were reseeded into 96-well plates. After single-cell cloning, the mutations present in each clone were examined by Sanger sequencing.
Plasmid constructs
The N-terminal FLAG-tagged FL BMPR2, BMPR2 mutants, and negative control were subcloned into pcDEF3 for overexpression experiments. The BMPR2 mutants were constructed by a PCR-based approach. The BMPR2 530X was constructed by inserting a FLAG coding sequence and stop codon between amino acids 529 and 530. The MBP-BMPR2-FLAG and MBP-ΔCTD-FLAG were constructed by replacing the extracellular domain of BMPR2 with MBP. pcDNA3.1-eGFP was constructed by inserting the eGFP sequence into the pcDNA3.1 vector.
siRNAs and adenovirus
The siRNAs targeting human, monkey, mouse, and rat FMR1 (5′-GGAUGAU-AAAGGGUGAGUUdTdT-3’ and 5′-GCUAGAAGCUUUCUCGAAUdTdT-3′) and mouse LIMK1 (5′-UAGUACUGGUGUGAAAGGGAGACdTdT-3′) were previously reported (73–75) and purchased from Dharmacon. The control siRNA was purchased from QIAGEN. Adenovirus vectors expressing FLAG-HA-FMRP isoform 1 and eGFP were prepared using AdEasy Adenoviral Vector System (Agilent Technologies). The adenovirus vectors that express FL and ΔCTD BMPR2 were previously reported (45).
qRT-PCR analysis
Total RNAs were isolated using TRIzol (Life Technologies), and real-time PCR was performed as described previously (76) using the oligodeoxynucleotide primers listed in table S1. The primers for human or mouse ID1 and GAPDH were reported previously (77).
Immunoblot analysis
Cells and mouse tissues were lysed in lysis buffer [tris-buffered saline (TBS) containing 1% NP-40, 1 µM NaF, and 1 mM EDTA). The frontal cortexes of FXS patients and controls were lysed in RIPA (radioimmunoprecipitation assay) buffer. Lysed samples were separated by SDS–polyacrylamide gel electrophoresis, transferred to nitrocellulose membrane (Millipore), immunoblotted with antibodies, and visualized using a LI-COR Imaging System. The antibodies used were directed against FLAG epitope tag (M2, Sigma), FMRP (ab17722, Abcam), BMPR2 KD (19087-1-AP, Proteintech Group), total SMAD1/5/8 (BMR 00479, Bio Matrix Research), phospho-SMAD1/5/8 (#9511, Cell Signaling), cofilin (#5175, Cell Signaling), phospho-cofilin (SC-12912-R, Santa Cruz), β-actin (A5441, Sigma), and GAPDH (MAB374, Millipore).
Phalloidin staining
Eight-well chamber slides (Nunc Lab-Tek) were coated with poly-D-lysine (0.1 mg/ml) overnight, followed by laminin (20 µg/ml) for 1 hour at 37°C. N1E cells (50,000) were seeded onto each chamber, transfected or infected with siRNAs or adenovirus vectors, and treated with BMP7 with or without inhibitors [LIMKi-3 (Millipore) and LDN] as indicated in the figure legends. The cells were fixed, permeabilized, stained with Alexa Fluor 568–conjugated phalloidin (Life Technologies), as described in the manufacturer’s protocols, and observed under a point scanning confocal microscope (Leica). The phalloidin signal on filopodia was traced and quantified using the ImageJ program by a blinded investigator.
Immunofluorescence microscopy
Primary cortical neurons were isolated from littermate FMR1−/− and wild-type P0 mice, and 80,000 cells were plated on 12-mm poly-d-lysine–coated coverslips (Corning). The cells were transfected with pcDNA3.1-eGFP plasmid using Lipofectamine 2000 (Life Technologies) after 3 days in vitro. Twenty-four hours after transfection, the cells were treated with LIMK-i for 12 hours, followed by BMP7 (10 ng/ml) treatments for 24 hours. The cells were fixed with 4% PFA/4% sucrose in phosphate-buffered saline (PBS) for 10 min, permeabilized with 0.1% Triton X-100 in PBS for 10 min, blocked with 3% bovine serum albumin/0.02% Triton X-100 in PBS, and stained with GFP antibody (AB13970, Abcam) overnight at 4°C, followed by incubation with an Alexa Fluor 488–conjugated secondary antibody. Confocal images were acquired using a point scanning confocal microscope (Leica). Images were from a projection of z sections.
RIP assay
RIP assay was performed as reported previously (11). Briefly, HEK293 cells were transfected with vectors expressing wild-type or mutant BMPR2. Forty-eight hours after transfection, the cells were washed with PBS containing cycloheximide (100 µg/ml), and the cell dishes were placed on an ice pack and ultraviolet (UV)–cross-linked in ice-cold PBS containing cycloheximide (100 mg/ml) for three times at 400 mJ/cm2 (254-nm UV light) using an UVC 500 cross-linker (Hoefer). The cells were collected, lysed in SDS-immunoprecipitation buffer, sonicated, and centrifuged at 15,000 rpm for 15 min at 4°C. The supernatant was heated to 90°C for 10 min with shaking and diluted nine times by volume with CSK buffer. The lysate was treated with ribonuclease T1 (5 U/ml) (Thermo Scientific) and deoxyribonuclease I (DNase I; 0.8 U/ml) (Roche) for 5 min at 37°C, and SUPERAse In (100 U/ml) (Life Technologies) was added to stop the reaction. After preclear with Protein A Dynabeads (Life Technologies), FMRP and RNA complexes were immunoprecipitated using Protein A Dynabeads bound to either FMRP antibody (ab17722, Abcam) or control Ig overnight at 4°C. After immunoprecipitation, the beads were washed with high-stringency buffer, high-salt buffer, low-salt buffer (twice), and NT2 buffer, followed by two washes with PK buffer. The beads were resuspended in PK buffer containing proteinase K (0.8 mg/ml) (Roche), incubated for 20 min at 37°C with shaking, mixed with an equal volume of PK-7 M urea buffer, and incubated for 20 min at 37°C with shaking. The RNAs were purified by acid phenol/chloroform extraction, followed by ethanol precipitation. The RNA solution was treated with DNase I (1U/ml) for 30 min at 37°C, purified by acid phenol/chloroform extraction and ethanol precipitation, and used for reverse transcription using SuperScript III Reverse Transcriptase (Life Technologies), followed by real-time PCR. The oligo-deoxynucleotide primers used for qRT-PCR are listed in table S2.
Larva immunofluorescence and image quantitation
Wandering late third instar larvae were dissected for analysis of NMJ phenotype. After dissection, the tissues were fixed with 8% formaldehyde in PBS for 10 min, washed with PBT (PBS containing 0.1% Triton X-100), and blocked with 5% normal donkey serum in PBT for 1 hour at room temperate. The larvae were incubated with DLG (4F3, Developmental Studies Hybridoma Bank) antibody overnight at 4°C, followed by incubation with Alexa Fluor 488– or Alexa Fluor 555–conjugated secondary antibodies against mouse Ig, Alexa Fluor 488–conjugated HRP antibody (123-545-021, Jackson ImmunoResearch), and Alexa Fluor 568–conjugated (A12380, Life Technologies) or Alexa Fluor 647–conjugated (#8940, Cell Signaling) phalloidin. Confocal images were acquired using a point scanning confocal microscope (Leica). Images used for quantification of NMJ bouton number and branch number were derived from a projection of z sections. For quantification of the branch number, branches with at least three boutons were counted blindly.
Golgi staining
The mice received LIMKi-3 (20 µg per pup) by intracerebroventricular injection at P1 and P4. At P7, the mice were anesthetized, and the brains were quickly removed and rinsed in distilled water to be used for Golgi’s staining. Golgi’s staining was performed as described in the manual (FD Rapid GolgiStain Kit, FD Neuro Technologies). Shortly, the brains were incubated in solution A + B for 3 weeks, followed by 3-day incubation in solution C. We stained 100-µm brain sections and performed spine density and morphological analyses under a microscope at ×100 magnification. Quantitation of dendritic spine number in 30 µm (>30 µm from cell body) was performed blindly.
FXS patient brain immunohistochemistry
FXS patient and control frontal cortexes were fixed by formalin, and the samples in paraffin were sectioned at 7 µm. Antigen retrieval was performed with 1× diva solution (heat-induced epitope retrieval buffer, Biocare Medical) at 110°C for 8 min, followed by 3% H2O2 treatment for 7 min. The samples were incubated with blocking buffer (TBS buffer containing 10% donkey serum and 0.025% Triton X-100) for 1 hour, primary antibodies (in TBS buffer containing 10% donkey serum and 0.0025% Tween 20) for 15 hours at 4°C, biotin-conjugated secondary antibodies (Jackson Immuno Research; in TBS buffer containing 10% donkey serum and 0.0025% Tween 20) for 1 hour at room temperature, and DAB Substrate (Vector Laboratories Inc.) for 5 min, followed by counterstaining with Harris hematoxylin (Thermo Scientific), dehydration with ethanol, clearing with xylene, and mounting with Permount (Fisher). An antibody against BMPR2 (Ab78422, Abcam) was used. The brain layers were categorized as upper (I and III), middle (III to V), and lower (V and VI) layers. To quantify the amount of protein in each tissue sample, the percentage of the stained area inside the cell was measured using Image J.
Statistical analysis
Statistical analysis was performed using the Prism 5.01 Graph Pad package and reviewed by M. Nojima at the University of Tokyo. Statistical test and significance are denoted in the figure legends.
Supplementary Material
Acknowledgments
We would like to dedicate this article to K. D. Bloch who passed in 2014. We thank K. Kawamura, A. Mundkur, and S. Manghise for technical supports. We also thank L. Jan for sharing FMR1-null mice and M. Nojima for reviewing the statistical analysis. Finally, we thank D. Hart, G. Davis, R. Edwards, and all members of the Hata Laboratory for the critical discussion and reading of the manuscript. Funding: R.K. is a recipient of a fellowship from Japan Society for the Promotion of Science. This work was supported by grants from the NIH (HL093154 and HL108317) and Foundation LeDucq to A.H., and from the NIH (HD040661) to P.J.H.
Footnotes
www.sciencesignaling.org/cgi/content/full/9/431/ra58/DC1
Fig. S1. Translational regulation of BMPR2 through the mRNA sequence encoding the CTD.
Fig. S2. FMRP binds BMPR2-CTDseq and suppresses translation.
Fig. S3. BMP4-SMAD1/5 signaling is increased, but TGFβ-SMAD2/3 signaling is not altered in FMR1-null cells.
Fig. S4. LIMK-i and LDN effectively inhibit LIMK1 and BMPR1 kinase activity in N1E cells.
Fig. S5. In vivo administration of LIMK-i inhibits phosphorylation of cofilin in mouse brain.
Table S1. qRT-PCR primers.
Table S2. RIP PCR primers.
Author contributions: R.K., S.R., S.K., J.L., Y.L., P.L., V.M.-C., and J.A.-T. designed and performed the experiments and interpreted the data. R.K., K.D.B., T.B.K., M.A., P.J.H., R.H., and G.L. designed the experiments, interpreted the data, and edited the manuscript. A.H. conceived the project, designed the experiments, interpreted the data, and wrote the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: The plasmids require a material transfer agreement from UCSF, USA.
REFERENCES
- 1.Hagerman R, Hagerman P. Advances in clinical and molecular understanding of the FMR1 premutation and fragile X-associated tremor/ataxia syndrome. Lancet Neurol. 2013;12:786–798. doi: 10.1016/S1474-4422(13)70125-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Santos AR, Kanellopoulos AK, Bagni C. Learning and behavioral deficits associated with the absence of the fragile X mental retardation protein: What a fly and mouse model can teach us. Learn. Mem. 2014;21:543–555. doi: 10.1101/lm.035956.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bagni C, Tassone F, Neri G, Hagerman R. Fragile X syndrome: Causes, diagnosis, mechanisms, and therapeutics. J. Clin. Invest. 2012;122:4314–4322. doi: 10.1172/JCI63141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Verkerk AJMH, Pieretti M, Sutcliffe JS, Fu Y-H, Kuhl DPA, Pizzuti A, Reiner O, Richards S, Victoria MF, Zhang F, Eussen BE, van Ommen G-JB, Blonden LAJ, Riggins GJ, Chastain JL, Kunst CB, Galjaard H, Caskey CT, Nelson DL, Oostra BA, Warren ST. Identification of a gene (FMR-1) containing a CGG repeat coincident with a breakpoint cluster region exhibiting length variation in fragile X syndrome. Cell. 1991;65:905–914. doi: 10.1016/0092-8674(91)90397-h. [DOI] [PubMed] [Google Scholar]
- 5.Kremer EJ, Pritchard M, Lynch M, Yu S, Holman K, Baker E, Warren ST, Schlessinger D, Sutherland GR, Richards RI. Mapping of DNA instability at the fragile X to a trinucleotide repeat sequence p(CCG)n. Science. 1991;252:1711–1714. doi: 10.1126/science.1675488. [DOI] [PubMed] [Google Scholar]
- 6.Kremer EJ, Yu S, Pritchard M, Nagaraja R, Heitz D, Lynch M, Baker E, Hyland VJ, Little RD, Wada M, Toniolo D, Vincent A, Rousseau F, Schlessinger D, Sutherland GR, Richards RI. Isolation of a human DNA sequence which spans the fragile X. Am. J. Hum. Genet. 1991;49:656–661. [PMC free article] [PubMed] [Google Scholar]
- 7.Vincent A, Hertz D, Petit C, Kretz C, Oberlé I, Mandel J-L. Abnormal pattern detected in fragile-X patients by pulsed-field gel electrophoresis. Nature. 1991;349:624–626. doi: 10.1038/349624a0. [DOI] [PubMed] [Google Scholar]
- 8.Darnell JC, Fraser CE, Mostovetsky O, Stefani G, Jones TA, Eddy SR, Darnell RB. Kissing complex RNAs mediate interaction between the Fragile-X mental retardation protein KH2 domain and brain polyribosomes. Genes Dev. 2005;19:903–918. doi: 10.1101/gad.1276805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Siomi H, Siomi MC, Nussbaum RL, Dreyfuss G. The protein product of the fragile X gene, FMR1, has characteristics of an RNA-binding protein. Cell. 1993;74:291–298. doi: 10.1016/0092-8674(93)90420-u. [DOI] [PubMed] [Google Scholar]
- 10.Ashley CT, Jr, Wilkinson KD, Reines D, Warren ST. FMR1 protein: Conserved RNP family domains and selective RNA binding. Science. 1993;262:563–566. doi: 10.1126/science.7692601. [DOI] [PubMed] [Google Scholar]
- 11.Darnell JC, Jensen KB, Jin P, Brown V, Warren ST, Darnell RB. Fragile X mental retardation protein targets G quartet mRNAs important for neuronal function. Cell. 2001;107:489–499. doi: 10.1016/s0092-8674(01)00566-9. [DOI] [PubMed] [Google Scholar]
- 12.Laggerbauer B, Ostareck D, Keidel E-M, Ostareck-Lederer A, Fischer U. Evidence that fragile X mental retardation protein is a negative regulator of translation. Hum. Mol. Genet. 2001;10:329–338. doi: 10.1093/hmg/10.4.329. [DOI] [PubMed] [Google Scholar]
- 13.Feng Y, Absher D, Eberhart DE, Brown V, Malter HE, Warren ST. FMRP associates with polyribosomes as an mRNP, and the I304N mutation of severe fragile X syndrome abolishes this association. Mol. Cell. 1997;1:109–118. doi: 10.1016/s1097-2765(00)80012-x. [DOI] [PubMed] [Google Scholar]
- 14.Corbin F, Bouillon M, Fortin A, Morin S, Rousseau F, Khandjian EW. The fragile X mental retardation protein is associated with poly(A)+ mRNA in actively translating polyribosomes. Hum. Mol. Genet. 1997;6:1465–1472. doi: 10.1093/hmg/6.9.1465. [DOI] [PubMed] [Google Scholar]
- 15.Pasciuto E, Bagni C. SnapShot: FMRP mRNA targets and diseases. Cell. 2014;158:1446–1446. doi: 10.1016/j.cell.2014.08.035. e1. [DOI] [PubMed] [Google Scholar]
- 16.Ascano M, Hafner M, Cekan P, Gerstberger S, Tuschl T. Identification of RNA-protein interaction networks using PAR-CLIP. Wiley Interdiscip. Rev.: RNA. 2012;3:159–177. doi: 10.1002/wrna.1103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.De Boulle K, Verkerk AJMH, Reyniers E, Vits L, Hendrickx J, Van Roy B, Van Den Bos F, de Graaff E, Oostra BA, Willems PJ. A point mutation in the FMR-1 gene associated with fragile X mental retardation. Nat. Genet. 1993;3:31–35. doi: 10.1038/ng0193-31. [DOI] [PubMed] [Google Scholar]
- 18.Hallahan BP, Craig MC, Toal F, Daly EM, Moore CJ, Ambikapathy A, Robertson D, Murphy KC, Murphy DGM. In vivo brain anatomy of adult males with Fragile X syndrome: An MRI study. Neuroimage. 2011;54:16–24. doi: 10.1016/j.neuroimage.2010.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Reiss AL, Abrams MT, Greenlaw R, Freund L, Denckla MB. Neurodevelopmental effects of the FMR-1 full mutation in humans. Nat. Med. 1995;1:159–167. doi: 10.1038/nm0295-159. [DOI] [PubMed] [Google Scholar]
- 20.Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: Maturation and pruning deficits. Proc. Natl. Acad. Sci. U.S.A. 1997;94:5401–5404. doi: 10.1073/pnas.94.10.5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nimchinsky EA, Oberlander AM, Svoboda K. Abnormal development of dendritic spines in FMR1 knock-out mice. J. Neurosci. 2001;21:5139–5146. doi: 10.1523/JNEUROSCI.21-14-05139.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ascano M, Jr, Mukherjee N, Bandaru P, Miller JB, Nusbaum JD, Corcoran DL, Langlois C, Munschauer M, Dewell S, Hafner M, Williams Z, Ohler U, Tuschl T. FMRP targets distinct mRNA sequence elements to regulate protein expression. Nature. 2012;492:382–386. doi: 10.1038/nature11737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Brown V, Jin P, Ceman S, Darnell JC, O’Donnell WT, Tenenbaum SA, Jin X, Feng Y, Wilkinson KD, Keene JD, Darnell RB, Warren ST. Microarray identification of FMRP-associated brain mRNAs and altered mRNA translational profiles in fragile X syndrome. Cell. 2001;107:477–487. doi: 10.1016/s0092-8674(01)00568-2. [DOI] [PubMed] [Google Scholar]
- 24.Miyashiro KY, Beckel-Mitchener A, Purk TP, Becker KG, Barret T, Liu L, Carbonetto S, Weiler IJ, Greenough WT, Eberwine J. RNA cargoes associating with FMRP reveal deficits in cellular functioning in Fmr1 null mice. Neuron. 2003;37:417–431. doi: 10.1016/s0896-6273(03)00034-5. [DOI] [PubMed] [Google Scholar]
- 25.Darnell JC, Van Driesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Darnell JC, Klann E. The translation of translational control by FMRP: Therapeutic targets for FXS. Nat. Neurosci. 2013;16:1530–1536. doi: 10.1038/nn.3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hogan BLM. Bone morphogenetic proteins: Multifunctional regulators of vertebrate development. Genes Dev. 1996;10:1580–1594. doi: 10.1101/gad.10.13.1580. [DOI] [PubMed] [Google Scholar]
- 28.Massagué J. TGF-b signaling in development and disease. FEBS Lett. 2012;586:1833. doi: 10.1016/j.febslet.2012.05.030. [DOI] [PubMed] [Google Scholar]
- 29.Lee-Hoeflich ST, Causing CG, Podkowa M, Zhao X, Wrana JL, Attisano L. Activation of LIMK1 by binding to the BMP receptor, BMPRII, regulates BMP-dependent dendritogenesis. EMBO J. 2004;23:4792–4801. doi: 10.1038/sj.emboj.7600418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Podkowa M, Zhao X, Chow C-W, Coffey ET, Davis RJ, Attisano L. Microtubule stabilization by bone morphogenetic protein receptor-mediated scaffolding of c-Jun N-terminal kinase promotes dendrite formation. Mol. Cell. Biol. 2010;30:2241–2250. doi: 10.1128/MCB.01166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee-Hoeflich ST, Zhao X, Mehra A, Attisano L. The Drosophila type II receptor, Wishful thinking, binds BMP and myoglianin to activate multiple TGFb family signaling pathways. FEBS Lett. 2005;579:4615–4621. doi: 10.1016/j.febslet.2005.06.088. [DOI] [PubMed] [Google Scholar]
- 32.Eaton BA, Davis GW. LIM Kinase1 controls synaptic stability downstream of the type II BMP receptor. Neuron. 2005;47:695–708. doi: 10.1016/j.neuron.2005.08.010. [DOI] [PubMed] [Google Scholar]
- 33.Goold CP, Davis GW. The BMP ligand Gbb gates the expression of synaptic homeostasis independent of synaptic growth control. Neuron. 2007;56:109–123. doi: 10.1016/j.neuron.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Piccioli ZD, Littleton JT. Retrograde BMP signaling modulates rapid activity-dependent synaptic growth via presynaptic LIM kinase regulation of cofilin. J. Neurosci. 2014;34:4371–4381. doi: 10.1523/JNEUROSCI.4943-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foletta VC, Lim MA, Soosairajah J, Kelly AP, Stanley EG, Shannon M, He W, Das S, Massagué J, Bernard O. Direct signaling by the BMP type II receptor via the cytoskeletal regulator LIMK1. J. Cell Biol. 2003;162:1089–1098. doi: 10.1083/jcb.200212060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bayat V, Jaiswal M, Bellen HJ. The BMP signaling pathway at the Drosophila neuromuscular junction and its links to neurodegenerative diseases. Curr. Opin. Neurobiol. 2011;21:182–188. doi: 10.1016/j.conb.2010.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Aberle H, Haghighi AP, Fetter RD, McCabe BD, Magalhães TR, Goodman CS. wishful thinking encodes a BMP type II receptor that regulates synaptic growth in Drosophila. Neuron. 2002;33:545–558. doi: 10.1016/s0896-6273(02)00589-5. [DOI] [PubMed] [Google Scholar]
- 38.Marqués G, Bao H, Haerry TE, Shimell MJ, Duchek P, Zhang B, O’Connor MB. The Drosophila BMP type II receptor Wishful Thinking regulates neuromuscular synapse morphology and function. Neuron. 2002;33:529–543. doi: 10.1016/s0896-6273(02)00595-0. [DOI] [PubMed] [Google Scholar]
- 39.Leyton PA, Beppu H, Pappas A, Martyn TM, Derwall M, Baron DM, Galdos R, Bloch DB, Bloch KD. Deletion of the sequence encoding the tail domain of the bone morphogenetic protein type 2 receptor reveals a bone morphogenetic protein 7-specific gain of function. PLOS One. 2013;8:e76947. doi: 10.1371/journal.pone.0076947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Siomi MC, Zhang Y, Siomi H, Dreyfuss G. Specific sequences in the fragile X syndrome protein FMR1 and the FXR proteins mediate their binding to 60S ribosomal subunits and the interactions among them. Mol. Cell. Biol. 1996;16:3825–3832. doi: 10.1128/mcb.16.7.3825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefani G, Fraser CE, Darnell JC, Darnell RB. Fragile X mental retardation protein is associated with translating polyribosomes in neuronal cells. J. Neurosci. 2004;24:7272–7276. doi: 10.1523/JNEUROSCI.2306-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keshishian H, Kim YS. Orchestrating development and function: Retrograde BMP signaling in the Drosophila nervous system. Trends Neurosci. 2004;27:143–147. doi: 10.1016/j.tins.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 43.Zhang YQ, Bailey AM, Matthies HJG, Renden RB, Smith MA, Speese SD, Rubin GM, Broadie K. Drosophila fragile X-related gene regulates the MAP1B homolog Futsch to control synaptic structure and function. Cell. 2001;107:591–603. doi: 10.1016/s0092-8674(01)00589-x. [DOI] [PubMed] [Google Scholar]
- 44.Guo X, Rueger D, Higgins D. Osteogenic protein-1 and related bone morphogenetic proteins regulate dendritic growth and the expression of microtubule-associated protein-2 in rat sympathetic neurons. Neurosci. Lett. 1998;245:131–134. doi: 10.1016/s0304-3940(98)00192-x. [DOI] [PubMed] [Google Scholar]
- 45.Chan MC, Nguyen PH, Davis BN, Ohoka N, Hayashi H, Du K, Lagna G, Hata A. A novel regulatory mechanism of the bone morphogenetic protein (BMP) signaling pathway involving the carboxyl-terminal tail domain of BMP type II receptor. Mol. Cell. Biol. 2007;27:5776–5789. doi: 10.1128/MCB.00218-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grossman AW, Elisseou NM, McKinney BC, Greenough WT. Hippocampal pyramidal cells in adult Fmr1 knockout mice exhibit an immature-appearing profile of dendritic spines. Brain Res. 2006;1084:158–164. doi: 10.1016/j.brainres.2006.02.044. [DOI] [PubMed] [Google Scholar]
- 47.McKinney BC, Grossman AW, Elisseou NM, Greenough WT. Dendritic spine abnormalities in the occipital cortex of C57BL/6 Fmr1 knockout mice. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2005;136B:98–102. doi: 10.1002/ajmg.b.30183. [DOI] [PubMed] [Google Scholar]
- 48.Irwin SA, Idupulapati M, Gilbert ME, Harris JB, Chakravarti AB, Rogers EJ, Crisostomo RA, Larsen BP, Mehta A, Alcantara CJ, Patel B, Swain RA, Weiler IJ, Oostra BA, Greenough WT. Dendritic spine and dendritic field characteristics of layer V pyramidal neurons in the visual cortex of fragile-X knockout mice. Am. J. Med. Genet. 2002;111:140–146. doi: 10.1002/ajmg.10500. [DOI] [PubMed] [Google Scholar]
- 49.Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb. Cortex. 2000;10:1038–1044. doi: 10.1093/cercor/10.10.1038. [DOI] [PubMed] [Google Scholar]
- 50.Ross-Macdonald P, de Silva H, Guo Q, Xiao H, Hung C-Y, Penhallow B, Markwalder J, He L, Attar RM, Lin T-a, Seitz S, Tilford C, Wardwell-Swanson J, Jackson D. Identification of a nonkinase target mediating cytotoxicity of novel kinase inhibitors. Mol. Cancer Ther. 2008;7:3490–3498. doi: 10.1158/1535-7163.MCT-08-0826. [DOI] [PubMed] [Google Scholar]
- 51.Yu PB, Deng DY, Lai CS, Hong CC, Cuny GD, Bouxsein ML, Hong DW, McManus PM, Katagiri T, Sachidanandan C, Kamiya N, Fukuda T, Mishina Y, Peterson RT, Bloch KD. BMP type I receptor inhibition reduces heterotopic [corrected] ossification. Nat. Med. 2008;14:1363–1369. doi: 10.1038/nm.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cuny GD, Yu PB, Laha JK, Xing X, Liu J-F, Lai CS, Deng DY, Sachidanandan C, Bloch KD, Peterson RT. Structure-activity relationship study of bone morphogenetic protein (BMP) signaling inhibitors. Bioorg. Med. Chem. Lett. 2008;18:4388–4392. doi: 10.1016/j.bmcl.2008.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tucker B, Richards RI, Lardelli M. Contribution of mGluR and Fmr1 functional pathways to neurite morphogenesis, craniofacial development and fragile X syndrome. Hum. Mol. Genet. 2006;15:3446–3458. doi: 10.1093/hmg/ddl422. [DOI] [PubMed] [Google Scholar]
- 54.Cruz-Martín A, Crespo M, Portera-Cailliau C. Delayed stabilization of dendritic spines in fragile X mice. J. Neurosci. 2010;30:7793–7803. doi: 10.1523/JNEUROSCI.0577-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.He CX, Portera-Cailliau C. The trouble with spines in fragile X syndrome: Density, maturity and plasticity. NeuroScience. 2013;251:120–128. doi: 10.1016/j.neuroscience.2012.03.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McBrayer ZL, Dimova J, Pisansky MT, Sun M, Beppu H, Gewirtz JC, O’Connor MB. Forebrain-specific loss of BMPRII in mice reduces anxiety and increases object exploration. PLOS One. 2015;10:e0139860. doi: 10.1371/journal.pone.0139860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen E, Joseph S. Fragile X mental retardation protein: A paradigm for translational control by RNA-binding proteins. Biochimie. 2015;114:147–154. doi: 10.1016/j.biochi.2015.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol. Cell. 2014;54:407–417. doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lucá R, Averna M, Zalfa F, Vecchi M, Bianchi F, La Fata G, Del Nonno F, Nardacci R, Bianchi M, Nuciforo P, Munck S, Parrella P, Moura R, Signori E, Alston R, Kuchnio A, Farace MG, Fazio VM, Piacentini M, De Strooper B, Achsel T, Neri G, Neven P, Evans DG, Carmeliet P, Mazzone M, Bagni C. The fragile X protein binds mRNAs involved in cancer progression and modulates metastasis formation. EMBO Mol. Med. 2013;5:1523–1536. doi: 10.1002/emmm.201302847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Heulens I, Suttie M, Postnov A, De Clerck N, Perrotta CS, Mattina T, Faravelli F, Forzano F, Kooy RF, Hammond P. Craniofacial characteristics of fragile X syndrome in mouse and man. Eur. J. Hum. Genet. 2013;21:816–823. doi: 10.1038/ejhg.2012.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hayano S, Komatsu Y, Pan H, Mishina Y. Augmented BMP signaling in the neural crest inhibits nasal cartilage morphogenesis by inducing p53-mediated apoptosis. Development. 2015;142:1357–1367. doi: 10.1242/dev.118802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Komatsu Y, Yu PB, Kamiya N, Pan H, Fukuda T, Scott GJ, Ray MK, Yamamura K-i, Mishina Y. Augmentation of Smad-dependent BMP signaling in neural crest cells causes craniosynostosis in mice. J. Bone Miner. Res. 2013;28:1422–1433. doi: 10.1002/jbmr.1857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Owens P, Pickup MW, Novitskiy SV, Giltnane JM, Gorska AE, Hopkins CR, Hong CC, Moses HL. Inhibition of BMP signaling suppresses metastasis in mammary cancer. Oncogene. 2015;34:2437–2449. doi: 10.1038/onc.2014.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Morrell NW. Role of bone morphogenetic protein receptors in the development of pulmonary arterial hypertension. Adv. Exp. Med. Biol. 2010;661:251–264. doi: 10.1007/978-1-60761-500-2_16. [DOI] [PubMed] [Google Scholar]
- 65.Edwards DC, Sanders LC, Bokoch GM, Gill GN. Activation of LIM-kinase by PAK1 couples Rac/Cdc42 GTPase signalling to actin cytoskeletal dynamics. Nat. Cell Biol. 1999;1:253–259. doi: 10.1038/12963. [DOI] [PubMed] [Google Scholar]
- 66.Say E, Tay H-G, Zhao Z-s, Baskaran Y, Li R, Lim L, Manser E. A functional requirement for PAK1 binding to the KH(2) domain of the fragile X protein-related FXR1. Mol. Cell. 2010;38:236–249. doi: 10.1016/j.molcel.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 67.Hayashi ML, Rao BSS, Seo J-S, Choi H-S, Dolan BM, Choi S-Y, Chattarji S, Tonegawa S. Inhibition of p21-activated kinase rescues symptoms of fragile X syndrome in mice. Proc. Natl. Acad. Sci. U.S.A. 2007;104:11489–11494. doi: 10.1073/pnas.0705003104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dolan BM, Duron SG, Campbell DA, Vollrath B, Rao BSS, Ko H-Y, Lin GG, Govindarajan A, Choi S-Y, Tonegawa S. Rescue of fragile X syndrome phenotypes in Fmr1 KO mice by the small-molecule PAK inhibitor FRAX486. Proc. Natl. Acad. Sci. U.S.A. 2013;110:5671–5676. doi: 10.1073/pnas.1219383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kumar R, Gururaj AE, Barnes CJ. p21-activated kinases in cancer. Nat. Rev. Cancer. 2006;6:459–471. doi: 10.1038/nrc1892. [DOI] [PubMed] [Google Scholar]
- 70.Pröschel C, Blouin MJ, Gutowski NJ, Ludwig R, Noble M. LIMK1 is predominantly expressed in neural tissues and phosphorylates serine, threonine and tyrosine residues in vitro. Oncogene. 1995;11:1271–1281. [PubMed] [Google Scholar]
- 71.Cuberos H, Vallee B, Vourc’h P, Tastet J, Andres CR, Benedetti H. Roles of LIM kinases in central nervous system function and dysfunction. FEBS Lett. 2015;589:3795–3806. doi: 10.1016/j.febslet.2015.10.032. [DOI] [PubMed] [Google Scholar]
- 72.Scott RW, Hooper S, Crighton D, Li A, König I, Munro J, Trivier E, Wickman G, Morin P, Croft DR, Dawson J, Machesky L, Anderson KI, Sahai EA, Olson MF. LIM kinases are required for invasive path generation by tumor and tumor-associated stromal cells. J. Cell Biol. 2010;191:169–185. doi: 10.1083/jcb.201002041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Khalil AM, Faghihi MA, Modarresi F, Brothers SP, Wahlestedt C. A novel RNA transcript with antiapoptotic function is silenced in fragile X syndrome. PLOS One. 2008;3:e1486. doi: 10.1371/journal.pone.0001486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zou K, Liu J, Zhu N, Lin J, Liang Q, Brown WT, Shen Y, Zhong N. Identification of FMRP-associated mRNAs using yeast three-hybrid system. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2008;147B:769–777. doi: 10.1002/ajmg.b.30678. [DOI] [PubMed] [Google Scholar]
- 75.Tursun B, Schlüter A, Peters MA, Viehweger B, Ostendorff HP, Soosairajah J, Drung A, Bossenz M, Johnsen SA, Schweizer M, Bernard O, Bach I. The ubiquitin ligase Rnf6 regulates local LIM kinase 1 levels in axonal growth cones. Genes Dev. 2005;19:2307–2319. doi: 10.1101/gad.1340605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Davis BN, Hilyard AC, Lagna G, Hata A. SMAD proteins control DROSHA-mediated microRNA maturation. Nature. 2008;454:56–61. doi: 10.1038/nature07086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lagna G, Ku MM, Nguyen PH, Neuman NA, Davis BN, Hata A. Control of phenotypic plasticity of smooth muscle cells by bone morphogenetic protein signaling through the myocardin-related transcription factors. J. Biol. Chem. 2007;282:37244–37255. doi: 10.1074/jbc.M708137200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.