Abstract
Altered metabolic phenotype has been recognized as a hallmark of tumor cells for many years, but this aspect of the cancer phenotype has come into greater focus in recent years. NOS2 (inducible nitric oxide synthase of iNOS) has been implicated as a component in many aggressive tumor phenotypes, including melanoma, glioblastoma and breast cancer. Nitric oxide has been well established as a modulator of cellular bioenergetics pathways, in many ways similar to the alteration of cellular metabolism observed in aggressive tumors. In this review we attempt to bring these concepts together with the general hypothesis that one function of NOS2 and NO in cancer is to modulate metabolic processes to facilitate increased tumor aggression. There are many mechanisms by which NO can modulate tumor metabolism, including direct inhibition of respiration, alterations in mitochondrial mass, oxidative inhibition of bioenergetic enzymes, and the stimulation of secondary signaling pathways. Here we review metabolic alterations in the context of cancer cells and discuss the role of NO as a potential mediator of these changes.
Keywords: Nitric Oxide, metabolism, cancer, energy, proliferation, mitochondria, glycolysis
Introduction
Nitric oxide (NO) has been implicated in cancer biology and tumor progression for many years. From discoveries of the mutagenic activities of nitrosamines to the effect of nitric oxide synthase inhibitors in various cancer models, there has been much activity in this area. This review focusses primarily on breast cancer and breast cancer models, and also on the old and yet emerging concept of bioenegetic and metabolic modulation. There is new and exciting evidence that inducible nitric oxide synthase (iNOS) is an important mediator of tumor aggressiveness in breast, and other, cancers. This review brings together this observation with other observations of the effect of NO on cellular bioenergetics to suggest the basic thesis that the modulation of cellular metabolism and bioenergetics by NO could be a component of its effect on cancer progression. While this review primarily focusses on iNOS, it should be noted that other NOS isoforms, particularly eNOS have also been implicated in tumor aggressiveness in breast cancer [1,2].
We first review metabolic alterations in cancer cell metabolism and bioenergetics, and then in the second part discuss how NO could modulate or mediate some of these changes.
Bioenergetics, Metabolism and Breast Cancer
Breast cancer, a malignant tumor originating from breast tissue, is the most common cancer diagnosed and the second leading cause of cancer-related mortality among American women [3]. With sustained investigation of the molecular mechanisms of breast cancer progression and development of anti-cancer therapies, the death-rate of breast cancer has dropped dramatically in the last decades. However, it is estimated that every woman in the United States has a 12 % lifetime risk of developing breast cancer. In addition, several types of breast cancers, for example triple negative breast cancer (which lacks expression of estrogen receptor, progesterone receptor, and HER2/neu receptor) either have, or develop, resistance to some of the most effective therapies, clearly indicated a need for targeted, and perhaps individualized, therapies for such pathologies. Therefore, understanding the mechanisms that underlie tumor aggressive and identifying the virulence factors for breast tumor progression are critical in the development of more effective and less toxic therapies.
Bioenergetics in cancer
Cancer is a disease caused by a heterogeneous collection of dysregulated cellular signaling processes involved in cell proliferation and homeostasis, arising in multiple tissues, and caused by a combination of genetic mutations and/or internal or external oncogenic stimuli [4,5]. Cancer etiology is a multi-step process and cancer cells acquire the following characteristics: uncontrolled growth in the absence of growth signals, resistance to anti-proliferative signals, evasion from apoptosis, limitless replication, development of new blood vessels (angiogenesis), and invasion to surrounding tissue and metastasis to distal organs. These six phenotypes were described as classic hallmarks of cancers by Hanahan and Weinberg in 2000 [6]. A decade later, the reprograming of energy metabolism and immune evasion have been considered as two additional emerging hallmarks of cancers [7]. It is well recognized that cancers have altered energy metabolism which fuels aberrant cancer cell proliferation [8–11]. Additionally, the reprogramming of energy metabolism has been associated with activated oncogenes or inactivated tumor suppressors, and contributes to aggressive cancer phenotypes, highlighting the important role of bioenergetic modulation in tumor progression [12–16].
Overview of energy metabolism
Glucose is a major energy substrate for cells to generate adenosine triphosphate (ATP) for multiple cellular processes in supporting cell functions, cell growth, and cell division. Cells take up glucose through glucose transporters (GLUT) and then it is metabolized to pyruvate by glycolytic enzymes. This process, called glycolysis, generates pyruvate, ATP, and reduced nicotinamide adenine dinucleotide (NADH). The pyruvate is then transported into mitochondria and converted to acetyl-CoA, which is directed to the tricarboxylic acid cycle (TCA cycle), to generate NADH and FADH2. These reduced nucleotides are oxidized through oxidative phosphorylation (OXPHOS) by the mitochondrial electron transport complexes to generate an electrochemical proton gradient that ultimately drives ATP production. The electrons donated by NADH or FADH2 reduce oxygen to water. The efficiency of ATP production by OXPHOS is almost 18 times higher than that by glycolysis [17], thus OXPHOS is the major cellular process of energy transduction in the presence of oxygen. Other metabolic substrates (lipids, amino acids) ultimately feed electrons into the electron transport chain and all require a functional electron transport chain to generate ATP. Consequently the presence of oxygen is essential to generate ATP from these substrates.
In the absence of oxygen ATP cannot be formed via mitochondria, so glycolysis becomes its major source. However glycolysis would soon be halted by the reduction of the NAD+/NADH redox couple as NAD+ is required in the pathway. Although the cytosolic and mitochondrial pools of NADH are isolated, cytosolic NADH electrons can still be fed into the electron transport chain via metabolic shuttles. Under anaerobic conditions, where the electron transport chain is not functioning, cytosolic NADH cannot be oxidized in this manner. Consequently an additional reaction is required to oxidize cytosolic NADH; namely the reduction of pyruvate to lactate catalyzed by lactate dehydrogenase. ATP production can thus be sustained at the expense of lactate production.
Metabolic alterations in cancers
Most cancer cells have increased reliance on glycolysis for ATP production and secrete significant glucose-derived carbon as lactate even in the presence of normal oxygen levels. This observation was first described by Otto Warburg in 1920s and the phenomena is known as the Warburg effect [18,19]. It was proposed that an impairment of OXPHOS in mitochondria caused the Warburg effect in tumor cells [20]; however, many tumor cells have no obvious impairment of OXPHOS [21] and mutations in mitochondrial proteins are rare in cancer. Furthermore, aerobic glycolysis is not unique to tumors. Studies reveled that in proliferating cells, such as proliferating lymphocytes, 90% of glucose was converted to lactate [22–24]. Even though there are similarities between tumor cells and normal proliferating cells with respect to bioenergetic pathways, there are also differences. Accumulating studies show that in addition to the up-regulation of glycolysis, many metabolic pathways are also modulated in tumors. Such metabolic alterations, caused by oncogenic mutations in combination with microenvironmental stimuli, strongly associate with cancer progression [25–27]. Here, several known alterations of metabolic processes in cancer cells will be discussed, including contributions from mutations of metabolic enzymes and oncogenic pathways. Several metabolic alterations in cancers will be discussed in the following sections, however, it is not appropriate to apply the list to all cancer cells, because individual cells may acquire different combination of metabolic modulations during tumorigenesis [28,29].
Why cancer cells exhibit altered metabolic rewiring remains an open question but there are several leading concepts. Much early thought on this was understandably ATP-centric. It has been suggested, for example, that although glycolysis is an inefficient way to generate ATP from glucose, it may be faster and be required for increased ATP demand during proliferation [30]. More recently the focus has shifted away from ATP to the idea that bioenergetics and metabolism are intricately linked, and the same metabolic processes that are used to make ATP are also used to generate non-essential metabolites used to build the carbon polymers essential for proliferation [30,31]. Ribose nucleotides, non-essential amino acids, heme, and other essential biomolecule synthesis pathways are all sub-branches of the basic bioenergetic tree, and the observed metabolic rewiring may facilitate the diversification of the cellular metabolome required for rapid proliferation. Put simplistically, carbon dioxide, the oxidation product of aerobic metabolism is not very useful to a proliferating cell.
It is also possible that cancer cells become glycolytic for the purpose of generating lactate. Local lactic-acidosis appears to contribute to tumor progression through multiple mechanisms including facilitation of matrix disruption and the increase of metastatic potential. The lactate export monocarboxylate transporters, are considered potential therapeutic targets. [32]
Modulations in glycolysis
Many tumors have higher rates of glucose uptake than non-transformed tissues, and this metabolic feature has been applied for clinical tumor imaging, such as 18F-2-deoxyglucose (FDG) accumulation detected by positron emission tomography (PET) [33]. Glucose is taken up through GLUTs and metabolized to pyruvate. Pyruvate produced from the glycolysis is either fed into the TCA cycle and OXPHOS or is converted to lactate by lactate dehydrogenase in the expense of NADH. The lactate is transported to the extracellular space through plasma membrane monocarboxylate transporters (MCTs). As mentioned above cancer cells have increased reliance on this latter pathway and increased expression of GLUTs, lactate dehydrogenase (LDH-A), and MCTs. Moreover, elevated expression of these proteins has been reported in in several cancers and is associated with cancer progression [34–38].
The first step of glycolysis is the phosphorylation of glucose to glucose-6-phosphate (G-6-P) catalyzed by hexokinases (HKs), consuming ATP. There are four isoforms of HK (HK I–IV) expressed in different tissues/cells and subcellular localizations [39]. The over-expression of HK II has been found in many tumors facilitating glycolysis and associated with cancer progression [40]. A predominant fraction of HK II binds to the voltage-dependent anion channel (VDAC) in the mitochondrial outer membrane [41] and this interaction increases the efficiency for G-6-P production in tumors by decreasing of the sensitivity of G-6-P product inhibition [42], increasing the accessibility to ATP generated by mitochondrial OXPHOS [43], and increasing the stability of HK II protein [44]. In addition, the binding of HK II decreases the available binding sites on VDAC for pro-apoptotic factors (such as Bax); thereby preventing cytochrome c release and apoptosis [45]. Therefore, the increased mitochondria-bound HK II contributes not only to the glycolytic phenotype of tumors but also to apoptosis resistance.
The third step of glycolysis involves phosphorylation of fructose-6-phosphate by phosphofructokinase. Quantitative and qualitative changes to phosphofructokinase-1 (PFK-1) are also linked to cancer progression. PFK-1 phosphorylates fructose-6-phosphate to fructose-1,6-bisphosphate in the glycolytic pathway. There are three human PFK-1 isoforms, PFK-M, PFK-L, and PFK-P [46] and increased PFK-1 activity and isoform switch have been associated to cancer malignancy [47,48]. The mechanisms of isoform switching in cancer progression are still unclear. PFK-1 activity can be regulated by many cellular metabolites. One of the potent activators of PFK-1 is fructose-2,6-bisphosphate (F-2,6-BP) generated by phosphofructokinase-2 (PFK-2) [49,50]. Increased expression of PFK-2 (also known as PFKFB3) has been shown in cultured cancer cell lines and primary tumor tissues [51]. Silencing of the PFK-2 gene or administration of PFK-2 inhibitors decrease cancer cell growth indicating the important regulatory role of PFK-2 in cancer metabolism [52,53] and its potential as a drug development target [54].
Pyruvate kinase (PK) is the other glycolytic enzyme linked to tumorigenesis [55]; however, its contribution to tumorigenesis via accelerating glycolysis is in debate. PK catalyzes the dephosphorylation of phosphoenolpyruvate with the production of ATP and pyruvate. Four isoforms exist in human: type L, type R, type M1, and type M2. PK-M2 is the dominant isoform expressed in tumor cells/tissues which enhances glycolysis and lactate production, and PK-M2 expression increased tumor growth in a xenograft mouse model indicating its role in tumor formation [56]. Conversely, recent PK-M2 knockdown studies showed that PK-M2 is not necessary for all tumor cell growth in vivo [57,58] suggesting other roles of PK-M2 beyond its glycolytic function may play important roles in tumorigenesis, such as diversion of glycolytic flux to the pentose phosphate pathway (PPP) supporting tumor cells survival under oxidative stress [59].
The PPP is the metabolic process leading to the generation of 5-carbon sugars and NADPH (reduced nicotinamide adenine dinucleotide phosphate) for anabolic processes. NADPH is not only the cofactor for lipid and nucleotide synthesis, but also for regeneration of glutathione (GSH) and maintenance of cellular redox balance. It has been shown that activation of the PPP prevents oxidative stress-induced cell death in tumors [60–62]. As mentioned previously, PK-M2 inactivation by reactive oxygen species (ROS) increased amounts of upstream metabolites in glycolysis beneficial for biosynthesis of amino acids and phospholipids, and increased shunting of G-6-P to the PPP for nucleotide biosynthesis and NADPH production [63,64]. Therefore, the up-regulation of glycolysis and the PPP has several advantages for cancers: increase of metabolic intermediates for biosynthetic reactions (e.g. fatty acid, nucleotides, and amino acids), rise in detoxification capacity for ROS, and evasion of apoptosis.
Metabolic alterations in the TCA cycle
The TCA cycle serves two important roles in cell metabolism: deriving maximal ATP production from oxidizable metabolites and generating metabolic intermediates for biosynthesis pathways. Proliferating cells and tumor cells generate substrates for biosynthesis of lipids and amino acids, and NADPH through the TCA cycle. During fatty acid synthesis, mitochondrial citrate, one of the intermediates in the TCA cycle, is first transported out to the cytosol and then converted to oxaloacetate (OAA) and acetyl-CoA by ATP citrate lyase (ACL). The acetyl-CoA is then used for fatty acid synthesis, while the OAA is converted to malate by cytosolic malate dehydrogenase at the expense of NADH. The malate can then be converted to pyruvate by malic enzyme, generating NADPH. Increased either ACL or fatty acid synthase (FASN) have been associated to tumor growth [65,66]. In addition, the citrate removed from the TCA cycle by fatty acid synthesis can be replenished by glutamine metabolism. Glutamine can be converted to α-ketoglutarate (α-KG) via glutaminolysis and α-KG can then be diverted into the TCA cycle and metabolized to citrate. Many tumors highly rely on glutamine and increased expression of enzymes involved in glutaminolysis have been identified in tumors [67–69]. In addition, glutaminolysis elevates the TCA intermediate malate which can then be transported to the cytosol and converted to pyruvate by cytosolic malic enzyme accompanied with NADPH production. The glutamine addiction phenomenon in tumors illustrates the important role of glutamine metabolism in fueling biosynthetic pathways in supporting tumor growth.
Generally, mutations of metabolic enzymes in the TCA cycle are rare in tumors. One such mutation related to tumorigenesis is in the enzyme isocitrate dehydrogenase (IDH). It has been shown that the mutations of genes encoded cytoplasmic and mitochondrial IDH (IDH1 and IDH2 respectively) are increased and associated with tumor progression, especially in glioblastoma and acute myeloid leukemia (AML) [70,71]. Wild type IDH converts its substrate, isocitrate, to α-KG; however, the IDH mutants lose this enzymatic activity. Instead, they gain the function of metabolizing α-KG to 2-hydroxyglutarate (2-HG), an oncometabolite [72]. 2-HG is a competitive inhibitor for α-KG-dependent oxygenases, including TET family proteins, which are involved in DNA demethylation processes [73,74]. This leads to global DNA hypermethylation in IDH-mutant AML [75]. IDH1/IDH2 mutations links metabolism and epigenetic modification to tumor formation [76].
Reductive carboxylation of glutamine-derived α-KG has been shown to support tumor cell growth with down-stream mitochondrial defects. This pathway converts α-KG to citrate via isocitratae dehydrogenase – effectively running the TCA cycle in reverse. Thus glutamine is not just an important source of amine groups for the proliferating cell, but also a source of citrate and subsequent fatty acid synthesis [77]. This pathway is elevating under hypoxia [78] and requires bidirectional, oxidative and reductive pathways of α-KG metabolism [79]
Oncogenic signaling and the regulation of cancer metabolism
Phosphoinositide 3-kinase (PI3K) pathway
The PI3K pathway is downstream of receptor tyrosine kinases activated by growth factors, and controls cell proliferation and survival in cooperation with energy metabolism. PI3K signaling is one of the most frequently altered pathways in cancers, and aberrant activation of this pathway by amplification or mutations in genes of its regulatory components has been reported in tumors [80–82]. This also contributes to the glycolytic phenotype in cancers [83]. PI3K activates one of its downstream effectors Akt (also known as protein kinase B), and Akt stimulates glycolysis by directly stimulating glycolytic enzyme activities [84,85] and/or increasing glycolytic enzyme expression via the serine/threonine kinase mammalian target of rapamycin (mTOR). Akt stimulates mTOR signaling by phosphorylation and inhibition of its inhibitor tuberous sclerosis 1/2 complex (TSC1/TSC2) leading to the translation of hypoxia inducible factor 1α (HIF-1α) [86]. HIF-1α is an important transcription factor regulating many glycolytic protein expressions and will be discussed later.
Increased fatty acid synthesis is associated with poor diagnosis in many cancers [87,88]. FASN expression can be activated by either PI3K/Akt or mitogen-activated protein kinase (MAPK) pathways [89–92]. Increased metabolic intermediates and the reducing equivalent NADPH from the PPP and glutaminolysis can be diverted to fatty acid synthesis. The substrate of FASN for fatty acid synthesis, acetyl-CoA can be generated by activation of ACL via Akt phosphorylation [93,94]. Taken together, activation of PI3K signaling directly affects glycolysis and biosynthetic pathways including fatty acid synthesis in cancers.
Transcription factors in metabolic regulation
HIF-1α
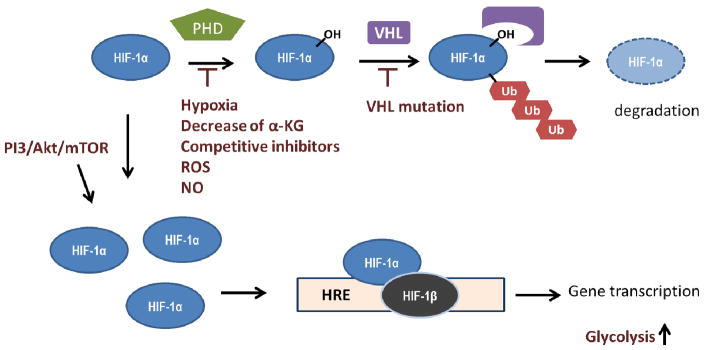
HIF1 signaling is one of the most common dysregulated pathway in cancers and the aberrant activation is associated with cancer progression [95]. HIF1, a heterodimer protein consisting of HIF-1α and HIF-1β, acts as a transcription factor, binding to hypoxia response elements (HREs) of promoters and activating downstream gene expression in response to hypoxic conditions. HIF1 signaling is mainly controlled at the level of HIF-1α. The stability of HIF-1α is controlled by oxygen tension. In the presence of oxygen, proline residues (at Pro-402/Pro-564) of HIF-1α are hydroxylated by prolyl hydroxylase domain proteins (PHD), which use oxygen and α-KG as substrates and produce CO2 and succinate. Prolyl hydroxylated HIF-1α is then recognized by the von Hippel-Lindau tumor suppressor (VHL), an E3 ubiqutin ligase, and subjected to proteasomal degradation [96–98]. Under hypoxic conditions, the PHD-dependent hydroxylation of HIF-1α is inhibited because of the lack of oxygen, and the stabilized HIF-1α can associate with HIF-1β leading to the transactivation of metabolic proteins responsible for the adaptation responses under hypoxia, including glucose transporters, glycolytic enzymes, LDH-A [99–102], MCT-4 [103], and PDK [104]. In cancers, hypoxia-independent activation of HIF1 signaling has three major etiologies: 1) the regulation of PHD activity 2) VHL loss-of-function 3) increase of HIF-1α expression. Functional mutations of the TCA enzymes, fumarate hydratase (FH) and succinate dehydrogenase (SDH), cause the accumulation of fumarate and succinate, which competitively inhibits PHD [105,106]. Interestingly, fumerate is able to react with glutathione to form succinated glutathione, which can impact mitochondrial superoxide production [107]. Loss-of-function in IDH1 mutants results in decrease of conversion of isocitrate to α-KG, leading to the reduction of α-KG which is required for PHD-dependent HIF-1α hydroxylation [108]. VHL mutations are found in renal carcinoma with the stabilization of HIF-1α [109]. ROS accumulation can also stabilize HIF-1α, partly caused by the inactivation of the nonheme iron(II) catalytic center of PHD [110,111]. Increased levels of nitric oxide (NO) have been associated with HIF-1α accumulation under normoxia [112]. In addition, the over-activation of PI3K/Akt/mTOR signaling increases HIF-1α expression and contributes to the glycolytic phenotype in cancers [113]. A short summary is shown in Figure 1.1.
Figure 1.1. Mechanisms of HIF-1α activation in cancers.
In normoxia, hypoxia-inducible factor (HIF)-1α is hydroxylated by prolyl hydroxylase domain proteins (PHD), which uses oxygen and α-KG as substrates. Hydroxylated HIF-1α is recognized by von Hippel-Lindau (VHL), which facilitates HIF-1α ubiquitination, and such modification subjects HIF-1α to proteasomal degradation. Upregulation of HIF-1 signaling has been observed in many cancers caused by aberrant activation of PI3K/Akt/mTOR pathway leading to an increase of HIF-1α translation. The inhibition of PHD by decrease of α-KG, increase of competitive inhibitors, and inactivation of enzyme activity mediated by reactive oxidative species (ROS) or NO have been observed. In addition, loss-of-function mutation of VHL also contributes to HIF-1α stabilization. HIF-1α associates with HIF-1β and binds to the hypoxia response element (HRE) of promoters resulting in transcription of target genes, including those involved in glycolysis regulation. Herein, HIF-1α stabilization contributes to tumor glycolytic phenotype.
Myc
c-Myc (Myc) is a transcription factor regulating genes involved in a broad range of cellular activities in cell proliferation, and also energy metabolism [114]. Constitutively activated Myc by gene amplification, translocation, and mutations have been found frequently in many cancers [115]. Myc directly transactivates the genes in glycolysis and contributes to the glycolytic phenotype [116]. Myc also increases PK-M2 expression via the up-regulation of RNA splicing factors, which promotes RNA splicing for the expression of PK-M2 [117] and leads to the increase of glycolytic intermediates and flux to the PPP for nucleotide synthesis and NADPH production [118]. Myc also contributes to the glutaminolytic phenotype in tumors via transactivation of the glutamine transporter ASCT2 and glutaminase [119]. Moreover, Myc stimulates mitochondrial biogenesis through direct activation of nuclear encoded genes involved in mitochondrial biogenesis [120–122]. Therefore, Myc is the metabolic regulator in boosting cellular energetic metabolism for cell proliferation and the constitutively active mutant contributes to tumor metabolic phenotypes.
p53
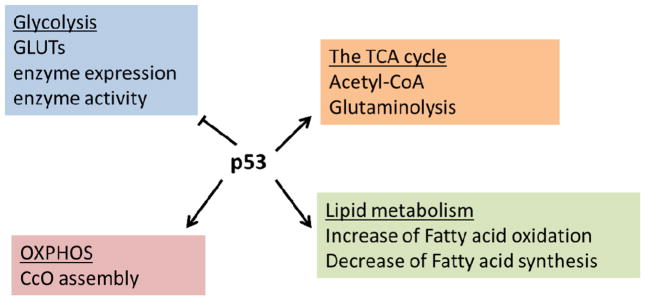
Transcription factor p53, a well-known tumor suppressor, participates in repairing or preventing cellular damage triggered by intrinsic or extrinsic stresses and controls cell fate. The function of p53 has also been extended to regulate energy metabolism, including glycolysis, glutaminolysis, OXPHOS, and fatty acid metabolism [123]. In contrast to HIF-1α and Myc, p53 generally functions as a negative regulator of glycolysis and fatty acid synthesis, and positive regulator for mitochondrial respiration and fatty acid oxidation that fits in its role as tumor suppressor. Actions on glycolysis inhibition by p53 include: 1) inhibition of GLUTs transcription [124], 2) downregulation of glycolytic enzymes including phosphoglycerate mutase, which converts 3-phosphoglycerate to 2-phosphoglycerate [125], 3) induction of the expression of TP53-induced glycolysis and apoptosis regulator (TIGAR), which acts as a bisphosphatase that decreases F-2,6-BP levels, a potent activator of PFK1, and dampens glycolytic flux [126,127], 4) downregulation of pyruvate dehydrogenase kinase 2 (PDK2) expression leading to decreased phosphorylation of pyruvate dehydrogenase complex (PDC) and increased PDC activity. PDC converts pyruvate to acetyl-CoA which then can be fed into the TCA cycle. In addition, p53 favors OXPHOS not only by increasing the levels of acetyl-CoA, but also by transcriptional activation of the SCO2 (synthesis of cytochrome c oxidase) which helps cytochrome c oxidase assembly to facilitate OXPHOS [128]. Glutamine metabolism is also controlled by p53 via the induction of the expression of mitochondrial glutaminase GLS2 which leads to increased cellular glutamate, α-KG, and mitochondrial respiration [129]. p53 is also an important regulator in lipid metabolism. Carnitine palmitoyltransferase 1C, a protein responsible for transporting fatty acids to mitochondria for fatty acid oxidation, can be upregulated by p53 resulting the increase of fatty acid oxidation and ATP production [130]. p53 suppresses fatty acid synthesis by negatively regulating sterol regulatory element-binding protein-1 (SREBP-1), a key transcriptional regulator of lipogenic enzymes including FASN and ACL [131]. The effects of p53 on cellular metabolism are enormously complicated and this is still a growing area for investigation. Generally, p53 acts like a reverse Warburg effect- decreases glycolysis and increase of OXPHOS (general summary in Figure 1.2); as one can imagine, the inactivation of p53 in many tumors contributes to the Warburg effect, at least in part.
Figure 1.2. Metabolic regulations by p53.
p53 inhibits glycolysis by downregulation of glycolytic enzyme, glucose transportor (GLUTs), and F-2,6-BP, a glycolysis activator. Mitochondrial oxidative phosphorylation (OXPHOS) is also maintained by p53 through facilitating cytochrome c oxidase (CcO) assembly. Increasing conversion of pyruvate to acetyl-CoA and transcription of glutaminase for glutaminolysis ultimately elevate the metabolites in the TCA cycle. p53 also inhibits fatty acid synthesis and activates fatty acid oxidation.
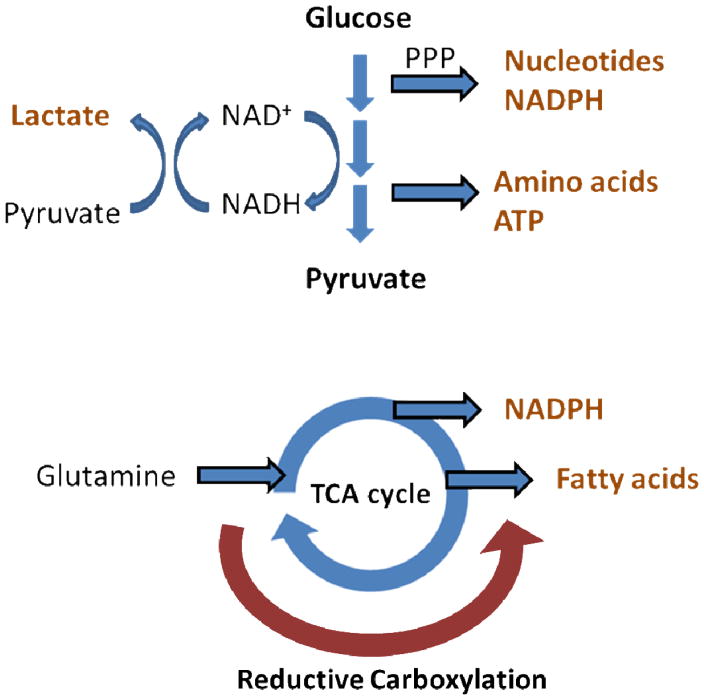
To summarize, the common features and benefits of altered metabolic pathways are illustrated in figure 1.3 in many cancers include: up-regulation of aerobic glycolysis for ATP production and biomass synthesis, suppression of mitochondrial OXPHOS, increase of metabolic flux to PPP for production of nucleotide precursors and NADPH, and utilization of glutaminolysis to replenish TCA intermediates for fatty acid synthesis and NADPH production. The metabolic reprogramming is optimized to meet the need for cell proliferation by stimulation of the carbon flux to biosynthesis pathways, and increase of NADPH for anabolism and oxidative defense [132].
Figure 1.3. Metabolic alterations in cancers.
Many cancers have increased glycolysis for ATP production and amino acid synthesis, and elevated metabolic flux to pentose phosphate pathway (PPP) for NADPH production and nucleotide synthesis. Pyruvate is converted to lactate for NAD+ regeneration to maintain glycolytic flux. Many cancers also rely on glutamine metabolism which replenishes the TCA cycle intermediates and further leads to NADPH production and fatty acid synthesis. Reductive carboxylation allows the use of glutamine for fatty acid synthesis and other metabolic needs even in the presence of hypoxia or mitochondrial dysfunction.
Nitric Oxide and Cancer Metabolism
In the second half of this review we will discuss how the intrinsic biological chemistry of NO is able to modulate cellular metabolism. We will first discuss biological targets of NO and mechanisms by which it is able to alter cellular bioenergetics and metabolism.
NO-dependent signaling mechanisms
The reactivity of NO
One of the most abundant and well disseminated myths concerning NO is that it is highly reactive. Granted, “highly reactive” is a relative term and in comparison with the majority of biomolecules this epithet may be somewhat justified, however, compared to many other free radicals NO is remarkably unreactive. Most importantly it does not react with itself to any appreciable amount. The NO dimer (NO)2 is only 1–2 kcal/mol lower in energy than NO {5938} and pure NO is a stable gaseous molecule at room temperature and pressure. Indeed when one contemplates relatively stable biologically-relevant free radicals, one has to consider much more complex organic structures, such as flavin radicals and protein radicals, which only exist stably in intricately defined environments. Other small molecule inorganic radicals that are often implicated in biological processes such as hydroxyl radical, chloride radical, and even superoxide, self-annihilate with rate constants in excess of 105 M−1s−1 {5939}, and in most cases close to the diffusion limit. The other important concept here is that “reactivity” is almost entirely defined by the magnitude of second order rate constants between two molecules. Nitric oxide reactivity spans the entire gamut of the second-order range constants giving NO its highly selective reactivity that allows it to diffuse unscathed through a sea of biomolecules until it collides with a molecule with which it can rapidly react.
Molecular targets of NO
In order for NO to act as a signal it must be sensed. Most signaling molecules are sensed through non-covalent interactions with receptor proteins, resulting in a conformation change in the receptor that subsequently triggers a down-stream response. The interactions between receptor and ligand usually take the form of hydrogen bonds and this interaction is often referred to as ‘binding’ as the dissolution of the hydrogen bonds allows release of the intact ligand. It has long been understood that metal ions make the best NO ‘receptors’ as the unpaired electron of NO can interact with a d-orbital electron to make an inorganic complex {4404}. A major biological target for NO appears to be ferrous heme iron. NO reacts with ferrous heme groups to form iron nitrosyls which generally are very stable species. However, as with hydrogen bonds, the d-orbital bond has a probability of spontaneously breaking, allowing release of NO, and so again this interaction could be termed ‘binding’. The iron nitrosyl of hemoglobin, for example, has a binding constant in excess of 1010 M, with an on rate of 5 × 107 M−1s−1 and an off rate less than 10−3 s−1 {4093}. Any ferrous heme protein with a vacant coordination site is likely a potential NO “receptor”. The classic example of this interaction occurs in the canonical pathway of NO signaling with soluble guanylyl cyclase (sGC) as the receptor {3985}, although studies indicate that this is not the whole story of NO-dependent sGC activation {5449}. In addition, the binding of NO to the heme/Copper center of the terminal electron transfer complex, cytochrome c oxidase, has been highlighted as a mechanism by which NO may alter cellular metabolism and consequently modulate a multiplicity of cellular processes {1099}. Other metallo-proteins, such as ferric iron, iron-sulfur clusters, copper and zinc ion-containing structures may also be targets for NO, but the underlying chemistry is more poorly resolved and no such proteins have been generally regarded as ‘NO receptors’.
The other major molecular targets of NO are other free radicals. Although relatively unreactive with itself, NO reacts with other free radicals with lightning speed. Perhaps the best studies such reaction is that between NO and superoxide which generates the strong biological oxidant, peroxynitrite {474}. Much has been explored and written concerning the biological fate and consequences of peroxynitrite formation, but suffice to say that this reaction combines two rather innocuous free radicals into a non-radical molecule with a much greater propensity for bio-molecule oxidation {5878}. Other NO-radical interaction include those with lipid-derived radicals, where NO can potently inhibit lipid oxidation but lead to the generation of nitro-lipid species {81}. Finally, NO may also react with molecular oxygen. This latter reaction generates nitrogen dioxide, a potent biological oxidant, but its role in the biological chemistry of NO is uncertain. This is because the reaction is kinetically third order, and very slow at physiological levels of NO and oxygen {288}{4148}. It should be highlighted that a major danger of using non-physiological levels of NO and oxygen in model systems is that the NO/oxygen reaction will be emphasized and the oxidative and damaging effects of NO, via nitrogen dioxide formation, may overwhelm more physiological pathways.
The formation of peroxynitrite and nitrogen dioxide lead into the domain of redox signaling which can be loosely (but perhaps not completely) defined as the propagation of cellular signals by the oxidation and reduction of protein cysteinyl thiols {5487}. Often vicinal thiols are oxidized to form a disulfide by a small-molecule oxidant, which then can be reduced by the cellular reducing machinery with electrons ultimately derived from NADPH. The oxidized and reduced forms of the protein then have different function that can act as a signal per se or modulate other signaling mechanisms. In addition to the thiol/disulfide switch, several different types of thiol modifications have been recognized and one of these, S-nitrosation, is NO specific {5458}{124}. There is an abundance of discussion on S-nitrosation as an NO-dependent post-translational thiol modification and the development and application of proteomic methods to detect this modification have identified numerous proteins that are susceptible to S-nitrosation {5940}. S-nitrosation is a very attractive mechanism by which the observed pleotropic nature of NO can be rationalized at the molecular level, and strong evidence exists for the participation of this post-translation modification in cellular signaling events.
The effect of NO on tumor biology
The effect of NO on cellular signaling appears to be dependent on the concentration and duration of NO exposure under the influence of the cellular microenvironment. The NO levels needed to elicit specific cellular responses correlate well with the observed biphasic effect of NO on tumor biology [133–135]; low concentrations of NO stimulate tumor proliferation and high NO levels elicit tumoricidal activity [136]. The potential molecular mechanisms by which NO promotes tumor progression in breast tumors have been demonstrated at various NO levels. Low to medium levels of NO (<300 nM) mediate cell proliferative and protective responses via cGMP signaling, mTOR activation, phosphorylation of Akt, and stabilization of HIF-1α, and these signaling events have been associated with the promotion of tumor proliferation, migration and angiogenesis [137–142]. NO-induced PI3K/Akt activation at NO concentrations of 100–400 nM has been shown to be mediated by phosphorylation and/or tyrosine nitration of TIMP-1 (tissue inhibitor matrix metalloproteinase-1), which activates pro-survival signaling through binding to the cell surface receptor CD63 complex [143]. NO at a level of 300 nM has been found to activate EGFR (epidermal growth factor receptor), Src kinase, and Ras by the mechanisms of nitrosation which leads to the suppression of tumor suppressor protein phosphatase 2A activity and/or the activation of oncogenic signaling pathways, including c-Myc, Akt, β-catenin, and Ets-1 [144,145]. As NO levels increase above 300 nM, the increased phosphorylation of p53, a tumor suppressor, is observed indicating a cytostatic and/or apoptotic response can be expected. That observation is correlated to tumoricidal effects of high NO levels [146–148]. Suppression of DNA syntheses, disruption of iron homeostasis, inhibition of mitochondrial respiration, inactivation of ERK and Akt by increased expression and activity of MAP kinase phosphatase-1 (MKP-1), decrease of protein synthesis mediated by activation of protein kinase R, and activation of intrinsic apoptotic signaling have also been attributed to tumoricidal effect of high NO levels [149–153]. In addition, the effect of NO is also influenced by the sensitivity of target proteins to NO. For example, in human mammary adenocarcinoma MCF7 cells, HIF-1α is activated by NO immediately as long as the NO levels reach 100–300 nM [154]. On the other hand, phosphorylation of p53 requires at least 2 hours of NO exposure and the activation lasts long after the NO exposure [155]. Therefore, the NO targets can be classified into immediate-transient, immediate-sustained, delayed-transient, and delayed-sustained [156] which implicates the duration of NO exposure can lead to different biological responses. Furthermore, the redox environment also affects the behavior of NO. Reactive oxygen species (ROS), such as superoxide and hydrogen peroxide, generated during cell metabolism are recognized as signaling molecules and plays important roles in cancer progression [157]. It has been shown that NO signaling can be attenuated by superoxide [158–160]; therefore, any factor altering superoxide levels can modulate NO signaling that may convert a growth-inhibition phenotype to a pro-growth phenotype and vice versa [161,162]. Herein, the versatile biological responses elicited by NO can be attributed to the concentrations of NO at local environments and the type of molecular targets which NO or NO derivatives interact with.
The effect of NO on bioenergetics
NO has great impact on energy metabolism [163,164]. NO increases the energy substrates and oxygen availability to cells and tissues by increasing blood flow. NO also alters cellular bioenergetics mediated by direct or indirect NO actions that modulate enzyme activities or signaling intermediates in metabolic pathways. Several known effects of NO on mitochondrial functions and the regulation of glycolysis will be discussed in the following sections.
Effect of NO on mitochondrial respiration
Mitochondrial respiration takes place in the mitochondrial inner membrane and is mediated by electron transport chain (ETC) complexes using the reducing equivalents (NADH and FADH2). Electrons donated from NADH or FADH2 to the ETC via NADH- ubiquinone oxidoreductase (complex I) or succinate-ubiquinone oxidoreductase (complex II) are transferred to ubiquinone-cytochrome c oxidoreductase (complex III) by ubiquinone, then passed to cytochrome c oxidase (CcO, complex IV) via cytochrome c. At CcO these electrons reduce oxygen, the terminal electron acceptor, to form water. The electron transporting processes are coupled to the pumping of proton from the mitochondrial matrix into the intermembrane space and the consequent proton electrochemical gradient is used to generate ATP by ATP synthase (complex V). Extensive experimental evidence has shown that NO inhibits mitochondrial respiration in multiple ways through either direct or indirect modification on mitochondrial complex activities [165–167] and NO-mediated mitochondrial inhibition is associated with pathological conditions [168–173]. CcO is very sensitive to NO inhibition compared to other mitochondrial complexes and it has been shown that nanomolar concentrations of NO at physiological levels (1–200 nM) can inhibit mitochondrial respiration at CcO [174–177]. NO reversibly inhibits mitochondrial oxygen consumption by competing with oxygen at the CcO heme iron:copper (a3/CuB) dinuclear center [178–182], and the inhibitory effect increases with the decrease of oxygen tension in respiring mitochondria. This illustrates the potential regulatory function of NO in modulating cellular respiration under physiological conditions [183–186]. It has also been reported that high levels of NO treatment or prolonged incubation can cause persistent inhibition of CcO caused by the S-nitrosation of cysteine residues in subunit II [187] and by decrease of CcO protein level [188,189].
Mitochondrial complex I can be inhibited by NO mainly through the indirect NO reactions of S-nitrosation [190,191] and/or tyrosine nitration [192,193]. NO-dependent inactivation of complex I is accelerated under hypoxia suggesting complex I inhibition may be enhanced under pathological conditions contributing to cell dysfunction [194]. Purified mitochondrial complex II can be inhibited by high levels of NO, possibly via the disruption of iron-sulfur complex [195]. Complex III activity can be reversibly inhibited by high levels of NO in purified enzymes or submitochondrial particles, but the mechanisms are unclear [196–198]. Tyrosine nitration in β subunit of complex V has been shown to inhibit the enzyme activity [199,200]. Taken together, mitochondrial respiration and ATP synthesis can be either reversibly or irreversibly inhibited by NO and NO derivatives. Persistently irreversible inhibition may be critical in pathological conditions; however, it has also been illustrated that the S-nitrosation of cysteine residues in complex I provides cytoprotective effects against heart ischemia-reperfusion injury by the reduction of reactive oxidants production [201,202].
NO and the TCA cycle
The TCA cycle involves a series of enzyme-catalyzed reactions in the mitochondrial matrix to generate reducing equivalents, NADH and FADH2, for ATP production by OXPHOS. In these reactions, the six-carbon molecule citrate is oxidized to the four-carbon molecule oxaloacetate (OAA) with the production of NADH and FADH2, and citrate is then regenerated using the acetyl group of acetyl-CoA and OAA Acetyl-coA is generated by pyruvate dehydrogenase complex (PDC) using pyruvate as the substrate; therefore, PDC couples glycolysis and the TCA cycle and it controls the flow of the TCA cycle. It has been shown that PDC can be inhibited by SIN-1, a peroxynitrite producing compound, in a concentration dependent manner and it is not reversible by treatment with dithiothreitol (DTT), a thiol reducing agent, suggesting that tyrosine nitration of PDC plays major role in the inhibition [203]. Aconitase is an iron-sulfur containing protein that catalyzes the isomerization of citrate to isocitrate and is a sensitive target of NO and NO-derivatives. NO binds to the iron center and reversibly inhibits aconitase [204–206]. Peroxynitrite is a potent aconitase inhibitor and the inhibition is reversible by DTT and iron supplement [207,208] suggesting the inhibition is mediated by the oxidation of cysteine residue bound to the iron-sulfur center which facilitates the removal of iron from [4Fe-4S]2+ cluster [209]. Tyrosine nitration on aconitase has been detected in purified protein treated with peroxynitrite or in animal models, but this modification seems not the major cause for enzyme inhibition [210–212]. S-Nitrosoglutathione (GSNO) can also inhibit aconitase but the reaction is irreversible and the mechanism is still unclear. S-Nitrosation reversibly inhibits aconitase activity in purified mitochondria treated with mitochondria-targeted S-nitrosothiol Mito-SNO [213]. Hence, the NO-dependent aconitase inhibition is caused by the disruption and/or removal of iron from the iron-sulfur cluster center, at least in part. Further down the TCA cycle, IDH is inhibited in LPS-stimulated murine macrophage RAW264.7 cells that may be a result of protein S-nitrosation [214]. α-KG dehydrogenase converts α-KG to succinyl-CoA and produces NADH. It is reversibly inhibited by S-nitrosation [215] and peroxynitrite treatment, and N-acetyl cysteine (NAC, cell permeable cysteine supplementation for GSH synthesis) reversed peroxynitrite-mediated inhibition indicating a reversible thiol modification involved in the inactivation [216].
NO and mitochondrial biogenesis
NO plays important roles in mitochondrial proliferation. Mitochondrial biogenesis is mainly controlled at the level of transcription and can be stimulated by various factors, including exercise, caloric restriction, hormones, cytokines, and cold [217,218]. Calorie-restricted mice had increased eNOS expression, mitochondrial proteins, and mitochondrial respiration [219]; in addition, lower mitochondrial density in many tissues of eNOS knockout mice compared to wild-type mice indicates NO generated from eNOS plays an important role in the regulation of mitochondrial biogenesis [220]. The mechanism by which NO up-regulates mitochondrial biogenesis appears to be mediated by the activation of sGC. In some cell-types in culture the increase of mitochondrial proteins and mitochondrial DNA (mtDNA) content by NO, from NO donors or eNOS expression, were accompanied by an increase of cGMP and key transcription factors responsible for mitochondrial biogenesis programming. These include PGC-1α (peroxisome proliferator-activated receptor gamma coactivator-1α), NRF-1 (nuclear respiratory factor-1), and mtTFA (mitochondrial transcription factor A) [221,222]. Mitochondrial biogenesis can be stimulated by the addition of the cGMP analog (8 Br-cGMP) and attenuated by ODQ (a sGC inhibitor) which indicates that NO-dependent mitochondrial biogenesis occurs via sGC-cGMP signaling. Additionally, NO-dependent PGC-1α activation and mitochondrial biogenesis were attenuated by eNOS knockdown [223]. NO-induced PGC-1α expression can also be mediated by AMPK activation in skeletal muscle cells exposed to nanomolar levels of NO [224,225] that may be caused by the AMPK-dependent phosphorylation of PGC-1α leading to an increase of PGC-1α transcription [226]. AMPK activation can also be mediated by mitochondrial ROS production driven by NO-dependent CcO inhibition which leads to the phosphorylation of CREB (cAMP-response-element-binding protein) and increase of PGC-1α transcription [227–231].
NO and glycolysis
A biphasic effect of NO on glycolysis has been reported, but such effects appear to be cell type-dependent. In general, low/physiological levels of NO stimulate glycolysis and high levels of NO inhibit it. NO stimulates glycolysis in astrocytes via mitochondrial inhibition, which results in AMPK activation, increase levels of F-2,6-BP, a potent PFK-1 activator, and glucose uptake [232,233]. This activation of glycolysis in astrocytes is cGMP-independent, and NO failed to stimulate glycolysis in neurons indicating the effect of NO is context-dependent [234]. NO-dependent stimulation of glucose uptake in insulin-sensitive skeletal muscle cells occurs through a different pathway that is cGMP-dependent [235,236]. Furthermore, HIF-1α plays a role in NO-dependent activation of glycolysis. It has been shown that, under hypoxia, NO decreases HIF-1α stability by redistribution of oxygen from mitochondria respiration to other uses, such as for PHD activity leading to HIF-1α hydroxylation and proteasomal degradation [237]; In contrast, NO can create “pseudohypoxia” for HIF-1 activation under normoxia, and it plays an important role for tumor progression [238–241]. However, the mechanisms of NO-dependent HIF-1α activation are still under investigation, and appear to depend on NO concentration and ROS generation [242–244]. Several mechanisms have been proposed including inhibition of PHD activity by NO interaction with the iron containing catalytic center, by the S-nitrosation of cysteine residues on PHD [245,246], and S-nitrosation of cysteine residues on HIF-1α resulting in stabilization of HIF-1α [247]. In addition, NO increases HIF-1α levels in astrocytes via PI3K/AKT/mTOR signaling leading to the up-regulation of glycolytic enzymes and enhancement of glycolysis [248]. On the other hand, excess amounts of NO inhibit glycolysis. High flux of NO, for example under inflammatory conditions, increases the generation of NO derivatives which leads to the inhibition of glycolytic enzymes, including glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and aldolase. GAPDH converts glyceraldehyde-3-phosphate to 1,3-bisphosphglycerate with the production of NADH. It has been shown that cells treated with NO donors or with S-nitrosothiols exhibited either reversible or irreversible inhibition of GAPDH activity mediated by S-glutathiolation or S-nitrosation of cysteine residues [249,250]. GAPDH is also sensitive to peroxynitrite resulting in the inhibition of GAPDH activity through thiol oxidation or tyrosine nitration [251,252]. Aldolase, also known as fructose-bisphosphate aldolase, splits fructose-1,6-bisphoaphate to glyceraldehyde-3-phosphate and dihydroxyacetone phosphate. Aldolase can be inhibited by peroxynitrite or high levels of GSNO through tyrosine nitration [253,254]. Overall, glycolysis is stimulated by NO through signaling triggered either by mitochondrial inhibition, NO-sGC-cGMP, or HIF-1α. Glycolysis inhibition generally occurs under high NO flux mediated by chemical modification of enzymes resulting in enzymatic inhibition.
Summary
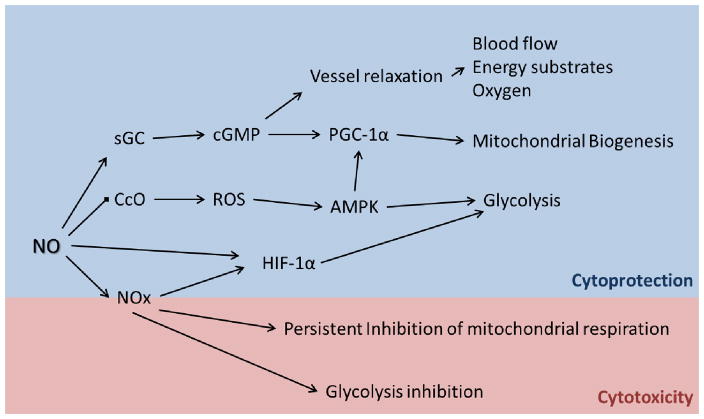
Figure 1.3 summarizes many of the pathways discussed above. NO plays an important role in maintaining cellular energetic balance in response to various environmental situations. Under physiological conditions, NO increases the availability of oxygen and carbon substrates for cells by sGC/cGMP-mediated vessel relaxation; stimulates mitochondrial biogenesis through transcriptional regulation; elevates glycolytic metabolism mediated by AMPK and HIF-1α signaling. The NO-dependent bioenergetic stimulatory effects are critical for tissue repair and other energy demanding conditions, for example inflammation and muscle contraction. In converse, NO can cause energetic suppression by persistent inhibition in mitochondrial respiration and glycolysis which is often observed under high NO flux.
Significance
Up-regulation of NOS2 expression in breast and other tumors has been associated with tumor proliferation, migration, and angiogenesis [255,256], and NOS2 expression is positively correlated with tumor size, decreased tumor differentiation (increasing tumor grade), and poor prognosis in aggressive breast cancers suggesting the NOS2 expression may be an early event, and probably necessary for tumor progression [257–261]. The signaling events by which NO activates and promotes breast tumor progression have been illustrated in breast tumors [262–266]; however, the potential role of NO in breast tumor progression is still under investigation. In addition, accumulating evidence shows that cancers have altered energy metabolism which is essential for tumor growth and survival, and such metabolic modulation is associated with aggressive breast cancers [267–270]. NO is a potent regulator of energy metabolism and NO signaling is intimately associated with cancer progression; so much so that it has recently been suggested to be a positive regulator of the Warburg effect in ovarian cancer {5941}. It is possible that the modulating effects of NO on cancer cell bioenergetics and metabolism can contribute to increased aggressiveness and that iNOS expression may be another diagnostic biomarker that could guide therapeutic strategy.
Figure 1.4. NO signaling and cellular bioenergetics.
Under physiological conditions, NO increases the availability of energy substrates and oxygen to cells and mitochondrial mass through sGC-cGMP signaling. The reversible inhibition of mitochondrial CcO by NO elevates oxidant (ROS) formation resulting in the activation of AMPK and upregulation of glycolysis. Glycolysis is also stimulated by HIF-1α signaling through direct or indirect NO activities. The boost of energetic pathways by NO in physiological conditions plays important roles in cytoprotection and tissue repair. In pathological conditions, usually characterized with high NO flux under inflammatory situations, NO derivatives (NOx) cause the irreversible inhibition of mitochondrial respiration and glycolysis leading to cell death.
Highlights.
We discuss metabolic alterations in cancer cells
We discuss how metabolic alterations can alter cancer aggressiveness
We summarize the chemical biology of Nitric Oxide
We discuss how Nitric Oxide can alter cancer cell metabolism
We link the effects of Nitric Oxide on bioenergetics and metabolism to aggressive cancer growth
Acknowledgments
This research was supported by the Redox Biology Program at the Medical College of Wisconsin (N.H.), the Wisconsin Breast Cancer Showhouse (N.H) an Interdisciplinary Cancer Research Post-Doctoral Fellowship from the Cancer Center of the Medical College of Wisconsin (A.R.D.), and National Institutes of Health grant R01-GM-55792 (N.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference List
- 1.Martin JH, Begum S, Alalami O, Harrison A, Scott KW. Endothelial nitric oxide synthase: correlation with histologic grade, lymph node status and estrogen receptor expression in human breast cancer. Tumour Biol. 2000;21:90–97. doi: 10.1159/000030114. [DOI] [PubMed] [Google Scholar]
- 2.Vakkala M, Paakko P, Soini Y. eNOS expression is associated with the estrogen and progesterone receptor status in invasive breast carcinoma. Int J Oncol. 2000;17:667–671. doi: 10.3892/ijo.17.4.667. [DOI] [PubMed] [Google Scholar]
- 3.DeSantis C, Ma J, Bryan L, Jemal A. Breast cancer statistics, 2013. CA A Cancer Journal for Clinicians. 2013 doi: 10.3322/caac.21203. n/a. [DOI] [PubMed] [Google Scholar]
- 4.Cantor JR, Sabatini DM. Cancer Cell Metabolism: One Hallmark, Many Faces. Cancer Discovery. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 6.Hanahan D, Weinberg RA. The Hallmarks of Cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 7.Hanahan D, Weinberg RA. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 8.Isidoro A, Martinez M, Fernaíndez PL, Ortega AD, Santamaria G, Chamorro M, Reed JC, Cuezva JM. Alteration of the bioenergetic phenotype of mitochondria is a hallmark of breast, gastric, lung and oesophageal cancer. Biochem J. 2004;378:17–20. doi: 10.1042/BJ20031541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Barger JF, Plas DR. Balancing biosynthesis and bioenergetics: metabolic programs in oncogenesis. Endocrine-Related Cancer. 2010;17:R287–R304. doi: 10.1677/ERC-10-0106. [DOI] [PubMed] [Google Scholar]
- 10.Cantor JR, Sabatini DM. Cancer Cell Metabolism: One Hallmark, Many Faces. Cancer Discovery. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ward PS, Thompson CB. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate (abstract) Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Contractor T, Harris CR. p53 Negatively Regulates Transcription of the Pyruvate Dehydrogenase Kinase Pdk2. Cancer Res. 2012;72:560–567. doi: 10.1158/0008-5472.CAN-11-1215. [DOI] [PubMed] [Google Scholar]
- 13.Dang CV. MYC, Metabolism, Cell Growth, and Tumorigenesis. Cold Spring Harbor Perspectives in Medicine. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of Glucose Transporter 1 and Glycolytic Gene Expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 15.Levine AJ, Puzio-Kuter AM. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 16.Ward PS, Thompson CB. Metabolic Reprogramming: A Cancer Hallmark Even Warburg Did Not Anticipate (abstract) Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Otto Warburg FWaEN. The metabolism of tumors in the body. J Gen Physiol 1927. 1927 Mar 7;8(6):519–530. doi: 10.1085/jgp.8.6.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11:325–337. doi: 10.1038/nrc3038. [DOI] [PubMed] [Google Scholar]
- 20.WEINHOUSE SIDN, WARBURG OTTO, BURK DEAN, SCHADE AL. On Respiratory Impairment in Cancer Cells. Science. 1956;124:267–272. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 21.Moreno-Sanchez R, Rodriguez-Enriquez S, Marin-Hernandez A, Saavedra E. Energy metabolism in tumor cells. FEBS Journal. 2007;274:1393–1418. doi: 10.1111/j.1742-4658.2007.05686.x. [DOI] [PubMed] [Google Scholar]
- 22.WANG TING, MARQUARDT CATH, FOKER JOHN. Aerobic glycolysis during lymphocyte proliferation. Nature. 1976;261:702–705. doi: 10.1038/261702a0. [DOI] [PubMed] [Google Scholar]
- 23.Hedeskov CJ. Early effects of phytohaemagglutinin on glucose metabolism of normal human lymphocytes. Biochem J. 1968;110:373–380. doi: 10.1042/bj1100373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brand K. Glutamine and glucose metabolism during thymocyte proliferation. Pathways of glutamine and glutamate metabolism. Biochem J. 1985;228:353–361. doi: 10.1042/bj2280353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vander Heiden MG, Cantley LC, Thompson CB. Understanding the Warburg Effect: The Metabolic Requirements of Cell Proliferation. Science. 2009;324:1029–1033. doi: 10.1126/science.1160809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cantor JR, Sabatini DM. Cancer Cell Metabolism: One Hallmark, Many Faces. Cancer Discovery. 2012;2:881–898. doi: 10.1158/2159-8290.CD-12-0345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dang CV. MYC, Metabolism, Cell Growth, and Tumorigenesis. Cold Spring Harbor Perspectives in Medicine. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Levine AJ, Puzio-Kuter AM. The Control of the Metabolic Switch in Cancers by Oncogenes and Tumor Suppressor Genes. Science. 2010;330:1340–1344. doi: 10.1126/science.1193494. [DOI] [PubMed] [Google Scholar]
- 30.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 31.Ward PS, Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell. 2012;21:297–308. doi: 10.1016/j.ccr.2012.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dhup S, Dadhich RK, Porporato PE, Sonveaux P. Multiple biological activities of lactic acid in cancer: influences on tumor growth, angiogenesis and metastasis. Curr Pharm Des. 2012;18:1319–1330. doi: 10.2174/138161212799504902. [DOI] [PubMed] [Google Scholar]
- 33.Gambhir SS. Molecular imaging of cancer with positron emission tomography. Nat Rev Cancer. 2002;2:683–693. doi: 10.1038/nrc882. [DOI] [PubMed] [Google Scholar]
- 34.Macheda ML, Rogers S, Best JD. Molecular and cellular regulation of glucose transporter (GLUT) proteins in cancer. J Cell Physiol. 2005;202:654–662. doi: 10.1002/jcp.20166. [DOI] [PubMed] [Google Scholar]
- 35.Kennedy KM, Dewhirst MW. Tumor metabolism of lactate: the influence and therapeutic potential for MCT and CD147 regulation. Future Oncology. 2009;6:127–148. doi: 10.2217/fon.09.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Medina RA, Owen GI. Glucose transporters: expression, regulation and cancer. Biol Res. 2002;35:9–26. doi: 10.4067/s0716-97602002000100004. [DOI] [PubMed] [Google Scholar]
- 37.Schwartz MK. Enzymes as prognostic markers and therapeutic indicators in patients with cancer. Clinica Chimica Acta. 1992;206:77–82. doi: 10.1016/0009-8981(92)90008-e. [DOI] [PubMed] [Google Scholar]
- 38.Grover-McKay M, Walsh SA, Seftor EA, Thomas PA, Hendrix MJ. Role for glucose transporter 1 protein in human breast cancer. Pathol Oncol Res. 1998;4:115–120. doi: 10.1007/BF02904704. [DOI] [PubMed] [Google Scholar]
- 39.Wilson JE. Isozymes of mammalian hexokinase: structure, subcellular localization and metabolic function. Journal of Experimental Biology. 2003;206:2049–2057. doi: 10.1242/jeb.00241. [DOI] [PubMed] [Google Scholar]
- 40.Mathupala SP, Ko YH, Pedersen PL. Hexokinase II: Cancer’s double-edged sword acting as both facilitator and gatekeeper of malignancy when bound to mitochondria. Oncogene. 2006;25:4777–4786. doi: 10.1038/sj.onc.1209603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakashima RA, Mangan PS, Colombini M, Pedersen PL. Hexokinase receptor complex in hepatoma mitochondria: evidence from N,N′-dicyclohexlycarbodiimide-labeling studies for the involvement of the pore-forming protein VDAC. Biochemistry. 1986;25:1015–1021. doi: 10.1021/bi00353a010. [DOI] [PubMed] [Google Scholar]
- 42.Bustamante E, Pedersen PL. High aerobic glycolysis of rat hepatoma cells in culture: Role of mitochondrial hexokinase. Proceedings of the National Academy of Sciences. 1977;74:3735–3739. doi: 10.1073/pnas.74.9.3735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Arora KK, Pedersen PL. Functional significance of mitochondrial bound hexokinase in tumor cell metabolism. Evidence for preferential phosphorylation of glucose by intramitochondrially generated ATP. J Biol Chem. 1988;263:17422–17428. [PubMed] [Google Scholar]
- 44.Rose IA, Warms JVB. Stability of hexokinase II in Vitro and in ascites tumor cells. Archives of Biochemistry and Biophysics. 1982;213:625–634. doi: 10.1016/0003-9861(82)90592-6. [DOI] [PubMed] [Google Scholar]
- 45.Pastorino JG, Shulga N, Hoek JB. Mitochondrial Binding of Hexokinase II Inhibits Bax-induced Cytochrome c Release and Apoptosis. J Biol Chem. 2002;277:7610–7618. doi: 10.1074/jbc.M109950200. [DOI] [PubMed] [Google Scholar]
- 46.Vora S, Halper JP, Knowles DM. Alterations in the Activity and Isozymic Profile of Human Phosphofructokinase during Malignant Transformation in Vivo and in Vitro: Transformation- and Progression-linked Discriminants of Malignancy. Cancer Res. 1985;45:2993–3001. [PubMed] [Google Scholar]
- 47.Wang G, Xu Z, Wang C, Yao F, Li J, Chen C, Sun S. Differential phosphofructokinase-1 isoenzyme patterns associated with glycolytic efficiency in human breast cancer and paracancer tissues. Oncol Lett. 2013;6:1701–1706. doi: 10.3892/ol.2013.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vora S, Halper JP, Knowles DM. Alterations in the Activity and Isozymic Profile of Human Phosphofructokinase during Malignant Transformation in Vivo and in Vitro: Transformation- and Progression-linked Discriminants of Malignancy. Cancer Res. 1985;45:2993–3001. [PubMed] [Google Scholar]
- 49.Van Schaftingen E, Jett MF, Hue L, Hers HG. Control of liver 6-phosphofructokinase by fructose 2,6-bisphosphate and other effectors. Proceedings of the National Academy of Sciences. 1981;78:3483–3486. doi: 10.1073/pnas.78.6.3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Okar DA, Lange AJ, Manzano A, Navarro-Sabate A, Riera L, Bartrons R. PFK-2/FBPase-2: maker and breaker of the essential biofactor fructose-2,6-bisphosphate. Trends in Biochemical Sciences. 2001;26:30–35. doi: 10.1016/s0968-0004(00)01699-6. [DOI] [PubMed] [Google Scholar]
- 51.Atsumi T, Chesney J, Metz C, Leng L, Donnelly S, Makita Z, Mitchell R, Bucala R. High Expression of Inducible 6-Phosphofructo-2-Kinase/Fructose-2,6-Bisphosphatase (iPFK-2; PFKFB3) in Human Cancers. Cancer Res. 2002;62:5881–5887. [PubMed] [Google Scholar]
- 52.Calvo MN, Bartrons R, Castano E, Perales JC, Navarro-Sabate A, Manzano A. PFKFB3 gene silencing decreases glycolysis, induces cell-cycle delay and inhibits anchorage-independent growth in HeLa cells. FEBS Letters. 2006;580:3308–3314. doi: 10.1016/j.febslet.2006.04.093. [DOI] [PubMed] [Google Scholar]
- 53.Clem B, Telang S, Clem A, Yalcin A, Meier J, Simmons A, Rasku MA, Arumugam S, Dean WL, Eaton J, Lane A, Trent JO, Chesney J. Small-molecule inhibition of 6-phosphofructo-2-kinase activity suppresses glycolytic flux and tumor growth. Molecular Cancer Therapeutics. 2008;7:110–120. doi: 10.1158/1535-7163.MCT-07-0482. [DOI] [PubMed] [Google Scholar]
- 54.Clem BF, O’Neal J, Tapolsky G, Clem AL, Imbert-Fernandez Y, Kerr DA, Klarer AC, Redman R, Miller DM, Trent JO, Telang S, Chesney J. Targeting 6-phosphofructo-2-kinase (PFKFB3) as a therapeutic strategy against cancer. Mol Cancer Ther. 2013;12:1461–1470. doi: 10.1158/1535-7163.MCT-13-0097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Seminars in Cancer Biology. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 56.Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, Fleming MD, Schreiber SL, Cantley LC. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. 2008;452:230–233. doi: 10.1038/nature06734. [DOI] [PubMed] [Google Scholar]
- 57.Cortes-Cros M, Hemmerlin C, Ferretti S, Zhang J, Gounarides JS, Yin H, Muller A, Haberkorn A, Chene P, Sellers WR, Hofmann F. M2 isoform of pyruvate kinase is dispensable for tumor maintenance and growth. Proceedings of the National Academy of Sciences. 2013;110:489–494. doi: 10.1073/pnas.1212780110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Israelsen WJ, Dayton TL, Davidson SM, Fiske BP, Hosios AM, Bellinger G, Li J, Yu Y, Sasaki M, Horner JW, Burga LN, Xie J, Jurczak MJ, DePinho RA, Clish CB, Jacks T, Kibbey RG, Wulf GM, Di Vizio D, Mills GB, Cantley LC, Vander Heiden MG. PKM2 Isoform-Specific Deletion Reveals a Differential Requirement for Pyruvate Kinase in Tumor Cells. Cell. 2013;155:397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang Jk, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Cellular Antioxidant Responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen EI, Hewel J, Krueger JS, Tiraby C, Weber MR, Kralli A, Becker K, Yates JR, Felding-Habermann B. Adaptation of Energy Metabolism in Breast Cancer Brain Metastases. Cancer Res. 2007;67:1472–1486. doi: 10.1158/0008-5472.CAN-06-3137. [DOI] [PubMed] [Google Scholar]
- 61.Schafer ZT, Grassian AR, Song L, Jiang Z, Gerhart-Hines Z, Irie HY, Gao S, Puigserver P, Brugge JS. Antioxidant and oncogene rescue of metabolic defects caused by loss of matrix attachment. Nature. 2009;461:109–113. doi: 10.1038/nature08268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Boada J, Roig T, Perez X, Gamez A, Bartrons R, Cascante M, Bermudez J. Cells overexpressing fructose-2,6-bisphosphatase showed enhanced pentose phosphate pathway flux and resistance to oxidative stress. FEBS Letters. 2000;480:261–264. doi: 10.1016/s0014-5793(00)01950-5. [DOI] [PubMed] [Google Scholar]
- 63.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang Jk, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Cellular Antioxidant Responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Seminars in Cancer Biology. 2005;15:300–308. doi: 10.1016/j.semcancer.2005.04.009. [DOI] [PubMed] [Google Scholar]
- 65.Milgraum LZ, Witters LA, Pasternack GR, Kuhajda FP. Enzymes of the fatty acid synthesis pathway are highly expressed in in situ breast carcinoma. Clin Cancer Res. 1997;3:2115–2120. [PubMed] [Google Scholar]
- 66.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 67.Souba WW. Glutamine and cancer. Ann Surg. 1993;218:715–728. doi: 10.1097/00000658-199312000-00004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Medina MA. Glutamine and Cancer. The Journal of Nutrition. 2001;131:2539S–2542S. doi: 10.1093/jn/131.9.2539S. [DOI] [PubMed] [Google Scholar]
- 69.DeBerardinis RJ, Mancuso A, Daikhin E, Nissim I, Yudkoff M, Wehrli S, Thompson CB. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proceedings of the National Academy of Sciences. 2007;104:19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, Fulton LA, Locke DP, Magrini VJ, Abbott RM, Vickery TL, Reed JS, Robinson JS, Wylie T, Smith SM, Carmichael L, Eldred JM, Harris CC, Walker J, Peck JB, Du F, Dukes AF, Sanderson GE, Brummett AM, Clark E, McMichael JF, Meyer RJ, Schindler JK, Pohl CS, Wallis JW, Shi X, Lin L, Schmidt H, Tang Y, Haipek C, Wiechert ME, Ivy JV, Kalicki J, Elliott G, Ries RE, Payton JE, Westervelt P, Tomasson MH, Watson MA, Baty J, Heath S, Shannon WD, Nagarajan R, Link DC, Walter MJ, Graubert TA, DiPersio JF, Wilson RK, Ley TJ. Recurring Mutations Found by Sequencing an Acute Myeloid Leukemia Genome. N Engl J Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yan H, Parsons DW, Jin G, McLendon R, Rasheed BA, Yuan W, Kos I, Batinic-Haberle I, Jones S, Riggins GJ, Friedman H, Friedman A, Reardon D, Herndon J, Kinzler KW, Velculescu VE, Vogelstein B, Bigner DD. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, Fantin VR, Jang HG, Jin S, Keenan MC, Marks KM, Prins RM, Ward PS, Yen KE, Liau LM, Rabinowitz JD, Cantley LC, Thompson CB, Vander Heiden MG, Su SM. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, Rao A. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim SH, Ito S, Yang C, Wang P, Xiao MT, Liu Lx, Jiang Wq, Liu J, Zhang Jy, Wang B, Frye S, Zhang Y, Xu Yh, Lei Qy, Guan KL, Zhao Sm, Xiong Y. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of alpha-Ketoglutarate-Dependent Dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Figueroa ME, bdel-Wahab O, Lu C, Ward PS, Patel J, Shih A, Li Y, Bhagwat N, Vasanthakumar A, Fernandez HF, Tallman MS, Sun Z, Wolniak K, Peeters JK, Liu W, Choe SE, Fantin VR, Paietta E, Lowenberg B, Licht JD, Godley LA, Delwel R, Valk PJM, Thompson CB, Levine RL, Melnick A. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prensner JR, Chinnaiyan AM. Metabolism unhinged: IDH mutations in cancer. Nat Med. 2011;17:291–293. doi: 10.1038/nm0311-291. [DOI] [PubMed] [Google Scholar]
- 77.Mullen AR, Wheaton WW, Jin ES, Chen PH, Sullivan LB, Cheng T, Yang Y, Linehan WM, Chandel NS, DeBerardinis RJ. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2012;481:385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wise DR, Ward PS, Shay JE, Cross JR, Gruber JJ, Sachdeva UM, Platt JM, DeMatteo RG, Simon MC, Thompson CB. Hypoxia promotes isocitrate dehydrogenase-dependent carboxylation of alpha-ketoglutarate to citrate to support cell growth and viability. Proc Natl Acad Sci U S A. 2011;108:19611–19616. doi: 10.1073/pnas.1117773108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Mullen AR, Hu Z, Shi X, Jiang L, Boroughs LK, Kovacs Z, Boriack R, Rakheja D, Sullivan LB, Linehan WM, Chandel NS, DeBerardinis RJ. Oxidation of alpha-ketoglutarate is required for reductive carboxylation in cancer cells with mitochondrial defects. Cell Rep. 2014;7:1679–1690. doi: 10.1016/j.celrep.2014.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Luo J, Manning BD, Cantley LC. Targeting the PI3K-Akt pathway in human cancer: Rationale and promise. Cancer Cell. 2003;4:257–262. doi: 10.1016/s1535-6108(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 81.Shaw RJ, Cantley LC. Ras, PI(3)K and mTOR signalling controls tumour cell growth. Nature. 2006;441:424–430. doi: 10.1038/nature04869. [DOI] [PubMed] [Google Scholar]
- 82.Yuan TL, Cantley LC. PI3K pathway alterations in cancer: variations on a theme. Oncogene. 2008;27:5497–5510. doi: 10.1038/onc.2008.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Elstrom RL, Bauer DE, Buzzai M, Karnauskas R, Harris MH, Plas DR, Zhuang H, Cinalli RM, Alavi A, Rudin CM, Thompson CB. Akt Stimulates Aerobic Glycolysis in Cancer Cells. Cancer Res. 2004;64:3892–3899. doi: 10.1158/0008-5472.CAN-03-2904. [DOI] [PubMed] [Google Scholar]
- 84.Deprez J, Vertommen D, Alessi DR, Hue L, Rider MH. Phosphorylation and Activation of Heart 6-Phosphofructo-2-kinase by Protein Kinase B and Other Protein Kinases of the Insulin Signaling Cascades. J Biol Chem. 1997;272:17269–17275. doi: 10.1074/jbc.272.28.17269. [DOI] [PubMed] [Google Scholar]
- 85.Gottlob K, Majewski N, Kennedy S, Kandel E, Robey RB, Hay N. Inhibition of early apoptotic events by Akt/PKB is dependent on the first committed step of glycolysis and mitochondrial hexokinase. Genes & Development. 2001;15:1406–1418. doi: 10.1101/gad.889901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) Signaling Increases the Rate of Hypoxia-Inducible Factor 1alpha (HIF-1alpha) Synthesis: Novel Mechanism for HIF-1-Mediated Vascular Endothelial Growth Factor Expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kuhajda FP. Fatty-acid synthase and human cancer: new perspectives on its role in tumor biology. Nutrition. 2000;16:202–208. doi: 10.1016/s0899-9007(99)00266-x. [DOI] [PubMed] [Google Scholar]
- 88.Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer. 2007;7:763–777. doi: 10.1038/nrc2222. [DOI] [PubMed] [Google Scholar]
- 89.Yoon S, Lee MY, Park SW, Moon JS, Koh YK, Ahn YH, Park BW, Kim KS. Up-regulation of Acetyl-CoA Carboxylase alpha and Fatty Acid Synthase by Human Epidermal Growth Factor Receptor 2 at the Translational Level in Breast Cancer Cells. J Biol Chem. 2007;282:26122–26131. doi: 10.1074/jbc.M702854200. [DOI] [PubMed] [Google Scholar]
- 90.Van de Sande T, De Schrijver E, Heyns W, Verhoeven G, Swinnen JV. Role of the Phosphatidylinositol 3-Kinase/PTEN/Akt Kinase Pathway in the Overexpression of Fatty Acid Synthase in LNCaP Prostate Cancer Cells. Cancer Res. 2002;62:642–646. [PubMed] [Google Scholar]
- 91.Yang YA, Han WF, Morin PJ, Chrest FJ, Pizer ES. Activation of Fatty Acid Synthesis during Neoplastic Transformation: Role of Mitogen-Activated Protein Kinase and Phosphatidylinositol 3-Kinase. Experimental Cell Research. 2002;279:80–90. doi: 10.1006/excr.2002.5600. [DOI] [PubMed] [Google Scholar]
- 92.Lupu R, Menendez JA. Targeting Fatty Acid Synthase in Breast and Endometrial Cancer: An Alternative to Selective Estrogen Receptor Modulators? Endocrinology. 2006;147:4056–4066. doi: 10.1210/en.2006-0486. [DOI] [PubMed] [Google Scholar]
- 93.Berwick DC, Hers I, Heesom KJ, Moule SK, Tavar+¬ JM. The Identification of ATP-citrate Lyase as a Protein Kinase B (Akt) Substrate in Primary Adipocytes. J Biol Chem. 2002;277:33895–33900. doi: 10.1074/jbc.M204681200. [DOI] [PubMed] [Google Scholar]
- 94.Bauer DE, Hatzivassiliou G, Zhao F, Andreadis C, Thompson CB. ATP citrate lyase is an important component of cell growth and transformation. Oncogene. 2005;24:6314–6322. doi: 10.1038/sj.onc.1208773. [DOI] [PubMed] [Google Scholar]
- 95.Zhong H, De Marzo AM, Laughner E, Lim M, Hilton DA, Zagzag D, Buechler P, Isaacs WB, Semenza GL, Simons JW. Overexpression of Hypoxia-inducible Factor 1alpha in Common Human Cancers and Their Metastases. Cancer Res. 1999;59:5830–5835. [PubMed] [Google Scholar]
- 96.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Current Opinion in Genetics & Development. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bruick RK, McKnight SL. A Conserved Family of Prolyl-4-Hydroxylases That Modify HIF. Science. 2001;294:1337–1340. doi: 10.1126/science.1066373. [DOI] [PubMed] [Google Scholar]
- 98.Kaelin J, Ratcliffe PJ. Oxygen Sensing by Metazoans: The Central Role of the HIF Hydroxylase Pathway. Molecular Cell. 2008;30:393–402. doi: 10.1016/j.molcel.2008.04.009. [DOI] [PubMed] [Google Scholar]
- 99.Semenza GL, Roth PH, Fang HM, Wang GL. Transcriptional regulation of genes encoding glycolytic enzymes by hypoxia-inducible factor 1. J Biol Chem. 1994;269:23757–23763. [PubMed] [Google Scholar]
- 100.Xia X, Lemieux ME, Li W, Carroll JS, Brown M, Liu XS, Kung AL. Integrative analysis of HIF binding and transactivation reveals its role in maintaining histone methylation homeostasis. Proceedings of the National Academy of Sciences. 2009;106:4260–4265. doi: 10.1073/pnas.0810067106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Mole DR, Blancher C, Copley RR, Pollard PJ, Gleadle JM, Ragoussis J, Ratcliffe PJ. Genome-wide Association of Hypoxia-inducible Factor (HIF)-1alpha and HIF-2alpha DNA Binding with Expression Profiling of Hypoxia-inducible Transcripts. J Biol Chem. 2009;284:16767–16775. doi: 10.1074/jbc.M901790200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Robey Ian F, Lien Anthony D, Welsh Sarah J, Baggett Brenda, Gillies Robert J. Hypoxia-inducible factor-1alpha and the glycolytic phenotype in tumors. Neoplasia. 2005;7:324–330. doi: 10.1593/neo.04430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Ullah MS, Davies AJ, Halestrap AP. The Plasma Membrane Lactate Transporter MCT4, but Not MCT1, Is Up-regulated by Hypoxia through a HIF-1alpha-dependent Mechanism. J Biol Chem. 2006;281:9030–9037. doi: 10.1074/jbc.M511397200. [DOI] [PubMed] [Google Scholar]
- 104.Kim Jw, Tchernyshyov I, Semenza GL, Dang CV. HIF-1-mediated expression of pyruvate dehydrogenase kinase: A metabolic switch required for cellular adaptation to hypoxia. Cell Metabolism. 2006;3:177–185. doi: 10.1016/j.cmet.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 105.Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, Pan Y, Simon MC, Thompson CB, Gottlieb E. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIFalpha prolyl hydroxylase. Cancer Cell. 2005;7:77–85. doi: 10.1016/j.ccr.2004.11.022. [DOI] [PubMed] [Google Scholar]
- 106.Isaacs JS, Jung YJ, Mole DR, Lee S, Torres-Cabala C, Chung YL, Merino M, Trepel J, Zbar B, Toro J, Ratcliffe PJ, Linehan WM, Neckers L. HIF overexpression correlates with biallelic loss of fumarate hydratase in renal cancer: Novel role of fumarate in regulation of HIF stability. Cancer Cell. 2005;8:143–153. doi: 10.1016/j.ccr.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 107.Sullivan LB, Martinez-Garcia E, Nguyen H, Mullen AR, Dufour E, Sudarshan S, Licht JD, DeBerardinis RJ, Chandel NS. The proto-oncometabolite fumarate binds glutathione to amplify ROS-dependent signaling. Mol Cell. 2013;51:236–248. doi: 10.1016/j.molcel.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhao S, Lin Y, Xu W, Jiang W, Zha Z, Wang P, Yu W, Li Z, Gong L, Peng Y, Ding J, Lei Q, Guan KL, Xiong Y. Glioma-Derived Mutations in IDH1 Dominantly Inhibit IDH1 Catalytic Activity and Induce HIF-1alpha. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Sudarshan S, Sourbier C, Kong HS, Block K, Romero VAV, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, Lee MJ, Gourlay CW, Trepel J, Linehan WM, Neckers L. Fumarate Hydratase Deficiency in Renal Cancer Induces Glycolytic Addiction and Hypoxia-Inducible Transcription Factor 1alpha Stabilization by Glucose-Dependent Generation of Reactive Oxygen Species. Mol Cell Biol. 2009;29:4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Sun W, Zhou S, Chang SS, McFate T, Verma A, Califano JA. Mitochondrial Mutations Contribute to HIF1alpha Accumulation via Increased Reactive Oxygen Species and Up-regulated Pyruvate Dehydrogenease Kinase 2 in Head and Neck Squamous Cell Carcinoma. Clin Cancer Res. 2009;15:476–484. doi: 10.1158/1078-0432.CCR-08-0930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Sudarshan S, Sourbier C, Kong HS, Block K, Romero VAV, Yang Y, Galindo C, Mollapour M, Scroggins B, Goode N, Lee MJ, Gourlay CW, Trepel J, Linehan WM, Neckers L. Fumarate Hydratase Deficiency in Renal Cancer Induces Glycolytic Addiction and Hypoxia-Inducible Transcription Factor 1alpha Stabilization by Glucose-Dependent Generation of Reactive Oxygen Species. Mol Cell Biol. 2009;29:4080–4090. doi: 10.1128/MCB.00483-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Brune B, Zhou J. Nitric oxide and superoxide: Interference with hypoxic signaling. Cardiovascular Research. 2007;75:275–282. doi: 10.1016/j.cardiores.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 113.Laughner E, Taghavi P, Chiles K, Mahon PC, Semenza GL. HER2 (neu) Signaling Increases the Rate of Hypoxia-Inducible Factor 1alpha (HIF-1alpha) Synthesis: Novel Mechanism for HIF-1-Mediated Vascular Endothelial Growth Factor Expression. Mol Cell Biol. 2001;21:3995–4004. doi: 10.1128/MCB.21.12.3995-4004.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Dang CV. MYC, Metabolism, Cell Growth, and Tumorigenesis. Cold Spring Harbor Perspectives in Medicine. 2013;3 doi: 10.1101/cshperspect.a014217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Conacci-Sorrell M, McFerrin L, Eisenman RN. An Overview of MYC and Its Interactome. Cold Spring Harbor Perspectives in Medicine. 2014;4 doi: 10.1101/cshperspect.a014357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Osthus RC, Shim H, Kim S, Li Q, Reddy R, Mukherjee M, Xu Y, Wonsey D, Lee LA, Dang CV. Deregulation of Glucose Transporter 1 and Glycolytic Gene Expression by c-Myc. J Biol Chem. 2000;275:21797–21800. doi: 10.1074/jbc.C000023200. [DOI] [PubMed] [Google Scholar]
- 117.David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Anastasiou D, Poulogiannis G, Asara JM, Boxer MB, Jiang Jk, Shen M, Bellinger G, Sasaki AT, Locasale JW, Auld DS, Thomas CJ, Vander Heiden MG, Cantley LC. Inhibition of Pyruvate Kinase M2 by Reactive Oxygen Species Contributes to Cellular Antioxidant Responses. Science. 2011;334:1278–1283. doi: 10.1126/science.1211485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, Nissim I, Daikhin E, Yudkoff M, McMahon SB, Thompson CB. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proceedings of the National Academy of Sciences. 2008;105:18782–18787. doi: 10.1073/pnas.0810199105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zhang H, Gao P, Fukuda R, Kumar G, Krishnamachary B, Zeller KI, Dang C, Semenza GL. HIF-1 Inhibits Mitochondrial Biogenesis and Cellular Respiration in VHL-Deficient Renal Cell Carcinoma by Repression of C-MYC Activity. Cancer Cell. 2007;11:407–420. doi: 10.1016/j.ccr.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 121.Kim J, Lee Jh, Iyer VR. Global Identification of Myc Target Genes Reveals Its Direct Role in Mitochondrial Biogenesis and Its E-Box Usage In Vivo. PLoS ONE. 2008;3:e1798. doi: 10.1371/journal.pone.0001798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Li F, Wang Y, Zeller KI, Potter JJ, Wonsey DR, O’Donnell KA, Kim Jw, Yustein JT, Lee LA, Dang CV. Myc Stimulates Nuclearly Encoded Mitochondrial Genes and Mitochondrial Biogenesis. Mol Cell Biol. 2005;25:6225–6234. doi: 10.1128/MCB.25.14.6225-6234.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Berkers C, Maddocks O, Cheung E, Mor I, Vousden K. Metabolic Regulation by p53 Family Members (abstract) Cell Metabolism. 2013;18:617–633. doi: 10.1016/j.cmet.2013.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Schwartzenberg-Bar-Yoseph F, Armoni M, Karnieli E. The Tumor Suppressor p53 Down-Regulates Glucose Transporters GLUT1 and GLUT4 Gene Expression. Cancer Res. 2004;64:2627–2633. doi: 10.1158/0008-5472.can-03-0846. [DOI] [PubMed] [Google Scholar]
- 125.Kondoh H, Lleonart ME, Gil J, Wang J, Degan P, Peters G, Martinez D, Carnero A, Beach D. Glycolytic Enzymes Can Modulate Cellular Life Span. Cancer Res. 2005;65:177–185. [PubMed] [Google Scholar]
- 126.Bensaad K, Tsuruta A, Selak MA, Vidal MNC, Nakano K, Bartrons R, Gottlieb E, Vousden KH. TIGAR, a p53-Inducible Regulator of Glycolysis and Apoptosis. Cell. 2006;126:107–120. doi: 10.1016/j.cell.2006.05.036. [DOI] [PubMed] [Google Scholar]
- 127.Li H, Jogl G. Structural and Biochemical Studies of TIGAR (TP53-induced Glycolysis and Apoptosis Regulator) J Biol Chem. 2009;284:1748–1754. doi: 10.1074/jbc.M807821200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Matoba S, Kang JG, Patino WD, Wragg A, Boehm M, Gavrilova O, Hurley PJ, Bunz F, Hwang PM. p53 Regulates Mitochondrial Respiration. Science. 2006;312:1650–1653. doi: 10.1126/science.1126863. [DOI] [PubMed] [Google Scholar]
- 129.Suzuki S, Tanaka T, Poyurovsky MV, Nagano H, Mayama T, Ohkubo S, Lokshin M, Hosokawa H, Nakayama T, Suzuki Y, Sugano S, Sato E, Nagao T, Yokote K, Tatsuno I, Prives C. Phosphate-activated glutaminase (GLS2), a p53-inducible regulator of glutamine metabolism and reactive oxygen species. Proceedings of the National Academy of Sciences. 2010;107:7461–7466. doi: 10.1073/pnas.1002459107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the Yeast Metabolic Cycle: Temporal Compartmentalization of Cellular Processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 131.Yahagi N, Shimano H, Matsuzaka T, Najima Y, Sekiya M, Nakagawa Y, Ide T, Tomita S, Okazaki H, Tamura Y, Iizuka Y, Ohashi K, Gotoda T, Nagai R, Kimura S, Ishibashi S, Osuga Ji, Yamada N. p53 activation in adipocytes of obese mice. J Biol Chem. 2003 doi: 10.1074/jbc.M302364200. [DOI] [PubMed] [Google Scholar]
- 132.Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi: 10.1038/nrc1478. [DOI] [PubMed] [Google Scholar]
- 133.Pervin S, Singh R, Chaudhuri G. Nitric oxide, N[omega]-hydroxy-l-arginine and breast cancer. Nitric Oxide. 2008;19:103–106. doi: 10.1016/j.niox.2008.04.016. [DOI] [PubMed] [Google Scholar]
- 134.Ridnour LA, Thomas DD, Switzer C, Flores-Santana W, Isenberg JS, Ambs S, Roberts DD, Wink DA. Molecular mechanisms for discrete nitric oxide levels in cancer. Nitric Oxide. 2008;19:73–76. doi: 10.1016/j.niox.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: Implications in cellular signaling. Free Radical Biology and Medicine. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ridnour LA, Thomas DD, Donzelli S, Espey MG, Roberts DD, Wink DA, Isenberg JS. The biphasic nature of nitric oxide responses in tumor biology. Antioxid Redox Signal. 2006;8:1329–1337. doi: 10.1089/ars.2006.8.1329. [DOI] [PubMed] [Google Scholar]
- 137.Roberts DD, Isenberg JS, Ridnour LA, Wink DA. Nitric Oxide and Its Gatekeeper Thrombospondin-1 in Tumor Angiogenesis. Clin Cancer Res. 2007;13:795–798. doi: 10.1158/1078-0432.CCR-06-1758. [DOI] [PubMed] [Google Scholar]
- 138.Thomas DD, Espey MG, Ridnour LA, Hofseth LJ, Mancardi D, Harris CC, Wink DA. Hypoxic inducible factor 1a, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Pervin S, Singh R, Hernandez E, Wu G, Chaudhuri G. Nitric Oxide in Physiologic Concentrations Targets the Translational Machinery to Increase the Proliferation of Human Breast Cancer Cells: Involvement of Mammalian Target of Rapamycin/eIF4E Pathway. Cancer Res. 2007;67:289–299. doi: 10.1158/0008-5472.CAN-05-4623. [DOI] [PubMed] [Google Scholar]
- 140.Jadeski LC, Chakraborty C, Lala PK. Nitric oxide-mediated promotion of mammary tumour cell migration requires sequential activation of nitric oxide synthase, guanylate cyclase and mitogen-activated protein kinase. Int J Cancer. 2003;106:496–504. doi: 10.1002/ijc.11268. [DOI] [PubMed] [Google Scholar]
- 141.Sanuphan Arpasinee, Chunhacha Preedakorn, Pongrakhananon Varisa, Chanvorachote Pithi. Long-Term Nitric Oxide Exposure Enhances Lung Cancer Cell Migration. BioMed Research International. 2013;2013 doi: 10.1155/2013/186972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 143.Ridnour LA, Barasch KM, Windhausen AN, Dorsey TH, Lizardo MM, Yfantis HG, Lee DH, Switzer CH, Cheng RYS, Heinecke JL, Brueggemann E, Hines HB, Khanna C, Glynn SA, Ambs S, Wink DA. Nitric Oxide Synthase and Breast Cancer: Role of TIMP-1 in NO-mediated Akt Activation. PLoS ONE. 2012;7:e44081. doi: 10.1371/journal.pone.0044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Switzer C, Cheng R, Ridnour L, Glynn S, Ambs S, Wink D. Ets-1 is a transcriptional mediator of oncogenic nitric oxide signaling in estrogen receptor-negative breast cancer. Breast Cancer Research. 2012;14:R125. doi: 10.1186/bcr3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Switzer CH, Glynn SA, Cheng RYS, Ridnour LA, Green JE, Ambs S, Wink DA. S-Nitrosylation of EGFR and Src Activates an Oncogenic Signaling Network in Human Basal-Like Breast Cancer. Molecular Cancer Research. 2012;10:1203–1215. doi: 10.1158/1541-7786.MCR-12-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Brune B, von Knethen A, Sandau KB. Nitric oxide and its role in apoptosis. European Journal of Pharmacology. 1998;351:261–272. doi: 10.1016/s0014-2999(98)00274-x. [DOI] [PubMed] [Google Scholar]
- 147.Thomas DD, Espey MG, Ridnour LA, Hofseth LJ, Mancardi D, Harris CC, Wink DA. Hypoxic inducible factor 1a, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Ridnour LA, Thomas DD, Mancardi D, Espey MG, Miranda KM, Paolocci N, Feelisch M, Fukuto J, Wink DA. The chemistry of nitrosative stress induced by nitric oxide and reactive nitrogen oxide species. Putting perspective on stressful biological situations. Biological Chemistry. 2004;385:1–10. doi: 10.1515/BC.2004.001. [DOI] [PubMed] [Google Scholar]
- 149.Pervin S, Singh R, Freije WA, Chaudhuri G. MKP-1-Induced Dephosphorylation of Extracellular Signal-Regulated Kinase Is Essential for Triggering Nitric Oxide-Induced Apoptosis in Human Breast Cancer Cell Lines: Implications in Breast Cancer. Cancer Res. 2003;63:8853–8860. [PubMed] [Google Scholar]
- 150.Stuehr DJ, Nathan CF. Nitric oxide. A macrophage product responsible for cytostasis and respiratory inhibition in tumor target cells. J Exp Med. 1989;169:1543–1555. doi: 10.1084/jem.169.5.1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Snyder CM, Shroff EH, Liu J, Chandel NS. Nitric Oxide Induces Cell Death by Regulating Anti-Apoptotic BCL-2 Family Members. PLoS ONE. 2009;4:e7059. doi: 10.1371/journal.pone.0007059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Lepoivre M, Flaman JM, Bobe P, Lemaire G, Henry Y. Quenching of the tyrosyl free radical of ribonucleotide reductase by nitric oxide. Relationship to cytostasis induced in tumor cells by cytotoxic macrophages. J Biol Chem. 1994;269:21891–21897. [PubMed] [Google Scholar]
- 153.Wink DA, Vodovotz Y, Laval J, Laval F, Dewhirst MW, Mitchell JB. The multifaceted roles of nitric oxide in cancer. Carcinogenesis. 1998;19:711–721. doi: 10.1093/carcin/19.5.711. [DOI] [PubMed] [Google Scholar]
- 154.Thomas DD, Espey MG, Ridnour LA, Hofseth LJ, Mancardi D, Harris CC, Wink DA. Hypoxic inducible factor 1a, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Thomas DD, Ridnour LA, Espey MG, Donzelli S, Ambs S, Hussain SP, Harris CC, DeGraff W, Roberts DD, Mitchell JB, Wink DA. Superoxide Fluxes Limit Nitric Oxide-induced Signaling. J Biol Chem. 2006;281:25984–25993. doi: 10.1074/jbc.M602242200. [DOI] [PubMed] [Google Scholar]
- 156.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: Implications in cellular signaling. Free Radical Biology and Medicine. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Acharya A, Das I, Chandhok D, Saha T. Redox regulation in cancer: a double-edged sword with therapeutic potential. Oxid Med Cell Longev. 2010;3:23–34. doi: 10.4161/oxim.3.1.10095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Brune B. The intimate relation between nitric oxide and superoxide in apoptosis and cell survival. Antioxid Redox Signal. 2005;7:497–507. doi: 10.1089/ars.2005.7.497. [DOI] [PubMed] [Google Scholar]
- 159.Herr B, Zhou J, Drsoe S, Brnne B. The interaction of superoxide with nitric oxide destabilizes hypoxia-inducible factor-1a. Cell Mol Life Sci. 2007;64:3295–3305. doi: 10.1007/s00018-007-7371-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Thomas DD, Ridnour LA, Espey MG, Donzelli S, Ambs S, Hussain SP, Harris CC, DeGraff W, Roberts DD, Mitchell JB, Wink DA. Superoxide Fluxes Limit Nitric Oxide-induced Signaling. J Biol Chem. 2006;281:25984–25993. doi: 10.1074/jbc.M602242200. [DOI] [PubMed] [Google Scholar]
- 161.Brune B. The intimate relation between nitric oxide and superoxide in apoptosis and cell survival. Antioxid Redox Signal. 2005;7:497–507. doi: 10.1089/ars.2005.7.497. [DOI] [PubMed] [Google Scholar]
- 162.Broniowska KA, Diers AR, Corbett JA, Hogg N. Effect of Nitric Oxide on Naphthoquinone Toxicity in Endothelial Cells: Role of Bioenergetic Dysfunction and Poly(ADP-ribose) Polymerase Activation. Biochemistry. 2013;52:4364–4372. doi: 10.1021/bi400342t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Bolanos JP, Almeida A. 5.2 Nitric Oxide in Regulation of Mitochondrial Function, Respiration, and Glycolysis. In: Lajtha A, Gibson G, Dienel G, editors. Handbook of Neurochemistry and Molecular Neurobiology. Springer; US: 2007. pp. 487–517. [Google Scholar]
- 164.Jobgen WS, Fried SK, Fu WJ, Meininger CJ, Wu G. Regulatory role for the arginine nitric oxide pathway in metabolism of energy substrates. The Journal of Nutritional Biochemistry. 2006;17:571–588. doi: 10.1016/j.jnutbio.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 165.Moncada S, Bolanos JP. Nitric oxide, cell bioenergetics and neurodegeneration. Journal of Neurochemistry. 2006;97:1676–1689. doi: 10.1111/j.1471-4159.2006.03988.x. [DOI] [PubMed] [Google Scholar]
- 166.Brown GC. Nitric oxide and mitochondrial respiration. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1999;1411:351–369. doi: 10.1016/s0005-2728(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 167.Brown GC, Borutaite V. Nitric oxide and mitochondrial respiration in the heart. Cardiovascular Research. 2007;75:283–290. doi: 10.1016/j.cardiores.2007.03.022. [DOI] [PubMed] [Google Scholar]
- 168.Haynes V, Traaseth NJ, Elfering S, Fujisawa Y, Giulivi C. Nitration of specific tyrosines in FoF1 ATP synthase and activity loss in aging. American Journal of Physiology - Endocrinology and Metabolism. 2010;298:E978–E987. doi: 10.1152/ajpendo.00739.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proceedings of the National Academy of Sciences. 2001;98:12056–12061. doi: 10.1073/pnas.221269198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Chen CL, Chen J, Rawale S, Varadharaj S, Kaumaya PPT, Zweier JL, Chen YR. Protein Tyrosine Nitration of the Flavin Subunit Is Associated with Oxidative Modification of Mitochondrial Complex II in the Post-ischemic Myocardium. J Biol Chem. 2008;283:27991–28003. doi: 10.1074/jbc.M802691200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Mongin AA, Dohare P, Jourd’heuil D. Selective vulnerability of synaptic signaling and metabolism to nitrosative stress. Antioxid Redox Signal. 2012;17:992–1012. doi: 10.1089/ars.2012.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Liu B, Tewari AK, Zhang L, Green-Church KB, Zweier JL, Chen YR, He G. Proteomic analysis of protein tyrosine nitration after ischemia reperfusion injury: Mitochondria as the major target. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2009;1794:476–485. doi: 10.1016/j.bbapap.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Brown GC. Nitric oxide and mitochondrial respiration. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1999;1411:351–369. doi: 10.1016/s0005-2728(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 174.Brown GC. Nitric oxide regulates mitochondrial respiration and cell functions by inhibiting cytochrome oxidase. FEBS Letters. 1995;369:136–139. doi: 10.1016/0014-5793(95)00763-y. [DOI] [PubMed] [Google Scholar]
- 175.Koivisto A, Matthias A, Bronnikov G, Nedergaard J. Kinetics of the inhibition of mitochondrial respiration by NO. FEBS Letters. 1997;417:75–80. doi: 10.1016/s0014-5793(97)01258-1. [DOI] [PubMed] [Google Scholar]
- 176.Brown GC, Cooper C. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Letters. 1994;356:295–298. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 177.Clementi E, Brown GC, Foxwell N, Moncada S. On the mechanism by which vascular endothelial cells regulate their oxygen consumption. Proceedings of the National Academy of Sciences. 1999;96:1559–1562. doi: 10.1073/pnas.96.4.1559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schweizer M, Richter C. Nitric Oxide Potently and Reversibly Deenergizes Mitochondria at Low Oxygen Tension. Biochemical and Biophysical Research Communications. 1994;204:169–175. doi: 10.1006/bbrc.1994.2441. [DOI] [PubMed] [Google Scholar]
- 179.Mason MG, Nicholls P, Wilson MT, Cooper CE. Nitric oxide inhibition of respiration involves both competitive (heme) and noncompetitive (copper) binding to cytochrome c oxidase. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:708–713. doi: 10.1073/pnas.0506562103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Brown GC, Cooper C. Nanomolar concentrations of nitric oxide reversibly inhibit synaptosomal respiration by competing with oxygen at cytochrome oxidase. FEBS Letters. 1994;356:295–298. doi: 10.1016/0014-5793(94)01290-3. [DOI] [PubMed] [Google Scholar]
- 181.Cleeter MWJ, Cooper JM, rley-Usmar VM, Moncada S, Schapira AHV. Reversible inhibition of cytochrome c oxidase, the terminal enzyme of the mitochondrial respiratory chain, by nitric oxide: Implications for neurodegenerative diseases. FEBS Letters. 1994;345:50–54. doi: 10.1016/0014-5793(94)00424-2. [DOI] [PubMed] [Google Scholar]
- 182.Brown GC. Nitric oxide and mitochondrial respiration. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1999;1411:351–369. doi: 10.1016/s0005-2728(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 183.Palacios-Callender M, Quintero M, Hollis VS, Springett RJ, Moncada S. Endogenous NO regulates superoxide production at low oxygen concentrations by modifying the redox state of cytochrome c oxidase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7630–7635. doi: 10.1073/pnas.0401723101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Koivisto A, Matthias A, Bronnikov G, Nedergaard J. Kinetics of the inhibition of mitochondrial respiration by NO. FEBS Letters. 1997;417:75–80. doi: 10.1016/s0014-5793(97)01258-1. [DOI] [PubMed] [Google Scholar]
- 185.Brookes PS, Kraus DW, Shiva S, Doeller JE, Barone MC, Patel RP, Lancaster JR, Drley-Usmar V. Control of Mitochondrial Respiration by NO., Effects of Low Oxygen and Respiratory State. J Biol Chem. 2003;278:31603–31609. doi: 10.1074/jbc.M211784200. [DOI] [PubMed] [Google Scholar]
- 186.Cooper C. Competitive, Reversible, Physiological? Inhibition of Mitochondrial Cytochrome Oxidase by Nitric Oxide. IUBMB Life. 2003;55:591–597. doi: 10.1080/15216540310001628663. [DOI] [PubMed] [Google Scholar]
- 187.Zhang J, Jin B, Li L, Block ER, Patel JM. Nitric oxide-induced persistent inhibition and nitrosylation of active site cysteine residues of mitochondrial cytochrome-c oxidase in lung endothelial cells. American Journal of Physiology - Cell Physiology. 2005;288:C840–C849. doi: 10.1152/ajpcell.00325.2004. [DOI] [PubMed] [Google Scholar]
- 188.Dranka BP, Hill BG, Darley-Usmar VM. Mitochondrial reserve capacity in endothelial cells: The impact of nitric oxide and reactive oxygen species. Free Radical Biology and Medicine. 2010;48:905–914. doi: 10.1016/j.freeradbiomed.2010.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Ramachandran A, Ceaser E, rley-Usmar VM. Chronic exposure to nitric oxide alters the free iron pool in endothelial cells: Role of mitochondrial respiratory complexes and heat shock proteins. Proceedings of the National Academy of Sciences. 2004;101:384–389. doi: 10.1073/pnas.0304653101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J. 2006;394:627–634. doi: 10.1042/BJ20051435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Clementi E, Brown GC, Feelisch M, Moncada S. Persistent inhibition of cell respiration by nitric oxide: Crucial role of S-nitrosylation of mitochondrial complex I and protective action of glutathione. Proceedings of the National Academy of Sciences of the United States of America. 1998;95:7631–7636. doi: 10.1073/pnas.95.13.7631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Yamamoto T, Maruyama W, Kato Y, Yi H, Shamoto-Nagai M, Tanaka M, Sato Y, Naoi M. Selective nitration of mitochondrial complex I by peroxynitrite: involvement in mitochondria dysfunction and cell death of dopaminergic SH-SY5Y cells. J Neural Transm. 2002;109:1–13. doi: 10.1007/s702-002-8232-1. [DOI] [PubMed] [Google Scholar]
- 193.Riobo NA, Clementi E, Melani M, Boveris A, Cadenas E, Moncada S, Poderoso JJ. Nitric oxide inhibits mitochondrial NADH:ubiquinone reductase activity through peroxynitrite formation. Biochem J. 2001;359:139–145. doi: 10.1042/0264-6021:3590139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Frost MT, Wang Q, Moncada S, Singer M. Hypoxia accelerates nitric oxide-dependent inhibition of mitochondrial complex I in activated macrophages. American Journal of Physiology - Regulatory, Integrative and Comparative Physiology. 2005;288:R394–R400. doi: 10.1152/ajpregu.00504.2004. [DOI] [PubMed] [Google Scholar]
- 195.Welter R, Yu L, Yu CA. The Effects of Nitric Oxide on Electron Transport Complexes. Archives of Biochemistry and Biophysics. 1996;331:9–14. doi: 10.1006/abbi.1996.0276. [DOI] [PubMed] [Google Scholar]
- 196.Poderoso JJ, Carreras M, Lisdero C, Riob+¦ N, Sch+¦pfer F, Boveris A. Nitric Oxide Inhibits Electron Transfer and Increases Superoxide Radical Production in Rat Heart Mitochondria and Submitochondrial Particles. Archives of Biochemistry and Biophysics. 1996;328:85–92. doi: 10.1006/abbi.1996.0146. [DOI] [PubMed] [Google Scholar]
- 197.Welter R, Yu L, Yu CA. The Effects of Nitric Oxide on Electron Transport Complexes. Archives of Biochemistry and Biophysics. 1996;331:9–14. doi: 10.1006/abbi.1996.0276. [DOI] [PubMed] [Google Scholar]
- 198.Brown GC. Nitric oxide and mitochondrial respiration. Biochimica et Biophysica Acta (BBA) - Bioenergetics. 1999;1411:351–369. doi: 10.1016/s0005-2728(99)00025-0. [DOI] [PubMed] [Google Scholar]
- 199.Haynes V, Traaseth NJ, Elfering S, Fujisawa Y, Giulivi C. Nitration of specific tyrosines in FoF1 ATP synthase and activity loss in aging. American Journal of Physiology - Endocrinology and Metabolism. 2010;298:E978–E987. doi: 10.1152/ajpendo.00739.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Aulak KS, Miyagi M, Yan L, West KA, Massillon D, Crabb JW, Stuehr DJ. Proteomic method identifies proteins nitrated in vivo during inflammatory challenge. Proceedings of the National Academy of Sciences. 2001;98:12056–12061. doi: 10.1073/pnas.221269198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Prime TA, Blaikie FH, Evans C, Nadtochiy SM, James AM, Dahm CC, Vitturi DA, Patel RP, Hiley CR, Abakumova I, Requejo R, Chouchani ET, Hurd TR, Garvey JF, Taylor CT, Brookes PS, Smith RAJ, Murphy MP. A mitochondria-targeted S-nitrosothiol modulates respiration, nitrosates thiols, and protects against ischemia-reperfusion injury. Proceedings of the National Academy of Sciences. 2009;106:10764–10769. doi: 10.1073/pnas.0903250106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Shiva S, Sack MN, Greer JJ, Duranski M, Ringwood LA, Burwell L, Wang X, MacArthur PH, Shoja A, Raghavachari N, Calvert JW, Brookes PS, Lefer DJ, Gladwin MT. Nitrite augments tolerance to ischemia/reperfusion injury via the modulation of mitochondrial electron transfer. The Journal of Experimental Medicine. 2007;204:2089–2102. doi: 10.1084/jem.20070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Richards EM, Rosenthal RE, Kristian T, Fiskum G. Postischemic hyperoxia reduces hippocampal pyruvate dehydrogenase activity. Free Radical Biology and Medicine. 2006;40:1960–1970. doi: 10.1016/j.freeradbiomed.2006.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 204.Drapier JC, Hibbs JB., Jr Murine cytotoxic activated macrophages inhibit aconitase in tumor cells. Inhibition involves the iron-sulfur prosthetic group and is reversible. J Clin Invest. 1986;78:790–797. doi: 10.1172/JCI112642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Castro L, Rodriguez M, Radi R. Aconitase is readily inactivated by peroxynitrite, but not by its precursor, nitric oxide. J Biol Chem. 1994;269:29409–29415. [PubMed] [Google Scholar]
- 206.Tortora V, Quijano C, Freeman B, Radi R, Castro L. Mitochondrial aconitase reaction with nitric oxide, S-nitrosoglutathione, and peroxynitrite: Mechanisms and relative contributions to aconitase inactivation. Free Radical Biology and Medicine. 2007;42:1075–1088. doi: 10.1016/j.freeradbiomed.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 207.Castro L, Rodriguez M, Radi R. Aconitase is readily inactivated by peroxynitrite, but not by its precursor, nitric oxide. J Biol Chem. 1994;269:29409–29415. [PubMed] [Google Scholar]
- 208.Tortora V, Quijano C, Freeman B, Radi R, Castro L. Mitochondrial aconitase reaction with nitric oxide, S-nitrosoglutathione, and peroxynitrite: Mechanisms and relative contributions to aconitase inactivation. Free Radical Biology and Medicine. 2007;42:1075–1088. doi: 10.1016/j.freeradbiomed.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 209.Han D, Canali R, Garcia J, Aguilera R, Gallaher TK, Cadenas E. Sites and Mechanisms of Aconitase Inactivation by Peroxynitrite: Modulation by Citrate and Glutathione. Biochemistry. 2005;44:11986–11996. doi: 10.1021/bi0509393. [DOI] [PubMed] [Google Scholar]
- 210.Castro L, Demicheli V, Tortora V, Radi R. Mitochondrial protein tyrosine nitration. Free Radical Research. 2010;45:37–52. doi: 10.3109/10715762.2010.516254. [DOI] [PubMed] [Google Scholar]
- 211.Liu B, Tewari AK, Zhang L, Green-Church KB, Zweier JL, Chen YR, He G. Proteomic analysis of protein tyrosine nitration after ischemia reperfusion injury: Mitochondria as the major target. Biochimica et Biophysica Acta (BBA) - Proteins and Proteomics. 2009;1794:476–485. doi: 10.1016/j.bbapap.2008.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Tortora V, Quijano C, Freeman B, Radi R, Castro L. Mitochondrial aconitase reaction with nitric oxide, S-nitrosoglutathione, and peroxynitrite: Mechanisms and relative contributions to aconitase inactivation. Free Radical Biology and Medicine. 2007;42:1075–1088. doi: 10.1016/j.freeradbiomed.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 213.Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RAJ, Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Yang ES, Richter C, Chun JS, Huh TL, Kang SS, Park JW. Inactivation of NADP+-dependent isocitrate dehydrogenase by nitric oxide. Free Radical Biology and Medicine. 2002;33:927–937. doi: 10.1016/s0891-5849(02)00981-4. [DOI] [PubMed] [Google Scholar]
- 215.Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RAJ, Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J. 2010;430:49–59. doi: 10.1042/BJ20100633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Shi Q, Xu H, Yu H, Zhang N, Ye Y, Estevez AG, Deng H, Gibson GE. Inactivation and Reactivation of the Mitochondrial alpha-Ketoglutarate Dehydrogenase Complex. J Biol Chem. 2011;286:17640–17648. doi: 10.1074/jbc.M110.203018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Piantadosi CA, Suliman HB. Redox regulation of mitochondrial biogenesis. Free Radical Biology and Medicine. 2012;53:2043–2053. doi: 10.1016/j.freeradbiomed.2012.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Bossy-Wetzel E, Lipton SA. Nitric oxide signaling regulates mitochondrial number and function. Cell Death Differ. 2003;10:757–760. doi: 10.1038/sj.cdd.4401244. [DOI] [PubMed] [Google Scholar]
- 219.Nisoli E, Tonello C, Cardile A, Cozzi V, Bracale R, Tedesco L, Falcone S, Valerio A, Cantoni O, Clementi E, Moncada S, Carruba MO. Calorie Restriction Promotes Mitochondrial Biogenesis by Inducing the Expression of eNOS. Science. 2005;310:314–317. doi: 10.1126/science.1117728. [DOI] [PubMed] [Google Scholar]
- 220.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial Biogenesis in Mammals: The Role of Endogenous Nitric Oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 221.Clementi E, Nisoli E. Nitric oxide and mitochondrial biogenesis: A key to long-term regulation of cellular metabolism. Comparative Biochemistry and Physiology Part A: Molecular & Integrative Physiology. 2005;142:102–110. doi: 10.1016/j.cbpb.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 222.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial Biogenesis in Mammals: The Role of Endogenous Nitric Oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 223.Nisoli E, Clementi E, Paolucci C, Cozzi V, Tonello C, Sciorati C, Bracale R, Valerio A, Francolini M, Moncada S, Carruba MO. Mitochondrial Biogenesis in Mammals: The Role of Endogenous Nitric Oxide. Science. 2003;299:896–899. doi: 10.1126/science.1079368. [DOI] [PubMed] [Google Scholar]
- 224.Lira VA, Brown DL, Lira AK, Kavazis AN, Soltow QA, Zeanah EH, Criswell DS. Nitric oxide and AMPK cooperatively regulate PGC-1a in skeletal muscle cells. The Journal of Physiology. 2010;588:3551–3566. doi: 10.1113/jphysiol.2010.194035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.McConell GK, Ng GPY, Phillips M, Ruan Z, Macaulay SL, Wadley GD. Central role of nitric oxide synthase in AICAR and caffeine-induced mitochondrial biogenesis in L6 myocytes. J Appl Physiol. 2010;108:589–595. doi: 10.1152/japplphysiol.00377.2009. [DOI] [PubMed] [Google Scholar]
- 226.Jager S, Handschin C, Pierre J, Spiegelman BM. AMP-activated protein kinase (AMPK) action in skeletal muscle via direct phosphorylation of PGC-1alpha. Proceedings of the National Academy of Sciences. 2007;104:12017–12022. doi: 10.1073/pnas.0705070104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 227.Colombo SL, Moncada S. AMPKalpha1 regulates the antioxidant status of vascular endothelial cells. Biochem J. 2009;421:163–169. doi: 10.1042/BJ20090613. [DOI] [PubMed] [Google Scholar]
- 228.Quintero M, Colombo SL, Godfrey A, Moncada S. Mitochondria as signaling organelles in the vascular endothelium. Proceedings of the National Academy of Sciences. 2006;103:5379–5384. doi: 10.1073/pnas.0601026103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zou MH, Kirkpatrick SS, Davis BJ, Nelson JS, Wiles WG, Schlattner U, Neumann D, Brownlee M, Freeman MB, Goldman MH. Activation of the AMP-activated Protein Kinase by the Anti-diabetic Drug Metformin in Vivo: ROLE OF MITOCHONDRIAL REACTIVE NITROGEN SPECIES. J Biol Chem. 2004;279:43940–43951. doi: 10.1074/jbc.M404421200. [DOI] [PubMed] [Google Scholar]
- 230.Zou MH, Hou XY, Shi CM, Kirkpatick S, Liu F, Goldman MH, Cohen RA. Activation of 5′-AMP-activated Kinase Is Mediated through c-Src and Phosphoinositide 3-Kinase Activity during Hypoxia-Reoxygenation of Bovine Aortic Endothelial Cells: ROLE OF PEROXYNITRITE. J Biol Chem. 2003;278:34003–34010. doi: 10.1074/jbc.M300215200. [DOI] [PubMed] [Google Scholar]
- 231.Palacios-Callender M, Quintero M, Hollis VS, Springett RJ, Moncada S. Endogenous NO regulates superoxide production at low oxygen concentrations by modifying the redox state of cytochrome c oxidase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7630–7635. doi: 10.1073/pnas.0401723101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 232.Cidad P, Almeida A, Bolanos JP. Inhibition of mitochondrial respiration by nitric oxide rapidly stimulates cytoprotective GLUT3-mediated glucose uptake through 5′-AMP-activated protein kinase. Biochem J. 2004;384:629–636. doi: 10.1042/BJ20040886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 233.Almeida A, Moncada S, Bolanos JP. Nitric oxide switches on glycolysis through the AMP protein kinase and 6-phosphofructo-2-kinase pathway. Nat Cell Biol. 2004;6:45–51. doi: 10.1038/ncb1080. [DOI] [PubMed] [Google Scholar]
- 234.Almeida A, Moncada S, Bolanos JP. Nitric oxide switches on glycolysis through the AMP protein kinase and 6-phosphofructo-2-kinase pathway. Nat Cell Biol. 2004;6:45–51. doi: 10.1038/ncb1080. [DOI] [PubMed] [Google Scholar]
- 235.Bergandi L, Silvagno F, Russo I, Riganti C, Anfossi G, Aldieri E, Ghigo D, Trovati M, Bosia A. Insulin Stimulates Glucose Transport Via Nitric Oxide/Cyclic GMP Pathway in Human Vascular Smooth Muscle Cells. Arterioscler Thromb Vasc Biol. 2003;23:2215–2221. doi: 10.1161/01.ATV.0000107028.20478.8e. [DOI] [PubMed] [Google Scholar]
- 236.McConell GK, Wadley GD. POTENTIAL ROLE OF NITRIC OXIDE IN CONTRACTION-STIMULATED GLUCOSE UPTAKE AND MITOCHONDRIAL BIOGENESIS IN SKELETAL MUSCLE. Clinical and Experimental Pharmacology and Physiology. 2008;35:1488–1492. doi: 10.1111/j.1440-1681.2008.05038.x. [DOI] [PubMed] [Google Scholar]
- 237.Hagen T, Taylor CT, Lam F, Moncada S. Redistribution of Intracellular Oxygen in Hypoxia by Nitric Oxide: Effect on HIF1alpha. Science. 2003;302:1975–1978. doi: 10.1126/science.1088805. [DOI] [PubMed] [Google Scholar]
- 238.Quintero M, Brennan PA, Thomas GJ, Moncada S. Nitric Oxide Is a Factor in the Stabilization of Hypoxia-Inducible Factor-1alpha in Cancer: Role of Free Radical Formation. Cancer Res. 2006;66:770–774. doi: 10.1158/0008-5472.CAN-05-0333. [DOI] [PubMed] [Google Scholar]
- 239.Sandau KB, Fandrey J, Brune B. Accumulation of HIF-1alpha under the influence of nitric oxide. Blood. 2001;97:1009–1015. doi: 10.1182/blood.v97.4.1009. [DOI] [PubMed] [Google Scholar]
- 240.Brune B, Zhou J. Nitric oxide and superoxide: Interference with hypoxic signaling. Cardiovascular Research. 2007;75:275–282. doi: 10.1016/j.cardiores.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 241.Semenza GL. HIF-1: upstream and downstream of cancer metabolism. Current Opinion in Genetics & Development. 2010;20:51–56. doi: 10.1016/j.gde.2009.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Quintero M, Brennan PA, Thomas GJ, Moncada S. Nitric Oxide Is a Factor in the Stabilization of Hypoxia-Inducible Factor-1alpha in Cancer: Role of Free Radical Formation. Cancer Res. 2006;66:770–774. doi: 10.1158/0008-5472.CAN-05-0333. [DOI] [PubMed] [Google Scholar]
- 243.Thomas DD, Espey MG, Ridnour LA, Hofseth LJ, Mancardi D, Harris CC, Wink DA. Hypoxic inducible factor 1a, extracellular signal-regulated kinase, and p53 are regulated by distinct threshold concentrations of nitric oxide. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:8894–8899. doi: 10.1073/pnas.0400453101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Brune B, Zhou J. Nitric oxide and superoxide: Interference with hypoxic signaling. Cardiovascular Research. 2007;75:275–282. doi: 10.1016/j.cardiores.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 245.Quintero M, Brennan PA, Thomas GJ, Moncada S. Nitric Oxide Is a Factor in the Stabilization of Hypoxia-Inducible Factor-1alpha in Cancer: Role of Free Radical Formation. Cancer Res. 2006;66:770–774. doi: 10.1158/0008-5472.CAN-05-0333. [DOI] [PubMed] [Google Scholar]
- 246.Chowdhury R, Godoy LC, Thiantanawat A, Trudel LJ, Deen WM, Wogan GN. Nitric Oxide Produced Endogenously Is Responsible for Hypoxia-Induced HIF-1alpha Stabilization in Colon Carcinoma Cells. Chem Res Toxicol. 2012;25:2194–2202. doi: 10.1021/tx300274a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Li F, Sonveaux P, Rabbani ZN, Liu S, Yan B, Huang Q, Vujaskovic Z, Dewhirst M, Li CY. Regulation of HIF-1alpha Stability through S-Nitrosylation. Molecular Cell. 2007;26:63–74. doi: 10.1016/j.molcel.2007.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Brix B, Mesters JR, Pellerin L, Johren O. Endothelial Cell-Derived Nitric Oxide Enhances Aerobic Glycolysis in Astrocytes via HIF-1alpha-Mediated Target Gene Activation. The Journal of Neuroscience. 2012;32:9727–9735. doi: 10.1523/JNEUROSCI.0879-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249.Mohr S, Hallak H, de Boitte A, Lapetina EG, Br++ne B. Nitric Oxide-induced S-Glutathionylation and Inactivation of Glyceraldehyde-3-phosphate Dehydrogenase. J Biol Chem. 1999;274:9427–9430. doi: 10.1074/jbc.274.14.9427. [DOI] [PubMed] [Google Scholar]
- 250.Broniowska KA, Hogg N. Differential mechanisms of inhibition of glyceraldehyde-3-phosphate dehydrogenase by S-nitrosothiols and NO in cellular and cell-free conditions. American Journal of Physiology - Heart and Circulatory Physiology. 2010;299:H1212–H1219. doi: 10.1152/ajpheart.00472.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Buchczyk DP, Grune T, Sies H, Klotz LO. Modifications of glyceraldehyde-3-phosphate dehydrogenase induced by increasing concentrations of peroxynitrite: early recognition by 20S proteasome. Biol Chem. 2003;384:237–241. doi: 10.1515/BC.2003.026. [DOI] [PubMed] [Google Scholar]
- 252.Souza JM, Radi R. Glyceraldehyde-3-Phosphate Dehydrogenase Inactivation by Peroxynitrite. Archives of Biochemistry and Biophysics. 1998;360:187–194. doi: 10.1006/abbi.1998.0932. [DOI] [PubMed] [Google Scholar]
- 253.Sekar Y, Moon TC, Slupsky CM, Befus AD. Protein Tyrosine Nitration of Aldolase in Mast Cells: A Plausible Pathway in Nitric Oxide-Mediated Regulation of Mast Cell Function. The Journal of Immunology. 2010;185:578–587. doi: 10.4049/jimmunol.0902720. [DOI] [PubMed] [Google Scholar]
- 254.Koeck T, Levison B, Hazen SL, Crabb JW, Stuehr DJ, Aulak KS. Tyrosine Nitration Impairs Mammalian Aldolase A Activity. Molecular & Cellular Proteomics. 2004;3:548–557. doi: 10.1074/mcp.M300141-MCP200. [DOI] [PubMed] [Google Scholar]
- 255.Vakkala M, Kahlos K, Lakari E, Paakko P, Kinnula V, Soini Y. Inducible Nitric Oxide Synthase Expression, Apoptosis, and Angiogenesis in in Situ and Invasive Breast Carcinomas. Clin Cancer Res. 2000;6:2408–2416. [PubMed] [Google Scholar]
- 256.Jenkins DC, Charles IG, Thomsen LL, Moss DW, Holmes LS, Baylis SA, Rhodes P, Westmore K, Emson PC, Moncada S. Roles of nitric oxide in tumor growth. Proc Natl Acad Sci U S A. 1995;92:4392–4396. doi: 10.1073/pnas.92.10.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Loibl S, Buck A, Strank C, von Minckwitz G, Roller M, Sinn HP, Schini-Kerth V, Solbach C, Strebhardt K, Kaufmann M. The role of early expression of inducible nitric oxide synthase in human breast cancer. European Journal of Cancer. 2005;41:265–271. doi: 10.1016/j.ejca.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 258.Bulut A, Erden E, Sak S, Doruk H, Kursun N, Dincol D. Significance of inducible nitric oxide synthase expression in benign and malignant breast epithelium: an immunohistochemical study of 151 cases. Virchows Arch. 2005;447:24–30. doi: 10.1007/s00428-005-1250-2. [DOI] [PubMed] [Google Scholar]
- 259.Vakkala M, Kahlos K, Lakari E, Paakko P, Kinnula V, Soini Y. Inducible Nitric Oxide Synthase Expression, Apoptosis, and Angiogenesis in in Situ and Invasive Breast Carcinomas. Clin Cancer Res. 2000;6:2408–2416. [PubMed] [Google Scholar]
- 260.Thomsen LL, Miles DW, Happerfield L, Bobrow LG, Knowles RG, Moncada S. Nitric oxide synthase activity in human breast cancer. Br J Cancer. 1995;72:41–44. doi: 10.1038/bjc.1995.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Glynn SA, Boersma BJ, Dorsey TH, Yi M, Yfantis HG, Ridnour LA, Martin DN, Switzer CH, Hudson RS, Wink DA, Lee DH, Stephens RM, Ambs S. Increased NOS2 predicts poor survival in estrogen receptor-negative breast cancer patients. J Clin Invest. 2010;120:3843–3854. doi: 10.1172/JCI42059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Ridnour LA, Thomas DD, Switzer C, Flores-Santana W, Isenberg JS, Ambs S, Roberts DD, Wink DA. Molecular mechanisms for discrete nitric oxide levels in cancer. Nitric Oxide. 2008;19:73–76. doi: 10.1016/j.niox.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Ridnour LA, Barasch KM, Windhausen AN, Dorsey TH, Lizardo MM, Yfantis HG, Lee DH, Switzer CH, Cheng RYS, Heinecke JL, Brueggemann E, Hines HB, Khanna C, Glynn SA, Ambs S, Wink DA. Nitric Oxide Synthase and Breast Cancer: Role of TIMP-1 in NO-mediated Akt Activation. PLoS ONE. 2012;7:e44081. doi: 10.1371/journal.pone.0044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Roberts DD, Isenberg JS, Ridnour LA, Wink DA. Nitric Oxide and Its Gatekeeper Thrombospondin-1 in Tumor Angiogenesis. Clin Cancer Res. 2007;13:795–798. doi: 10.1158/1078-0432.CCR-06-1758. [DOI] [PubMed] [Google Scholar]
- 265.Switzer CH, Glynn SA, Ridnour LA, Cheng RYS, Vitek MP, Ambs S, Wink DA. Nitric oxide and protein phosphatase 2A provide novel therapeutic opportunities in ER-negative breast cancer. Trends in Pharmacological Sciences. 2011;32:644–651. doi: 10.1016/j.tips.2011.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Thomas DD, Ridnour LA, Isenberg JS, Flores-Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. The chemical biology of nitric oxide: Implications in cellular signaling. Free Radical Biology and Medicine. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267.Gatenby RA, Smallbone K, Maini PK, Rose F, Averill J, Nagle RB, Worrall L, Gillies RJ. Cellular adaptations to hypoxia and acidosis during somatic evolution of breast cancer. Br J Cancer. 2007;97:646–653. doi: 10.1038/sj.bjc.6603922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Isidoro A, Casado E, Redondo A, Acebo P, Espinosa E, Alonso AM, Cejas P, Hardisson D, Fresno Vara JA, Belda-Iniesta C, Gonzaílez-Baron M, Cuezva JM. Breast carcinomas fulfill the Warburg hypothesis and provide metabolic markers of cancer prognosis. Carcinogenesis. 2005;26:2095–2104. doi: 10.1093/carcin/bgi188. [DOI] [PubMed] [Google Scholar]
- 269.Jerby L, Wolf L, Denkert C, Stein GY, Hilvo M, Oresic M, Geiger T, Ruppin E. Metabolic Associations of Reduced Proliferation and Oxidative Stress in Advanced Breast Cancer. Cancer Res. 2012;72:5712–5720. doi: 10.1158/0008-5472.CAN-12-2215. [DOI] [PubMed] [Google Scholar]
- 270.McCleland ML, Adler AS, Shang Y, Hunsaker T, Truong T, Peterson D, Torres E, Li L, Haley B, Stephan JP, Belvin M, Hatzivassiliou G, Blackwood EM, Corson L, Evangelista M, Zha J, Firestein R. An Integrated Genomic Screen Identifies LDHB as an Essential Gene for Triple-Negative Breast Cancer. Cancer Res. 2012;72:5812–5823. doi: 10.1158/0008-5472.CAN-12-1098. [DOI] [PubMed] [Google Scholar]