Abstract
A better understanding of HIV-1 transmission is critical for developing preventative strategies. To that end, we analyzed 524 full-length env sequences of SIVmac251 at 6 and 10 days post intrarectal infection of rhesus macaques. There was no tissue compartmentalization of founder viruses across plasma, rectal and distal lymphatic tissues for most animals; however one animal has evidence of virus tissue compartmentalization. Despite identical viral inoculums, founder viruses were animal-specific, primarily derived from rare variants in the inoculum, and have a founder virus signature that can distinguish dominant founder variants from minor founder or untransmitted variants in the inoculum. Importantly, the sequences of post-transmission defective viruses were phylogenetically associated with competent viral variants in the inoculum and were mainly converted from competent viral variants by frameshift rather than APOBEC mediated mutations, suggesting the converting the transmitted viruses into defective viruses through frameshift mutation is an important component of rectal transmission bottleneck.
Introduction
During human immunodeficiency virus type 1 (HIV-1) mucosal transmission, a complex quasispecies of viruses is restricted through a transmission bottleneck into a single or limited number of founder viruses that establish a systemic infection [1-4]. This phenomenon has been observed in vaginal [5], rectal [6,7], and penile transmission [8].
To establish a systemic infection through mucosal transmission, HIV-1 has to overcome both mucosal physical and immunological barriers [9,10], such as innate and intrinsic immunity [11,12], as well as the availability and susceptibility of target cells within the lamina propria mucosa [13-15]. Moreover, it has been well documented that both viral and host factors can impact the transmission bottleneck [16]. For example, HIV-1 using the CCR5 (R5) coreceptor is preferentially transmitted over virus using the CXCR4 (X4) coreceptor [14,17-21]. Previous reports have also shown that HIV-1 that is resistant to type I interferon has an increased chance to become a founder virus [13,22]. Consistent with the in situ observation that CD4+ T cells rather than macrophages are the major infected cell type in early mucosal transmission [23,24], infectious molecular clones reconstructed from founder HIV-1 infect CD4+ T cells more efficiently than monocytes and macrophages [14,20]. Viral load and virus composition of the transmission donor also impacts mucosal transmission [25]. It has also been demonstrated that infrequent or rare HIV-1 variants, rather than high-abundant variants from the donors, can become the founder viruses in recipients [3,26]. A recent study compared the amino acid sequences of HIV-1 gag genes in heterosexual linked transmission pairs and found that founder virus has a selection bias [27], and sequence analyses of HIV-1 founder viruses in men who have sex with men and in heterosexual transmission have also revealed a selection bottleneck [28,29]. However, in human studies it is difficult to unambiguously define the composition of the HIV-1 population present on mucosal surfaces of the transmission recipient, to sample and analyze founder viruses at or near the time of transmission, or to compare the founder viruses in different recipients who have been exposed to an identical virus population to determine if founder viruses in one individual also have an advantage in other individuals.
While the transmission bottleneck is well documented, the underlying mechanisms remain controversial or largely unknown. For example, are founder viruses in the mucosa, at the draining or distal lymphatic tissues identical or different? Are founder viruses preferentially derived from rare or high- abundant variants from the inoculum? Do founder viruses in one individual also have an advantage in other individuals? Through what mechanisms is viral diversity constrained by the host in early rectal transmission?
To answer these questions and to overcome the previously mentioned limitations, we studied very early virological events, prior to the emergence of adaptive immunity, in the SIV-rhesus macaque model. SIV-rhesus macaques are the best available model to study early events of rectal transmission where the composition and dosage of viral variants in the inoculum and the time of infection can be experimentally controlled. We simultaneously analyzed the sequences of SIV in rectum, axillary lymph nodes, and plasma from each animal in the context of a well-defined SIVmac251 inoculum at 6 and 10 days post intra-rectal inoculation (dpi). We found that founder viruses are preferentially derived from low abundant variants in inoculum. During very early rectal transmission of SIV, there is no tissue compartmentalization of founder viruses for the most animals; however one animal has evidence of virus tissue compartmentalization. Strikingly, the sequences of post-transmission defective viruses were phylogenetically associated with competent viral variants in the inoculum and were mainly converted from competent viral variants by frameshift mutations rather than APOBEC derived mutations.
Results
The genetic make-up of the inoculum provides an adequate diversity of virus variants with varying abundance
To study the early virological events of SIV rectal transmission, it is imperative that the virus inoculum contains diverse virus variants with varying abundance. To confirm this, full-length env sequences of SIVmac251 from the inoculum were analyzed. The env nucleotide (nt) sequences were further analyzed at the amino acid (AA) level in order to distinguish synonymous and non-synonymous mutations. A functional Env should contain only one stop codon, and Env AA sequences containing more than one stop codon were considered as defective variants. Of the 68 full-length env sequences (Fig.1A and 1B) in the inoculum, there are 59 intact AA sequences with only one stop codon (Fig. 1C), which have quite different abundances. 26 AA sequences clustered with more than 99.5% identity, whereas others dispersed as rare variants (Fig. 1C). Thus, the inoculum presented broad viral diversity and an obvious stratum of variant abundance.
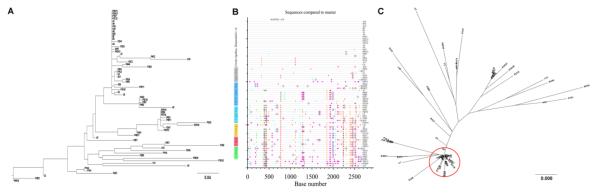
Fig 1. The genetic make-up of SIVmac251 inoculum.
Maximum likelihood tree (A) and highlighter plots (B) of full-length env nucleotide sequences after SGA and Sanger sequencing. Nucleotide polymorphisms in the highlighter plots are indicated by a colored mark. Thymine is represented in red, guanine in orange, adenine in green, cytosine in blue, pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown by gray in the highlighter plots. (C) Maximum likelihood tree of full-length Env AA sequences. The red circle indicates the dominant virus variant group composed of 26 AA sequences with 99.5% identity; other variants were dispersed as rare variants. Bar length indicates 0.008 AA substitutions per site.
There is no tissue compartmentalization of founder viruses in most of studied animals
To gauge the relative contribution of the rectal mucosa and the secondary lymphatic tissues (LTs) to the transmission bottleneck and to determine whether there is tissue compartmentalization for founder viruses during very early rectal transmission, nt and AA sequences of virus variants from rectal, axillary lymphoid tissue, and plasma were analyzed. As shown in Fig S1 to S3, 262 full-length env nt sequences were obtained from three macaques at 6 dpi (Rh4809, Rh5052 and Rh5053). And 194 nt sequences were obtained from three macaques at 10 dpi (Rh4976, Rh4978 and Rh4979). Their phylogenetic tree based on nt sequences were shown in Fig 2 and S4 to S5. The env nt sequences were further analyzed at AA level in order to distinguish the synonymous and non-synonymous mutations and identify Env AA signatures of founder viruses.
Fig 2. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 10 dpi macaque Rh4976.
The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Based on the reverse transcription error rate of 1 mutation per 104 nucleotides and the 1.2 day duration of the HIV-1 life cycle in vivo, the founder virus variants post transmission would have limited (<0.5%) AA changes within such a short period of time [30]. Thus the AA sequences within 99.5% identity were clustered as a single group, since variants in the same cluster may derive from the same variant in inoculum or a certain variant may derive from any variants within the same cluster in inoculum. We also analyzed our data with 99.9% identify as the cutoff for the clustering; however that does not change our conclusion.
To distinguish dominant and minor founder variant clusters, the founder virus variants in each animal were clustered based on 99.5% AA identity. The clusters were normalized as described in the experimental procedures and normalized clusters were pooled. As shown in Fig S6, the abundance of founder virus variant clusters clearly separated the dominant and minor founder variant clusters.
The variants of dominant founder virus groups highlighted by red circles were dispersed in all three tissue compartments (Fig. 3 and Fig. S7 to S11), although few founder virus variants were detected only in a single tissue site. As shown in Table 2, statistical analyses using three independent methods all indicate there is no virus tissue compartmentalization for most of studied animals except an animal of 6 dpi, suggesting overall there is no virus tissue compartmentalization at very early infection. As expected, only a small fraction of inoculum virus variants was detected in the rectal tissues, lymph node tissues, and plasma in each macaque, supporting the notion of presence of a transmission bottleneck in SIV rectal infection.
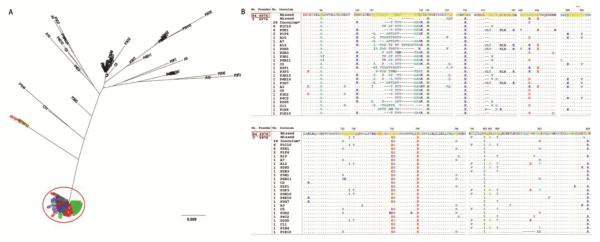
Fig 3. The relationship between founder and inoculum variants as well as the founder virus signature (FVS) in 10 dpi macaque Rh4976.
(A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red circle indicates the dominant variant group of founder viruses with greater than 99.5% AA identity. Bar length indicates 0.009 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Table 2.
The distribution of functional and defective variants and the causes of defects
| Animal ID | Total Sequences |
Functional Variants (%) |
Defective Variants (%) |
Defective Caused by Frameshift (%) |
Defective Caused by APOBEC (%) |
|
|---|---|---|---|---|---|---|
| 4809 | 74 | 75.68 | 24.32 | 22.97 | 1.35 | |
| 6 dpi | 5052 | 118 | 69.49 | 30.51 | 29.66 | 0.85 |
| 5053 | 70 | 50.00 | 50.00 | 48.57 | 1.43 | |
| 4976 | 105 | 82.86 | 17.14 | 14.29 | 2.86 | |
| 10 dpi | 4978 | 53 | 94.34 | 5.66 | 0.00 | 5.66 |
| 4979 | 36 | 94.44 | 5.56 | 0.00 | 5.56 |
Founder viruses are mainly derived from rare variants in the inoculum
We next sought to understand the mechanism underlying the transmission bottleneck. With a typical sampling depth, SGA cannot capture the entire sequence space present in a population of viruses. Therefore, there were rare variants in the inoculum that were only detected once or missed by SGA, but later they became founder variants. It is intractable to estimate their abundance within the inoculum. In contrast, high-abundant variants were repeatedly detected and their abundance can be estimated based on their frequency to the total variants number. Based on whether variants can be repeatedly detected and whether their frequency can be estimated, variants in the inoculum are divided into abundant and rare clusters. In the inoculum, 4 abundant clusters were detected 26, 6, 4, and 2 times respectively, whereas other rare variants were only detected once or were not detected (Fig. 1C and 3B). The abundant variants take up more than 64.4% of all 59 functional AA sequences, whereas rare variants take up less than 35.6% of abundance.
As shown in panel A of Fig 3 and Fig S7 to S11, the red circles on the AA phylogenetic tree for each animal indicate the dominant (solid) and subdominant (dotted) founder variant groups. For macaques at 10 dpi, all of the dominant founder variant groups were phylogenetically colocalized with rare variants in the inoculum. Rh4976 at 10 dpi had only one dominant founder group with 84 AA sequences that were phylogenetically colocalized with a missed variant in the inoculum (Fig. 3). In Rh4978 at 10 dpi, there was only one dominant founder group with 47 AA sequences that were phylogenetically colocalized with a rare variant A10 (Fig. S7). In Rh4979 at 10 dpi, both dominant founder groups were located far from the high-abundant variants in the inoculum (Fig. S8). The 6 dpi macaques displayed mixed picture with dominant founder groups corresponding to both abundant and rare variants in the inoculum. For macaque Rh4809 at 6 dpi, there were two dominant founder groups; the larger group contained 21 AA sequences that corresponded to a missed variant in the inoculum, while the smaller group containing 14 AA sequences corresponded to the most abundant group in the inoculum (Fig. S9). In another 6 dpi animal (Rh5052), a founder variant group containing 62 AA sequences corresponded to the most abundant group in the inoculum, whereas another founder variant group, comprised of 15 AA sequences, corresponded to a missed variant in the inoculum (Fig. S10). In macaque Rh5053 at 6 dpi, the only dominant founder variant group, comprised of 35 AA sequences, corresponded to a highly-abundant cluster in the inoculum (Fig. S11).
For all six macaques, nine dominant founder virus groups were identified. Six dominant founder virus groups came from rare variants in the inoculum (66.6% derived from 35.6%), whereas only three dominant founder viruses came from the high-abundant group in the inoculum (33.3% derived from 64.4%). Our statistical analyses show that the abundance of founder viruses for each animal is highly independent of their ancestor variant proportion in the inoculum (p=3.3E-16, Supplemental statistical analysis). Our analyses at the nt level were consistent with the AA analysis results (Fig. 2 and S1-S5). Taken together, our data suggest that founder viruses are preferentially derived from rare variants.
Founder viruses are animal-specific with founder virus signature (FVS)
To test whether founder viruses are shared among different macaques inoculated with the same viral inoculum, we evaluated the sequences of founder viruses from all macaques. The locations and composition of founder virus sequences among macaques were not phylogenetically shared (Fig. S12). There are significant differences in the composition of founder variants among animals at 6 dpi (p=1.4e-8), among animals at 10 dpi (p<2.2e-16), and between the two groups of macaques at 6 and 10 dpi (p<2.2e-16, Supplemental statistical analysis). Together, these results indicate that the transmission bottleneck is animal-specific and that founder viruses in one host do not have a predetermined advantage in another host.
To examine whether there is a FVS that can distinguish the dominant founder variant group from the minor transmitted variants or untransmitted variants in the inoculum, the consensus AA sequence of the dominant founder virus group was compared with sequences of the minor transmitted variants and untransmitted variants. Panel B of Fig 3 and Fig S7 to S11 show the variable region MUSCLE alignments of the subdominant, minor transmitted variants, and untransmitted functional variants, with the consensus AA sequence of the dominant founder group for each animal. The dominant founder variant group of each animal has an animal-specific FVS that comprises a combination of several unique AA sequences, which are highlighted in yellow. We identified the FVS that can distinguish dominant founder variants from minor founder variants and untransmitted variants in the inoculum for each animal (Fig. S6). To better understand the animal-specific FVS, we mapped and annotated FVS positions on SIV GP120 and GP41 in a 3D structure based on published SIV and HIV-1 homolog AA sequences [31]. As shown in Fig 4, although sequences of FVS are animal-specific, the 3D positions of most FVS are shared among the six monkeys (Fig. 4B).
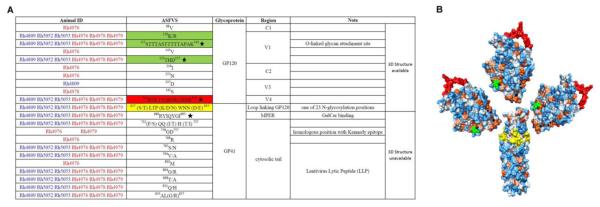
Fig 4. Summary of the founder virus signature (FVS) from all animals.
(A) The feature of FVS and their corresponding animals are listed, of which some are shared among animals in amino acid spatial position but are unique in sequence. A star indicates a particular sequence chosen from a group of sequence with similar position. A blue animal ID indicates 6 dpi and red indicates 10 dpi. The highlighted FVS are also depicted using the same colors on a 3D structure in panel B. (B) Localization of FVS on available 3D structures of SIV Env. The clipped V1V2 region is highlighted in green (truncated for easier crystallization), the V4 region is highlighted in red, and the FVS region on GP41 is highlighted in yellow.
Sequences of post-transmission defective virus variants were mainly caused by frameshift rather than APOBEC mediated mutations
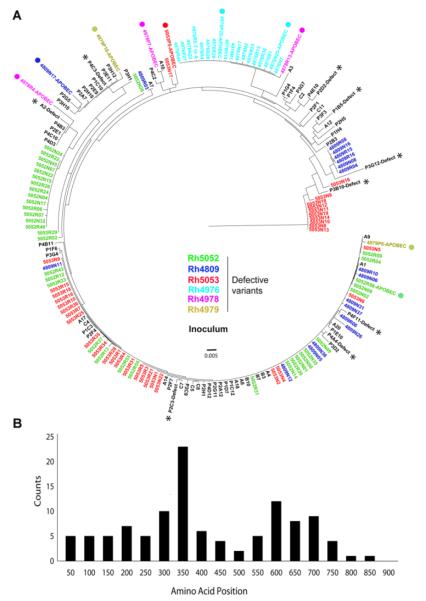
When we analyzed AA sequences, as shown in Table 2, we found that many variants contained more than one stop codon, indicating defective variants. Strikingly, these post-transmission defective variants were phylogenetically colocalized with functional variants in the inoculum (Fig. 5A), suggesting those defective variants were converted from functional variants. Surprisingly, we found that defective variants had mainly been converted by frameshift mutations, but not APOBEC mediated mutations (Fig. 5A & Table 2). We then mapped the position of the first introduced stop codon and found most defective variants were truncated at the beginning or middle of Env, indicating a dysfunctional Env. The peak location of the first introduced stop codon is located near the V3 region and only few are located within the cytoplasmic tail of gp41, which may still be functional (Fig. 5B).
Fig 5. Defective post-transmission variants were primarily caused by frameshift.
(A) The phylogenetic relationship of defective post-transmission variants with all the variants in the inoculum. Defective post-transmission variants are color coded by animal ID and are phylogenetically associated with inoculum functional rather than defective variants. The majority of post-transmission defective variants are caused by frameshift. The APOBEC mediated mutations are labeled with solid circles. All variants in the inoculum are shown in black with defective variants labeled with stars. (B) The distribution of the first introduced stop codon in post-transmission defective variants. The peak of the first stop codon location is near the V3 region and very few are located within the cytoplasmic tail of GP41.
As shown in Table 2, the ratio of defective to functional founder variants significantly decreased from 6 dpi (34.9±13.4%) to 10 dpi (9.5±6.6%) (p=1.1e-7, S13_Statistical analysis). Frameshift mutations were detected in the majority of defective variants at 6 dpi, but were significantly decreased at 10 dpi (p=6.0e-10, S13_Statistical analysis) corresponding with the overall decrease of defective variants. Further, we observed an increase in the frequency of APOBEC derived defective variants from 6 dpi to 10 dpi; however, they are not significantly different (p=0.15, Pearson’s chi-squared test, S13_Statistical analysis) due to a limited number of APOBEC derived defective variants at both 6 and 10 dpi. These results indicate that transmitted functional variants were converted into defective variants mainly through frameshift rather than APOBEC mediated mutations during very early rectal transmission.
Discussion
Transmission bottlenecks play a pivotal role in HIV-1 mucosal transmission. A better understanding the nature and mechanism of transmission bottleneck may inform HIV-1 vaccine design. To better understand the earliest virological events of HIV-1 rectal transmission, we chose samples from 6 dpi, which was the first time point when systemic viremia could be detected in our cohort, and 10 dpi, which was before emergence of the adaptive immune response [32]. To our knowledge this is the first study of founder viruses at multiple tissue sites during very early rectal transmission.
We found that the founder virus variants were enriched into one or two dominant variant groups, indicating there is a transmission bottleneck as has been reported [6,7]. When analyzed at a tissue-specific level, we did not find evidence of tissue compartmentalization of founder viruses in most of studied animals during early rectal transmission. However, in one animal there is evidence of virus tissue compartmentalization, indicating tissue compartmentalization can happen at very early infection. We do not know the exact underlying mechanism of this outlier animal with tissue compartmentalization at early infection. We speculate that tissue microenvironments may play a critical role for determination of virus local fitness, however further studies are needed to specifically address this question.
An ideal study to test the random versus selection hypotheses of HIV-1 rectal transmission is to look at founder viruses in a large group of genetically identical humans or macaques infected with an identical swarm of viruses. However, such a study is lacking. Therefore, to test these two competing hypotheses, counterexamples are often used, where repeated observations of small probability events are against the purely random hypothesis. We found that all of the dominant founder variant groups in 10 dpi macaques corresponded to rare variants in the inoculum, which provides counterexamples to the purely random hypothesis. The dominant founder variant groups in the 6 dpi animals corresponded either to rare variants or to abundant variants in the inoculum. However, due to our limited samples size, we fell that further study is needed to reach a definitive conclusion.
We then sought to investigate whether macaques inoculated with identical viruses would lead to shared founder variants by analyzing all of the animals together. As shown in Fig S12, each macaque has its own founder variants located at different positions on the phylogenetic tree, demonstrating that the transmission bottleneck is animal-specific. As discussed in the introduction, there are many factors that may impact the HIV-1 transmission. Each rhesus macaque has a distinct genetic background associated with differences in intrinsic and innate immunity, which may be one of the underlying reasons why the dominant founder variant group of each monkey is unique even though each animal has been inoculated with the same virus stock at the same dosage.
To better understand the relationship of post-transmission defective variants, functional variants, and the host anti-viral immunity during very early rectal transmission, we analyzed the defective variants in the inoculum and the post-transmission defective variants in tissues. We did not find any post-transmission defective variants that were identical with defective variants in the inoculum; therefore, post-transmission defective variants in tissues are most likely converted from transmitted functional variants. Furthermore, even the defective variants in the inoculum may be able to cross the mucosal barrier but they are unable to replicate and amply themselves. Strikingly, the post-transmission defective variants were mainly derived from frameshift rather than APOBEC mediated mutations (Fig. 5B & Table 2). For the APOBEC mediated mutation rate (0.85%-5.7%) we observed in the very early rectal transmission in this study, it may just reflect limited virus replication in such early infection. Importantly, the decline of defective variants paralleled the decline of frameshift mutations, suggesting that frameshift mutations may play an important role in rectal transmission through reducing viral diversity or even preventing infection. We did not find post-transmission recombinant variants in our analyses, which may be due to a limited number of founder variants and short duration of infection at this very early stage.
The Env of founder viruses may play a critical role in transmission [16]. Recently, pseudoviruses reconstructed from the Env of founder viruses were found to be enriched for higher Env content [13,33]. Another study identified different HIV-1 Env AA signatures through comparing viruses from acute and chronic infection [34]. We therefore compared the Env sequences of dominant founder variants with untransmitted variants in the inoculum. We found that the dominant founder virus group has a founder virus signature (FVS).
Collectively, our data supports a new model of rectal transmission. After viruses gain entry through the rectal mucosa, the host further reduces viral diversity by converting some of the transmitted functional viruses into defective viruses. These conversions were mainly caused by an unidentified frameshift mechanism. Moreover, founder viruses are primarily deriving from rare variants in the inoculum and have an animal-specific FVS. Future elucidation of the role of the animal-specific FVS and how transmitted functional viruses are converted into defective variants may uncover the underlying mechanism of transmission bottleneck and help to inform the design of new strategies to prevent HIV-1 transmission.
Materials and Methods
Ethics Statement
This study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) at the University of Nebraska-Lincoln (protocol number 559) and BIOQUAL, INC. (Protocol number 10-0000-01). BIOQUAL was accredited by the Association for the Assessment and Accreditation of Laboratory Animal Care (AAALAC file #000624), and holds an assurance on file with the National Institute of Health, Office for Protection of Research Risks as required by the US Public Health Service Policy on Humane Care and Use of Laboratory Animals. BIOQUAL was also certified by the PHS Animal Welfare Assurance (file number A3086-01) and the USDA certification. The rhesus macaque work was conducted at BIOQUAL, Inc. and the care and husbandry of all non-human primates were provided in compliance with federal laws and guidelines as well as in accordance with recommendations provided in the NIH guide and other accepted standards of laboratory animal care and use. The comprehensive veterinary care program includes: disease detection and surveillance, prevention, diagnosis, treatment, and resolution; monitoring and promoting physical and psychological well-being of all animals. Monkeys were housed socially, if possible, and had access to food, water, light at 12- hour cycle and were provided with enrichment toys. Animals were sedated with ketamine for all technical procedures and were fully anesthetized for SIV inoculation. Animals were euthanized by exsanguinations under deep (surgical plane) anesthesia using under directions of the attending veterinarian.
Rhesus macaques, SIVmac251 rectal inoculation, and tissue collection
Male adult rhesus macaques (Macaca mulatta) of Indian origin were housed at Bioqual Inc. All animals enrolled in this study were negative for HIV-2, SIV, type-D retrovirus, and simian T cell lymphotropic virus-1. Animals were intrarectally inoculated with cell-free SIVmac251 at a dose of 3.4 ×104 TCID50 provided by Dr. Nancy Miller, and euthanized at 6 dpi (n=3, Rh4809, Rh5052 and Rh5053) and 10 dpi (n=3, Rh4976, Rh4978 and Rh4979). During animal euthanasia, plasma, anorectal tissues, and axillary lymph node (LN) tissues were collected.
Viral RNA isolation and cDNA synthesis
Total RNA was extracted from homogenized tissues using TRIzol (Invitrogen) and purified with an RNeasy Mini Kit (Qiagen). Viral RNA from plasma or inoculum was isolated using the QIAamp Ultrasens Viral Kit (QIAGEN) according to the manufacturer’s protocol. RNA quality was verified by Nanodrop (Thermo Scientific). The cDNA synthesis from extracted RNA was conducted by following published protocols with minor modifications [5,10]. Briefly, RNA was reversely transcribed into cDNA using SuperscriptIII reagents (Invitrogen) with gene-specific primers.
Single genome amplification (SGA) of SIV full-length env and Sanger sequencing
Full-length SIVmac251 env was amplified from cDNA using nested PCR by following published protocols [5,10]. Briefly, cDNA was serially diluted to obtain less than 30% positivity in the total PCR reactions. PCR amplification was performed using Q5 DNA polymerase (NEB) or High Fidelity Platinum Taq polymerase (Invitrogen) with gene-specific primers. The amplicons were checked by agarose gel. All of the PCR procedures were performed with safeguards against sample contamination, including pre-aliquoting all reagents, use of dedicated equipment, and physical separation of sample processing from pre- and post-PCR amplification steps. Sanger sequencing was performed on amplicons of full-length env from a single genome with eight overlapping primers at Sequetech (Mountain View, CA).
Primers used in this study
RT primer: 5′-TGTAATAAATCCCTTCCAGTCCCCCC-3′. The first round PCR was performed with forward primer SIVsm/macEnvF1 5′-CCT CCC CCT CCA GGA CTA GC-3′ and reverse primer SIVsm/macEnvR1 5′-TGT AAT AAA TCC CTT CCA GTC CCC CC-3′. The second round PCR was conducted using the forward primer SIVEnvF2 5′-TAT AAT AGA CAT GGA GAC ACC CTT GAG GGA GC-3′ and reverse primer SIVEnvR2 5′-ATG AGA CAT RTC TAT TGC CAA TTT GTA-3′. The eight sequencing primers are: MS.1-CTT GGT TTG GCT TTA ATG, MS.3-TAT TTG CCT CCA AGA GAG, MS.5-TGG CCA AAT GCA AGT CTA, MS.A-TTA GAC TTG CAT TTG GCC, MS.C-ACT TTA TGC CAA GTG TTG, MS.E-GCC AAA CCA AGT AGA AGT, MS.F-AGC TTT GAT GCT TGG GAG, and MS.RTTG CTG AAT AGC CAA GTC.
Sequence analysis
To assure that the env sequences were derived from a single genome, chromatograms were manually examined for multiple peaks, indicating the presence of amplicons from multiple variant templates. Thus, the env sequences containing more than one peak or other ambiguous sites were excluded from further analysis. Then the sequences were assembled to the full-length env using Sequencher 5.0 (Gene Codes Inc., Ann Arbor, Michigan). Nucleotide (nt) sequences were aligned using MUSCLE 3.8 [35]. The assembled env nt sequences were translated into amino acid (AA) sequences and the defective AA sequences that contain more than one stop codon were separately analyzed. Aligned sequences were output in PHYLIP format and the maximum iteration of alignments was used in order to obtain the highest accuracy. Phylogenetic analysis on both nt and AA sequences of founder viruses and viruses in the inoculum were performed using PHYML 3.0 [36] with the maximum likelihood method.
Compartmentalization analysis was carried out using Hyphy package with distance based methods FST and Snn [37]. Three different calculations were used in FST: HSM (Hudson, Slatkin, and Maddison), S (Slatkin), and HBK (Hudson, Boos, and Kaplan) [38-40]. Snn used the nearest neighbor calculation [41]. One thousand permutation tests were used to randomly allocating sequences into subpopulations and tabulating the distribution of various statistics. Sample was defined as compartmentalized if all tests show a probability value of < 0.05.
The Env AA sequences of each individual animal were clustered using CD-HIT 4.6 with a cluster threshold of 0.995 and word size of 5.0 [42]. To distinguish dominant and minor founder variant clusters, the clusters in each animal were normalized using the formula, , where is the ith normalized cluster size, Ci is the ith cluster, and sums the cluster sizes. The normalized clusters were then pooled together to make a histogram as shown in Fig S6. This Figure shows that an abundance level of 0.2 clearly separates the dominant and minor founder variant clusters.
Consensus sequences were generated from the clusters using majority votes. The motifs extracted from the alignment reflect the differences between the dominant and minor Env sequences. Motifs of dominant groups were computed by comparing the aligned consensus sequence with sequences from inoculum corresponding with minor or undetected groups. The motifs were visualized using UCSF Chimera. PDB files of SIV gp120 and gp41 were downloaded from the RCSB Protein Data Bank as 3FUS and 1QBZ respectively [43-45].
Statistical Analysis
Pearson’s chi-squared test was used to determine whether there were significant differences between inoculum and samples, within different tissue types, between 6 dpi and 10 dpi groups, and among individual macaques. The tests were performed using R statistical software (http://www.r-project.org). Pearson’s chi-squared test was performed to test whether the counts of founder variants were randomly sampled with similar proportions of virus variants in the inoculum. For variants sequenced in the inoculum sample, the proportion is simply the sequence number for that variant divided by the total number of sequences. For rare variants that were missed by SGA detection in the inoculum, the proportion is set to be less than 0.043. The rationale for doing so is based on probability theory. There is 95% likelihood that the proportion of a missing variant is <1 − 0.051/n, where n (68 in our case) is the number of sequences sampled [46]. Using 0.043 for the proportion of the rare variants will lead to a conservative estimate of the p-value. After the p-value was obtained for each monkey, Fisher’s method was used to combine the results from the six monkeys to form a single p-value for the test. For the tissue compartmentalization analysis, we used the following method. Suppose a proportion q of variants found in a particular tissue compartment also exists in other tissues. By classical probability theory, the number of variants (x, say) found in this compartment and also in other tissues follows a binomial distribution with the parameters being n (the total number of variants found in this tissue compartment) and q. As the value of q is unknown, we use the lower bound of the one-sided confidence interval for q as a conservative estimate for q with confidence level 95%. All other supporting statistical details are included in the S13_Statistical analysis.
Data availability
All 524 full-length SIV env sequence data in this study have been deposited into GenBank under accession numbers KM212303 - KM212826.
Supplementary Material
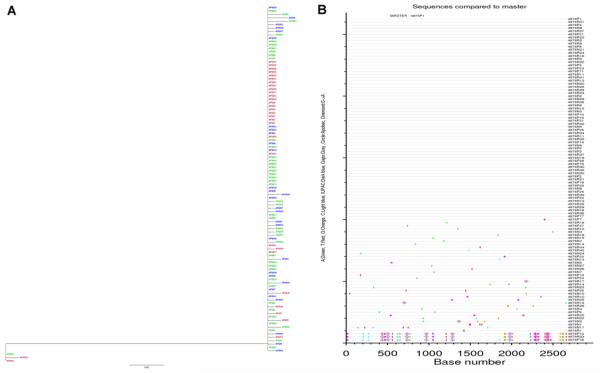
Fig S1. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 6 dpi macaque Rh4809. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S6. Separation of dominant and minor founder variant clusters based on abundance. Founder variant clusters from each animal were pooled together to create a histogram that shows the difference between minor and dominant founder variant clusters. A majority of clusters have an abundance of less than 0.2 and are designated as minor founder variant clusters. A smaller number of clusters have greater abundances (> 0.2) and are designated as major founder variant clusters.
Fig S7. The relationship between founder and inoculum variants and the founder virus signature (FVS) in 10 dpi macaque Rh4978. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red circle indicates the dominant variant group of founder viruses with greater than 99.5% AA identity. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S8. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 10 dpi macaque Rh4979. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red solid-line circle and dotted-line circle indicates the dominant and subdominant variant group of founder viruses with greater than 99.5% AA identity, respectively. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S9. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 6 dpi macaque Rh4809. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red solid-line circle and dotted-line circle indicates the dominant and subdominant variant group of founder viruses with greater than 99.5% AA identity, respectively. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S10. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 6 dpi macaque Rh5052. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red solid-line circle and dotted-line circle indicates the dominant and subdominant variant group of founder viruses with greater than 99.5% AA identity, respectively. Bar length indicates 0.008 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S11. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 6 dpi macaque Rh5053. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red circle indicates the dominant variant group of founder viruses with greater than 99.5% AA identity. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S12. The distribution of founder virus variants from 6 macaques and the variants in the inoculum on an AA phylogenetic tree. Different colors depict the variants derived from different macaques and the inoculum.
Supplemental statistical analysis.
Fig S2. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 6 dpi macaque Rh5052. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S3. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 6 dpi macaque Rh5053. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S4. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 10 dpi macaque Rh4978. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S5. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 10 dpi macaque Rh4979. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Table 1.
Test of tissue compartmentalization.
| Subjects | Days post inoculation (dpi) |
Test of Compartmentalization |
Compartmentalization | |||
|---|---|---|---|---|---|---|
| Fst | Snn | |||||
|
| ||||||
| HSM | S | HBK | ||||
| 4809 N vs P | 6 | 0.482 | 0.482 | 0.482 | 0.354 | No |
| 4809 N vs R | 6 | 0.993 | 0.993 | 0.993 | 0.02 | No |
| 4809 P vs R | 6 | 0.117 | 0.117 | 0.117 | 0.113 | No |
|
| ||||||
| 4976 N vs P | 10 | 0.729 | 0.729 | 0.729 | 0.162 | No |
| 4976 N vs R | 10 | 0.396 | 0.396 | 0.396 | 0.039 | No |
| 4976 P vs R | 10 | 0.168 | 0.168 | 0.168 | 0.762 | No |
|
| ||||||
| 4978 N vs P | 10 | 0.385 | 0.385 | 0.385 | 0.002 | No |
| 4978 N vs R | 10 | 0.458 | 0.448 | 0.422 | 0.299 | No |
| 4978 P vs R | 10 | 0.642 | 0.642 | 0.641 | 0.104 | No |
|
| ||||||
| 5053 N vs P | 6 | 0.086 | 0.086 | 0.086 | 0.131 | No |
| 5053 N vs R | 6 | 0 | 0 | 0 | 0 | Yes |
| 5053 P vs R | 6 | 0 | 0 | 0 | 0 | Yes |
N – lymph node, P - plasma, and R - rectal; HSM – Hudson, Slatkin and Maddison, S – Slatkin, and HBK – Hudson, Boos and Kaplan; 5052 or 4979 sequences from some tissue type cannot reach statistical power
Significance.
Anorectal receptive intercourse is a common route of HIV-1 transmission and a better understanding of the transmission mechanisms is critical for developing HIV-1 preventative strategies. Here, we report that there is no tissue compartmentalization of founder viruses during very early rectal transmission of SIV in the majority of rhesus macaques and founder viruses are preferentially derived from rare variant in the inoculum. We also found that founder viruses are animal-specific despite identical viral inoculums. After viruses cross the mucosal barriers, the host further reduces viral diversity by converting some of the transmitted functional viruses into defective viruses through frameshift rather than APOBEC derived mutations. To our knowledge, this is the first study of founder viruses at multiple tissue sites during very early rectal transmission.
Highlights.
Founder viruses are animal-specific in SIV rectal transmission in rhesus macaques
Founder viruses are preferentially derived from rare variants in the inoculum.
No tissue compartmentalization of founder viruses for the majority of macaques
Post-transmission defective viruses were derived by frameshift mediated mutations
Acknowledgments
The authors thank Dr. Brandon Keele for SGA technical support and discussion, Dr. James Mullins, Lance Daharsh, and Dr. John West for discussion and critical reading of the manuscript. The authors also thank Dr. Nancy Miller for providing the SIVmac251 stock. The funding sources: R01 DK087625-01 (Q. Li), R01 AI111862-01 (Q. Li and J. Guo), NCRR COBRE 15635.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Edwards C, Holmes E, Wilson D, Viscidi R, Abrams E, et al. Population genetic estimation of the loss of genetic diversity during horizontal transmission of HIV-1. BMC Evolutionary Biology. 2006;6:28. doi: 10.1186/1471-2148-6-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abrahams M-R, Anderson JA, Giorgi EE, Seoighe C, Mlisana K, et al. Quantitating the Multiplicity of Infection with Human Immunodeficiency Virus Type 1 Subtype C Reveals a Non-Poisson Distribution of Transmitted Variants. Journal of Virology. 2009;83:3556–3567. doi: 10.1128/JVI.02132-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boeras DI, Hraber PT, Hurlston M, Evans-Strickfaden T, Bhattacharya T, et al. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proceedings of the National Academy of Sciences. 2011;108:E1156–E1163. doi: 10.1073/pnas.1103764108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keele BF, Estes JD. Barriers to mucosal transmission of immunodeficiency viruses. Blood. 2011;118:839–846. doi: 10.1182/blood-2010-12-325860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stone M, Keele BF, Ma Z-M, Bailes E, Dutra J, et al. A Limited Number of Simian Immunodeficiency Virus (SIV) env Variants Are Transmitted to Rhesus Macaques Vaginally Inoculated with SIVmac251. Journal of Virology. 2010;84:7083–7095. doi: 10.1128/JVI.00481-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu J, Keele BF, Li H, Keating S, Norris PJ, et al. Low-Dose Mucosal Simian Immunodeficiency Virus Infection Restricts Early Replication Kinetics and Transmitted Virus Variants in Rhesus Monkeys. Journal of Virology. 2010;84:10406–10412. doi: 10.1128/JVI.01155-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li H, Bar KJ, Wang S, Decker JM, Chen Y, et al. High Multiplicity Infection by HIV-1 in Men Who Have Sex with Men. PLoS Pathog. 2010;6:e1000890. doi: 10.1371/journal.ppat.1000890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma ZM, Keele BF, Qureshi H, Stone M, Desilva V, et al. SIVmac251 is inefficiently transmitted to rhesus macaques by penile inoculation with a single SIVenv variant found in ramp-up phase plasma. AIDS Res Hum Retroviruses. 2011;27:1259–1269. doi: 10.1089/aid.2011.0090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Derdeyn CA, Hunter E. Viral characteristics of transmitted HIV. Curr Opin HIV AIDS. 2008;3:16–21. doi: 10.1097/COH.0b013e3282f2982c. [DOI] [PubMed] [Google Scholar]
- 10.Haaland RE, Hawkins PA, Salazar-Gonzalez J, Johnson A, Tichacek A, et al. Inflammatory Genital Infections Mitigate a Severe Genetic Bottleneck in Heterosexual Transmission of Subtype A and C HIV-1. PLoS Pathogens. 2009;5:e1000274. doi: 10.1371/journal.ppat.1000274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Lehner T. Induction of innate immunity in control of mucosal transmission of HIV. Curr Opin HIV AIDS. 2011;6:398–404. doi: 10.1097/COH.0b013e3283499df7. [DOI] [PubMed] [Google Scholar]
- 12.Yan N, Chen ZJ. Intrinsic antiviral immunity. Nat Immunol. 2012;13:214–222. doi: 10.1038/ni.2229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, et al. Phenotypic properties of transmitted founder HIV-1. Proceedings of the National Academy of Sciences. 2013;110:6626–6633. doi: 10.1073/pnas.1304288110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keele BF, Giorgi EE, Salazar-Gonzalez JF, Decker JM, Pham KT, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc Natl Acad Sci U S A. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma D, Jasinska AJ, Feyertag F, Wijewardana V, Kristoff J, et al. Factors associated with siman immunodeficiency virus transmission in a natural African nonhuman primate host in the wild. J Virol. 2014;88:5687–5705. doi: 10.1128/JVI.03606-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Joseph SB, Swanstrom R, Kashuba AD, Cohen MS. Bottlenecks in HIV-1 transmission: insights from the study of founder viruses. Nat Rev Microbiol. 2015;13:414–425. doi: 10.1038/nrmicro3471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harouse JM, Buckner C, Gettie A, Fuller R, Bohm R, et al. CD8+ T cell-mediated CXC chemokine receptor 4-simian/human immunodeficiency virus suppression in dually infected rhesus macaques. Proceedings of the National Academy of Sciences. 2003;100:10977–10982. doi: 10.1073/pnas.1933268100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moore JP, Kitchen SG, Pugach P, Zack JA. The CCR5 and CXCR4 Coreceptors—Central to Understanding the Transmission and Pathogenesis of Human Immunodeficiency Virus Type 1 Infection. AIDS Research and Human Retroviruses. 2004;20:111–126. doi: 10.1089/088922204322749567. [DOI] [PubMed] [Google Scholar]
- 19.Grivel J-C, Shattock R, Margolis L. Selective transmission of R5 HIV-1 variants: where is the gatekeeper? Journal of Translational Medicine. 2011;9:1–17. doi: 10.1186/1479-5876-9-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar-Gonzalez JF, Salazar MG, Keele BF, Learn GH, Giorgi EE, et al. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. The Journal of Experimental Medicine. 2009;206:1273–1289. doi: 10.1084/jem.20090378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ochsenbauer C, Edmonds TG, Ding H, Keele BF, Decker J, et al. Generation of Transmitted/Founder HIV-1 Infectious Molecular Clones and Characterization of Their Replication Capacity in CD4 T Lymphocytes and Monocyte-Derived Macrophages. Journal of Virology. 2012;86:2715–2728. doi: 10.1128/JVI.06157-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology. 2013;10:146. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z-Q, Schuler T, Zupancic M, Wietgrefe S, Staskus KA, et al. Sexual Transmission and Propagation of SIV and HIV in Resting and Activated CD4+ T Cells. Science. 1999;286:1353–1357. doi: 10.1126/science.286.5443.1353. [DOI] [PubMed] [Google Scholar]
- 24.Li Q, Estes JD, Schlievert PM, Duan L, Brosnahan AJ, et al. Glycerol monolaurate prevents mucosal SIV transmission. Nature. 2009;458:1034–1038. doi: 10.1038/nature07831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gray RH, Wawer MJ, Brookmeyer R, Sewankambo NK, Serwadda D, et al. Probability of HIV-1 transmission per coital act in monogamous, heterosexual, HIV-1-discordant couples in Rakai, Uganda. The Lancet. 2001;357:1149–1153. doi: 10.1016/S0140-6736(00)04331-2. [DOI] [PubMed] [Google Scholar]
- 26.Wolinsky SM, Wike CM, Korber BT, Hutto C, Parks WP, et al. Selective transmission of human immunodeficiency virus type-1 variants from mothers to infants. Science. 1992;255:1134–1137. doi: 10.1126/science.1546316. [DOI] [PubMed] [Google Scholar]
- 27.Carlson JM, Schaefer M, Monaco DC, Batorsky R, Claiborne DT, et al. Selection bias at the heterosexual HIV-1 transmission bottleneck. Science. 2014;345 doi: 10.1126/science.1254031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tully DC, Ogilvie CB, Batorsky RE, Bean DJ, Power KA, et al. Differences in the Selection Bottleneck between Modes of Sexual Transmission Influence the Genetic Composition of the HIV-1 Founder Virus. PLoS Pathog. 2016;12:e1005619. doi: 10.1371/journal.ppat.1005619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gonzalez MW, DeVico AL, Lewis GK, Spouge JL. Conserved molecular signatures in gp120 are associated with the genetic bottleneck during simian immunodeficiency virus (SIV), SIV-human immunodeficiency virus (SHIV), and HIV type 1 (HIV-1) transmission. J Virol. 2015;89:3619–3629. doi: 10.1128/JVI.03235-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Perelson AS, Neumann AU, Markowitz M, Leonard JM, Ho DD. HIV-1 dynamics in vivo: virion clearance rate, infected cell life-span, and viral generation time. Science. 1996;271:1582–1586. doi: 10.1126/science.271.5255.1582. [DOI] [PubMed] [Google Scholar]
- 31.Franchini G, Gurgo C, Guo HG, Gallo RC, Collalti E, et al. Sequence of simian immunodeficiency virus and its relationship to the human immunodeficiency viruses. Nature. 1987;328:539–543. doi: 10.1038/328539a0. [DOI] [PubMed] [Google Scholar]
- 32.Goulder PJR, Watkins DI. HIV and SIV CTL escape: implications for vaccine design. Nat Rev Immunol. 2004;4:630–640. doi: 10.1038/nri1417. [DOI] [PubMed] [Google Scholar]
- 33.Fenton-May AE, Dibben O, Emmerich T, Ding H, Pfafferott K, et al. Relative resistance of HIV-1 founder viruses to control by interferon-alpha. Retrovirology. 2013;10:1742–4690. doi: 10.1186/1742-4690-10-146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, et al. Recurrent Signature Patterns in HIV-1 B Clade Envelope Glycoproteins Associated with either Early or Chronic Infections. PLoS Pathog. 2011;7:e1002209. doi: 10.1371/journal.ppat.1002209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guindon S, Dufayard JF, Lefort V, Anisimova M, Hordijk W, et al. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 37.Pond SL, Frost SD, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 38.Hudson RR, Slatkin M, Maddison WP. Estimation of levels of gene flow from DNA sequence data. Genetics. 1992;132:583–589. doi: 10.1093/genetics/132.2.583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Slatkin M. Isolation by Distance in Equilibrium and Non-Equilibrium Populations. Evolution. 1993;47:264–279. doi: 10.1111/j.1558-5646.1993.tb01215.x. [DOI] [PubMed] [Google Scholar]
- 40.Hudson RR, Boos DD, Kaplan NL. A statistical test for detecting geographic subdivision. Mol Biol Evol. 1992;9:138–151. doi: 10.1093/oxfordjournals.molbev.a040703. [DOI] [PubMed] [Google Scholar]
- 41.Hudson RR. A new statistic for detecting genetic differentiation. Genetics. 2000;155:2011–2014. doi: 10.1093/genetics/155.4.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li W, Jaroszewski L, Godzik A. Clustering of highly homologous sequences to reduce the size of large protein databases. Bioinformatics. 2001;17:282–283. doi: 10.1093/bioinformatics/17.3.282. [DOI] [PubMed] [Google Scholar]
- 43.Chen B, Vogan EM, Gong H, Skehel JJ, Wiley DC, et al. Structure of an unliganded simian immunodeficiency virus gp120 core. Nature. 2005;433:834–841. doi: 10.1038/nature03327. [DOI] [PubMed] [Google Scholar]
- 44.Chen X, Lu M, Poon BK, Wang Q, Ma J. Structural improvement of unliganded simian immunodeficiency virus gp120 core by normal-mode-based X-ray crystallographic refinement. Acta Crystallogr D Biol Crystallogr. 2009;65:339–347. doi: 10.1107/S0907444909003539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang Z-N, Mueser TC, Kaufman J, Stahl SJ, Wingfield PT, et al. The Crystal Structure of the SIV gp41 Ectodomain at 1.47 Å Resolution. Journal of Structural Biology. 1999;126:131–144. doi: 10.1006/jsbi.1999.4116. [DOI] [PubMed] [Google Scholar]
- 46.Salazar-Gonzalez JF, Bailes E, Pham KT, Salazar MG, Guffey MB, et al. Deciphering Human Immunodeficiency Virus Type 1 Transmission and Early Envelope Diversification by Single-Genome Amplification and Sequencing. Journal of Virology. 2008;82:3952–3970. doi: 10.1128/JVI.02660-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 6 dpi macaque Rh4809. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S6. Separation of dominant and minor founder variant clusters based on abundance. Founder variant clusters from each animal were pooled together to create a histogram that shows the difference between minor and dominant founder variant clusters. A majority of clusters have an abundance of less than 0.2 and are designated as minor founder variant clusters. A smaller number of clusters have greater abundances (> 0.2) and are designated as major founder variant clusters.
Fig S7. The relationship between founder and inoculum variants and the founder virus signature (FVS) in 10 dpi macaque Rh4978. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red circle indicates the dominant variant group of founder viruses with greater than 99.5% AA identity. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S8. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 10 dpi macaque Rh4979. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red solid-line circle and dotted-line circle indicates the dominant and subdominant variant group of founder viruses with greater than 99.5% AA identity, respectively. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S9. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 6 dpi macaque Rh4809. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red solid-line circle and dotted-line circle indicates the dominant and subdominant variant group of founder viruses with greater than 99.5% AA identity, respectively. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S10. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 6 dpi macaque Rh5052. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red solid-line circle and dotted-line circle indicates the dominant and subdominant variant group of founder viruses with greater than 99.5% AA identity, respectively. Bar length indicates 0.008 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S11. The relationship between founder and inoculum variants and the animal-specific founder virus signature (FVS) in 6 dpi macaque Rh5053. (A) Maximum likelihood tree of Env AA sequences of founder virus variants. Sequences derived from plasma are represented in red, rectum in green, lymph node in blue. Functional variants in inoculum are represented in black. The red circle indicates the dominant variant group of founder viruses with greater than 99.5% AA identity. Bar length indicates 0.01 AA substitutions per site. (B) The animal-specific founder virus signature (FVS) of the dominant founder variant group (top, highlighted in yellow). The variant nomenclature rule: the number represents the variant number of the cluster/group from which the consensus sequence was derived, followed by a space and the variant ID. The montage of MUSCLE alignment of Env AA sequences of minor founder variants and undetected functional variants in the inoculum are compared with the consensus sequence of the dominant founder variant group (red box).
Fig S12. The distribution of founder virus variants from 6 macaques and the variants in the inoculum on an AA phylogenetic tree. Different colors depict the variants derived from different macaques and the inoculum.
Supplemental statistical analysis.
Fig S2. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 6 dpi macaque Rh5052. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S3. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 6 dpi macaque Rh5053. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S4. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 10 dpi macaque Rh4978. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Fig S5. Maximum likelihood tree (A) and Highlighter plots (B) of SGA-derived env nucleotide sequences from 10 dpi macaque Rh4979. The sequences from rectum (green), axillary lymph node tissue (blue), plasma (red), or inoculum (black) are depicted by different colors. Nucleotide polymorphisms in the Highlighter plots are depicted by a colored tic mark (thymine in red, guanine in orange, adenine in green, cytosine in blue). Pink filled circles denote APOBEC signatures, open diamonds represent G-to-A conversions, and gaps are shown in gray).
Data Availability Statement
All 524 full-length SIV env sequence data in this study have been deposited into GenBank under accession numbers KM212303 - KM212826.