Summary
Single nucleotide polymorphisms (SNPs) in the vitamin D pathway genes have been implicated in cutaneous melanoma (CM) risk, but their role in CM disease-specific survival (DSS) remains obscure. We conducted comprehensively survival analysis of 2,669 common SNPs in the vitamin D pathway using data from a published genome-wide association study (GWAS) at The University of Texas M.D. Anderson Cancer Center (MDACC), followed by a replication GWAS from the Nurses’ Health Study and Health Professionals Follow-up Study. Among the 2,669 SNPs, 203 were significantly associated with DSS in MDACC dataset (P<0.05 and false positive report probability<0.2), of which 18 were the tag SNPs. In the replication, 2 of these 18 SNPs showed nominal significance: the VDBP rs12512631 T>C was associated with a better DSS [combined hazards ratio (HR)=0.66]; and the same for RXRA rs7850212 C>A (combined HR=0.38). Further bioinformatics analyses indicated that these loci may modulate corresponding gene methylation status.
Keywords: cutaneous melanoma, vitamin D pathway, disease specific survival, single nucleotide polymorphisms, Cox regression
Introduction
Vitamin D is a fat-soluble steroid hormone, 25(OH)D3 is the most accepted measure of the body stores of vitamin D, and 1α,25(OH)2D3 is an active metabolite of vitamin D (Holick, 1996). The first step in the vitamin D synthesis is the formation of vitamin D3 in the skin through stimulation by ultraviolet irradiation. Endogenous or dietary vitamin D3 is hydroxylated by 25OHase (encoded by CYP27A1) to 25(OH)D3 in the liver and then 1α-hydroxylated via 1α-OHase (encoded by CYP27B1) to 1α,25(OH)2D3 in the kidney. The 24-hydroxylation of 25(OH)D3 and 1α,25(OH)2D3 by 24OHase (encoded by CYP24A1) is the rate-limiting step for the vitamin D catabolism (Hewison et al., 2000). The vitamin D-binding protein (DBP) encoded by the VDBP gene facilitates vitamin D actions by carrying vitamin D metabolites to various sites of action, and polymorphic DBP proteins differ in their affinity for 1,25(OH)2D (Pani et al., 2002). Classical action of 1α,25(OH)2D3 is mediated by the binding of vitamin D receptor (VDR)-9-cis-retinoid X receptor (RXR) complex at the vitamin-D response elements (VDREs), resulting in activation of gene expression of specific target genes (Carlberg et al., 1993). Vitamin D may inhibit the MAPK signaling by suppressing the epidermal growth factor receptor (EGFR) pathway and insulin-like growth factor, but it may also promote apoptosis via the IGFR1/PI3K-Akt signaling pathway and by inhibiting telomerase (Deeb et al., 2007). Overall, vitamin D exerts pleiotropic effects in regulating cell proliferation, growth modulation, differentiation, apoptosis and immune modulation, which have an influence on both carcinogenesis of normal cells and metastatic potential of cancer cells (Deeb et al., 2007).
There is a myriad of epidemiological evidence associating vitamin D with mortality rates of several cancers (Chowdhury et al., 2014). For example, higher vitamin D levels were found to be associated with a lower risk of death in patients with colorectal cancer (Ng et al., 2008), prostate cancer (Tretli et al., 2009), and non-small cell lung cancer with stage IB/IIB (Zhou et al., 2007), while a deficient vitamin D status in patients with early breast cancer was associated with a worse disease-free survival, compared with those with an adequate vitamin D status (Goodwin et al., 2009). Preclinical studies also suggested that 1α,25(OH)2D3 potentiates anticancer activity of some chemotherapeutic agents (Hershberger et al., 2002), and attempts have been made to translate these findings to clinical application of 1α,25(OH)2D3 across several tumor types (Fakih et al., 2007; Lappe et al., 2007; Wactawski-Wende et al., 2006).
Tumor Breslow thickness remains the most important single prognostic factor for patients with cutaneous melanoma (CM) (Balch et al., 2009). In general, CM patients with thin tumors have a much longer survival than those with thick tumors (Balch et al., 2009). Serum vitamin D levels have been tested as a potential modifier of CM prognosis associated with Breslow thickness. For example, in 1043 cases from the first Leeds case-control study of CM, a single estimation of serum vitamin D level taken at recruitment was inversely correlated with Breslow thickness (Randerson-Moor et al., 2009). In another study of 271 patients with CM, higher vitamin D levels were found to be associated with lower Breslow thickness at diagnosis and were positively associated with relapse and death (Newton-Bishop et al., 2009). A subsequent small study in patients with stage IV CM reported that lower vitamin D levels were associated with poorer survival (Nurnberg et al., 2009). A more recent study of CM patients in Germany revealed that lower 25(OH)D levels were significantly associated with greater Breslow thickness and later disease stages (Gambichler et al., 2013).
Data on the role of single nucleus polymorphisms (SNPs) in the vitamin D pathway genes in CM outcome are sparse (Davies et al., 2014; Hutchinson et al., 2000; Santonocito et al., 2007; Schafer et al., 2012). Most of the studies used Breslow thickness as a proxy of survival, and the results have been conflicting. In addition, only a handful of SNPs have generally been assessed, as identified by the tagging SNPs in certain genes. Finally, most studies have been performed in single hospital or cancer center, and frequently lack either the scientific rigor or the external validity required to account for widespread changes in practice. To the best of our knowledge, a systematic, multicenter evaluation of genetic variants of vitamin D-related genes and CM specific survival is still lacking. In the present study, to identify prognostic SNPs in the vitamin D pathway, we conducted Cox proportional hazards regression analyses in a previously published large genome-wide association study (GWAS) dataset conducted at the University of M.D. Anderson Cancer Center (MDACC), and subsequently validate the significant SNPs in another GWAS dataset from Harvard University (Amos et al., 2011; Song et al., 2012). Both of these GWASs had available long-term follow-up data. We comprehensively evaluated whether the selected SNPs in CYP24A1, CYP27A1, CYP27B1, CYP2R1, VDR, VDBP, RXRA, RXRB, RXRG, NCOA1, NCOA2, NCOA3, NCOR1 and SNW1 contribute to disease specific survival (DSS) in CM patients. These 14 genes were selected based on their biological roles in the vitamin D metabolism and signaling in cancer (Deeb et al., 2007).
Results
Study populations
Overall, 858 patients from MDACC and 409 patients from Harvard University were included in the analyses (Table 1). All patients with primary CM were non-Hispanic white. Harvard patients had a relatively longer median follow-up time (MFT, 179 months) as compared with MDACC patients (81 months). During the follow-up, there were 95 (11.1%) and 48 (11.5%) patients who died of CM in MDACC and Harvard patients, respectively. Mean ages of MDACC and Harvard patients were 52.4 and 60.1 years old, respectively.
Table 1.
Characteristics of the study populations at the time of analysis.
| Parameter | MDACC
|
Harvard
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Patients | Death (%) | MFT | HR (95%CI)* | P* | Patients | Death (%) | MFT | HR (95%CI)* | P* | |
| Total | 858 | 95 (11.1) | 81.0 | 409 | 48 (11.5) | 179.0 | ||||
| Age (years old) | ||||||||||
| ≤50 | 371 | 31 (8.4) | 85.8 | 1.00 | 72 | 3 (4.2) | 352.5 | 1.00 | ||
| >50 | 487 | 64 (13.1) | 78.1 | 1.69 (1.10 – 2.59) | 0.017 | 337 | 45 (13.4) | 167.0 | 4.04 (1.25 – 13.06) | 0.020 |
| Sex | ||||||||||
| Male | 496 | 69 (13.9) | 77.8 | 1.00 | 138 | 17 (12.3) | 198.0 | 1.00 | ||
| Female | 362 | 26 (7.2) | 85.9 | 0.48 (0.31 – 0.76) | 0.002 | 271 | 31 (11.4) | 155.5 | 1.16 (0.64 – 2.10) | 0.622 |
MDACC = MD Anderson Cancer Center, MFT = median follow-up time (months);
univariate analysis.
Survival-analysis
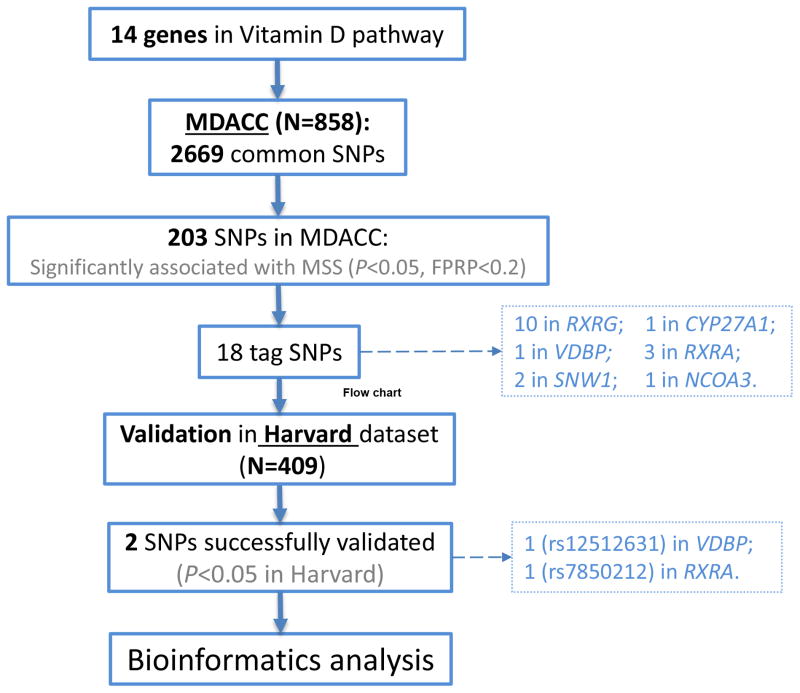
In the discovery analysis of the MDACC dataset for 2,669 SNPs that passed strict quality control measures, we found that 203 SNPs met our selection criteria (i.e., FPRP<0.2; among them, 81 with P< 10−3). As shown in Supplementary Table 1, these 203 SNPs included 111 SNPs of CYP27A1, six of VDBP, two of NCOA3, 20 of RXRA, 61 of RXRG, three of SNW1. Next, with r2>0.6 between SNPs in the same gene as the cut-off value, 18 SNPs were chosen as the tagging SNPs (Table 2 and Supplementary Figure 1).
Table 2.
Survival results of 18 tag SNPs (r-square >0.6) with FPRP < 0.2 in the MDACC dataset and their replication in the Harvard dataset.
| SNP | Gene | Chr | EA | MDACC
|
Harvard
|
Meta
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| MAF | HR (95%CI) | P* | FPRP | MAF | HR (95%CI) | P** | HR(95%CI) | P | I2 | Phet | ||||
| rs6426914 | RXRG | 1 | G | 0.06 | 2.04 (1.18 – 3.53) | 1.10×10−2 | 0.171 | 0.06 | 0.82 (0.32–2.07) | 0.669 | 1.40 (0.58 – 3.30) | 0.453 | 63.60 | 0.099 |
| rs283695 | RXRG | 1 | A | 0.45 | 1.68 (1.24 – 2.29) | 9.00×10−4 | 0.011 | 0.44 | 0.87 (0.57–1.31) | 0.492 | 1.23 (0.65 – 2.34) | 0.534 | 84.18 | 0.013 |
| rs285480 | RXRG | 1 | G | 0.22 | 1.47 (1.06 – 2.04) | 2.05×10−2 | 0.165 | 0.22 | 1.14 (0.72–1.80) | 0.581 | 1.35 (1.03 – 1.76) | 0.027 | 0.00 | 0.376 |
| rs3767332 | RXRG | 1 | A | 0.24 | 0.59 (0.39 – 0.89) | 1.10×10−2 | 0.120 | 0.24 | 0.91 (0.57–1.46) | 0.687 | 0.72 (0.47 – 1.10) | 0.127 | 46.23 | 0.175 |
| rs157861 | RXRG | 1 | G | 0.21 | 1.51 (1.09 – 2.08) | 1.23×10−2 | 0.099 | 0.19 | 0.97 (0.59–1.60) | 0.914 | 1.26 (0.83 – 1.93) | 0.285 | 53.64 | 0.144 |
| rs115079560 | RXRG | 1 | C | 0.16 | 1.65 (1.18 – 2.30) | 3.08×10−3 | 0.031 | 0.14 | 0.87 (0.48–1.58) | 0.654 | 1.26 (0.68 – 2.34) | 0.467 | 70.55 | 0.066 |
| rs3753896 | RXRG | 1 | G | 0.38 | 1.58 (1.15 – 2.15) | 4.35×10−3 | 0.034 | 0.35 | 0.88 (0.57–1.34) | 0.541 | 1.20 (0.68 – 2.13) | 0.531 | 78.92 | 0.030 |
| rs3820367 | RXRG | 1 | G | 0.21 | 1.51 (1.11 – 2.04) | 7.95×10−3 | 0.063 | 0.21 | 0.87 (0.52–1.46) | 0.597 | 1.19 (0.70 – 2.04) | 0.514 | 69.19 | 0.071 |
| rs2194899 | RXRG | 1 | A | 0.37 | 1.51 (1.13 – 2.01) | 5.18×10−3 | 0.042 | 0.34 | 0.72 (0.46–1.14) | 0.159 | 1.07 (0.52 – 2.20) | 0.863 | 86.20 | 0.007 |
| rs3753898 | RXRG | 1 | G | 0.21 | 1.44 (1.06 – 1.95) | 1.84×10−2 | 0.144 | 0.20 | 0.84 (0.49–1.43) | 0.515 | 1.15 (0.69 – 1.94) | 0.593 | 66.11 | 0.086 |
| rs1529382 | CYP27A1 | 2 | T | 0.49 | 1.70 (1.26 – 2.31) | 6.00×10−4 | 0.007 | 0.50 | 1.25 (0.83–1.89) | 0.282 | 1.50 (1.12 – 2.02) | 0.007 | 28.86 | 0.238 |
| rs12512631 | VDBP | 4 | C | 0.36 | 0.70 (0.51 – 0.95) | 2.41×10−2 | 0.168 | 0.35 | 0.58 (0.36–0.93) | 0.025 | 0.66 (0.51 – 0.86) | 1.88×10−3 | 0.00 | 0.516 |
| rs7850212 | RXRA | 9 | A | 0.11 | 0.41 (0.21 – 0.78) | 7.15×10−3 | 0.179 | 0.09 | 0.31 (0.10–0.97) | 0.043 | 0.38 (0.22 – 0.68) | 9.54×10−4 | 0.00 | 0.676 |
| rs67965144 | RXRA | 9 | A | 0.12 | 1.52 (1.07 – 2.16) | 2.04×10−2 | 0.158 | 0.13 | 0.79 (0.43–1.48) | 0.469 | 1.15 (0.61 – 2.17) | 0.656 | 68.86 | 0.071 |
| rs62576319 | RXRA | 9 | T | 0.11 | 1.71 (1.17 – 2.49) | 5.12×10−3 | 0.055 | 0.12 | 1.00 (0.55–1.82) | 0.992 | 1.38 (0.82 – 2.31) | 0.222 | 54.59 | 0.137 |
| rs12895681 | SNW1 | 14 | T | 0.32 | 1.42 (1.06 – 1.90) | 1.81×10−2 | 0.142 | 0.35 | 0.92 (0.59–1.43) | 0.719 | 1.18 (0.78 – 1.80) | 0.439 | 61.72 | 0.109 |
| rs12890375 | SNW1 | 14 | G | 0.26 | 0.66 (0.45 – 0.95) | 2.45×10−2 | 0.197 | 0.27 | 1.10 (0.71–1.72) | 0.664 | 0.84 (0.51 – 1.39) | 0.490 | 66.68 | 0.084 |
| rs77513685 | NCOA3 | 20 | T | 0.21 | 1.50 (1.07 – 2.12) | 2.01×10−2 | 0.170 | 0.21 | 1.52 (0.98–2.35) | 0.061 | 1.51 (1.15 – 1.97) | 0.003 | 0.00 | 0.963 |
EA = effect allele; MAF=minor allele frequency; HR = hazards ratio; LHR = lower hazards ratio; UHR = upper hazards ratio; FPRP = false positive report probability; Phet = P heterogeniety;
A2 is the effect allele;
Adjusted by age, sex, tumor stage, Breslow thickness, Clark level and ulceration of tumor;
Adjusted by age, sex.
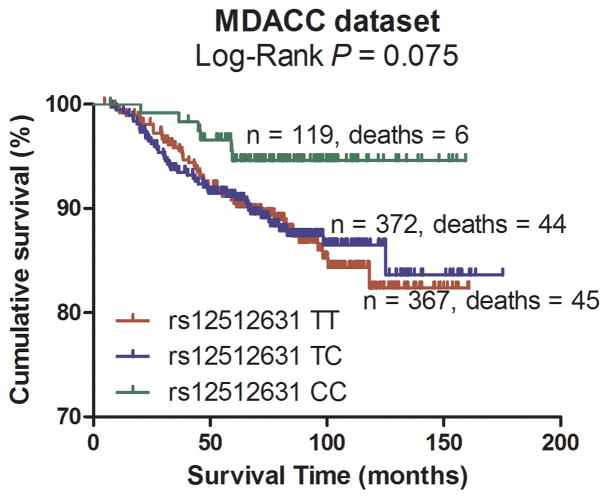
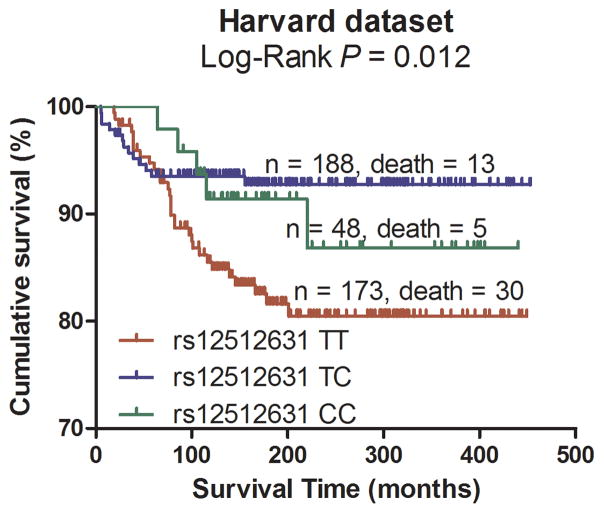
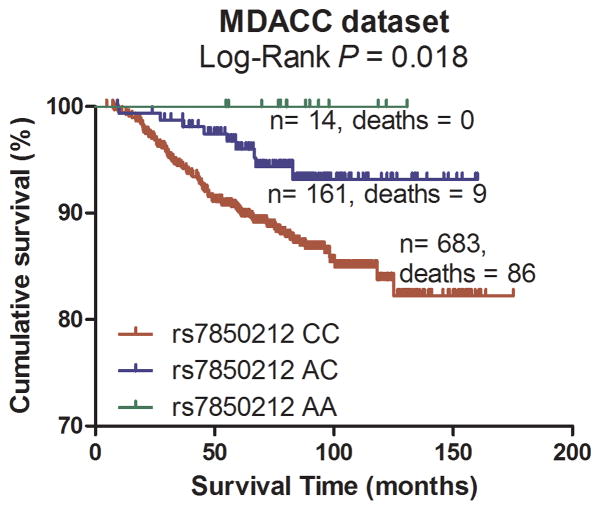
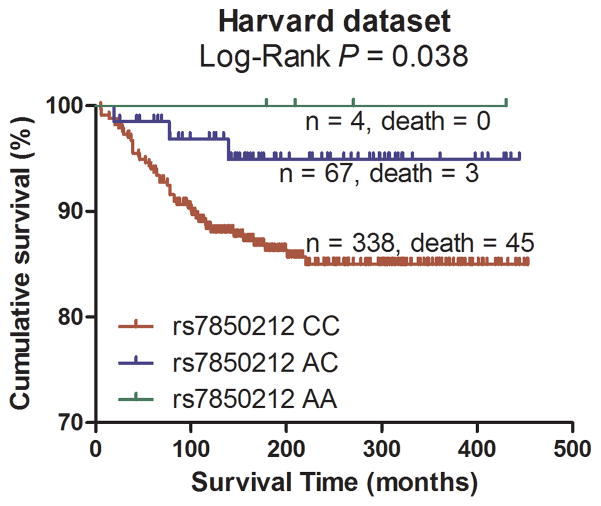
Subsequently, the significant association between the 18 tagging SNPs and CM DSS were validated in the CM patient population from Harvard University following the same eligibility criteria as used in the discovery population. As summarized in Table 2, under an additive model, two SNPs were statistically significantly associated with DSS and had the same direction of effects with the MDACC dataset. One SNP was located in the VDBP gene, while the other was mapped to the RXRA gene (VDBP rs12512631 T>C and RXRA rs7850212 C>A).
Pooling data from each of the datasets, we derived the joint HR and 95%CI under a conservative random-effects model for each SNP and the associated per-allele P values. As shown in Table 2 for detailed results of the survival analysis, VDBP rs12512631 (T>C) was associated with a better DSS in both MDACC and Harvard datasets, with an HR of 0.70 and 0.58, respectively. Similarly, the A allele of rs7850212 C>A was associated a decreased risk of death (HR = 0.41 in MDACC patients and HR = 0.31 in Harvard patients). As shown in Figure 2 with the Kaplan-Meier curves, patients with the VDBP rs12512631 TT genotype had a poorer survival compared with those with TC and CC genotypes; likewise, the rs7850212 CC genotype was also associated with an unfavorable DSS. In the meta-analysis, none of the effects of the two SNPs were significantly heterogeneous among the studies (rs12512631, Pheterogeneity = 0.516; and rs7850212, Pheterogeneity = 0.676). The VDBP rs12512631 C allele was significantly associated with the survival of CM patients (HR=0.66, 95%CI= 0.51 – 0.86, P = 1.88×10−3). For RXRA rs7850212, there was a significant association between the variant allele A and a better DSS (HR= 0.38, 95%CI = 0.22–0.68, P = 9.54×10−4). Additionally, we also combined the HRs and 95%CIs without any adjustment, and the results were similar with those with the adjustment (Supplementary Figure 2).
Figure 2.
Kaplan-Meier plots of specific survival of CM patients with different genotypes of rs12512631 (panel A, B) and rs7850212 (panel C, D).
In silica functional validation
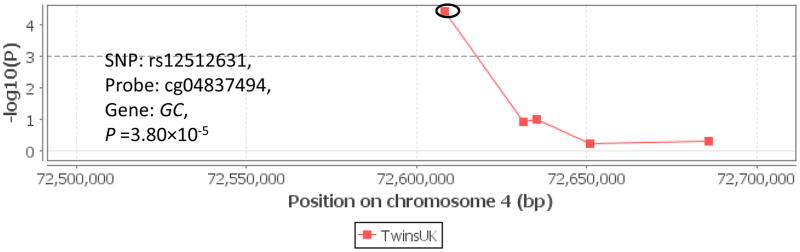
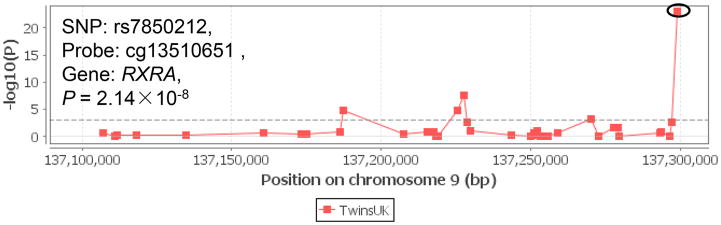
To further understand how the germline genetic variation influences the vitamin D pathway in tumor progress, we examined meQTL associations in the Multiple Tissue Human Expression Resource (MuTHER) project. As shown in Figure 3, there was evidence for some cismeQTL effects, including rs12512631 (VDBP, probe ID = cg04837494, P = 3.80×10−5) and rs7850212 (RXRA, probe ID cg13510651, P = 2.14×10−8). In detail, the variant alleles of these two SNPs were associated with methylation status of corresponding genes, and then potentially affect gene transcription and phenotypic variation.
Figure 3.
meQTL associations of VDBP rs12512631 (panel A), RXRA rs7850212 (panel B) with corresponding genes. The meQTL associations was assessed by Genevar on adipose tissue from a population of 428 female twin-pairs (856 individuals), collected as a part of the MuTHER Project.
Discussion
In the present study, we comprehensively evaluated the effects of SNPs from some major vitamin D-related genes on DSS of CM patients. We identified that two SNPs in VDBP and RXRA genes were protective and prolonged CM DSS across two different GWAS datasets. These results are biologically plausible and supported by previous experimental studies. For example, vitamin D has been reported to inhibit growth of malignant melanocytes both in vitro and in vivo (Colston et al., 1981). Additionally, vitamin D was also identified as inducing differentiation (Danielsson et al., 1998) and inhibiting invasiveness (Yudoh et al., 1999) in melanoma cell lines. Other in vivo trials found that vitamin D can suppress melanoma proliferation and inhibit metastasis in immune suppressed rodents (Eisman et al., 1987; Yudoh et al., 1999). Similarly, in the laboratory-based trials, 1,25(OH)2D molecule was shown to induce apoptosis in human melanoma cell lines in vitro (Seifert et al., 2004).
In the present study, VDBP rs12512631 and RXRA rs7850212 were found to be associated with CM survival; in addition, these two SNPs were associated with the methylation status of their corresponding genes. There is an existing evidence that VDBP may contribute to the variation of serum vitamin D levels in healthy populations (Biernacka et al., 2009; Bu et al., 2010; Wacholder et al., 2004) and cancer patients (Laddha et al., 2014). In Prostate, Lung, Colorectal and Ovarian Cancer Screening Trial performed in 2009, 150 control participants were randomly selected and associations between Vitamin D-related genes and serum vitamin D concentrations were tested; the results showed that rs12512631 was associated with modest differences in serum 25(OH)D (Laddha et al., 2014). Meanwhile, rs12512631 was statistically significantly associated with serum 25(OH)D concentrations in healthy Danish children and adults (Pani et al., 2002). A similar result was also found in the subset of 404 individuals with colorectal adenomas from the Ursodeoxycholic Acidtrial (Song et al., 2012). Therefore, it is likely that rs12512631 may modulate the risk of death by affecting corresponding gene’s function or the vitamin D level, providing a biological basis for the observed associations. The evidence for the role of rs7850212 of RXRA in CM survival is relatively rare. However, there was a report that RXRA SNPs were associated with poorer disease-free survival in breast cancer patients (Pande et al., 2013); additionally, a gene-level association was observed for RXRA and colon adenoma recurrence (Egan et al., 2010). Meanwhile, the representative tagging SNPs are also associated with corresponding gene methylation status, providing an additional biological support.
In the literature, some VDR SNPs (e.g., Apa1 rs7975232, Fok1 rs2228570, Bsm1 rs1544410, and Taq1 rs731236) are the vitamin D-related SNPs that have been mostly studied for an association with CM prognosis (Hutchinson et al., 2000; Santonocito et al., 2007; Schafer et al., 2012); however, the reported results have been inconclusive. For example, it was reported that Taq1 rs731236 and Fok1 rs2228570 were associated with Breslow thickness (Hutchinson et al., 2000). However, in another study of 101 CM patients, a significant association between Bsm1 rs1544410 and Breslow thickness was found, but not for the Fok1 rs2228570 (Santonocito et al., 2007), Taq1 rs731236 and Apa1 rs7975232 (Schafer et al., 2012). In the present study, we found that Apa1 rs7975232 and Bsm1 rs1544410 were marginally associated with MSS with P values of 0.045 and 0.043 in the MDACC dataset, respectively, which did not reach the significant threshold in the Harvard dataset (P values of 0.481 and 0.452, respectively). The discrepancies between the results of these GWAS studies may attribute to differences in populations recruited as well as study sizes and designs; especially we used melanoma-specific survival instead of Breslow thickness. Meanwhile, the VDBP rs2282679 genotype, the strongest genetic determinant of serum vitamin D levels reported in a previous GWAS study (Wang et al., 2010), was not significantly associated with CM survival in the MDACC dataset (P = 0.852), which is consistent with the report of Davies et al. (Davies et al., 2014). Similarly, in the breast cancer study reported by Mala et al., the association of rs2282679 with survival did not reach the threshold of significance (Pande et al., 2013).
The major strength of the present study is that we used DSS as the end point for CM outcomes. Although Breslow thickness works well in predicting CM prognosis and overall survival is similar to DSS in most cases, DSS is the most clinically critical outcome measurement for CM survival analyses. Moreover, a pathway-based analysis, rather than single-gene or single-SNP studies, can assist in identifying biologically meaningful prognostic SNPs from the available high-dimensional data and also can improve detection of the combined effects of these SNPs on survival.
One short coming of the present study is that serum vitamin D levels were not measured because only the historical data were made available to us for the analysis. Another potential limitation could be the differences in tumor characteristics or treatment regimens between the two study sites (i.e., MDACC and Harvard University), which might confound specific survival differences. To minimize the effect, we combined the results using the conservative random-effects model. It should be noted that we used less stringent FPRP method instead of false discovery rate (Benjamini and Yekutieli, 2001) or Bonferroni correction (Bland and Altman, 1995) to control for multiple comparisons in the discovery MDACC dataset. Although this may lead to higher probability of false positive, considering the consistent effects of those identified SNPs between the discovery and validation datasets, and their potential functions in regulating gene methylation, it is less likely that our findings are due to false discovery.
In conclusion, our meta-analysis suggests a role of VDBP rs12512631 T>C and RXRA rs7850212 C>A in CM DSS as assessed in two independent GWAS datasets. Considering the important role of vitamin D throughout the cancer continuum, these genetic variants may represent promising prognostic biomarkers in decision making for CM clinical management.
Materials & Methods
Study populations and genotyping
MDACC GWAS dataset
The MDACC CM GWAS has been previously reported (Amos et al., 2011). The characteristic details of the patient subjects in the survival analysis have also been recently described (Yin et al., 2014; Yuan et al., 2015; Zhang et al., 2015). Briefly, the present study included 858 histologically confirmed CM patients who were non-Hispanic white, had complete information for selected prognostic variables and were enrolled at MDACC between March 1998 and August 2008. For each of the patients, a DNA sample was extracted from the whole blood, and demographic, prospective clinical and pathological data were collected from a standard life-style questionnaire and/or extracted from patient medical charts. Tumor stages were classified according to the American Joint Committee on Cancer (AJCC) for the melanoma staging system (Balch et al., 2009). The follow-up was conducted using the standardized guidelines (Gershenwald and Ross, 2011). Stage of the disease and length of the follow-up were determined from the date of diagnosis. All individuals provided a written informed consent under an Institutional Review Board-approved protocol.
The genotype data in the present study can be accessed by using the National Center for Biotechnology Information (NCBI) Database of Genotypes and Phenotypes (dbGaP; http://www.ncbi.nlm.nih.gov/gap), with the study accession number phs000187.v1.p1 (Mailman et al., 2007; Tryka et al., 2014). Briefly, the genomic DNA extracted from the whole blood was genotyped with the Illumina HumanOmni-Quad_v1_0_B array, and the genotypes were called using the BeadStudio algorithmat the John Hopkins University Center for Inherited Disease Research (CIDR). The detailed genotyping information and data quality control can be found in the previously described GWAS (Amos et al., 2011). Genome-wide imputation was performed by using the MACH software based on the 1000 Genomes project, phase I V2 CEU data (Li et al., 2010), in which around 6.78 SNPs were included, after the quality control, if they had a minor allele frequency ≥ 0.05, genotyping rate ≥ 95%, Hardy-Weinberg equilibrium p-value ≥ 0.00001, or imputation r2 ≥ 0.8.
Harvard GWAS dataset
The Harvard CM GWAS dataset consisted of two studies: Nurses’ Health Study (NHS) and Health Professionals Follow-up Study (HPFS). Sampling, genotyping and quality control procedures have been described previously (Song et al., 2012). In brief, eligible cases in both the NHS and HPFS cohorts were participants with histopathologically confirmed invasive melanoma, diagnosed at any time after baseline up to the 2008 follow-up cycle for both cohorts. All subjects were US non-Hispanic white.
Genotyping was performed using the Illumina HumanHap610 array. Based on the genotyped SNPs and haplotype information from the 1000 Genomes Project [Utah Residents with Northern and Western European Ancestry (CEU) data, phase I v3, March 2012), genotypes for over 5.5 million SNPs were imputed using the program MACH (Biernacka et al., 2009). Only SNPs with imputation quality r2 ≥ 0.8 and minor allele frequency ≥ 0.05 in each study were used in the final analysis. In the final analysis, only 409 patients were kept in the data after quality control.
Gene and SNP selection
We selected variants of 14 genes in the vitamin D pathway based on their roles in the vitamin D metabolism and signaling in cancer (Deeb et al., 2007): CYP24A1, CYP27A1, CYP27B1, CYP2R1, VDR, VDBP, RXRA, RXRB, RXRG, NCOA1, NCOA2, NCOA3, NCOR1 and SNW1. Genotyped or imputed common SNPs within these genes or their ± 20-kb flanking regions were extracted. As a result, 2,669 (428 genotyped or 2241 imputed) common SNPs in the vitamin D pathway were extracted from the MDACC GWAS dataset.
Statistical methods
In the follow-up, causes of death other than CM were considered censored. We assessed an additive genetic model for each SNP in the MDACC discovery dataset by multivariable Cox proportional hazards regression analysis by using GenABEL package of R software (Aulchenko et al., 2007); the adjusted variables included age at diagnosis, sex, clinical stage, Breslow thickness, Clark level and ulceration of tumor in the MDACC dataset. False positive report probability (FPRP) was calculated to assess the false-positive association findings (Wacholder et al., 2004). For all the significant results, we assigned a prior probability of 0.1 to detect a HR of 1.5 for an association with genotypes and alleles of each SNP. Only those results with an FPRP value < 0.2 were considered as a noteworthy association (Wacholder et al., 2004). Kaplan-Meier survival curves and log-rank tests were used to evaluate the effects of genetic variants on the cumulative probability of death (Kaplan and Meier, 1958). For SNPs showing statistically significant differences in DSS with an FPRP vale <0.2 in the discovery, we selected the tagging SNPs based on pairwise r2>0.6, using LD information from the latest 1000 Genomes Project for CEU populations (Genomes Project et al., 2012). Next, the prognostic roles of tagging SNPs were validated in the Harvard GWAS dataset, with adjustment for age at diagnosis and sex in multivariable Cox analysis. Pooled hazards ratios (HRs) and 95% confidence intervals (95%CIs) were calculated for the meta-analysis using a conservative random-effects model, and inter-study heterogeneity was assessed with Cochrane’s Q test. Finally, the methylation quantitative trait loci (meQTL) association was assessed by Genevar on adipose tissue from a population of 428 female twin-pairs (856 individuals), collected as a part of the MuTHER Project and publically available (Grundberg et al., 2012).
All statistical analyses were carried out by R software (version 3.0.2; The R Foundation for Statistical Computing, Vienna, Austria), Stata v14.0 (Stata College, Texas, US) and Statistical Analysis System software (version 9.1.3; SAS Institute, Cary, NC, USA). Figure 1 provides the flow chart, illustrating procedures of analyses in this study.
Figure 1.
Flow chart of this study.
Supplementary Material
Supplementary Figure 1. Linkage disequilibrium map of 203 SNPs in 6 genes with FPRP < 0.2 in the MDACC dataset [Panel A (CYP27A1, 111 SNPs), Panel B (VDBP, six SNPs), Panel C (NCOA3, two SNPs), Panel D (RXRA, 20 SNPs), Panel E (RXRG, 61 SNPs) and Panel F (SNW1, three SNPs)]. The linkage disequilibrium structure was created using genotype data from the1000 Genomes project for CEU populations (Utah Residents with Northern and Western European Ancestry).
Supplementary Figure 2. Forest plot of meta-analysis of rs12512631 and rs7850212 in univariate and multivariate Cox proportional hazards regression model.
Association of SNPs in vitamin D pathway and CM specific survival in the MDACC dataset.
Significance.
Our study suggests a role of VDBPrs12512631 T>C and RXRA rs7850212 C>A in CM DSS as assessed in two independent GWAS datasets. Considering the important role of vitamin D throughout the cancer continuum, these genetic variants may represent promising prognostic biomarkers in decision making for CM clinical management.
Acknowledgments
Financial support:
This work was partially supported by the National Institutes of Health (National Cancer Institute) Grants R01 CA100264 and 2P50CA093459; the Marit Peterson Fund for Melanoma Research; the Start-Up funds from Duke University and support from the Duke Cancer Institute as part of the P30 Cancer Center Support Grant (Grant ID: NIH CA014236); and the Harvard cohort grants (UM1 CA186107, P01 CA87969, R01 CA49449, UM1 CA167552). Additional funding was provided by philanthropic contributions to The University of Texas M. D. Anderson Cancer Center Moon Shots Program, the Miriam and Jim Mulva Melanoma Research Fund and the Marit Peterson Fund for Melanoma Research. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
We thank the individuals who participated in this project. We thank the John Hopkins University Center for Inherited Disease Research for conducting high-throughput genotyping for this study. Jieyun Yin was sponsored by the China Scholarship Council for studying at Duke University. We would like to thank the participants and staff of the Nurses’ Health Study (NHS)and Health Professionals Follow-up Study (HPFS) for their valuable contributions as well as the following state cancer registries for their help: AL, AZ, AR, CA, CO, CT, DE, FL, GA, ID, IL, IN, IA, KY, LA, ME, MD, MA, MI, NE, NH, NJ, NY, NC, ND, OH, OK, OR, PA, RI, SC, TN, TX, VA, WA, WY. The authors assume full responsibility for analyses and interpretation of these data.
Abbreviation
- CM
cutaneous melanoma
- SNPs
single nucleotide polymorphisms
- DSS
disease specific survival
- HR
Hazards Ratio
- 95%CI
95% confident interval
Footnotes
Conflict of interest:
The authors state no conflict of interest.
References
- Amos CI, Wang LE, Lee JE, Gershenwald JE, Chen WV, Fang S, Kosoy R, Zhang M, Qureshi AA, Vattathil S, et al. Genome-wide association study identifies novel loci predisposing to cutaneous melanoma. Human molecular genetics. 2011;20:5012–23. doi: 10.1093/hmg/ddr415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aulchenko YS, Ripke S, Isaacs A, Van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23:1294–6. doi: 10.1093/bioinformatics/btm108. [DOI] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, et al. Final version of 2009 AJCC melanoma staging and classification. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:6199–206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of statistics. 2001:1165–1188. [Google Scholar]
- Biernacka JM, Tang R, Li J, Mcdonnell SK, Rabe KG, Sinnwell JP, Rider DN, De Andrade M, Goode EL, Fridley BL. Assessment of genotype imputation methods. BMC proceedings. 2009;3(Suppl 7):S5. doi: 10.1186/1753-6561-3-s7-s5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bland JM, Altman DG. Multiple significance tests: the Bonferroni method. Bmj. 1995;310:170. doi: 10.1136/bmj.310.6973.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bu FX, Armas L, Lappe J, Zhou Y, Gao G, Wang HW, Recker R, Zhao LJ. Comprehensive association analysis of nine candidate genes with serum 25-hydroxy vitamin D levels among healthy Caucasian subjects. Human genetics. 2010;128:549–56. doi: 10.1007/s00439-010-0881-9. [DOI] [PubMed] [Google Scholar]
- Carlberg C, Bendik I, Wyss A, Meier E, Sturzenbecker LJ, Grippo JF, Hunziker W. Two nuclear signalling pathways for vitamin D. Nature. 1993;361:657–60. doi: 10.1038/361657a0. [DOI] [PubMed] [Google Scholar]
- Chowdhury R, Kunutsor S, Vitezova A, Oliver-Williams C, Chowdhury S, Kiefte-De-Jong JC, Khan H, Baena CP, Prabhakaran D, Hoshen MB, et al. Vitamin D and risk of cause specific death: systematic review and meta-analysis of observational cohort and randomised intervention studies. Bmj. 2014;348:g1903. doi: 10.1136/bmj.g1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colston K, Colston MJ, Feldman D. 1,25-dihydroxyvitamin D3 and malignant melanoma: the presence of receptors and inhibition of cell growth in culture. Endocrinology. 1981;108:1083–6. doi: 10.1210/endo-108-3-1083. [DOI] [PubMed] [Google Scholar]
- Danielsson C, Fehsel K, Polly P, Carlberg C. Differential apoptotic response of human melanoma cells to 1 alpha,25-dihydroxyvitamin D3 and its analogues. Cell death and differentiation. 1998;5:946–52. doi: 10.1038/sj.cdd.4400437. [DOI] [PubMed] [Google Scholar]
- Davies JR, Field S, Randerson-Moor J, Harland M, Kumar R, Anic GM, Nagore E, Hansson J, Hoiom V, Jonsson G, et al. An inherited variant in the gene coding for vitamin D-binding protein and survival from cutaneous melanoma: a BioGenoMEL study. Pigment cell & melanoma research. 2014;27:234–43. doi: 10.1111/pcmr.12193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deeb KK, Trump DL, Johnson CS. Vitamin D signalling pathways in cancer: potential for anticancer therapeutics. Nature reviews Cancer. 2007;7:684–700. doi: 10.1038/nrc2196. [DOI] [PubMed] [Google Scholar]
- Egan JB, Thompson PA, Ashbeck EL, Conti DV, Duggan D, Hibler E, Jurutka PW, Leroy EC, Martinez ME, Mount D, et al. Genetic polymorphisms in vitamin D receptor VDR/RXRA influence the likelihood of colon adenoma recurrence. Cancer research. 2010;70:1496–504. doi: 10.1158/0008-5472.CAN-09-3264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisman JA, Barkla DH, Tutton PJ. Suppression of in vivo growth of human cancer solid tumor xenografts by 1,25-dihydroxyvitamin D3. Cancer research. 1987;47:21–5. [PubMed] [Google Scholar]
- Fakih MG, Trump DL, Muindi JR, Black JD, Bernardi RJ, Creaven PJ, Schwartz J, Brattain MG, Hutson A, French R, et al. A phase I pharmacokinetic and pharmacodynamic study of intravenous calcitriol in combination with oral gefitinib in patients with advanced solid tumors. Clinical cancer research : an official journal of the American Association for Cancer Research. 2007;13:1216–23. doi: 10.1158/1078-0432.CCR-06-1165. [DOI] [PubMed] [Google Scholar]
- Gambichler T, Bindsteiner M, Hoxtermann S, Kreuter A. Serum 25-hydroxyvitamin D serum levels in a large German cohort of patients with melanoma. The British journal of dermatology. 2013;168:625–8. doi: 10.1111/j.1365-2133.2012.11212.x. [DOI] [PubMed] [Google Scholar]
- Genomes Project C. Abecasis GR, Auton A, Brooks LD, Depristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, Mcvean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gershenwald JE, Ross MI. Sentinel-lymph-node biopsy for cutaneous melanoma. The New England journal of medicine. 2011;364:1738–45. doi: 10.1056/NEJMct1002967. [DOI] [PubMed] [Google Scholar]
- Goodwin PJ, Ennis M, Pritchard KI, Koo J, Hood N. Prognostic effects of 25-hydroxyvitamin D levels in early breast cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:3757–63. doi: 10.1200/JCO.2008.20.0725. [DOI] [PubMed] [Google Scholar]
- Grundberg E, Small KS, Hedman AK, Nica AC, Buil A, Keildson S, Bell JT, Yang TP, Meduri E, Barrett A, et al. Mapping cis- and trans-regulatory effects across multiple tissues in twins. Nature genetics. 2012;44:1084–9. doi: 10.1038/ng.2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hershberger PA, Mcguire TF, Yu WD, Zuhowski EG, Schellens JH, Egorin MJ, Trump DL, Johnson CS. Cisplatin potentiates 1,25-dihydroxyvitamin D3-induced apoptosis in association with increased mitogen-activated protein kinase kinase kinase 1 (MEKK-1) expression. Molecular cancer therapeutics. 2002;1:821–9. [PubMed] [Google Scholar]
- Hewison M, Zehnder D, Bland R, Stewart PM. 1alpha-Hydroxylase and the action of vitamin D. Journal of molecular endocrinology. 2000;25:141–8. doi: 10.1677/jme.0.0250141. [DOI] [PubMed] [Google Scholar]
- Holick MF. Vitamin D and bone health. The Journal of nutrition. 1996;126:1159S–64S. doi: 10.1093/jn/126.suppl_4.1159S. [DOI] [PubMed] [Google Scholar]
- Hutchinson PE, Osborne JE, Lear JT, Smith AG, Bowers PW, Morris PN, Jones PW, York C, Strange RC, Fryer AA. Vitamin D receptor polymorphisms are associated with altered prognosis in patients with malignant melanoma. Clinical cancer research : an official journal of the American Association for Cancer Research. 2000;6:498–504. [PubMed] [Google Scholar]
- Kaplan EL, Meier P. Nonparametric estimation from incomplete observations. Journal of the American statistical association. 1958;53:457–481. [Google Scholar]
- Laddha SV, Ganesan S, Chan CS, White E. Mutational landscape of the essential autophagy gene BECN1 in human cancers. Molecular cancer research : MCR. 2014;12:485–90. doi: 10.1158/1541-7786.MCR-13-0614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lappe JM, Travers-Gustafson D, Davies KM, Recker RR, Heaney RP. Vitamin D and calcium supplementation reduces cancer risk: results of a randomized trial. The American journal of clinical nutrition. 2007;85:1586–91. doi: 10.1093/ajcn/85.6.1586. [DOI] [PubMed] [Google Scholar]
- Li Y, Willer CJ, Ding J, Scheet P, Abecasis GR. MaCH: using sequence and genotype data to estimate haplotypes and unobserved genotypes. Genetic epidemiology. 2010;34:816–34. doi: 10.1002/gepi.20533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailman MD, Feolo M, Jin Y, Kimura M, Tryka K, Bagoutdinov R, Hao L, Kiang A, Paschall J, Phan L. The NCBI dbGaP database of genotypes and phenotypes. Nature genetics. 2007;39:1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newton-Bishop JA, Beswick S, Randerson-Moor J, Chang YM, Affleck P, Elliott F, Chan M, Leake S, Karpavicius B, Haynes S, et al. Serum 25-hydroxyvitamin D3 levels are associated with breslow thickness at presentation and survival from melanoma. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2009;27:5439–44. doi: 10.1200/JCO.2009.22.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng K, Meyerhardt JA, Wu K, Feskanich D, Hollis BW, Giovannucci EL, Fuchs CS. Circulating 25-hydroxyvitamin d levels and survival in patients with colorectal cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2008;26:2984–91. doi: 10.1200/JCO.2007.15.1027. [DOI] [PubMed] [Google Scholar]
- Nurnberg B, Graber S, Gartner B, Geisel J, Pfohler C, Schadendorf D, Tilgen W, Reichrath J. Reduced serum 25-hydroxyvitamin D levels in stage IV melanoma patients. Anticancer research. 2009;29:3669–74. [PubMed] [Google Scholar]
- Pande M, Thompson PA, Do KA, Sahin AA, Amos CI, Frazier ML, Bondy ML, Brewster AM. Genetic variants in the vitamin D pathway and breast cancer disease-free survival. Carcinogenesis. 2013;34:587–94. doi: 10.1093/carcin/bgs369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani MA, Regulla K, Segni M, Hofmann S, Hufner M, Pasquino AM, Usadel KH, Badenhoop K. A polymorphism within the vitamin D-binding protein gene is associated with Graves’ disease but not with Hashimoto’s thyroiditis. The Journal of clinical endocrinology and metabolism. 2002;87:2564–7. doi: 10.1210/jcem.87.6.8562. [DOI] [PubMed] [Google Scholar]
- Randerson-Moor JA, Taylor JC, Elliott F, Chang YM, Beswick S, Kukalizch K, Affleck P, Leake S, Haynes S, Karpavicius B, et al. Vitamin D receptor gene polymorphisms, serum 25-hydroxyvitamin D levels, and melanoma: UK case-control comparisons and a meta-analysis of published VDR data. European journal of cancer. 2009;45:3271–81. doi: 10.1016/j.ejca.2009.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santonocito C, Capizzi R, Concolino P, Lavieri MM, Paradisi A, Gentileschi S, Torti E, Rutella S, Rocchetti S, Di Carlo A, et al. Association between cutaneous melanoma, Breslow thickness and vitamin D receptor BsmI polymorphism. The British journal of dermatology. 2007;156:277–82. doi: 10.1111/j.1365-2133.2006.07620.x. [DOI] [PubMed] [Google Scholar]
- Schafer A, Emmert S, Kruppa J, Schubert S, Tzvetkov M, Mossner R, Reich K, Berking C, Volkenandt M, Pfohler C, et al. No association of vitamin D metabolism-related polymorphisms and melanoma risk as well as melanoma prognosis: a case-control study. Archives of dermatological research. 2012;304:353–61. doi: 10.1007/s00403-012-1243-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seifert M, Rech M, Meineke V, Tilgen W, Reichrath J. Differential biological effects of 1, 25-dihydroxyVitamin D3 on melanoma cell lines in vitro. The Journal of steroid biochemistry and molecular biology . 2004;89–90:375–9. doi: 10.1016/j.jsbmb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- Song F, Qureshi AA, Zhang J, Amos CI, Lee JE, Wei Q, Han J. Exonuclease 1 (EXO1) gene variation and melanoma risk. DNA repair. 2012;11:304–9. doi: 10.1016/j.dnarep.2011.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tretli S, Hernes E, Berg JP, Hestvik UE, Robsahm TE. Association between serum 25(OH)D and death from prostate cancer. British journal of cancer. 2009;100:450–4. doi: 10.1038/sj.bjc.6604865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tryka KA, Hao L, Sturcke A, Jin Y, Wang ZY, Ziyabari L, Lee M, Popova N, Sharopova N, Kimura M. NCBI’s Database of Genotypes and Phenotypes: dbGaP. Nucleic acids research. 2014;42:D975–D979. doi: 10.1093/nar/gkt1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacholder S, Chanock S, Garcia-Closas M, El Ghormli L, Rothman N. Assessing the probability that a positive report is false: an approach for molecular epidemiology studies. Journal of the National Cancer Institute. 2004;96:434–42. doi: 10.1093/jnci/djh075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wactawski-Wende J, Kotchen JM, Anderson GL, Assaf AR, Brunner RL, O’sullivan MJ, Margolis KL, Ockene JK, Phillips L, Pottern L, et al. Calcium plus vitamin D supplementation and the risk of colorectal cancer. The New England journal of medicine. 2006;354:684–96. doi: 10.1056/NEJMoa055222. [DOI] [PubMed] [Google Scholar]
- Wang TJ, Zhang F, Richards JB, Kestenbaum B, Van Meurs JB, Berry D, Kiel DP, Streeten EA, Ohlsson C, Koller DL, et al. Common genetic determinants of vitamin D insufficiency: a genome-wide association study. Lancet. 2010;376:180–8. doi: 10.1016/S0140-6736(10)60588-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin J, Liu H, Liu Z, Wang LE, Chen WV, Zhu D, Amos CI, Fang S, Lee JE, Wei Q. Genetic Variants in Fanconi Anemia Pathway Genes BRCA2 and FANCA Predict Melanoma Survival. The Journal of investigative dermatology. 2014 doi: 10.1038/jid.2014.416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan H, Liu H, Liu Z, Zhu D, Amos CI, Fang S, Lee JE, Wei Q. Genetic variants in Hippo pathway genes YAP1, TEAD1 and TEAD4 are associated with melanoma-specific survival. International journal of cancer. Journal international du cancer. 2015 doi: 10.1002/ijc.29429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yudoh K, Matsuno H, Kimura T. 1alpha,25-dihydroxyvitamin D3 inhibits in vitro invasiveness through the extracellular matrix and in vivo pulmonary metastasis of B16 mouse melanoma. The Journal of laboratory and clinical medicine. 1999;133:120–8. doi: 10.1016/s0022-2143(99)90004-5. [DOI] [PubMed] [Google Scholar]
- Zhang W, Liu H, Liu Z, Zhu D, Amos CI, Fang S, Lee JE, Wei Q. Functional Variants in Notch Pathway Genes NCOR2, NCSTN and MAML2 Predict Survival of Patients with Cutaneous Melanoma. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology. 2015 doi: 10.1158/1055-9965.EPI-14-1380-T. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou W, Heist RS, Liu G, Asomaning K, Neuberg DS, Hollis BW, Wain JC, Lynch TJ, Giovannucci E, Su L, et al. Circulating 25-hydroxyvitamin D levels predict survival in early-stage non-small-cell lung cancer patients. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2007;25:479–85. doi: 10.1200/JCO.2006.07.5358. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure 1. Linkage disequilibrium map of 203 SNPs in 6 genes with FPRP < 0.2 in the MDACC dataset [Panel A (CYP27A1, 111 SNPs), Panel B (VDBP, six SNPs), Panel C (NCOA3, two SNPs), Panel D (RXRA, 20 SNPs), Panel E (RXRG, 61 SNPs) and Panel F (SNW1, three SNPs)]. The linkage disequilibrium structure was created using genotype data from the1000 Genomes project for CEU populations (Utah Residents with Northern and Western European Ancestry).
Supplementary Figure 2. Forest plot of meta-analysis of rs12512631 and rs7850212 in univariate and multivariate Cox proportional hazards regression model.
Association of SNPs in vitamin D pathway and CM specific survival in the MDACC dataset.