Abstract
The spliceosomal protein SF3b49, a component of the splicing factor 3b (SF3b) protein complex in the U2 small nuclear ribonucleoprotein, contains two RNA recognition motif (RRM) domains. In yeast, the first RRM domain (RRM1) of Hsh49 protein (yeast orthologue of human SF3b49) reportedly interacts with another component, Cus1 protein (orthologue of human SF3b145). Here, we solved the solution structure of the RRM1 of human SF3b49 and examined its mode of interaction with a fragment of human SF3b145 using NMR methods. Chemical shift mapping showed that the SF3b145 fragment spanning residues 598–631 interacts with SF3b49 RRM1, which adopts a canonical RRM fold with a topology of β1‐α1‐β2‐β3‐α2‐β4. Furthermore, a docking model based on NOESY measurements suggests that residues 607–616 of the SF3b145 fragment adopt a helical structure that binds to RRM1 predominantly via α1, consequently exhibiting a helix–helix interaction in almost antiparallel. This mode of interaction was confirmed by a mutational analysis using GST pull‐down assays. Comparison with structures of all RRM domains when complexed with a peptide found that this helix–helix interaction is unique to SF3b49 RRM1. Additionally, all amino acid residues involved in the interaction are well conserved among eukaryotes, suggesting evolutionary conservation of this interaction mode between SF3b49 RRM1 and SF3b145.
Keywords: nuclear magnetic resonance, RNA recognition motif, SF3b49, SF3b145, U2 snRNP
Short abstract
PDB Code(s): 5GVQ
Abbreviations
- HSQC
heteronuclear single quantum coherence spectroscopy
- RRM
RNA recognition motif
- SF3b
splicing factor 3b
- snRNP
small nuclear ribonucleoprotein
- TALOS
torsion angle likelihood obtained from shift and sequence similarity
Introduction
Pre‐mRNA splicing occurs in the nucleus through two successive trans‐esterification reactions, in which the U2 small nuclear ribonucleoprotein (snRNP) binds to the branch point sequence in pre‐mRNA for the formation of the active center of splicing machinery.1, 2 The recognition of the branch point sequence involves splicing factor 3b (SF3b) protein complex, which is an essential component of the U2 snRNP.3, 4, 5, 6, 7, 8 In human, SF3b is a large protein complex composed of seven proteins: SF3b155, SF3b145, SF3b130, SF3b49, SF3b14b, SF3b10, and p14.4, 5, 8
The SF3b49 protein interacts with the SF3b145 protein, forming a protein complex that is thought to play a role in tethering U2 snRNP to the branch site.3, 5 Human SF3b49, composed of 424 amino acid residues, contains two consecutive RNA recognition motif (RRM) domains (first RRM domain [RRM1] for residues 15–86 and RRM2 for residues 102–174), while human SF3b145, composed of 895 residues, contains two domains of unknown function and structure, according to the Pfam database [Fig. 1(A)]. One of the human SF3b145 domains is DUF382 (domain of unknown function, residues 549–597) and the other is the PSP domain (proline‐rich domain, residues 605–657), a domain type often found in spliceosome‐associated proteins [Fig. 1(A)]. The region including DUF382 and PSP is termed the Cus1 domain. The sequence and domain architecture of both SF3b49 and SF3b145 are well conserved among eukaryotes ranging from yeast to human [Fig. 1(B,C)].
Figure 1.
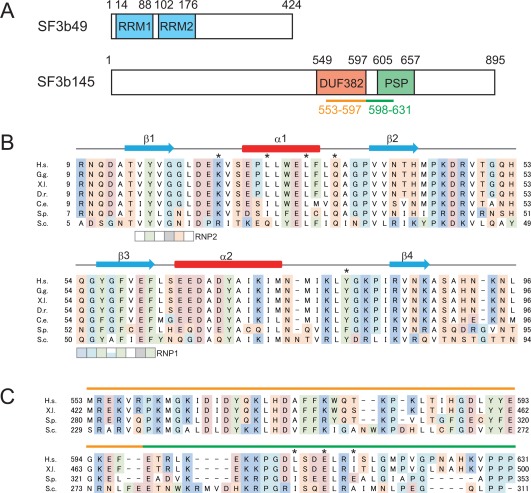
Primary structures of two SF3b component proteins: SF3b49 and SF3b145. (A) Schematic diagrams of human SF3b49 (upper) and SF3b145 (lower). SF3b49 has two RRM domains, RRM1 and RRM2, while SF3b145 has two well‐conserved domains of unknown function, DUF382 and PSP. In the yeast counterparts, the successive regions underlined in orange and green are supposed to interact with SF3b49 RRM1. (B) Multiple sequence alignment of the RRM1 of SF3b49 proteins from various eukaryotes. The SF3b49 sequences from Homo sapiens (H.s., UniProt accession No. Q15427), Gallus gallus (G.g., H9L019), Xenopus laevis (X.l., Q7ZX30), Danio rerio (D.r., Q6NWB3), Caenorhabditis elegans (C.e., Q09442), Schizosaccharomyces pombe (S.p., O14102), and Saccharomyces cerevisiae (S.c., Q99181) were aligned with ClustalX.9 Alignments are colored as follows: cyan: glycine (G) and proline (P); white: hydrophobic amino acids (A, V, L, I, M); red: negatively charged amino acids (D and E); blue: positively charged amino acids (K and R); green: aromatic amino acids (F, Y, W); and orange: hydroxyl and amine amino acids (S, T, N, Q, H, C). Secondary structure elements of the RRM1 structure determined in this study are depicted with cyan arrows (β‐sheet) and red boxes (α‐helix). The conserved signature sequences of RNP1 and RNP2 are indicated by the boxes. Asterisks indicate the amino acid residues shown in this study to interact with SF3b145. (C) Alignment of sequences of the putative SF3b145 region interacting with SF3b49 RRM1. The orange and green lines above the sequences correspond to those shown in (A). The sequences from H. sapiens (H.s., Q13435), X. laevis (X.l., Q52KT1), S. pombe (S.p., Q9UUI3), and S. cerevisiae (S.c., Q02554) were aligned with ClustalX.9 Asterisks indicate the amino acid residues shown in this study to interact with SF3b49.
An in vitro systematic evolution of ligands by exponential enrichment system showed that SF3b49 from Caenorhabditis elegans possesses specific RNA‐binding activity that resides in RRM2, but not or rarely in RRM1.10 Additionally, yeast two‐hybrid screening and pull‐down assays showed that RRM1 of Hsh49 protein (yeast orthologue of human SF3b49) binds to the Cus1 domain of Cus1 protein (yeast orthologue of human SF3b145).11, 12 Thus, SF3b49 RRM1 is considered to be required for the interaction with SF3b145. However, no further detailed information about SF3b49 RRM1 interactions, particularly in humans, have been reported.
An RRM domain is one of the most abundant protein domains in eukaryotes and was first identified as the protein module that plays important roles in sequence‐specific RNA binding.13 The canonical RRM domain consists of a four‐stranded antiparallel β‐sheet packed against two α‐helices with a β1‐α1‐β2‐β3‐α2‐β4 topology.14 It contains two well‐conserved consensus sequences termed RNP1 ([K/R]‐G‐[F/Y]‐[G/A]‐[F/Y]‐[I/L/V]‐X‐[F/Y]) and RNP2 ([I/L/V]‐[F/Y]‐[I/L/V]‐X‐N‐L), which are included in the β3 and β1 strands, respectively.15, 16 The RRM domain typically interacts with RNA on its β‐sheet surface through specific intermolecular hydrogen bonds and stacking interactions mediated by the conserved and exposed aromatic rings located on the β1 and β3 strands.17 However, several RRMs have since been revealed to function as protein–protein interaction modules.18 Some RRMs interact with a protein via the β‐sheet surface, as in the case of RNA recognition, while others interact via the opposite α‐helical surface. For example, among splicing factors, the RRM domain of p14, a component of the SF3b complex, is classified into the former group and that of the small subunit of U2 snRNP auxiliary factor (U2AF35) is classified into the latter. The RRM domains of both types have characteristic amino acid residues required in the protein–protein interaction. The interaction with p14 RRM involves Gly and Thr laying side‐by‐side in β2 and β3, respectively, and Ile and Val laying side‐by‐side in β1 and β3, respectively; this combination of four residues is unique to the p14 RRM.19, 20 In contrast, the U2AF RRM is classified into a novel class of protein recognition motif, and this unusual RRM is termed U2AF homology motif (UHM). The UHM family proteins always have a D/E‐X‐X‐D/E motif in α1 and a R/K‐W/X‐F motif in the region followed by α2.21 However, the RRM1 of SF3b49 has none of the characteristic amino acid residues or motifs that have been previously identified as being required for RRM–protein interaction.
In this study, using multidimensional NMR spectroscopy, we solved the solution structure of the human SF3b49 RRM1 and examined its mode of interaction with a fragment of human SF3b145. Intermolecular NOE and mutational analyses indicated a limited number of residues crucial for this interaction, which led to the establishment of a model of docking between the RRM domain and the fragment. Our findings also reveal a novel mode of interaction with SF3b49 RRM1 in that, on binding, part of the SF3b145 fragment adopts a helical structure and contacts an α‐helix of the RRM domain.
Results
Structure determination of human SF3b49 RRM1
Chemical shift assignments for human SF3b49 RRM1 were performed by standard methods using a combination of triple resonance NMR experiments. All of the main‐chain chemical shifts, except for Gly55 and 90.9% of the side‐chain chemical shifts, were assigned in residues Thr14–Lys88 of SF3b49 RRM1. We solved the solution structures using NOE distance restraints obtained from 15N‐ and 13C‐edited NOESY spectra.
The structure calculations were performed using CYANA 2.0.17.22 An average of 25 restraints per residue were utilized for these calculations (Supporting Information Table S1). The structure composed of Thr14–Lys88 was well defined by the NMR data (Supporting Information Table S1 and Fig. S1). SF3b49 RRM1 adopts a canonical RRM fold with the topology of βαββαβ [Fig. 2(A)].13 Residues Thr14–Gly18 (β1), Val39–His43 (β2), Tyr56–Phe61 (β3), and Arg85–Lys88 (β4) constitute a four‐stranded antiparallel β‐sheet [Fig. 1(B)]. Helix 1 (α1, Glu26–Phe33) and helix 2 (α2, Glu64–Met74) connect β1–β2 and β3–β4, respectively [Fig. 1(B)]. An electrostatic potential surface of SF3b49 RRM1 is also shown in Figure 2(B). Furthermore, a characteristic feature of SF3b49 RRM1 is found in the α‐helical surface, where there is a rather hydrophobic region in the space between α1 and α2. This region appears to form a concave structure, as often seen in other RRMs.
Figure 2.
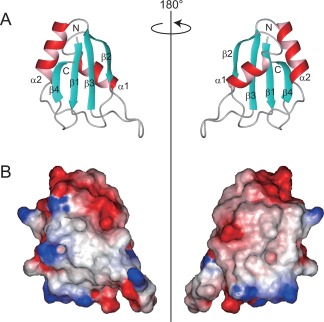
Solution structure of human SF3b49 RRM1. (A) Ribbon representations of SF3b49 RRM1, with views of the β‐sheet surface (left) and of the α‐helical surface (right). Only the well‐converged region ranging from Thr14 to Ala90 is presented. The β‐strands and α‐helical structures are shown in cyan and red, respectively. (B) Electrostatic‐potential surfaces of SF3b49 RRM1. Red and blue indicate negative and positive charges, respectively. The views of the left and right representations correspond to the ribbon representations shown in (A). Electrostatic potentials were calculated, and all of the structural representations were prepared with the software MOLMOL.23
Interaction analysis between SF3b49 RRM1 and the SF3b145 fragment
RRM1 of yeast SF3b49 reportedly interacts with Cus1, which corresponds to yeast SF3b145, in two‐hybrid experiments.11 Another report using two‐hybrid experiments showed that yeast SF3b49 interacts with the fragment spanning residues 229–437 of yeast SF3b145, which corresponds to residues 553–788 in the human counterpart [Fig. 1(C)], but does not interact with either of the two parts into which the fragment was divided (residues 229–311 and residues 312–437).12 Additionally, a pull‐down assay showed that yeast SF3b49 binds to the SF3b145 fragment spanning residues 171–311.12 However, expression of the SF3b145 fragment spanning residues 229–311 in Escherichia coli was undetectable. The authors therefore suggested that the region spanning SF3b145 residues 229–311 includes a binding site of yeast SF3b49, although the peptide composed of only this region may be instable or fold incorrectly.12 We examined if human SF3b49 RRM1 can bind to the human SF3b145 fragment spanning residues 553–631, which corresponds to residues 229–311 in the yeast counterpart [Fig. 1(C)]. We constructed a corresponding N‐terminal GST‐tagged fragment [termed the GST‐SF3b145(553–631) fragment] and purified the fused protein as well as the GST protein as a control. However, we could not remove the GST tag from the GST‐SF3b145(553–631) fragment because the GST‐cleaved fragment becomes aggregated easily (data not shown). This would be consistent with the reported failure of expression of the region spanning residues 229–311 in the yeast counterpart. We then compared a series of [1H,15N]‐heteronuclear single quantum coherence spectroscopy (HSQC) titration spectra of 15N‐labeled SF3b49 RRM1 with GST with those of 15N‐labeled SF3b49 RRM1 with the GST‐SF3b145(553–631) fragment [Fig. 3(A,B)]. No obvious chemical shift changes were observed following the addition of GST to 15N‐labeled SF3b49 RRM1 [Fig. 3(A)]. In contrast, when 15N‐labeled SF3b49 RRM1 was titrated with the GST‐SF3b145(553–631) fragment, distinct chemical shift changes were observed [Fig. 3(B)], suggesting that human SF3b49 RRM1 interacts with the fragment of human SF3b145.
Figure 3.
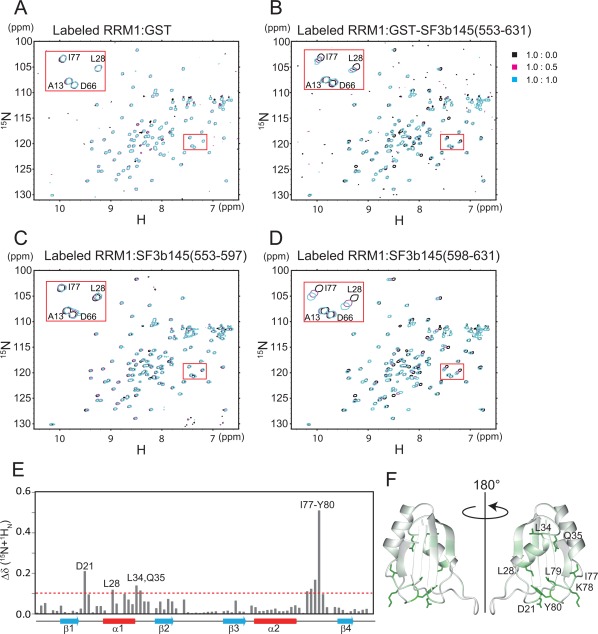
NMR chemical shift perturbations between labeled SF3b49 RRM1 and the non‐labeled SF3b145 fragment. (A) [1H,15N]‐HSQC spectra of SF3b49 RRM1 in the absence or presence of GST (control). (B) [1H,15N]‐HSQC spectra of SF3b49 RRM1 in the absence or presence of the GST‐SF3b145(553–631) fragment. (C) [1H,15N]‐HSQC spectra of SF3b49 RRM1 in the absence or presence of the SF3b145(553–597) fragment. (D) [1H,15N]‐HSQC spectra of SF3b49 RRM1 in the absence or presence of the SF3b145(598–631) fragment. (E) Quantification of the chemical shift perturbation values of SF3b49 RRM1 on binding to the SF3b145(598–631) fragment. The perturbation values were obtained from the [1H,15N]‐HSQC spectrum. The absolute values of the chemical shift change Δδ (15N + 1HN) are shown. Δδ (15N + 1HN) was calculated as follows: Δδ (15N + 1HN) = [(δ15N/6.5)2 + δ1H 2]1/2. Perturbation values greater than the average (0.039 ppm) plus the standard deviation (0.065 ppm) were defined as significant perturbations (i.e., the significance level is 0.104 ppm, indicated by a dotted line). Residues with significant chemical shift changes are shown. The cross‐peaks of Asp12, Trp30, Leu62, and Asn93 disappeared after the addition of the SF3b145(598–631) fragment. (F) Ribbon representation of SF3b49 RRM1 colored according to the chemical shift perturbation values of the main‐chain amides. The color is coded from white (0 ppm) to green (0.509 ppm). The side chains of the residues with significant side‐chain chemical shift perturbations on SF3b145 binding are shown in green.
To clarify the interaction region of SF3b145, we divided the fragment into two parts: a fragment spanning residues 553–597 [termed the SF3b145(553–597) fragment], which is part of DUF382, and a fragment spanning residues 598–631 [the SF3b145(598–631) fragment], which consists of part of the PSP domain and the linker region between the two domains [Fig. 1(A)]. We then titrated 15N‐labeled SF3b49 RRM1 with each fragment. When the SF3b145(598–631) fragment was added to 15N‐labeled SF3b49 RRM1, the chemical shift changes that occurred [Fig. 3(D)] were almost the same as those that occurred on the addition of the GST‐SF3b145(553–631) fragment [Fig. 3(B)]. In contrast, the addition of the SF3b145(553–597) fragment to 15N‐labeled SF3b49 RRM1 did not cause any obvious chemical shift changes [Fig. 3(C)]. These findings show that the region ranging from residue 598 to 631 in SF3b145 is required for interaction with SF3b49 RRM1. Additionally, mapping of the chemical shift changes on the SF3b49 RRM1 structure suggested that a hydrophobic region in the α‐helical surface is involved in the interaction with the SF3b145 fragment [Fig. 3(E,F)].
Furthermore, to confirm the interaction between SF3b49 RRM1 and the SF3b145 fragment, we prepared the 15N‐labeled SF3b145(598–631) fragment and performed the reverse titration experiments. In the [1H,15N]‐HSQC spectra of the15N‐labeled SF3b145(598–631) fragment alone, the 1H chemical shifts of these resonances were observed within a narrow range between approximately 8.0 and 8.5 ppm [Fig. 4(A), red cross‐peaks], indicating that this SF3b145 fragment predominantly adopts a random coil structure. In contrast, on addition of SF3b49 RRM1 to the 15N‐labeled fragment, distinct chemical shift changes were observed in the [1H,15N]‐HSQC spectrum [Fig. 4(A)], showing that the fragment becomes structural on its binding to the RRM domain. Within the SF3b145 fragment, the most significant chemical shift changes occurred in residues Leu609–Val621, suggesting that these residues contribute to the interaction with SF3b49 RRM1 [Fig. 4(B)]. Based on the chemical shift changes, we estimate that the exchange rate between free and bound states is in the fast–medium range in the NMR time‐scale (estimated K d value is nearly 50 µM).
Figure 4.
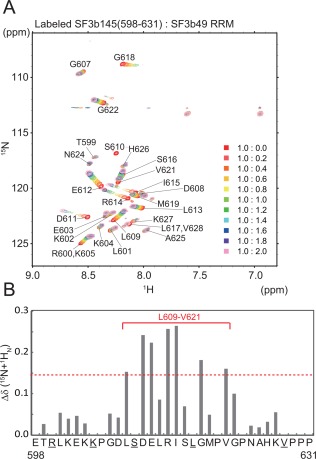
NMR chemical shift perturbations between non‐labeled SF3b49 RRM1 and the labeled SF3b145 fragment. (A) [1H,15N]‐HSQC spectra of the labeled SF3b145(598–631) fragment in the absence or presence of SF3b49 RRM1. (B) Quantification of the chemical shift perturbation values of the SF3b145(598–631) fragment on binding to SF3b49 RRM1. The perturbation values were obtained from the [1H,15N]‐HSQC spectrum. The absolute values of the chemical shift change Δδ (15N + 1HN) were calculated as described in Figure 3(E). The red solid line indicates the region in which amino acid residues with significant chemical shift changes were concentrated. The cross‐peaks of Ser610 (asterisk) disappeared after the addition of SF3b49 RRM. Because the cross‐peaks of Arg600 and Lys605 and of Leu617 and Val628 (underlined) overlap, we could not calculate the chemical shift change.
NOESY measurements and docking model of SF3b49 RRM1 with the SF3b145 fragment
To obtain information about the direct interaction between SF3b49 RRM1 and the SF3b145 fragment, we obtained 13C‐ and 15N‐edited NOESY spectra for a mixture of the labeled and non‐labeled proteins. Observation of the intramolecular NOEs of the SF3b145(598–631) fragment in the presence of SF3b49 RRM1 indicate that the region spanning residues Gly607–Ser616 adopts a helical structure (Supporting Information Fig. S2). Additionally, we observed 15 intermolecular NOEs between Leu609, Glu612, and Ile615 in the SF3b145(598–631) fragment and Lys23, Leu28, Leu32, Gln35, and Tyr80 of SF3b49 RRM1 (Supporting Information Table S2). The most obvious NOEs were confirmed between the aromatic ring protons of Tyr80 of SF3b49 RRM1 and the Hγ protons of Glu612 and Hγ and Hδ protons of Ile615 of the SF3b145 fragment (Supporting Information Fig. S3). The formation of a helical structure within the SF3b145 fragment allows Leu609, Glu612, and Ile615 to adopt a linear arrangement. This positioning of the three residues is supported by the observed intermolecular NOEs from the three residues. These findings suggest that one side of the helical structure of the SF3b145(598–631) fragment contacts SF3b49 RRM1.
We then established a docking model based on the intermolecular NOE distance restraints (Supporting Information Table S2). The structure calculations were performed using CYANA 2.0.17.22 The 15 intermolecular restraints and 85 intramolecular NOEs for the SF3b145(598–631) fragment were utilized to create a model structure (Supporting Information Fig. S2 and Table S2). Because the region spanning residues Gly607–Ser616 in the SF3b145(598–631) fragment is helical, dihedral angle restraints for the φ and ψ angles of the residues were introduced into the structure calculations. Additionally, we assumed that the intensities of the NOEs between the consecutive amide protons, dNN, in this helical region correspond to a distance of 3.6 Å and that the intensities of dαβ(i, i + 3) correspond to 4.4 Å in the 15N‐ and 13C‐edited NOESY experiments for the labeled peptide.24 Based on the intensities of the sequential NOEs, we classified the intermolecular NOEs into two groups with the upper distance bounds of 4.0 Å and 5.0 Å (Supporting Information Table S2). The calculated structures were then refined in 1500‐step energy minimizations in a vacuum to avoid bad steric contacts using the AMBER9 program.25 An ensemble of 20 NMR conformers and a ribbon representation of the lowest energy structure are shown in Supporting Information Figure S4 and Figure 5(A), respectively. In the ensemble of 20 NMR conformers, the region spanning residues 608–616 in the SF3b145 fragment converged well against SF3b49 RRM1 (Supporting Information Fig. S4). Thus, we used the calculated structure as a model for the interaction between SF3b49 RRM1 and the SF3b145 fragment.
The modeled complex structure demonstrated that the helical structure of the SF3b145(598–631) fragment mainly binds to α1 of SF3b49 RRM1 rather than the concave between α1 and α2 [Fig. 5(A)]. This binding mode is consistent with the results of the perturbation experiments [Fig. 5(B)]. In SF3b49 RRM1, Lys23 in the β1‐α1 loop, Leu28 and Leu32 in α1, Gln35 in the α1‐β2 loop, and Tyr80 in the α2‐β4 loop are arranged in a straight line, interacting with one side of the helical structure of the SF3b145 fragment. In particular, Leu28, Leu32 in α1, and Tyr80 form a hydrophobic patch [Fig. 5(C)]. The side chain of Leu32 and the aromatic ring of Tyr80 contact the side chains of Leu609 and Ile615 in the SF3b145 fragment, respectively, and the side chain of Leu28 contacts the alkyl side chain of Glu612 [Fig. 5(A)]. Additionally, it seems likely that Hη of the aromatic ring of Tyr80 hydrogen‐bonds with Oɛ of the side chain of Glu612, although only three of the 20 calculated structures show this interaction. Notably, the residues involved in this interaction are well conserved among various eukaryotes [Fig. 1(B,C)].
Figure 5.
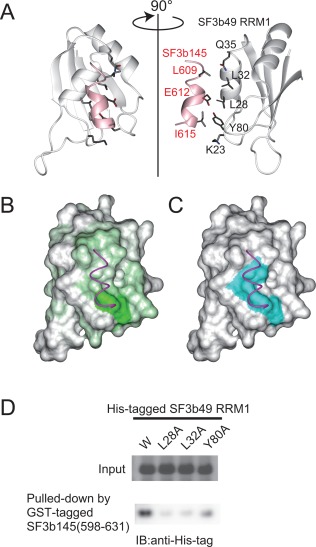
Model structure of the complex between SF3b49 RRM1 and the SF3b145 fragment. (A) Ribbon representation of the model complex of SF3b49 (white) and the SF3b145 fragment (pink). Well‐converged regions of SF3b49 RRM1 (Thr14–Ala90) and SF3b145 (Gly607–Leu617) are shown. Residues directly involved in the interaction between SF3b49 RRM1 and the SF3b145 fragment are shown in stick models (dark gray, carbon; red, oxygen; blue, nitrogen), as described in the text. In all panels, the complex models are presented from the same view. (B) Surface representation of SF3b49 RRM1 colored according to the chemical shift perturbation values of the main‐chain amides as shown in Figure 3(F). The main chain of the SF3b145 fragment corresponding to the region Gly607–Leu617 is illustrated by a magenta wire. (C) Surface representation of SF3b49 RRM1 showing hydrophobic residues, Leu28, Leu32, and Tyr80, involved in the interaction, as described in the text. The three residues, all colored cyan, form a hydrophobic patch. (D) GST pull‐down assay of SF3b49 RRM1 and SF3b145(598‐631). The cell lysates of E. coli expressing wild‐type or mutant His‐tagged SF3b49 RRM1 proteins were analyzed by SDS‐PAGE to confirm the expression level of each the protein (upper panel). Each of the lysates was mixed with the cell lysate of E. coli expressing the GST‐tagged SF3b145(598‐631) or glutathione sepharose, and the protein fractions bound to the resin were analyzed by western blotting using an anti‐His‐tag antibody (lower panel).
Mutation analysis of SF3b49 RRM1
According to the model structure, Leu28, Leu32, and Tyr80 in SF3b49 RRM1 directly contact the SF3b145(598–631) fragment. To confirm these contacts, we prepared three mutants of His‐tagged SF3b49 RRM1 into which Ala was introduced in one of the three residues, and performed a GST pull‐down assay against the GST‐SF3b145(598–631) fragment. The circular dichroic spectra of the mutants were similar to those of the wild‐type, showing that the mutations do not have significant effects on the structure (Supporting Information Fig. S5). Western blotting using an anti‐His‐tag antibody detected His‐tagged SF3b49 RRM1 in the purified fraction of the GST‐F3b145(598–631) fragment. This confirms that SF3b49 RRM1 associates with the SF3b145(598–631) fragment. In contrast, the replacement of Leu28 or Leu32 with an Ala residue severely abolished the detected band, and that of Tyr80 significantly decreased the band intensity [Fig. 5(D)]. These results confirm that Leu28, Leu32, and Tyr80 in SF3b49 RRM1 directly contribute to complex formation with the SF3b145 fragment.
Discussion
The present study shows that the region spanning residues Gly607–Ser616 of SF3b145 adopts a helical structure on binding to the α1 of SF3b49 RRM1 mainly via a hydrophobic interaction. All of the residues involved in this interaction are well preserved among eukaryotes [Fig. 1(C)], suggesting evolutionary conservation of this mode of interaction between SF3b49 RRM1 and the fragment of SF3b145.
In many RRMs with peptide‐binding activity, the α‐helical surface is used as a ligand‐binding site. Figure 6 shows schematic diagrams of the relative positions of the two α‐helices in an RRM domain and a bound protein fragment that are derived from the complex structures that have been determined thus far. The RRM interaction mode can be simply classified into two types, depending on whether the concave between α1 and α2 primarily contributes to the interaction. In the ligand complex with UHM family proteins21 and RNPS1–Acinus,26 the ligand in an extended conformation binds to the concave between α1 and α2, where an invariant Trp residue of the ligand is inserted into a hydrophobic pocket formed by α1 and α2 residues. Similarly, in the complex of PTB–Raver1, instead of Trp, a pair of Leu play a similar role to that of Trp.27 In contrast, in the other RRM interaction mode type, each ligand peptide either crosses over both α1 and α2 (Snu17p–Bud13p),28 binds mainly to α2 (ALYREF–ICP27 and ALYREF–ORF57),29, 30 or binds only to α1 (Snu17p–Pml1p).28 Additionally, Snu17p–Bud13p, ALYREF–ICP27, and ALYREF–ORF57 each have a Trp residue that interacts with α1 residues, while Snu17p–Pml1p has Phe and Leu residues instead of a Trp residue that interact with residues of α1. Furthermore, the bound fragments of Bud13p, ICP27, and Pml1p form an extended conformation, while that of ORF57 forms a helical conformation. We found that in the complex of SF3b49 RRM1–SF3b145, the bound ligand forms a helical conformation and crosses over α1 without interacting with α2, resulting in a helix–helix interaction in almost antiparallel (Fig. 6). Thus, a comparison among the known RRM binding modes indicates that this interaction mode is unique to SF3b49 RRM1. This example also reflects the diversity of RRM–protein interactions. It seems likely that because various splicing factors have multiple RRM domains each binding to different proteins, the existence of such distinct binding modes for RRM domains may be effective at preventing unwanted crosstalk during the complicated interactions of protein–protein and RNA–protein that occur at various stages of the splicing process.
Figure 6.
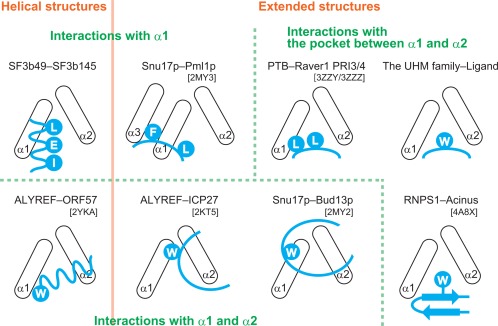
Comparison of schematic diagrams of the relative positions of the two α‐helices in an RRM domain (black) and a bound protein fragment (cyan). The UHM family contains U2AF65‐SF1 [protein data bank code: 1OPI], SPF45‐SF3b155 [PDB 2PEH], U2AF35‐U2AF65 [PDB 1JMT], RBM39‐U2AF65 [PDB 5CXT], and CAPER‐SF3b155 [PDB 4OZ0]. The left abbreviation indicates the RRM‐containing protein, and the right one indicates the bound protein. A Trp residue is indicated by the letter “W,” and the other amino acid residues are shown using one‐letter codes.
The region within the SF3b145(598–631) fragment that is involved in the interaction with SF3b49 is located in the PSP domain, the function of which has remained unknown [Fig. 1(A)]. Thus, the present results are the first to show that in humans the PSP domain of SF3b145 has a protein–protein interaction site. However, it is not clear from this study if the free forms of the region spanning residues 598–631 and the PSP domain each form a stable structure. In humans, according to the Pfam database, only two PSP domains have been found: one in SF3b145 and one in the Zinc finger CCHC domain‐containing protein 8 (ZCCHC8). Given that ZCCHC8 is thought to be a spliceosome‐associated protein, PSP domains may be generally related to protein–protein interactions during the splicing process.
It was reported that Vpr, an accessory gene product of human immunodeficiency virus type 1 (HIV‐1), associates with SF3b145.31, 32 Vpr interferes with the formation of the SF3b49–SF3b145 complex and inhibits the splicing of cellular pre‐mRNA, therefore having multiple biological influences on both viral and cellular proliferation. In light of our results, a potential candidate for the Vpr‐binding site is a region in the SF3b145 PSP domain that includes residues 598–631 because Vpr binding here could mask the site of interaction with SF3b49 RRM1.
The structure of a yeast‐activated spliceosome including the U2 snRNP was very recently determined by cryo‐electron microscopy. Although the electron density of Hsh49 (yeast orthologue of SF3b49) is so weak in the EM structure that there is no available information on the detailed molecular interactions, the RRM1 of Hsh49 obviously interacts with Cus1.33 There appears to be a helix–helix interaction between the RRM1 and Cus1, which is consistent with the result obtained in our present study. Additionally, the low binding affinity between SF3b49 RRM1 and the SF3b145 fragment may be related to the weak electron density of Hsh49. In fact, a similar weak binding affinity between structural domains is found in some spliceosomal protein complexes with dynamic properties. For example, the third RRM of RBM39 and the fragment of U2AF65, components of splicing machinery, showed a K d value of ∼20 µM.
SF3b49 RRM1 is clearly involved in a type of protein‐binding, but this structural study cannot rule out the possibility that the RRM1 can also bind to some RNA because the SF3b49 RRM1 has a structural feature typical of RNA‐binding capacity. The β‐sheet surface of SF3b49 RRM1 has both negatively and positively charged clusters around the hydrophobic patch formed by the two well‐conserved aromatic amino acid residues on β1 and β3 [Tyr16 and Phe58 in the RNP motifs, Fig. 1(B)].13 It is possible that the RNA‐binding activity of RRM1 is exhibited only in the complex form of two RRM domains that arises at a stage during the conformational changes of SF3b49 that occur in the course of the splicing process. Further studies will be required to obtain a comprehensive understanding of the roles of the SF3b49 RRM domains.
Materials and Methods
Protein expression and purification
For structure determination, the DNA encoding the RRM1 (Pro5–Leu96) of human SF3b49 (GenBank: BAD97042.1) was subcloned by PCR from the human cDNA clone with the ID RIKEN cDNA hss001003928. Note that the amino acid sequence of the expressed RRM protein differs only in position 75 (Asn vs. Asp) from that of the protein of SwissProt accession no. Q15427. The changed position is located at the C‐terminal end of α2, which is located in a side nearly opposite from the interaction site in the helical region.
The DNA fragment was cloned into the expression vector pCR2.1 (Invitrogen, Carlsbad, CA) as a fusion with an N‐terminal native His affinity tag and a TEV protease cleavage site. The 15N, 13C ‐labeled fusion protein was synthesized using a cell‐free protein expression system from E. coli.34, 35 The resulting tagged protein was purified by a 5‐ml His Trap column (GE Healthcare, Little Chalfont, Buckinghamshire, UK) with an imidazole gradient from 12 to 500 mM. After tag removal, the tag‐free protein was further purified by HiTrap SP and HiTrap Q column chromatography (GE Healthcare).
For the NMR titration experiments and the pull‐down assay, the RRM1 (Pro5–Leu96) of human SF3b49 was cloned into the Nde I/Bam HI sites of pET‐15b (Novagen). Each of the fragments of SF3b145 (Q13435, 553–631, and 598–631), was cloned into the Eco RI/Sal I sites of pGEX6P‐1 (GE Healthcare). In all constructs, a TEV protease cleavage site was placed between the tag and the protein sequences. E. coli strain BL21 (DE3) cells with the recombinant plasmids were grown at 37°C in LB medium supplemented with 50 mg/L of ampicillin for the non‐labeled samples and in modified minimal medium36 supplemented with 50 mg/L ampicillin for the 15N, 13C‐labeled samples. After isopropyl β‐D‐1‐thiogalactopyranoside induction (1 mM), the harvested cells were lyzed, and the lysates were applied to a His Accept column (Nacalai Tesque) or a Glutathione Sepharose 4 Fast Flow column (GE Healthcare) eluted by the addition of imidazole or glutathione. The tag‐free SF3b49 RRM1 was further purified by RESOURCE Q column chromatography (GE Healthcare). The GST‐tagged SF3b145(553–631) and the tag‐free SF3b145(598–631) were further purified by gel‐filtration column chromatography (GE Healthcare). The peptides corresponding to the two non‐labeled SF3b145 fragments [SF3b145(553–597) and SF3b145(598–631)] were purchased from Toray Research Center.
NMR spectroscopy and resonance assignments
The NMR samples were concentrated to approximately 1.0 mM in 20 mM d11‐Tris‐HCl buffer (pH 7.0), containing 100 mM NaCl, 1 mM 1,4‐dl‐dithiothreitol‐d 10 (d‐DTT), and 0.02% NaN3 (in 90% H2O/10% D2O), using an Amicon Ultra‐15 (5000 MWCO; Millipore). NMR experiments were performed at 25°C on 700 and 800 MHz spectrometers (Bruker AV700 and AV800) equipped with xyz‐pulsed field gradients. Backbone and side‐chain assignments were obtained by standard triple resonance experiments.37 All assignments were checked for consistency with three‐dimensional 15N‐ and 13C‐edited NOESY‐HSQC spectra. Three‐dimensional NOESY spectra were recorded with mixing times of 80–150 ms. The NMR data were processed with the program NMRPipe.38 Spectra were analyzed with the programs NMRView,39 KUJIRA,40 and SPARKY (T. D. Goddard and D. G. Kneller, SPARKY 3, University of California, San Francisco, CA).
Structure calculations
The three‐dimensional structure was determined by combined automated NOESY cross‐peak assignment41 and structure calculations with torsion angle dynamics42 implemented in the program CYANA 2.0.17.22 Dihedral angle constraints for φ and ψ were obtained from the main‐chain and 13Cα chemical shift values using the program TALOS (torsion angle likelihood obtained from shift and sequence similarity),43 and by analyzing the NOESY spectra. Structure calculations started from 100 randomized conformers and used the standard CYANA simulated annealing schedule42 with 20,000 torsion angle dynamic steps per conformer. The 20 conformers with the lowest overall energy were subjected to restrained energy refinement with the program AMBER9, using the Generalized Born model.25 PROCHECK‐NMR was used to validate the final structures.44 Structure figures were prepared with the program MOLMOL.23
NMR titration experiments
For the chemical shift titration experiments, GST‐SF3b145(553–631) was dissolved in the same buffer as the SF3b49 RRM1 sample to a concentration of 1 mM, and each of the two non‐labeled SF3b145 fragments, SF3b145(553–597) and SF3b145(598–631), was dissolved to a concentration of 5 mM. The SF3b145(553–597) fragment often precipitated in the buffer utilized for the structure determination of SF3b49 RRMs. Thus, we added choline‐O‐sulfate [2‐(trimethylammonio)ethyl sulfate, COS] to each of the solutions to achieve a final concentration of 250 mM for protein solubilization and stabilization.45, 46 Two‐dimensional [1H,15N]‐HSQC spectra were recorded while increasing the concentrations of the GST‐fused protein or of each of the SF3b145 fragments relative to that of SF3b49 RRM1 (0.2 mM) to a final ratio of 1:2 of SF3b49 RRM1:GST‐fused protein or each of the fragments. The chemical shift titration experiments between the 15N‐labeled SF3b145(598–631) fragment and the non‐labeled SF3b49 RRM1 were performed in the same way, except for the number of increments of the titrations.
Intermolecular NOE between SF3b49 RRM1 and the SF3b145(598–631) fragment
We prepared 13C‐ and 15N‐labeled SF3b145(598–631) and measured the 13C‐ and 15N‐filtered NOESY spectra in the presence of the non‐labeled SF3b49 RRM1 [2.0:1.0 molar ratio of SF3b49 RRM1:SF3b145(598–631)]. Simultaneously, we performed the NOESY experiment with the 13C‐ and 15N‐labeled SF3b49 RRM1 and non‐labeled SF3b145(598–631) to identity the intermolecular NOEs [1.0:2.0 molar ratio of SF3b49 RRM1:SF3b145(598–631)].
Pull‐down assay and immunoblotting
His‐tagged SF3b49 RRM1 and the GST‐tagged SF3b145(598–631) fragment were expressed in E. coli cells, as described above. Cell lysates at volumes of 200 µL and 1 mL containing SF3b49 RRM1 and the SF3b145(598–631) fragment, respectively, were mixed, to which 25 µL of the glutathione sepharose resin was added. The resultant mixture was incubated for 30 min at 4°C, and then the resin was washed five times with PBS containing 0.05% Nonidet P‐40 (Nacalai Tesque). The proteins bound to the resin were analyzed by SDS‐PAGE and transferred to a PVDF membrane (Bio‐Rad). His‐tagged proteins were immunoblotted by Anti‐His‐tag mAb‐HRP‐DirecT (MBL). The protein was visualized using ImageQuant LAS‐4000 (GE Healthcare).
Accession codes
The atomic coordinates for the ensemble of 20 energy‐refined NMR conformers that represent the solution structures of the first RRM of SF3b49 have been deposited in the Protein Data Bank with accession code PDB 5GVQ.
Supporting information
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Figure 4.
Supporting Information Figure 5.
Supporting Information Table 1.
Supporting Information Table 2.
Acknowledgment
The authors have no conflict of interest to report.
Contributor Information
Kanako Kuwasako, Email: kanameki@musashino-u.ac.jp.
Yutaka Muto, Email: ymuto@musashino-u.ac.jp.
References
- 1. Nelson KK, Green MR (1989) Mammalian U2 snRNP has a sequence‐specific RNA‐binding activity. Genes Dev 3:1562–1571. [DOI] [PubMed] [Google Scholar]
- 2. Pan ZQ, Prives C (1989) U2 snRNA sequences that bind U2‐specific proteins are dispensable for the function of U2 snRNP in splicing. Genes Dev 3:1887–1898. [DOI] [PubMed] [Google Scholar]
- 3. Champion‐Arnaud P, Reed R (1994) The prespliceosome components SAP 49 and SAP 145 interact in a complex implicated in tethering U2 snRNP to the branch site. Genes Dev 8:1974–1983. [DOI] [PubMed] [Google Scholar]
- 4. Das BK, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R (1999) Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol 19:6796–6802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Gozani O, Feld R, Reed R (1996) Evidence that sequence‐independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev 10:233–243. [DOI] [PubMed] [Google Scholar]
- 6. Gozani O, Potashkin J, Reed R (1998) A potential role for U2AF‐SAP 155 interactions in recruiting U2 snRNP to the branch site. Mol Cell Biol 18:4752–4760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. MacMillan AM, Query CC, Allerson CR, Chen S, Verdine GL, Sharp PA (1994) Dynamic association of proteins with the pre‐mRNA branch region. Genes Dev 8:3008–3020. [DOI] [PubMed] [Google Scholar]
- 8. Will CL, Urlaub H, Achsel T, Gentzel M, Wilm M, Luhrmann R (2002) Characterization of novel SF3b and 17S U2 snRNP proteins, including a human Prp5p homologue and an SF3b DEAD‐box protein. EMBO J 21:4978–4988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Thompson JD, Gibson TJ, Higgins DG (2002) Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics Chapter 2:Unit 2 3. [DOI] [PubMed]
- 10. Tanaka Y, Ohta A, Terashima K, Sakamoto H (1997) Polycistronic expression and RNA‐binding specificity of the C. elegans homologue of the spliceosome‐associated protein SAP49. J Biochem 121:739–745. [DOI] [PubMed] [Google Scholar]
- 11. Igel H, Wells S, Perriman R, Ares M, Jr (1998) Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA 4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 12. Pauling MH, McPheeters DS, Ares M Jr. (2000) Functional Cus1p is found with Hsh155p in a multiprotein splicing factor associated with U2 snRNA. Mol Cell Biol 20:2176–2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Oubridge C, Ito N, Evans PR, Teo CH, Nagai K (1994) Crystal structure at 1.92 A resolution of the RNA‐binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature 372:432–438. [DOI] [PubMed] [Google Scholar]
- 14. Nagai K, Oubridge C, Ito N, Avis J, Evans P (1995) The RNP domain: a sequence‐specific RNA‐binding domain involved in processing and transport of RNA. Trends Biochem Sci 20:235–240. [DOI] [PubMed] [Google Scholar]
- 15. Bandziulis RJ, Swanson MS, Dreyfuss G (1989) RNA‐binding proteins as developmental regulators. Genes Dev 3:431–437. [DOI] [PubMed] [Google Scholar]
- 16. Burd CG, Dreyfuss G (1994) Conserved structures and diversity of functions of RNA‐binding proteins. Science 265:615–621. [DOI] [PubMed] [Google Scholar]
- 17. Perez‐Canadillas JM, Varani G (2001) Recent advances in RNA‐protein recognition. Curr Opin Struct Biol 11:53–58. [DOI] [PubMed] [Google Scholar]
- 18. Clery A, Blatter M, Allain FH (2008) RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol 18:290–298. [DOI] [PubMed] [Google Scholar]
- 19. Kuwasako K, Dohmae N, Inoue M, Shirouzu M, Taguchi S, Guntert P, Seraphin B, Muto Y, Yokoyama S (2008) Complex assembly mechanism and an RNA‐binding mode of the human p14‐SF3b155 spliceosomal protein complex identified by NMR solution structure and functional analyses. Proteins 71:1617–1636. [DOI] [PubMed] [Google Scholar]
- 20. Schellenberg MJ, Edwards RA, Ritchie DB, Kent OA, Golas MM, Stark H, Luhrmann R, Glover JN, MacMillan AM (2006) Crystal structure of a core spliceosomal protein interface. Proc Natl Acad Sci USA 103:1266–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kielkopf CL, Lucke S, Green MR (2004) U2AF homology motifs: protein recognition in the RRM world. Genes Dev 18:1513–1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Güntert P, Buchner L (2015) Combined automated NOE assignment and structure calculation with CYANA. J Biomol NMR 62:453–471. [DOI] [PubMed] [Google Scholar]
- 23. Koradi R, Billeter M, Wüthrich K (1996) MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph 14:51–55, 29–32. [DOI] [PubMed] [Google Scholar]
- 24. Kurt Wüthrich (1986) NMR of proteins and nucleic acids. New York: Wiley. [Google Scholar]
- 25. Case DA, Cheatham TE, III , Darden T, Gohlke H, Luo R, Merz KM, Jr , Onufriev A, Simmerling C, Wang B, Woods RJ (2005) The Amber biomolecular simulation programs. J Comput Chem 26:1668–1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Murachelli AG, Ebert J, Basquin C, Le Hir H, Conti E (2012) The structure of the ASAP core complex reveals the existence of a Pinin‐containing PSAP complex. Nat Struct Mol Biol 19:378–386. [DOI] [PubMed] [Google Scholar]
- 27. Joshi A, Coelho MB, Kotik‐Kogan O, Simpson PJ, Matthews SJ, Smith CW, Curry S (2011) Crystallographic analysis of polypyrimidine tract‐binding protein‐Raver1 interactions involved in regulation of alternative splicing. Structure 19:1816–1825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wysoczanski P, Becker S, Zweckstetter M (2015) Structures of intermediates during RES complex assembly. Sci Rep 5:12545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Tunnicliffe RB, Hautbergue GM, Kalra P, Jackson BR, Whitehouse A, Wilson SA, Golovanov AP (2011) Structural basis for the recognition of cellular mRNA export factor REF by herpes viral proteins HSV‐1 ICP27 and HVS ORF57. PLoS Pathog 7:e1001244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tunnicliffe RB, Hautbergue GM, Wilson SA, Kalra P, Golovanov AP (2014) Competitive and cooperative interactions mediate RNA transfer from herpesvirus saimiri ORF57 to the mammalian export adaptor ALYREF. PLoS Pathog 10:e1003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hashizume C, Kuramitsu M, Zhang X, Kurosawa T, Kamata M, Aida Y (2007) Human immunodeficiency virus type 1 Vpr interacts with spliceosomal protein SAP145 to mediate cellular pre‐mRNA splicing inhibition. Microbes Infect 9:490–497. [DOI] [PubMed] [Google Scholar]
- 32. Terada Y, Yasuda Y (2006) Human immunodeficiency virus type 1 Vpr induces G2 checkpoint activation by interacting with the splicing factor SAP145. Mol Cell Biol 26:8149–8158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Yan C, Wan R, Bai R, Huang G, Shi Y (2016) Structure of a yeast activated spliceosome at 3.5 A resolution. Science 353:904–911. [DOI] [PubMed] [Google Scholar]
- 34. Kigawa T, Yabuki T, Matsuda N, Matsuda T, Nakajima R, Tanaka A, Yokoyama S (2004) Preparation of Escherichia coli cell extract for highly productive cell‐free protein expression. J Struct Funct Genomics 5:63–68. [DOI] [PubMed] [Google Scholar]
- 35. Kigawa T, Yabuki T, Yoshida Y, Tsutsui M, Ito Y, Shibata T, Yokoyama S (1999) Cell‐free production and stable‐isotope labeling of milligram quantities of proteins. FEBS Lett 442:15–19. [DOI] [PubMed] [Google Scholar]
- 36. Kuwasako K, He F, Inoue M, Tanaka A, Sugano S, Güntert P, Muto Y, Yokoyama S (2006) Solution structures of the SURP domains and the subunit‐assembly mechanism within the splicing factor SF3a complex in 17S U2 snRNP. Structure 14:1677–1689. [DOI] [PubMed] [Google Scholar]
- 37. Clore GM, Gronenborn AM (1994) Multidimensional heteronuclear nuclear magnetic resonance of proteins. Methods Enzymol 239:349–363. [DOI] [PubMed] [Google Scholar]
- 38. Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293. [DOI] [PubMed] [Google Scholar]
- 39. Johnson BA (2004) Using NMRView to visualize and analyze the NMR spectra of macromolecules. Methods Mol Biol 278:313–352. [DOI] [PubMed] [Google Scholar]
- 40. Kobayashi N, Iwahara J, Koshiba S, Tomizawa T, Tochio N, Güntert P, Kigawa T, Yokoyama S (2007) KUJIRA, a package of integrated modules for systematic and interactive analysis of NMR data directed to high‐throughput NMR structure studies. J Biomol NMR 39:31–52. [DOI] [PubMed] [Google Scholar]
- 41. Herrmann T, Güntert P, Wüthrich K (2002) Protein NMR structure determination with automated NOE assignment using the new software CANDID and the torsion angle dynamics algorithm DYANA. J Mol Biol 319:209–227. [DOI] [PubMed] [Google Scholar]
- 42. Güntert P, Mumenthaler C, Wüthrich K (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J Mol Biol 273:283–298. [DOI] [PubMed] [Google Scholar]
- 43. Cornilescu G, Delaglio F, Bax A (1999) Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR 13:289–302. [DOI] [PubMed] [Google Scholar]
- 44. Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM (1996) AQUA and PROCHECK‐NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486. [DOI] [PubMed] [Google Scholar]
- 45. Hagihara M, Takei A, Ishii T, Hayashi F, Kubota K, Wakamatsu K, Nameki N (2012) Inhibitory effects of choline‐O‐sulfate on amyloid formation of human islet amyloid polypeptide. FEBS Open Bio 2:20–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xiang L, Ishii T, Hosoda K, Kamiya A, Enomoto M, Nameki N, Inoue Y, Kubota K, Kohno T, Wakamatsu K (2008) Interaction of anti‐aggregation agent dimethylethylammonium propane sulfonate with acidic fibroblast growth factor. J Magn Reson 194:147–151. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure 1.
Supporting Information Figure 2.
Supporting Information Figure 3.
Supporting Information Figure 4.
Supporting Information Figure 5.
Supporting Information Table 1.
Supporting Information Table 2.


