Abstract
Type 2 diabetes (T2D) is an increasing health problem worldwide with particularly high occurrence in specific subpopulations and ancestry groups. The high prevalence of T2D is caused both by changes in lifestyle and genetic predisposition. A large number of studies have sought to identify the genetic determinants of T2D in large, open populations such as Europeans and Asians. However, studies of T2D in population isolates are gaining attention as they provide several advantages over open populations in genetic disease studies, including increased linkage disequilibrium, homogeneous environmental exposure, and increased allele frequency. We recently performed a study in the small, historically isolated Greenlandic population, in which the prevalence of T2D has increased to more than 10%. In this study, we identified a common nonsense variant in TBC1D4, which has a population-wide impact on glucose-stimulated plasma glucose, serum insulin levels, and T2D. The variant defines a specific subtype of non-autoimmune diabetes characterized by decreased post-prandial glucose uptake and muscular insulin resistance. These and other recent findings in population isolates illustrate the value of performing medical genetic studies in genetically isolated populations. In this review, we describe some of the advantages of performing genetic studies of T2D and related cardio-metabolic traits in a population isolate like the Greenlandic, and we discuss potentials and perspectives for future research into T2D in this population.
Keywords: type 2 diabetes, population isolate, linkage disequilibrium, GWAS, genetic drift, loss of function
Abbreviations: ADCY5 - adenylate cyclase 5; APOC3 - apolipoprotein C 3; B99 - Greenland Population Study 1999; CDKN2B - cyclin-dependent kinase inhibitor 2B; CVD - cardiovascular disease; DGKB - diacylglycerol kinase beta; G6PC2 - glucose-6-phosphatase catalytic subunit 2; GCK - glucokinase; GCKR - glucokinase (hexokinase 4) regulator; GLUT4 - glucose transporter 4; GTPase - guanosine triphosphatase; GWAS - genome-wide association studies; HNF1A - hepatocyte nuclear factor 1 alpha; IHIT - Inuit Health in Transition; LD - Linkage disequilibrium; LDL - low-density lipoprotein; MAF - minor allele frequency; MODY - maturity-onset diabetes of the young; mRNA - messenger ribonucleic acid; MTNR1B - melatonin receptor 1B; OGTT - oral glucose tolerance test; OR - odds ratio; PCSK9 - proprotein convertase subtilisin/kexin type 9; RAF - risk allele frequency; SD - standard deviation; SLC16A11 - solute carrier family 16 member 11; SLC30A8 - solute carrier family 30 member 8; T2D - type 2 diabetes; TBC1D4 - TBC1 domain family member 4; TCF7L2 - transcription factor 7 like 2; WHO - World Health Organization
1. Prevalence of type 2 diabetes
Over the past three decades, the global prevalence of type 2 diabetes (T2D) has increased dramatically [1]. Projections estimate that 592 million people will have T2D in 2035 [2]. Thus, T2D is a serious worldwide health problem. In countries such as India, Brazil, and China, the prevalence is increasing dramatically along with major changes in socioeconomic status of large proportions of the population [2]. A major cause for T2D increment is the complex societal change consisting of urbanization and lifestyle change characterized by physical inactivity, increased food intake, and changed food composition. Furthermore, genetic and epigenetic factors contribute to the epidemic, probably in a complex manner interacting with lifestyle and environmental factors [1].
Some populations and ancestry groups feature specific subtypes of T2D [3]. Pima Indians in Arizona represent a classic and well-studied example with a very high prevalence of T2D. Clinically, their diabetes appears to be a subtype of T2D, characterized by obesity, insulin resistance, and a relative insulin deficiency [4, 5]. The increase in prevalence among Pima Indians has occurred along with a transition from a traditional lifestyle with limited food supply and high physical activity to a modern, sedentary lifestyle with a consistent food supply. The Pima Indian population is a population isolate, i.e. the population is derived from a small number of individuals who became isolated, and have remained isolated to a certain degree for many generations. In general, such populations may show a unique profile of rare diseases [6], and their prevalence of common diseases, such as T2D, may also be significantly different from larger, open populations, such as Europeans and Asians. This is especially true when the environment and lifestyle of the isolated population change rapidly under the influence of the modern western lifestyle. As in other populations, some diseases in isolated populations are due to specific genetic variants causing monogenic disease. Other diseases are of a more complex nature, but well-defined molecular subtypes of common diseases are more likely to be present in isolated populations.
The Greenlandic population is another population isolate. As defined by the World Health Organization (WHO) 1999 criteria [7], the prevalence of T2D has increased in Greenland from a very low level in the 1960s to more than 10%, as estimated from two population-based investigations conducted in 1999-2010 [8, 9]. As such, the prevalence of T2D is on the same level as in India and twice as high as in Denmark [2], a country that is politically and culturally linked to Greenland. The increase in T2D prevalence in Greenland can be attributed to several factors. Importantly, the rapid societal change leading to a major change in lifestyle probably plays an important role. However, while urbanization in most other populations is associated with an increased T2D risk, the prevalence of T2D in Greenland has been found to be higher in rural than in urban areas [10]. Furthermore, the results of studies of dietary intake are counterintuitive as Greenlanders eating a traditional Inuit diet with a high content of marine mammals and fish have the highest prevalence of T2D [11]. Changes in traditional lifestyle risk factors therefore do not fully explain the high T2D prevalence. Genetic risk factors may cause some of the high risk of T2D in the Greenlandic population, and may possibly explain some of the surprising epidemiological findings.
2. Genetic characteristics of the Greenlandic population
Despite being the largest island in the world, Greenland has historically been sparsely populated because of its inhospitable environment, with the result that the current population consists of less than 60,000 individuals only. Greenland was one of the last areas on Earth to be populated by humans, and the Inuit ancestors of modern Greenlanders did not arrive in Greenland until less than 1,000 years ago [12]. They migrated to Greenland from the northern part of Canada [12, 13], and lived in the Arctic for thousands of years before reaching Greenland. It is likely that they have adapted both culturally and genetically to the extreme environmental conditions of this region [14]. Historically, both before and after they reached Greenland, they were a small population that lived in isolation. However, in recent centuries, they have admixed with Europeans, which has led to a relatively high proportion of genetic European ancestry in modern Greenlanders. The average proportion of European ancestry is 25%, and more than 80% of modern Greenlanders have some European ancestry [13]. However, the amount of recent European ancestry varies across Greenland, and is far smaller in the more historically isolated areas in the north and east and in the small villages in the south of Greenland [13].
The fact that the Greenlandic population is a historically small and isolated population has the following consequences for its genetic characteristics and thereby for genetic mapping of disease-associated variation:
1. Linkage disequilibrium (LD) is markedly higher in the Greenlanders than in Europeans, although the recent European admixture has reduced LD significantly, except in the ancestral Inuit population [13]. This higher LD makes indirect association mapping more efficient.
2. The Greenlandic population harbors less DNA nucleotide variability than larger open populations, and most variants are either common or absent. The reasons for this are founder effects and increased genetic drift. Founder effects lead to loss of genetic variation. They appear when a new population is founded by a very small number of individuals separated from a larger population. In contrast, increased genetic drift results from historic isolation and small size. Importantly, this implies that relatively few rare alleles are observed in the Greenlandic population. Therefore, disease-causing variants, if not absent in Inuits, will have a higher probability of being common and thereby of holding increased statistical power of association mapping in the Greenlandic population compared to large, open populations. In other words, the history of the Greenlandic population provides several advantages in genetic disease studies including genome-wide association studies (GWAS). These advantages are, at least to some extent, shared with other population isolates.
3. GWAS in population isolates
In the past decade, GWAS in large, open populations such as Europeans and Asians have proven highly successful in identifying common variants associated with complex diseases and traits, among them T2D and related metabolic traits [15-20]. Developments in genotyping and sequencing technologies have further enabled the study of low-frequency and rare genetic variation [18, 21, 22]. However, given that most loci contribute only modest effects in the studied populations, the sample sizes required to obtain sufficient statistical power are enormous, reaching >500,000 European or Asian individuals. Disease-causing variants that are rare in large populations may have higher allele frequencies in population isolates like the Greenlandic population. Studying population isolates may thus provide sufficiently high statistical power to detect the variants by using a much lower number of individuals. This phenomenon has been observed in several association studies performed in population isolates. For instance, a nonsense variant (p.Arg19Ter) in APOC3 has been shown to associate with decreased circulating triglyceride levels and lower risk of cardiovascular disease in Europeans [23, 24] and in a Greek isolate [25], and with decreased circulating triglyceride levels in an Amish population isolate living in the mid-west USA [26]. Importantly, while the variant is rare in Europeans, 3-5% of individuals in the two isolates are heterozygous carriers, which increases statistical power for association in these populations enormously. In fact, while more than 75,000 individuals were necessary to establish the effect in Europeans, less than 1,500 individuals were needed to detect the effect in the Greek population isolate.
Similar observations have been reported in studies of T2D. In the Mexican and Mexican-American population, variants in SLC16A11, which are common in these populations, but rare in Europeans, were found to be associated with a modest increase in T2D prevalence [27]. Furthermore, a whole exome sequencing-based study identified a rare missense variant, which has a high impact on T2D (OR ~5) [28]. This variant is located in HNF1A, a gene in which damaging mutations are known to cause MODY subtype 3 [29]. The variant was also observed at an allele frequency of 2.1% in T2D cases and 0.36% in control individuals in the Mexican population [28]. In comparison, it was found only in 2 of 32,990 non-Finnish Europeans sequenced as part of the Exome Aggregation Consortium [30]. In studies of European isolates, genetic associations with T2D have also been detected. Large-scale sequencing and genotyping studies have been performed in the Icelandic founder population [31], and these efforts have led to the identification of numerous associations of sequence variants with complex diseases, among them both common and rare variants associated with T2D [32, 33]. Many of these findings can be extrapolated to the general European population because of genetic similarity, although population-specific rare alleles have also been detected [33].
Together, these studies show that GWAS in population isolates can be a powerful approach to identify disease-causing variants that are rare in the more commonly studied large, open European and Asian populations. This phenomenon also applies to the study of the Greenlandic Inuit population, which is a founder population, and shares some advantages with other founder populations. However, it is important to note that the Greenlandic population is different from well-studied founder populations (such as the Finnish and the Icelandic) in at least one way: the well-studied founder populations are all genetically comparable to at least one large population, whereas the Greenlandic Inuit population does not have a close genetic relation to a large population. Let us consider the genetic relations between Greenlanders, Europeans, and Han Chinese. Based on Fst, a common measure for population differentiation, Han Chinese represent one of the large populations that is genetically closest related to Greenlandic Inuit, but they are even more closely related to Europeans than to Greenlandic Inuit [13]. A key reason for this genetic differentiation is the fact that the Greenlandic population has been small and isolated for a much longer period than the other populations.
4. The p.Arg684Ter nonsense variant TBC1D4 identified in Greenland imposes high effects on glycemic traits
As a part of population-based health surveys, more than 4,000 Greenlanders were investigated in two cohorts, the Inuit Health in Transition (IHIT, n = 2,733) and the Greenland Population Study 1999 (B99, n = 1,331) [8, 9]. In these surveys, T2D-related phenotypes were collected, including data from an oral glucose tolerance test (OGTT) in most individuals. In initial studies to discover specific risk variants associated with T2D and T2D-defining phenotypes, we performed a genetic association study of variation on the MetaboChip [34] in relation to fasting and 2-h plasma glucose and serum insulin during an OGTT. Using data from the IHIT cohort, we discovered an intronic variant in TBC1D4, which was associated with plasma glucose and serum insulin 2 h after an oral glucose load, and we were able to replicate these associations in the B99 cohort [35]. Exome sequencing in a small subset of the population revealed that the identified associated variant was in high LD with a nonsense TBC1D4 p.Arg684Ter variant not included in the MetaboChip [35]. After genotyping of the TBC1D4 p.Arg684Ter variant in both cohorts, we showed that it was almost certainly the causal variant, and that it mainly imposes increased phenotypic values for homozygous carriers. Approximately 4% of the Greenlanders have two copies of p.Arg684Ter. On average, these individuals had 3.8 mmol/l higher 2-h plasma glucose and 160 pmol/l higher 2-h serum insulin during the OGTT.
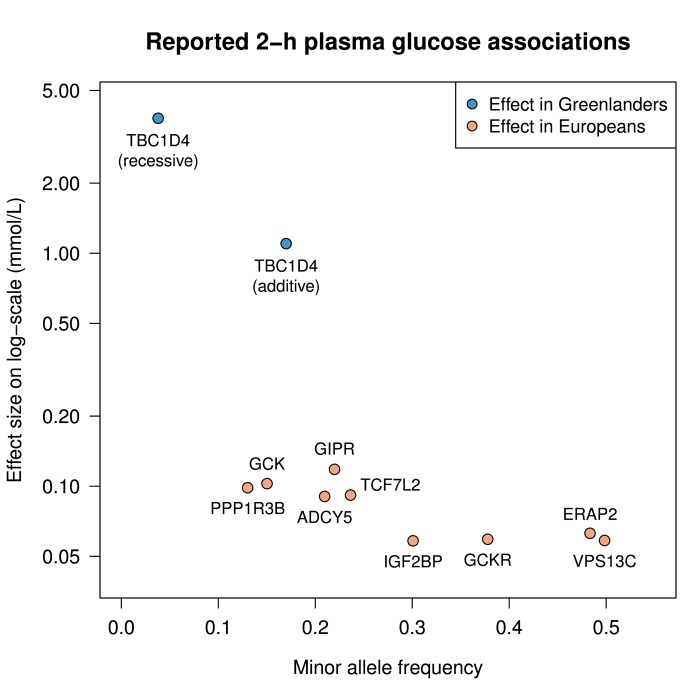
Interestingly, the effect of TBC1D4 p.Arg684 Ter observed on 2-h plasma glucose―1.1 mmol/l (equal to 0.35 SD) per allele in an additive model and 3.8 mmol/l (equal to 1.2 SD) in a recessive model―is higher than previously reported for common variants on complex traits and several times higher than previously seen for 2-h plasma glucose. Figure 1 compares the effect of TBC1D4 with variants previously reported for 2-h plasma glucose in European ancestry individuals [19, 36], and shows that the effect of the TBC1D4 nonsense variant is approximately 10- and 40-fold higher than previously established 2-h plasma glucose loci in an additive and a recessive model, respectively.
Figure 1. The effect of the TBC1D4 p.Arg684Ter variant on 2-h plasma glucose in Greenlanders compared to the effect of reported variants in Europeans.
The figure shows the effects of the reported variants on 2-h plasma glucose [19, 36] in mmol/l as a function of minor allele frequency (as a function of risk genotype frequency, assuming a recessive model). The effect sizes are reported on a log-scale. The TBC1D4 variant is plotted assuming both an additive and a recessive model.
5. The TBC1D4 p.Arg684Ter variant defines a subtype of type 2 diabetes characterized by isolated peripheral insulin resistance
We performed additional association studies showing that the prevalence of T2D (diagnosed according to WHO 1999 guidelines [7]) in carriers of two copies was much higher than in the group of heterozygous carriers and non-carriers (53.1% vs. 9.5%, OR 10.3). This large effect size made it possible to achieve genome-wide statistical significance for association with T2D both in the IHIT discovery cohort with only 223 T2D cases (p = 1.6 x 10-24) and in the replication B99 cohort using only 79 T2D cases (p = 4.6 x 10-12) [35]. Also, homozygous carriers of p.Arg684Ter had a 1 SD decreased insulin sensitivity estimated by the Gutt insulin sensitivity index [37]. Interestingly, while 2-h plasma glucose and 2-h serum insulin were increased, carriers of the p.Arg684Ter variant had decreased fasting plasma glucose and serum insulin, although with markedly smaller effect sizes than for the glucose-stimulated levels. The TBC1D4 p.Arg684Ter nonsense variant is located in an exon that is spliced out in the short isoform of TBC1D4, and therefore theoretically it affects the long isoform only. The long isoform is primarily expressed in skeletal muscle, and homozygous carriers of the variant have nearly immeasurable mRNA and protein expression levels of the long isoform of TBC1D4. Furthermore, these individuals had decreased GLUT4 protein expression [35].
TBC1D4 encodes a protein that acts as a mediator of insulin-stimulated glucose uptake in the cell through its GTPase-activating domain and interactions with Rab proteins. The protein increases GLUT4 translocation when its GTPase activity is inhibited by Akt phosphorylation [38]. It has previously been shown that Tbc1d4-/- knockout mice have decreased basal plasma glucose levels, are resistant to insulin-stimulated glucose uptake in muscle and adipose tissue, and have overall markedly lower GLUT4 levels compared with wild-type littermates [39, 40]. TBC1D4 may also play a role in the insulin-sensitizing effects of exercise [41], as exercise training has been shown to restore TBC1D4 phosphorylation in skeletal muscle from T2D patients [42]. In conclusion, our findings suggest that TBC1D4 p.Arg684Ter in homozygous carriers imposes a reduced insulin-stimulated translocation of GLUT4 to the cell membranes of skeletal muscle, which leads to post-prandial hyperinsulinemia and hyperglycemia. Thus, the variant causes isolated peripheral insulin resistance and eventually T2D.
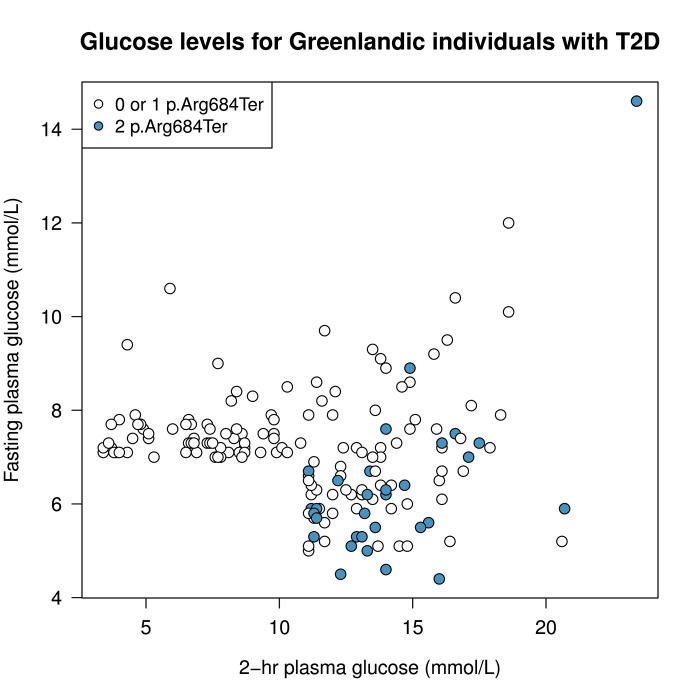
Constitutive recycling of GLUT4 causing an elevated level of GLUT4 in the muscle cell membrane in the fasting state is probably responsible for the decreased fasting levels of glucose and insulin. Whereas regular T2D is characterized by pathophysiologic defects in multiple organs, each contributing to overt disease, the identified homozygous carriers of the TBC1D4 variant seem to suffer from a subtype of T2D featuring a tissue-specific molecular defect. Figure 2 shows fasting and 2-h plasma glucose levels of p.Arg684Ter allele carriers with T2D in the Greenlandic cohorts. These data show that homozygous TBC1D4 p.Arg684Ter carriers are almost always diagnosed with T2D―because of elevated 2-h plasma glucose levels―compared with homozygous p.Arg684Ter non-carriers and heterozygous carriers. Also, it is remarkable that many of the homozygous carriers with T2D have rather low fasting plasma glucose levels, although patients with simultaneously elevated 2-h and fasting plasma glucose also exist (Figure 2). These observations underline the specific nature of this T2D subtype. Given the large effect size of the variant (88% of homozygous carriers above 60 years of age have T2D), it acts almost as a monogenic Mendelian disease-causing variant, which is not rare but quite common, with ~4% of the population being homozygous carriers.
Figure 2. Fasting and 2-h plasma glucose levels in p.Arg684Ter allele carriers with T2D.
Homozygous TBC1D4 p.Arg684Ter variant carriers are shown in blue; homozygous p.Arg684Ter variant non-carriers and heterozygotes are shown in white. Data are from the 172 individuals with screen-detected T2D from the IHIT cohort for whom genotype information for this specific variant was available. T2D was diagnosed according to WHO 1999 criteria [7] based on fasting and 2-h plasma glucose values.
The discovery of such a T2D subtype has the following implications:
Defining a subtype of T2D may have specific implications for treatment and prognosis of the disease, although such knowledge is still lacking.
Given the single molecular defect in patients with this T2D subtype, this finding enables detailed studies of biological and physiological mechanisms in humans.
Epidemiological investigations of the long-term risks associated with this subtype will help to elucidate the relationship between intermediary risk factors, such as isolated postprandial hyperglycemia, and disease endpoints.
T2D is known to impose a risk of cardiovascular disease (CVD), but it has been discussed whether fasting plasma glucose levels, 2-h OGTT plasma glucose, or HbA1c are better predictors for CVD risk [43-45]. Homozygous carriers of the TBC1D4 p.Arg684Ter variant have isolated post-prandial hyperglycemia. Their cardiovascular risk profile can therefore be regarded as empirical evidence for the risk associated with lifelong elevated post-prandial glucose.
6. TBC1D4 p.Arg684Ter – the effect of genetic drift or positive selection
Given the large effect of the variant in TBC1D4 on 2-h plasma glucose, 2-h serum insulin, insulin sensitivity, and T2D it is questionable how and why the variant has a frequency as high as ~17%, with ~4% of the Greenlandic population even being homozygous carriers. According to estimates, the allele frequency in the ancestral Inuit population was even higher (~23%), while the variant was found as a single copy in only 3 out of ~60,000 individuals in the data set of the Exome Aggregation Consortium [30].
There are several possible explanations for how the variant could reach such a high frequency:
It is plausible that the high frequency of the variant is due to random genetic drift, which in small populations, like Greenlandic Inuit, has a rather strong impact on allele frequencies. As described, few variants segregate at low frequency in this population; variants are either lost or driven to high frequency [13].
The high allele frequency of the TBC1D4 variant could also be due to effects of positive selection favoring the variant. While the current interpretation of the TBC1D4 p.Arg684Ter variant is that it is associated with risk of insulin resistance and T2D, the impact on disease endpoints may well have been different in the context of the traditional Inuit diet. Since the variant decreases glucose uptake into skeletal muscle cells, and causes post-prandial hyperglycemia, it may have been an advantage to have the variant when living on a diet high in protein and fat, but low in carbohydrates. Therefore, the variant could have increased in frequency over generations because of positive selection, even though in modern Greenlandic society, it poses a high risk of T2D. We presented suggestive evidence that this is actually the case by showing decreased variability at this locus [34]. However, future studies of selection and interaction between genotype and diet are needed to clarify this question.
7. Genetics of glycemic traits – similarities between the European, Asian, and isolated Greenlandic population
Over the past decade, GWAS with a high number of individuals have detected numerous genetic loci associated with T2D and glycemic traits related to T2D in the European population [15-17, 19, 20]. These loci are primarily represented by common variants, which have rather modest individual effects on diseases and traits. Furthermore, the causal genetic variant and gene have not been identified for most of the detected loci. Previous studies have shown that variants associated with T2D in Europeans are largely transferable to other large populations such as the East and South Asian [17]. However, it is unknown whether the same alleles are associated with metabolic phenotypes in the Greenlandic population. Thus far, ~45 variants have been shown in GWAS to associate with fasting plasma glucose [19, 20, 46] or with 2-h plasma glucose after OGTT [20, 36].
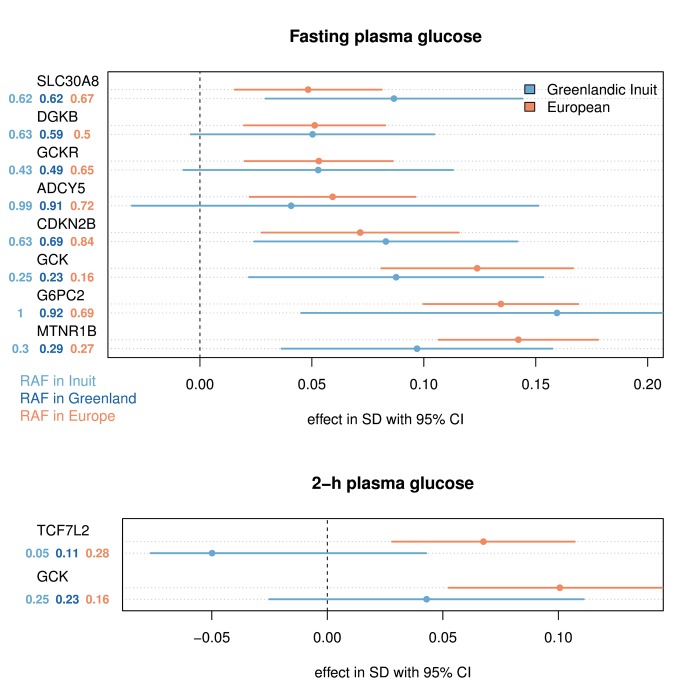
Figure 3 shows the effects of the variants associated with fasting glucose and 2-h glucose seen in patients from the Danish Inter99 cohort (n ≈ 5,700), which is a European cohort, and the Greenlandic IHIT cohort (n ≈ 2,900). Only variants associated with the trait are included in the plot, assuming an additive genetic model (p < 0.01, for joint analysis of Greenlandic IHIT and Danish Inter99). The effect of each variant was assessed, using a method that provides effect size estimates for Europeans on the one hand and unmixed Greenlandic Inuit on the other [47]. These data show that most loci have comparable effects in the European ancestry component and the Greenlandic Inuit population (Figure 3). Variants in the MTNR1B, GCK, DGKB, GCKR, and G6PC2 loci, which are the most impactful fasting glucose loci in Europeans [46], show comparable effects in the Greenlandic Inuit population. However, even though their effect per allele is comparable, their impact on the population, i.e. the amount of heritability explained by the variant (which is proportional to statistical power), is very different in several cases. For example, the CDKN2B and GCKR variants have much higher minor allele frequencies (MAF) in Greenlandic Inuit than in Europeans, and thus have a much greater impact on the Greenlandic population. The opposite is true for the ADCY5 and TCF7L2 variants, which have a higher MAF in Europeans than in Greenlandic Inuit. For variants associated with 2-h plasma glucose, the risk variant in TCF7L2 has effect estimates pointing in the opposite direction in the two populations. However, the two effects are not significantly different (Figure 3). Thus, previously described glycemic risk variants seem to have comparable effects per allele in Greenlandic Inuit and Danish Europeans, but their impact on the two populations differs.
Figure 3. The effect of variants reported to associate with glycemic traits in Europeans and Greenlandic Inuit.
The effects were estimated for the ancestral population using AsaMap [47]. Only variants associated with the trait in an additive model (p < 0.01, for joint analysis of Greenlandic IHIT and B99 (n = 3693 and n = 3437) and Danish Inter99 (n = 6116 and n = 5774)) for fasting and 2-h plasma glucose respectively were included in the plot. No assumptions of similar effects of the rare allele in the two populations were made in this test, and therefore opposite effects in the two populations can still provide a statistical significant association. The effect sizes are given in standard deviation (SD) for the reported risk allele, following a rank-based inverse normal transformation. The risk allele frequency (RAF) is shown for Europeans, Greenlandic Inuit, and the admixed Greenlandic population. The variants included in the plot are: SLC30A8 rs11558471 (A/G), DGKB rs2191349 (T/G), GCKR rs780094 (C/T), ADCY5 rs11708067 (A/G), CDKN2B rs10811661 (T/C), GCK rs4607517 (G/A), G6PC2 rs560887 (G/A), MTNR1B rs10830963 (C/G) and TCF7L2 rs7903146 (C/T). In parentheses are the major/minor alleles mapped on the plus strand with the risk allele in bold face.
8. Characterization of human loss-of-function variants in isolated populations
Homozygosity of the nonsense variant in TBC1D4 is similar to a tissue-specific knockout of the long isoform of TBC1D4, as evidenced by the measurement of mRNA and protein levels in skeletal muscle. In rodent animal models, creating homozygous knockouts has long been a key procedure in understanding the biological and physiological role of specific genes and proteins. While this remains an important tool, differences between mice and men may disrupt biological translation to humans. In the light of this problem, finding humans with homozygous loss-of-function variants is very valuable, because detailed physiological investigations of such individuals can shed light on gene functions in humans, and potentially pave the way for novel treatment targets in human diseases.
A prime example of successful identification and translation of loss-of-function variants is the PSCK9 locus. Individuals with very low LDL-cholesterol levels were found to be homozygous carriers of loss-of-function variants in PCSK9 [48], which led to the development of a novel class of cholesterol-lowering agents as alternatives or adjuvants to statins. In fact, clinical trials have shown that a significant LDL cholesterol-lowering effect is induced by the treatment with monoclonal antibodies against PCSK9 [49].
Although whole-genome and whole-exome sequencing of a large number of European individuals show that each individual harbors approximately 400 loss-of-function variants, most of these disruptive alleles are rare in Europeans [50], making investigations of homozygous human loss-of-function mutation carriers particularly challenging. For example, 40,000 individuals will have to be investigated to identify one homozygous carrier of a loss-of-function allele, which is carried by 1% of the population (MAF ~0.5%). If the frequency of the loss-of-function allele decreases, the required sample size to identify a homozygous loss-of-function allele carrier increases exponentially.
While investigations of heterozygous loss-of-function variants may be of interest, as exemplified on several occasions [51], it adds significant complexity to the biological interpretation since the functioning allele may rescue or alter gene function. One way to address the problem of the extremely low frequency of homozygous loss-of-function carriers is to study population isolates, where founder effects and genetic drift can make such individuals occur more frequently. Indeed, a large set of rare homozygous loss-of-function carriers in the Icelandic population was recently reported, showing that 7.7% of genotyped Icelanders are homozygote or compound heterozygote carriers of loss-of-function variants with a MAF below 2% in 1,171 genes [52]. Similarly, mapping of loss-of-function mutations in Finns showed that, despite fewer variable sites, the average Finn has more low-frequency loss-of-function variants and complete gene knockouts than non-Finnish Europeans [53].
Given the previously described differences between the Icelandic, Finnish, and Greenlandic population, we expect that the frequencies of such variants may be even higher in the Greenlandic population. This can be explained by the higher likelihood of disruptive alleles occurring at higher frequencies in populations that have been small and isolated for a long time. For this reason, the Greenlandic population may be of great value in the context of loss-of-function studies. A forthcoming endeavor will be to map systematically the potential loss-of-function alleles present at sufficient frequency in this population. During this procedure, homozygous carriers can be found, characterized after recall-by-genotype principles, and physiologically evaluated with statistical confidence.
9. Perspectives
Studies of population isolates like the Greenlandic are most suitable to contribute important new discoveries in the era of sequence-based association studies and loss-of-function studies. The advantages of studying population isolates include increased statistical power due to increased LD and higher allele frequencies. These advantages have been well-documented, and have recently been demonstrated through successful identification of complex trait loci.
While the effect of some of these identified loci can be readily translated to other populations, other findings are population-specific. Notably, such loci can still identify the potential pathways involved in the pathogenesis of clinical traits and diseases, as illustrated by the TBC1D4 variant identified in Greenland. This can in turn generate new knowledge of general disease mechanisms, and may eventually lead to new drug targets. Therefore, we believe that future large-scale efforts to combine genome-wide data from a number of isolated populations will add to the interpretation of the genetic impact on metabolic diseases and T2D across isolated populations.
Acknowledgments
Acknowledgments
The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent research center at the University of Copenhagen, partially funded by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk). A.A. is supported by the Villum foundation. I.M. is funded by a grant from the Danish Council for Independent Research, reference number DFF-4090-00244.
Disclosures
The authors report no conflict of interests.
References
- 1.Chen L, Magliano DJ, Zimmet PZ. The worldwide epidemiology of type 2 diabetes mellitus - present and future perspectives. Nat Rev Endocrinol. 2012;8(4):228–236. doi: 10.1038/nrendo.2011.183. [DOI] [PubMed] [Google Scholar]
- 2.IDF Diabetes Atlas, Sixth Edition. http://www.idf.org/diabetesatlas.
- 3.Diamond J. The double puzzle of diabetes. Nature. 2003;423(6940):599–602. doi: 10.1038/423599a. [DOI] [PubMed] [Google Scholar]
- 4.Schulz LO, Chaudhari LS. High-Risk Populations: The Pimas of Arizona and Mexico. Curr Obes Rep. 2015;4(1):92–98. doi: 10.1007/s13679-014-0132-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baier L, Hanson R. Genetic studies of the etiology of type 2 diabetes in Pima Indians: hunting for pieces to a complicated puzzle. Diabetes. 2004;53:1181–1186. doi: 10.2337/diabetes.53.5.1181. [DOI] [PubMed] [Google Scholar]
- 6.Arcos-Burgos M, Muenke M. Genetics of population isolates. Clin Genet. 2002;61(4):233–247. doi: 10.1034/j.1399-0004.2002.610401.x. [DOI] [PubMed] [Google Scholar]
- 7.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15(7):539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 8.Jorgensen ME, Borch-Johnsen K, Stolk R, Bjerregaard P. Fat distribution and glucose intolerance among Greenland Inuit. Diabetes Care. 2013;36(10):2988–2994. doi: 10.2337/dc12-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bjerregaard P, Curtis T, Borch-Johnsen K, Mulvad G, Becker U, Andersen S, Backer V. Inuit health in Greenland: a population survey of life style and disease in Greenland and among Inuit living in Denmark. Int J Circumpolar Health. 2003;62(Suppl 1):3–79. doi: 10.3402/ijch.v62i0.18212. [DOI] [PubMed] [Google Scholar]
- 10.Jorgensen ME, Borch-Johnsen K, Witte DR, Bjerregaard P. Diabetes in Greenland and its relationship with urbanization. Diabet Med. 2012;29(6):755–760. doi: 10.1111/j.1464-5491.2011.03527.x. [DOI] [PubMed] [Google Scholar]
- 11.Jeppesen C, Bjerregaard P, Jorgensen ME. Dietary patterns in Greenland and their relationship with type 2 diabetes mellitus and glucose intolerance. Public Health Nutr. 2014;17(2):462–470. doi: 10.1017/S136898001300013X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gullov HC. Gronlands forhistorie. Gyldendal; 2004. [Google Scholar]
- 13.Moltke I, Fumagalli M, Korneliussen TS, Crawford JE, Bjerregaard P, Jorgensen ME, Grarup N, Gullov HC, Linneberg A, Pedersen O. et al. Uncovering the genetic history of the present-day Greenlandic population. Am J Hum Genet. 2015;96(1):54–69. doi: 10.1016/j.ajhg.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fumagalli M, Moltke I, Grarup N, Racimo F, Bjerregaard P, Jorgensen ME, Korneliussen TS, Gerbault P, Skotte L, Linneberg A. et al. Greenlandic Inuit show genetic signatures of diet and climate adaptation. Science. 2015;349(6254):1343–1347. doi: 10.1126/science.aab2319. [DOI] [PubMed] [Google Scholar]
- 15.Grarup N, Sandholt CH, Hansen T, Pedersen O. Genetic susceptibility to type 2 diabetes and obesity: from genome-wide association studies to rare variants and beyond. Diabetologia. 2014;57(8):1528–1541. doi: 10.1007/s00125-014-3270-4. [DOI] [PubMed] [Google Scholar]
- 16.Morris AP, Voight BF, Teslovich TM, Ferreira T, Segre AV, Steinthorsdottir V, Strawbridge RJ, Khan H, Grallert H, Mahajan A. et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44(9):981–990. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mahajan A, Go MJ, Zhang W, Below JE, Gaulton KJ, Ferreira T, Horikoshi M, Johnson AD, Ng MC, Prokopenko I. et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46(3):234–244. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Albrechtsen A, Grarup N, Li Y, Sparso T, Tian G, Cao H, Jiang T, Kim SY, Korneliussen T, Li Q. et al. Exome sequencing-driven discovery of coding polymorphisms associated with common metabolic phenotypes. Diabetologia. 2013;56(2):298–310. doi: 10.1007/s00125-012-2756-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scott RA, Lagou V, Welch RP, Wheeler E, Montasser ME, Luan Ja, Magi R, Strawbridge RJ, Rehnberg E, Gustafsson S. et al. Large-scale association analyses identify new loci influencing glycemic traits and provide insight into the underlying biological pathways. Nat Genet. 2012;44(9):991–1005. doi: 10.1038/ng.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manning AK, Hivert MF, Scott RA, Grimsby JL, Bouatia-Naji N, Chen H, Rybin D, Liu CT, Bielak LF, Prokopenko I. et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44(6):659–669. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lohmueller KE, Sparso T, Li Q, Andersson E, Korneliussen T, Albrechtsen A, Banasik K, Grarup N, Hallgrimsdottir I, Kiil K. et al. Whole-exome sequencing of 2,000 Danish individuals and the role of rare coding variants in type 2 diabetes. Am J Hum Genet. 2013;93(6):1072–1086. doi: 10.1016/j.ajhg.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mahajan A, Sim X, Ng HJ, Manning A, Rivas MA, Highland HM, Locke AE, Grarup N, Im HK, Cingolani P. et al. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. Plos Genet. 2015;11(1):e1004876. doi: 10.1371/journal.pgen.1004876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jorgensen AB, Frikke-Schmidt R, Nordestgaard BG, Tybjaerg-Hansen A. Loss-of-function mutations in APOC3 and risk of ischemic vascular disease. N Engl J Med. 2014;371(1):32–41. doi: 10.1056/NEJMoa1308027. [DOI] [PubMed] [Google Scholar]
- 24.Crosby J, Peloso GM, Auer PL, Crosslin DR, Stitziel NO. Loss-of-function mutations in APOC3, triglycerides, and coronary disease. N Engl J Med. 2014;371(1):22–31. doi: 10.1056/NEJMoa1307095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tachmazidou I, Dedoussis G, Southam L, Farmaki AE, Ritchie GR, Xifara DK, Matchan A, Hatzikotoulas K, Rayner NW, Chen Y. et al. A rare functional cardioprotective APOC3 variant has risen in frequency in distinct population isolates. Nat Commun. 2013;4:2872. doi: 10.1038/ncomms3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pollin TI, Damcott CM, Shen H, Ott SH, Shelton J, Horenstein RB, Post W, McLenithan JC, Bielak LF, Peyser PA. et al. A null mutation in human APOC3 confers a favorable plasma lipid profile and apparent cardioprotection. Science. 2008;322(5908):1702–1705. doi: 10.1126/science.1161524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The Sigma Type Diabetes Consortium. Williams AL, Jacobs SB, Moreno-Macias H, Huerta-Chagoya A, Churchhouse C, Marquez-Luna C, Garcia-Ortiz H, Gomez-Vazquez MJ, Burtt NP, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506(7486):97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.The Sigma Type Diabetes Consortium. Estrada K, Aukrust I, Bjorkhaug L, Burtt NP, Mercader JM, Garcia-Ortiz H, Huerta-Chagoya A, Moreno-Macias H, Walford G, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311(22):2305–2314. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCarthy MI, Hattersley AT. Learning from molecular genetics: novel insights arising from the definition of genes for monogenic and type 2 diabetes. Diabetes. 2008;57(11):2889–2898. doi: 10.2337/db08-0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.The Sigma Type Diabetes Consortium. Cambridge, MA: http://exac.broadinstitute.org. [Google Scholar]
- 31.Gudbjartsson DF, Helgason H, Gudjonsson SA, Zink F, Oddson A, Gylfason A, Besenbacher S, Magnusson G, Halldorsson BV, Hjartarson E. et al. Large-scale whole-genome sequencing of the Icelandic population. Nat Genet. 2015;47(5):435–444. doi: 10.1038/ng.3247. [DOI] [PubMed] [Google Scholar]
- 32.Grant S, Thorleifsson G, Reynisdottir I, Benediktsson R, Manolescu A, Sainz J, Helgason A, Stefansson H, Emilsson V, Helgadottir A. et al. Variant of transcription factor 7-like 2 (TCF7L2) gene confers risk of type 2 diabetes. Nat Genet. 2006;38:320–323. doi: 10.1038/ng1732. [DOI] [PubMed] [Google Scholar]
- 33.Steinthorsdottir V, Thorleifsson G, Sulem P, Helgason H, Grarup N, Sigurdsson A, Helgadottir HT, Johannsdottir H, Magnusson OT, Gudjonsson SA. et al. Identification of low-frequency and rare sequence variants associated with elevated or reduced risk of type 2 diabetes. Nat Genet. 2014;46(3):294–298. doi: 10.1038/ng.2882. [DOI] [PubMed] [Google Scholar]
- 34.Voight BF, Kang HM, Ding J, Palmer CD, Sidore C, Chines PS, Burtt NP, Fuchsberger C, Li Y, Erdmann J. et al. The metabochip, a custom genotyping array for genetic studies of metabolic, cardiovascular, and anthropometric traits. Plos Genet. 2012;8(8):e1002793. doi: 10.1371/journal.pgen.1002793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moltke I, Grarup N, Jorgensen ME, Bjerregaard P, Treebak JT, Fumagalli M, Korneliussen TS, Andersen MA, Nielsen TS, Krarup NT. et al. A common Greenlandic TBC1D4 variant confers muscle insulin resistance and type 2 diabetes. Nature. 2014;512(7513):190–193. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 36.Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, Vollenweider P, Lyssenko V, Bouatia-Naji N, Dupuis J, Jackson AU. et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42(2):142–148. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gutt M, Davis CL, Spitzer SB, Llabre MM, Kumar M, Czarnecki EM, Schneiderman N, Skyler JS, Marks JB. Validation of the insulin sensitivity index (ISI(0,120)): comparison with other measures. Diabetes Res Clin Pract. 2000;47(3):177–184. doi: 10.1016/s0168-8227(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 38.Sano H, Kane S, Sano E, Mieinea CP, Asara JM, Lane WS, Garner CW, Lienhard GE. Insulin-stimulated Phosphorylation of a Rab GTPase-activating Protein Regulates GLUT4 Translocation. J Biol Chem. 2003;278(17):14599–14602. doi: 10.1074/jbc.C300063200. [DOI] [PubMed] [Google Scholar]
- 39.Wang HY, Ducommun S, Quan C, Xie B, Li M, Wasserman DH, Sakamoto K, Mackintosh C, Chen S. AS160 deficiency causes whole-body insulin resistance via composite effects in multiple tissues. Biochem J. 2013;449(2):479–489. doi: 10.1042/BJ20120702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lansey MN, Walker NN, Hargett SR, Stevens JR, Keller SR. Deletion of Rab GAP AS160 modifies glucose uptake and GLUT4 translocation in primary skeletal muscles and adipocytes and impairs glucose homeostasis. Am J Physiol Endocrinol Metab. 2012;303(10):E1273–E1286. doi: 10.1152/ajpendo.00316.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Treebak JT, Frosig C, Pehmoller C, Chen S, Maarbjerg SJ, Brandt N, MacKintosh C, Zierath JR, Hardie DG, Kiens B. et al. Potential role of TBC1D4 in enhanced post-exercise insulin action in human skeletal muscle. Diabetologia. 2009;52(5):891–900. doi: 10.1007/s00125-009-1294-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vind BF, Pehmoller C, Treebak JT, Birk JB, Hey-Mogensen M, Beck-Nielsen H, Zierath JR, Wojtaszewski JF, Hojlund K. Impaired insulin-induced site-specific phosphorylation of TBC1 domain family, member 4 (TBC1D4) in skeletal muscle of type 2 diabetes patients is restored by endurance exercise-training. Diabetologia. 2011;54(1):157–167. doi: 10.1007/s00125-010-1924-4. [DOI] [PubMed] [Google Scholar]
- 43.Sarwar N, Aspelund T, Eiriksdottir G, Gobin R, Seshasai SR, Forouhi NG, Sigurdsson G, Danesh J, Gudnason V. Markers of dysglycaemia and risk of coronary heart disease in people without diabetes: Reykjavik Prospective Study and systematic review. Plos Med. 2010;7(5):e1000278. doi: 10.1371/journal.pmed.1000278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faerch K, Vistisen D, Johansen NB, Jorgensen ME. Cardiovascular risk stratification and management in pre-diabetes. Curr Diab Rep. 2014;14(6):493. doi: 10.1007/s11892-014-0493-1. [DOI] [PubMed] [Google Scholar]
- 45.Kodama S, Saito K, Tanaka S, Horikawa C, Fujiwara K, Hirasawa R, Yachi Y, Sone Y, Tada Iida K, Shimano H. et al. Fasting and post-challenge glucose as quantitative cardiovascular risk factors: a meta-analysis. J Atheroscler Thromb. 2012;19(4):385–396. doi: 10.5551/jat.10975. [DOI] [PubMed] [Google Scholar]
- 46.Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, Jackson AU, Wheeler E, Glazer NL, Bouatia-Naji N, Gloyn AL. et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Skotte L, Korneliussen TS, Moltke I, Albrechtsen A. Ancestry specific association mapping in admixed populations. 2015. http://dx.doi.org/10.1101/014001. [DOI] [PubMed]
- 48.Cohen J, Pertsemlidis A, Kotowski IK, Graham R, Garcia CK, Hobbs HH. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat Genet. 2005;37(2):161–165. doi: 10.1038/ng1509. [DOI] [PubMed] [Google Scholar]
- 49.Blom DJ, Hala T, Bolognese M, Lillestol MJ, Toth PD, Burgess L, Ceska R, Roth E, Koren MJ, Ballantyne CM. et al. A 52-week placebo-controlled trial of evolocumab in hyperlipidemia. N Engl J Med. 2014;370(19):1809–1819. doi: 10.1056/NEJMoa1316222. [DOI] [PubMed] [Google Scholar]
- 50.MacArthur DG, Balasubramanian S, Frankish A, Huang N, Morris J, Walter K, Jostins L, Habegger L, Pickrell JK, Montgomery SB. et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335(6070):823–828. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Flannick J, Thorleifsson G, Beer NL, Jacobs SB, Grarup N, Burtt NP, Mahajan A, Fuchsberger C, Atzmon G, Benediktsson R. et al. Loss-of-function mutations in SLC30A8 protect against type 2 diabetes. Nat Genet. 2014;46(4):357–363. doi: 10.1038/ng.2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sulem P, Helgason H, Oddson A, Stefansson H, Gudjonsson SA, Zink F, Hjartarson E, Sigurdsson GT, Jonasdottir A, Jonasdottir A. et al. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47(5):448–452. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 53.Lim ET, Würtz P, Havulinna AS, Palta P, Tukiainen T, Rehnström K, Esko T, Mägi R, Inouye M, Lappalainen T. et al. Distribution and Medical Impact of Loss-of-Function Variants in the Finnish Founder Population. Plos Genet. 2014;10(7):e1004494. doi: 10.1371/journal.pgen.1004494. [DOI] [PMC free article] [PubMed] [Google Scholar]