Abstract
Interaction between the acidic motif (AM) of protein kinase WNK4 and the Kelch domain of KLHL3 are involved in the pathogenesis of pseudohypoaldosteronism type II, a hereditary form of hypertension. This interaction is disrupted by some disease‐causing mutations in either WNK4 or KLHL3, or by angiotensin II‐ and insulin‐induced phosphorylation of KLHL3 at serine 433, which is also a site frequently mutated in patients. However, the mechanism by which this phosphorylation disrupts the interaction is unclear. In this study, we approached this problem using molecular dynamics simulation with structural, dynamical and energetic analyses. Results from independent simulations indicate that when S433 was phosphorylated, the electrostatic potential became more negative in the AM binding site of KLHL3 and therefore was unfavorable for binding with the negatively charged AM. In addition, the intermolecular hydrogen bond network that kept the AM stable in the binding site of KLHL3 was disrupted, and the forces for the hydrophobic interactions between the AM of WNK4 and KLHL3 were also reduced. As a result, the weakened interactions were no longer capable of holding the AM of WNK4 at its binding site in KLHL3. In conclusion, phosphorylation of KLHL3 at S433 disrupts the hydrogen bonds, hydrophobic and electrostatic interactions between the Kelch domain of KLHL3 and the AM of WNK4. This study provides a key molecular understanding of the KLHL3‐mediated regulation of WNK4, which is an integrative regulator of electrolyte homeostasis and blood pressure regulation in the kidney.
Significances Statement: WNK4 is an integrative regulator of electrolyte homeostasis, which is important in the blood pressure regulation by the kidney. Interaction between WNK4 and KLHL3 is a key physiological process that is impaired in a hereditary form of hypertension. This study provides substantial new insights into the role of phosphorylation of KLHL3 in regulating the interaction with WNK4, and therefore advances our understanding of molecular pathogenesis of hypertension and the mechanism of blood pressure regulation.
Keywords: KLHL3, WNK4, phosphorylation, molecular dynamics simulation
Abbreviations
- AM
acidic motif
- CUL3
Cullin 3
- KLHL3
kelch like family member 3
- MM/GBSA
molecular mechanics/generalized Born surface area
- NCC
Na+–Cl− cotransporter
- OSR1
oxidative stress‐responsive kinase 1
- PDB
protein data bank; Rg, radius of gyration
- RMSD
root mean square deviation
- RMSF
root mean square fluctuation
- SPAK
STE20/SPS1‐related proline/alanine‐rich kinase
- WNK4
with‐no‐lysine [K] kinase 4
Introduction
Pseudohypoaldosteronism type II (PHAII), also known as familial hyperkalemic hypertension or Gordon's syndrome, is a hereditary disease characterized by hypertension and hyperkalemia.1, 2 Mutations in genes encoding With‐No‐lysine [K] (WNK) kinases WNK1 and WNK4, were first identified in PHAII patients.3 As protein kinases, WNK1 and WNK4 phosphorylate and activate oxidative stress‐responsive kinase 1 (OSR1) and STE20/SPS1‐related proline/alanine‐rich kinase (SPAK),4, 5, 6 which in turn, phosphorylate and activate the thiazide‐sensitive Na+–Cl− cotransporter (NCC) and possibly other targets.7, 8, 9, 10, 11, 12, 13, 14 It has been shown that disease‐causing mutations in WNK4 gene lead to an elevation of the level of kinase protein and/or activity.15, 16, 17, 18, 19 This in turn augments their effects on ion transport proteins, resulting in increased Na+ reabsorption and decreased K+ secretion in the kidney, leading to PHAII.
In addition to WNK1 and WNK4, mutations in KLHL3 (kelch like family member 3), and CUL3 (Cullin 3) genes were recently identified in PHAII patients.20, 21 The newly identified PHAII related proteins are involved in regulating WNK1 and WNK4 protein degradation. KLHL3 and Cullin 3 are components of KLHL3–Cullin 3 ubiquitin E3 ligase complex, and WNK1 and WNK4 are substrates of this ligase complex. As the substrate adaptor of this complex, KLHL3 interacts with WNK1 and WNK4 at the “acidic motif” (AM), a stretch of sequence rich in negatively charged residues glutamate and aspartate. Most PHAII‐causing mutations of WNK4 are located in the AM. We previously found that the mutations in AM disrupted the calcium‐dependent regulation of WNK4 kinase activity.16 Recent findings indicate these mutations also disrupt the binding between KLHL3 and WNK4, and therefore, prevent the degradation of WNK4 protein.17, 18, 19, 22
Structurally, KLHL3 constitutes of three domains: an N‐terminal BTB domain, a BACK domain, and a C‐terminal Kelch domain that interacts with substrates. Recently, the crystal structure (PDB ID: 4CH9) of KLHL3 Kelch domain in complex with the AM of WNK4 was reported.23 Similar to the other members of BTB–BACK−Kelch family,24, 25, 26 the Kelch domain of KLHL3 has a β‐propeller structure which is formed by six Kelch repeats, each composed of four antiparallel β‐strands. The AM (557EPEEPEADQ565) of WNK4 is nested in the binding interface spanning all six Kelch repeats.23 Most PHAII‐causing mutations in KLHL3 are located in the Kelch domain, highlighting its significance in the interaction with WNK4.
As a key step in WNK kinases degradation, the interaction between KLHL3 and WNK4 likely responds to physiological cues. One of the mechanisms mediating the physiological response is phosphorylation. Serine 433 (S433), a key residue situated in the Kelch domain, was found to be phosphorylated by protein kinase A (cyclic AMP‐dependent protein kinase), protein kinase B (also known as Akt), and protein kinase C (PKC) isozymes PKC‐α and PKC‐βI.27, 28 Phosphorylation of S433 reduces the interaction between KLHL3 and AM, and in turn, prevents WNK4 protein from degradation.27, 28 This is a physiologically important process, as it can be a part of angiotensin II, insulin, and/or vasopressin signaling on WNK kinases.27, 28 Mutation at S433 (S433N) was also frequently found in PHAII patients,20, 21 suggesting S433 is critical in the interaction between KLHL3 and WNK4. However, it is unclear how phosphorylation at S433 affects the interaction between the AM of WNK4 and the Kelch domain in KLHL3. To gain a better understanding of this mechanism, we performed molecular dynamics (MD) simulations using the structure of KLHL3 Kelch domain in complex with the AM. Results from structural, dynamical and electrostatic analyses of the MD simulations suggest that phosphorylation at S433 of KLHL3 alter the hydrogen bonds, electrostatic and hydrophobic interactions between KLHL3 and WNK4.
Results and Discussion
To investigate the effect of phosphorylation of S433 in the Kelch domain of KLHL3 on its interaction with the AM of WNK4, two systems were set up based on the crystal structure of the Kelch domain of KLHL3 in complex with the AM of WNK4 (PDB ID 4CH9): one with unphosphorylated S433 (S433) and the other with phosphorylated S433 (pS433). In the initial structure of system with phosphorylated S433, the phosphate group of pS433 was located in the pocket formed by P561, E562, and A563 in the AM of WNK4 (Fig. 1). For each system, three independent 100 ns MD simulations were performed and the results are denoted as S433‐1 to S433‐3 and pS433‐1 to pS433‐3, respectively.
Figure 1.
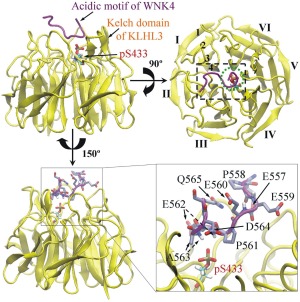
The initial positions of modeled phosphorylated S433 (pS433) in the Kelch domain of KLHL3 (residues 300–585, in yellow) and residues in the acidic motif (AM, residues 557 − 565, in purple) of WNK4. In the top view (upper right), the six Kelch repeats in the Kelch domain of KLHL3 are labeled as I–VI, and four antiparallel β‐strands in repeat I are labeled as 1–4. The black dashed rectangle represents the AM‐binding site on Kelch domain of KLHL3, and the green dot circle represents the location of pS433 in the top view. For clarity, residues of the AM in WNK4 and pS433 of KLHL3 are enlarged (lower right). The pS433 residue is located in the pocket formed by the side chain of P561, main chain of E562 and both main and side chains of A563 in the AM of WNK4. For simplicity, the hydrogen atoms in the side chain of AM are not shown.
The equilibration of the simulations was verified by calculating the values of RMSD and Rg of the Kelch domain of KLHL3 (residues 300–585) and the AM of WNK4 (residues 557–565) prior to data analysis. The time‐dependent changes of RMSD and Rg relative to the initial structure are presented in Figure 2. In both S433 and pS433 systems, the RMSD and Rg values for Kelch domain were stable (RMSD = 1.03 ± 0.11 Å; Rg = 17.13 ± 0.06 Å), implying the tertiary structure of the Kelch domain in KLHL3 did not vary significantly during the simulations. On the other hand, the RMSD and Rg values for the AM of WNK4 were low in the S433 systems whereas they were much higher in the pS433 systems. The RMSD and Rg curves indicated that the simulations reached equilibration before 60 ns, therefore, data from the last 40 ns (from 60 to 100 ns) simulation trajectories were used for following analyses.
Figure 2.
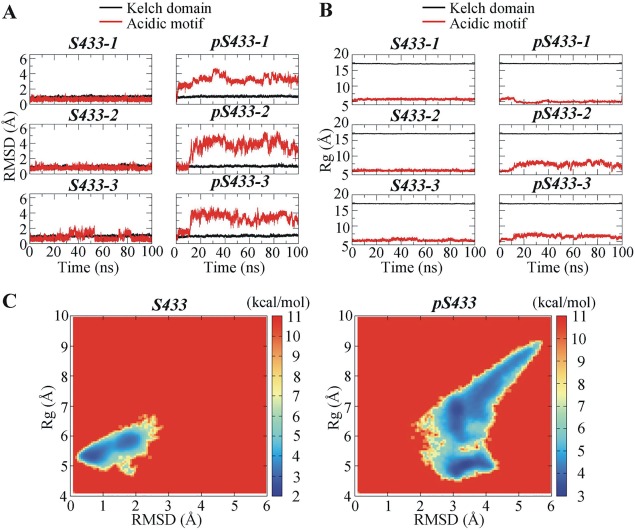
Time‐dependent structural alterations of the Kelch domain of KLHL3 and the acidic motif (AM) of WNK4. (A) Root mean square deviation (RMSD) as a function of simulation time. (B) Radius of gyration (Rg) as a function of simulation time; (C) Free energy landscape for the AM of WNK4 with respect to RMSD and Rg. In (A) and (B), three independent simulations for the Kelch domain−AM system containing unphosphorylated S433 (S433‐1 to S433‐3) and phosphorylated S433 (p433‐1 to pS433‐3) were performed.
Phosphorylation of S433 does not induce dramatic structural and dynamical changes in the Kelch domain of KLHL3
The RMSD and Rg values for the Kelch domain [shown black lines in Fig. 2(A,B), respectively] are not significantly different between the S433 and pS433 systems in three simulations. Since these variables represent the averaged distance of backbone carbon (Cα) atoms in the Kelch domain relative to their initial positions (RMSD) or to the center of mass of the Kelch domain (Rg), these results implied that phosphorylation of S433 in KLHL3 did not induce significant structural change in the Kelch domain of KLHL3. To further investigate the changes induced by phosphorylation of KLHL3 at S433, secondary structural and RMSF analyses were performed (Supporting Information Fig. S1). The values for β sheet occupancy averaged from the three independent trajectories for the S433 and pS433 systems are not significantly different [Supporting Information Fig. S1(A), 47.08 and 48.06%, respectively]. Furthermore, the RMSF values, which represent the atomic mobility for each residue in the Kelch domain over time, were very close between S433 and pS433 systems [Supporting Information Fig. S1(B)] These results indicate that the phosphorylation at S433 did not induce significant changes in the structure and dynamics in the Kelch domain of KLHL3.
Phosphorylation of S433 in KLHL3 results in conformational change in WNK4 acidic motif
In contrast to the Kelch domain of KLHL3, the RMSD and Rg values for the AM of WNK4 [red lines in Fig. 2(A,B), respectively] suggested that phosphorylation of S433 in KLHL3 induced robust structural change in the AM of WNK4. To further illustrate the conformational change of WNK4 AM, the free energy landscape with respect to RMSD and Rg was calculated from three independent trajectories [Fig. 2(C)]. The free energy landscape reflects the conformational space explored by WNK4 AM. The high free energy areas (deep red) were seldom explored whereas the low free energy areas (deep blue) were frequently explored by AM. In S433 systems, there were two major populations of conformations characterized by RMSD ∼ 0.7 Å/Rg ∼ 5.3 Å and RMSD ∼ 1.8 Å/Rg ∼ 5.8 Å. In pS433 systems, the position of the major population shifted to RMSD ∼ 3.2 Å/Rg ∼ 4.9 Å and RMSD ∼ 3.2 Å/Rg ∼ 6.9 Å. In addition, compared to S433 systems, the area explored by AM was broader in the pS433 systems, indicating that the AM in pS433 systems had more deviated structures and explored more conformations than that in S433 systems.
Phosphorylation of S433 changes the electrostatic potential for the WNK4 binding site on the Kelch domain of KLHL3
The AM of WNK4 (557EPEEPEADQ565), which is rich in glutamate/aspartate (E/D) residues, carries negative charges. To accommodate this motif, the surface electrostatic potential of the AM binding site on KLHL3 is expected to be positive. To test this proposition, the electrostatic potential maps for the initial structures of S433 and pS433 were calculated (Fig. 3). As expected, the electrostatic potential in the AM binding site of S433 was positive in order to attract the negatively charged WNK4 AM. Specially, the electrostatic potential for residue S433 in the initial S433 system was favorable for AM binding. As the phosphate group of residue pS433 brought two negative charges to the Kelch domain, the electrostatic potential shifted towards negative in the AM binding site on Kelch domain (Fig. 3). This resultant negative potential may not favor the binding of the negatively charged AM of WNK4. The alteration in the electrostatic potential of the Kelch domain by the phosphorylation of S433 was not only observed in the initial structures but also in the equilibrated structures. The representative equilibrated structures of S433 and pS433 systems, each from three independent simulations, were extracted using the clustering analysis. In each of the simulations, the electrostatic potential was more negative in the AM binding region of the Kelch domain for pS433 than that for S433 (Supporting Information Fig. S2). This further confirmed that the electrostatic potential in the Kelch domain became more negative upon phosphorylation of the S433 residue.
Figure 3.
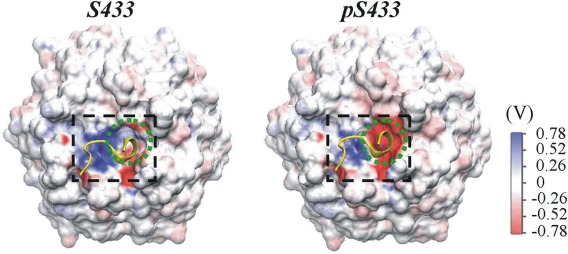
Electrostatic potential for the initial structure of KLHL3 Kelch domain with unphosphorylated (left) or phosphorylated S433 (right). The surface electrostatic potential for the acidic motif‐binding site (in black dashed rectangle) and that for S433/pS433 (in the green dashed circle) are shown. The initial structure of acidic motif (in yellow) is also indicated.
Phosphorylation of S433 disrupts the interaction network formed between the Kelch domain and the acidic motif
In the crystal structure of KLHL3 Kelch domain in complex with the AM (PDB ID: 4CH9), R339, R360 and R528 of KLHL3 form hydrogen bonds (H‐bonds) with E559, E557 and D564, respectively, in the AM of WNK4.23 Together with S432 of KLHL3, S433 forms an H‐bond with A563 of WNK4 through a water molecule. S432 also forms an H‐bond with E562 of WNK4. By assessing the interaction energy for each residues in the KLHL3 Kelch domain [Fig. 4(A), left panels], it is clear that the R339, R360, and R528 are among the residues with lowest interaction energies. Negative interaction energy indicates the residue favors interaction, thus R339, R360, and R528 are important in the interaction with the AM.
Figure 4.
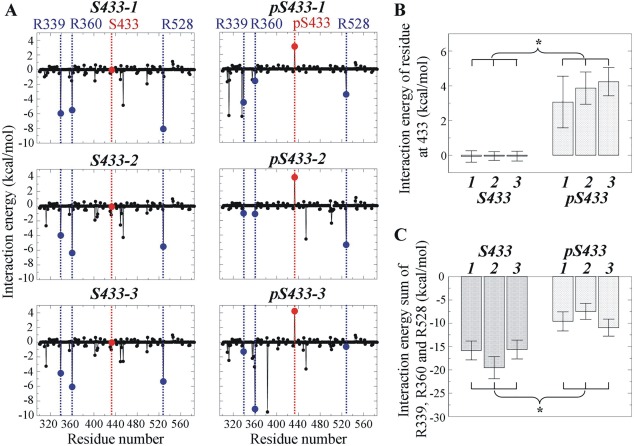
Interaction energy for residues of KLHL3 involved in the formation of intermolecular hydrogen bonds between acidic motif and Kelch domain. (A) Interaction energy for residues R339, R360, and R528 in Kelch domain containing S433 or pS433. Blue dash lines indicate the positions of these key interaction residues and red dash lines indicate the positions of S433 or pS433. (B) Interaction energy at S433 rose significantly after phosphorylation. (C) The sum of interaction energy for R339, R360, and R528 decreased significantly upon phosphorylation at S433. For (B and C), three simulations (labeled as 1, 2, and 3) were performed in the presence and in the absence of phosphorylation at S433, respectively. * indicates that the difference is statistically significant based on the mean and standard deviation of the analyzed variable (Student's t test, P < 0.05).
Upon phosphorylation, the interaction energy for S433 was raised from −0.07 to 4.23 kcal/mol [Fig. 4(B)]. Thus, the phosphorylation of S433 is unfavorable for interaction with the AM of WNK4. This was accompanied by the raised interaction energies for residues participating in H‐bond formation [Fig. 4(A,C)]. The raised interaction energy, together with a more negative surface electrostatic potential as a result of phosphorylation of S433, ultimately resulted in the disruption of the H‐bond network between the Kelch domain and the AM.
A visual comparison of the H‐bond network between the Kelch domain–AM systems in the presence and in the absence of the S433 phosphorylation is shown in Figure 5. In the three simulations with unphosphorylated S433, the AM peptide was fairly stable and no much difference in H‐bond network was observed among the simulations. One exception was that in two simulations (S433‐2 and S433‐3), S432 formed H‐bond with E562 in the AM; in one simulation (S433‐1), it formed H‐bond with the main chain of A563 in the AM through a water molecule (WAT) in the system [Fig. 5(A), left panel].
Figure 5.
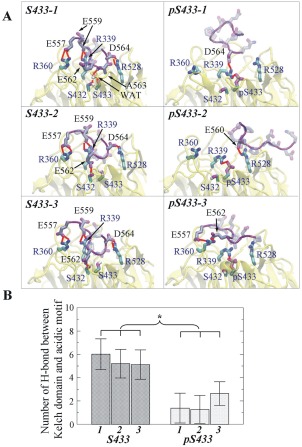
Hydrogen bonds between acidic motif and the Kelch domain containing unphosphorylated or phosphorylated S433. (A) Hydrogen bond (H‐bond) network formed in three simulations of Kelch domain‐acidic motif complex containing unphosphorylated (labeled as S433‐1, −2, and −3, left panels) or phosphorylated S433 (labeled as pS433‐1, −2, and −3, right panels). For clarity, the H‐bonds are labeled in red, and the hydrogen atoms for AM are not shown. (B) H‐bond number averaged from the equilibrated simulations. The numbers of H‐bonds in the three simulations with acidic motif and the Kelch domain containing pS433 are significantly lower than those in the simulations with systems containing S433. Asterisk indicates that the difference is statistically significant based on the mean and standard deviation of the analyzed variable (Student's t test, P < 0.05).
In contrast, when S433 was phosphorylated, pS433 formed an intramolecular H‐bond with R339 of Kelch domain in all three pS433 simulations [Fig. 5(A), right panels]. This disrupted the formation of intermolecular H‐bond between R339 of Kelch domain and E559 of AM. Possibly, this was the initial event that disrupted the formation of other intermolecular H‐bonds, including R360–E557 and R528–D564, as observed in the unphosphorylated S433 simulations [Fig. 5(A)]. As a result, fewer intermolecular H‐bonds were observed when S433 was phosphorylated. In the first simulation [Fig. 5(A), pS433‐1], only R339 of Kelch domain formed an H‐bond with D564 of AM; in the second simulation [Fig. 5(A), pS433‐2], only R528 of Kelch domain formed an H‐bond with E560 of AM; and in the third simulation [Fig. 5(A,) pS433‐3], only R360 of Kelch domain formed H‐bond with E557 and E562 of AM. To provide an estimate of the change in H‐bonds between Kelch domain and AM induced by the phosphorylation of S433, the time‐dependent formation of intermolecular H‐bonds among the three key residues of Kelch domain and AM were calculated (Supporting Information Fig. S3). The numbers of intermolecular hydrogen bonds, averaged from the equilibrated 40 ns of simulations, were significantly lower in the pS433 simulations than those in the S433 simulations [Fig. 5(B)], indicating the phosphorylation of S433 resulted in the disruption of the key H‐bond network between KLHL3 and WNK4.
In addition to the H‐bonds, residues F355, F402, G404, Y449, G451, and H498 in the Kelch domain exhibit hydrophobic interactions with AM of WNK4.23 The sum of interaction energy of F355, F402, Y449, and H498 exhibited a significant decrease upon the phosphorylation of S433 as well (Supporting Information Fig. S4), this likely lead to a reduction in the hydrophobic interactions between the Kelch domain and the AM.
The acidic motif moves away from the Kelch domain upon the phosphorylation of Ser433
The RMSD and Rg values of AM shown in Figure 2 indicate that the AM of WNK4 undergo robust conformational changes upon phosphorylation at S433. To further assess to what extent it stays around the initial binding position, the distance between the center of mass (COM) of Kelch domain and that of AM was calculated for both S433 and pS433 systems (Fig. 6, Supporting Information Fig. S5). The snapshots of one of the three simulations with systems containing phosphorylated S433 (pS433‐1) at 10, 40, and 100 ns were represented in Figure 6(B). For comparison, the corresponding snapshots of one simulation in the system with unphosphorylated S433 (S433‐1) were also shown in Figure 6(A). Compared to the relative stable distance between the AM and the Kelch domain with unphosphorylated S433, that with pS433 varied significantly, indicating that the AM shifted away from its initial binding site on the Kelch domain during the course of simulation. Similar results were observed in the other two pairs of simulations as well (Supporting Information Fig. S5). To give another view of the positions of the AM relative to the Kelch domain during simulations, three representative structures for the Kelch domain–AM complex were extracted by clustering analysis [Fig. 6(C)]. The position of AM was fairly stable when S433 was unphosphorylated during the course of simulation whereas it changed significantly when S433 was phosphorylated, indicating it no longer bound to the Kelch domain.
Figure 6.
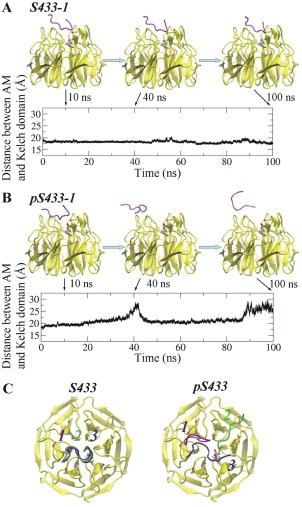
Distance between the acidic motif and the Kelch domain as a function of time. Time‐dependent change in the distance between the acidic motif and the Kelch domain with unphosphorylated S433 (A: S433‐1) or phosphorylated S433 (B: pS433‐1). Shown are results from one of the three simulations in each system. Snapshots of the acidic motif at 10, 40, and 100 ns of are shown. (C) Representative structures of the AM relative to the Kelch domain containing unphosphorylated or phosphorylated S433. The AM in the three independent simulations are shown in purple, green, and blue, respectively.
Phosphorylation of S433 adds a phosphate group to the residue. The phosphate group not only introduces two negative charges to S433, but also increases the size of its side chain. Thus, the unstable interaction between the AM and the Kelch domain containing pS433 during MD simulations could be due to either an electrostatic effect or a steric effect or both. To examine whether the unstable binding between the Kelch domain and AM is caused by the steric effect of the large side chain of pS433, the electroneutral protonated pS433 was modeled in the structure of the Kelch domain and the MD simulation was carried out. The results were compared with those with phosphorylated and unphosphorylated S433 (Supporting Information Fig. S6). The simulation snapshots at 100 ns show that similar to the situation with unphosphorylated S433 [Supporting Information Fig. S6(A)], the nearby residues P561 and A563 of the AM did not move significantly in the presence of electroneutral protonated pS433 [Supporting Information Fig. S6(C)], whereas they drifted away in the presence of negative charged pS433 [Supporting Information Fig. S6(B)]. In addition, the distance between the Kelch domain and AM was stable during the simulation in the presence of electroneutral protonated pS433, similar to that in the presence of unphosphorylated S433. Thus, the large phosphate group of the protonated pS433 did not result in unstable interaction between the Kelch domain and the AM. These data indicate that it is the negative charge rather than the size of the side chain of pS433 leads to the unstable binding between the Kelch domain and the AM of WNK4.
Conclusion and Outlook
The results of this study provide a theoretical explanation why the phosphorylation modification at S433 of the Kelch domain disrupts the interaction between KLHL3 and WNK4. The phosphorylation of S433 in the center of the Kelch domain did not induce drastic conformational or dynamical changes in the Kelch domain. However, this modification results in two alterations that ultimately disrupted the interaction between the Kelch domain and the AM of WNK4. Firstly, the two negative charges of pS433 increased the negative surface electrostatic potential that is unfavorable for the Kelch domain to interact with the negatively charged AM of WNK4. Secondly, phosphorylation of S433 resulted in the formation of an intramolecular H‐bond between pS433 and R339 in the Kelch domain, which ultimately lead to the disruption of the intermolecular H‐bond network between the Kelch domain and the AM. The disrupted H‐bonds and the reduced hydrophobic forces for interactions were unable to hold the AM to the Kelch domain. This could explain the experimental results that phosphomimetic mutation at S433 results in the reduction of interaction between KLHL3 and WNK4 and in turn, a decrease in WNK degradation.27 The MD simulation results of this study are consistent with the experimental observations,27, 28 indicating that MD simulation approach may be utilized to understand the mechanism by which the individual disease‐causing mutations in the Kelch lead to dysregulation of WNK4 that result in disease.
The results of our study do not identify which of the two major alterations cause by S433 phosphorylation is more important in disrupting the interaction between KLHL3 and WNK4. It is likely the phosphorylation at S433 only occur when KLHL3 is not occupied by WNK4, as WNK4 may block the accessibility of protein kinases to S433. In reality, phosphorylation of S433 will not disrupt the existing H‐bonds between KLHL3 and WNK4; rather these H‐bonds may not form in the first place. A possible scenario is that after phosphorylation of KLHL3 at S433, the negative electrostatic potential at the Kelch domain surface will repel the negatively charged AM of WNK4. Even though interaction may occur randomly, stable hydrogen bonds could not form between the Kelch domain and the AM. Thus, when S433 is phosphorylated, KLHL3 may not be able to hold WNK4 in place for ubiquitination.
The significance of S433 residue in KLHL3 has been confirmed that S433N mutation associates with PHAII.19, 23 The position of S433 in the center of Kelch domain surface makes it a perfect target for phosphorylation regulation. This position is accessible to protein kinases, and it has been shown that S433 can be phosphorylated by protein kinase A, B and C.27, 28 This makes S433 a suitable mediator of the effects of angiotensin II and insulin, as indicated recently.27, 28 The phosphorylation of S433 will lead to the reduction of interaction between KLHL3 and WNK4, and in turn an increase of the level of WNK4 protein, thus will sustain or increase the effects of WNK4 on ion transport proteins and ultimately lead to retention of both Na+ and K+.
Materials and Methods
System modeling
The structure of KLHL3 Kelch domain in complex with the acidic motif (AM) of WNK4 was obtained from the Protein Data Bank (PDB) with the PDB ID of 4CH9. The main chain of phosphorylated S433 (pS433) is modeled using the structure of S433 in the crystal structure, and the phosphate group atoms of pS433 were formed automatically using the LEAP module of AMBER12.29 In the modeled structure, the phosphate group of pS433 is located in the pocket formed by P561, E562, and A563 of WNK4 (Fig. 1). The two systems without and with the phosphorylation of KLHL3 at S433 are denoted as S433 and pS433, respectively. In the initial model of pS433, residues P561 and A563 of the AM of WNK4 are close to the side chain of pS433 in the Kelch domain. To examine whether a steric effect between the increased side chain of pS433 and the two residues causes the unstable interaction between the Kelch domain and the AM of WNK4, electroneutral protonated pS433 were also modeled.
Each system was placed in a box filled with 11,570 transferable intermolecular potential 3P (TIP3P) water molecules.30 All the crystallographic waters were kept in the systems. The size of the water sphere was chosen so that the distance between every atom in the protein and the boundary of the water was at least 10 Å. Na+ and Cl‐ ions were added to the system to neutralize it and maintain a 150 mM NaCl concentration. The Amber ff99SB force field31 was applied to all the residues and ions including Na+ and Cl‐ ions. The force field parameter for pS433 (with a −2 charge)32 was obtained from the AMBER parameter database.
Molecular dynamics simulations
To investigate the effect of phosphorylation of KLHL3 at S433 on the interaction between KLHL3 and WNK4, three independent 100 ns molecular dynamic (MD) simulations were performed for the two systems S433 and pS433, respectively, using the AMBER12 simulation package.29 Before production simulations, the two systems were further refined. Each system was relaxed by 2000 steps of steepest descent followed by 2000 steps of conjugate gradient minimization. After energy minimization of the whole system, the water molecules were equilibrated for 20 ps, while the protein and ions were restrained at a constant number–pressure–temperature (NpT) at 50 K and 1 atm by applying a force constant of 100 kcal mol−1 Å−1. Then the system was gradually heated to 300 K via six 100 ps constant number–volume−temperature (NVT) MD simulations, still maintaining the restraint on the proteins. The restraints were gradually reduced to zero in the subsequent 200 ps equilibration simulation at NpT of 300 K and 1 atm. Lastly, 100 ns production simulations were carried out using Berendsen temperature and pressure coupling33 without any restraint. The SHAKE constraints34 were applied to all hydrogen heavy bonds to permit a dynamics time step of 2 fs. Electrostatic interactions were calculated by the particle‐mesh Ewald method (PME)35, 36 with grid spacing of 0.12 nm and interpolation of order four. Both of the direct space PME and Lennard–Jones cutoffs were set at 10 Å. The data for the MD simulations were collected every 2 ps. The simulations were performed on the AMD Opterons cluster in the Alabama Supercomputer Center.
Structural and dynamical analysis and electrostatic potential calculation
With the equilibrated MD simulation trajectories, we performed secondary structural analysis, dynamical fluctuation analysis and electrostatic potential calculation to better understand the change of KLHL3 caused by the phosphorylation of S433. Root mean square deviation (RMSD) and radius of gyration (Rg) of protein backbone carbon (Cα) atoms were analyzed to check the system's equilibration tendencies and the structural change of KLHL3 and WNK4. To characterize the conformational change of WNK4 AM, the free energy landscape ΔG respect to RMSD and Rg was calculated using the following equation:
where k B is the Boltzmann constant, and P(RMSD, Rg) is the probability distribution obtained from the MD data at temperature T.
We calculated the β strand occupancy for KLHL3 to evaluate secondary structural changes caused by the phosphorylation. The DSSP method was used to determine whether an amino acid residue belonged to an β strand.37 The occupancy of each residue in a β strand was determined on the basis of the percentage of time that the residue existed in the β strand over the simulation. The occupancy percentage for each system was calculated via dividing the sum of total occupancy percentage of each residue by the total residue number.
Clustering analysis using the MMTSB toolset38 was performed to get the representative structure for the systems. The K‐means algorithm39 based on the RMSD similarity of the structures was used in the clustering analysis. A centroid structure was generated by averaging the PDB structures of KLHL3. The structure that had the lowest RMSD from the centroid structure was obtained as the representative structure. To study the conformational fluctuation for each residue, root‐mean‐square fluctuations (RMSF) for the backbone carbon (Cα) atoms of KLHL3 were calculated on a residue‐by‐residue basis and averaged over the equilibrated simulations. All these analyses were performed using the equilibrated simulation trajectories by the ptraj program of AMBER. The electrostatic potential for KLHL3 was calculated by APBS.40 To calculate the electrostatic potential, dielectric constants of 1 and 78.54 were used separately for the protein and solvent, respectively. Similar to our previous studies,41, 42 an ion concentration of 150 mM was used in the APBS calculation. The electrostatic potential was mapped onto the molecular surface of KLHL3 using VMD.43
Interaction energy contribution calculations
To investigate which residue in KLHL3 plays a key role in binding the AM of WNK4, the contribution of each KLHL3 residue to the interaction energy were calculated using the molecular mechanics/generalized Born surface area (MM/GBSA) approach.44 The MM/GBSA energy consists of molecular mechanics energy (including the internal energy, the electrostatic energy and the van der Waals energy) and solvation free energy (including a polar part and a nonpolar part). The molecular mechanics energy was calculated by summing the contributions of internal energy (including bond, angle, and torsional angle energies), electrostatic energy, and van der Waals energy from the gas‐phase energies of MD simulation. The polar solvation free energy was computed with the GBOBC model.45 A dielectric constant of 80 was used for the solvent, and that of 1 was used for the solute. The nonpolar solvation free energy was estimated by a solvent accessible surface area (SASA)‐dependent term using ICOSA method. The MM/GBSA energy was then decomposed based on the contributions per residue. All counterions and water molecules were stripped during the calculation. For the equilibrated MD simulation trajectories, frames at an interval of 20 ps were used for the calculation of all of the components of MM/GBSA energy.
Statistical methods
To investigate the change of hydrogen bond between KLHL3 and the AM of WNK4 caused by the phosphorylation, the average values and the standard deviations for hydrogen bond number were calculated. Because the adjacent snapshots from the MD trajectory have the tendency to be correlated with each other, the autocorrelation time46, 47 for the studied variables were obtained to resample the trajectories into statistically independent periods in order to calculate the standard deviations for the studied variables. With the obtained decorrelation times, bootstrap analysis was performed following steps similar to Chen and Pappu48 and our previous study.49, 50 Significant differences in the mean and standard deviations for the studied variables were determined using the Student's t test51 with 95% confidence.
Supporting Information
This provides a detailed description of secondary structural and RMSF analysis for KLHL3 (Supporting Information Fig. S1), electrostatic potential of KLHL3 for the equilibrated simulations (Supporting Information Fig. S2), time‐dependent hydrogen bonds formed between the key residues of KLHL3 and the acidic motif of WNK4 (Supporting Information Fig. S3), comparison of the interaction energy for residues of KLHL3 involved in hydrophobic interactions between Kelch domain and acidic motif of WNK4 (Supporting Information Fig. S4), and distance between the acidic motif and the Kelch domain (Supporting Information Fig. S5) as well as a comparison between the distances in the presence of pS433 and protonated pS433 (Supporting Information Fig. S6).
Supporting information
Supporting Information
ACKNOWLEDGMENTS
We thank the Alabama Supercomputer Center and Supercomputer facility at the University of Alabama at Birmingham for providing computational resources.
REFERENCES
- 1. Gordon RD (1986) Syndrome of hypertension and hyperkalaemia with normal glomerular filtration rate. Hypertension 8:93–102. [DOI] [PubMed] [Google Scholar]
- 2. Achard JM, Disse‐Nicodeme S, Fiquet‐Kempf B, Jeunemaitre X (2001) Phenotypic and genetic heterogeneity of familial hyperkalaemic hypertension (Gordon syndrome). Clin Exp Pharmacol Physiol 28:1048–1052. [DOI] [PubMed] [Google Scholar]
- 3. Wilson FH, Disse‐Nicodeme S, Choate KA, Ishikawa K, Nelson‐Willams C, Desitter I, Gunel M, Milford DV, Lipkin GW, Achard JM, Feely MP, Dussol B, Berland Y, Unwin RJ, Mayan H, Simon DB, Farfel Z, Jeunemaitre X, Lifton RP (2001) Human hypertension caused by mutations in WNK kinases. Science 293:1107–1112. [DOI] [PubMed] [Google Scholar]
- 4. Moriguchi T, Urushiyama S, Hisamoto N, Iemura SI, Uchida S, Natsume T, Matsumoto K, Shibuya H (2005) WNK1 regulates phosphorylation of cation‐chloride‐coupled cotransporters via the STE20‐related kinases, SPAK and OSR1. J Biol Chem 280:42685–42693. [DOI] [PubMed] [Google Scholar]
- 5. Vitari AC, Deak M, Morrice NA, Alessi DR (2005) The WNK1 and WNK4 protein kinases that are mutated in Gordon's hypertension syndrome phosphorylate and activate SPAK and OSR1 protein kinases. Biochem J 391:17–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Pathare G, Hoenderop JG, Bindels RJ, San‐Cristobal P (2013) A molecular update on pseudohypoaldosteronism type II. Am J Physiol Renal Physiol 305:1513–1520. [DOI] [PubMed] [Google Scholar]
- 7. Susa K, Kita S, Iwamoto T, Yang SS, Lin SH, Ohta A, Sohara E, Rai T, Sasaki S, Alessi DR, Uchida S (2012) Effect of heterozygous deletion of WNK1 on the WNK‐OSR1/SPAK‐NCC/NKCC1/NKCC2 signal cascade in the kidney and blood vessels. Clin Exp Nephrol 16:530–538. [DOI] [PubMed] [Google Scholar]
- 8. Castaneda‐Bueno M, Cervantes‐Perez LG, Vazquez N, Uribe N, Kantesaria S, Morla L, Bobadilla NA, Doucet A, Alessi DR, Gamba G (2012) Activation of the renal Na+:Cl− cotransporter by angiotensin II is a WNK4‐dependent process. Proc Natl Acad Sci USA 109:7929–7934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Yang SS, Morimoto T, Rai T, Chiga M, Sohara E, Ohno M, Uchida K, Lin SH, Moriguchi T, Shibuya H, Kondo Y, Sasaki S, Uchida S (2007) Molecular pathogenesis of pseudohypoaldosteronism type II: Generation and analysis of a Wnk4D561A/+ knockin mouse model. Cell Metab 5:331–344. [DOI] [PubMed] [Google Scholar]
- 10. Chiga M, Rafiqi FH, Alessi DR, Sohara E, Ohta A, Rai T, Sasaki S, Uchida S (2011) Phenotypes of pseudohypoaldosteronism type II caused by the WNK4 D561A missense mutation are dependent on the WNK–OSR1/SPAK kinase cascade. J Cell Sci 124:1391–1395. [DOI] [PubMed] [Google Scholar]
- 11. Bergaya S, Faure S, Baudrie V, Rio M, Escoubet B, Bonnin P, Henrion D, Loirand G, Achard JM, Jeunemaitre X, Hadchouel J (2011) WNK1 regulates vasoconstriction and blood pressure response to α1‐adrenergic stimulation in mice. Hypertension 58:439. [DOI] [PubMed] [Google Scholar]
- 12. Vidal‐Petiot E, Elvira‐Matelot E, Mutig K, Soukaseum C, Baudrie V, Wu S, Cheval L, Huc E, Cambillau M, Bachmann S, Doucet A, Jeunemaitre X, Hadchouel J (2013) WNK1‐related familial hyperkalemic hypertension results from an increased expression of L‐WNK1 specifically in the distal nephron. Proc Natl Acad Sci USA 110:14366–14371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rafiqi FH, Zuber AM, Glover M, Richardson C, Fleming S, Jovanovic S, Jovanovic A, O'Shaughnessy KM, Alessi DR (2010) Role of the WNK‐activated SPAK kinase in regulating blood pressure. EMBO Mol Med 2:63–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ohta A, Rai T, Yui N, Chiga M, Yang SS, Lin SH, Sohara E, Sasaki S, Uchida S (2009) Targeted disruption of the Wnk4 gene decreases phosphorylation of Na‐Cl cotransporter, increases Na excretion and lowers blood pressure. Hum Mol Genet 18:3978–3986. [DOI] [PubMed] [Google Scholar]
- 15. Na T, Wu GJ, Zhang W, Dong WJ, Peng JB (2013) Disease‐causing R1185C mutation of WNK4 disrupts a regularoty mechanism involving calmodulin binding and SGK1 phosphorylation sites. Am J Physiol Renal Physiol 304:F8–F18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Na T, Wu GJ, Peng JB (2012) Disease‐causing mutations in the acidic motif of WNK4 impair the sensitivity of WNK4 kinase to calcium ions. Biochem Biophys Res Commun 419:293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu GJ, Peng JB (2013) Disease‐causing mutations in KLHL3 impair its effect on WNK4 degradation. FEBS Lett 587:1717–1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Shibata S, Zhang J, Puthumana J, Stone KL, Lifton RP (2013) Kelch‐like 3 and Cullin 3 regulate electrolyte homeostasis via ubiquitination and degradation of WNK4. Proc Natl Acad Sci USA 110:7838–7843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ohta A, Schumacher FR, Mehellou Y, Johnson C, Knebel A, Macartney TJ, Wood NT, Alessi DR, Kurz T (2013) The CUL3–KLHL3 E3 ligase complex mutated in Gordon's hypertension syndrome interacts with and ubiquitylates WNK isoforms: disease‐causing mutations in KLHL3 and WNK4 disrupt interaction. Biochem J 451:111–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Boyden LM, Choi M, Choate KA, Nelson‐Williams CJ, Farhi A, Toka HR, Tikhonova IR, Bjornson R, Mane SM, Colussi G (2012) Mutations in kelch‐like 3 and cullin 3 cause hypertension and electrolyte abnormalities. Nature 482:98–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Louis‐Dit‐Picard H, Barc J, Trujillano D, Miserey‐Lenkei S, Bouatia‐Naji N, Pylypenko O, Beaurain G, Bonnefond A, Sand O, Simian C, Vidal‐Petiot E, Soukaseum C, Mandet C, Broux F, Chabre O, Delahousse M, Esnault V, Fiquet B, Houillier P, Bagnis CI, Koenig J, Konrad M, Landais P, Mourani C, Niaudet P, Hadchouel J, Schott JJ, Jeunemaitre X (2012) KLHL3 mutations cause familial hyperkalemic hypertension by impairing ion transport in the distal nephron. Nat Genet 44:456–460. [DOI] [PubMed] [Google Scholar]
- 22. Wakabayashi M, Mori T, Isobe K, Sohara E, Susa K, Araki Y, Chiga M, Kikuchi E, Nomura N, Mori Y, Matsuo H, Murata T, Nomura S, Asano T, Kawaguchi H, Nonoyama S, Rai T, Sasaki S, Uchida S (2013) Impaired KLHL3‐mediated ubiquitination of WNK4 causes human hypertension. Cell Rep 3:858–868. [DOI] [PubMed] [Google Scholar]
- 23. Schumacher F, Sorrell FJ, Alessi DR, Bullock AN, Kurz T (2014) Structural and biochemical characterization of the KLHL3–WNK kinase interaction important in blood pressure regulation. Biochem J 460:237–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lo SC, Li XC, Henzl MT, Beamer LJ, Hannink M (2006) Structure of the Keap1:Nrf2 interface provides mechanistic insight into Nrf2 signaling. EMBO J 25:3605–3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Canning P, Cooper CDO, Krojer T, Murray JW, Pike ACW, Chaikuad A, Keates T, Thangaratnarajah C, Hojzan V, Marsden BD, Gileadi O, Knapp S, von Deft F, Bullock AN (2013) Structural basis for Cul3 assembly with the BTB‐Kelch family of E3 ubiquitin ligases. J Biol Chem 288:7803–7814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Li XC, Zhang D, Hannink M, Beamer LJ (2004) Crystal structure of the Kelch domain of human Keap1. J Biol Chem 279:54750–54758. [DOI] [PubMed] [Google Scholar]
- 27. Shibata S, Arroyo JP, Castaneda‐Bueno M, Puthumana J, Zhang J, Uchida S, Stone KL, Lam TT, Lifton RP (2014) Angiotensin II signaling via protein kinase C phosphorylates Kelch‐like 3, preventing WNK4 degradation. Proc Natl Acad Sci USA 111:15556–15561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Yoshizaki Y, Mori Y, Tsuzaki Y, Mori T, Nomura N, Wakabayashi M, Takahashi D, Zeniya M, Kikuchi E, Araki Y, Ando F, Isobe K, Nishida H, Ohta A, Susa K, Inoue Y, Chiga M, Rai T, Sasaki S, Uchida K, Sohara E (2015) Impaired degradation of WNK by Akt and PKA phosphorylation of KLHL3. Biochem Biophys Res Commun 467:229–234. [DOI] [PubMed] [Google Scholar]
- 29. Case DA, Darden TA, Cheatham ITE, Simmerling CL, Wang J, Duke RE, Luo R, Walker RC, Zhang W, Merz KM, Roberts B, Hayik S, Roitberg A, Seabra G, Swails J, Gotz AW, Kolossváry I, Wong KF, Paesani F, Vanicek J, Wolf RM, Liu J, Wu X, Brozell SR, Steinbrecher T, Gohlke H, Cai Q, Ye X, Wang J, Hsieh MJ, Cui G, Roe DR, Mathew DH, Seetin MG, Salomon‐Ferrer R, Sagui C, Babin V, Luchko T, Gusarov S, Kovalenko A, Kollman PA (2012) AMBER 12, University of California, San Francisco. [Google Scholar]
- 30. Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML (1983) Comparison of simple potential functions for simulating liquid water. J Chem Phys 79:926–935. [Google Scholar]
- 31. Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C (2006) Comparison of multiple amber force fields and development of improved protein backbone parameters. Proteins 65:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Homeyer N, Horn AHC, Lanig H, Sticht H (2006) AMBER force‐field parameters for phosphorylated amino acids in different protonation states: phosphoserine, phosphothreonine, phosphotyrosine, and phosphohistidine. J Mol Model 12:281–289. [DOI] [PubMed] [Google Scholar]
- 33. Berendsen HJC, Postma JPM, van Gunsteren WF, DiNola A, Haak JR (1984) Molecular dynamics with coupling to an external bath. J Chem Phys 81:3684–3690. [Google Scholar]
- 34. Ryckaert J‐P, Ciccotti G, Berendsen HJC (1977) Numerical integration of the Cartesian equations of motion of a system with constraints: molecular dynamics of n‐alkanes. J Comput Phys 23:327–341. [Google Scholar]
- 35. Darden T, York D, Pedersen L (1993) Particle mess Ewald: an N•log (N) method for Ewald sums in large systems. J Chem Phys 98:10089–10092. [Google Scholar]
- 36. Essmann U, Perera L, Berkowitz ML, Darden T, Lee H, Pedersen L (1995) A smooth particle mess Ewald method. J Chem Phys 103:8577–8593. [Google Scholar]
- 37. Kabsch W, Sander C (1983) Dictionary of protein secondary structure: pattern recognition of hydrogen‐bonded and geometrical features. Biopolymers 22:2577–2637. [DOI] [PubMed] [Google Scholar]
- 38. Feig M, Karanicolas J, Brooks I, C. L. (2001) MMTSB Tool set, MMTSB NIH Research Resource, The Scripps Research Institute.
- 39. MacQueen JB (1967) Some methods for classification and analysis of multivariate observations. Proceedings of 5‐th Berkeley Symposium on Mathematical Statistics and Probability, Vol. 1. Berkeley: University of California Press, pp 281–297.
- 40. Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA (2001) Electrostatics of nanosystems: Application to microtubules and the ribosome. Proc Natl Acad Sci USA 98:10037–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Fancy RM, Wang L, Napier T, Lin J, Jing G, Lucius AL, McDonald JM, Zhou T, Song Y (2014) Characterization of calmodulin‐Fas death domain interaction: an integrated experimental and computational study. Biochemistry 53:2680–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fancy RM, Wang L, Zeng Q, Wang H, Zhou T, Buchsbaum DJ, Song Y (2016) Characterization of the interactions between calmodulin and death receptor 5 in triple‐negative and estrogen receptor positive breast cancer cells: an integrated experimental and computational study. J Biol Chem M116:727727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graphics Model 14:33–38. [DOI] [PubMed] [Google Scholar]
- 44. Gohlke H, Kiel C, Case DA (2003) Insights into protein‐protein binding by binding free energy calculation and free energy decomposition for the Ras–Raf and Ras–RalGDS complexes. J Mol Biol 330:891–913. [DOI] [PubMed] [Google Scholar]
- 45. Onufriev A, Bashford D, Case DA (2004) Exploring protein native states and large‐scale conformational changes with a modified generalized Born model. Proteins 55:383–394. [DOI] [PubMed] [Google Scholar]
- 46. Allen MP, Tildesley DJ (1987) Computer simulation of liquids. New York: Oxford University Press. [Google Scholar]
- 47. Lee SJ, Song Y, Baker NA (2008) Molecular dynamics simulations of asymmetric NaCl and KCl solutions separated by phosphatidylcholine bilayers: potential drops and structural changes induced by strong Na+–lipid interactions and finite size effects. Biophys J 94:3565–3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Efron B, Tibshirani RJ (1998) An introduction to the bootstrap. New York: Chapman & Hall. [Google Scholar]
- 49. Wang L, Murphy‐Ullrich JE, Song Y (2014) Molecular insight for the effect of lipid bilayer environments on thrombospondin‐1 and calreticulin interactions. Biochemistry 53:6309–6322. [DOI] [PubMed] [Google Scholar]
- 50. Wang L, Homes RP, Peng JB (2016) Molecular modeling of the structural and dynamical changes in calcium channel TRPV5 induced by the African‐specific A563T variation lin interactions. Biochemistry 55:1254–1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Bailey NTJ (1995) Statistical methods in biology, 3rd edition ed. New York: Cambridge University Press. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


