Abstract
Colorectal cancer (CRC) is the second leading cause of cancer deaths in the United States. It arises from loss of intestinal epithelial homeostasis and hyperproliferation of the crypt epithelium. In order to further understand the pathogenesis of CRC it is important to further understand the factors regulating intestinal epithelial proliferation and more specifically, regulation of the intestinal epithelial stem cell compartment. Here, we investigated the role of the RNA binding protein RBM3 in stem cell homeostasis in colorectal cancers. Using a doxycycline (Dox) inducible RBM3 overexpressing cell lines HCT 116 and DLD-1, we measured changes in side population (SP) cells that have high xenobiotic efflux capacity and increased capacity for self-renewal. In both cell lines, RBM3 induction showed significant increases in the percentage of side population cells. Additionally, we observed increases in spheroid formation and in cells expressing DCLK1, LGR5 and CD44Hi. As the Wnt/β-catenin signaling pathway is important for both physiologic and cancer stem cells, we next investigated the effects of RBM3 overexpression on β-catenin activity. RBM3 overexpression increased levels of nuclear β-catenin as well as TCF/LEF transcriptional activity. In addition, there was inactivation of GSK3β leading to decreased β-catenin phosphorylation. Pharmacologic inhibition of GSK3β using (2′Z,3′E)-6-Bromoindirubin-3′-oxime (BIO) also recapitulates the RBM3 induced β-catenin activity. In conclusion, we see that RNA binding protein RBM3 induces stemness in colorectal cancer cells through a mechanism involving suppression of GSK3β activity thereby enhancing β-catenin signaling.
Keywords: cancer stem cells, Wnt signaling, GSK3β, DCLK1, LGR5, CD44
INTRODUCTION
The RNA binding motif containing protein 3 (RBM3) was originally discovered through cDNA selection in yeast artificial chromosomes and was found to be a cold shock response protein [1,2]. Further studies revealed that RBM3 had cytoprotective effects and that exogenous overexpression of RBM3 impaired polyglutamine repeat induced cell death in both neural and non-neural cells [3]. The cytoprotective effects were further expanded to include chemical inducers of apoptosis, hypothermia, and serum deprivation [4–6]. Indeed, it was shown that RBM3 expression is necessary for appropriate cell cycle progression and loss of RBM3 induced mitotic catastrophe [6,7]. Studies of RBM3 expression in human tumors showed that it is upregulated in many solid tumors including colon, prostate, and breast cancers, and more specifically it is upregulated in a stage dependent manner in human colon cancers [7]. RBM3 has been shown to be expressed at higher levels in poorly differentiated, more aggressive prostate cancer and it has been shown to be an independent prognostic factor predicting early recurrence [8,9]. RBM3 expression has also been shown to be correlated with high grade astrocytoma compared to low grade or normal tissue [10]. Furthermore, we have previously shown that overexpression of the protein results in transformation of NIH-3T3 cells [7]. We have previously reported that RBM3 is upregulated in a stage dependent manner in colon cancer, and that upregulation of RBM3 is capable of inducing oncogenic transformation [7]. Mechanistically, we have demonstrated that RBM3 is an RNA binding proteins, which when overexpressed increases the stability and translation of rapidly degraded mRNAs such as cyclooxygenase 2 (COX-2), interleukin-8 (IL-8), and vascular endothelial growth factor (VEGF) [7]. Downregulating RBM3 expression has also shown to increase chemotherapeutic susceptibility in prostate cancer cell lines [11].
Paradoxically, some studies have shown that RBM3 expression levels correlate with decreased tumor progression and recurrence. RBM3 was found to be upregulated in breast cancer tissues compared to normal controls, but high RBM3 expression was correlated with increased disease free survival [12]. Similarly, it was found that in a prospective cohort study of melanoma, the majority of tumors strongly expressed RBM3, though the tumors with low RBM3 expression levels correlated with increased aggressiveness [13]. One study postulated that RBM3 may serve to maintain genomic integrity in ovarian cancer cell lines treated with genotoxic agents [14]. However, these studies are largely observational, it remains unclear why this discrepancy exists. Importantly, the role for RBM3 in tumor aggressiveness remains contested. We have designed these studies in an attempt to elucidate the effects of ectopic RBM3 overexpression and reveal a mechanistic role of RBM3 in the pathogenesis of colorectal carcinoma. In doing so, we hope to begin understanding the role of RBM3 expression on tumor aggressiveness.
Colon cancer is the second leading cause of cancer related deaths in the United States [15]. Akey pathway that is dysregulated in colon cancer is initiated is the Wnt (Wnt/β-catenin) pathway [16]. In the absence of stimuli, β-catenin is subjected to high turnover at the Adenomatous Polyposis Coli (APC)/Axin/Casein Kinase 1α (CK1α)/GSK3β destruction complex [17]. Upon exogenous stimuli, such as presence of a Wnt ligand or through the actions of phosphorylated AKT, the destruction complex is inactivated resulting in the cytoplasmic accumulation and nuclear translocation of β-catenin, where it associates with a member of the Transcription factor/Lymphoid enhancer binding factor (TCF/LEF) and alters the transcriptional state of the cell [17,18]. Importantly, GSK3β kinase activity can be modulated by phosphorylation at the Serine 9 position resulting in the inability to phosphorylate β-catenin for degradation [19]. Wnt/β-catenin signaling has also been heavily implicated in the maintenance of the stem cell compartment within the normal adult intestine and in regulating the stem cell (CSC) population in colorectal cancers [20]. CSCs are a small subset of cells within the heterogeneous tumor that have the capacity to resist classical chemotherapeutics, survive under stressful conditions and regenerate the tumor following radiation or chemotherapeutic intervention [21].
Previous studies have shown that RNA binding proteins are capable of affecting the cancer stem cell population [22,23]. Given that RBM3 is able to induce properties shared by cancer stem cells, namely resistance to chemotherapy, hypoxia and serum deprivation, we investigated the role of RBM3 in affecting the cancer stem cell population. We chose to overexpress RBM3 in the colon cancer cell lines HCT 116 and DLD-1 primarily because these cell lines have different genotypes allowing for modulation of β-catenin signaling. DLD-1 cells contain an inactivating mutation of APC, while HCT 116 cells contain a mutation in the CTNNB1 gene which prevents one copy of β-catenin from being phosphorylated for degradation [24]. We show that RBM3 overexpression results in increased stemness in colon cancer cells as measured by side population, spheroid formation capacity and expression of the stem cell markers. Additionally, RBM3 overexpression results the inactivation of GSK3 by phosphorylation at Ser9, thereby enhancing β-catenin signaling activity the colorectal cancer cells.
MATERIALS AND METHODS
Cell Culture and Reagents
HCT 116 and DLD-1 cells were obtained from American Type Culture Collections (Manassas, VA) and grown in Dulbecco’s modified eagle medium (DMEM) with 10% heat inactivated fetal bovine serum (Sigma–Aldrich, St. Louis, MO) and 2% penicillin/streptomycin/amphotericin B solution (Mediatech Inc., Manassas, VA). Cells were grown in a humidified incubator at 37°C with 5% CO2. Tetracycline inducible RBM3 overexpressing plasmids were generated using the pLVX Tet-On Advanced plasmid system (ClonTech Laboratories Inc., Mountain View, CA). Lentiviral particles were generated using Lenti-X cells transfected with the pGIPZ set of packaging plasmids generously donated by Roy Jensen (University of Kansas Medical Center, Kansas City, KS). Both HCT 116 and DLD-1 cells were cotranduced with pLVX-Tet-On and pLVX-Tight plasmids followed by selection with 1 mg/mL G418 (Mediatech Inc.) and 2 µg/mL puromycin (Life Technologies, Grand Island, NY) for 7 d. Cells were consistently maintained in 500 µg/mL G418 and 1 µg/mL puromycin following selection unless otherwise noted.
Western Blots
Cell extracts were separated by poly-acrylamide gel electrophoresis using a Miniprotean Tetracell apparatus (BioRad, Hercules, CA) followed by transfer on to 0.45 µm pore size Immobilon polyvinyl difluoride membrane (Millipore, Bedford, MA) using a mini Transblot module (BioRad). Specific proteins were detected by the enhanced chemiluminescence system (GE Health Care, Piscataway, NJ). Nuclear cytoplasmic extraction was generated through NE-PER kit (Thermo Fisher Scientific, Rockford, IL) according to manufacturer recommendations. Antibodies for RBM3 were obtained from AbCam (AbCam, Cambridge, MA) or custom generated through Fisher (Thermo Fisher Scientific). Antibodies for β-catenin and phospho-β-catenin were obtained from Cell Signaling (Cell Signaling Technology, Danvers, MA) or BD Biosciences (BD Biosciences, San Jose, CA). Antibodies for GSK3β and phosphor-GSK3β were obtained from Cell Signaling (Cell Signaling Technology).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total cellular RNA was isolated using TRIzol reagent followed by reverse transcription using SuperScript II in the presence of random hexonucleotide primers (Life Technologies). cDNA was then analyzed by realtime PCR using Jumpstart Taq polymerase (Sigma–Aldrich) and SYBR Green nucleic acid stain (Life Technologies). Threshold crossing values for each gene were normalized to glyceraldehyde phosphate dehydrogenase (GAPDH) gene expression. mRNA expression was then normalized to fold change relative to uninduced controls. Primers used in this study are shown in Figure S5.
Side Population
Side population assay was performed as previously described with minor modifications [25]. In brief, cells were plated at 100 000 cells/mL into 6 well plates with relevant treatments. Following 72 h of treatment cells were trypsinized and resuspended at 1 million cells/mL in DMEM and pre-warmed for 15 min at 37°C. Cells were then given 50 µM verapamil (Sigma–Aldrich), as a pan efflux inhibitor, or vehicle for 30 min. Cells were then incubated with 10 µM Vybrant DyeCycle Violet (DCV) (Life Technologies) for 90 min at 37°C. Cells were then resuspended in ice cold SP buffer containing 2% FBS, 2 mM HEPES, 1 µg/mL 7-amino actinomycin D (7AAD) (Life Technologies) in Hank’s balanced salt solution and analyzed on a BD LSR II for side population. DCV was excited using a violet diode laser (408 nm) and emission was measured with a 450/50 nm bandpass filter (DCV Blue) and 630 nm longpass, 660/40 nm bandpass filter (DCV Red). Viability gating was established with 7-amino actinomycin D staining (7AAD) with excitation at 552 nm and emission monitored with a 630 nm longpass, 660/40 nm bandpass filter. Cells were initially gated for appropriate size using the side scatter (SSC) and forward scatter (FSC). Cells were then gated for viability using 7-AAD uptake to eliminate non-viable cells. Finally, the location of the SP gate was established using the verapamil treated sample.
Stem Cell Marker Flow Cytometry
Cells were treated with Dox to induce expression of GFP or RBM3 for 72 h. Cells were then trypsinized then resuspended at 1 × 106 cells. For DCLK1 and LGR5, cells were stained with Alexafluor 647 conjugated LGR5 antibody (BD Biosciences) or PE conjugated DCLK1 antibody (AbCam) according to manufacturer recommendations. PE conjugation was performed using the Zenon PE labelling kit (Life Technologies) according to manufacturer recommendations. For the CD44/CD24 experiment, cells were stained using allophycocyanin (APC) conjugated CD44 antibody and phycoerythrin (PE) conjugated CD24 antibody according to manufacturer recommendations with the exception of using half the recommended concentration of APC-CD44 antibody (BD Biosciences). Compensation was performed using standardized fluorescent beads.
Proliferation Assay
Cells were induced with Dox 2 for 72 h. Following induction of RBM3 or GFP vector control, cells were plated at 50 000 cells/mL into 96 well plates and allowed to adhere overnight followed by treatment with paclitaxel or doxorubicin at varying concentrations. Following 48 h of treatment, cells were assayed for proliferation using the hexoseaminidase assay as previously described [26]. Briefly, cell culture medium was removed followed by addition of 75 µL hexoseaminidase substrate buffer (7.5 mM sodium citrate, pH 5.0, 3.75 mM 4-nitrophenyl-N-acetyl-β-D-glucosaminide, 0.25% Triton) per well and incubation at 37°C for 30 min. The reaction was quenched using 112.5 µL of hexoseaminidase developer solution (50 mM glycine, 5 mM EDTA, pH 10.4) and absorbance at 405 nm was read on a plate reader. Percentage proliferation was calculated as absorbance relative to untreated control and curves for inhibitory response were fit using GraphPad Prism software (GraphPad, La Jolla, CA).
Spheroid Formation
Cells were appropriately treated in 2D culture, trypsinized followed by a wash in phosphate buffered saline (PBS) to remove residual serum. Cells were then resuspended in spheroid media containing 1× B27 supplement, 20 ng/mL basic recombinant fibroblast growth factor (bFGF) and 20 ng/mL recombinant epidermal growth factor (EGF) (Life Technologies) in DMEM. Cells were passed through a 35 µm sieve, counted and plated onto ultra-low attachment surface plates (Corning, Tewksbury, MA). Spheroid formation was allowed to proceed for 10–14 d followed by spheroid quantification and analysis using a Celigo Cyntellect embryoid body counter.
siRNA transfection
siRNA to CTNNB1 and scrambled siRNA were both purchased from Life Technologies (Life Technologies). Transfection was performed using Lipofectamine RNAi-Max and OptiMEM according to manufacturer recommendations (Life Technologies). Briefly, cells were plated at 400 000 cells per well into 6 well plates and transfected using either siSCR (Cat# S28186) or siCTNNB1 (Cat# S436) using 25 pmol of siRNA into each well. Six hours following transfection, cells were treated with indicated concentrations of Dox. Seventy-two hours following siRNA transfection, cells were then assayed for side population, flow cytometry or western blot as indicated above.
TOPFlash Luciferase Assay
Cells were plated in 24 well plates in antibiotic/antimycotic free medium and without selection antibiotics. Each well was transfected using 2 µL of Lipofectamine 2000 complexed with 900 ng of TOP-Flash luciferase plasmid and 100 ng of CMV renilla luciferase plasmid. Eight hours following transfection, the media was replaced with normal growth medium containing selection antibiotics and appropriate treatments. Luciferase activity was then read using the dual-luciferase reporter assay system (Promega, Madison, WI) on a plate reader. TOPFlash luciferase activity was normalized to CMV renilla luciferase activity.
Immunofluorescence and Immunohistochemistry
For immunofluorescence, cells were plated on sterile glass cover slips with appropriate treatments. Cells were then fixed using neutral buffered formalin then permeablized using 0.1% Triton. Antigen blocking was achieved using 1% bovine serum albumin in phosphate buffered saline (PBS). Cells were incubated with primary antibody overnight at 4°C followed by staining with fluorophore conjugated secondary antibody for 1 h at room temperature. Coverslips were mounted using Prolong Gold with DAPI (Life Technologies). For immunohistochemistry, spheroids were embedded in paraffin blocks and slides were obtained with 5 µm sections. Slides were deparaffinized then boiled for 20 min in antigen retrieval buffer (10 mM sodium citrate, pH6, 0.05% Tween-20) followed by peroxidase quenching using Peroxabolish (Biocare Medical, Concord, CA). Slides were then stained using Histostain SP kit (Life Technologies) as per manufacturer recommendations. Antibodies used were the same as those used for western blots.
Statistics
Results are expressed with standard error of mean. Statistical analysis was performed using Student’s t-test using GraphPad Prism software (GraphPad, La Jolla, CA). Values of P < 0.05 were considered statistically significant.
RESULTS
RBM3 Overexpression Increases Side Population in Colon Cancer Cells
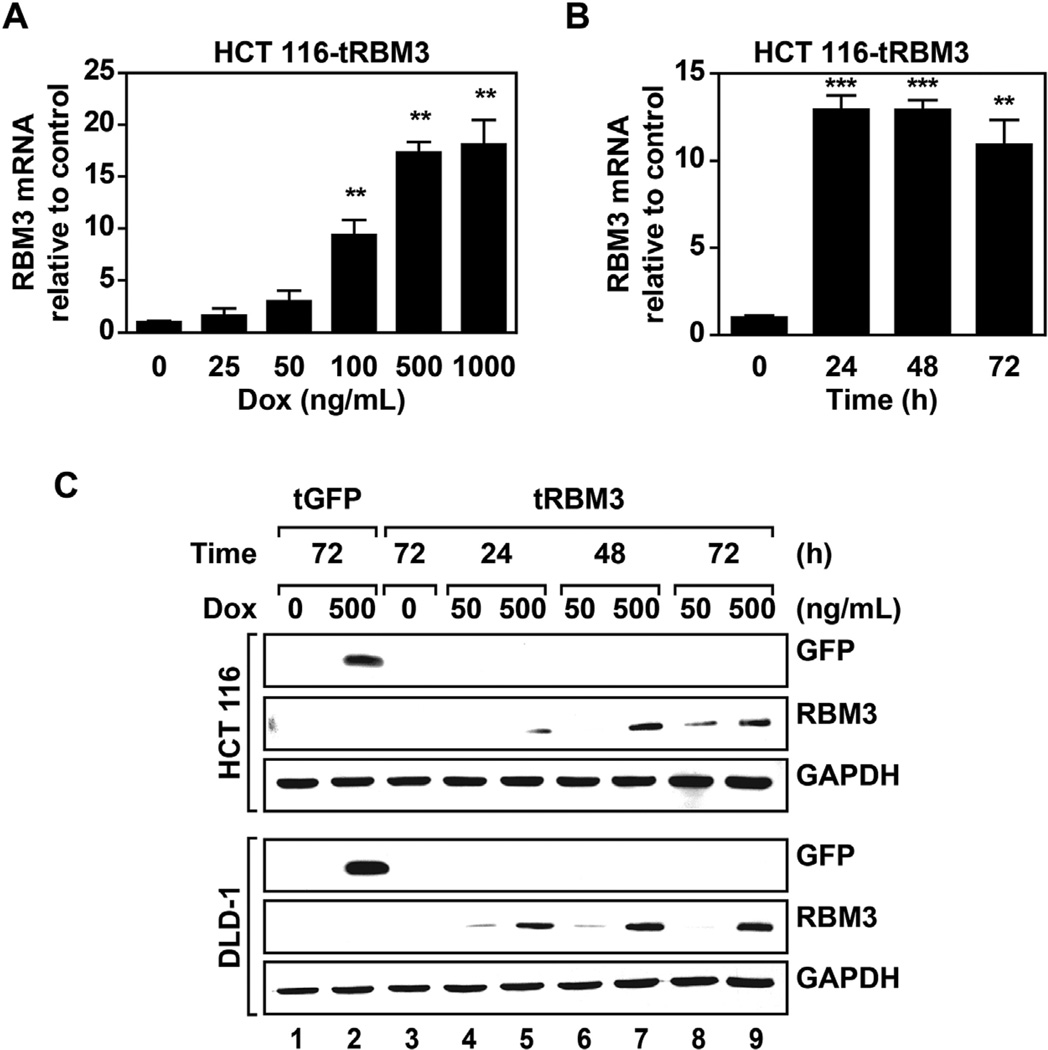
We first developed HCT 116 and DLD-1 colon cancer cell lines with stable doxycycline (Dox) inducible RBM3 overexpression. There was a dose-dependent increase in RBM3 mRNA expression with maximal induction seen with 500 ng/mL Dox (Figure 1A). Additionally, RBM3 mRNA and levels reached maximal induction within 24 and 48 h, respectively (Figure 1B,C). We also validated that the GFP vector control had similar Dox-mediated induction (Figure 1C). Importantly, there is very low baseline expression of both RBM3 and GFP in the absence of Dox.
Figure 1.
RBM3 overexpression in colorectal cancer cells. (A) Real time PCR analysis of RBM3 mRNA in HCT 116 cells following 72 h induction with indicated concentrations of doxycycline. mRNA fold change is normalized to uninduced control. (B) Real time PCR analysis of RBM3 mRNA following 500 ng/mL Dox induction with indicated durations of induction. mRNA fold change is normalized to uninduced control. Graphs show mean fold change with SEM. (C) Western blot analysis of RBM3 or GFP overexpression in DLD-1 and HCT 116 inducible GFP or RBM3 cells with indicated concentrations of and times of Dox induction. Data shows significant dose- and time-dependent induction of RBM3 in the two cell lines. Graphs show mean fold change with SEM. *P < 0.05, **P < 0.01, ***P <0.001 (Student's t-test).
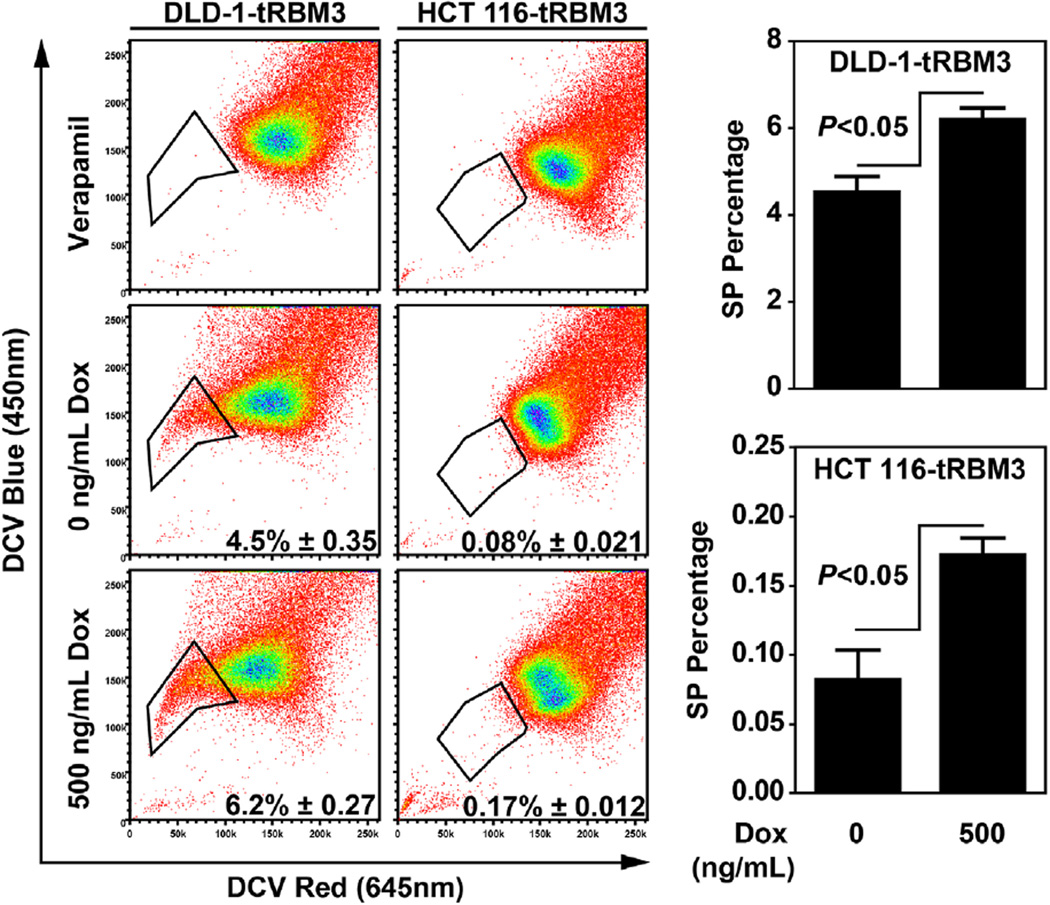
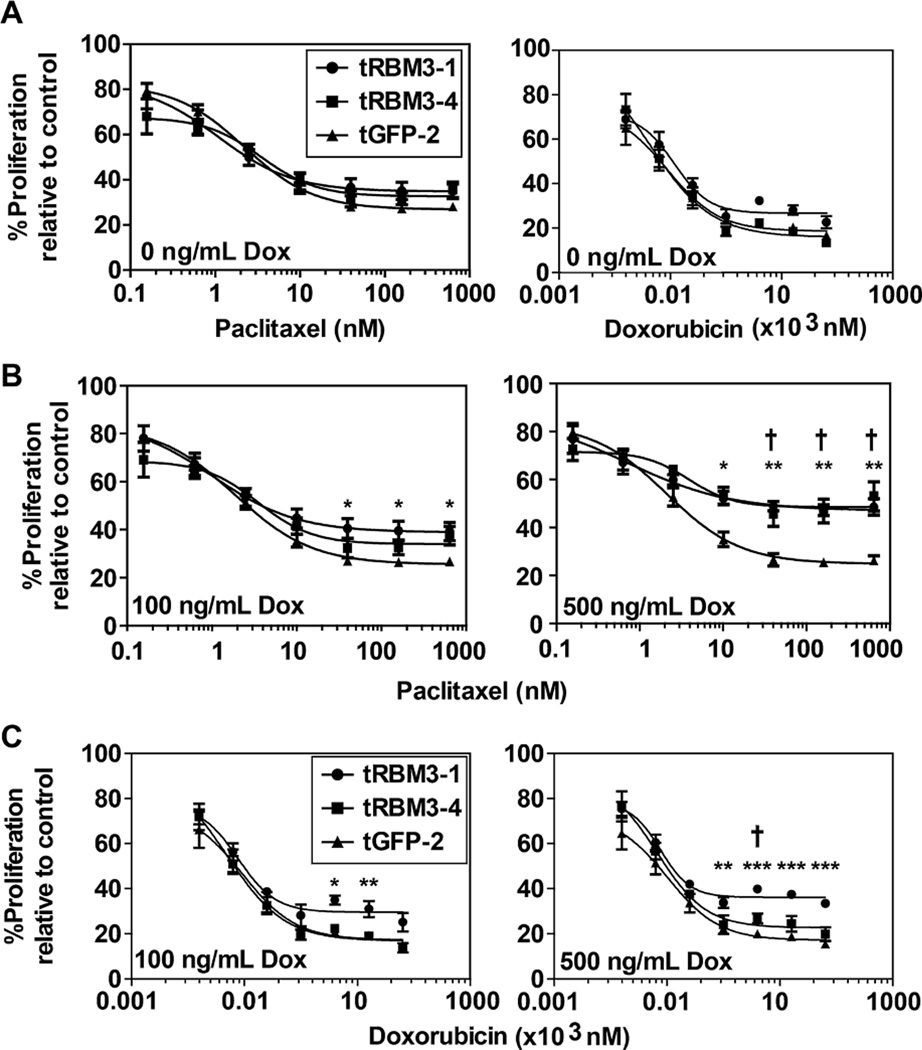
To measure changes in the stem cell population, we first determined whether there was a change in the number of cells inside population (SP) following RBM3 induction. SP population, originally described in mouse bone marrow cells, represent a small fraction of cells with enhanced capacity to efflux therapeutic agents, as measured by intracellular fluorescent dye, such as Hoescht 33342, and have the capacity to self-renew and differentiate [27]. SP cells have also been shown to be upregulated in many primary tumors and cancer cell lines [28]. Following RBM3 induction, both DLD-1 and HCT 116 cells showed increased numbers of DyeCycle Violet negative SP cells compared to uninduced controls (Figure 2). Verapamil was used as a pan efflux inhibitor to establish the negative control for locating the SP fraction (Figure 2). As mentioned above, SP cells have the increased capacity to efflux chemotherapeutic agents. We next hypothesized that with a higher side population, RBM3 expression should also confer increased resistance to chemotherapies that are substrates of efflux mechanisms. For this, we measured cell proliferation following treatment with paclitaxel and doxorubicin, both being of which are substrates for ATP-binding cassette (ABC) transporters [29]. HCT 116 Tet-RBM3 clones were induced for 72 h with 500 ng/mL Dox and then treated with increasing concentrations of the two compounds. There was significantly higher level of survival of RBM3 overexpressing cells, especially at higher concentrations of paclitaxel and doxorubicin (Figure 3). As expected, uninduced RBM3 cells showed no appreciable difference between GFP vector controls or parental cells (Figure S1). These data suggest that RBM3 overexpression increases stem cells within colon cancer cells and alters sensitivity to chemotherapies. The studies are further confirmation of previous studies that correlated RBM3 knockdown with loss of resistance to doxorubicin and cisplatin [11].
Figure 2.
RBM3 overexpression increases side population. Flow cytometry analyses of side population following 72 h treatment with 0 or 500 ng/mL Dox. Side population negative gate was set with 50 µM verapamil treated control. One representative experiment is shown with dot plots with bar graphs showing mean of three replicates. Shown in dot plots are percentage of side population with respective 95% confidence intervals.
Figure 3.
RBM3 induction increases chemotherapy resistance. (A) Comparison of proliferation of HCT 116 Tet-GFP and Tet-RBM3 clones with varying concentrations of paclitaxel or doxorubicin with 0 ng/mL Dox induction. The two cell lines show no difference in proliferation rate. (B) Comparison of hexoseaminidase proliferation assay of HCT 116 Tet-GFP and Tet-RBM3 clones treated with varying concentrations of paclitaxel following 72 h of 100 or 500 ng/mL Dox induction. In the presence of RBM3, the proliferation inhibition by paclitaxel is reduced. (C) Comparison of hexoseaminidase proliferation assay of HCT 116 Tet-GFP and Tet-RBM3 clones treated with varying concentrations of doxorubicin following 72 h of 100 or 500 ng/mL Dox induction. Again there was significant reduction in doxorubicin-mediated proliferation inhibition in the presence of RBM3. Graphs show SEM. *P < 0.05, **P < 0.01, ***P < 0.001 for RBM3-1, †P < 0.05, ††P < 0.01, †††P < 0.001 for RBM3-4 (Student's t-test).
RBM3 Increases Spheroid Formation
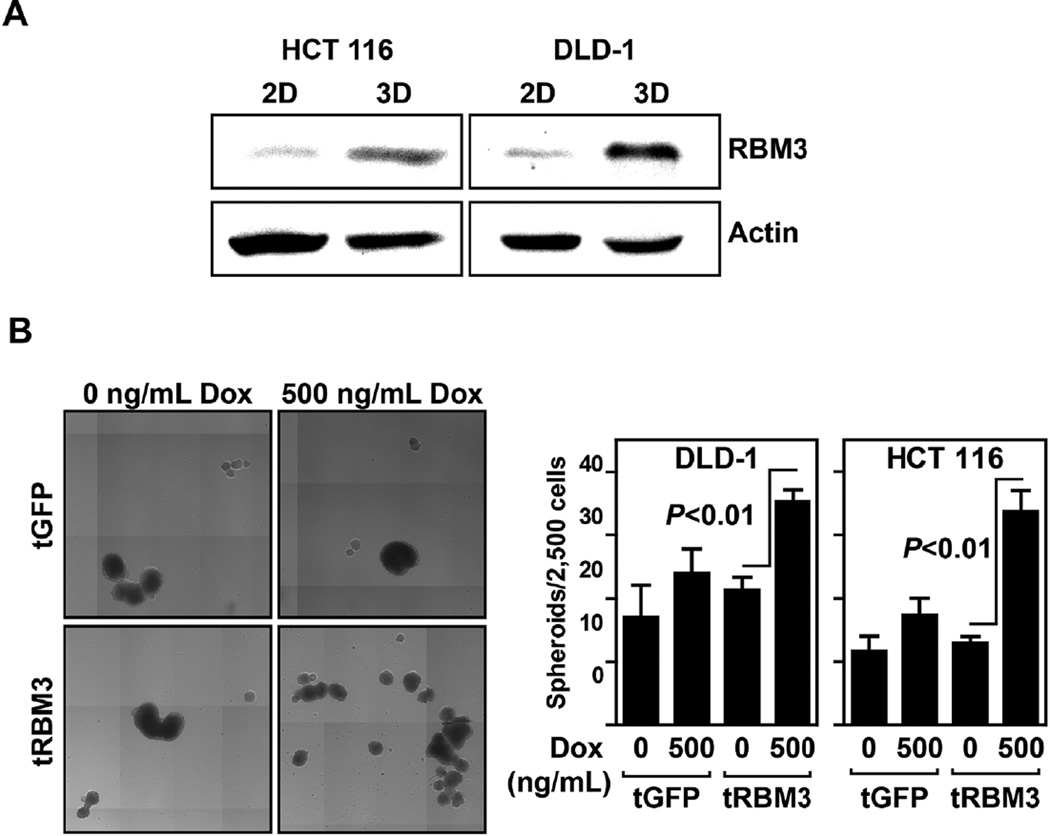
Spheroid forming capacity of cells is a marker of stemness [30,31]. Cells cultured as spheroids have been shown to have significant increases in SP and in self-renewal and differentiation capacity [32]. We first compared levels of RBM3 protein within spheroids to levels within monolayer cell cultures and found that there were significantly higher levels of RBM3 in the spheroids. This was observed in both DLD-1 and HCT 116 cells (Figure 4A). We next determined whether increasing RBM3 levels in the cells affects spheroid formation. Following 72 h Dox induction in both DLD-1 and HCT 116 cells expressing vector control or RBM3, cells were plated into ultra-low attachment conditions and allowed to form spheroids. Both DLD-1 RBM3 and HCT 116 RBM3 overexpressing cells showed increase in spheroid formation compared to vector or RBM3 uninduced controls (Figure 4B). These data suggest that colorectal cancer cells increase RBM3 expression in the process of spheroid formation and that RBM3 is capable of enhancing spheroid formation capacity. Taken together, these data suggest that RBM3 overexpression enhances stemness in cancer cells.
Figure 4.
RBM3 overexpression increases spheroid formation. (A) Western blot analyses for RBM3 from HCT 116 and DLD-1 cells grown in regular tissue culture dish (2D culture) and ultra-low attachment plates (3D culture). Data shows that cells grown in 3D cultures have higher levels of RBM3 compared to 2D cultures. (B) Representative composite image of HCT 116 GFP or RBM3 inducible cells induced for 72 h with 0–500 ng/mL of Dox then plated in ultra-low attachment plates with colonosphere media and allowed to form spheroids for 14 d. Quantification of HCT 116 and DLD-1 spheroids was performed by Celligo embryoid body counting system. Bar graph shows spheroid number from 2500 cells.
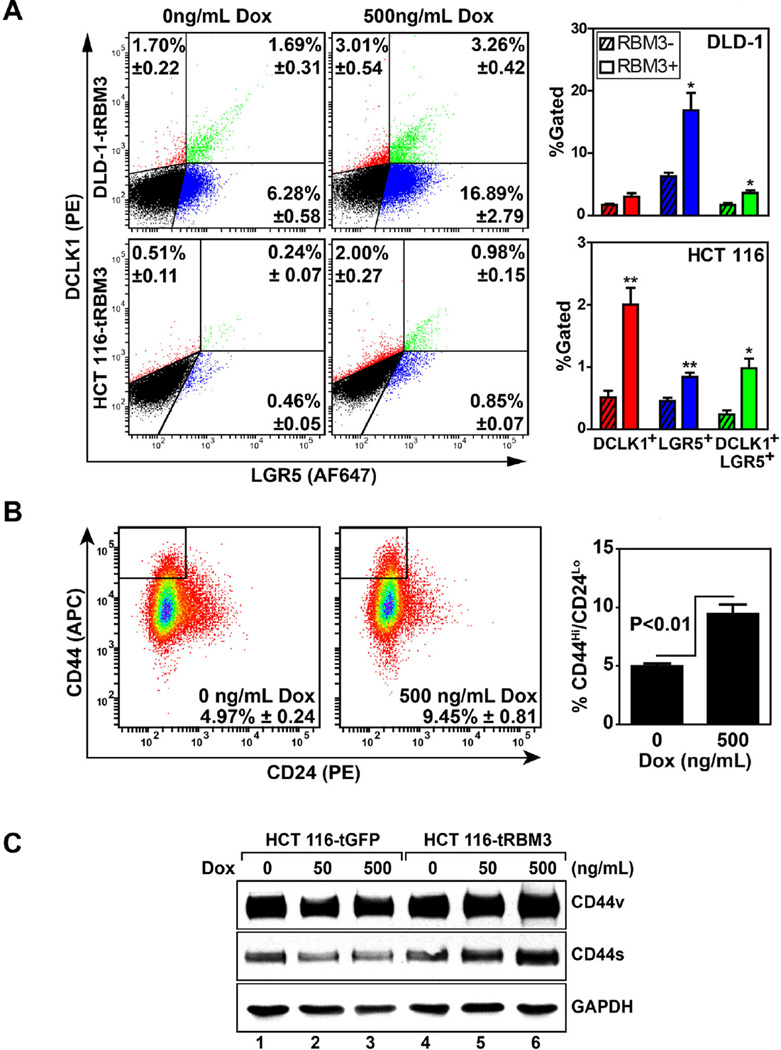
RBM3 Overexpression Increases Stem Cell Marker Expression
Two putative markers elucidated for the intestinal epithelial stem cell is doublecortin-like calmodulin kinase 1 (DCLK1) and leucine-rich-repeat containing G-protein-coupled receptor 5 (LGR5) [33,34]. Interestingly, both markers have also been implicated in the process of tumorigenesis and may serve as important markers in the context of both physiologic and cancer stem cells [35,36]. To further validate the model of RBM3 induced colon cancer stem cells, we next determined the expression of both DCLK1 and LGR5 in HCT 116 and DLD-1 cells. Following induction of RBM3 with 500 ng/mL Dox for 72 h, there was significant increase in the DCLK1+ (0.51 ± 0.11 vs. 2.00 ± 0.27 in control and RBM3 overexpressing cells, respectively), LGR5+ (0.46 ± 0.05 vs. 0.85 ± 0.07 in control and RBM3 overexpressing cells, respectively) and double positive (0.24 ± 0.07 vs. 0.98 ± 0.15 in control and RBM3 overexpressing cells, respectively) populations of HCT 116 when compared to uninduced control (Figure 5A). Similar increase in LGR5+ and double positive populations with only a modest increase in DCLK1+ (P¼0.08) in DLD-1 cells when compared to uninduced controls (Figure 5A). The modest increase in DCLK1+ LGR5− cell population in DLD1 is compensated for by the higher percentage of DCLK1+LGR5+ cells.
Figure 5.
Increased stem cell marker expression in HCT 116 and DLD-1 cells overexpressing RBM3. (A) Representative flow cytometry dot plot of HCT 116 and DLD-1 cells stained for DCLK1 (phycoerythrin-PE) and LGR5 (AlexaFluor 647-AF647). Bar graphs on right represent percentage of DCLK1+, LGR5+, and DCLK1+/LGR5+ double positive populations. Error bars represent SEM. P values shown are obtained with Student's t-test. *P < 0.05, **P < 0.01, ***P < 0.001 (Student's t-test). (B) Representative flow cytometry dot plot of HCT 116 cells stained for CD44 (allophycocyanin-APC) and CD24 (PE). Bar graphs on right represent percentage of CD44hi/CD24lo population of cells. Error bars represent SEM. (C) Western blot analyses for the two variants of CD44 of extracts from HCT 116 cells expressing GFP or RBM3 after induction with increasing doses (0–500 ng/mL) of doxorubicin. Data shows increased expression of both isoforms of CD44 following RBM3 overexpression.
As an additional marker of stem cell population, we next measured changes in the CD44Hi/CD24Lo population. Previous studies have demonstrated that CD44Hi/CD24Lo population of cells display characteristics of cancer stem cells [37]. Following RBM3 induction, HCT 116 Tet-RBM3 cells showed an increase in CD44Hi/CD24Lo compared uninduced controls (4.987 ± 0.24 vs. 9.45 ± 0.81 in control and RBM3 overexpressing cells, respectively) (Figure 5B). GFP vector controls showed no appreciable difference following Dox induction (data not shown). Previous reports have also shown that RBM3 overexpression induces alternative splicing of CD44 increasing the tumor suppressing variant of CD44 (CD44s) while suppressing levels of the pro-oncogenic CD44 variants (CD44v) in prostate cancer cells [38]. In HCT 116 Tet-RBM3 cells, RBM3 induction showed significant increases in both major isoforms of CD44 with no significant changes in GFP vector controls (Figure 5C). Additional studies are required to determine whether the CD44Hi/CD24Lo cells also overexpress DCLK1 and LGR5. Nevertheless, taken together, these data imply that RBM3 overexpression is capable of increasing the cancer stem cell population.
RBM3 increases β-Catenin Signaling Activity
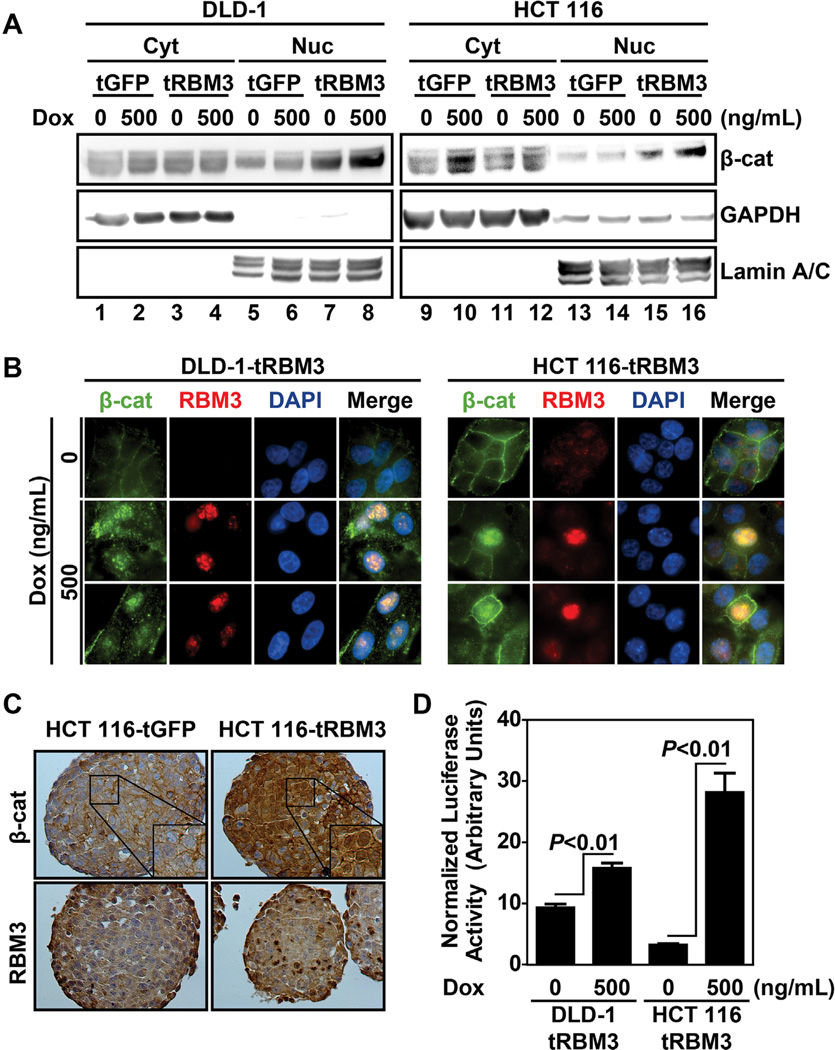
The Wnt/β-catenin signaling pathway is known to be active in stem cells of the embryo and of the adult as well as in cancer stem cells [39]. During embryogenesis, β-catenin activation is required for maintenance and transition from a pluripotent state. In cancers, the pathway is believed to contribute to the self-renewal of cancer stem cells [17]. Furthermore, both CD44 and LGR5 have been identified as β-catenin transcriptional targets [30,33,40]. Moreover, β-catenin signaling activity has been shown to be essential for SP cells and spheroid formation [30,41]. Since, RBM3 overexpression increased stemness and expression of stem cell marker protein LGR5, we next determined whether the Wnt/β-catenin pathway is affected. Following RBM3 induction in DLD-1 and HCT 116 cells, there was only a modest increase in total β-catenin levels (Figure S2A). As β-catenin transcriptional activity depends upon its localization to the nucleus, we next examined changes in β-catenin levels in the cytoplasm and nucleus. RBM3 overexpression did not significantly affect cytoplasmic levels of β-catenin. However, nuclear levels of β-catenin were increased upon RBM3 overexpression in both DLD-1 and HCT 116 compared to uninduced or vector controls (Figure 6A). Additionally, immunofluorescence labeling revealed nuclear accumulation of β-catenin in RBM3 overexpressing cells, while membrane localization of β-catenin showed no significant difference (Figure 6B). This effect was further replicated with immunohistochemistry of spheroids (Figure 6C). Finally, to validate that the increase in nuclear β-catenin levels correlates with increased transcriptional activity, we examined TOPFlash luciferase assay. Following 48 h RBM3 induction, TOPFlash luciferase activity, normalized to CMV/Renilla luciferase was significantly increased in both cell lines with HCT 116 cells showing an average of 28.3 ± 3.1 compared to 3.3 ± 0.13 in uninduced controls (Figure 6D). Similarly, in DLD-1 cells, luciferase levels were observed to be at 15.9 ± 0.79 RBM3 overexpressing cells compared to an average of 9.4 ± 0.52 in uninduced controls (Figure 6D). As expected, DLD-1 cells show a higher basal TOPFlash activity because it encodes mutated APC whereas HCT 116 maintains intact wild type APC and at least one wild type β-catenin allele [24]. Further confirmation of increased β-catenin transcriptional activity was observed through elevated levels of its target genes c-Myc, LGR5, and CD44 [33,40,42] (Figure S2B). These data demonstrate that upon RBM3 overexpression, β-catenin transcriptional activity is increased resulting in higher DCLK1+ and LGR5+ stem cell population thereby enhancing side population and spheroid formation.
Figure 6.
RBM3 overexpression increases β-catenin signaling. (A) β-catenin western blot of DLD-1 and HCT 116 inducible GFP or RBM3 cell lines following nuclear and cytoplasmic fractionation. Lamin A/C shown as loading control for nuclear fraction and GAPDH shown as loading control for cytoplasmic fraction. Cells were treated with indicated concentration of Dox for 72 h. (B) Immunofluorescence of DLD-1 and HCT 116 RBM3 cells treated with 0 or 500 ng/mL Dox for 72 h. Both β-catenin and RBM3 are nuclear following RBM3 induction. (C) Immunohistochemistry of HCT 116 GFP and RBM3 overexpressing spheroids. Cells were treated with 500 ng/mL for 72 h Dox in monolayer culture followed by 14 d growth in spheroid culture. Again, there is increased nuclear levels of β-catenin and RBM3 following induction. (D) TOPFlash luciferase assay of DLD-1 and HCT 116 RBM3 inducible cells. Cells were treated with indicated concentrations of Dox for 48 h following transfection with TOPFlash and CMV renilla luciferase plasmid. Bar graphs represent mean values of TOPFlash firefly luciferase luminescence normalized to CMV renilla luciferase luminescence. There was increased luciferase activity following RBM3 induction. Bar graphs show SEM. P values were obtained from Student's t-test.
RBM3 Mediated Stem Cell Induction Is Dependent Upon Increased β-catenin Activity
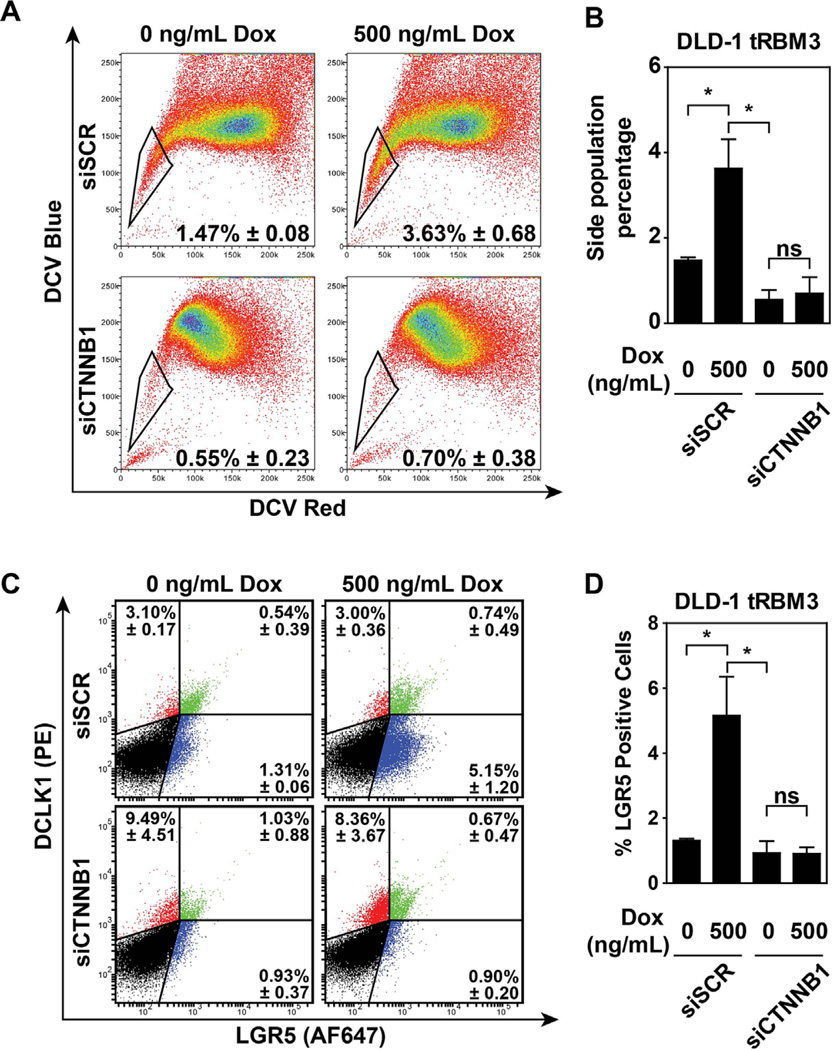
We next sought to determine whether the initial observations of increased SP percentages and stem cell marker expression were dependent upon the β-catenin signaling pathway. In order to validate this, we transiently knocked down CTNNB1 expression using siRNA (siCTNNB1) in DLD-1 tRBM3 cells (Figure S3). We observed that following transfection of siCTNNB1, there was attenuation in the SP percentages (0.55 ± 0.23) compared to scrambled siRNA (siSCR) transfected controls (1.47 ± 0.08) (Figure 7A). As expected, RBM3 overexpression in siSCR transfected cells showed similar increases in both SP. Moreover, RBM3 overexpression in siCTNNB1-transfected cells was unable to rescue either the loss in SP (Figure 7A,B). We also determined the effect of the knockdown on LGR5 and DCLK1 expressing cells. LGR5+ only cells were significantly suppressed upon β-catenin suppression (5.15 ± 1.2 in siSCR vs. 0.90 ± 0.12 siCTNNB1-transfected cells, respectively) (Figure 7C). As expected, overexpression of RBM3 did not overcome β-catenin knockdown (Figure 7C,D). In addition, there was a significant effect on DCLK+LGR5+ cells (0.54 ± 0.39 vs. 1.03 ± 0.88 in siSCR vs. 0.90 ± 0.12 siCTNNB1-transfected cells, respectively) (Figure 7C). Surprisingly, however, there was no effect on DCLK1-positive LGR-negative cells between siSCR and siCTNNB1- transfected cells in the absence (3.10 ± 0.17 vs. 9.49 ± 4.51, respectively) or presence of RBM3 overexpression (3.00 ± 0.36 vs. 8.36 ± 3.67, respectively) (Figure 7C). These data suggest that the ability of RBM3 overexpression to modulate SP and LGR5 expression, but not DCLK1 expression is dependent on β-catenin levels.
Figure 7.
β-catenin is necessary for RBM3 mediated stemness. (A) Flow cytometry analysis of SP in DLD-1 cells overexpressing RBM3 transfected with scrambled (siSCR) or β-catenin-specific (siCTNNB1) siRNA and induced with 0 or 500 ng/mL Dox. (B) Bar graph representations of the SP percentages following the indicated treatments. (C) Flow cytometry analysis of LGR5 and DCLK1 expression in DLD-1 cells overexpressing RBM3 transfected with siSCR or siCTNNB1 and induced with 0 or 500 ng/mL Dox. (D) Bar graph representations of the LGR5 and DCLK1 expression following the indicated treatments. *P < 0.05, **P < 0.01, ***P < 0.001, ns P > 0.05 (Student's t-test).
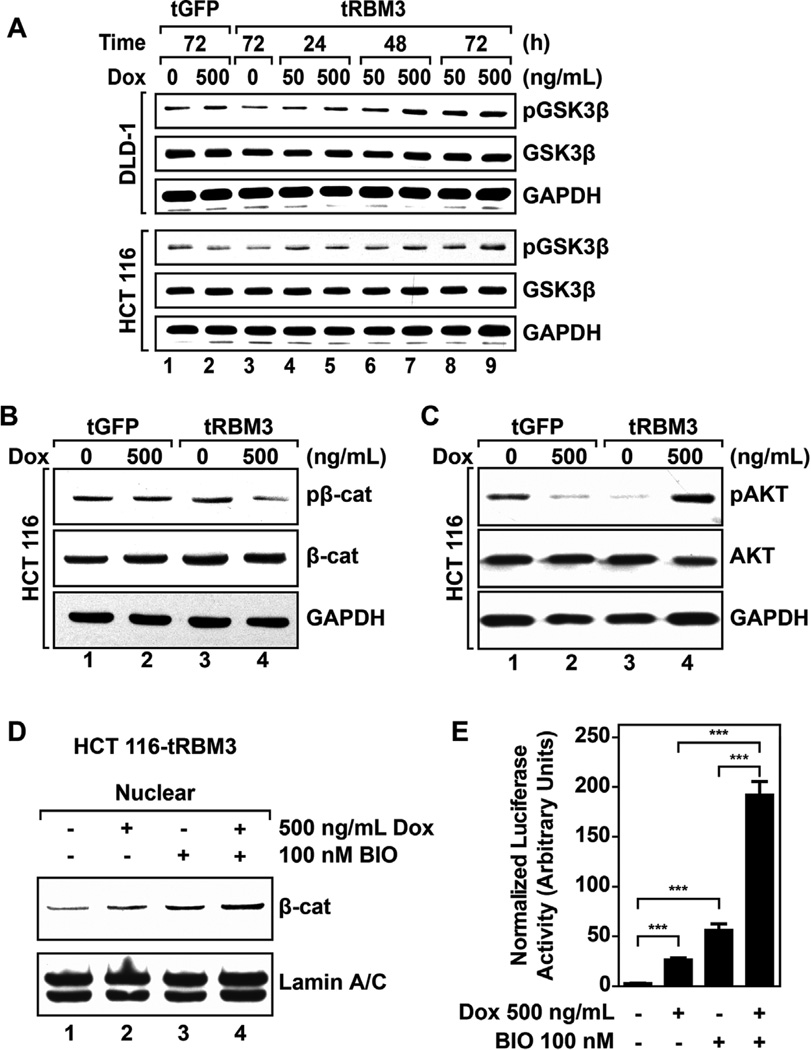
GSK3β Activity Is Decreased Following RBM3 Overexpression
Marked increases in nuclear β-catenin levels and activity in HCT 116 RBM3 cells suggests that degradation of the protein might be affected. Under normal conditions,β-catenin targeted by the APC/Axin/GSK-3β degradation complex in the cytoplasm to degradation by the ubiquitination-proteasomal degradation pathway. We chose to investigate GSK3β as a target for RBM3 mediated β-catenin activity because previous reports show that GSK3β is inactivated during hypoxia, and RBM3 expression is also induced [43,44]. Western blot analyses showed that there is an increase in the phosphorylation of the protein at serine position 9 in both HCT 116 and DLD-1 cells (Figure 8A). Phosphorylation at serine 9 (Ser9) inhibits GSK-3β activity [45]. As expected, GFP vector control showed no appreciable difference in GSK3β phosphorylation in either cell line (Figure 8A). We next validated this increase in GSK3β phosphorylation by determining β-catenin phosphorylation at serines 33 and 37, and threonine at position 41 because phosphorylated GSK-3β cannot phosphorylate β-catenin in these residues. There was reducedβ-catenin phosphorylation in HCT 116 cells (Figure 8B). These data suggest that GSK3β is inactivated. GSK3β phosphorylation can occur through either induction of Wnt signaling or through the action of activated AKT kinase [18]. In previous studies, we have demonstrated that RBM3 induces the production of Interleukin 8 and Prostaglandin E2, both of which activate AKT by phosphorylation [7,46]. Accordingly, we determined whether AKT activity may explain attenuated GSK3β activity. RBM3 overexpression showed significant increases in AKT phosphorylation at serine 473, whereas no appreciable increase was observed in vector controls or uninduced RBM3 cells (Figure 8C). To confirm that the increasedβ-catenin activity is a result of inactivating GSK3β, we used a specific inhibitor, BIO, a compound previously shown to upregulate β-catenin signaling and increase the expression of ABC proteins [47,48]. HCT 116 cells treated with BIO showed increase in both total and nuclear β-catenin levels similar to that of RBM3 overexpression(Figures 8D, S4A, S4B). Additionally, BIO treatment induced c-myc expression in both GFP and RBM3 overexpressing cells similar to RBM3 overexpression (Figure S4C). Moreover, the combination of RBM3 overexpression and BIO treatment generated an even more robust increase inβ-catenin signaling as measured the levels of nuclear β-catenin, and increased c-Myc protein expression (Figures 8D, S4B,C). Furthermore, there was significantly higher TOPFlash luciferase activity following treatment with both RBM3 and BIO (Figure 8E). These data imply that RBM3 overexpression increases β-catenin activity in part by inhibiting GSK3β through the actions of activated AKT kinase.
Figure 8.
Cells overexpressing RBM3 show attenuated GSK3β activity. (A) Western blots for Ser9 phosphorylation in GSK3β in DLD-1 and HCT 116 GFP or RBM3 inducible cells treated with Dox for 72 h Data shows higher levels of Ser9 phosphorylation in RBM3 overexpressing cells in both cell lines. (B) Western blots for β-catenin at Ser33/37 and Thr41 in HCT 116 GFP or RBM3 inducible cells following 72 h treatment with indicated concentration of Dox followed by 6 h of treatment with 5 µM MG132. Data shows reduced phosphorylation of β-catenin following RBM3 induction. (C) Western blots for phosphorylation of Akt at Ser473 in HCT116 GFP or RBM3 inducible cells following 72 h treatment with indicated Dox. Data shows increased phosphorylation of Akt in RBM3 overexpressing cells, while a decreased phosphorylation is observed in GFP overexpressing cells. (D) Western blot of nuclear extracts of HCT 116 RBM3 inducible cells following treatment with indicated concentrations of Dox and BIO for 72 h. Data shows increased β-catenin levels in the nucleus of following RBM3 induction, which was further increased in the presence of BIO. (E) TOPFlash luciferase assay of DLD-1 and HCT 116 RBM3 inducible cells. Cells were treated with indicated concentrations of Dox or BIO for 48 h following transfection with TOPFlash and CMV renilla luciferase plasmid. Bar graphs represent mean values of TOPFlash firefly luciferase luminescence normalized to CMV renilla luciferase luminescence. Data shows increased TOPFlash luciferase activity in the presence of RBM3 and BIO. Bar graphs show SEM. P values obtained from Student's t-test.
DISCUSSION
Previously, our studies suggested that RBM3 might be working as a protooncogene because it was able to transform NIH3T3 cells [7]. However, its role in tumor progression is yet to be carefully studied. Previously, we had discerned in a small sample of tissues that there might be a correlation in the level of RBM3 expression and tumor stage, suggesting that the protein may play a role in progression of the disease. Indeed previous studies have demonstrated that stem cells have the capacity to contribute to tumor progression [49]. Interestingly, much of these have been with mesenchymal stem cells or stromal cells, which have been shown to suppress immune responses directly by producing immunomodulatory molecules [50]. Mesenchymal stem cells have also been shown to modulate epithelial-to-mesenchymal transition (EMT) followed by induction of tumor metastasis and cancer-initiating stem cells, as well as angiogenesis followed by promotion of tumor growth [51,52]. In this case, RBM3 is inducing stemness in the cells, suggesting that this can circumvent the need for this aspect of mesenchymal stem cells. Further studies are required to determine whether RBM3 overexpression affects EMT, angiogenesis or tumor metastatic phenotype.
RBM3 has been previously demonstrated to confer resistance to classical chemotherapeutic agents cisplatin and doxorubicin [11]. However, no clear mechanism was demonstrated regarding this chemoresistance. Our current study further confirms RBM3-induced chemoresistance to these agents and to paclitaxel. Our study further reveals a potential mechanism by which RBM3 is capable of increasing chemoresistance by inducing a side population of cells with high xenobiotic efflux capacity. However, what is also interesting is that RBM3 might be conferring chemoresistance to the cells by multiple mechanisms, one of which would be through the induction of ATP-binding cassette (ABC) transporters. Both, doxorubicin and paclitaxel are both substrates of the ABC transporters [53], and therefore cells overexpressing RBM3 might be able to acquire the resistance to these compounds through the expression of these transporters. However, the mechanism for resistance to cisplatin in the RBM3-overexpressing cells is currently unknown, but cisplatin is not a substrate for the ABC transporters. Understanding the resistance to cisplatin may be complex and may involve multiple pathways. One such mechanism involves the upregulation of enzymes such as γ-glutamylcysteine synthetase or glutathione S-transferase that catalyze the production of GSH, or conjugates the compound to GSH, respectively [54]. While we do not know whether any of the proteins are upregulated in the cells, this and expression of the drug resistance genes such as MRP2 would be an interesting new direction to understand RBM3 function.
Studies have demonstrated a link between hypoxia and its effects on GSK3β [43]. Our study further demonstrates that increased RBM3 expression affects GSK3β activity thereby enhancing β-catenin signaling. Additionally, our report suggests that there must be a secondary mechanism by which RBM3 activates β-catenin signaling as a concurrent overexpression of RBM3 as well as inhibition of GSK3β through use of BIO was capable of increasing nuclear β-catenin and TOP-Flash activity beyond that of either one alone. Additionally, we show that RBM3 overexpression is capable of increasing β-catenin signaling irrespective of their APC and β-catenin status. There was increased signaling in cell lines with intact APC as well as those with mutant APC that show the inability to generate phospho-β-catenin species through GSK3β activity or to induce its degradation [24]. These findings reveal that while RBM3 overexpression results in phosphorylation of GSK3β, thereby attenuating β-catenin degradation, this is only a partial mechanism and there is an as yet unknown role that RBM3 plays in modulatingβ-catenin signaling. Although previous studies have reported that hypoxia is capable of effecting the stem cell population in various cancers, these studies focused on the hypoxia inducible factor 1α (HIF1α) as the primary mechanism of modulating stem cell signaling pathways such as WNT/β-catenin [55–57]. As RBM3 is also regulated by hypoxia in a HIF1α independent mechanism this provides a novel target to further examine as a potential mediator of hypoxia induced stem cell signaling.
β-catenin signaling pathway is dysregulated in the majority of colorectal cancers and is a critical factor governing colorectal cancer aggressiveness and cancer stem cells [17]. Since RBM3 levels are increased in many solid tumors with more aggressive tumors showing higher expression, it is important to understand the effects that RBM3 confers upon the cells and further understand how this affects colorectal cancers [7,9,10]. The studies reported here reveal a novel mechanism by which the β-catenin signaling pathway can be regulated through alterations in expression of RBM3. Further studies are required to determine whether the effect of RBM3 overexpression affects tumor aggressiveness in vivo through changes in stem cells, a focus of our future publications.
Supplementary Material
Acknowledgments
This work was funded by the grants DK094532 (Venugopal A.) and CA135559, CA182872, and CA190291 (Anant S.) from the National Institutes of Health. Support for the studies were also obtained through a pilot project grant supported by National Cancer Institute Cancer Support Grant P30 CA168524. S. Anant is an Eminent Scholar of Kansas Bioscience Authority. The authors would also like to thank members of Dr. Andrew Godwin’s group for technical assistance with the side population experiments and from the Anant and Umar labs for their scientific input regarding this study.
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found in the online version of this article at the publisher’s web-site.
REFERENCES
- 1.Derry JM, Kerns JA, Francke U. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum Mol Genet. 1995;4:2307–2311. doi: 10.1093/hmg/4.12.2307. [DOI] [PubMed] [Google Scholar]
- 2.Danno S, Nishiyama H, Higashitsuji H, et al. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236:804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- 3.Kita H, Carmichael J, Swartz J, et al. Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Hum Mol Genet. 2002;11:2279–2287. doi: 10.1093/hmg/11.19.2279. [DOI] [PubMed] [Google Scholar]
- 4.Ferry AL, Vanderklish PW, Dupont-Versteegden EE. Enhanced survival of skeletal muscle myoblasts in response to overexpression of cold shock protein RBM3. Am J Physiol Cell Physiol. 2011;301:C392–C402. doi: 10.1152/ajpcell.00098.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chip S, Zelmer A, Ogunshola OO, et al. The RNA-binding protein RBM3 is involved in hypothermia induced neuroprotection. Neurobiol Dis. 2011;43:388–396. doi: 10.1016/j.nbd.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 6.Wellmann S, Truss M, Bruder E, et al. The RNA-binding protein RBM3 is required for cell proliferation and protects against serum deprivation-induced cell death. Pediatr Res. 2009;67:35–41. doi: 10.1203/PDR.0b013e3181c13326. [DOI] [PubMed] [Google Scholar]
- 7.Sureban SM, Ramalingam S, Natarajan G, et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27:4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Grupp K, Wilking J, Prien K, et al. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur J Cancer. 2014;50:852–861. doi: 10.1016/j.ejca.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Shaikhibrahim Z, Lindstrot A, Ochsenfahrt J, Fuchs K, Wernert N. Epigenetics-related genes in prostate cancer: Expression profile in prostate cancer tissues, androgen-sensitive and -insensitive cell lines. Int J Mol Med. 2013;31:21–25. doi: 10.3892/ijmm.2012.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang HT, Zhang ZW, Xue JH, et al. Differential expression of the RNA-binding motif protein 3 in human astrocytoma. Chin Med J (Engl) 2013;126:1948–1952. [PubMed] [Google Scholar]
- 11.Zeng Y, Kulkarni P, Inoue T, Getzenberg RH. Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. J Cell Biochem. 2009;107:179–188. doi: 10.1002/jcb.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jogi A, Brennan DJ, Ryden L, et al. Nuclear expression of the RNA-binding protein RBM3 is associated with an improved clinical outcome in breast cancer. Mod Pathol. 2009;22:1564–1574. doi: 10.1038/modpathol.2009.124. [DOI] [PubMed] [Google Scholar]
- 13.Jonsson L, Bergman J, Nodin B, et al. Low RBM3 protein expression correlates with tumour progression and poor prognosis in malignant melanoma: An analysis of 215 cases from the malmo diet and cancer study. J Transl Med. 2011;9:114–122. doi: 10.1186/1479-5876-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ehlen A, Brennan DJ, Nodin B, et al. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J Transl Med. 2010;8:78–89. doi: 10.1186/1479-5876-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Cancer Society. Cancer Facts & Figures 2013. American Cancer Society: Atlanta, GA; 2013. [Google Scholar]
- 16.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61:759–767. doi: 10.1016/0092-8674(90)90186-i. [DOI] [PubMed] [Google Scholar]
- 17.Fodde R, Brabletz T. Wnt/beta-catenin signaling in cancer stemness and malignant behavior. Curr Opin Cell Biol. 2007;19:150–158. doi: 10.1016/j.ceb.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 18.Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat Rev Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- 19.Welsh GI, Proud CG. Glycogen synthase kinase-3 is rapidly inactivated in response to insulin and phosphorylates eukaryotic initiation factor eIF-2B. Biochem J. 1993;294:625–629. doi: 10.1042/bj2940625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brabletz S, Schmalhofer O, Brabletz T. Gastrointestinal stem cells in development and cancer. J Pathol. 2009;217:307–317. doi: 10.1002/path.2475. [DOI] [PubMed] [Google Scholar]
- 21.Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: Accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- 22.Okano H, Kawahara H, Toriya M, Nakao K, Shibata S, Imai T. Function of RNA-binding protein Musashi-1 in stem cells. Exp Cell Res. 2005;306:349–356. doi: 10.1016/j.yexcr.2005.02.021. [DOI] [PubMed] [Google Scholar]
- 23.King CE, Wang L, Winograd R, et al. LIN28B fosters colon cancer migration, invasion and transformation through let-7-dependent and -independent mechanisms. Oncogene. 2011;30:4185–4193. doi: 10.1038/onc.2011.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang J, Zhang W, Evans PM, Chen X, He X, Liu C. Adenomatous polyposis coli (APC) differentially regulates beta-catenin phosphorylation and ubiquitination in colon cancer cells. J Biol Chem. 2006;281:17751–17757. doi: 10.1074/jbc.M600831200. [DOI] [PubMed] [Google Scholar]
- 25.Mathew G, Timm EA, Jr, Sotomayor P, et al. ABCG2-mediated DyeCycle Violet efflux defined side population in benign and malignant prostate. Cell Cycle. 2009;8:1053–1061. doi: 10.4161/cc.8.7.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramaniam D, May R, Sureban SM, et al. Diphenyl difluoroketone: A curcumin derivative with potent in vivo anticancer activity. Cancer Res. 2008;68:1962–1969. doi: 10.1158/0008-5472.CAN-07-6011. [DOI] [PubMed] [Google Scholar]
- 27.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirschmann-Jax C, Foster AE, Wulf GG, et al. A distinct ”side population” of cells with high drug efflux capacity in human tumor cells. Proc Natl Acad Sci U S A. 2004;101:14228–14233. doi: 10.1073/pnas.0400067101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- 30.Kanwar SS, Yu Y, Nautiyal J, Patel BB, Majumdar AP. The Wnt/beta-catenin pathway regulates growth and maintenance of colonospheres. Mol Cancer. 2010;9:212–224. doi: 10.1186/1476-4598-9-212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ricci-Vitiani L, Lombardi DG, Pilozzi E, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007;445:111–115. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 32.Dontu G, Abdallah WM, Foley JM, et al. In vitro propagation and transcriptional profiling of human mammary stem/ progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Barker N, van Es JH, Kuipers J, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 34.May R, Riehl TE, Hunt C, Sureban SM, Anant S, Houchen CW. Identification of a novel putative gastrointestinal stem cell and adenoma stem cell marker, doublecortin and CaM kinase-like-1, following radiation injury and in adenomatous polyposis coli/multiple intestinal neoplasia mice. Stem Cells. 2008;26:630–637. doi: 10.1634/stemcells.2007-0621. [DOI] [PubMed] [Google Scholar]
- 35.Barker N, Ridgway RA, van Es JH, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 36.Nakanishi Y, Seno H, Fukuoka A, et al. Dclk1 distinguishes between tumor and normal stem cells in the intestine. Nat Genet. 2013;45:98–103. doi: 10.1038/ng.2481. [DOI] [PubMed] [Google Scholar]
- 37.Mani SA, Guo W, Liao MJ, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zeng Y, Wodzenski D, Gao D, et al. Stress-response protein RBM3 Attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res. 2013;73:4123–4133. doi: 10.1158/0008-5472.CAN-12-1343. [DOI] [PubMed] [Google Scholar]
- 39.Klaus A, Birchmeier W. Wnt signalling and its impact on development and cancer. Nat Rev Cancer. 2008;8:387–398. doi: 10.1038/nrc2389. [DOI] [PubMed] [Google Scholar]
- 40.Wielenga VJ, Smits R, Korinek V, et al. Expression of CD44 in Apc and Tcf mutant mice implies regulation by the WNT pathway. Am J Pathol. 1999;154:515–523. doi: 10.1016/S0002-9440(10)65297-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chikazawa N, Tanaka H, Tasaka T, et al. Inhibition of Wnt signaling pathway decreases chemotherapy-resistant side-population colon cancer cells. Anticancer Res. 2010;30:2041–2048. [PubMed] [Google Scholar]
- 42.He TC, Sparks AB, Rago C, et al. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 43.Mottet D, Dumont V, Deccache Y, et al. Regulation of hypoxia-inducible factor-1 alpha protein level during hypoxic conditions by the phosphatidylinositol 3-kinase/Akt/glycogen synthase kinase 3beta pathway in HepG2 cells. J Biol Chem. 2003;278:31277–31285. doi: 10.1074/jbc.M300763200. [DOI] [PubMed] [Google Scholar]
- 44.Wellmann S, Buhrer C, Moderegger E, et al. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 45.van Weeren PC, de Bruyn KM, de Vries-Smits AM, van Lint J, Burgering BM. Essential role for protein kinase B (PKB) in insulin-induced glycogen synthase kinase 3 inactivation. Characterization of dominant-negative mutant of PKB. J Biol Chem. 1998;273:13150–13156. doi: 10.1074/jbc.273.21.13150. [DOI] [PubMed] [Google Scholar]
- 46.Knall C, Young S, Nick JA, Buhl AM, Worthen GS, Johnson GL. Interleukin-8 regulation of the Ras/Raf/mitogen-activated protein kinase pathway in human neutrophils. J Biol Chem. 1996;271:2832–2838. doi: 10.1074/jbc.271.5.2832. [DOI] [PubMed] [Google Scholar]
- 47.Meijer L, Skaltsounis AL, Magiatis P, et al. GSK-3-selective inhibitors derived from Tyrian purple indirubins. Chem Biol. 2003;10:1255–1266. doi: 10.1016/j.chembiol.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 48.Liu KP, Luo F, Xie SM, et al. Glycogen Synthase Kinase 3beta Inhibitor (2'Z,3'E)-6-Bromo-indirubin-3'-Oxime Enhances Drug Resistance to 5-Fluorouracil Chemotherapy in Colon Cancer Cells. Chin J Cancer Res. 2012;24:116–123. doi: 10.1007/s11670-012-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Behnan J, Isakson P, Joel M, et al. Recruited brain tumor-derived mesenchymal stem cells contribute to brain tumor progression. Stem Cells. 2014;32:1110–1123. doi: 10.1002/stem.1614. [DOI] [PubMed] [Google Scholar]
- 50.Kudo-Saito C. Cancer-associated mesenchymal stem cells aggravate tumor progression. Front Cell Dev Biol. 2015;3:23–28. doi: 10.3389/fcell.2015.00023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Uccelli A, Moretta L, Pistoia V. Mesenchymal stem cells in health and disease. Nat Rev Immunol. 2008;8:726–736. doi: 10.1038/nri2395. [DOI] [PubMed] [Google Scholar]
- 52.Yang X, Hou J, Han Z, et al. One cell, multiple roles: Contribution of mesenchymal stem cells to tumor development in tumor microenvironment. Cell Biosci. 2013;3:5–14. doi: 10.1186/2045-3701-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sikic BI, Fisher GA, Lum BL, Halsey J, Beketic-Oreskovic L, Chen G. Modulation and prevention of multidrug resistance by inhibitors of P-glycoprotein. Cancer Chemother Pharmacol. 1997;40:S13–S19. doi: 10.1007/s002800051055. [DOI] [PubMed] [Google Scholar]
- 54.Galluzzi L, Vitale I, Michels J, et al. Systems biology of cisplatin resistance: Past, present and future. Cell Death Dis. 2014;5:e1257–e1274. doi: 10.1038/cddis.2013.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yoshida T, Hashimura M, Mastumoto T, et al. Transcriptional upregulation of HIF-1 alpha by NF-kappaB/p65 and its associations with beta-catenin/p300 complexes in endometrial carcinoma cells. Lab Invest. 2013;93:1184–1193. doi: 10.1038/labinvest.2013.111. [DOI] [PubMed] [Google Scholar]
- 56.Mazumdar J, O'Brien WT, Johnson RS, et al. O2 regulates stem cells through Wnt/beta-catenin signalling. Nat Cell Biol. 2010;12:1007–1013. doi: 10.1038/ncb2102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kaidi A, Williams AC, Paraskeva C. Interaction between betacatenin and HIF-1 promotes cellular adaptation to hypoxia. Nat Cell Biol. 2007;9:210–217. doi: 10.1038/ncb1534. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.