Abstract
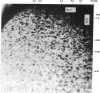
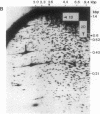
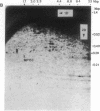
We have developed a powerful genomic scanning method, termed "restriction landmark genomic scanning," that is useful for analysis of the genomic DNA of higher organisms using restriction sites as landmarks. Genomic DNA is radioactively labeled at cleavage sites specific for a rare cleaving restriction enzyme and then size-fractionated in one dimension. The fractionated DNA is further digested with another more frequently occurring enzyme and separated in the second dimension. This procedure gives a two-dimensional pattern with thousands of scattered spots corresponding to sites for the first enzyme, indicating that the genome of mammals can be scanned at approximately 1-megabase intervals. The position and intensity of a spot reflect its locus and the copy number of the corresponding restriction site, respectively, based on the nature of the end-labeling system. Therefore, this method is widely applicable to genome mapping or detection of alterations in a genome.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Avner P., Amar L., Dandolo L., Guénet J. L. Genetic analysis of the mouse using interspecific crosses. Trends Genet. 1988 Jan;4(1):18–23. doi: 10.1016/0168-9525(88)90123-0. [DOI] [PubMed] [Google Scholar]
- Bingham P. M., Levis R., Rubin G. M. Cloning of DNA sequences from the white locus of D. melanogaster by a novel and general method. Cell. 1981 Sep;25(3):693–704. doi: 10.1016/0092-8674(81)90176-8. [DOI] [PubMed] [Google Scholar]
- Blin N., Stafford D. W. A general method for isolation of high molecular weight DNA from eukaryotes. Nucleic Acids Res. 1976 Sep;3(9):2303–2308. doi: 10.1093/nar/3.9.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brilliant M. H., Gondo Y., Eicher E. M. Direct molecular identification of the mouse pink-eyed unstable mutation by genome scanning. Science. 1991 Apr 26;252(5005):566–569. doi: 10.1126/science.1673574. [DOI] [PubMed] [Google Scholar]
- Garrels J. I. The QUEST system for quantitative analysis of two-dimensional gels. J Biol Chem. 1989 Mar 25;264(9):5269–5282. [PubMed] [Google Scholar]
- Russell L. B., Hunsicker P. R., Cacheiro N. L., Bangham J. W., Russell W. L., Shelby M. D. Chlorambucil effectively induces deletion mutations in mouse germ cells. Proc Natl Acad Sci U S A. 1989 May;86(10):3704–3708. doi: 10.1073/pnas.86.10.3704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Uitterlinden A. G., Slagboom P. E., Knook D. L., Vijg J. Two-dimensional DNA fingerprinting of human individuals. Proc Natl Acad Sci U S A. 1989 Apr;86(8):2742–2746. doi: 10.1073/pnas.86.8.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]