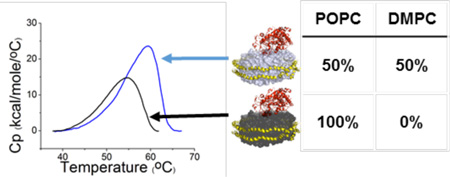
Abstract
Cytochrome P4503A4 (CYP3A4) is a peripheral membrane protein that plays a major role in enzymatic detoxification of many drugs and toxins. CYP3A4 has an integral membrane N-terminal helix and a localized patch comprised of the G′ and F′ helix regions that are embedded in the membrane, but the effects of membrane composition on CYP3A4 function are unknown. Here, circular dichroism and differential scanning calorimetry were used to compare the stability of CYP3A4 in lipid bilayer nanodiscs with varying ratios of 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine to 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC). These lipids differ in the acyl-chain length and their degree of unsaturation. The thermal denaturation of CYP3A4 in nanodiscs occurs in a temperature range distinct from that of the nanodisc denaturation so it can be monitored calorimetrically. Melting temperatures (Tm), heat capacities (ΔCp), and calorimetric enthalpies (ΔHcal) for denaturation of CYP3A4 each increased with an increasing fraction of DMPC, with a maximum at 50% DMPC, before decreasing at 75% DMPC. Addition of the inhibitor ketoconazole results in increased thermal stability, and larger ΔCp and ΔHcal values, with different sensitivities to lipid composition. Effects of lipid composition on ligand binding dynamics were also studied. Equilibrium binding affinities of both ketoconazole (KTZ) and testosterone (TST) were minimally affected by lipid composition. However, stopped-flow analyses indicate that the rates of KTZ binding reach a maximum in membranes containing 50% DMPC, whereas the rate of TST binding decreases continuously with an increasing DMPC concentration. These results indicate that CYP3A4 is highly sensitive to the acyl-chain composition of the lipids and fluidity of the membrane in which it is embedded.
Graphical abstract
The cytochrome P450 monooxygenases (CYPs) dominate enzymatic detoxification and drug metabolism in humans.1–4 Among the hepatic membrane-bound CYPs, CYP3A4 is the isoform that contributes most to drug clearance and its substrate promiscuity results in drug–drug inter-actions.5,6 The resolution of several X-ray crystal structures of various CYP isoforms has provided mechanistic details about the CYP catalytic cycle, as well as a basis for understanding their overlapping, but distinct, substrate selectivities.7–11 However, the effects of membrane composition, and lipid acyl-chain structure in particular, on the structure and function of CYPs are unknown.
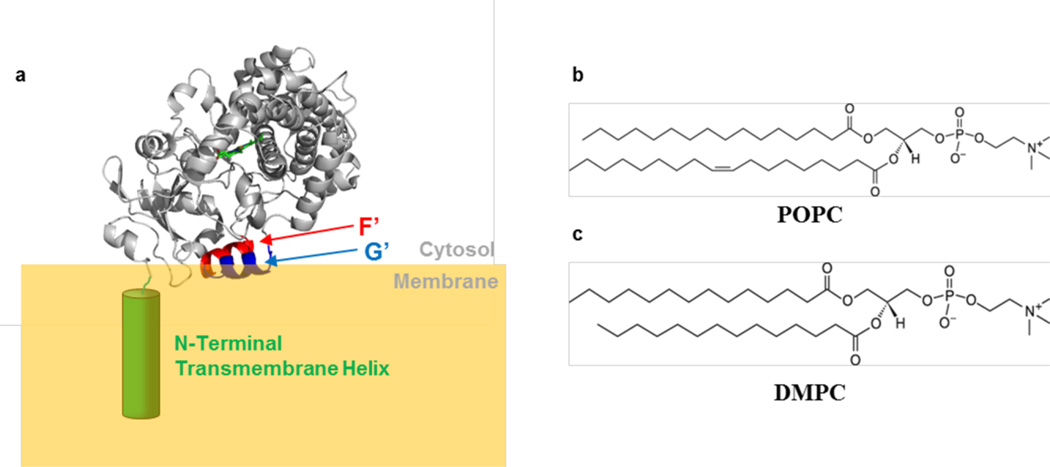
Interactions of CYP3A4 with the membrane are likely to be a critical determinant of enzyme function and stability. On the basis of H/D exchange mass spectrometry and molecular dynamics (MD) simulations, CYP3A4 adopts an orientation on the membrane with the N-terminal membrane anchor deeply embedded, and with the F′ and G′ helices inserted less deeply into the lipid phase (Figure 1a).12–14 Most of the remainder of the protein appears to interact only with the membrane surface or bulk solution. Several putative access or egress channels for substrates or products have been suggested, and it is expected that substrates and inhibitors access the active site through the membrane.14,15 However, experimental methods that report on the CYP3A4-membrane interactions are not well developed. Furthermore, the thermal stabilities of membrane-bound CYPs, and membrane-bound proteins in general, have been difficult to assess in the presence of a lipid bilayer.
Figure 1.
(a) Putative orientation of CYP3A4 in the lipid bilayer. This representation is consistent with many MD simulations and H/DX mass spectrometry. The N-terminal helix is expected to insert deeply into the membrane, whereas the F′ and G′ helices insert less deeply (Protein Data Bank entry 1TQN11). The structures of (b) POPC and (c) DMPC are also shown.
Lipid nanodiscs make up a model membrane platform that allows for the interrogation of CYP3A4 properties in a lipid bilayer.16–20 Because of their narrow size distribution (~10 nm diameter), nanodiscs make possible the study of monomeric CYPs, or CYPs in complex with associated redox partner proteins, in a membrane of defined lipid composition. In contrast, it is often more difficult to control the size distribution and lamellarity of liposomes.17 Consequently, it is difficult to study the monomeric form of membrane proteins in liposomes, without the potential for transient or persistent, and poorly controlled, protein–protein interactions. We recently reported a calorimetric study of the stability of CYP3A4 in POPC lipid nanodiscs, which demonstrated a very significant membrane-induced stabilization.21 Here we extend those studies to nanodiscs containing varying ratios of POPC and DMPC (Figure 1a,b). The results suggest correlations among the lipid acyl-chain structure or membrane fluidity, the thermal stability of CYP3A4, substrate binding kinetics, and the heat capacity of the folded CYP. The results, therefore, emphasize the sensitivity of the stability and function of CYP3A4 to its lipid environment.
MATERIALS AND METHODS
Protein Expression and Nanodisc Self-Assembly
CYP3A4 and MSP1D1 expression and purification and CYP3A4 nanodisc assembly were performed as described previously with routine use of disc forming buffer (DFB).21 Briefly, CYP3A4 was expressed in Escherichia coli C41(DE3) cells transformed with pCWOri+ encoding ampicillin resistance and a CYP3A4 gene with an N-terminal deletion of residues 3–12 and further modified with a C-terminal His tag. The protein also includes a serine to phenylalanine substitution at position 18. Most of the membrane anchor residues, residues 1, 2, and 13–27 of full length CYP3A4, are contained in the construct used here for all experiments. The sequence of the N-terminus of the wild type is NH2-MALIPDLAMETWLLLAVSLVLL…, where the underlined residues are deleted in the construct used here and the bold residue is a Phe to Ser replacement. For Laurdan-containing nanodiscs, 1% (w/w) Laurdan was included in the lipid film prior to initiation of the nanodisc self-assembly process. Measurement of Laurdan generalized polarization (LGP) was done with an Aminco AB2 fluorimeter with 5 mm excitation and emission slits at room temperature. The Laurdan generalized polarization was determined from
where I440 and I490 are the relative fluorescence emission intensities at 440 and 490 nm, respectively.
The total lipid:MSP1D1 ratios for nanodiscs containing a mixture of POPC and DMPC were empirically optimized; nanodisc formation was validated using size-exclusion chromatography prior to CYP3A4 nanodiscs, and the percent yields for each CYP3A4-nanodisc lipid composition was ~60% for pure POPC, ~70% for a 3:1 POPC:DMPC ratio, ~90% for a 1:1 POPC:DMPC ratio, and ~35% for a 1:3 POPC:DMPC ratio. The lipid:MSP1D1 ratios that yielded optimal nanodisc formation for each lipid composition are summarized in Table S1.
Circular Dichroism (CD)
Circular dichroism (CD) spectroscopy was performed using a Jasco 720 circular dichroism spectrophotometer (Jasco, Inc., Easton, MD). The instrument was allowed to pre-equilibrate at 25 °C for 10 min prior to the acquisition of data. To prevent the dynode from exceeding 700 V, CYP3A4 and CYP3A4 nanodiscs were diluted to a total CYP3A4 concentration of 1 µM with CD buffer [25 mM potassium phosphate and 50 mM sodium fluoride (pH 7.4)] prior to analysis. Phosphate and chloride ions are known to absorb in the far-UV region of interest, and as such, it was necessary to lower the phosphate concentration as well as substitute sodium chloride in our buffer with sodium fluoride. The sample was loaded into a 0.1 cm path length quartz cuvette and allowed to equilibrate inside the CD spectrophotometer for 5 min at 25 °C prior to the collection of data. The instrument was set to record data from 25 to 85 °C with increments of 10 °C. Eight replicate scans were collected from 195 to 270 nm in continuous scanning mode for each temperature, with a scan speed of 100 nm/min and a 2 nm bandwidth. The temperature slope was set to 1 °C/min, and samples were allowed to equilibrate for an additional 5 min after the instrument reached a new temperature increment prior to the collection of data.
CD spectra were smoothed, buffer-subtracted, and corrected for CYP3A4 concentration. For CYP3A4 nanodiscs, to correct for the contribution of the MSP1D1 from the nanodisc, CD spectra were collected with empty nanodiscs in the same manner as described above. Assuming that CYP3A4 does not significantly alter the MSP1D1 CD signal, the nanodisc raw spectra at each temperature were then scaled for concentration and subtracted from the raw spectrum of the CYP3A4 nanodisc at the corresponding temperature.
At 85 °C, the state of CYP3A4 is not defined by this approach. However, the fractional conversion to this thermally generated, but undefined, state was used to compare empirically CYP3A4 in buffer versus that in nanodiscs. Specifically, the fractional change at 222 nm versus temperature was considered as the fraction of denatured protein by fitting to a logistical function (eq 1). This is a general equation frequently used to determine the midpoint of a sigmoidal curve.
| (1) |
where T is the temperature, Tm is the melting temperature, and k is a measure of the cooperativity of unfolding, with larger values indicating higher cooperativity.
The secondary structural content was estimated from far-UV CD spectra recorded at 25 C using CONTINLL, CDSSTR, and SELCON3 from the CDPro software package.22,23
Ligand Binding Titrations
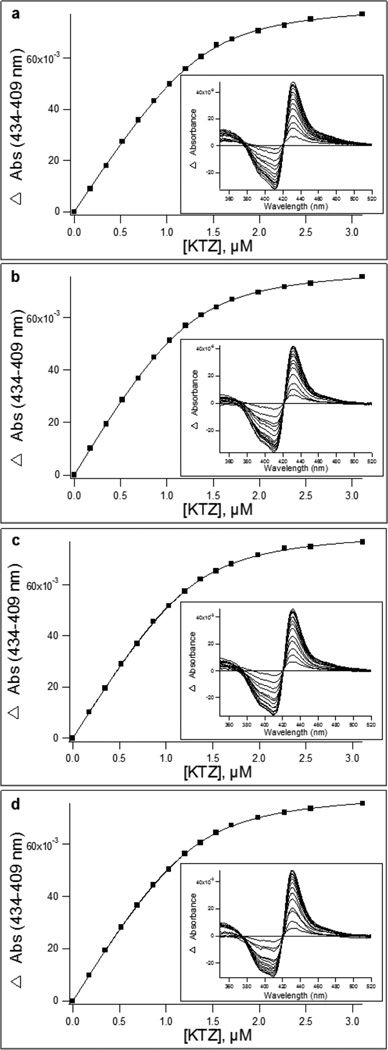
Binding titrations were performed at 25 °C and a scan range of 350–700 nm using an Aminco DW2a spectrophotometer (Olis, Inc., Norcross, GA). Ketoconazole (KTZ) was purchased from Sigma-Aldrich (St. Louis, MO), and 75 and 125 µM stocks were made in 50% (v/v) methanol. A sample cuvette was filled with CYP3A4 nanodiscs diluted to 1.4 µM with DFB. A reference cuvette was filled with an equivalent concentration of empty nanodiscs in DFB to best compensate for any potential effects from binding of ligand to the nanodisc membrane. Cuvettes were allowed to equilibrate to temperature for 10 min. Prior to the addition of KTZ, a ligand-free absolute absorbance spectrum was recorded. Binding of KTZ to the enzyme was then monitored by titrating equal volumes of ligand into each cuvette and recording absolute spectra after each addition of ligand. The final concentration of methanol in both cuvettes did not exceed 1% (v/v). Data were baseline-corrected and corrected for the change in volume, and difference spectra were calculated by subtracting the ligand-free spectrum for CYP3A4 nanodiscs from the spectra recorded from each subsequent addition of ligand. Binding isotherms were generated by plotting the difference between the absorbance maximum and minimum of the spectra as a function of ligand concentration. To determine the dissociation constant, KD, IgorPro (Wavemetrics, Inc.) was used to fit data using a quadratic equation for tight binding ligands (eq 2):
| (2) |
where ΔAbs is the change in peak-to-trough absorbance, Absmax is the maximal absorbance difference at infinite ligand concentrations, [E] is the total enzyme concentration, [L] is the KTZ concentration, and KD is the dissociation constant.
Binding titrations with testosterone (TST) were carried out in a fashion similar to that described above. For the TST titrations, 13 and 4 mM stocks of TST were prepared in 70% methanol. The final methanol concentration never exceeded 0.5% (v/v). Titrations, data collection, and data correction were performed as described above for KTZ, and an apparent KD was determined by fitting the data to a Hill equation (eq 3):
| (3) |
where KD,app is the apparent dissociation constant, n is the Hill coefficient, and all other parameters are the same as in eq 2.
Differential Scanning Calorimetry
Differential scanning calorimetry (DSC) experiments were performed using a MicroCal Capillary DSC System (MicroCal, Northampton, MA). CYP3A4 nanodiscs or empty nanodisc samples were dialyzed overnight against DFB and diluted to a final concentration of 2.5 µM. KTZ-bound CYP3A nanodisc samples were prepared in the same manner as described above, and KTZ (dissolved in methanol) was added to a final concentration of 50 µM immediately prior to the DSC scans being performed [the final methanol cosolvent concentration did not exceed 1% (v/v)]. As recommended for DSC, the instrument was “conditioned” by performing three consecutive scans with DFB alone prior to the analysis of a single sample. For KTZ-containing samples, the instrument was “conditioned” using DFB supplemented with 50 µM KTZ. CYP3A4 endotherms were measured by scanning samples from 10 to 130 °C at a scan rate of 60 °C/h. CYP3A4 endotherms were baseline corrected, and transition temperatures (Tm), changes in heat capacity (ΔCp), calorimetric enthalpies (ΔHcal), and van’t Hoff enthalpies (ΔHvH) were determined by fitting the data using the software package Origin (MicroCal).
Stopped-Flow Spectroscopy
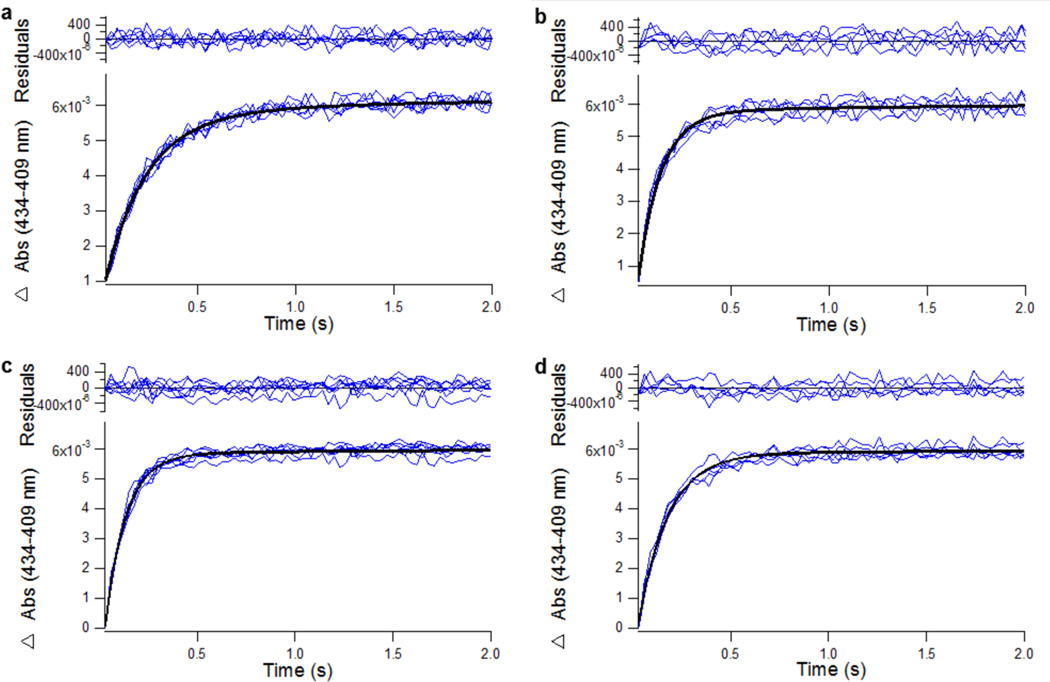
Stopped-flow spectroscopy was performed using an Applied Photophysics (Surrey, U.K.) SX-20 stopped-flow apparatus. All stopped-flow experiments were performed at 25 °C using a 1:10 mixing setup. A 2 mM stock of KTZ dissolved in methanol was diluted to 55 µM using DFB and loaded into a 250 µL syringe, and a 2.5 mL syringe was loaded with 0.55 µM CYP3A4 nanodiscs of varying lipid composition in DFB. This results in final KTZ and CYP3A4 nanodisc concentrations of 5 and 0.5 µM after mixing, respectively. The contents of both syringes were rapidly mixed, and absorbance data were collected using a photodiode array detector. For each lipid composition, seven repeat injections were performed and 1000 data points were collected for each injection. The absorbance difference between 434 and 409 nm was plotted as a function of time (t) and fit to an equation for a biphasic process (eq 4):
| (4) |
where Absmax is the maximal absorbance change, t0 is the earliest time point that is fit to the equation, Afast and Aslow are the magnitudes of the fast and slow phases of the kinetic trace, respectively, and kfast and kslow are the observed rates for the fast and slow phases, respectively.
Stopped-flow spectroscopy with TST was performed with a protocol similar to that described above. The initial concentration of CYP3A4 nanodiscs was kept at 0.55 µM (final concentration of 0.5 µM), and the initial concentration of TST was 2.2 mM (final concentration of 200 µM). Injections were performed and data collected as described above for KTZ. The absorbance difference between 387 and 418 nm was plotted as a function of time, and data were fit to an equation for a monophasic process (eq 5):
| (5) |
where kobs is the observed rate constant for the reaction, A is the magnitude of the kinetic trace, and all other parameters are the same as in eq 4.
RESULTS
Circular Dichroism
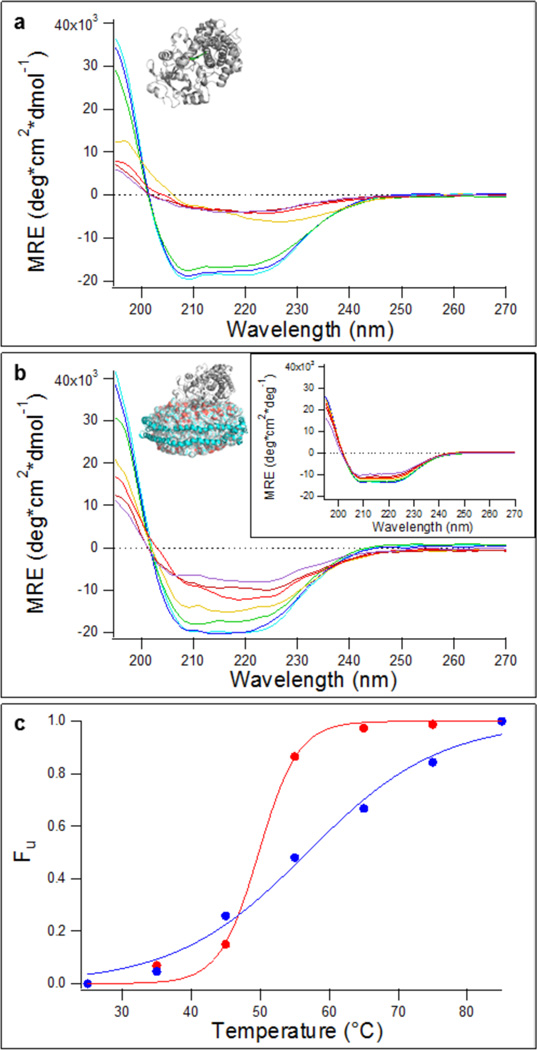
To compare the sensitivity of the secondary structure of CYP3A4 in buffer versus in POPC nanodiscs, CD spectroscopy was performed at varying temperatures between 25 and 85 °C. This temperature increase was expected to result in CYP3A4 denaturation with a negligible contribution from the nanodisc. Previous DSC results with CYP3A4 in POPC nanodiscs have indicated that CYP3A4 denatures with a complex non-two-state behavior, with a Tm for the major transition at ~56 °C. In this temperature range, there is a negligible enthalpy change for the nanodisc, which yielded a Tm of >75 °C. Thus, the DSC transitions in the 56 °C region were assigned to CYP3A4. The resolution of CYP3A4 transitions from POPC nanodisc transitions allows for more detailed analysis of the structural changes of CYP3A4 in the membrane. Figure 2 shows CD spectra recorded at varying temperatures for CYP3A4 in buffer and CYP3A4 incorporated into POPC nanodiscs. Also shown are CD spectra for empty POPC nanodiscs (Figure 2b, inset). The mean residue ellipticity of empty nanodiscs changes minimally over the temperature range analyzed, so the contribution of MSP1D1 to changes in CD signal is small relative to that of CYP3A4.
Figure 2.
Circular dichroism spectroscopy for samples collected at varying temperatures. CD spectra were recorded at temperatures between 25 and 80 °C for (a) CYP3A4 in DFB and (b) CYP3A4 incorporated into 100% POPC nanodiscs (inset, CD spectra for empty POPC nanodiscs). (c) Fractional change in ellipticity at 222 nm (Fu) at increasing temperatures: red for CYP3A4 in buffer and blue for CYP3A4 in 100% POPC nanodiscs. Solid lines are fits to a logistic function to empirically estimate Tm. The specific temperatures used to measure CD spectra were 25, 35, 45, 55, 65, 75, and 85 °C.
As expected, at room temperature both CYP3A4 in buffer and CYP3A4 nanodiscs yield double minima characteristic of a protein that is largely α-helical in structure. Secondary structure analysis results in an α-helix content of 63.7 ± 1.5% and a β-sheet content of 3.5 ± 1.0% for CYP3A4 in buffer. While the α-helix content of CYP3A4 nanodiscs remained unchanged (64.0 ± 1.8%), the β-sheet content showed a slight increase to 5.0 ± 0.5%, relative to that of CYP3A4 in buffer. Available crystal structures of CYP3A4 indicate that it is approximately 50% α-helical and 8% β-sheet in secondary structure.11,25–28 This is in good agreement with our experimental results, and the minor discrepancies in secondary structure content may likely be explained, in part, by the fact that the available crystal structures do not include the crystallographically disordered N-terminal helix in the calculation of secondary structure.
For CYP3A4 and CYP3A4 in nanodiscs, as the temperature increases, the mean residue ellipticity at 222 nm increases (becomes less negative), corresponding to an apparent loss of α-helix content. Plots of fraction unfolded versus temperature indicate that denaturation of CYP3A4 in buffer is highly cooperative, whereas denaturation of CYP3A4 in POPC nanodiscs is less cooperative (Figure 2c). An estimate of Tm for CYP3A4 in POPC nanodiscs based on the CD spectra is 56.8 ± 1.1 °C, versus 49.8 ± 0.5 °C for CYP3A4 in buffer, in excellent agreement with the previously reported Tm values based on DSC.21 In separate samples, the UV-vis spectrum of the CYP3A4 in nanodiscs was monitored after they had been heated to 85 °C at the same rate as in the CD experiments, and a CO difference spectrum was obtained (not shown). The intensity of the spectrum was reduced by ~10-fold compared to that of an unheated sample of CYP3A4 in nanodiscs, and the remaining hemoprotein was nearly completely converted to P420 (>95%). Apparently, a significant fraction of the protein lost the heme, and the remainder had a highly denatured active site. No heme spectrum was apparent in the corresponding CYP3A4 heated in buffer.
The plots in Figure 2c should not be interpreted as evidence that the unfolding of CYP3A4 in either environment is a two-state process; denaturation is clearly highly complex, and the protein likely populates multiple states. Because of this complexity, we are cautious not to interpret the date in terms of an unfolding mechanism. Also, the final CD spectrum of CYP3A4 in buffer at 85 °C is very low intensity and cannot be analyzed for secondary structure with confidence. In contrast, the final spectrum of CYP3A4 in nanodiscs retains some residual structure, and spectral analysis indicates it retains approximately 22.8% helix. Speculatively, the remaining helicity partially reflects the N-terminal membrane helix that is anchored in the membrane. Because there is slightly more residual helical structure at the highest temperature for CYP3A4 in nanodiscs than for CYP3A4 in buffer, the normalized fractional changes may slightly underestimate the stabilization provided by the nanodisc. That is, the magnitude of the total spectral change is larger for the CYP3A4 in buffer, but this difference in magnitude is lost upon internal normalization of each data set.
Together, the CD results confirm that CYP3A4 in POPC nanodiscs is thermally more stable than CYP3A4 in buffer and that the denaturation of CYP3A4 in POPC nanodiscs is resolved from the higher-temperature denaturation of the nanodisc complex. This temperature resolution further suggested the possibility of measuring the thermodynamic effects of varying lipid composition on the thermal unfolding of CYP3A4 in membranes of different composition. The previously reported calorimetric signal in the range of 45–65 °C is nearly completely associated with changes in secondary structure of CYP3A4 with a minimal contribution from changes in the MSP1D1 structure.
Differential Scanning Calorimetry in Lipid Mixtures
Thermodynamic parameters for CYP3A4 denaturation in pure POPC nanodiscs by DSC were demonstrated previously.21 In that study, we reported the total calorimetric enthalpy as well as the van’t Hoff enthalpy/calorimetric enthalpy ratio, ΔHvH/ΔHcal, for several individual endotherms recovered from the deconvolution of the complex endotherm. For the CYP3A4 POPC nanodiscs, three transitions were observed, with Tm values of 46, 53, and 56 °C, which span most of the transition observed in CD.
To determine the effects of lipid composition on the stability of CYP3A4 in a membrane, calorimetric parameters for unfolding were determined by DSC, using CYP3A4 nanodiscs with varying POPC:DMPC ratios. POPC and DMPC have identical headgroups but differ in the length and saturation of the acyl chains (Figure 1b,c). Specifically, DMPC is completely saturated and has an acyl chain shorter that that of POPC, which also contains a double bond in one of its acyl chains. Of course, a limitation of this approach is the requirement for lipids with gel–liquid transitions that do not overlap with the denaturation of CYP3A4, and it is, therefore, impossible to exactly model biological membranes from specific cells or organelles. As shown below, POPC and DMPC satisfy this requirement. In addition, the identity of the lipid headgroup could have an impact on the stability and function of CYPs. However, to focus on a single chemical property of the lipids in this initial demonstration of membrane effects, we limited these initial studies to the variable acyl chain.
CYP3A4 nanodiscs were prepared with pure POPC, or a POPC:DMPC molar ratio of 3:1 (25% DMPC by mole content), 1:1 (50% DMPC by mole content), and 1:3 (75% DMPC by mole content). To achieve this, it was necessary to optimize the lipid:MSP1D1 ratio used for assembly of nanodiscs with each different lipid mixture composition. The results of this optimization are shown in the Discussion, and the data are compiled in Table S1; however, here it is noted that the results emphasize that different combinations of lipids pack with different densities, resulting in variation in the optimal lipid:MSP1D1 ratio. Pure DMPC empty nanodiscs were also prepared. However, attempts to incorporate CYP3A4 into DMPC nanodiscs were unsuccessful in maintaining the integrity of the enzyme and thus could not be analyzed.
To ensure that the nanodiscs prepared with different lipid mixtures had different bilayer properties, their relative fluidity was compared using Laurdan generalized polarization measurements. Laurdan is a fluorescent probe that is commonly used to study the lateral dynamics of membrane components.29,30 The Laurdan fluorescence emission maximum is primarily dependent upon the dynamics of water molecules at the lipid-water interface of the membrane near the excited state dipole of the naphthalene moiety of the probe, and this in turn is linked to dynamics of the lipid molecules. Upon excitation at 340 nm, the Laurdan fluorescence displays a maximum at 440 nm when the membrane is in the gel phase. This emission maximum shifts to 490 nm in the liquid phase, as the water molecules and lipids are less spatially restricted and more dynamic. Laurdan generalized polarization measurements, which monitor the ratio of emission at 440 nm to that at 490 nm, suggest that the membrane surface near the aqueous boundary becomes increasingly ordered with an increasing DMPC content. This is further elaborated in the Discussion. The specific values for the Laurdan polarization are included in Table S2. Combined with the optimal lipid:MSP1D1 ratio at varying POPC:DMPC ratios, these data suggest a complex relationship between membrane order and packing density, and they demonstrate that the different lipid mixtures used here yield bilayers with different fluidities.
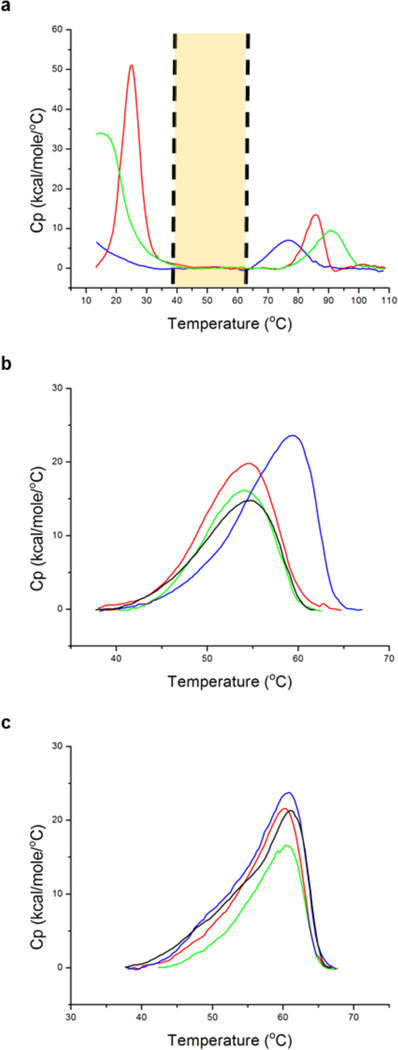
To ensure that the thermal lipid transitions in mixed lipid nanodiscs were resolved from CYP3A4, as in pure POPC nanodiscs, DSC of the empty nanodiscs was performed. The DSC thermograms are shown in Figure 3a. Each lipid composition has a characteristic low-temperature transition (<35 °C) that corresponds to the gel-to-liquid phase transition of the lipids and a high-temperature transition (~65–100 °C) due to nanoparticle denaturation. Most importantly, it is clear that for each lipid composition, the DSC transitions of the empty nanodiscs do not occur in the range of 35–65 °C (Figure 3).
Figure 3.
DSC endotherms for empty nanodiscs and CYP3A4 nanodiscs with varying POPC:DMPC ratios. (a) DSC of empty nanodiscs indicates that for each lipid composition there is a negligible DSC signal in the range of 35–65 °C: blue for 100% POPC, red for 100% DMPC, and green for a 50% POPC/50% DMPC mixture. The highlighted area represents the temperature range of CYP3A4 unfolding. DSC thermograms of ligand-free CYP3A4 nanodiscs (b) and KTZ-bound CYP3A4 nanodiscs of varying lipid composition (c) are shown: black for pure POPC, red for a 3:1 POPC:DMPC ratio, blue for a 1:1 POPC:DMPC ratio, and green for a 1:3 POPC:DMPC ratio. Melting temperatures, enthalpies, and heat capacity changes for the CYP3A4 endotherms are summarized in Table 1.
The CYP3A4 nanodisc DSC thermograms are shown for each composition in the region of 35–65 °C, in the absence of ligand (Figure 3b), as well as in the presence of a saturating concentration of KTZ (Figure 3c), a well-documented tight binding inhibitor of CYP3A4.31–34 To ensure that the concentration of KTZ used was saturating for each lipid composition, the equilibrium binding affinity of KTZ was determined in each nanodisc (Figure 5). The KD for KTZ varied from 127 to 138 nM among the CYP3A4 nanodiscs with different lipid compositions; the KD was not altered significantly by the lipid composition. DSC of the ligand-free samples showed an apparent increase in the CYP3A4 endotherm maximum for 50% DMPC nanodiscs. Further increasing the DMPC content of the nanodiscs to 75% results in the CYP3A4 endotherm shifting back to a lower temperature. Upon addition of a saturating concentration of KTZ, there are not significant differences in the DSC in different lipid compositions. Regardless of the lipid composition, KTZ shifts the CYP3A4 endotherms to a similar melting temperature. The calorimetric enthalpy, heat capacity, and melting temperature values for each CYP3A4 endotherm are listed in Table 1. The endotherms were further analyzed by deconvolution into the sum of multiple components (Figure S1), and the Tm, the van’t Hoff enthalpy (ΔHvH), the calorimetric enthalpy (ΔHcal), and the ΔHvH/ΔHcal ratio for each are reported in Table S3. It is apparent for each lipid composition that the DSC endotherm is complex and unfolding is a non-two-state process (Figure 4).
Figure 5.
Stopped-flow kinetic analysis of binding of KTZ to CYP3A4 nanodiscs composed of (a) pure POPC, (b) a 3:1 POPC:DMPC ratio, (c) a 1:1 POPC:DMPC ratio, and (d) a 1:3 POPC:DMPC ratio. Solid black lines are the result of fitting the kinetic data to eq 4. The saturating KTZ concentration was 5 µM. Kinetic parameters for KTZ binding are summarized in Table 2.
Table 1.
Melting Temperatures, Calorimetric Enthalpies, and Heat Capacities from DSC Analysis of CYP3A4 Nanodiscs
|
Tm (°C) |
total ΔHcal (cal/mol) |
ΔCp (cal mol−1 °C−1) |
||||
|---|---|---|---|---|---|---|
| system | without KTZ | with KTZa | without KTZ | with KTZa | without KTZ | with KTZa |
| 0% DMPCb | 54.8 | 61.2 | 1.5 × 105 | 2.4 × 105 | 3.3 × 103 | 1.1 × 104 |
| 25% DMPCb | 54.3 | 60.1 | 2.0 × 105 | 2.2 × 105 | 5.0 × 103 | 8.1 × 103 |
| 50% DMPCb | 59.5 | 60.8 | 2.4 × 105 | 2.6 × 105 | 1.2 × 104 | 1.8 × 104 |
| 75% DMPCb | 54.0 | 60.5 | 1.5 × 105 | 1.6 × 105 | 1.8 × 103 | 2.0 × 103 |
In 50 µM ketoconazole.
Percentage of DMPC in POPC nanodiscs.
Figure 4.
Binding isotherms for binding of KTZ to CYP3A4 in nanodiscs composed of (a) pure POPC, (b) 3:1 POPC/DMPC, (c) 1:1 POPC/DMPC, and (d) 1:3 POPC/DMPC CYP3A4 nanodiscs. The inset shows the corresponding difference absorbance spectra. Equilibrium binding parameters are summarized in Table 2.
Ligand Binding Kinetics
The kinetics of binding of two ligands were also examined using stopped-flow spectroscopy at a single saturating concentration of each. The first ligand was KTZ, which is a tight binding inhibitor as noted above. KTZ drives the ferric heme to a greater fraction of the low-spin state as a result of its coordination to the heme iron via an imidazole group. The second ligand was testosterone (TST), which binds with an apparent 3:1 TST:CYP3A4 stoichiometry, with modest positive cooperativity.35,36 TST pushes the ferric equilibrium toward the high-spin form. Thus, the two ligands studied span several different behaviors for CYP3A4: low spin versus high spin, and single versus multiple binding.
When KTZ was mixed with CYP3A4 nanodiscs, there was a clear time-dependent increase in the fraction of low-spin CYP3A4 (Figure 5). The approach to equilibrium for each nanodisc composition was fit to a double exponential with the recovered parameters listed in Table 2. There is a clear increase in the rate of KTZ binding with an increase in the nanodisc DMPC content to 25%, relative to 0% DMPC. Further increasing the DMPC content to 50% resulted in a further increase in rate; however, increasing the DMPC content to 75% resulted in a decrease in the rate of binding. This is elaborated further in the Discussion.
Table 2.
Dissociation Constants and Observed Rates of Binding of KTZ to CYP3A4 Nanodiscs
| system | KD (µM) | kfast (s−1) | kslow (s−1) |
|---|---|---|---|
| 0% DMPCa | 0.127 ± 0.010 | 4.80 ± 0.18 | 0.939 ± 0.141 |
| 25% DMPCa | 0.138 ± 0.008 | 8.73 ± 0.28 | 0.363 ± 0.044 |
| 50% DMPCa | 0.130 ± 0.009 | 9.51 ± 0.16 | 0.349 ± 0.033 |
| 75% DMPCa | 0.134 ± 0.006 | 6.61 ± 0.13 | 0.172 ± 0.019 |
Percentage of DMPC in POPC nanodiscs.
For TST, the binding isotherms were fit to a Hill model as done previously by many investigators (Figure S2). We note that for the case of multiple-ligand binding, the spectral change may not correlate precisely with the extent of binding if sequential binding steps differentially perturb the spin state.35,36 Regardless, the binding isotherms reveal relevant binding parameters. The equilibrium S0.5 value and Hill coefficient are ~40 µM and 1.4, respectively, in close agreement with previously published values.35,36 As with KTZ, the equilibrium binding isotherm with TST was not significantly affected by the lipid composition. However, in contrast to KTZ, the rate of TST binding decreases continuously with an increasing fraction of DMPC in the nanodisc (Table 3). The stopped-flow data are found in Figure S3. The rate constant for binding in POPC nanodiscs is ~4-fold faster than the rate in 25% POPC/75% DMPC nanodiscs. This is elaborated further in the Discussion.
Table 3.
Apparent Dissociation Constants, Hill Coefficients, and Observed Rate Constants for Binding of TST to CYP3A4 Nanodiscs
| system | KD,App (µM)b | nc | Kobs (s−1) |
|---|---|---|---|
| 0% DMPCa | 43.6 ± 0.9 | 1.51 ± 0.04 | 1256.7 ± 96.9 |
| 25% DMPCa | 33.4 ± 0.5 | 1.47 ± 0.03 | 731.7 ± 74.2 |
| 50% DMPCa | 40.7 ± 0.7 | 1.50 ± 0.03 | 524.7 ± 54.5 |
| 75% DMPCa | 38.3 ± 0.6 | 1.48 ± 0.03 | 398.6 ± 46.0 |
Percentage of DMPC in POPC nanodiscs.
Determined using the Hill equation.
Hill coefficient.
DISCUSSION
The lipid nanodisc system used here provides a platform for interrogating the effects of membrane structure or composition on membrane proteins embedded within them. A limitation of this system for calorimetry is the fact that some lipids yield calorimetric signals from their gel transitions in the temperature range that overlaps with that of protein unfolding.37–39 Therefore, we have limited our studies to mixtures of POPC and DMPC, which conveniently lack DSC signals in the temperature range in which CYP3A4 denatures and which allow for the specific interrogation of the effect of acyl-chain composition on CYP3A4 properties (Figure 3). The studies reported here specifically demonstrate a significant sensitivity of CYP3A4 thermal stability, thermodynamic parameters for thermal denaturation, and ligand binding kinetics to the lipid acyl-chain composition or fluidity in mixed lipid bilayers. POPC and DMPC differ only in the presence of a single double bond in one acyl chain and two additional carbons in POPC (Figure 1a,b). It is well established that membrane fluidity is influenced by the degree of acyl-chain saturation, with membranes containing unsaturated lipids being more fluid than membranes containing fully saturated lipids. Furthermore, it has been demonstrated that lipid acyl-chain length also modulates membrane fluidity, with longer chain lengths decreasing the fluidity of membranes.38–40 The lipids used in this work differ in both acyl-chain length and degree of saturation, resulting in their two contrasting phase transition temperatures (−2 °C for POPC, 25 °C for DMPC) as well as in the optimal lipid:MSP1D1 ratio for each of these lipid mixtures. Interestingly, in this work, we observed that the optimal lipid:MSP1D1 ratio required for nanodisc self-assembly deviated from a linear trend, and this suggests that fewer lipids are incorporated per nanodisc in the lipid mixtures than in the pure POPC nanodiscs or pure DMPC nanodiscs. The different properties of the mixed lipid nanodiscs were also evident in the Laurdan generalized polarization, which clearly indicates increasing fluidity with increasing DMPC content. These properties are discussed further below.
The CD and DSC results further extend our previous demonstration that CYP3A4 undergoes thermal denaturation at temperatures lower than that of the nanodiscs, allowing for thermal resolution of CYP3A4 unfolding. The thermal stability of CYP3A4 in nanodiscs is sensitive to even modest changes in lipid acyl-chain composition and is more stable in the POPC/DMPC mixtures than in pure POPC (Figure 3). This is interesting because CYP3A4 is thought to have only a single structural element, the N-terminal helix, which penetrates deeply into the membrane. The N-terminal helix, which has 12 amino acids truncated from the wild-type sequence in the engineered variant used here, is expected to penetrate ~15–20 Å into the membrane, based on modeling studies with several CYP isoforms, including CYP3A4.41,42 The F′ and G′ helices have been suggested to penetrate much less deeply, and the remainder of the CYP3A4 is solvent-exposed, including the highly plastic F and G helices and interconnecting loop that are thought to control access to the active site.13,41,42
The POPC/DMPC lipid mixture appears to provide a hydrocarbon environment less densely packed than that of pure POPC or pure DMPC nanodiscs, based on the optimal lipid:protein ratios. Presumably, this favors deeper insertion of the N-terminal helix into the membrane. Recently, crystal structures of full length Saccharomyces cerevisiae lanosterol 14α-demethylase (CYP51) incorporated into N-decyl β-d-maltoside micelles were obtained by Monk and co-workers.43 On the basis of these structures, they suggest a possible interaction of the N-terminal membrane helix with the catalytic domain of the enzyme, which could have potential effects on enzyme conformation and ligand binding dynamics. A similar interaction may be occurring with CYP3A4 in nanodiscs, wherein the N-terminal helix interacts with the catalytic domain more efficiently in mixed lipid nanodiscs than in pure POPC nanodiscs. This in turn could promote deeper insertion of CYP3A4 into the membrane, resulting in an increase in its thermal stability and in ΔHcal and ΔCp for denaturation, as observed here (Table 1). Above 50% DMPC, the lipid packing density increases, potentially limiting the interaction of the N-terminal anchor with the catalytic domain and resulting in our observed decreases in thermal stability, ΔHcal, and ΔCp for denaturation. This could also explain why attempts to incorporate CYP3A4 into pure DMPC nanodiscs were unsuccessful in maintaining enzyme structural integrity. Alternatively, the lipid mixtures could perturb the boundary lipid layer that forms an ordered phase on the periphery of the nanodisc, and this could alter the location or orientation of CYP3A4 within the bilayer.43–47 Regardless, it is clear that the lipid acyl-chain composition modulates the thermodynamics of irreversible CYP3A4 denaturation, but this effect is limited to the ligand-free enzyme; the thermal stability was nearly independent of lipid composition when KTZ was bound.
The effects of the lipid acyl chain on the rate of KTZ binding or TST binding are interesting and not necessarily surprising, but such effects also have not been previously demonstrated. They clearly demonstrate that lipid acyl-chain composition affects the kinetics of binding in a ligand-dependent manner. It is instructive to consider possible correlations between membrane properties and ligand binding for the cases we studied here.
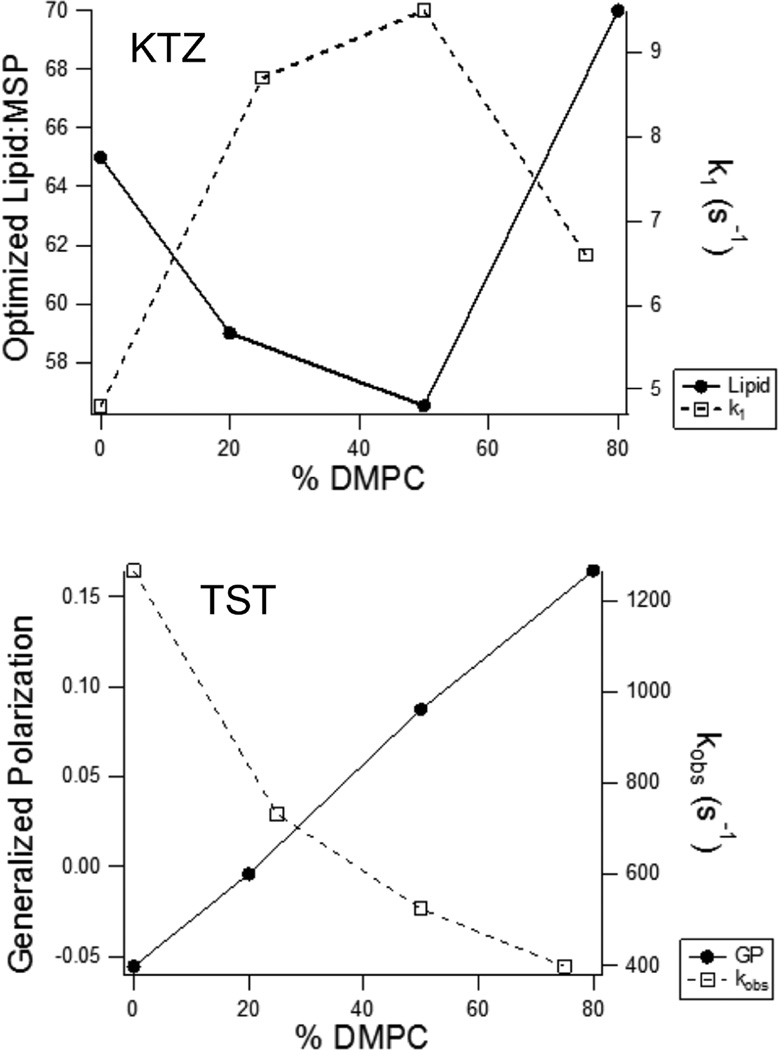
For KTZ binding, which is complex and multiphasic, the rate constant for the fast phase increases with an increasing DMPC content initially but reaches a maximum at 50% DMPC before decreasing. Interestingly, this correlates inversely with the optimal lipid:MSP1D1 ratio for the different lipid compositions, which reflects the lipid packing density. The lipid packing density and rate constant for the fast binding phase of KTZ are plotted as a function of lipid composition in Figure 6 (top). These plots suggest the interesting possibility that the KTZ binds fastest when the lipid packing density is lowest. In contrast, the rate of TST binding decreases steadily as the fraction of DMPC increases, and thus, the binding rate correlates inversely with the Laurdan generalized polarization or the membrane headgroup fluidity. As the lipid headgroups become less fluid, TST binds less rapidly (Figure 6, bottom). Although the detailed molecular bases for these observations remain unknown, the results emphasize the importance of lipid acyl chain in ligand interactions with CYP3A4, possibly resulting from differences in lipid packing and headgroup fluidity.
Figure 6.
Correlations of ligand binding kinetics with membrane properties of CYP3A4 nanodiscs. (Top) Optimized total lipid:MSP ratio for mixed lipid nanodiscs (● and solid line) and the rate constants for the fast phase of KTZ binding obtained by stopped-flow spectroscopy (○ and dashed line) plotted as a function of the mole percentage of DMPC in the nanodisc membrane. (Bottom) Laurdan generalized polarization (● and solid line) for mixed lipid nanodiscs and the observed rate constants for TST binding from stopped-flow spectroscopy (○ and dashed line), plotted as a function of the mole percentage of DMPC in the nanodisc membrane.
While it is widely assumed that hydrophobic substrates and ligands for CYP3A4 bind to the active site via the membrane, there are few data that directly demonstrate this. The modulation of KTZ or TST binding and dissociation kinetics with a change in lipid composition is consistent with this, although the results do not directly prove that ligands must enter the active site from the membrane. Speculatively, both ligand diffusion in the lipid bilayer and protein movement within the bilayer could be facilitated in the less densely packed lipid phase with the mixed lipids. An increased level of diffusion of the ligand and protein elements would be expected to increase rates of binding and dissociation. Also, lipid–CYP3A4 interactions near the aqueous boundary could alter ligand binding kinetics, and the lipid mixture clearly alters the relative order in this region of the membrane, based on the Laurdan polarization.
These effects could be dependent on the CYP isoform, as well. Presumably, different CYPs with different F′ and G′ helix region sequences would be differentially affected by a changing lipid composition. In fact, Das and co-workers have demonstrated that mutations in the F and G helix regions of CYP2J2 result in changes in penetration of the protein into the lipid bilayer of nanodiscs.48 Together, the data suggest the likelihood that both the CYP sequence at the membrane interface and the lipid composition are likely to contribute to functional properties of these enzymes.
A related aspect of CYP function that should be acknowledged is the possibility that, in vivo, various CYP isoforms more readily localize to different regions of the endoplasmic reticulum, based on the lipid composition of the membrane.47–49 This could have important effects on both the catalytic properties and substrate selectivity of membrane-bound CYPs. While this study focuses on the effects of lipid composition on CYP3A4 stability and function, in principle the mixed lipid nanodisc system could be extended to other CYP isoforms, which have been demonstrated to have differential lipid domain preferences.50,51
In conclusion, we have demonstrated the differential thermal stability of CYP3A4 in nanodiscs containing varying POPC:DMPC ratios. The increase in thermal stability, enthalpy of unfolding, and heat capacity of unfolding using a 1:1 POPC/DMPC mixture suggests a greater degree of interaction between the enzyme and the membrane in optimal lipid mixtures, which may in turn facilitate the binding of some hydrophobic substrates. In fact, the maximum that is reached for the ΔCp for denaturation, the ΔHcal for denaturation, and the rate constant for the fast phase of KTZ binding coincides in each case with the lowest packing density of lipids in the 50:50 POPC/DMPC mixture, highlighting the role of membrane dynamics or packing density in the structure and function of CYP3A4 (Figure 6). Obviously, TST binding is controlled by other properties, such as membrane fluidity near the head-groups. In the absence of crystal structures of mammalian CYPs in a membrane environment, DSC in combination with the use of phospholipid bilayer nanodiscs serves as a powerful method for investigating the membrane topology and conformational dynamics of mammalian CYPs under varying lipid conditions.
Supplementary Material
Acknowledgments
DSC was performed in the Analytical Biopharmacy Core facility of the Department of Medicinal Chemistry and Molecular Engineering and Sciences Institute (http://depts.washington.edu/cidb4bio/index.shtml).
Funding
This work was supported by National Institutes of Health Grants R01GM098457 (W.M.A.) and T32GM007750 (W.D.M.).
ABBREVIATIONS
- CD
circular dichroism
- CYP
cytochrome P450
- DFB
disc forming buffer
- DMPC
1,2-dimyristoyl-sn-glycero-3-phospho-choline
- POPC
1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocho-line
- DSC
differential scanning calorimetry
- KTZ
ketoconazole
- MD
molecular dynamics
- MSP
membrane scaffold protein
- MSP1D1
molecular scaffold protein 1D1
- ND
nanodisc
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem.6b00715.
Tabulated results for lipid:MSP1D1 ratios for different lipid mixtures in nanodiscs, tabulated results for ketoconazole binding kinetics and thermodynamics at varying lipid compositions, tabulated results for testosterone binding kinetics and thermodynamics at varying lipid compositions, binding isotherms and stopped-flow results, and deconvolution of the DSC thermograms at varying lipid compositions and the recovered thermodynamic parameters for each fitted transition (PDF)
Author Contributions
W.D.M. prepared all proteins and nanodiscs and performed DSC, with assistance from J.P.S., and CD with assistance from M.S. L.P. assisted with thermal denaturation studies. W.M.A. directed the research and wrote part of the manuscript.
The authors declare no competing financial interest.
REFERENCES
- 1.Guengerich FP. In: Cytochrome P450: Structure, mechanism, and biochemistry. 3rd. Montellano Ortiz de P, editor. New York: Kluwer Academic/Plenum Publishers; 2005. pp. 423–430. [Google Scholar]
- 2.Wrighton SA, Stevens JC. The human hepatic cytochromes P450 involved in drug metabolism. Crit. Rev. Toxicol. 1992;22:1–21. doi: 10.3109/10408449209145319. [DOI] [PubMed] [Google Scholar]
- 3.Anzenbacher P, Anzenbacherova E. Cytochromes P450 and metabolism of xenobiotics. Cell. Mol. Life Sci. 2001;58:737–747. doi: 10.1007/PL00000897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nebert DW, Russell DW. Clinical importance of the cytochromes P450. Lancet. 2002;360:1155–1162. doi: 10.1016/S0140-6736(02)11203-7. [DOI] [PubMed] [Google Scholar]
- 5.Guengerich FP. Cytochrome P-450 3A4: regulation and role in drug metabolism. Annu. Rev. Pharmacol. Toxicol. 1999;39:1–17. doi: 10.1146/annurev.pharmtox.39.1.1. [DOI] [PubMed] [Google Scholar]
- 6.Haddad A, Davis M, Lagman R. The pharmacological importance of cytochrome CYP3A4 in the palliation of symptoms: review and recommendations for avoiding adverse drug interactions. Support Care Cancer. 2007;15:251–257. doi: 10.1007/s00520-006-0127-5. [DOI] [PubMed] [Google Scholar]
- 7.Scott EE, Halpert JR. Structures of cytochrome P450 3A4. Trends Biochem. Sci. 2005;30:5–7. doi: 10.1016/j.tibs.2004.11.004. [DOI] [PubMed] [Google Scholar]
- 8.Rowland P, Blaney FE, Smyth MG, Jones JJ, Leydon VR, Oxbrow AK, Lewis CJ, Tennant MG, Modi S, Eggleston DS, Chenery RJ, Bridges AM. Crystal structure of human cytochrome P450 2D6. J. Biol. Chem. 2006;281:7614–7622. doi: 10.1074/jbc.M511232200. [DOI] [PubMed] [Google Scholar]
- 9.Williams PA, Cosme J, Ward A, Angove HC, Matak Vinkovic D, Jhoti H. Crystal structure of human cytochrome P450 2C9 with bound warfarin. Nature. 2003;424:464–468. doi: 10.1038/nature01862. [DOI] [PubMed] [Google Scholar]
- 10.Williams PA, Cosme J, Vinkovic DM, Ward A, Angove HC, Day PJ, Vonrhein C, Tickle IJ, Jhoti H. Crystal structures of human cytochrome P450 3A4 bound to metyrapone and progesterone. Science. 2004;305:683–686. doi: 10.1126/science.1099736. [DOI] [PubMed] [Google Scholar]
- 11.Yano JK, Wester MR, Schoch GA, Griffin KJ, Stout CD, Johnson EF. The structure of human microsomal cytochrome P450 3A4 determined by X-ray crystallography to 2.05-A resolution. J. Biol. Chem. 2004;279:38091–38094. doi: 10.1074/jbc.C400293200. [DOI] [PubMed] [Google Scholar]
- 12.Black SD. Membrane Topology of the Mammalian P450-Cytochromes. FASEB J. 1992;6:680–685. doi: 10.1096/fasebj.6.2.1537456. [DOI] [PubMed] [Google Scholar]
- 13.Baylon JL, Lenov IL, Sligar SG, Tajkhorshid E. Characterizing the Membrane-Bound State of Cytochrome P450 3A4: Structure, Depth of Insertion, and Orientation. J. Am. Chem. Soc. 2013;135:8542–8551. doi: 10.1021/ja4003525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Berka K, Paloncyova M, Anzenbacher P, Otyepka M. Behavior of Human Cytochromes P450 on Lipid Membranes. J. Phys. Chem. B. 2013;117:11556–11564. doi: 10.1021/jp4059559. [DOI] [PubMed] [Google Scholar]
- 15.Berka K, Hendrychova T, Anzenbacher P, Otyepka M. Membrane Position of Ibuprofen Agrees with Suggested Access Path Entrance to Cytochrome P450 2C9 Active Site. J. Phys. Chem. A. 2011;115:11248–11255. doi: 10.1021/jp204488j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Denisov IG, Grinkova YV, Lazarides AA, Sligar SG. Directed self-assembly of monodisperse phospholipid bilayer Nanodiscs with controlled size. J. Am. Chem. Soc. 2004;126:3477–3487. doi: 10.1021/ja0393574. [DOI] [PubMed] [Google Scholar]
- 17.Nath A, Atkins WM, Sligar SG. Applications of phospholipid bilayer nanodiscs in the study of membranes and membrane proteins. Biochemistry. 2007;46:2059–2069. doi: 10.1021/bi602371n. [DOI] [PubMed] [Google Scholar]
- 18.Bayburt TH, Sligar SG. Membrane protein assembly into Nanodiscs. FEBS Lett. 2010;584:1721–1727. doi: 10.1016/j.febslet.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ritchie TK, Grinkova YV, Bayburt TH, Denisov IG, Zolnerciks JK, Atkins WM, Sligar SG. Reconstitution of Membrane Proteins in Phospholipid Bilayer Nanodiscs. Methods Enzymol. 2009;464:211–231. doi: 10.1016/S0076-6879(09)64011-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Denisov IG, Sligar SG. Cytochromes P450 in nanodiscs. Biochim. Biophys. Acta, Proteins Proteomics. 2011;1814:223–229. doi: 10.1016/j.bbapap.2010.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Treuheit NA, Redhair M, Kwon H, McClary WD, Guttman M, Sumida JP, Atkins WM. Membrane Interactions, Ligand-dependent Dynamics, and Stability of Cyto-chrome P4503A4 in Lipid Nanodiscs. Biochemistry. 2016;55:1058–1069. doi: 10.1021/acs.biochem.5b01313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 23.Sreerama N, Woody RW. On the analysis of membrane protein circular dichroism spectra. Protein Sci. 2004;13:100–112. doi: 10.1110/ps.03258404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nath A, Grinkova YV, Sligar SG, Atkins WM. Ligand binding to cytochrome P450 3A4 in phospholipid bilayer nanodiscs - The effect of model membranes. J. Biol. Chem. 2007;282:28309–28320. doi: 10.1074/jbc.M703568200. [DOI] [PubMed] [Google Scholar]
- 25.Kaur P, Chamberlin AR, Poulos TL, Sevrioukova IF. Structure-Based Inhibitor Design for Evaluation of a CYP3A4 Pharmacophore Model. J. Med. Chem. 2016;59:4210–4220. doi: 10.1021/acs.jmedchem.5b01146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sevrioukova IF, Poulos TL. Dissecting cytochrome P450 3A4-ligand interactions using ritonavir analogues. Biochemistry. 2013;52:4474–4481. doi: 10.1021/bi4005396. [DOI] [PubMed] [Google Scholar]
- 27.Ekroos M, Sjogren T. Structural basis for ligand promiscuity in cytochrome P450 3A4. Proc. Natl. Acad. Sci. U. S. A. 2006;103:13682–13687. doi: 10.1073/pnas.0603236103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sevrioukova IF, Poulos TL. Pyridine-substituted desoxyritonavir is a more potent inhibitor of cytochrome P450 3A4 than ritonavir. J. Med. Chem. 2013;56:3733–3741. doi: 10.1021/jm400288z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bagatolli LA. To see or not to see: lateral organization of biological membranes and fluorescence microscopy. Biochim. Biophys. Acta, Biomembr. 2006;1758:1541–1556. doi: 10.1016/j.bbamem.2006.05.019. [DOI] [PubMed] [Google Scholar]
- 30.Parasassi T, De Stasio G, d’Ubaldo A, Gratton E. Phase fluctuation in phospholipid membranes revealed by Laurdan fluorescence. Biophys. J. 1990;57:1179–1186. doi: 10.1016/S0006-3495(90)82637-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moody DE, Walsh SL, Rollins DE, Neff JA, Huang W. Ketoconazole, a cytochrome P450 3A4 inhibitor, markedly increases concentrations of levo-acetyl-alpha-methadol in opioid-naive individuals. Clin. Pharmacol. Ther. 2004;76:154–166. doi: 10.1016/j.clpt.2004.04.004. [DOI] [PubMed] [Google Scholar]
- 32.Pearson JT, Hill JJ, Swank J, Isoherranen N, Kunze KL, Atkins WM. Surface plasmon resonance analysis of antifungal azoles binding to CYP3A4 with kinetic resolution of multiple binding orientations. Biochemistry. 2006;45:6341–6353. doi: 10.1021/bi0600042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Katz HI. Drug interactions of the newer oral antifungal agents. Br. J. Dermatol. 1999;141(Suppl. 56):26–32. doi: 10.1046/j.1365-2133.1999.00011.x. [DOI] [PubMed] [Google Scholar]
- 34.Greenblatt DJ, Zhao Y, Venkatakrishnan K, Duan SX, Harmatz JS, Parent SJ, Court MH, von Moltke LL. Mechanism of cytochrome P450-3A inhibition by ketoconazole. J. Pharm. Pharmacol. 2011;63:214–221. doi: 10.1111/j.2042-7158.2010.01202.x. [DOI] [PubMed] [Google Scholar]
- 35.Roberts AG, Campbell AP, Atkins WM. The thermodynamic landscape of cytochrome P450 3A4: ligand binding and spin state equilibria. Biochemistry. 2005;44:1353–1365. doi: 10.1021/bi0481390. [DOI] [PubMed] [Google Scholar]
- 36.Denisov IG, Baas BJ, Grinkova YV, Sligar SG. Cooperativity in cytochrome P450 3A4: linkages in substrate binding, spin state, uncoupling, and product formation. J. Biol. Chem. 2006;282:7066–7076. doi: 10.1074/jbc.M609589200. [DOI] [PubMed] [Google Scholar]
- 37.Denisov IG, McLean MA, Shaw AW, Grinkova YV, Sligar SG. Thermotropic phase transition in soluble nanoscale lipid bilayers. J. Phys. Chem. B. 2005;109:15580–15588. doi: 10.1021/jp051385g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shaw AW, McLean MA, Sligar SG. Phospholipid phase transitions in homogeneous nanometer scale bilayer discs. FEBS Lett. 2004;556:260–264. doi: 10.1016/s0014-5793(03)01400-5. [DOI] [PubMed] [Google Scholar]
- 39.Marsh D. Structural and thermodynamic determinants of chain-melting transition temperatures for phospholipid and glycolipids membranes. Biochim. Biophys. Acta, Biomembr. 2010;1798:40–51. doi: 10.1016/j.bbamem.2009.10.010. [DOI] [PubMed] [Google Scholar]
- 40.Lande MB, Donovan JM, Zeidel ML. The relationship between membrane fluidity and permeabilities to water, solutes, ammonia, and protons. J. Gen. Physiol. 1995;106:67–84. doi: 10.1085/jgp.106.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ruan KH, So SP, Zheng W, Wu J, Li D, Kung J. Solution structure and topology of the N-terminal membrane anchor domain of a microsomal cytochrome P450: prostaglandin I2 synthase. Biochem. J. 2002;368:721–728. doi: 10.1042/BJ20021001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Szczesna-Skorupa E, Kemper B. The Signal-Anchor Sequence of CYP2C1 Inserts into the Membrane as a Hairpin Structure. Biochem. Biophys. Res. Commun. 2011;415:405–409. doi: 10.1016/j.bbrc.2011.10.082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Skar-Gislinge N, Kynde SA, Denisov IG, Ye X, Lenov I, Sligar SG, Arleth L. Small-angle scattering determination of the shape and localization of human cytochrome P450 embedded in a phospholipid nanodisc environment. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2015;71:2412–2421. doi: 10.1107/S1399004715018702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mustafa G, Nandekar PP, Yu X, Wade RC. On the application of the MARTINI coarse-grained model to immersion of a protein in a phospholipid bilayer. J. Chem. Phys. 2015;143:243139. doi: 10.1063/1.4936909. [DOI] [PubMed] [Google Scholar]
- 45.Monk BC, Tomasiak TM, Keniya MV, Huschmann FU, Tyndall JDA, O’Connell JD, Cannon RD, McDonald JG, Rodriguez A, Finer-Moore JS, Stroud RM. Architecture of a single membrane spanning cytochrome P450 suggests constraints that orient the catalytic domain relative to a bilayer. Proc. Natl. Acad. Sci. U. S. A. 2014;111:3865–3870. doi: 10.1073/pnas.1324245111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tall AR, Small DM, Deckelbaum RJ, Shipley GG. Structure and thermodynamic properties of high density lipoprotein recombinants. J. Biol. Chem. 1977;252:4701–4711. [PubMed] [Google Scholar]
- 47.Edwards WL, Bush SF, Mattingly TW, Weisgraber KH. Raman spectroscopic study of boundary lipid in 1,2-dipalmitoylphosphatidylcholine/apolipoprotein A—I recombinants. Spectrochim Acta A Mol. Biomol Spectrosc. 1993;49:2027–2038. [Google Scholar]
- 48.McDougle DR, Baylon JL, Meling DD, Kambalyal A, Grinkova YV, Hammernik J, Tajkhorshid E, Das A. Incorporation of charged residues in the CYP2J2 F-G loop disrupts CYP2J2-lipid bilayer interactions. Biochim. Biophys. Acta, Biomembr. 2015;1848:2460–2470. doi: 10.1016/j.bbamem.2015.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brignac-Huber L, Reed JR, Backes WL. Organization of NADPH-cytochrome P450 reductase and CYP1A2 in the endoplasmic reticulum-microdomain localization affects monooxygenase function. Mol. Pharmacol. 2011;79:549–557. doi: 10.1124/mol.110.068817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brignac-Huber LM, Reed JR, Eyer MK, Backes WL. Relationship between CYP1A2 Localization and Lipid Mcrodomain Formation as a Function of Lipid Composition. Drug Metab. Dispos. 2013;41:1896–1905. doi: 10.1124/dmd.113.053611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Park JW, Reed JR, Brignac-Huber LM, Backes WL. Cytochrome P450 system proteins reside in different regions of the endoplasmic reticulum. Biochem. J. 2014;464:241–249. doi: 10.1042/BJ20140787. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.