Abstract
Obtaining well‐ordered crystals remains a significant challenge in protein X‐ray crystallography. Carrier‐driven crystallization can facilitate crystal formation and structure solution of difficult target proteins. We obtained crystals of the small and highly flexible SPX domain from the yeast vacuolar transporter chaperone 4 (Vtc4) when fused to a C‐terminal, non‐cleavable macro tag derived from human histone macroH2A1.1. Initial crystals diffracted to 3.3 Å resolution. Reductive protein methylation of the fusion protein yielded a new crystal form diffracting to 2.1 Å. The structures were solved by molecular replacement, using isolated macro domain structures as search models. Our findings suggest that macro domain tags can be employed in recombinant protein expression in E. coli, and in carrier‐driven crystallization.
Keywords: carrier‐driven crystallization, crystallization tag, macro domain, histone macroH2A, recombinant protein expression
Short abstract
PDB Code(s): 5LNC
Abbreviations
- GST
glutathione‐S‐transferase
- MBP
maltose binding protein
- SEC
size‐exclusion chromatography
- TTM
triphosphate tunnel metalloenzyme
- VTC
vacuolar transporter chaperone
Introduction
Carrier‐driven crystallization,1, 2 or chaperone‐assisted crystallization,3 describes the crystallization of a target protein by fusing it to a well‐behaving protein tag, which may contribute to the formation of the crystal lattice. A small selection of well‐characterized proteins has thus far been used as crystallization tags: Short fragments of fibrinogen and α‐actin have been crystallized by fusing them to E. coli glutathione‐S‐transferase (GST)4 or the catalytic domain of myosin II,5 respectively. The β2 adrenergic G protein‐coupled receptor crystallized after inserting T4 lysozyme into a flexible loop region.6 Several structures obtained by carrier‐driven crystallization contain E. coli maltose binding protein (MBP) as a fusion tag3: the ectodomain of the human T cell leukemia virus type 1 gp21,7 the yeast mating regulator MATa1 homeodomain,8 segments of the amyloid‐forming α‐synuclein9 and the fungal Kar3 kinesin motor domain.10 Corsini et al. demonstrated the importance of using small rigid linkers between the target protein and the fusion tag, when crystallizing the U2AF homology motif of splicing factor Pfu60 fused to E. coli thioredoxin A.11
Given that there are high resolution structures available for most of the fusion proteins used thus far, the fusion tag, besides assisting in the formation of diffracting crystals, can facilitate structure solution by molecular replacement. In addition, an increased stability of the fusion protein is frequently observed.12, 13
We have recently reported that SPX domains of previously unknown biochemical function are sensors for inositol pyrophosphate signaling molecules, controlling phosphate (P i) homeostasis in fungi, plants and animals.14 The name SPX originates from the yeast SYG1 and Pho81 and the mammalian XPR1 proteins, all of which contain SPX domains (https://www.ebi.ac.uk/interpro/entry/IPR004331). Comparing different eukaryotic organisms, we located this small, α‐helical domain at the N‐termini of proteins involved in P i transport15, 16 and P i signaling,17, 18 and as a single‐domain protein in plants.14, 19, 20
Previously, we mapped three sets of domains in the vacuolar transporter chaperone (VTC) complex, a multi‐subunit protein assembly embedded in the vacuolar membrane in yeast cells.21, 22 The central catalytic triphosphate tunnel metalloenzyme (TTM) domain synthesizes inorganic polyphosphate chains from ATP22, 23 and a trans‐membrane pore translocates the growing polymer into the vacuole where inorganic polyphosphate represents an important P i store.22, 24 VTC subunits Vtc2, 3 and 4 contain additional ∼180 amino‐acid SPX domains at their N‐termini. We could solubly express and purify S. cerevisiae SPXScVtc 2 (residues 1‐182) and SPXScVtc 4 (residues 1‐178) as well as other SPX domains from various fungal and plant species, but could neither obtain crystals from the purified proteins nor readily interpretable NMR spectra.14 In addition, carrier driven crystallization using the established thioredoxin A11 and maltose‐binding protein3 tags were unsuccessful.
Results
SPX domains can be crystallized as macro domain fusion proteins
The VTC SPX domains contain highly conserved N‐termini and are connected to the catalytic TTM domain by a short linker22 (Fig. 1). We attempted to crystallize SPXScVtc 2 and SPXScVtc 4 fused to their respective catalytic TTM domains (residues 1‐553 and 1‐480 in ScVtc2 and ScVtc4, respectively). We obtained crystals for SPXScVtc4‐TTM‐6xHis, diffracting to 4.5 Å resolution and showing clear signs of perfect merohedral twinning. We replaced the TTM domain by thioredoxin A,11 but this construct again failed to yield well‐diffracting crystals [Fig. 2(A)]. Next, we engineered SPXScVtc 4 (residues 1‐178) with a C‐terminal macro domain tag (residues 181‐366), connected via a short Ala‐Gly‐Ser linker to decrease inter‐domain flexibility11 [Fig. 2(B); see Methods]. We chose the macro domain of human histone variant macroH2A1.1 as fusion tag as it (1) expresses to high levels in E. coli, (2) crystallizes in many and very different crystal lattices25, 26, 27 (PDB‐IDs 1YD9, 1ZR3, 1ZR5, 2FXK, 3IID, 3IIF), and (3) has solvent accessible N‐ and C‐termini that allow for the addition of a fusion protein. Importantly, macro domains bind ADP‐ribose, and ligand binding results in small conformational changes that alter the crystallization properties of the domain.26, 27 Our construct provides a C‐terminal, non‐cleavable 6xHis tag for metal affinity purification.
Figure 1.
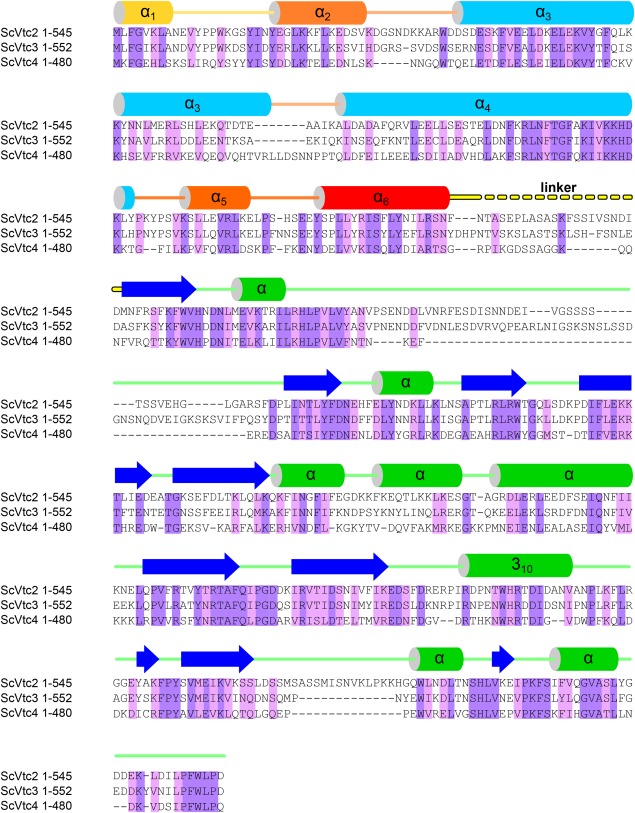
The VTC complex harbors N‐terminal SPX domains. Structure based sequence alignment of VTC subunits ScVtc2 (S. cerevisiae Vtc2; UniProt Acc. P43585), ScVtc3 (S. cerevisiae Vtc3; Q02725) and ScVtc4 (S. cerevisiae Vtc4; P47075) and including a secondary structure assignment calculated with the program DSSP48 and based on PDB entry 5IIG.14 Invariant and conserved residues are highlighted in dark‐ and light‐purple, respectively. SPXScVtc 2 shows 62% and 32% sequence identity with SPXScVtc 3 and SPXScVtc 4, respectively. The two long core helices of the SPX domain are colored in light blue, surrounding helices are colored from yellow to red. The C‐terminal catalytic TTM domain (α‐helices in green, β‐strands in blue) is connected to the SPX domain via a variable linker (in yellow).
Figure 2.
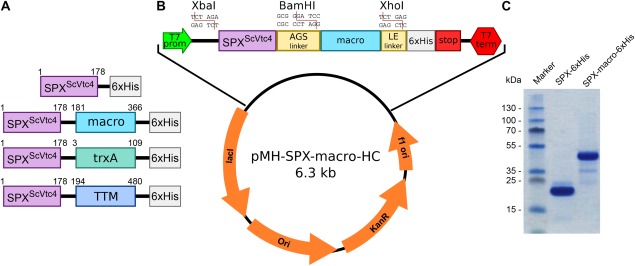
A stable SPXScVtc4‐macro fusion protein can be recombinantely expressed in E. coli. (A) Four SPXScVtc 4 constructs were assayed for crystallization, all of which contained a C‐terminal 6xHis tag and either no fusion protein, a macro tag (macro domain of the human histone macroH2A1.126, 27), trxA (E. coli thioredoxinA11) or the catalytic TTM domain22, 23 of ScVtc4. (B) Overview of the constructed macro domain expression plasmid providing a macro tag (human macroH2A1.1 residues 181‐366) connected to the N‐terminal SPX domain via a Ala‐Gly‐Ser linker and followed by a non‐cleavable 6xHis tag. Target proteins can be inserted using the restriction enzymes XbaI/BamHI. (C) SDS‐PAGE analysis of purified SPXScVtc4−6xHis and SPXScVtc4‐macro‐6xHis fusion proteins expressed in E. coli.
The SPXScVtc4‐6xHis and SPXScVtc4‐macro‐6xHis fusion proteins expressed to similar levels in E. coli and both could be purified to homogeneity, while N‐terminal fusion constructs were insoluble14 [Fig. 2(C)]. The fusion protein displayed increased protein stability and yielded needle shaped crystals in more than 25 different crystallization conditions, after screening 10 different 96‐well grid screens using the sitting drop vapor diffusion method. In parallel, screening of a reductively methylated SPXScVtc4‐macro fusion protein28 produced crystals of a different morphology.
The macro domain can be utilized to solve the phase problem
The non‐methylated SPXScVtc4‐macro fusion protein yielded orthorhombic crystals diffracting to 3.3 Å resolution. Structural superposition of the existing macroH2A1.1 models suggests that the N‐ (residues 180‐185) and C‐termini (residues 354‐368) of the macro domain can adopt different orientations [core r.m.s.d.'s are between 0.6 Å and 1.0 Å comparing 169 corresponding Cα atoms; Fig. 3(A)]. We thus used all available macroH2A1.1 structures as search models in molecular replacement searches as implemented in the program PHASER.29 The best solution comprises two macro domains derived from PDB entry 1YD925 in the asymmetric unit, which are related by a pseudo two‐fold axis [Fig. 3(B,C)]. We refined this crystallographic dimer, which accounts for ∼50% of protein atoms in the asymmetric unit in the program autoBUSTER (see Methods) and used the resulting phase information as starting phases for density modification (solvent content is ∼0.65). The resulting density modified map at 3.3 Å revealed the presence of long segments of electron density, which could be modeled as two long α‐helices forming the core of the SPX domain [Fig. 3(C)]. Towards the end of refinement, an α‐helical hairpin motif and two C‐terminal helical segments could be modeled, which complete the SPX domain fold [Fig. 3(C), Table 1].14
Figure 3.
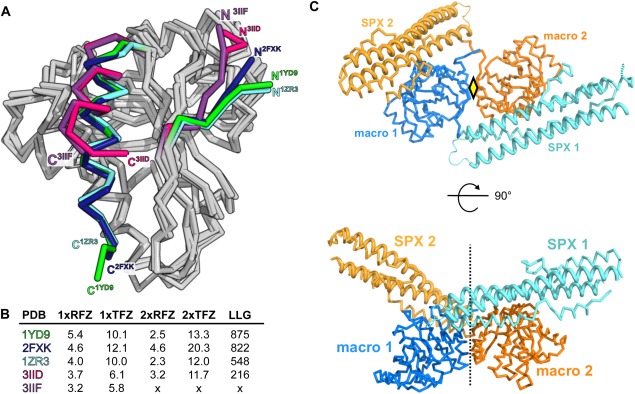
The known macro domain structures allow for structure solution of SPXScVtc 4 by molecular replacement. (A) Structural superposition of different, previously described macro H2A1.1 macro domain structures indicates that N‐ and C‐termini are flexible. R.m.s.d. is between 0.7‐2.6 Å comparing 180 corresponding Cα atoms. (B) Table summary of molecular replacement calculations using the different known macroH2A1.1 structures. 1xRFZ: rotation function Z‐score for the 1st molecule to be placed. 1xTFZ: translation function Z‐score for the 1st molecule to be placed. 2xRFZ: rotation function Z‐score for the 2nd molecule to be placed. 2xTFZ: translation function Z‐score for the 2nd molecule. LLG: Log Likelihood Gain of the refined solution. (C) Cα traces of two SPXScVtc4‐macro molecules (SPX domain are shown in light‐blue and yellow, the C‐terminal macro domains in dark‐blue and orange, respectively) related by a pseudo 2‐fold axis in the asymmetric unit of the I212121 crystal form. The central core helices of the SPX domains are highlighted (ribbon diagrams).
Table 1.
Crystallographic Data Collection and Refinement for Non‐ and Reductively Methylated SPXScVtc 4‐Macro Fusion Proteins
| SPXScVtc4‐macro (non‐methylated) | SPXScVtc 4‐macro (reproduced from 14) (red. methylated) | |
|---|---|---|
| Data collection | ||
| Beam‐line | SLS PXII | ESRF ID29 |
| Wavelength (Å) | 0.97963 | 0.976251 |
| Space‐group | I 21 21 21 | P 21 |
| Resolution (Å) | 48.90–3.29 (3.38–3.29) | 46.86–2.13 (2.26–2.13) |
| Unit cell dimensions, a, b, c (Å) | 114.02, 130.05, 158.33 | 105.64, 67.92, 129.68 |
| Unit cell dimensions, α,β,γ (°) | 90, 90, 90 | 90, 93.32, 90 |
| Molecules per AU | 2 | 4 |
| No. reflections: total | 126,657 (5,814) | 688,776 (103,122) |
| No. reflections: unique | 17,957 (1,103) | 101,489 (15,910) |
| Completeness (%) | 98.4 (82.3) | 99.1 (96.7) |
| Multiplicity | 7.1 (5.3) | 6.8 (6.5) |
| I/σIa | 15.4 (0.6) | 15.5 (1.9) |
| CC(1/2)a (%) | 100 (25.3) | 99.0 (80.9) |
| Rmeas a (%) | 7.2 (289.1) | 8.4 (111.6) |
| Wilson B‐factor (Å2) | 193.0 | 51.0 |
| Refinement | ||
| No. atoms/No. atoms test set | 17,957/898 | 101,454/5,074 |
| Resolution (Å) | 48.90–3.29 | 46.86–2.13 |
| Rcryst/Rfree b (%) | 27.9/30.0 | 21.1/24.7 |
| R.m.s. deviations: bond distancesb (Å) | 0.01 | 0.004 |
| R.m.s. deviations: bond anglesb (°) | 1.1 | 0.75 |
| Structure/Stereochemistry | ||
| No. atoms: protein | 4,673 | 11,352 |
| No atoms: water | 183 | |
| Average B‐factors: protein (Å2) | 179.5 | 58.4 |
| Average B‐factors: water (Å2) | 52.2 | |
| Ramachandran plot: most favored regionsc (%) | 95.71 | 98.95 |
| Ramachandran plot. outliers (%) | 1.1 | 0 |
| MolProbity scorec | 1.30 | 1.00 |
| Protein Data Bank ID | 5LNC | 5IIT |
Monoclinic crystals of the reductively methylated SPXScVtc4‐macro fusion protein diffracted to 2.1 Å (PDB‐ID 5IIT,14 Table 1). The structure was solved using the macro domain PDB entry 1ZR326 and the two SPX core helices from the low resolution SPXScVtc4‐macro structure as search models in molecular replacement calculations with PHASER,29 as described.14 There are four molecules in the asymmetric unit, with the SPX and macro domains making extensive interactions [Fig. 4(A)]. While the two long core helices of the non‐methylated and methylated SPXScVtc 4 domain structures closely align (r.m.s.d. is ∼1.0 Å comparing 71 corresponding Cα atoms), their N‐ (residues 2‐22) and C‐termini (154‐177) adopt different orientations in our structures: The very C‐terminal helix in SPX does not engage in the formation of a three helix bundle [Fig. 4(B)], but is pointed outwards [shown in red in Fig. 4(A,C)], establishing contacts with a neighboring molecule [Fig. 4(A)]. Remarkably, we found the N‐terminal α‐helical hairpin motif initially located in our 3.3 Å map, to be disordered in the 2.1 Å structure [Fig. 4(B,C)]. Importantly, the Ala‐Gly‐Ser linker is well‐defined by electron density in the 2.1 Å structure [Fig. 4(C)]. We used this high‐resolution SPXScVtc4‐macro model to confirm the topology of the SPX domain and the directionality of α‐helices in our low resolution maps. The refined models revealed that SPX domains fold into three‐helix bundles with two central ∼ 80 Å long core helices and two shorter C‐terminal helices, connected by short loops. The N‐terminus appears flexible but can fold into an α‐helical hairpin motif, which provides a binding site for inositol polyphosphate signaling molecules14 [Fig. 4(B,C)].
Figure 4.
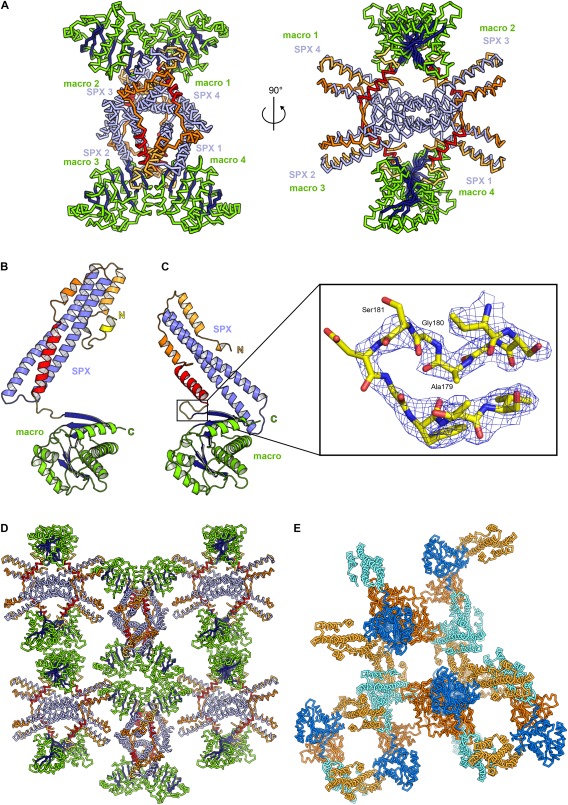
Reductively and non‐methylated SPXScVtc4‐macro protein adopts different conformations. (A) Cα trace of the SPXScVtc4‐macro tetramer in the asymmetric unit of the P21 crystal form. The C‐terminal helix of the SPX domain is highlighted in red. The C‐terminal helix of human SPXHsXPR1 has a similar configuration, again contacting a symmetry‐related molecule (PDB‐ID 5IJH14). Comparison of (B) non‐methylated and (C) reductively methylated SPXScVtc4‐macro fusion proteins (in ribbon representation). The zoom‐in shows the connecting Ala‐Gly‐Ser linker in bonds representation (colored in yellow) and including a 2Fo‐Fc omit electron density map contoured at 1.0 σ (blue mesh). (D) Reductively methylated and (E) non‐methylated SPXScVtc4‐macro fusion proteins form different crystal lattices, involving SPX–SPX, macro–macro and SPX–macro interactions. Colors are as in (A) and Figure 3(C).
Crystal packing involves SPX and macro domain protein–protein interactions
We next analyzed the arrangement of the SPXScVtc 4 and macroH2A1.1 domains in our two crystal forms (solvent content is ∼0.65 and ∼0.55 for the non‐methylated and methylated forms, respectively): Crystal lattice formation in our low‐resolution structure of non‐methylated SPXScVtc4‐macro is achieved by SPX–SPX (interface area: ∼1050 Å2), by macro–macro (interface area: ∼870 Å2) and by SPX–macro domain interactions (interface area ∼770 Å2), as calculated with the program PISA30 [Fig. 4(D)]. Similar results were obtained for the crystal form of the reductively methylated SPXScVtc4‐macro fusion protein, although very different crystal contacts are established in the two crystal forms [Fig. 4(D,E)]. In both cases, SPX–SPX interactions are not sufficient to build‐up a three‐dimensional crystal lattice [Fig. 4(D,E)] suggesting that the additional interactions formed by the macro domain allowed for the crystallization of the fusion protein.
Discussion
Our experiments suggest that the macro domains are suitable tags for recombinant protein expression and carrier‐driven crystallization. Based on our previous work on the histone variant macroH2A1.1,26, 27 we employed this human macro domain as fusion tag as it (1) stably expresses to high levels in E. coli, (2) can be easily purified, (3) is very stable in minimal buffers, (4) crystallizes in many different lattice combinations25, 26, 27 (PDB‐IDs 1YD9, 1ZR3, 1ZR5, 2FXK, 3IID, 3IIF), and (5) provides solvent accessible N‐ and C‐termini for the design of both N‐ and C‐terminal (this study) protein fusions. It is of note that macro domains bind ADP‐ribose with nano‐ to micromolar affinity.26, 31 Thus, ADP‐ribose could be used as an additive in carrier‐driven crystallization. In our previous co‐crystallization experiments with the isolated macroH2A1.1 macro domain, ADP‐ribose induced the formation of new crystal forms (compared to the existing ‘apo’ structures25, 26), as ligand‐binding induces a structural re‐orientation of the C‐terminal helix.26, 27 Notably, the macro ADP‐ribose binding pocket can accommodate a MES buffer molecule (e.g., PDB‐ID 1ZR326), and we could only obtain crystals for the SPXScVtc4‐macro fusion when using MES‐based protein storage buffers. In addition to ADP‐ribose, macro domains can also bind ADP, albeit with lower affinity.26, 31 It might thus be possible to soak or co‐crystallize macro fusion proteins with halogenated nucleotide variants, in order to introduce heavy atoms at defined positions in macro fusion protein crystal lattices for experimental phasing.
Our work has focused on the use of the macroH2A1.1 macro domain, but many alternative macro domain structures have been reported from human (PDB‐IDs 2X47,32 2L8R,33 4IQY,34 3VFQ,35 4J5S36), trypanosomal (PDB‐ID 5FSY37), archaeal (PDB‐IDs 1HJZ,38 2BFQ31), bacterial (5CB3,39 5KIV40) and viral (PDB‐IDs 3EJF,41 3EWP,42 3GPO,43 5DUS44) proteins. Given that all these different macro domains produced well‐diffracting crystals (n = 61 were refined at a resolution between 1.5 and 2.5 Å), one interesting approach to carrier‐driven crystallization would be to screen target proteins in fusion with different macro domains and in the pre‐ or absence of ADP‐ribose. In addition, it may be worthwhile to test linkers of different length and composition between the protein of interest and the macro fusion tag.
Linker length is an important factor in carrier‐driven crystallization as it determines the molecular dynamics between the target protein and the fusion tag. While too short linkers may cause expression and folding problems, long linkers may hinder crystal formation.11 In the present study a short, Ala‐Gly‐Ser linker was used, which allowed for high‐level expression of the SPXScVtc4‐macro fusion protein and which has well‐defined electron density in our 2.1 Å structure [Figs. 2(C), 4(C)].
Taken together, crystallization of SPXScVtc 4 as a macro fusion protein allowed us to define the SPX domain fold and borders, which later on enabled the successful crystallization and structure solution of other fungal and human SPX domains.14 Importantly, two crystal structures of ScVtc41‐480, containing both the SPX and TTM domains [Figs. 1, 22(A)], revealed that addition of a macro tag does not induce large structural changes in SPXScVtc 4 (r.m.s.d. is ∼1.5 and 2.2 Å comparing 179 corresponding Cα atoms, vs. PDB‐IDs 5IIG, 5IIQ, respectively14). Based on our findings, we suggest that macro domains could be interesting fusion tags for recombinant protein expression and for the carrier‐driven crystallization of ‘difficult' target proteins.
Materials and Methods
Cloning and purification of proteins
The S. cerevisiae SPXScVtc 4 (amino acids 1‐178) was cloned into plasmid pMH‐HC using BspHI and XhoI restrictions sites. The plasmid provided a C‐terminal non‐cleavable 6xHis tag. For carrier‐driven crystallization, a new plasmid was constructed which provides a C‐terminal macro tag (amino‐acids 181‐366 of human histone macroH2A1.1) followed by 6xHis tag [pMH‐macroHC, Fig. 2(B)]. SPXScVtc 4 and the macro domain are connected by an Ala‐Gly‐Ser linker. Target proteins can be cloned utilizing the XbaI/BamHI restriction sites.
For recombinant protein expression, plasmids were transformed into E. coli BL21 (DE3) RIL cells and selected on LB‐Agar plates containing kanamycin and chloramphenicol. 3 l terrific broth medium containing 30 mg/mL kanamycin and 34 mg/mL chloramphenicol were inoculated and cells grown to OD600nm = 0.6 at 37°C. Then, the temperature was reduced to 16°C and protein expression was induced by adding 300 μM Isopropyl‐β‐D‐thiogalactopyranoside. After 16 hours, cells were harvested by centrifugation for 20 min at 4,000g at 4°C. The pellet was washed with PBS buffer, resuspended in a small volume of lysis buffer (50 mM Tris/HCl pH 7.8, 500 mM NaCl, 2 mM β‐mercaptoethanol [β‐ME]) and snap‐frozen in liquid nitrogen.
For protein purification, pellets from 3 l of bacterial culture were thawed and lysis buffer, supplemented with 0.1% (v/v) IGEPAL, 1 mM MgCl2, 500 units TurboNuclease (BioVision) and 2 tablets Protease Inhibitor Cocktail (Roche), was added to a final volume of 300 mL. Cells were lysed using an EmulsiFlex‐C3 (Avestin) and the cell debris was removed by centrifugation for 1 hour at 7,000g at 4°C. Proteins were purified by Ni2+ affinity chromatography using a 5 mL HisTrapHP column (GE Healthcare). After dialysing proteins against size‐exclusion chromatography (SEC) buffer (20 mM Mes pH 6.5, 300 mM NaCl, 0.5 mM TCEP) overnight at 4°C, monomeric SPXScVtc 4 or SPXScVtc4‐macro peak fractions were isolated on a Superdex 75 HR 26/60 column (GE healthcare). Reductive methylation of SPXScVtc4‐macro was carried out as described previously.28 Methylated protein sample was re‐purified by an additional SEC step. Purified proteins were concentrated and immediately used for crystallization.
Crystallization and data collection
SPXScVtc4‐macro (16 mg/mL in 20 mM MES pH6.5, 300 mM NaCl, 0.5 mM TCEP) crystals were grown in 2.75M NaCl, 8.75% (v/v) PEG 6,000. Crystals were cryo‐protected by serial transfer through 10 μL drops containing crystallization buffer supplemented with increasing concentrations of ethylene glycol (final concentration: 8% [v/v]), and diffracted to ∼3.3 Å at beam‐line PXII of the Swiss Light Source (SLS), Villigen, Switzerland. The reductively methylated SPXScVtc4‐macro protein (10 mg/mL in 20 mM MES pH 6.5, 300 mM, 0.5 mM TCEP) crystallized in 19% (v/v) PEG 3,350, 0.1M (NH4)2SO4, 0.1M MES pH 6.5 as described.14 Data processing and scaling was done with XDS (version: May, 2016).45
Crystallographic structure solution and refinement
The structure of the non‐methylated SPXScVtc4‐macro domain was solved using the molecular replacement method as implemented in the program PHASER,29 and using the isolated macro domain of human histone macroH2A1.1 (PDB‐ID 1YD925) as search model. The solution comprised a dimer in the asymmetric unit, which was refined in autoBUSTER (Global Phasing Limited, version 2.10.3). The resulting phases were used as starting phases for density modification as implemented in PHENIX.RESOLVE.46 The structure was completed in alternating cycles of manual model building in COOT25 and restrained TLS refinement in autoBUSTER (Global Phasing Limited), using external reference restraints based on the high‐resolution structures of the macroH2A1.1 macro (PDB‐ID 1ZR326) and SPXScVtc 4 domains (PDB‐ID 5IIG14), respectively. The side‐chains of most amino acids in the SPX domain could not be modeled with certainty and were thus truncated to Ala. The quality of the refined structure was validated using the program MolProbity47 (Table 1) and structural presentations were prepared in PyMOL (Molecular Graphics System, Version 1.7 Schrödinger, LLC).
Acknowledgments
Atomic coordinates and structure factors have been deposited with the Protein Data Bank with accession number 5LNC. The pMH‐macroHC plasmid is available from Addgene (ID 82337). Diffraction experiments were performed at beam lines PXII of the Swiss Light Source, Paul Scherrer Institute, Villigen, Switzerland.
Statement of Significance: A novel fusion tag for recombinant protein expression and carrier driven crystallization is presented.
References
- 1. Donahue JP, Patel H, Anderson WF, Hawiger J (1994) Three‐dimensional structure of the platelet integrin recognition segment of the fibrinogen gamma chain obtained by carrier protein‐driven crystallization. Proc Natl Acad Sci USA 91:12178–12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lim K, Ho JX, Keeling K, Gilliland GL, Ji X, Rüker F, Carter DC (1994) Three‐dimensional structure of Schistosoma japonicum glutathione S‐transferase fused with a six‐amino acid conserved neutralizing epitope of gp41 from HIV. Protein Sci 3:2233–2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Waugh DS (2016) Crystal structures of MBP fusion proteins. Protein Sci 25:559–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ware S, Donahue JP, Hawiger J, Anderson WF (1999) Structure of the fibrinogen gamma‐chain integrin binding and factor XIIIa cross‐linking sites obtained through carrier protein driven crystallization. Protein Sci 8:2663–2671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kliche W, Fujita‐Becker S, Kollmar M, Manstein DJ, Kull FJ (2001) Structure of a genetically engineered molecular motor. EMBO J 20:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rosenbaum DM, Cherezov V, Hanson MA, Rasmussen SGF, Thian FS, Kobilka TS, Choi H‐J, Yao X‐J, Weis WI, Stevens RC, Kobilka BK (2007) GPCR engineering yields high‐resolution structural insights into beta2‐adrenergic receptor function. Science 318:1266–1273. [DOI] [PubMed] [Google Scholar]
- 7. Kobe B, Center RJ, Kemp BE, Poumbourios P (1999) Crystal structure of human T cell leukemia virus type 1 gp21 ectodomain crystallized as a maltose‐binding protein chimera reveals structural evolution of retroviral transmembrane proteins. Proc Natl Acad Sci USA 96:4319–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Ke A, Wolberger C (2003) Insights into binding cooperativity of MATa1/MATα2 from the crystal structure of a MATa1 homeodomain‐maltose binding protein chimera. Protein Sci 12:306–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhao M, Cascio D, Sawaya MR, Eisenberg D (2011) Structures of segments of α‐synuclein fused to maltose‐binding protein suggest intermediate states during amyloid formation. Protein Sci 20:996–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Delorme C, Joshi M, Allingham JS (2012) Crystal structure of the Candida albicans Kar3 kinesin motor domain fused to maltose‐binding protein. Biochem Biophys Res Commun 428:427–432. [DOI] [PubMed] [Google Scholar]
- 11. Corsini L, Hothorn M, Scheffzek K, Sattler M, Stier G (2008) Thioredoxin as a fusion tag for carrier‐driven crystallization. Protein Sci 17:2070–2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Zhou P, Lugovskoy AA, Wagner G (2001) A solubility‐enhancement tag (SET) for NMR studies of poorly behaving proteins. J Biomol NMR 20:11–14. [DOI] [PubMed] [Google Scholar]
- 13. Hiller S, Kohl A, Fiorito F, Herrmann T, Wider G, Tschopp J, Grütter MG, Wüthrich K (2003) NMR structure of the apoptosis‐ and inflammation‐related NALP1 pyrin domain. Structure 11:1199–1205. [DOI] [PubMed] [Google Scholar]
- 14. Wild R, Gerasimaite R, Jung J‐Y, Truffault V, Pavlovic I, Schmidt A, Saiardi A, Jessen HJ, Poirier Y, Hothorn M, Mayer A (2016) Control of eukaryotic phosphate homeostasis by inositol polyphosphate sensor domains. Science 352:986–990. [DOI] [PubMed] [Google Scholar]
- 15. Hürlimann HC, Pinson B, Stadler‐Waibel M, Zeeman SC, Freimoser FM (2009) The SPX domain of the yeast low‐affinity phosphate transporter Pho90 regulates transport activity. EMBO Rep 10:1003–1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Giovannini D, Touhami J, Charnet P, Sitbon M, Battini J‐L (2013) Inorganic phosphate export by the retrovirus receptor XPR1 in metazoans. Cell Rep 3:1866–1873. [DOI] [PubMed] [Google Scholar]
- 17. Ogawa N, DeRisi J, Brown PO (2000) New components of a system for phosphate accumulation and polyphosphate metabolism in Saccharomyces cerevisiae revealed by genomic expression analysis. Mol Biol Cell 11:4309–4321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park BS, Seo JS, Chua N‐H (2014) Nitrogen limitation adaptation recruits phosphate2 to target the phosphate transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26:454–464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duan K, Yi K, Dang L, Huang H, Wu W, Wu P (2008) Characterization of a sub‐family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J 54:965–975. [DOI] [PubMed] [Google Scholar]
- 20. Secco D, Wang C, Arpat BA, Wang Z, Poirier Y, Tyerman SD, Wu P, Shou H, Whelan J (2012) The emerging importance of the SPX domain‐containing proteins in phosphate homeostasis. New Phytol 193:842–851. [DOI] [PubMed] [Google Scholar]
- 21. Müller O, Bayer MJ, Peters C, Andersen JS, Mann M, Mayer A (2002) The Vtc proteins in vacuole fusion: coupling NSF activity to V0 trans‐complex formation. embo J 21:259–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hothorn M, Neumann H, Lenherr ED, Wehner M, Rybin V, Hassa PO, Uttenweiler A, Reinhardt M, Schmidt A, Seiler J, Ladurner AG, Herrmann C, Scheffzek K, Mayer A (2009) Catalytic core of a membrane‐associated eukaryotic polyphosphate polymerase. Science 324:513–516. [DOI] [PubMed] [Google Scholar]
- 23. Martinez J, Truffault V, Hothorn M (2015) Structural determinants for substrate binding and catalysis in triphosphate tunnel metalloenzymes. J Biol Chem 290:23348–23360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Gerasimaitė R, Sharma S, Desfougères Y, Schmidt A, Mayer A (2014) Coupled synthesis and translocation restrains polyphosphate to acidocalcisome‐like vacuoles and prevents its toxicity. J Cell Sci 127:5093–5104. [DOI] [PubMed] [Google Scholar]
- 25. Chakravarthy S, Gundimella SKY, Caron C, Perche P‐Y, Pehrson JR, Khochbin S, Luger K (2005) Structural characterization of the histone variant macroH2A. Mol Cell Biol 25:7616–7624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kustatscher G, Hothorn M, Pugieux C, Scheffzek K, Ladurner AG (2005) Splicing regulates NAD metabolite binding to histone macroH2A. Nat Struct Mol Biol 12:624–625. [DOI] [PubMed] [Google Scholar]
- 27. Timinszky G, Till S, Hassa PO, Hothorn M, Kustatscher G, Nijmeijer B, Colombelli J, Altmeyer M, Stelzer EHK, Scheffzek K, Hottiger MO, Ladurner AG (2009) A macrodomain‐containing histone rearranges chromatin upon sensing PARP1 activation. Nat Struct Mol Biol 16:923–929. [DOI] [PubMed] [Google Scholar]
- 28. Shaw N, Cheng C, Liu Z‐J (2007) Procedure for reductive methylation of protein to improve crystallizability. Protocol Exchange. Available at: http://dx.doi.org/10.1038/nprot.2007.287. [Google Scholar]
- 29. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Krissinel E, Henrick K (2007) Inference of macromolecular assemblies from crystalline state. J Mol Biol 372:774–797. [DOI] [PubMed] [Google Scholar]
- 31. Karras GI, Kustatscher G, Buhecha HR, Allen MD, Pugieux C, Sait F, Bycroft M, Ladurner AG (2005) The macro domain is an ADP‐ribose binding module. embo J 24:1911–1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen D, Vollmar M, Rossi MN, Phillips C, Kraehenbuehl R, Slade D, Mehrotra PV, von Delft F, Crosthwaite SK, Gileadi O, Denu JM, Ahel I (2011) Identification of macrodomain proteins as novel O‐acetyl‐ADP‐ribose deacetylases. J Biol Chem 286:13261–13271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Peterson FC, Chen D, Lytle BL, Rossi MN, Ahel I, Denu JM, Volkman BF (2011) Orphan macrodomain protein (human C6orf130) is an O‐acyl‐ADP‐ribose deacylase: solution structure and catalytic properties. J Biol Chem 286:35955–35965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG (2013) A family of macrodomain proteins reverses cellular mono‐ADP‐ribosylation. Nat Struct Mol Biol 20:508–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forst AH, Karlberg T, Herzog N, Thorsell A‐G, Gross A, Feijs KLH, Verheugd P, Kursula P, Nijmeijer B, Kremmer E, Kleine H, Ladurner AG, Schüler H, Lüscher B (2013) Recognition of mono‐ADP‐ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 21:462–475. [DOI] [PubMed] [Google Scholar]
- 36. Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I (2013) Deficiency of terminal ADP‐ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. embo J 32:1225–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Haikarainen T, Lehtiö L (2016) Proximal ADP‐ribose hydrolysis in trypanosomatids is catalyzed by a macrodomain. Sci Rep 6:24213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Allen MD, Buckle AM, Cordell SC, Löwe J, Bycroft M (2003) The crystal structure of AF1521 a protein from Archaeoglobus fulgidus with homology to the non‐histone domain of macroH2A. J Mol Biol 330:503–511. [DOI] [PubMed] [Google Scholar]
- 39. Zhang W, Wang C, Song Y, Shao C, Zhang X, Zang J (2015) Structural insights into the mechanism of Escherichia coli YmdB: a 2′‐O‐acetyl‐ADP‐ribose deacetylase. J Struct Biol 192:478–486. [DOI] [PubMed] [Google Scholar]
- 40. Appel CD, Feld GK, Wallace BD, Williams RS (2016) Structure of the sirtuin‐linked macrodomain SAV0325 from Staphylococcus aureus. Protein Sci doi: 10.1002/pro.2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Piotrowski Y, Hansen G, Boomaars‐van der Zanden AL, Snijder EJ, Gorbalenya AE, Hilgenfeld R (2009) Crystal structures of the X‐domains of a Group‐1 and a Group‐3 coronavirus reveal that ADP‐ribose‐binding may not be a conserved property. Protein Sci 18:6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Xu Y, Cong L, Chen C, Wei L, Zhao Q, Xu X, Ma Y, Bartlam M, Rao Z (2009) Crystal structures of two coronavirus ADP‐ribose‐1″‐monophosphatases and their complexes with ADP‐Ribose: a systematic structural analysis of the viral ADRP domain. J Virol 83:1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Malet H, Coutard B, Jamal S, Dutartre H, Papageorgiou N, Neuvonen M, Ahola T, Forrester N, Gould EA, Lafitte D, Ferron F, Lescar J, Gorbalenya AE, de Lamballerie C, Canard B (2009) The crystal structures of Chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol 83:6534–6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Cho C‐C, Lin M‐H, Chuang C‐Y, Hsu C‐H (2016) Macro domain from Middle East Respiratory Syndrome Coronavirus (MERS‐CoV) is an efficient ADP‐ribose binding module: crystal structure and biochemical studies. J Biol Chem 291:4894–4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kabsch W (1993) Automatic processing of rotation diffraction data from crystals of initially unknown symmetry and cell constants. J Appl Cryst 26:795–800. [Google Scholar]
- 46. Terwilliger TC (2003) SOLVE and RESOLVE: automated structure solution and density modification. Methods Enzymol 374:22–37. [DOI] [PubMed] [Google Scholar]
- 47. Davis IW, Leaver‐Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, Snoeyink J, Richardson JS, Richardson DC (2007) MolProbity: all‐atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res 35:W375–W383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Holm L, Sander C (1993) Protein structure comparison by alignment of distance matrices. J Mol Biol 233:123–138. [DOI] [PubMed] [Google Scholar]


