Abstract
Genetic studies in large outbred populations have documented a complex, highly polygenic basis for type 2 diabetes (T2D). Most of the variants currently known to be associated with T2D risk have been identified in large studies that included tens of thousands of individuals who are representative of a single major ethnic group such as European, Asian, or African. However, most of these variants have only modest effects on the risk for T2D; identification of definitive 'causal variant' or 'causative loci' is typically lacking. Studies in isolated populations offer several advantages over outbred populations despite being, on average, much smaller in sample size. For example, reduced genetic variability, enrichment of rare variants, and a more uniform environment and lifestyle, which are hallmarks of isolated populations, can reduce the complexity of identifying disease-associated genes. To date, studies in isolated populations have provided valuable insight into the genetic basis of T2D by providing both a deeper understanding of previously identified T2D-associated variants (e.g. demonstrating that variants in KCNQ1 have a strong parent-of-origin effect) or providing novel variants (e.g. ABCC8 in Pima Indians, TBC1D4 in the Greenlandic population, HNF1A in Canadian Oji-Cree). This review summarizes advancements in genetic studies of T2D in outbred and isolated populations, and provides information on whether the difference in the prevalence of T2D in different populations (Pima Indians vs. non-Hispanic Whites and non-Hispanic Whites vs. non-Hispanic Blacks) can be explained by the difference in risk allele frequencies of established T2D variants.
Keywords: type 2 diabetes, GWAS, imputation, SNP, meta-analysis, ethnic group, functional variant, risk
Abbreviations: ABCC8 – ATP-binding cassette, sub-family C (CFTR/MRP), member 8; ADAMTS9 – ADAM metallopeptidase with thrombospondin type 1 motif, 9; ADCY5 – adenylate cyclase 5; APOE – apolipoprotein E; ARL15 – ADP-ribosylation factor-like 15; BMI – body mass index; C2CD4A – C2 calcium-dependent domain containing 4A; CACNA1E – calcium channel, voltage-dependent, R type, alpha 1E subunit; CAPN10 – calpain 10; CCND2 – cyclin D2; CDKAL1 – CDK5 regulatory subunit associated protein 1-like 1; C0DKN2B – cyclin-dependent kinase inhibitor 2B (p15, inhibits CDK4); CGAS – candidate-gene association studies; DGKB – diacylglycerol kinase, beta 90kDa; DIAGRAM – Diabetes Genetics Replication and Meta-analysis; DNER – delta/notch-like EGF repeat containing; DUSP9 – dual specificity phosphatase 9; EXT2 – exostosin glycosyltransferase 2; EXT2 – exostosin glycosyltransferase 2; FTO – fat mass and obesity associated; FUSION – Finland-United States Investigation Of NIDDM Genetics; GAF – genetic attributable fraction; GCK – glucokinase; GCKR – glucokinase (hexokinase 4) regulator; GLUT4 – glucose transporter 4; GRB10 – growth factor receptor-bound protein 10; GRS – genetic risk score; GWAS – genome-wide association study; HbA1c – glycated hemoglobin; HHEX – hematopoietically expressed homeobox; HNF1A – HNF1 homeobox A; HNF1B – HNF1 homeobox B; HNF4A – hepatocyte nuclear factor 4, alpha; HSL – hormone-sensitive lipase; IGF2BP2 – insulin-like growth factor 2 mRNA binding protein 2; IRS1 – insulin receptor substrate 1; IRX3 – iroquois homeobox 3; KCNJ11 – potassium channel, inwardly rectifying subfamily J, member 11; KCNQ1 – potassium channel, voltage gated KQT-like subfamily Q, member 1; KLF14 – Kruppel-like factor 14; LAMA1 – laminin alpha 1; LD – linkage disequilibrium; LIPE – hormone sensitive lipase; LPP – LIM domain containing preferred translocation partner in lipoma; MAGIC – Meta-Analyses of Glucose and Insulin-related traits Consortium; MBL2 – mannose-binding lectin; MOB2 – MOB kinase activator 2; MODY – maturity onset diabetes of the young; MPHOSPH9 – M-phase phosphoprotein 9; MTNR1B – melatonin receptor 1B; MYBPC3 – myosin binding protein C, cardiac; NDSR – National diabetes statistics report; OGTT – oral glucose tolerance test; OR – ODDs ratio; PAM – peptidylglycine alpha-amidating monooxygenase; PDX1 – pancreatic and duodenal homeobox 1; POU5F1 – POU class 5 homeobox 1; PPARG – peroxisome proliferator-activated receptor gamma; PROX1 – prospero homeobox 1; PTP1B – protein tyrosine phosphatase, non-receptor type 1; PTPRD – protein tyrosine phosphatase, receptor type, D; SIGMA – Slim Initiative in Genomic Medicine for the Americas; SLC16A11 – solute carrier family 16, member 11; SLC30A8 – solute carrier family 30 (zinc transporter), member 8; SNP – single nucleotide polymorphism; T2D – type 2 diabetes; TBC1D4 – TBC1 domain family, member 4; TCF19 – transcription factor 19; TCF7L2 – transcription factor 7 like 2; TMEM154 – transmembrane protein 154; TREH – trehalase (brush-border membrane glycoprotein); WFS1 – Wolfram syndrome 1; ZFAND3 – zinc finger, AN1-type domain 3
1. Type 2 diabetes - an introduction
Diabetes is a metabolic condition defined by elevated serum glucose levels. Recent data from the International Diabetes Federation show that 387 million people worldwide suffer from diabetes; among these individuals, 90% have T2D. The etiology of T2D is heterogeneous, where inefficient insulin use (insulin resistance) and/or reduced insulin availability are predictive of the disease [1]. The recent increase in incidence and prevalence of T2D in developed and developing countries worldwide coincides with changes in the inhabitants' dietary habits and physical activity, suggesting that lifestyle/environment has a major impact on this disease. Indeed, T2D is frequently associated with obesity and sedentary life style, which can increase insulin resistance and further disease progression [2]. However, not all individuals with T2D are overweight or obese, and conversely, many obese people do not develop T2D. Therefore, an obesogenic environment cannot fully explain the risk of T2D.
2. Type 2 diabetes is heritable
Several lines of evidence, as detailed below, have documented a genetic basis for T2D.
2.1 Familial aggregation
Siblings of affected individuals are at increased risk of T2D. The diabetes risk odds ratio (OR) for an individual with one affected parent is 3.4-3.5. The OR increases to 6.1 when both parents are affected [3].
2.2 Twin studies
Multiple twin concordance studies have been conducted in T2D. The concordance rate is higher in monozygotic twins (0.29-1.00) than in dizygotic twins (0.10-0.43), indicating a significant genetic component of the disease [4-6].
2.3 Heritability of related traits
Usually, decreased insulin sensitivity and impaired insulin production precede the onset of T2D [1]. Data from multiple studies document that these prediabetic metabolic traits are also familial [7-8].
2.4 Different prevalence in different ethnic groups
The large variance in the prevalence of T2D in different ethnic groups living in a similar environment also indicates a genetic burden for T2D. According to the National Diabetes Statistics Report (Center for Disease Control, 2014), the prevalence of T2D among individuals in the United States varies from 7.6%-15.9% depending on race/ethnic background. The lowest prevalence is among non-Hispanic whites (7.6%) and highest prevalence is among American Indians/Alaskan Natives (15.9%). Among American Indians, the Pima Indians of Arizona have a very high prevalence of T2D; approximately 50% of adults above 35 years of age have T2D [9].
3. Delineating the heritability of T2D
Identifying genetic factors that influence the pathophysiology of T2D has proved to be challenging. Initially, success was primarily limited to the identification of variants explaining rare, monogenic forms of T2D. However, as knowledge of the human genome sequence became available, genomic technologies advanced, and the costs of new technologies declined. Powerful new methodologies have heralded unparalleled success in uncovering new disease genes. The plethora of studies to identify DNA variation that affects risk of T2D in diverse populations worldwide has led us to a better appreciation of the complexity of this disease. Both candidate-gene association studies (CGAS) and genome-wide association studies (GWAS) have highlighted the highly polygenic nature of T2D, and proven the early proposal of an oligogenic model as overly optimistic (reviewed in [10]).
Prior to the "genomic era" it was often argued that the primary defect in T2D was in skeletal muscle glucose uptake. However, genome-wide studies and a multitude of physiological studies have documented the importance of pancreatic beta-cell insulin secretion, adipocytes as a secretory organ, the central nervous system, and the liver in T2D (reviewed in [11]). Genetic studies have also shown that the genetic etiology of T2D can differ in diverse populations, highlighting the importance of identifying risk factors for the disease in different populations. In particular, studies in isolated populations with less variability among subjects can provide valuable information that could be missed in studies of heterogeneous populations.
3.1 Physiologic candidate gene studies
The earliest genetic studies focused on the analysis of coding regions of candidate genes selected because of their known role in T2D physiology. Most of the early candidate gene studies were underpowered, and independent replication was rare. Few of the early reported signals (KCNJ11, PPARG, HNF1B, WFS1, and IRS1) have been confirmed by subsequent GWASs [12-16]. Among those that were subsequently verified, the KCNJ11 (E23K) and PPARG (P12A) variants [12-13] have yielded therapeutic targets. PPARG codes for a type II nuclear receptor, peroxisome proliferator-activated receptor gamma, which regulates fatty acid storage and glucose metabolism [13]. KCNJ11 encodes one of the subunits (Kir6.2) of the ATP-sensitive potassium channel that regulates glucose-stimulated insulin secretion from pancreatic beta-cells [12]. The proteins encoded by both these genes are targets for two classes of widely used diabetes drugs (thiazolidinediones and sulphonylureas).
3.2 Family-based linkage studies
The first genomic studies to identify variants that increase risk for T2D based on their chromosomal position were family-based linkage studies. Linkage analyses were extremely successful in identifying genes responsible for highly penetrant Mendelian forms of T2D such as maturity onset diabetes of the young (MODY). One of these MODY genes, HNF4A, has subsequently been shown to have a role in common polygenic T2D as well [17]. The first gene to be identified via linkage studies for a common form of T2D was CAPN10 that encodes the cysteine protease calpain 10 [18].
Linkage was initially reported among families of Mexican American descent; however, replication in other ethnic groups has been inconsistent [19-24]. The most widely replicated gene for T2D identified via linkage studies is TCF7L2, which began as a modest linkage signal on chromosome 10q in an Icelandic population [25-26]. Common variation in this gene has been shown to associate robustly with T2D in nearly all ethnic groups, with the exception of Pima Indians [27]. These variants have the largest effect on T2D among individuals of European descent. However, despite the progress mentioned above, linkage studies have overall been largely unsuccessful in identifying genes that contribute to T2D.
3.3 Genome-wide association studies (GWASs)
Completion of the Human Genome and the 1000 Genomes sequencing projects, paired with knowledge that neighboring single nucleotide polymorphisms (SNPs) are often in high linkage disequilibrium, and humans inherit large stretches of the genome as haplotypes, has led to the use of tag SNPs to capture genotype information across many other SNPs. Microarrays or "chips" were manufactured, containing a million such tag SNPs which, in theory, capture genotypic information across the entire genome. This microarray technology enabled the advent of the GWAS era, and with it came the discovery of dozens of tag SNPs that associate with T2D. The first GWAS for T2D reported the identification of variants in/near two new "loci" for T2D. The loci were defined by the closest gene/genes in the genomic region captured by an associated variant. These two established loci were HHEX and SLC30A8 (a third locus EXT2-ATX4 has not been confirmed in subsequent studies). This study was followed by several others which confirmed both previous associations and identified additional gene regions [28-31]. Perhaps the most important value of GWAS was the repeated identification of the same top signals in different studies, thus further increasing its credibility. This success resulted in choosing GWAS as the "breakthrough of the year" by Science 2007 [29-31].
3.4 GWAS imputation and meta-analysis
A major challenge of these GWAS analyses was the modest effect size of individual common susceptibility variants. To identify additional variants with similar or smaller effect sizes, larger samples were required. A cost- and time-effective way to increase sample size is to combine data from different GWASs. However, a uniform catalogue of genotyped SNPs across studies was limited, which was in part due to different arrays used for genotyping in different studies. Therefore, the SNP coverage in each study was extended by imputing genotypes for missing SNPs based on patterns of haplotype variation from HapMap [32].
Imputation provided a uniform catalog of SNP genotypes across studies, allowing for ease of data combination, and a novel application of the statistical strategy called meta-analysis, which combined data from different cohorts, at the same time controlling for confounding factors [32, 33]. The major goal of these meta-analyses was to increase the statistical power of the association studies, leading to the identification of robust signals. This required the formation of consortia where different research groups can work together and effectively increase the sample size to increase the statistical power of the study.
Large consortia such as Diabetes Genetics Replication and Meta-Analysis (DIAGRAM) and Meta-Analyses of Glucose and Insulin-related Traits Consortium (MAGIC) have identified numerous loci that associate with diabetes and diabetes-related traits by performing a meta-analysis of data from different GWAS studies, largely in individuals of European descent. In addition to analyzing data from microarrays designed to capture SNPs positioned across the genome, these consortia used data from the custom-designed CardioMetabo chip (Illumina) which contains ~200,000 SNPs previously shown to associate with a metabolic disorder (T2D, lipid disorder, obesity, or cardiovascular disease). Data from the CardioMetabo chip led to the identification of ten new loci associated with T2D in European populations [34].
3.5 GWAS imputation and meta-analysis in diverse ethnic groups
In parallel, a number of GWASs and GWAS meta-analyses of both genotyped and imputed SNPs were also performed in other ethnic groups (East-Asians, South-Asians, African-Americans, North-Indians, American Indians, Mexican Americans, etc.) [35-47]. The majority of SNPs that associated with T2D in one ethnic group showed some evidence of association in other ethnic groups as well. These trans-ethnic findings provided further evidence of the reliability and robustness of GWASs. Imputation was carried out using either HapMap dataset or the 1000 genome dataset and provided dense genotyping that led to the identification of six new T2D loci in Japanese, African Americans, and Mexican and Latin Americans [48-51]. The availability of imputed genetic data in different ancestry groups also enabled meta-analyses using multiple ancestry groups, including one trans-ethnic meta-analysis that analyzed data from European, East Asian, South Asian, Mexican, and Mexican American studies to identify seven new T2D loci [52]. Trans-ancestry meta-analyses also enabled fine-mapping of regions associated with T2D because of differences in linkage disequilibrium across ethnically diverse groups.
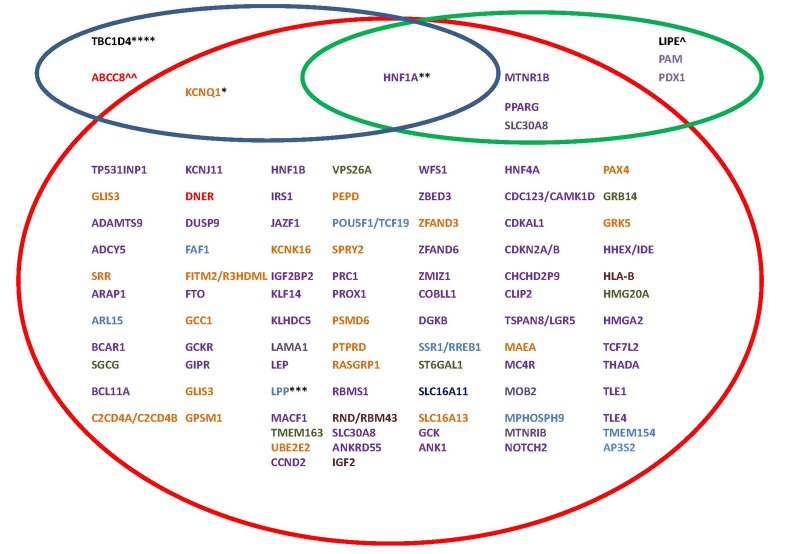
To date, more than 100 variants have been associated with T2D, mapping to 93 loci. These variants have been identified by single GWASs, meta-analyses, and trans-ancestry meta-analyses, with many more likely to be discovered in the future as sample size and diversity increases. Genes reported in these studies are represented in Figure 1 (red circle).
Figure 1. Loci that harbor variants associated with type 2 diabetes [10, 47, 68, 87, 135, 138, 145, 147].
Red circle: Loci harboring common variants with small effect size (OR < 1.5) primarily identified by GWASs. Green circle: Loci harboring low frequency variants with large effect size (OR > 1.8). Blue circle: Loci harboring common variants with large effect size (OR > 1.8). Also indicated are the populations in which the loci were initially detected: Caucasian European, East Asian, South Asian, African American, Pima Indians, Mexican, trans-ancestry meta-analysis (see color code). * Variants in KCNQ1 were first identified in Japanese; subsequently a strong parent-of-origin effect for KCNQ1 variants has been independently shown in family studies of Icelandic and Pima Indian populations. In Pima Indians, one of the KCNQ1 variants explained ~4% of the observed variance in T2D (OR > 1.9). ** A common variant (G319S) in HNF4A has been identified in the Oji-Cree population with an OR of 4.00 when comparing homozygous individuals for either allele. Rare variants in HNF4A have been identified in a Mexican population which increases T2D risk 5-fold. *** Variants in LPP were initially identified by a trans-ancestry meta-analysis; however studies in Pima Indians identified an independent variant associated with T2D. **** A Greenlandic study identified a common nonsense variant in TBC1D4 which increases T2D risk by ~10 fold. ^ A rare nonsense variant in LIPE was identified in the Old Order Amish which substantially increases risk for T2D (OR = 1.8). ^^ A common (3% of the population) missense variant was identified in ABCC8 in Pima Indians that associated with a 2-fold increased risk for T2D.
4. What have we learned about the heritability of T2D?
GWASs and the various strategies to expand the use of GWAS data (meta-analyses, imputation, and trans-ancestry studies) have been enormously successful in identifying new loci for T2D. Only 4 loci identified prior to the "GWAS era" (around the year 2007) meet the current definition of "established" T2D loci (i.e. required significance of p ≤ 5 x 10-8). After the year 2007, more than 100 variants have been identified and "established" according to this criterion, largely as a result of GWAS technology.
4.1 Many variants with modest effects
Historically, the field of complex disease genetics was dominated by the "common disease-common variant" hypothesis. However, based on GWAS data where common variants do not explain the high prevalence of the disease, other mechanisms have been proposed that include:
The infinitesimal model: a large number of small-effect common variants.
The rare allele model: a large number of large-effect rare variants.
The broad sense heritability model: some combination of genotypic, environmental, and epigenetic interaction.
The pros and cons of these models have been reviewed by Greg Gibson in reference [53]. As mentioned before, GWASs for T2D have uncovered multiple variants, but all have modest effect sizes (OR < 1.15) except the TCF7L2 variant which has an OR of roughly 1.50 in non-Hispanic Whites. However, all of these variants in combination explain less than 15% of the genetic component of the disease. Therefore, additional approaches to establish causality at GWAS loci have been created. These approaches include direct sequencing of genes located near GWAS signals in a large number of individuals to identify potential rare coding variants. They led to the identification of very rare coding mutations (frequency <0.1%) in MTNR1B (a previously identified GWAS locus) strongly associated with T2D (OR of 3.31) [54].
Functional studies of these rare variants identified 14 variants that were deleterious. In aggregate, these loss-of-function mutations yielded a 5.5 fold risk for T2D. The neutral variants had no effect on T2D risk. Yet another study identified rare coding variants in PPARG by sequencing the coding exons in ~20,000 multiethnic T2D cases/controls [55]. The authors used an adipocyte differentiation assay to identify functional variants among the observed coding variants, and reported a greater than 7 fold T2D risk in aggregation for the functional variants. As was observed in the study of MTNR1B, the neutral variants in PPARG had no effect on T2D risk in aggregation [55]. If indeed rare variants (near established GWAS loci or within unidentified loci) with large effect sizes contribute to the heritability of T2D, large sample sizes will be required to detect the individual effects of such variants at genome-wide or exome-wide significance.
4.2 Association does not imply causality
Despite the fact that GWASs have been highly successful in identifying new loci for T2D, the translatability of these disease loci for use in disease prediction has been disappointing since the ORs are typically too small to be clinically meaningful (most T2D loci, with the exception of TCF7L2, have an OR < 1.15). A translation for therapeutic advances is also lacking since most of these loci are not known to function in diabetes-related pathways [48]. The majority of SNPs identified by GWASs are intronic or intergenic, and most have been assigned to the nearest gene, even though there is no clear molecular mechanism linking the SNP to the gene or the gene to T2D pathophysiology [48]. For example, obesity-related variants within an intron of the FTO gene were assumed to affect obesity via an effect on the FTO. However, it has recently been shown that these variants affect expression of IRX3, a neighboring gene [56-57]. Conversely, the GWAS-implicated gene may actually be causal in nature, but its role in T2D pathophysiology has not yet been identified by molecular and functional studies. For example, TCF7L2 had no known role in T2D when intronic SNPs were first found to be associated with this disease. Subsequent expression and functional studies have now identified a key role for this gene in pancreatic beta-cell function [58-59].
Another key to the causal variants is to study diverse populations with different haplotype patterns. The majority of GWASs for T2D have been done in European populations, yet a number of observations suggest that studying the genetics of complex diseases in diverse populations can advance our understanding (reviewed in reference [60]). The following aspects are to be considered with this procedure:
Specific risk alleles might occur in specific populations only (e.g. MYBPC3 locus for cardiomyopathy [61]).
If the same variant occurs in different populations, the allele frequency may differ, as has been documented for TCF7L2 and KCNQ1 [27, 62].
The differences in recombination, mutation, and divergence of genealogical lineages can influence how easily one variant can be detectable in one population compared to other.
Disease prevalence may vary among populations, which may affect power, making detection in some populations more likely than in others.
The risk variants can have different effect sizes in different populations (e.g. the APOE locus for Alzheimer's disease [63]).
4.3 Most of the heritability remains unexplained
Although GWASs have identified many loci associated with T2D, the effect size of each variant is quite small, and in aggregate, these variants explain less than 15% of familial aggregation of T2D [34, 48]. To date, there is little explanation for this "unexplained heritability" of T2D. Some have proposed that the unexplained heritability is attributable to a large number of undiscovered common variants with low additive effects, and the disease represents the extremes of the normal distribution [10, 64]. Others have suggested that the unexplained heritability may be clarified by the detection of rare variants with large effect size. It has also been questioned whether established common variants are simply capturing information on nearby rare variants which are the true causal variants [10, 65-66].
Recent advances in next-generation sequencing technology allow the rare variant hypotheses to be directly tested. For example, a recent study in ~2000 Danes uncovered two coding variants in two novel genes associated with T2D [67]. The study was primarily characterized by low-coverage whole exome sequencing. With the rapid progress in next-generation sequencing, which has occurred since 2009, it is likely that additional rare variants with large effects on T2D will be identified, and may account for a larger proportion of the heritability of this disease.
Another source of the unexplained heritability could be structural variants (large insertions and deletions) and copy number variations which can affect numerous loci. GWASs typically capture simple variations such as SNPs, but no well-powered studies have been conducted to analyze structural or copy number variation. Similarly, large-scale epigenetic studies for T2D are lacking. Epigenetic modifications can be stable and heritable across generations and manifest as "parent-of-origin" effect, as observed for variations in KCNQ1 and KLF14, which will be discussed later [62, 68].
4.4 Limited overlap between variants influencing glucose- and insulin-related traits and T2D
As mentioned above in section 4.2, the functional significance of most of the T2D susceptibility variants identified by GWASs is not known. GWASs, such as those conducted in MAGIC, to identify variants influencing glucose- and insulin-related traits can provide some guidance as to how T2D susceptibility genes may affect T2D pathogenesis. Although some of the variants associated with glucose- and insulin-related traits were also found to be associated with T2D (e.g., variants in/near MTNR1B, DGKB, PROX1, ADCY5, GCKR etc.), there was a surprisingly limited overlap between those influencing glucose- or insulin-related traits and those influencing T2D [48]. For example, a meta-analysis of 21 GWASs informative for glucose- and insulin-related traits identified 17 variants associated with fasting glucose or HOMA-B, and found genome-wide significant associations with T2D for only 8 of these variants [8]. Interestingly, two of the variants, in/near ADCY5 and MADD, had similar allele frequency and effect size for association with fasting glucose, but only the variant in ADCY5 was robustly associated with T2D, whereas the one in MADD was not associated [8]. This led to the suggestion that genes regulating physiological levels of these glycemic traits are different from those that regulate the pathophysiological levels that lead to T2D, generating the hypothesis that the mechanism of elevation in fasting glucose rather than elevation itself may be more detrimental for T2D pathogenesis [8]. In accordance with this conclusion, a recent study observed that a combination of variants that increase fasting glucose levels was associated with impaired fasting glucose over a 9-year follow-up, but did not predict the incidence of overt T2D [69]. Conversely, as outlined in a recent review by Bonnefond et al., only ~50% of the known T2D susceptibility genes have been related to either insulin resistance or insulin secretion; the molecular mechanism of the remaining T2D susceptibility genes is generally unknown [48].
5. Using isolated populations to study complex diseases
Family-based association studies in population isolates have been successful in identifying loci associated with Mendelian traits [70]. However, in the study of polygenic complex diseases, where the disease does not necessarily segregate with a single variant, achieving associations with genome-wide significance helps to discern correct from incorrect findings. Studies in population isolates often involve smaller sample sizes than studies in outbred populations, and many lack the power to identify variants with moderate effects. Also, variants identified to be causal may be private or limited to the studied population, and may not be present in other populations, ruling out replication studies. However, now that large-scale outbred populations have failed in explaining the heritability of most complex diseases, and studies are more focused on the role of rare variation which could have larger effect sizes, there is renewed interest in performing studies in genetic isolates [71]. Genetically isolated populations are a powerful resource for the discovery of loci which may be common in the isolate, but rare and difficult to capture in outbred populations [71]. The unique genetic, social, and environmental characteristics of isolated populations can be used to expedite the discovery of disease-associated loci.
5.1 Population isolates
Isolated populations are formed when a subpopulation is derived from a small number of founders as a result of events such as famine, war, infectious disease epidemics, settlement in a new territory, social and cultural barriers, environmental disruption, etc., and when this subpopulation remains secluded for many generations [72-73]. The resulting geographical or cultural isolation has important consequences. Decreased gene flow from neighboring populations and increased endogamy in population isolates often result in increased genomic homogeneity. Population isolates also tend to have environmental and cultural homogeneity [72-73]. Thus, the individuals tend to share a common lifestyle, and have similar exposure to environmental factors. Many isolates also experience multiple population bottlenecks (a marked reduction in sample size followed by survival and expansion of a small random sample of the original population) which alternates with periods of rapid growth, resulting in independent branching and formation of regional subisolates [72]. For instance, in Finland both younger and older population isolates have appeared within one geographical region [72]. As an isolate recovers from the bottleneck, mating occurs between individuals who share a common ancestor. This reduces genetic variability, and affects allele frequencies in the isolated population. Another important aspect of population isolates is that they tend to have very different demographical histories which are influenced by a number of factors such as the total number of founders, number and intensity of bottlenecks, and age and duration of isolation [73]. Detailed genealogical records are frequently available for isolated populations.
5.2 Genetic consequences of isolation
Isolation and population bottlenecks result in genetic drift (random fluctuations in allele frequencies as genes are transmitted from one generation to another) which influences the genetic make-up of a population isolate. Both disease-associated alleles and neutral alleles are subjected to genetic drift in a population isolate [72]. As a result, some disease alleles are lost, whereas others become enriched. Consequently, every population isolate has specific recessive diseases, which are expressed at a higher prevalence in one isolate, but which may be near absent in others [74]. For example, the Pima Indians of Arizona have a very high prevalence of T2D (~50%), but a near absence of type 1 diabetes [9, 75-76]. Certain alleles reach fixation or extinction at a particular locus, which is due to genetic drift. Similarly, diseases causing variants which were rare in the founder population can drift to a much higher frequency in the subsequent generation [72]. This reduced genetic variability and higher frequency of disease-associated alleles can help in the identification of these variants with smaller sample sizes in isolated populations.
Isolation and subsequent genetic drift also affect the haplotype complexity [73]. HapMap and 1000 genome projects have significantly contributed to the understanding of linkage disequilibrium in outbred populations. In contrast to outbred populations, studies of isolated populations have identified extended genomic regions of linkage disequilibrium [77-78]. As expected, older population isolates tend to have shorter regions of linkage disequilibrium than younger population isolates. However, relatively extended regions of linkage disequilibrium can be observed in population isolates with a small number of founder individuals; this may result in a slower rate of growth following a bottleneck [79]. Genetic drift affects rare markers and rare haplotypes in the same way as it affects rare disease alleles. Some rare marker haplotypes can drift up in frequency while others are completely lost. Common marker haplotypes are rarely lost unless the number of founder individuals is very small [72].
6. Advantages of studying population isolates for complex diseases
6.1 Less genetic variability
Isolation, population bottlenecks, and consanguinity lead to a more uniform genetic background. Population isolates have extended regions of linkage disequilibrium and longer haplotypes, requiring fewer markers for GWASs and empowering imputation approaches that depend on predicting haplotype structure.
6.2 Enrichment of rare variants
As rare variants are more recent in origin, they are more likely to be geographically localized. It is also possible that these variants may drift up in frequency in geographically isolated, high-risk populations, which makes the detection of such variants easier. Alternatively, rare alleles could drift down and reach extinction, reducing the extent of genetic variability.
6.3 More uniform environment and lifestyle
Variability within a sample of individuals can be a consequence of environmental and lifestyle influences, and can also lead to differences in the genetic predisposition to T2D. For example, it has been shown that the T2D-predisposing alleles have a higher genetic effect in lean cases than in obese cases [80]. Stratification of GWAS meta-analysis data in Europeans by BMI led to the identification of a new T2D association signal in LAMA1, where a SNP within this gene was associated with T2D only in lean cases, with p = 8.4 × 10-9 and OR = 1.13 in lean cases compared to p = 0.04 and OR = 1.03 in obese cases [80]. Also, for 36 known T2D-associated variants analyzed in this study, 29 had a higher OR for T2D in lean cases, and the overall weighted per-risk allele OR was higher in lean cases than in obese cases [80]. Similarly, the Slim Initiative in Genomic Medicine for the Americas (SIGMA) consortium identified an association between a 5 SNP haplotype in SLC16A11 and T2D in Mexican and Latin Americans, in whom the association was stronger in younger and leaner individuals [50]. This finding was confirmed in a study of 13,267 American Indians who exhibited a significant SNP genotype x BMI interaction, such that the effect of the T2D risk allele was stronger in leaner individuals [81].
6.4 Enrichment of disease prevalence
Genetic drift following a population bottleneck results in the enrichment of certain diseases which may have been rare in the original population. As disease prevalence affects statistical power for genetic association studies, it follows that fewer subjects are required to study a disease with a high population-based frequency, as compared to a low-frequency disease.
6.5 More uniform clinical measures
Variability in association studies may be due to differences in phenotype definition. Smaller population size allows for standardized assessment and characterization of phenotypes, leading to limited variability.
6.6 Better access to patients and their families
Many isolates have good genealogical records due to less migration and more intact families. This provides opportunities for studying a cohort longitudinally, recalling subjects based on genotype, obtaining health records, and conducting family-based association studies.
7. Population Isolates studied for T2D
Research groups have studied the heritability of T2D in different isolated populations, and have provided valuable insights into the genetic pathophysiology of T2D. Below are listed some of the isolated populations in whom genetic studies of T2D have resulted in increased understanding of the disease.
7.1 The Pima Indians of Arizona
The ancestors of American Indians who now live in the Gila River Indian Community in Arizona were the first people to arrive in the Americas 30,000 years ago. They have continued to live in the Sonoran desert near the Gila River in Southern Arizona for more than 2,000 years. These early Americans called themselves “O’Odham”, the river people, and those with whom they intermarried, "Tohono O'Odham", the desert people. Exploring Spaniards called them Pima Indians. Archaeological finds suggest that the modern day Pima Indians descended from the Hohokam, "those who have gone", prehistoric people who originated in Mexico. Migrating from Mexico, they settled in the land up to where the Gila River and the Salt River meet, in what is now Arizona [82-83]. Modern day Pima Indians living in Arizona have minimal European admixture [84].
The Pima Indians were master weavers and farmers, and established a sophisticated system of irrigation to help with the farming needs [85]. Subsequent settlement of the west by people of European ancestry led to the disruption of the irrigation system, effecting Pima agriculture. This resulted in curtailment of subsistence farming, and led to fundamental changes in the lifestyle of Pima Indians from the Gila River. Most of the people from the tribe became dependent on government-issued foods. Early description of Pima Indians from the 1900's suggested that diabetes was either rare or not diagnosed at that time [86-87]. In late 1930's, Joslin identified 21 Pima Individuals with diabetes by reviewing medical records from hospitals serving the Pima population [88]. A survey of rheumatoid arthritis among Pimas in 1963 conducted by the National Institutes of Health led to the discovery of an extremely high rate of diabetes among Pima Indians. Two years later, the National Institutes of Health, the Indian Health Service, and the Pima community started a cooperative effort to understand the etiology of this disease. From 1965 to 2008, a systematic longitudinal study of Pima Indians from the Gila River Community was performed, focused on diabetes research. Individuals, predominantly Pima or the closely related Tohono O'Odham, greater than 5 years of age, were invited to participate in a biennial health examination that included a 75 g oral glucose tolerance test (OGTT) for assessing diabetes status, measurement of BMI, information on pregnancy and health of children, and assessment of diabetes-related complications [9]. Genealogical information was also documented.
This longitudinal study identified an extremely high rate of T2D in Pima Indians. Although the precise reasons underlying the high prevalence and incidence of T2D is not known, genetic factors and higher rates of obesity due to life style changes are suspected as major culprits [9, 89]. Indeed, adult Pima Indians (age >20 years) are more obese than adult non-Hispanic whites, non-Hispanic Blacks, or Mexican Americans living in the United States [9, 90]. This rapid rise in T2D prevalence in Pima Indians living in Arizona has been followed by a relatively stable incidence rate [91]. However, the onset of diabetes has shifted to a younger age. During the past years, the incidence of diabetes among Pima Indians less than 15 years of age has increased nearly six-fold. This shift to a younger age of onset may be a consequence of increasing rates of obesity in children and young adults [92]. Despite diabetes being diagnosed in Pima children as young as 3 years of age, diabetes in this American Indian tribe is exclusively T2D. Diabetes in Pima Indians lacks characteristics of type 1 diabetes such as insulin dependence, low levels of islet cells, and glutamic acid decarboxylase antibodies [93-94]. The absence of type 1 diabetes, a relatively younger onset of diabetes, and minimal admixture with European populations may indicate a more homogenous background for the disease. Additionally, the relatively young age of onset of diabetes in Pima Indians allows for a better estimate of "affected" vs. "unaffected" status for a given individual.
A subset of the Pima Indian population living in Arizona has also volunteered for patient metabolic trait studies in the Clinical Research Center of the Phoenix Epidemiology and Clinical Research Branch of the National Institutes of Health [1]. These examinations included assessing the determinants of T2D when the individual was non-diabetic. Physiologic tests included a 75 g OGTT with measures of insulin at fasting, 30, 60, and 120 minutes. Body composition was measured by underwater weighing and later by dual X-ray absorptiometry. Acute insulin release was measured in response to a 25 g intravenous bolus of glucose and insulin action determined by a euglycemic-hyperinsulinemic clamp [1]. DNA for genetic analysis was obtained from participants of the longitudinal study and the inpatient study.
7.2 The Amish of the Lancaster County
The Amish, named after Jacob Ammann, immigrated to the United States from Western Europe (primarily Switzerland) to escape religious persecution. Earliest immigrants settled in Pennsylvania, whereas latter groups settled in other Midwestern states. Approximately 200 families settled in Lancaster County and are considered founders of the current Lancaster Amish Community. As of the year 2000, the Amish population in and around Lancaster has exceeded 30,000. In 1995, the Amish Family Diabetes Study was initiated in Lancaster to identify genetic determinants of T2D and related traits [95-97].
The Amish represent a religious isolate. Members of this faith are conservative Christians whose rural lives are guided by Ordnung, which promotes religious devotion and family and community cohesion. The primary livelihood is farming, although modern agricultural machinery is not allowed. Use of other technology such as automobiles is also not allowed. The influence of the surrounding culture is minimal. Marriages from outside the sect are not allowed and individuals rarely immigrate [98]. Marriages between first cousins are not allowed, but on average, married couples are more closely related than second cousins once removed, but less related than second cousins [99]. Family genealogies are well documented, dating back to 1700s [95]. The Old Order Amish also have extensive genealogical records which is useful in genetic studies.
Although the Amish are very physically active because of their abstinence from modern technologies, Snitker et al. found Amish adults had a similar mean BMI as the general US non-Hispanic population (BMI = 27.9 kg/m2 in Amish compared with 27.0 kg/m2 in non-Hispanic whites) [100]. They also observed that the Amish were as likely to have an abnormal OGTT compared to non-Hispanic Whites, yet had half the rate of T2D. Higher BMI is a risk factor for the development of diabetes. However, a lower rate of conversion from impaired glucose tolerance to overt T2D in Amish suggest that physical activity may protect against T2D independent of weight loss [100].
7.3 Isolated populations from Finland, Iceland, and Greenland
The vast majority of Finns descended from two immigration waves occurring 4,000 and 2,000 years ago [72]. The earlier were eastern Uralic speakers, and the later were Indo-European speakers from the south. Both Y chromosomal haplotype and mitochondrial sequencing confirm the low genetic diversity among Finns compared to other European populations. The size of the founding population is not known, but as late as the twelfth century, the population was estimated to be only 50,000 [72]. The population size reached 400,000 before the great famine of 1696-1698 wiped out one third of the population. Since then, the Finnish population has expanded to more than 5,000,000. In the sixteenth century, during the reign of Swedish king Gustavus of Vasa (1523-1560), internal migrations created regional subisolates. This time period also saw the establishment of a national system of population records which served as an important resource for later genetic studies. Approximately 30 recessive diseases are highly enriched in Finland, whereas diseases like phenylketonuria and cystic fibrosis are almost nonexistent [72]. According to the Finnish Diabetes Association, at least one third of Finns have a genetic predisposition to develop T2D, and 10-20% have impaired glucose tolerance. In the year 2000, there were 200,000 people with diabetes in Finland. An estimated increase by 70% was predicted for the year 2010. This rapid increase in T2D was mostly due to unhealthy eating habits and reduced physical activity [101].
The population of Iceland was founded in the ninth and tenth century by a limited number of settlers from Norway, Ireland, and Scotland [72]. The country has experienced little immigration over the past centuries, and most of the current residents are descendants of the early settlers. Y-chromosomal haplotypes suggest that 20-25% of Icelandic founding males were from Gaelic origin and 75-80% of Scandinavian origin. A tradition of keeping family trees provides a unique resource to track the heredity of many diseases over hundreds of years [72]. deCODE genetics, a subsidiary of Amgen, has merged DNA from the population of Iceland, their extensive genealogical database, and health records from Iceland's national health-care system to study the genetic basis of a number of complex and rare Mendelian diseases [72].
Greenland was believed to be settled by descendants of the present Inuit culture, who identify the island as Kalaalit Nunaat, "land of the people". It is believed that Greenland’s first inhabitants arrived on the island about 4,500-5,000 years ago, most likely from the island of Ellesmere. But these early Inuit people disappeared from the land about 3,000 years ago for unknown reasons. These were followed by the "Dorset" and the "Thule". In 985 AD, the "Norse", who arrived from Iceland, settled along the east coast of Greenland [102]. In the 18th century, Europeans returned to Greenland, predominantly from Norway and Denmark, and in 1775 Denmark claimed the island as a colony. In 1979, a popular referendum gave Greenland "home rule" status as a distinct nation within the kingdom of Denmark. A hard-to-access geography along with the harsh climate may have made the settlement of the island difficult. The relative isolation from other regions and successive waves of migration may have created conditions for strong bottleneck and other effects of genetic drift. Today, almost 90% of Greenland's population is of Inuit or mixed Inuit/Danish heritage [103]. There is a certain amount of European influence in Greenland culture, but the island nonetheless features unique Inuit and European cultures that are distinct from one another.
7.4 The Natives of Kosrae
Kosrae is an island in the Federated States of Micronesia which was settled by a small number of Micronesian founders ~2,000 years ago. Previous studies using genotyping data from 30 Kosrean trios and ~110,000 genome-wide SNPs showed that Kosraens exhibit strikingly reduced haplotype diversity and extended LD, likely resulting from a strong founder effect and repeated population bottlenecks [104-106]. Indeed, they found that these features were much more dramatic than the "founder" populations of Finland and Iceland. Studies have also reported an increased prevalence of obesity and T2D in natives from Kosrae, similar to that seen in other indigenous populations [107].
7.5 Population from the Adriatic coast of Croatia
This Croatian island population is primarily of Slavic descent, originating from people who emigrated from the mainland at successive time periods [108-109]. This population has remained largely isolated mainly because of geographic separation with minimal immigration from the mainland. Although they share a common descent with Europeans, they are different culturally, practicing a traditional life style based on agricultural subsistence in a rural setting and living on a typical "Mediterranean diet" [109]. However, studies have shown an increased prevalence of obesity and hypertension in this population, suggesting factors other than lifestyle contributing to these conditions [110].
7.6 Oji-Cree population from Canada
The Oji-Cree is a native Canadian population residing in a narrow band extending from the Missinabi river region in Northeastern Ontario at the east and Lake Winnipeg at the west. The Oji-Cree people are descended from historical intermarriage between the Ojibwa and Cree cultures, but are considered a distinct nation from either parent groups. The Oji-Cree population has a very high prevalence of T2D (~40%). Consequently, this population has been studied to understand the genetic factors responsible for this disease [111].
8. Results of studies of T2D in isolated populations
Studies in isolated populations to delineate the heritability of T2D followed much the same pattern as studies in outbred populations. Initial work focused on candidate gene association studies, family-based linkage studies, and GWASs. Recent studies have also utilized whole genome or whole exome sequence data to identify potentially functional variants that were not captured in commercially available genotyping arrays. Some studies in isolated populations have led to several important observations which are either highly significant for the studied population or have advanced our understanding of the pathophysiology of diabetes. Several examples are detailed below.
8.1 HNF1A G319S is a major determinant of T2D in Canadian Oji-Cree population
The Oji-Cree population of Canada has a very high prevalence of T2D (~40%) [111]. In 1999, Hegele et al. reported a common (MAF > 0.05) Glycine319Serine mutation in HNF1A (hepatocyte nuclear factor 1 alpha) that was strongly associated with T2D in Oji-Cree [112]. The Ser319Ser homozygotes and Gly319Ser heterozygotes had an OR of 4.00 and 1.97 respectively compared to Gly319Gly homozygotes [112]. The HNF1A G319S heterozygosity was associated with increased odds of T2D in various subgroups divided by age, gender, and body mass. Among the subgroups examined, the association was strongest in adolescent individuals [112]. Furthermore, this mutation was reported to have specificity and predictive value of 97% and 95% respectively for developing T2D by age 50 [113]. Even though Oji-Cree subjects without this variant display characteristic features of T2D that include obesity, high plasma insulin levels, and insulin resistance, the diabetic phenotype of G319S carriers is more consistent with a defect in insulin secretion, with an earlier onset of diabetes, less obesity, and lower insulin levels, as compared to non-carriers [111].
The mutation was identified using a focused candidate gene approach which analyzed genes that code for proteins known to play a role in diabetes pathophysiology and genes in which mutations were reported to cause MODY (reviewed in [111]). Mutations in HNF1A are known to cause MODY3. Therefore, this gene was studied for sequence variation, leading to the discovery of the Gly319Ser mutation which associated with T2D. To date, the Gly319Ser mutation has not been detected in other ethnic groups. HNF1A belongs to the homeobox gene family of transcription factors and is expressed in the pancreas. It acts as a transcription activator for many genes including insulin [111]. In-vitro functional studies have shown that the G319S mutation does not affect DNA binding or dimerization of HNF1A, but significantly reduces its capacity to transactivate gene expression. This is in contrast to HNF1A mutations in MODY3, which imparted total loss of HNF1A function. This difference in functional impact may explain a reduced penetrance seen with the G319S mutation resulting in T2D [114]. Recent functional studies have also shown that the G319S variant produces two abnormal transcripts that alter the relative ratio of normal splicing products. This combination of abnormal splicing and reduced activity of the G319S protein are the underlying cause of the increase in diabetes susceptibility associated with this mutation [115]. Variations associated with T2D in other genes have also been reported in Oji-Cree. Variants in genes including PTP1B and PPARG associate with T2D in the Oji-Cree; however, HNF1A remains the strongest determinant in this isolated population [116, 117].
8.2 A null mutation in hormone-sensitive lipase (LIPE) increases risk for T2D in the Old Order Amish
Lipolysis plays an important role in energy homeostasis. A study in the Old Order Amish analyzed sequence data from 12 lipolytic-pathway genes in subjects whose fasting triglyceride levels were at the extremes of the distribution, and identified a 19-bp frameshift deletion in exon 9 of LIPE, a key enzyme for lipolysis [118]. LIPE encodes hormone-sensitive lipase (HSL), and primarily hydrolyzes stored triglycerides to free fatty acids in adipose tissue and heart, whereas in steroidogenic tissues, it converts cholesteryl esters to free cholesterol for steroid hormone production. Analysis of the 19-bp frameshift deletion in 2,738 Amish subjects identified an effect on T2D (OR = 1.80 for subjects heterozygous for the deletion compared to subjects without deletion despite similar BMI), dyslipidemia, hepatic steatosis, and systemic insulin resistance [118]. All four subjects who were homozygous for the deletion were diagnosed with T2D before the age of 50 years [118]. Analysis of adipose tissue from study participants who were homozygous for the deletion suggests that the mutation results in the complete absence of HSL protein and a downregulation of transcription factors responsive to PPARG and downstream target genes. Small adipocytes, impaired lipolysis, insulin resistance, and inflammation were also noted [118]. It was recently shown that Pima Indians have different missense mutations in LIPE that also associated with T2D (unpublished data, [119]). These studies highlight the importance of lipolysis in systemic lipid and glucose homeostasis, and the important role of HSL in this process [118]. Apart from variation in LIPE, a GWAS for T2D in Old Order Amish using the Affymetrix 100K SNP array identified significant associations for SNPs in GRB10 (growth factor receptor-bound protein 10) [120].
8.3 A loss-of-function mutation in ABCC8 increases the risk for T2D in Pima Indians
Whole genome sequencing in Pima Indians led to the identification of several novel variants in ABCC8 which encodes the SUR1 subunit of the KATP channel [121]. One novel R1420H variant was significantly associated with T2D in Pima Indians where heterozygous carriers had twice the risk for T2D as non-carriers (OR = 2.02, p = 3.6 × 10-5) [121]. In a community-based study of Pima Indians, 3.3% of the population carried this mutation, and the mean age of diabetes onset was, on average, 7 years earlier for carriers compared with non-carriers (HR = 2.05). Among the 7,528 individuals genotyped, only one individual was homozygous for this mutation. The clinical course of this individual included hypoglycemia, with seizures at the age of 4 months due to hyperinsulinemia, and a diagnosis of diabetes at 3.5 years of age [121]. The R1420H also associated with birth weight where newborns who carried this mutation were approximately 170 grams heavier than non-carriers [121]. This is consistent with the hypothesis of fetal hyperinsulinemia, resulting in increased birth weight. The mutation was functionally characterized, and in vitro studies showed that it reduced KATP channel activity [121].
Mutations in ABCC8 have been previously identified in individuals diagnosed with MODY [122, 123]. Other groups have reported that individuals with congenital hyperinsulinemia due to inactivating ABCC8 mutations eventually developed early- and adult-onset diabetes [124, 125]. Identification of a loss-of-function variant in ABCC8 with a carrier frequency of 3.3% suggested that homozygous carriers of this mutation are not uncommon in this American Indian population. This may have an impact on clinical practice since homozygous carriers may develop hyperinsulinemic hyperglycemia in infancy [121].
8.4 DNER, which functions in the NOTCH signaling pathway, is a T2D gene in Pima Indians
A GWAS of T2D in the Pima Indian population identified DNER as a new gene for T2D [47]. The DNER SNP (rs1861612) was not significantly associated with T2D in Europeans, and there was significant heterogeneity in effect between Europeans and American Indians (p = 1.6 × 10-6; I2 = 95.6%) [47]. However, a recent report identified a directionally consistent, significant association of the DNER SNP (rs1861612) with T2D risk in Han Chinese [126]. Functional studies implicate that DNER regulates notch signaling pathway genes in pancreatic beta-cells [47].
8.5 Variation in GCK can contribute to type 2 diabetes by affecting non-insulin secretory mechanisms in Pima Indians
It has previously been shown that rare mutations in GCK (encodes glucokinase) can cause MODY as a consequence of reduced glucose-stimulated insulin secretion. Glucokinase is the main glucose-phosphorylating enzyme in liver and pancreatic beta-cells, where it converts glucose to G6P as a first and rate-limiting step in glycolysis. In a study of Pima Indians, a novel variant in the 3'UTR of GCK was associated with a lower rate of carbohydrate oxidation, lower 24-hr energy expenditure, and higher risk of T2D [127]. The finding of a lower rate of glucose oxidation, observed post-absorptively, during insulin stimulation, and after a diet of mixed consumption, is consistent with a role of GCK in glycolysis. This variant was not associated with non-oxidative glucose disposal, suggesting that glucose storage (glycogen synthesis) was not affected [127].
8.6 A null mutation in TBC1D4 increases the risk for T2D in the Greenlandic population
Large-scale array-based genotyping and exome sequencing in individuals from Greenland led to the identification of a common nonsense mutation (p.Arg684Ter) in TBC1D4 (frequency = 17%) [128]. TBC1D4 codes for AS160 (AKT substrate of 160 kDa). Homozygous carriers of the mutation have markedly higher concentrations of plasma glucose (beta = 3.8 mmol/l) and serum insulin (beta = 165 pmol/l) two hours after an oral glucose load and a 10-fold increased risk of T2D [128]. The effect size for TBC1D4 on the risk of T2D among individuals in Greenland is much larger than any other gene for T2D. This further highlights the importance of studying diverse populations. Additional details of this study are included in the next Chapter of this RDS Special Edition.
9. Comparison of T2D loci in isolated vs. outbred populations
As detailed above, identification of disease alleles for complex diseases may be simplified by studies in isolated populations because of increased genomic, environmental, and cultural homogeneity [72, 73]. However, this advantage must be balanced with the fact that much of the currently available technology has been customized from outbred populations. For example, commercially available SNP arrays were primarily designed using haplotype and sequence information from outbred populations. Consequently, the SNPs included in these arrays may be less informative for isolated populations because of the difference in linkage disequilibrium and failure to capture variants that are either novel to a population or that occur at a much higher frequency than in outbred populations. Therefore, in addition to performing GWASs, many studies in isolated populations have directly assessed risk alleles identified in outbred populations to determine their effect in isolated populations.
9.1 Analysis of established T2D variants in isolated population
In an early study of established T2D variants (those identified by GWAS prior to 2008), SNPs in/near CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, CDKN2B, and TCF7L2 did not associate with T2D in Pima Indians; only the FTO locus provided evidence for replication [27, 129]. More recently, assessment of seven SNPs identified by genome-wide trans-ancestry meta-analysis for T2D (by the DIAGRAM consortium) in Pima Indians identified nominal associations with T2D for rs6813195 near TMEM154 and rs3130501 near POU5F1-TCF19 [130]. These variants were further associated with diabetes-related traits, including adiposity (rs6813195), 2-hr glucose, and insulin resistance (rs3130501) [130]. Importantly, this study identified an independent variant at the LPP locus which was not reported in the trans-ancestry analysis, highlighting the importance of studying a diverse population for genetics of complex traits [130].
Recently, Hanson et al. reported a comprehensive study of 63 established T2D-associated SNPs in Pima Indians [131]. Eight of these variants were nearly monomorphic in Pima Indians, and nine SNPs showed significant heterogeneity in effect between Pima Indians and Europeans, which may be explained by different patterns of linkage disequilibrium [131]. Among the remaining SNPs, only 9 were nominally significant and directionally consistent between Pima Indians and non-Hispanic Whites. A relatively smaller sample size available for the study of isolated populations may be one of the reasons that T2D-associated variants identified in outbred populations do not achieve statistical significance in isolated populations. However, a genetic risk score derived from all established loci was strongly associated with T2D and with lower insulin secretion in Pima Indians [131]. In a similar study of the role of established T2D and BMI loci on metabolic traits measured in an island population from Croatia, a significant association of TCF7L2 variants with fasting plasma glucose and HbA1c levels was reported [132].
9.2 Allele frequency differences at established T2D loci do not explain the higher diabetes prevalence in Pima Indians (Figures {f2} and {f3})
Several studies have investigated whether the difference in the prevalence of T2D among different populations is attributable to population differences in the frequencies of T2D risk alleles [133, 134]. Recent reports suggest that the difference in allele frequency at established T2D loci between major continental populations is greater than expected, given the genetic distance between the major continental populations. A gradient in genetic risk for T2D has also been proposed, with risk alleles having highest frequencies in Africans and those of lowest frequencies in East Asians [131, 133, 134]. Such divergence in allele frequency at disease-associated loci may represent an effect of natural selection along the course of the evolutionary history of these populations [131]. Given the very high prevalence of T2D in Pima Indians, one might expect higher frequencies of established T2D risk alleles in this isolated population.
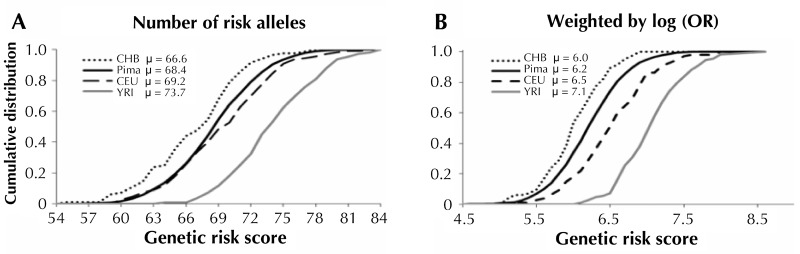
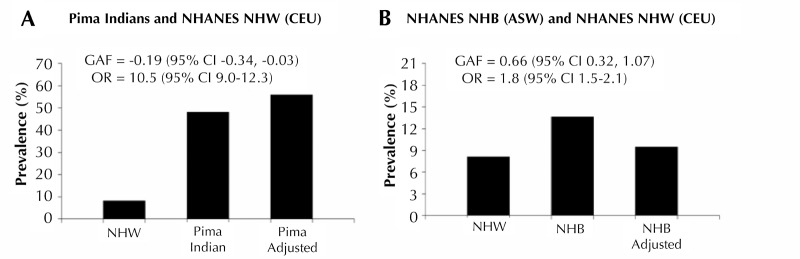
A recent study by Hanson et al. compared the allele frequencies at established T2D loci among Pima Indians, Europeans, Africans, and East Asians. The authors formally tested the significance of the difference in risk allele frequency between Pimas and other major HapMap populations [131]. A mean genetic risk score was used to determine whether T2D risk alleles were systematically higher in one population than another. The mean genetic risk score in Pimas was significantly lower than in Europeans and Africans, but higher than in East Asians (Figure 2). The genetic distances calculated as fixation index (FST) across the T2D loci between Pimas and other continental populations were not significantly different than distances derived using randomly selected markers. This observation suggests that the difference in allele frequency at T2D loci between Pima Indians and other populations is not as different as expected, despite the large genetic distance [131]. The genetic attributable fraction was also calculated; it is defined as the proportion of excess T2D prevalence in a "high-risk population" in relation to a "reference population" that is due to difference in risk allele frequency. The calculation suggested that the differences in allele frequency at established T2D loci account for little of the increased diabetes prevalence in Pimas compared to Europeans (Figure 3). However, a high proportion (66%) of the excess prevalence in non-Hispanic blacks compared to non-Hispanic whites was found to be attributable to differences in allele frequency at these loci [131].
Figure 2. Cumulative distribution of the genetic risk score (GRS) for type 2 diabetes in Pimas and in each of the HapMap populations (Han Chinese in Beijing (CHB), Utah residents with ancestry from northern and western Europe (CEU) and Yoruba in Ibadan (YRI)).
In the left panel (A), the GRS was calculated as the sum of the number of risk alleles across 63 type 2 diabetes loci, while in the right panel (B) it is the sum of the number of risk alleles multiplied by log (OR), as determined in Europeans. Differences in the mean GRS (µ) between populations were compared in a mixed model in which population was a fixed effect and sibship was a random effect. Modified from [131].
Figure 3. Prevalence of T2D risk in Pima Indians compared with non-Hispanic whites from the United States National Health and Nutrition Examination Survey (NHANES, A), and NHANES non-Hispanic blacks compared with non-Hispanic whites before and after adjusting for risk allele frequency in non-Hispanic whites (B).
A: The age-sex-adjusted prevalence of T2D was significantly higher in Pima Indians than in non-Hispanic whites (48.2% compared to 8.2%, OR = 10.5). When adjusted for the frequency of the risk allele in non-Hispanic whites (Pima adjusted), the prevalence in Pimas was slightly higher (55.9%), resulting in a genetic attributable fraction (GAF) of -0.19 (95% CI, -0.34, -0.03); the low value of GAF reflects the lower value of the genetic risk score in Pimas. B: The diagram shows the comparison between non-Hispanic whites and non-Hispanic blacks in NHANES. The "adjusted" value represents the age-sex-adjusted prevalence for the target population adjusted to the frequency of the risk alleles in the reference population across all 63 loci. Modified from [131].
10. Exploring an additional heritable mechanism: parent of origin
Genomic imprinting is a mechanism by which certain genes are expressed in a parent-of-origin-specific manner. If the paternally-derived allele is imprinted, then it is silenced, and the gene is expressed only from the maternally-derived allele and vice versa. Some of the genetic loci associated with T2D map to regions of the genome that are imprinted. However, knowledge of the parental genotypes is required to determine whether variants in imprinted regions have a parent-of-origin effect on disease risk.
An Icelandic study was the first to report a parent-of-origin effect for several loci using a combination of genealogy and long-range phasing to determine the parental origin of alleles [62]. For example, maternal transmission of the C allele of rs2237892 in the KCNQ1 gene was significantly associated with T2D (OR = 1.30, p = 0.008), but the paternal transmission showed no association with disease (p = 0.71) [61]. A second variant in KCNQ1, rs231362, which is not substantially correlated with rs2237892, also showed a maternal inheritance effect on T2D [62]. Similar results were seen for rs4731702 at the KLF14 locus where the effect was again restricted to maternally inherited allele [62]. Parent-of-origin analysis also identified a new variant for T2D, rs2334499 in MOB2, where the T allele was strongly associated with an increased risk of T2D when inherited paternally (OR = 1.35), whereas it was protective when inherited maternally (OR = 0.86). Strikingly, this SNP had a genome-wide significant effect on T2D when parent of origin was taken into account (OR = 1.35, p = 4.7 × 10-10 for the paternal inheritance), but only a nominal association was seen when using the standard case control analysis (OR = 1.08, p = 0.03).
Six SNPs were also analyzed for parent-of-origin effects in relation to T2D and insulin secretory function in Pima Indians [68]. The strongest effect was seen at the KCNQ1 locus. Three SNPs at the KCNQ1 locus (rs2237892, rs2273895, rs2299620) had risk allele frequencies of ~0.50, were in strong linkage disequilibrium, and provided the strongest evidence for association with T2D [68]. In Pimas, the C allele of rs2299620 increased T2D risk when maternally inherited (OR = 1.92), but not when paternally inherited. The maternally derived C allele also associated with a 28% decrease in insulin secretion (p = 0.002), and was found to account for 4% of the risk for T2D in Pima Indians [68]. This represents one of the largest single SNP contributions to T2D risk reported in any population, and demonstrates the importance of collecting family-based data and DNA, which is often facilitated by studying isolated populations where family members have not dispersed.
11. Going forward
Although efforts in the past decade have greatly advanced our understanding of DNA variation contributing to T2D, this knowledge currently has very limited predictive and translational value. Most of the loci identified to date have no known established function in the pathophysiology of T2D; they have been designated as "T2D loci" solely based on their proximity to a GWAS signal. A clear demonstration of causality at the gene and variant level will be important to increase the usefulness of these results in both basic science and translational studies. Whole genome sequencing studies using next generation sequencing, which are currently being explored, may help to achieve these aims.
As genomic studies move farther and farther away from hypothesis-based efforts, and instead aim to explore all genomic information, the complexity of T2D can be daunting. As suggested in a recent review by Froguel et al., experimental biologists and geneticists should not be overwhelmed by the massive amounts of new data, but rather formulate strategies to validate and exploit the new information towards a better understanding of the exact biochemical and molecular pathways involved in T2D [48].
Acknowledgments
Disclosures
The authors report no conflict of interests.
References
- 1.Lillioja S, Mott DM, Spraul M, Ferraro R, Foley JE, Ravussin E, Knowler WC, Bennett PH, Bogardus C. Insulin resistance and insulin secretory dysfunction as precursors of non-insulin-dependent diabetes mellitus. Prospective studies of Pima Indians. N Engl J Med. 1993;329(27):1988–1992. doi: 10.1056/NEJM199312303292703. [DOI] [PubMed] [Google Scholar]
- 2.Mayer-Davis EJ, Costacou T. Obesity and sedentary lifestyle: modifiable risk factors for prevention of type 2 diabetes. Curr Diab Rep. 2001;1(2):170–176. doi: 10.1007/s11892-001-0030-x. [DOI] [PubMed] [Google Scholar]
- 3.Meigs JB, Cupples LA, Wilson PW. Parental transmission of type 2 diabetes: the Framingham Offspring Study. Diabetes. 2000;49(12):2201–2207. doi: 10.2337/diabetes.49.12.2201. [DOI] [PubMed] [Google Scholar]
- 4.Barnett AH, Eff C, Leslie RD, Pyke DA. Diabetes in identical twins. A study of 200 pairs. Diabetologia. 1981;20(2):87–93. doi: 10.1007/BF00262007. [DOI] [PubMed] [Google Scholar]
- 5.Newman B, Selby JV, King MC, Slemenda C, Fabsitz R, Friedman GD. Concordance for type 2 (non-insulin-dependent) diabetes mellitus in male twins. Diabetologia. 1987;30(10):763–768. doi: 10.1007/BF00275741. [DOI] [PubMed] [Google Scholar]
- 6.Poulsen P, Kyvik KO, Vaag A, Beck-Nielsen H. Heritability of type II (non-insulin-dependent) diabetes mellitus and abnormal glucose tolerance - a population-based twin study. Diabetologia. 1999;42(2):139–145. doi: 10.1007/s001250051131. [DOI] [PubMed] [Google Scholar]
- 7.Hanson RL, Imperatore G, Narayan KM, Roumain J, Fagot-Campagna A, Pettitt DJ, Bennett PH, Knowler WC. Family and genetic studies of indices of insulin sensitivity and insulin secretion in Pima Indians. Diabetes Metab Res Rev. 2001;17(4):296–303. doi: 10.1002/dmrr.213. [DOI] [PubMed] [Google Scholar]
- 8.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Knowler WC, Bennett PH, Hamman RF, Miller M. Diabetes incidence and prevalence in Pima Indians: a 19-fold greater incidence than in Rochester, Minnesota. Am J Epidemiol. 1978;108(6):497–505. doi: 10.1093/oxfordjournals.aje.a112648. [DOI] [PubMed] [Google Scholar]
- 10.Prasad RB, Groop L. Genetics of type 2 diabetes-pitfalls and possibilities. Genes (Basel) 2015;6(1):87–123. doi: 10.3390/genes6010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das SK, Elbein SC. The genetic basis of type 2 diabetes. Cellscience. 2006;2(4):100–131. doi: 10.1901/jaba.2006.2-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hani EH, Boutin P, Durand E, Inoue H, Permutt MA, Velho G, Froguel P. Missense mutations in the pancreatic islet beta cell inwardly rectifying K+ channel gene (KIR6.2/BIR): a meta-analysis suggests a role in the polygenic basis of Type II diabetes mellitus in Non-Hispanic Whites. Diabetologia. 1998;41(12):1511–1515. doi: 10.1007/s001250051098. [DOI] [PubMed] [Google Scholar]
- 13.Deeb SS, Fajas L, Nemoto M, Pihlajamäki J, Mykkänen L, Kuusisto J, Laakso M, Fujimoto W, Auwerx J. A Pro12Ala substitution in PPARgamma2 associated with decreased receptor activity, lower body mass index and improved insulin sensitivity. Nat Genet. 1998;20(3):284–287. doi: 10.1038/3099. [DOI] [PubMed] [Google Scholar]
- 14.Winckler W, Weedon MN, Graham RR, McCarroll SA, Purcell S, Almgren P, Tuomi T, Gaudet D, Boström KB, Walker M. et al. Evaluation of common variants in the six known maturity-onset diabetes of the young (MODY) genes for association with type 2 diabetes. Diabetes. 2007;56(3):685–693. doi: 10.2337/db06-0202. [DOI] [PubMed] [Google Scholar]
- 15.Sandhu MS, Weedon MN, Fawcett KA, Wasson J, Debenham SL, Daly A, Lango H, Frayling TM, Neumann RJ, Sherva R. et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Genet. 2007;39(8):951–953. doi: 10.1038/ng2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almind K, Bjørbaek C, Vestergaard H, Hansen T, Echwald S, Pedersen O. Aminoacid polymorphisms of insulin receptor substrate-1 in non-insulin-dependent diabetes mellitus. Lancet. 1993;342(8875):828–832. doi: 10.1016/0140-6736(93)92694-o. [DOI] [PubMed] [Google Scholar]
- 17.Silander K, Mohlke KL, Scott LJ, Peck EC, Hollstein P, Skol AD, Jackson AU, Deloukas P, Hunt S, Stavrides G. et al. Genetic variation near the hepatocyte nuclear factor-4 alpha gene predicts susceptibility to type 2 diabetes. Diabetes. 2004;53(4):1141–1149. doi: 10.2337/diabetes.53.4.1141. [DOI] [PubMed] [Google Scholar]
- 18.Horikawa Y, Oda N, Cox NJ, Li X, Orho-Melander M, Hara M, Hinokio Y, Lindner TH, Mashima H, Schwarz PE. et al. Genetic variation in the gene encoding calpain-10 is associated with type 2 diabetes mellitus. Nat Genet. 2000;26(2):163–175. doi: 10.1038/79876. [DOI] [PubMed] [Google Scholar]
- 19.Baier LJ, Permana PA, Yang X, Pratley RE, Hanson RL, Shen GQ, Mott D, Knowler WC, Cox NJ, Horikawa Y. et al. A calpain-10 gene polymorphism is associated with reduced muscle mRNA levels and insulin resistance. J Clin Invest. 2000;106(7):R69–R73. doi: 10.1172/JCI10665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hegele RA, Harris SB, Zinman B, Hanley AJ, Cao H. Absence of association of type 2 diabetes with CAPN10 and PC-1 polymorphisms in Oji-Cree. Diabetes Care. 2001;24(8):1498–1499. doi: 10.2337/diacare.24.8.1498-a. [DOI] [PubMed] [Google Scholar]
- 21.Evans JC, Frayling TM, Cassell PG, Saker PJ, Hitman GA, Walker M, Levy JC, O'Rahilly S, Rao PV, Bennett AJ. et al. Studies of association between the gene for calpain-10 and type 2 diabetes mellitus in the United Kingdom. Am J Hum Genet. 2001;69(3):544–552. doi: 10.1086/323315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stumvoll M, Fritsche A, Madaus A, Stefan N, Weisser M, Machicao F, Häring H. Functional significance of the UCSNP-43 polymorphism in the CAPN10 gene for proinsulin processing and insulin secretion in nondiabetic Germans. Diabetes. 2001;50(9):2161–2163. doi: 10.2337/diabetes.50.9.2161. [DOI] [PubMed] [Google Scholar]
- 23.Tsai HJ, Sun G, Weeks DE, Kaushal R, Wolujewicz M, McGarvey ST, Tufa J, Viali S, Deka R. Type 2 diabetes and three calpain-10 gene polymorphisms in Samoans: no evidence of association. Am J Hum Genet. 2001;69(6):1236–1244. doi: 10.1086/324646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lynn S, Evans JC, White C, Frayling TM, Hattersley AT, Turnbull DM, Horikawa Y, Cox NJ, Bell GI, Walker M. Variation in the calpain-10 gene affects blood glucose levels in the British population. Diabetes. 2002;51(1):247–250. doi: 10.2337/diabetes.51.1.247. [DOI] [PubMed] [Google Scholar]
- 25.Reynisdottir I, Thorleifsson G, Benediktsson R, Sigurdsson G, Emilsson V, Einarsdottir AS, Hjorleifsdottir EE, Orlygsdottir GT, Bjornsdottir GT, Saemundsdottir J. et al. Localization of a susceptibility gene for type 2 diabetes to chromosome 5q34-q35.2. Am J Hum Genet. 2003;73(2):323–335. doi: 10.1086/377139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grant SF, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38(3):320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 27.Guo T, Hanson RL, Traurig M, Muller YL, Ma L, Mack J, Kobes S, Knowler WC, Bogardus C, Baier LJ. TCF7L2 is not a major susceptibility gene for type 2 diabetes in Pima Indians: analysis of 3,501 individuals. Diabetes. 2007;56(12):3082–3088. doi: 10.2337/db07-0621. [DOI] [PubMed] [Google Scholar]
- 28.Sladek R, Rocheleau G, Rung J, Dina C, Shen L, Serre D, Boutin P, Vincent D, Belisle A, Hadjadj S. et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445(7130):881–8815. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 29.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT; Lund University; Novartis Institutes of BioMedical Research. Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, Roix JJ, Kathiresan S, Hirschhorn JN, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316(5829):1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 30.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU. et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316(5829):1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zeggini E, Scott LJ, Saxena R, Voight BF, Marchini JL, Hu T, de Bakker PI, Abecasis GR, Almgren P, Andersen G. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40(5):638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, Welch RP, Zeggini E, Huth C, Aulchenko YS, Thorleifsson G. et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579–589. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cho YS, Chen CH, Hu C, Long J, Ong RT, Sim X, Takeuchi F, Wu Y, Go MJ, Yamauchi T. et al. Meta-analysis of genome-wide association studies identifies eight new loci for type 2 diabetes in east Asians. Nat Genet. 2011;44(1):67–72. doi: 10.1038/ng.1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kooner JS, Saleheen D, Sim X, Sehmi J, Zhang W, Frossard P, Been LF, Chia KS, Dimas AS, Hassanali N. et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43(10):984–989. doi: 10.1038/ng.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li H, Gan W, Lu L, Dong X, Han X, Hu C, Yang Z, Sun L, Bao W, Li P. et al. A genome-wide association study identifies GRK5 and RASGRP1 as type 2 diabetes loci in Chinese Hans. Diabetes. 2013;62(1):291–298. doi: 10.2337/db12-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ma RC, Hu C, Tam CH, Zhang R, Kwan P, Leung TF, Thomas GN, Go MJ, Hara K, Sim X. et al. Genome-wide association study in a Chinese population identifies a susceptibility locus for type 2 diabetes at 7q32 near PAX4. Diabetologia. 2013;56(6):1291–1305. doi: 10.1007/s00125-013-2874-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Palmer ND, McDonough CW, Hicks PJ, Roh BH, Wing MR, An SS, Hester JM, Cooke JN, Bostrom MA, Rudock ME. et al. A genome-wide association search for type 2 diabetes genes in African Americans. Plos One. 2012;7(1):e29202. doi: 10.1371/journal.pone.0029202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saxena R, Saleheen D, Been LF, Garavito ML, Braun T, Bjonnes A, Young R, Ho WK, Rasheed A, Frossard P. et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes. 2013;62(5):1746–1755. doi: 10.2337/db12-1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shu XO, Long J, Cai Q, Qi L, Xiang YB, Cho YS, Tai ES, Li X, Lin X, Chow WH. et al. Identification of new genetic risk variants for type 2 diabetes. Plos Genet. 2010;6(9):e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tabassum R, Chauhan G, Dwivedi OP, Mahajan A, Jaiswal A, Kaur I, Bandesh K, Singh T, Mathai BJ, Pandey Y. et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes. 2013;62(3):977–986. doi: 10.2337/db12-0406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tsai FJ, Yang CF, Chen CC, Chuang LM, Lu CH, Chang CT, Wang TY, Chen RH, Shiu CF, Liu YM. et al. A genome-wide association study identifies susceptibility variants for type 2 diabetes in Han Chinese. Plos Genet. 2010;6(2):e1000847. doi: 10.1371/journal.pgen.1000847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Unoki H, Takahashi A, Kawaguchi T, Hara K, Horikoshi M, Andersen G, Ng DP, Holmkvist J, Borch-Johnsen K, Jørgensen T. et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat Genet. 2008;40(9):1098–1102. doi: 10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 45.Yamauchi T, Hara K, Maeda S, Yasuda K, Takahashi A, Horikoshi M, Nakamura M, Fujita H, Grarup N, Cauchi S. et al. A genome-wide association study in the Japanese population identifies susceptibility loci for type 2 diabetes at UBE2E2 and C2CD4A-C2CD4B. Nat Genet. 2010;42(10):864–868. doi: 10.1038/ng.660. [DOI] [PubMed] [Google Scholar]
- 46.Yasuda K, Miyake K, Horikawa Y, Hara K, Osawa H, Furuta H, Hirota Y, Mori H, Jonsson A, Sato Y. et al. Variants in KCNQ1 are associated with susceptibility to type 2 diabetes mellitus. Nat Genet. 2008;40(9):1092–1097. doi: 10.1038/ng.207. [DOI] [PubMed] [Google Scholar]
- 47.Hanson RL, Muller YL, Kobes S, Guo T, Bian L, Ossowski V, Wiedrich K, Sutherland J, Wiedrich C, Mahkee D. et al. A genome-wide association study in American Indians implicates DNER as a susceptibility locus for type 2 diabetes. Diabetes. 2014;63(1):369–376. doi: 10.2337/db13-0416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bonnefond A, Froguel P. Rare and common genetic events in type 2 diabetes: what should biologists know? Cell Metab. 2015;21(3):357–368. doi: 10.1016/j.cmet.2014.12.020. [DOI] [PubMed] [Google Scholar]
- 49.Hara K, Fujita H, Johnson TA, Yamauchi T, Yasuda K, Horikoshi M, Peng C, Hu C, Ma RC, Imamura M. et al. Genome-wide association study identifies three novel loci for type 2 diabetes. Hum Mol Genet. 2014;23(1):239–246. doi: 10.1093/hmg/ddt399. [DOI] [PubMed] [Google Scholar]
- 50.SIGMA Type 2 Diabetes Consortium. Williams AL, Jacobs SB, Moreno-Macias H, Huerta-Chagoya A, Churchhouse C, Marquez-Luna C, Garcia-Ortiz H, Gomez-Vazquez MJ, Burtt NP, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506(7486):97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ng MC, Shriner D, Chen BH, Li J, Chen WM, Guo X, Liu J, Bielinski SJ, Yanek LR, Nalls MA. et al. Meta-analysis of genome-wide association studies in African Americans provides insights into the genetic architecture of type 2 diabetes. Plos Genet. 2014;10(8):e1004517. doi: 10.1371/journal.pgen.1004517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I. et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gibson G. Rare and common variants: twenty arguments. Nat Rev Genet. 2012;13(2):135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bonnefond A, Clement N, Fawcett K, Yengo L, Vaillant E, Guillaume JL, Dechaume A, Payne F, Roussel R, Czernichow S. et al. Rare MTNR1B variants impairing melatonin receptor 1B function contribute to type 2 diabetes. Nat Genet. 2012;44(3):297–301. doi: 10.1038/ng.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Majithia AR, Flannick J, Shahinian P, Guo M, Bray MA, Fontanillas P, Gabriel SB et al. GoT2D Consortium; NHGRI JHS/FHS Allelic Spectrum Project; SIGMA T2D Consortium. Rare variants in PPARG with decreased activity in adipocyte differentiation are associated with increased risk of type 2 diabetes. Proc Natl Acad Sci U S A. 2014;111(36):13127–13132. doi: 10.1073/pnas.1410428111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ragvin A, Moro E, Fredman D, Navratilova P, Drivenes O, Engström PG, Alonso ME, de la Calle Mustienes E, Gomez Skarmeta JL, Tavares MJ. et al. Long-range gene regulation links genomic type 2 diabetes and obesity risk regions to HHEX, SOX4, and IRX3. Proc Natl Acad Sci U S A. 2010;107(2):775–780. doi: 10.1073/pnas.0911591107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF. et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mitchell RK, Mondragon A, Chen L, Mcginty JA, French PM, Ferrer J, Thorens B, Hodson DJ, Rutter GA, Da Silva Xavier G. Selective disruption of Tcf7l2 in the pancreatic beta cell impairs secretory function and lowers beta cell mass. Hum Mol Genet. 2015;24(5):1390–1399. doi: 10.1093/hmg/ddu553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Takamoto I, Kubota N, Nakaya K, Kumagai K, Hashimoto S, Kubota T, Inoue M, Kajiwara E, Katsuyama H, Obata A. et al. TCF7L2 in mouse pancreatic beta cells plays a crucial role in glucose homeostasis by regulating beta cell mass. Diabetologia. 2014;57(3):542–553. doi: 10.1007/s00125-013-3131-6. [DOI] [PubMed] [Google Scholar]
- 60.Rosenberg NA, Huang L, Jewett EM, Szpiech ZA, Jankovic I, Boehnke M. Genome-wide association studies in diverse populations. Nat Rev Genet. 2010;11(5):356–366. doi: 10.1038/nrg2760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dhandapany PS, Sadayappan S, Xue Y, Powell GT, Rani DS, Nallari P, Rai TS, Khullar M, Soares P, Bahl A. et al. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat Genet. 2009;41(2):187–191. doi: 10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kong A, Steinthorsdottir V, Masson G, Thorleifsson G, Sulem P, Besenbacher S, Jonasdottir A, Sigurdsson A, Kristinsson KT, Jonasdottir A. et al. Parental origin of sequence variants associated with complex diseases. Nature. 2009;462(7275):868–874. doi: 10.1038/nature08625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tang MX, Stern Y, Marder K, Bell K, Gurland B, Lantigua R, Andrews H, Feng L, Tycko B, Mayeux R. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA. 1998;279(10):751–755. doi: 10.1001/jama.279.10.751. [DOI] [PubMed] [Google Scholar]
- 64.Plomin R, Haworth CM, Davis OS. Common disorders are quantitative traits. Nat Rev Genet. 2009;10(12):872–878. doi: 10.1038/nrg2670. [DOI] [PubMed] [Google Scholar]
- 65.McClellan J, King MC. Genetic heterogeneity in human disease. Cell. 2010;141(2):210–217. doi: 10.1016/j.cell.2010.03.032. [DOI] [PubMed] [Google Scholar]
- 66.Mitchell KJ. What is complex about complex disorders? Genome Biol. 2012;13(1):237. doi: 10.1186/gb-2012-13-1-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Albrechtsen A, Grarup N, Li Y, Sparso T, Tian G, Cao H, Jiang T, Kim SY, Korneliussen T, Li Q. et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56(2):298–310. doi: 10.1007/s00125-012-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanson RL, Guo T, Muller YL, Fleming J, Knowler WC, Kobes S, Bogardus C, Baier LJ. Strong parent-of-origin effects in the association of KCNQ1 variants with type 2 diabetes in American Indians. Diabetes. 2013;62(8):2984–2991. doi: 10.2337/db12-1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Vaxillaire M, Yengo L, Lobbens S, Rocheleau G, Eury E, Lantieri O, Marre M, Balkau B, Bonnefond A, Froguel P. Type 2 diabetes-related genetic risk scores associated with variations in fasting plasma glucose and development of impaired glucose homeostasis in the prospective DESIR study. Diabetologia. 2014;57(8):1601–1610. doi: 10.1007/s00125-014-3277-x. [DOI] [PubMed] [Google Scholar]
- 70.Sheffield VC, Stone EM, Carmi R. Use of isolated inbred human populations for identification of disease genes. Trends Genet. 1998;14(10):391–396. doi: 10.1016/s0168-9525(98)01556-x. [DOI] [PubMed] [Google Scholar]
- 71.Zeggini E. Using genetically isolated populations to understand the genomic basis of disease. Genome Med. 2014;6(10):83. doi: 10.1186/s13073-014-0083-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Peltonen L, Palotie A, Lange K. Use of population isolates for mapping complex traits. Nat Rev Genet. 2000;1(3):182–190. doi: 10.1038/35042049. [DOI] [PubMed] [Google Scholar]
- 73.Hatzikotoulas K, Gilly A, Zeggini E. Using population isolates in genetic association studies. Brief Funct Genomics. 2014;13(5):371–377. doi: 10.1093/bfgp/elu022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arcos-Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61(4):233–247. doi: 10.1034/j.1399-0004.2002.610401.x. [DOI] [PubMed] [Google Scholar]
- 75.Baier LJ, Hanson RL. Genetic studies of the etiology of type 2 diabetes in Pima Indians: hunting for pieces to a complicated puzzle. Diabetes. 2004;53(5):1181–1186. doi: 10.2337/diabetes.53.5.1181. [DOI] [PubMed] [Google Scholar]
- 76.Dabelea D, Hanson RL, Bennett PH, Roumain J, Knowler WC, Pettitt DJ. Increasing prevalence of Type II diabetes in American Indian children. Diabetologia. 1998;41(8):904–910. doi: 10.1007/s001250051006. [DOI] [PubMed] [Google Scholar]
- 77.Service SK, Ophoff RA, Freimer NB. The genome-wide distribution of background linkage disequilibrium in a population isolate. Hum Mol Genet. 2001;10(5):545–551. doi: 10.1093/hmg/10.5.545. [DOI] [PubMed] [Google Scholar]
- 78.Devlin B, Roeder K, Otto C, Tiobech S, Byerley W. Genome-wide distribution of linkage disequilibrium in the population of Palau and its implications for gene flow in Remote Oceania. Hum Genet. 2001;108(6):521–528. doi: 10.1007/s004390100511. [DOI] [PubMed] [Google Scholar]
- 79.Kruglyak L. Prospects for whole-genome linkage disequilibrium mapping of common disease genes. Nat. Genet;22(2):139–144. doi: 10.1038/9642. [DOI] [PubMed] [Google Scholar]
- 80.Perry JR, Voight BF, Yengo L, Amin N, Dupuis J, Ganser M, Grallert H, Navarro P, Li M, Qi L. et al. Stratifying type 2 diabetes cases by BMI identifies genetic risk variants in LAMA1 and enrichment for risk variants in lean compared to obese cases. Plos Genet. 2012;8(5):e1002741. doi: 10.1371/journal.pgen.1002741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Traurig M, Hanson RL, Marinelarena A, Kobes S, Piaggi P, Cole S, Curran JE, Blangero J, Göring H, Kumar S, Nelson RG. et al. Analysis of SLC16A1 variants in 12,811 American Indians: genotype-obesity interaction for type 2 diabetes and an association with RNASEK expression. Diabetes. 2015 doi: 10.2337/db15-0571. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.The Pima Indians: Pathfinders for Health. http://diabetes.niddk.nih.gov/dm/pubs/pima/index.htm.
- 83.Ravussin E, Valencia ME, Esparza J, Bennett PH, Schulz LO. Effects of a traditional lifestyle on obesity in Pima Indians. Diabetes Care. 1994;17(9):1067–1074. doi: 10.2337/diacare.17.9.1067. [DOI] [PubMed] [Google Scholar]
- 84.Williams RC, Steinberg AG, Knowler WC, Pettitt DJ. Gm 3;5,13,14 and stated-admixture: independent estimates of admixture in American Indians. Am J Hum Genet. 1986;39(3):409–413. [PMC free article] [PubMed] [Google Scholar]
- 85.Knowler WC, Pettitt DJ, Saad MF, Bennett PH. Diabetes mellitus in the Pima Indians: incidence, risk factors and pathogenesis. Diabetes Metab Rev. 1990;6(1):1–27. doi: 10.1002/dmr.5610060101. [DOI] [PubMed] [Google Scholar]
- 86.Hrdlicka A. The Bureau of American Ethnology: physiological and medical observations among the Indians of Southwestern United States and Northern Mexico. Washington, DC, U.S. Government Printing Office, 1908. Bulletin. 1908;34:1–347. [Google Scholar]
- 87.Russell F. The Pima Indians. In Twenty-Sixth Annual Report of the Bureau of American Ethnology to Secretary of Smithsonian Institution. U.S. Government Printing Office; Washington, DC: 1908. pp. 3–389. [Google Scholar]
- 88.Joslin EP. The universality of diabetes. JAMA. 1940;115:2033–2038. [Google Scholar]
- 89.Schulz LO, Bennett PH, Ravussin E, Kidd JR, Kidd KK, Esparza J, Valencia ME. Effects of traditional and western environments on prevalence of type 2 diabetes in Pima Indians in Mexico and the U.S. Diabetes Care. 2006;29(8):1866–1871. doi: 10.2337/dc06-0138. [DOI] [PubMed] [Google Scholar]
- 90.Ogden CL, Carroll MD, Curtin LR, McDowell MA, Tabak CJ, Flegal KM. Prevalence of overweight and obesity in the United States, 1999-2004. JAMA. 2006;295(13):1549–1555. doi: 10.1001/jama.295.13.1549. [DOI] [PubMed] [Google Scholar]
- 91.Pavkov ME, Hanson RL, Knowler WC, Bennett PH, Krakoff J, Nelson RG. Changing patterns of type 2 diabetes incidence among Pima Indians. Diabetes Care. 2007;30(7):1758–1763. doi: 10.2337/dc06-2010. [DOI] [PubMed] [Google Scholar]
- 92.Dabelea D, Hanson RL, Bennett PH, Roumain J, Knowler WC, Pettitt DJ. Increasing prevalence of type II diabetes in American Indian children. Diabetologia. 1998;41(8):904–910. doi: 10.1007/s001250051006. [DOI] [PubMed] [Google Scholar]
- 93.Savage PJ, Bennett PH, Senter RG, Miller M. High prevalence of diabetes in young Pima Indians: evidence of phenotypic variation in a genetically isolated population. Diabetes. 1979;28(10):937–942. doi: 10.2337/diab.28.10.937. [DOI] [PubMed] [Google Scholar]
- 94.Dabelea D, Palmer JP, Bennett PH, Pettitt DJ, Knowler WC. Absence of glutamic acid decarboxylase antibodies in Pima Indian children with diabetes mellitus. Diabetologia. 1999;42(10):1265–1266. doi: 10.1007/s001250051303. [DOI] [PubMed] [Google Scholar]
- 95.Hsueh WC, Mitchell BD, Aburomia R, Pollin T, Sakul H, Gelder Ehm M, Michelsen BK, Wagner MJ, St Jean PL, Knowler WC. et al. Diabetes in the Old Order Amish: characterization and heritability analysis of the Amish Family Diabetes Study. Diabetes Care. 2000;23(5):595–601. doi: 10.2337/diacare.23.5.595. [DOI] [PubMed] [Google Scholar]
- 96.Cross HE. Population studies and the old Order Amish. Nature. 1976;262:17–20. doi: 10.1038/262017a0. [DOI] [PubMed] [Google Scholar]
- 97.Gordonsville PA. In association with Katie Beiler. Church Directory of the Lancaster County Amish. Pequea publishers; 1996. 1:320pp, 2:322pp. [Google Scholar]
- 98.McKusick VA. Medical genetic studies of the Amish. Johns Hopkins University; Baltimore, MD: 1978. [Google Scholar]
- 99.Khoury MJ, Cohen BH, Diamond EL, Chase GA, McKusick VA. Inbreeding and prereproductive mortality in the Old Order Amish. I. Genealogic epidemiology of inbreeding. Am J Epidemiol. 1987;125(3):453–461. doi: 10.1093/oxfordjournals.aje.a114551. [DOI] [PubMed] [Google Scholar]
- 100.Snitker S, Mitchell BD, Shuldiner AR. Physical activity and prevention of type 2 diabetes. Lancet. 2003;361:87–88. doi: 10.1016/S0140-6736(03)12156-3. [DOI] [PubMed] [Google Scholar]
- 101.Finnish Diabetes Association. Programme for the prevention of type 2 diabetes in Finland 2003-2010. [Google Scholar]
- 102.Moltke I, Fumagalli M, Korneliussen TS, Crawford JE, Bjerregaard P, Jorgensen ME, Grarup N, Gullov HC, Linneberg A, Pedersen O. et al. Uncovering the genetic history of the present-day Greenlandic population. Am J Hum Genet. 2015;96(1):54–69. doi: 10.1016/j.ajhg.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jorgensen ME, Bjeregaard P, Borch-Johnsen K. Diabetes and impaired glucose tolerance among the inuit population of Greenland. Diabetes Care. 2002;25(10):1766–1771. doi: 10.2337/diacare.25.10.1766. [DOI] [PubMed] [Google Scholar]
- 104.Bonnen PE, Pe'er I, Plenge RM, Salit J, Lowe JK, Shapero MH, Lifton RP, Breslow JL, Daly MJ, Reich DE. et al. Evaluating potential for whole-genome studies in Kosrae, an isolated population in Micronesia. Nat Genet. 2006;38(2):214–217. doi: 10.1038/ng1712. [DOI] [PubMed] [Google Scholar]
- 105.Jakobsson M, Scholz SW, Scheet P, Gibbs JR, VanLiere JM, Fung HC, Szpiech ZA, Degnan JH, Wang K, Guerreiro R. et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature. 2008;451(7181):998–1003. doi: 10.1038/nature06742. [DOI] [PubMed] [Google Scholar]
- 106.Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, Cann HM, Barsh GS, Feldman M, Cavalli-Sforza LL. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319(5866):1100–1104. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 107.Shmulewitz D, Auerbach SB, Lehner T, Blundell ML, Winick JD, Youngman LD, Skilling V, Heath SC, Ott J, Stoffel M. et al. Epidemiology and factor analysis of obesity, type II diabetes, hypertension, and dyslipidemia (syndrome X) on the Island of Kosrae, Federated States of Micronesia. Hum Hered. 2001;51(1-2):8–19. doi: 10.1159/000022953. [DOI] [PubMed] [Google Scholar]
- 108.Rudan P, Roberts DF, Sujoldzic A, Macarol B, Zuskin E, Kastelan A. Strategy of anthropological research on the island of Hvar. Coll Anthropologicum. 1982;6:39–46. [Google Scholar]
- 109.Rudan P, Sujoldzic , Simic D, et al. Population structure in eastern Adriatic: the influence of historical processes, migration patterns, isolation and ecological pressures, and their interaction. In: Roberts DF, Fujiki N, Torizuka K, editors. Isolation, Migration and Health. Cambridge University Press; Cambridge (SSHB): 1992. pp. 204–218. [Google Scholar]
- 110.Smolej-Narancic N, Zagar I. Overweight and fatness in Dalmatia, Croatia: comparison with the US population reference. Coll Antropol. 2000;24:411–421. [PubMed] [Google Scholar]
- 111.Hegele RA, Zinman B, Hanley AJ, Harris SB, Barrett PH, Cao H. Genes, environment and Oji-Cree type 2 diabetes. Clin Biochem. 2003;36(3):163–170. doi: 10.1016/s0009-9120(03)00004-3. [DOI] [PubMed] [Google Scholar]
- 112.Hegele RA, Cao H, Harris SB, Hanley AJ, Zinman B. The hepatic nuclear factor-1alpha G319S variant is associated with early-onset type 2 diabetes in Canadian Oji-Cree. J Clin Endocrinol Metab. 1999;84(3):1077–1082. doi: 10.1210/jcem.84.3.5528. [DOI] [PubMed] [Google Scholar]
- 113.Hegele RA, Cao H, Hanley AJ, Zinman B, Harris SB, Anderson CM. Clinical utility of HNF1A genotyping for diabetes in aboriginal Canadians. Diabetes Care. 2000;23(6):775–778. doi: 10.2337/diacare.23.6.775. [DOI] [PubMed] [Google Scholar]
- 114.Triggs-Raine BL, Kirkpatrick RD, Kelly SL, Norquay LD, Cattini PA, Yamagata K, Hanley AJ, Zinman B, Harris SB, Barrett PH. et al. HNF-1alpha G319S, a transactivation-deficient mutant, is associated with altered dynamics of diabetes onset in an Oji-Cree community. Proc Natl Acad Sci U S A. 2002;99(7):4614–4619. doi: 10.1073/pnas.062059799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Harries LW, Sloman MJ, Sellers EA, Hattersley AT, Ellard S. Diabetes susceptibility in the Canadian Oji-Cree population is moderated by abnormal mRNA processing of HNF1A G319S transcripts. Diabetes. 2008;57(7):1978–1982. doi: 10.2337/db07-1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Mok A, Cao H, Zinman B, Hanley AJ, Harris SB, Kennedy BP, Hegele RA. A single nucleotide polymorphism in protein tyrosine phosphatase PTP-1B is associated with protection from diabetes or impaired glucose tolerance in Oji-Cree. J Clin Endocrinol Metab. 2002;87(2):724–727. doi: 10.1210/jcem.87.2.8253. [DOI] [PubMed] [Google Scholar]
- 117.Hegele RA, Cao H, Harris SB, Zinman B, Hanley AJ, Anderson CM. Peroxisome proliferator-activated receptor-gamma2 P12A and type 2 diabetes in Canadian Oji-Cree. J Clin Endocrinol Metab. 2000;85(5):2014–2019. doi: 10.1210/jcem.85.5.6610. [DOI] [PubMed] [Google Scholar]
- 118.Albert JS, Yerges-Armstrong LM, Horenstein RB, Pollin TI, Sreenivasan UT, Chai S, Blaner WS, Snitker S, O'Connell JR, Gong DW. et al. Null mutation in hormone-sensitive lipase gene and risk of type 2 diabetes. N Engl J Med. 2014;370(24):2307–2315. doi: 10.1056/NEJMoa1315496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Baier LJ, Muller YL, Huang K, Nair AK, Hsueh WC, Chen P, Piaggi P, Knowler WC, Kobes S, Hanson RL. Use of whole genome sequence data to design a custom genotyping chip for American Indians. 75th American Diabetes Association Scientific Sessions; Boston. Jun 5-9, 2015. [Google Scholar]
- 120.Rampersaud E, Damcott CM, Fu M, Shen H, McArdle P, Shi X, Shelton J, Yin J, Chang YP, Ott SH. et al. Identification of novel candidate genes for type 2 diabetes from a genome-wide association scan in the Old Order Amish: evidence for replication from diabetes-related quantitative traits and from independent populations. Diabetes. 2007;56(12):3053–3062. doi: 10.2337/db07-0457. [DOI] [PubMed] [Google Scholar]
- 121.Baier LJ, Muller YL, Remedi MS, Traurig M, Wiessner G, Paolo P, Huang K, Stacy A, Kobes S, Krakoff J, Bennett PH. et al. ABCC8 R1420H loss of function variant in a Southwest American Indian community: association with increased birth weight and doubled risk of type 2 diabetes. Diabetes. 2015 doi: 10.2337/db15-0459. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Bowman P, Flanagan SE, Edghill EL, Damhuis A, Shepherd MH, Paisey R, Hattersley AT, Ellard S. Heterozygous ABCC8 mutations are a cause of MODY. Diabetologia. 2012;55:123–127. doi: 10.1007/s00125-011-2319-x. [DOI] [PubMed] [Google Scholar]
- 123.Johansson S, Irgens H, Chudasama KK, Molnes J, Aerts J, Roque FS, Jonassen I, Levy S, Lima K, Knappskog PM. et al. Exome sequencing and genetic testing for MODY. Plos One. 2012;7:e38050. doi: 10.1371/journal.pone.0038050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Abdulhadi-Atwan M, Bushman J, Tornovsky-Babaey S, Perry A, Abu-Libdeh A, Glaser B, Shyng SL, Zangen DH. Novel de novo mutation in sulfonylurea receptor 1 presenting as hyperinsulinism in infancy followed by overt diabetes in early adolescence. Diabetes. 2008;57:1935–1940. doi: 10.2337/db08-0159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Vieira TC, Bergamin CS, Gurgel LC, Moises RS. Hyperinsulinemic hypoglycemia evolving to gestational diabetes and diabetes mellitus in a family carrying the inactivating ABCC8 E1506K mutation. Pediatr. Diabetes;11:505–508. doi: 10.1111/j.1399-5448.2009.00626.x. [DOI] [PubMed] [Google Scholar]
- 126.Deng Z, Shen J, Ye J, Shu Q, Zhao J, Fang M, Zhang T. Association between single nucleotide polymorphisms of delta/notch-like epidermal growth factor (EGF)-related receptor (DNER) and Delta-like 1 Ligand (DLL 1) with the risk of type 2 diabetes mellitus in a Chinese Han population. Cell Biochem Biophys. 2015;71(1):331–335. doi: 10.1007/s12013-014-0202-3. [DOI] [PubMed] [Google Scholar]
- 127.Muller YL, Piaggi P, Hoffman D, Huang K, Gene B, Kobes S, Thearle MS, Knowler WC, Hanson RL, Baier LJ. et al. Common genetic variation in the glucokinase gene (GCK) is associated with type 2 diabetes and rates of carbohydrate oxidation and energy expenditure. Diabetologia. 2014;57(7):1382–1390. doi: 10.1007/s00125-014-3234-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moltke I, Grarup N, Jorgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, Korneliussen TS, Andersen MA, Nielsen TS, Krarup NT. et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512(7513):190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 129.Rong R, Hanson RL, Ortiz D, Wiedrich C, Kobes S, Knowler WC, Bogardus C, Baier LJ. Association analysis of variation in/near FTO, CDKAL1, SLC30A8, HHEX, EXT2, IGF2BP2, LOC387761, and CDKN2B with type 2 diabetes and related quantitative traits in Pima Indians. Diabetes. 2009;58(2):478–488. doi: 10.2337/db08-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Nair AK, Muller YL, McLean NA, Abdussamad M, Piaggi P, Kobes S, Weil EJ, Curtis JM, Nelson RG, Knowler WC. et al. Variants associated with type 2 diabetes identified by the transethnic meta-analysis study: assessment in American Indians and evidence for a new signal in LPP. Diabetologia. 2014;57(11):2334–2338. doi: 10.1007/s00125-014-3351-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hanson RL, Rong R, Kobes S, Muller YL, Weil EJ, Curtis JM, Nelson RG, Baier LJ. The Role of Established Type 2 Diabetes-Susceptibility Genetic Variants in a High-Prevalence American Indian Population. Diabetes. 2015;64(7):2646–2657. doi: 10.2337/db14-1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Karns R, Zhang G, Jeran N, Havas-Augustin D, Missoni S, Niu W, Indugula SR, Sun G, Durakovic Z, Narancic NS. et al. Replication of genetic variants from genome-wide association studies with metabolic traits in an island population of the Adriatic coast of Croatia. Eur J Hum Genet. 2011;19(3):341–346. doi: 10.1038/ejhg.2010.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Chen R, Corona E, Sikora M, Dudley JT, Morgan AA, Moreno-Estrada A, Nilsen GB, Ruau D, Lincoln SE, Bustamante CD. et al. Type 2 diabetes risk alleles demonstrate extreme directional differentiation among human populations, compared to other diseases. Plos Genet. 2012;8(4):e1002621. doi: 10.1371/journal.pgen.1002621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Corona E, Chen R, Sikora M, Morgan AA, Patel CJ, Ramesh A, Bustamante CD, Butte AJ. Analysis of the genetic basis of disease in the context of worldwide human relationships and migration. Plos Genet. 2013;9(5):e1003447. doi: 10.1371/journal.pgen.1003447. [DOI] [PMC free article] [PubMed] [Google Scholar]