Abstract
In response to intracellular stress events ranging from starvation to pathogen invasion, the cell activates one or more forms of macroautophagy. The key event in these related pathways is the de novo formation of a new organelle called the autophagosome, which surrounds and sequesters either random portions of the cytoplasm or selectively targets individual intracellular challenges. Thus the autophagosome is a flexible membrane platform with dimensions that ultimately depend upon the target cargo. The intermediate membrane, termed the phagophore or isolation membrane, is a cup-like structure with a clear concave face and a highly curved rim. The phagophore is largely devoid of integral membrane proteins, thus its shape and size are governed by peripherally-associated membrane proteins and possibly by the lipid composition of the membrane itself. Growth along the phagophore rim marks the progress of both organelle expansion and ultimately of organelle closure around a particular cargo. These two properties, a reliance on peripheral membrane proteins and a structurally-distinct membrane architecture, suggest that the ability to target or manipulate membrane curvature might be an essential activity of proteins functioning in this pathway. In this review, we discuss the extent to which membranes are naturally curved at each of the cellular sites believed to engage in autophagosome formation, review basic mechanisms used to sense this curvature and then summarize the existing literature concerning which autophagy proteins are capable of curvature recognition.
Keywords: membrane curvature, amphipathic helix, BAR domain, reticulon domain
Graphical Abstract
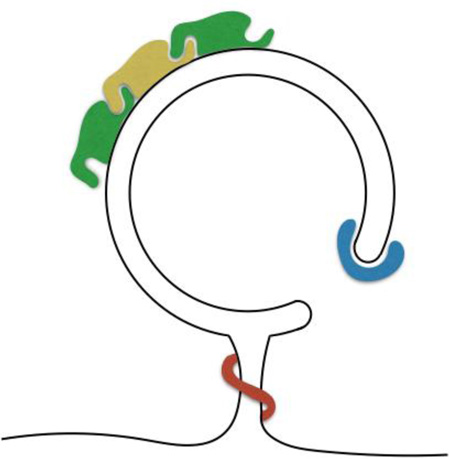
Membrane curvature on the growing autophagosome. Subdomains of the growing autophagosome present unique membrane architectures that likely recruit or activate subsets of macroautophagy proteins.
The phagophore is an autophagic intermediate with a unique membrane architecture
The mechanistic details of autophagosome formation are only beginning to emerge, but the unique morphologies of the organelle and its intermediates have long been apparent by electron microscopy (EM). Mature autophagosomes were readily observed as early as the 1960’s and defined as double-membrane vesicles filled with cytoplasmic material (e.g.1). Conditions that lead to enrichment of these structures such as starvation, also lead to the accumulation of other objects that were inferred to be autophagosome intermediates. The most striking structural intermediate is a cup-shaped double membrane called the phagophore or isolation membrane (IM; Figure 1). In EM sections the phagophore appears as a semi-circular cisterna from 0.3 to 3µm in diameter. The wall of the structure is composed of two membrane bilayers in very close apposition; continuity of the convex- and concave-facing phagophore membranes generates a subregion that we call the phagophore rim (Figure 2).
Figure 1. Sources of membrane curvature associated with macroautophagy in the mammalian cell.
Autophagosome biogenesis involves many structures that present strident positive curvature to the cytoplasm including very small vesicles, ER tubules, tubular protrusions from very small recycling endosome and golgi vesicles, and the rim of the expanding phagophore (see Table I). Proteins associated with each step in the organelle’s maturation have been shown to possess membrane curvature sensing in vitro and to rely on these motifs for proper function in vivo.
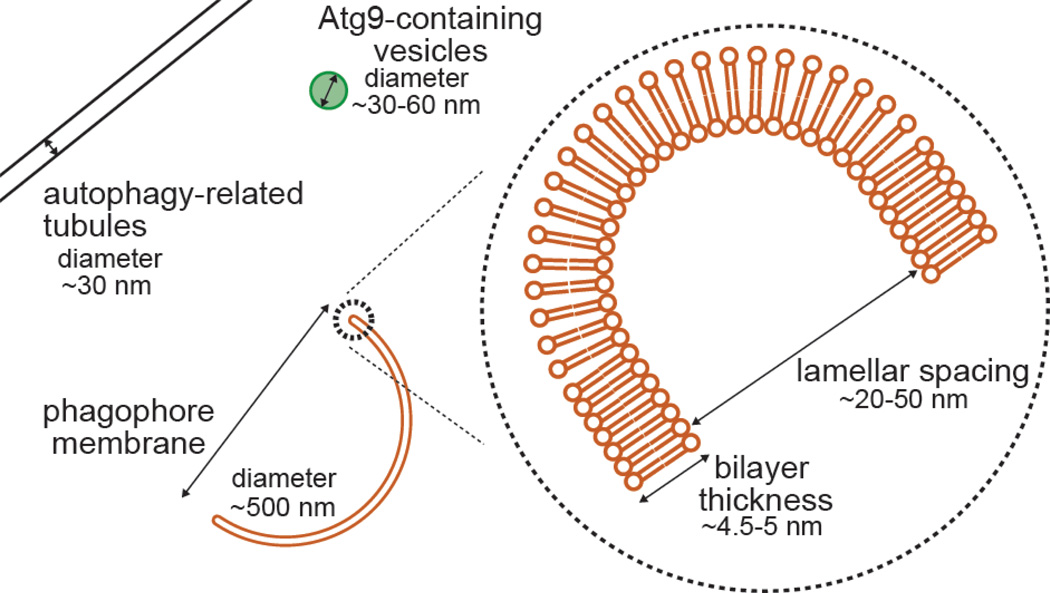
Figure 2. Phagophore dimensions.
The phagophore or isolation membrane is a cup-shaped intermediate in autophagosome development. It includes a concave inner and convex outer surface as well as a highly curved rim running along the open edge of the cup. Labeled are the diameters of several autophagy-related membrane structures exhibiting high curvature. In addition, we note the typical distances between the bilayers in the growing isolation membrane or the mature autophagosome (lamellar spacing). Although this distance does not directly represent a curvature, it sets a kind of lower bound for the radius of curvature connecting the two bilayers at the rim. In practice, actual measures of rim curvature are mostly not available. We have surveyed a representative set of articles in the literature to generate the range of measures in Table I.
Topologically, this arrangement is reminiscent of multi-vesicular bodies during luminal vesicle budding events; however the very close apposition of the two autophagosome bilayers suggests that the curvature at the phagophore rim is particularly dramatic. We have conducted a survey of the autophagosome literature in which electron microscopy was used to image these structures arising from a wide range of different model systems in an attempt to estimate typical dimensions of the phagophore (Table I). EM from many groups using varying fixation and preservation strategies suggests that the separation between membranes in the luminal space of the phagophore is less than 30 nm (the limit of measure we can easily make in reproduced images from publications) and in many cases is essentially not detectable (ex.2). Thus, the radius of curvature at the rim in these same structures may also be less than 30 nm (Figure 2) and often appears to be at or below the limiting radius of curvature for a protein-free bilayer (Table I and 3).
Table I. Sources of membrane curvature in cellular macroautophagy.
This Table represents a non-comprehensive survey of the literature on macroautophagy spanning the earliest electron microscopy measurements through more current cutting-edge approaches including cryo-electron tomography. Our objective is to illustrate first that the autophagosome itself, and the body of the growing phagophore or isolation membrane is not highly curved. This can be inferred by considering the size of the object and assuming a smooth curvature. Thus the diameter of the autophagosome, or the diameter of the phagophore volume (when considering the distance between its furthest points as in figure 2) reflects the relative curvature of the structure. In contrast, the phagophore rim, or other features of very high definition like small tubules, will present much more strident curvatures. For simplicity, we only considered phagophores that had already adopted a cup-like shape (i.e. the very early stages of phagophore growth are not represented here). It is currently impractical to measure the rim curvature in most electron microscopy images. Thus we present the lamellar spacing between the inner and outer membranes of the growing phagophore as a proxy for the lower limit of the radius of curvature (as shown in figure 2). Notably, this spacing is consistently less than 50 nm and usually much smaller, independent of model organism or the laboratory collecting the images.
Where possible, we have cited the actual measurements made by the authors of a publication. In most cases however, we have made these measurements ourselves from published images and presented the value as an upper limit for the dimension (in each case noted as “≤”). To be conservative, any spacing smaller than 30 nm was listed as ≤ 30 nm, even in cases where the two bilayers appear to touch and have essentially no separation. Thus, in many cases, “≤ 30 nm” represents a much smaller distance.
| Structure | Measurement | Method of observation |
Model organism | Reference |
|---|---|---|---|---|
| Autophagosome | lamellar spacing / diameter |
|||
| Autophagosomes | ≤30 nm /≤0.9 µm |
Freeze- substitution fixation for EM |
Carbon- or nitrogen- starved S. cerevisiae |
91 Fig. 1–8 |
| Autophagosomes | ≤30 nm /≤0.9 µm |
Freeze- fractured EM |
Nitrogen-starved S cerevisiae |
92 Fig. 3, 4 |
| Autophagosomes | ≤30 nm /≤2.5 µm |
TEM | Sucrose-starved A thaliana T87 cells |
93 Fig. 4, 5 |
| Autophagosomes | ≤50 nm / NM | Immunogold EM |
Differentiating root epidermal cells of Arabidopsis seedlings |
27 Fig. 6 |
| Autophagosomes | ≤ 50 nm / ≤ 1 µm |
EM | Isolated rat liver |
94 Fig. 1 |
| Autophagosomes | ≤ 60 nm / ≤ 1 µm |
Immunogold EM |
Starved rat kidney cells |
5 Fig. 2 |
| Purified autophagosomes (subcellular fractionation) |
≤ 50 nm / ≤ 2 µm |
TEM | Vinblastine-treated hepatocytes isolated from starved rats |
26 Fig. 1–3 |
| Autophagosomes | ≤60 nm /≤1.5 µm |
Silver-enhanced immunogold EM |
GFP-LC3 mouse liver, heart muscle, pancreatic exocrine, and stromal epithelial cells |
95 Fig. 3–6 |
| Cytolysomes | ≤ 50 nm / ≤ 1 µm |
TEM | Fat body of instar larva of C. ethlius |
96 Fig. 7, 8 |
| Mitophagosomes | ≤ 30 nm / ≤ 1 µm |
TEM | Valinomycin-treated HCT116 (Human colo- rectal cancer) cells |
97 Fig. 2 |
|
Phagophore & Omegasome |
lamellar spacing / diameter |
|||
| Autophagosomes & phagophores |
≤ 50 nm / ≤ 1 µm |
EM | Isolated rat liver |
98 Fig. 1, 6–9, 11 |
| Phagophore | ≤50 nm /≤ 1.5 µm |
Silver-enhanced immunogold EM |
Embryonic stem cells expressing GFP-Apg16L |
29 Fig. 7 |
| Autophagosomes and phagophores |
≤ 50 nm / ≤ 1 µm |
Silver-enhanced immunogold EM |
F9 teratocarcinoma cells |
99 Fig. 7 |
| Phagophores, ER-IM complexes |
≤ 50 nm / ≤ 2 µm |
Immunogold EM and electron tomography |
NIH3T3 cells and MEFS |
2 Fig. 1–4, S2, S4 |
| Phagophores | ≤30 nm /≤0.5 µm |
Immuno EM | Atg9-6xHA ypt7Λ S cerevisiae |
30 Fig. 5 |
| Phagophores | ≤30 nm / NM | Immuno EM and CLEM |
Atg3+/+ and Atg3−/− MEFs |
4 Fig. 2 |
| Omegasomes/punctate structures |
NM / ≤ 0.5 µm | Confocal microscopy |
HEK293 cells |
100 Fig. 4 |
| Omegasomes | NM / ≤ 1 µm | Cryo-CLXM | Starved HEK293 cells |
101 Fig. 6, 7 |
| Cvt vesicle | lamellar spacing / diameter |
|||
| Cvt vesicle | ≤30 nm /≤0.5 µm |
Freeze- substitution fixation for EM |
S. cerevisiae (vegetative growth) |
102 Fig. 2–4 |
| Tubules | diameter | |||
| ER tubules | ≤30 nm | EM and electron tomography |
High pressure frozen S cerevisiae |
103 Fig. 1 |
| ER-like membranes associate with autophagosomal structures |
≤50 nm | Immunogold EM |
Differentiating root epidermal cells of Arabidopsis seedlings |
27 Fig. 6 |
| RFP-LC3-positive Tubules |
40 – 65 nm | CLEM | Human Embryonic Kidney cell line over- expressing certain protocadherins |
104 Fig. 9 |
| Golgi membrane fragmentations and tubulations |
≤30 nm | TEM | Bif-1 shRNA-expressing HeLa cells |
43 Fig. 3 |
| GFP-LC3-decorated Tubules |
≤100 nm | Immunogold EM |
HeLa over-expressing SNX18 |
81 Fig. 8 |
| Anti-Kar2 ER double- ring structures and ER tubules |
≤100 nm | Immunogold EM |
S. cerevisiae treated with rapamycin |
12 Fig. 3 |
| Immuno-isolated GFP- LC3-positive tubulo- vesicular structures (tubule form) |
≤50 nm | EM | HEK293 and HCT116 stably expressing GFP- LC3 |
42 Fig. 3 |
| Vesicles | diameter | |||
| Spherical particles clustered around the cvt vesicle |
35 – 50 nm | Freeze- substitution fixation for EM |
S. cerevisiae (vegetative growth) |
102 Fig. 2, 4 |
| Clustered Atg9-GFP- positive vesicles at the phagophore assembly site |
30 – 40 nm | Immunogold EM |
S. cerevisiae (vegetative growth) |
32 Fig. 2, S1 |
| Atg9-GFP-positive vesicles purified from lysate |
30 – 60 nm | Single-particle tracking, dynamic light scattering, and TEM |
Rapamycin-treated S cerevisiae |
30 Fig. 1, 2 |
| Clustered vesicles around the autophagosomes |
100 – 300 nm | TEM | Sucrose-starved A thaliana T87 cells |
93 Fig. 4, 5 |
| Immuno-isolated GFP- LC3-positive tubulo- vesicular structures (vesicle form) |
≤50 nm | EM | HEK293 and HCT116 stably expressing GFP- LC3 |
42 Fig. 3, 7 |
| mRFP-Atg9-positive vesicles and/or tubules |
≤100 nm | CLEM | WIPI2-depleted HEK293 cell line |
31 Fig. 4 |
| GFP-DFCP1-positive clustered tubulo- vesicular structures at the edge of isolation membrane |
30 – 35 nm | Immuno EM and CLEM |
Atg3+/+ and Atg3−/− MEFs |
4 Fig. 3 |
| Other organelles: | diameter | |||
| Autophagic bodies | ≤0.8 µm | Freeze- fractured EM |
Nitrogen-starved S cerevisiae |
92 Fig. 3, 4 |
| Autophagic vacuoles (amphisome or autophagosome), phagophore |
≤1.2 µm | Freeze- fractured EM |
Isolated rat hepatocyte |
105 Fig. 3 |
| COPII-containing ER exit site vesicular and tubular structures |
≤50 nm | Electron tomography |
HepG2 mammalian cells |
106 Fig. 3, 4 |
| COPI and COPII Vesicles |
60 – 100 nm | Immunogold EM and CLEM |
MPR- and GGA- expressing HeLa cells |
107 Fig. 1–3 |
| Tubular-vesicular structures |
≤150 nm | Immunogold EM and CLEM |
MPR- and GGA- expressing HeLa cells |
107 Fig. 3–6 |
| Lysosomes | ≤ 300 nm | Freeze- fractured EM |
Isolated rat hepatocyte |
105 Fig. 4 |
| Lysosomes | 200 – 600 nm | Freeze- fractured EM |
Isolated rat liver |
108 Fig. 4 |
| Mitochondria | ≤ 500 nm | Freeze- fractured EM |
Isolated rat hepatocyte |
105 Fig. 4 |
| Endosomes | ≤ 250 nm | Freeze- fractured EM |
Isolated rat hepatocyte |
105 Fig. 5 |
≤ : dimensions are not given in the text by the authors and were measured from the published images.
NM: Not measured
EM: Electron microscopy
CLEM: Correlative light electron microscopy
TEM: Transmission electron microscopy
Cryo-CLXM: Cryo-correlative light and X-ray microscopy
MEF: Mouse Embryonic Fibroblasts
More recent studies using electron tomography have unambiguously confirmed the phagophore’s bowl-like structure. In addition to corroborating previous observations, these studies also identify short, narrow tubular connections between the endoplasmic reticulum (ER) and the phagophore, termed “IM-associated tubular/vesicular structures” (IMATs) 4. Like the rim of the phagophore, these membrane structures are also highly curved (∼ 30 nm diameter). Their direct connection between the ER and the phagophore suggests that phagophore expansion may be mediated by membrane flow from the ER2, 4, 5, possibly suggesting that specific lipids or proteins are sorted into the phagophore via this intermediary.
Eventually the phagophore closes upon itself, and in higher eukaryotes, moves away from its point of origin. These mature autophagosomes are spherical, but are large enough that the cytoplasm-facing membranes do not exhibit the kind of significant curvature ordinarily recognized by single proteins --diameters vary from ∼300 nm in yeast to several hundred in mammals; Figure 1 and Table I. Thus during maturation, this organelle proceeds from a morphologically diverse mixture of sub-regions exhibiting very strong positive and negative curvature to a closed sphere with homogeneous membranes that are read as “flat” at the single molecule level. In addition, a number of other organelles in the cell also contribute to autophagosome growth, each with their own natural capacity to adopt some level of local membrane bending (Figure 1). Thus how and whether proteins recognize membrane curvature is likely to play a critical role in the activation or recruitment of autophagy-relevant factors.
Peripheral proteins control autophagosome growth
Despite the apparent continuity of the ER and phagophore membrane6 in mammals, ER-derived transmembrane proteins are generally excluded from the autophagosome. Indeed, the paucity of integral membrane proteins in the autophagosome was predicted early on from the smooth appearance of autophagosomes in freeze-fracture EM, a feature used to uniquely identify autophagosomes prior to the discovery of any autophagy-specific protein markers7–10. Among proteins identified from the first yeast screens for atg genes, sequence analysis shows only Atg9 and Atg27 contain transmembrane domains, but whether either protein so much as transiently resides within the phagophore membrane remains controversial 11. Recent studies also identify putative transmembrane domains in S. cerevisiae Atg39 and Atg4012; however these integrate into membranous autophagic cargo rather than the phagophore or autophagosome membrane itself. In fact, it is possible that the structure of the junction joining the ER and isolation membrane actively excludes integral membrane proteins. For example, from immuno-EM and immunofluorescence 9, 10, 13, it is clear that the PI(3)P-binding autophagy protein WIPI and the PI(3)P sensor, GFP-FYVE, bind the entire phagophore surface. The addition of a transmembrane domain to GFP-FYVE relegates the sensor to a ring around nascent phagophores, suggesting it is actively excluded. Studies ascribing a mitochondrial membrane origin for phagophore biogenesis describe a similar exclusion of transmembrane domains, even as single leaflet anchors are efficiently delivered 14.
Thus, the absence and potential exclusion of integral membrane proteins suggests that instead peripheral proteins comprising the autophagic machinery must directly recognize features of the nascent membrane. These features will include direct detection of individual lipids (especially PI(3)P) but also likely rely upon recognition of membrane architecture including the strident curvature of the rim and of the junctions with donor organelles.
Sensing membrane curvature or lipid composition
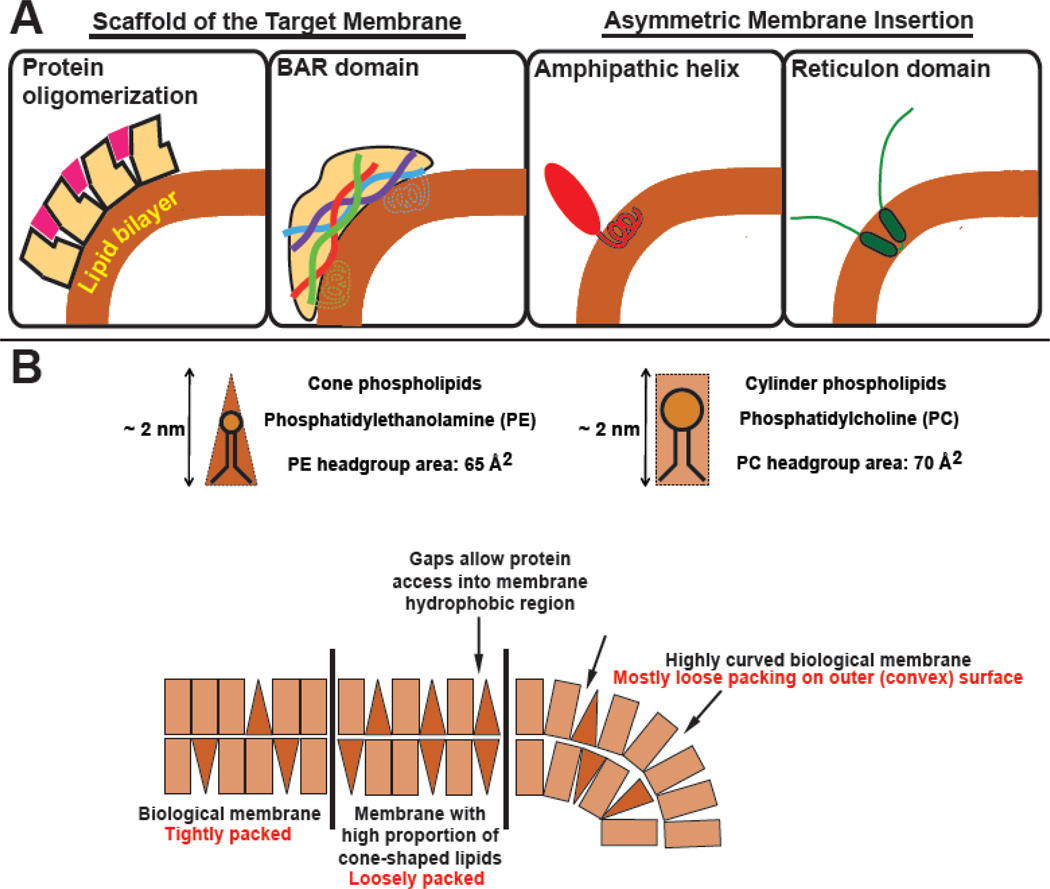
In the following sections we will briefly discuss protein motifs involved in sensing or inducing membrane curvature. For the most part, we use the term “curvature-sensing” even when in vitro work, such as the tubulation of small vesicles, could be interpreted as an example of curvature induction. In practice, the functional distinction is often a matter of protein concentration and ionic conditions, and whether curvature induction is physiologically relevant must be established by other methods. Two general mechanisms of curvature sensing are well-described (Figure 3). These rely on the protein’s (1) scaffolding of the target membrane or (2) asymmetric membrane insertion. These mechanisms have been extensively reviewed elsewhere (see for example 15–17 and 18), so here we explain them only briefly.
Figure 3. Mechanisms for proteins to recognize and interact with highly curved membranes.
A) Peripheral proteins either recognize the shape of the membrane primarily through engagement with phospholipid headgroups or interrogate the hydrophobic core of the bilayer with membrane insertion sequences. (BAR domains often include both scaffolding and amphipathic helix motifs). B) Membrane insertion of poorly hydrophobic motifs, such as those commonly associated with curvature-sensing, relies on poor lipid packing in the bilayer to favor partitioning of protein motifs into the membrane. If we consider phospholipids as cylinders or cones, with phospholipid headgroups’ and acyl chains’ cross sectional areas defining the base 17, 18, we can see that on planar surfaces, the packing of conical lipid headgroups becomes less ideal. In order to minimize exposed hydrophobic surface area of the acyl chains, membranes can bend or inverted conical lipids or protein insertion motifs can sort to these areas of the bilayer.
Scaffolding is the most intuitive and relies predominantly on direct interaction with the phospholipid headgroups. If a protein or protein complex adopts a shape that mirrors the organization of the bilayer and interacts with the lipid head groups of this bilayer weakly, this protein will accumulate on membranes exhibiting the same shape (Figure 3). Proteins that contain BAR (Bin/amphiphysin/Rvs) domains are the proto-typical example; BAR domains include long helices that assemble into crisscrossed dimers with a second BAR-domain containing protein. These dimers adopt an arc-like configuration and the degree of bending in the arc mirrors the curvature in the target membrane, even including membranes with a negative (concave) curvature (e.g. 18, 19). Proteins can also oligomerize into larger scaffolds that adopt a curved organization and thus will be found on similarly curved membranes. In both cases, as the affinity of the protein for the membrane increases, protein-dependent remodeling of the bilayer can instead be favored (where flat membranes are forced to adopt the curvature of the adhering protein complex).
In contrast, asymmetric membrane insertion relies upon the engagement of protein motifs with both the phospholipid headgroups as well as with the hydrophobic core of the bilayer. The degree to which a protein motif will be inserted into a bilayer is related to its partitioning coefficient, such that more hydrophobic structures will partition into the membrane more effectively. For example, classic membrane-binding amphipathic helices have one surface that is rich in highly hydrophobic amino acids including especially the aromatics tryptophan, phenylalanine and tyrosine. This surface is often bracketed by basic amino acids. Together these features are naturally recruited to biological membranes. In contrast, curvature-sensing amphipathic helices are a little less effective at binding membranes. They may have a polar surface that is missing any charged amino acids that would otherwise contribute to electrostatic engagement of the membrane or they can exhibit much less hydrophobicity on their membrane-apposing face; in either event, these proteins have a lower partitioning energy. In this case efficient membrane binding only becomes a possibility when the membrane is destabilized, usually with phospholipid compositions that do not naturally pack well into planar lamellar arrays, such as those rich in conical phospholipids (Figure 3B). At membranes that are very stridently curved, the packing of the lipids themselves can be compromised in a way that resembles conical-phospholipid rich membranes. At these sites, insertion of protein motifs like curvature-sensing amphipathic helices is often favored (Figure 4).
Figure 4. Highly curved membranes allow efficient interaction and activation of autophagic machinery.
Multiple protein sequence motifs, whether autophagy specific or not, have been shown to recognize structural deformation of target membranes. A common mechanism involves an alpha helix, consisting of hydrophobic residues flanked by hydrophilic residues, to penetrate the membrane. The distribution of these amino acids reflect the lipidic composition of the target membranes. For examples ALPS motifs were originally identified at the golgi (i.e. ArfGAP1) and are thought to be commonly associated membrane events early in the secretory cascade, while more charged amino acids are often involved in peripheral membrane targeting. In autophagy, proteins that propagate the nucleation, elongation, and completion stages of the autophagosome have each been described with curvature-dependent amphipathic helices including those that mimic ALPS motifs (human Atg14L) and those with more charged faces (human Atg3).
Additional membrane specificity can also be provided by interactions between charged lipid headgroups and the alpha helix’s hydrophilic face. The prevalence of charge on this face is often an indication that the helix will target a strongly anionic membrane and thus is likely used at peripheral structures (the plasma membrane or some endosomes). In contrast, Bruno Antonny and colleagues have described a mostly charge-free amphipathic helix termed the ALPS motif that is instead rich in polar serines and threonines 20; e.g. the helices on ArfGAP1 in Figure 4). This sort of helix has been implicated in targeting intracellular, charge-poor membranes like the golgi or ER. In autophagy, curvature-sensitive amphipathic helices have now been identified on multiple proteins operating both at the growing isolation membrane and also on distal compartments contributing to autophagosome growth and include both ALPS motifs (Atg14L) and charged helices (Atg3), perhaps consistent with biogenesis models that implicate multiple membrane sources.
Asymmetric insertion is also the curvature-sensing mechanism used by the reticulon fold (21 and Figure 3A), a pair of membrane-embedded helices that assemble into a protein wedge and naturally concentrate at highly curved regions of the endoplasmic reticulum. Reticulon motifs have not been implicated in autophagosome biogenesis, but likely do play a role in the recognition and targeting of certain cargo (described below).
Recruitment to the phagophore rim
An intriguing model for the utilization of curvature sensing during autophagosome biogenesis is that the proteins are recruited specifically to the phagophore rim (22 and Figure 4B). This targeting could serve two purposes: 1) to sculpt and stabilize this stridently bent membrane and 2) to temporally limit the protein activity to developing phagophores because this curvature will only exist on immature autophagosomes. Closed mature autophagosomes have membrane curvatures that appear flat at the single molecule or single protein level. Thus there is tremendous interest in defining the set of proteins unique to or at least enriched at the phagophore rim. However, despite extensive microscopy on autophagic structures, evidence for specific targeting remains limited.
The first demonstrated marker of the rim was actually not a protein, but rather a set of glycosylations typically found at the ER, Golgi and the late secretory compartment. Employing lectin immunochemistry on EM sections, Tashiro and colleagues showed that at the phagophore, lectins are enriched at or adjacent to the phagophore rim 23. They speculate this may represent a local pool of donor membrane sourced from the secretory pathway.
Fluorescence microscopy approaches have generally been unable to describe sub-domains of the phagophore, because in most cases, phagophores are less than ∼500 nm across (Figure 1) and thus very near the resolution limit of conventional microscopy. However, when selective autophagy targets very large cargoes, the phagophore must necessarily grow to larger dimensions. In a series of clever experiments, the Ohsumi group exploited this principle to begin to map subdomains in yeast. They developed a frustrated cytoplasm-to-vacuole targeting system (CVT) in which they force the CVT cargo ApeI to grow into a non-physiological extremely large oligomer 24. Under these conditions, phagophores growing around the oligomers extend out to a few micrometers and are ultimately unable to complete engulfment. In fluorescence images, these frustrated phagophores appear as cup-like structures. Some proteins including Atg8 decorate the whole of the structure while others define punctate subdomains, one along the broad convex surface that appears to delimit autophagosome-vacuole interaction sites and a second domain that is near the rim. Proteins localizing to the rim include Atg2, Atg18 and Atg9. How this targeting is determined is not yet clear, these proteins do not have described curvature-sensing motifs. Possibly they are targeted via interactions with other molecules. To this end, it is interesting that this site is also immediately adjacent to ER exit site (ERES) protein complexes on the ER and so perhaps organelle-organelle junctions are also playing a critical organizing role. Whether these protein localizations are physically on the rim and whether they are present along the entirety of the rim are not clear; the two-dimensional nature of conventional fluorescence microscopy simply resolves them as puncta. Future studies will hopefully include fluorescence nanoscopy approaches such as PALM/STORM25 and STED in three dimensions.
Purified autophagosomes from one particular species can have a relatively narrow distribution of sizes (e.g. 26), but it is clear that the typical autophagosome size varies dramatically across different species and cell types. Thus, it may also be possible to resolve rim-like distributions in species with naturally large phagophores. For example, Bianchi and co-workers have shown that the growing phagophore in the plant species Arabidopsis is sufficiently large to distinguish Atg8 and Atg5 labeling 27. Further, Atg5 demarcates a toroidal ring that first grows with the phagophore and then shrinks before disappearing altogether. In three-dimensional reconstructions they establish that this toroid is likely facing the ER throughout the phagophore growth, and they postulate that the toroid is in fact the limiting rim of the membrane. Consistent with Atg5’s known role as an E3-like complex for Atg8 lipidation, this localization could put the complex at the right place to recruit or activate local Atg3 (see below). Whether this result is generalizable to other systems is unclear however. Arabidopsis Atg5’s localization appears different from that of its mammalian counterparts, as both mouse Atg528 and Atg16L2 localize uniformly along the phagophore surface by immuno-EM. Likewise, live fluorescence imaging of Atg16L also suggests a continuous cup-shaped distribution29. A plausible alternative interpretation then could be that Arabidopsis Atg5 is forming a ring someplace else, for example around a region of membrane that marks the site of autophagosome efflux from the ER and includes most of the nascent cup-shaped membrane. Ultimately imaging conditions that reveal both the lipids and the proteins will be necessary to firmly establish the morphological context of this label.
Curvature sensing proteins in autophagy
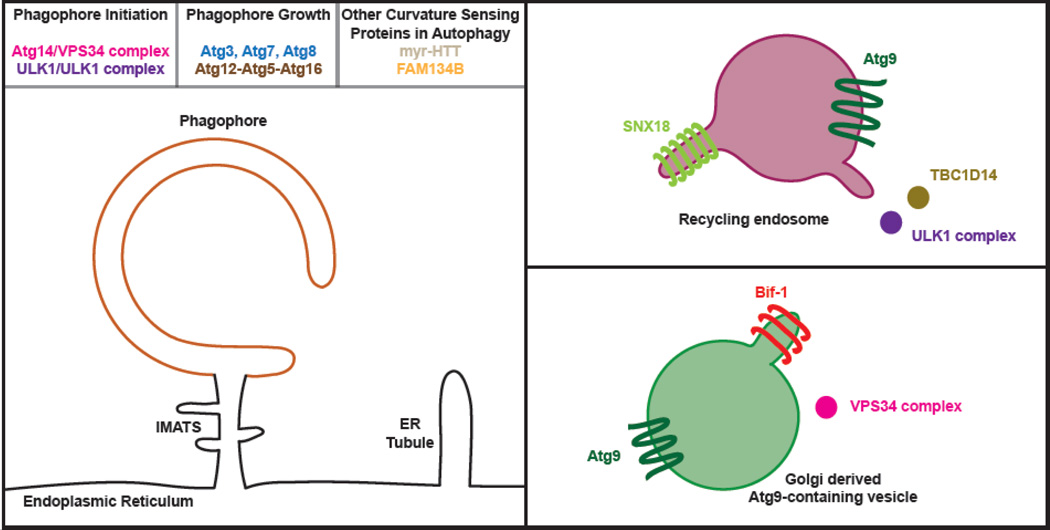
The majority of conserved autophagy-related (Atg) proteins were identified by genetic screens in the 1990’s, and comprise at least five functional complexes. In both yeast and mammalian cells, autophagy is initiated by the Atg1 kinase complex (ULK1 in mammals), which consists of Atg proteins 13, 29, 31, 17 and the eponymous Atg1. PI(3)P is generated locally by the Vps34 complex which includes Atg14, Vps15 and Atg6/Beclin. Phagophore biogenesis also requires the recruitment of Atg9-positive membranes. As the phagophore forms, two ubiquitin-related complexes become active; first the Atg5-Atg12/Atg16 complex scaffolds the maturing phagophore and second this complex promotes the recruitment and activation of the Atg7/Atg3 proteins driving the local lipidation of Atg8. Intriguingly, each of these complexes has one or more components that have been implicated in curvature sensing. In the following sections, we’ll discuss these examples including especially the in vitro mechanistic data that supports a role for these proteins at membrane curvature sites.
Atg9 vesicles and the Atg1 kinase complex
Atg9 is an integral membrane protein with six transmembrane alpha helices11. The protein localizes to several classes of membrane structures within the cell including especially upon clusters of very small vesicles and tubules30–33. These vesicles are highly mobile and in yeast appear to be the source of membrane used in the initial biogenesis of the phagophore assembly site30. In mammals, the direct role of the vesicles is less certain, but loss of Atg9-positive vesicles leads to a block in isolation membrane formation (e.g. 31). The origin of these vesicles appears to vary across species. In yeast, Atg9-GFP vesicles are enriched near the mitochondria or Golgi30, 32, while in HEK293 cells, transmission electron microscopy suggests mRFP-Atg9 containing vesicles and tubules emerge from many organelles including multivesicular structures and recycling endosomes (RE) 31, 34. Traffic out of the RE is largely controlled by Rab-specific tubular protrusions. In the case of Atg9, this involves the Rab GAP TBC1D14, which itself promotes tubulation34 and which also engages the TRAPP III complex to ensure trafficking of Atg9 back to peri-Golgi compartments35, ahead of delivery to the nascent autophagosome. Through a variety of imaging and biophysical approaches, Atg9-positive vesicles have generally been measured at about 30 nm in diameter, smaller than most other transport vesicles. This suggests Atg9 is present upon and/or may generate a highly curved vesicular intermediate important during phagophore growth.
The pentameric Atg1 complex in yeast, or the equivalent ULK complexes in mammals, are directly involved in the recruitment of Atg9 vesicles during early autophagosome membrane biogenesis. 36, 37. Atg1 itself has been shown to have curvature-dependent membrane binding 38 and derives at least in part from a motif termed the EAT domain (Early Autophagy Targeting/Tethering)39. In addition, the Atg1-binding protein Atg13 binds membranes of high curvature38. Atg1/Atg13 binds to Atg17/Atg29/Atg31 to form the pentameric complex which can assemble into a higher-order dimer39. In vitro, this complex also binds membranes of high curvature and is competent to tether liposomes together, including Atg9-bearing liposomes, but whether membrane-binding or only protein-protein interactions are principally involved in this tethering is unclear. In yeast, one consequence of tethering might be to promote the fusion of Atg9 vesicles, which appears to be the first step in phagophore formation30, however in mammals, correlative light electron microscopy and cryoimmuno-EM of HEK293 cells suggest mammalian Atg9s remain at tubular and vesicular structures and may not be integrated on autophagosome membranes31.
The biogenesis of these very small vesicles is only poorly understood. In principle, large trans-membrane proteins can influence the size and shape of the compartments into which they are delivered (e.g. 40, but see also 18 for review of mechanisms in which trans-membrane proteins influence membrane shape). In fact, overexpression of Atg9 in yeast causes the accumulation of small tubular-vesicular compartments30, 32. Likewise, Atg9 has a natural propensity to homo-oligomerize which appears to be required to support autophagy 41 and would further constrain the set of membrane architectures that could accomodate this protein. Atg9 is also enriched at LC3-decorated tubular vesicular structures in HEK293 cells42. Thus it is possible that this protein has a natural propensity for curved structures, but in practice, establishing such a property for a large integral membrane protein is very challenging.
Bif-1, also known as endophilin b1 or SH3P2, is an N-BAR domain containing protein that regulates Atg9 vesicle formation and trafficking 43. Takahashi et al. observed that Bif-1 requires its N-BAR domain to remain bound to Golgi or TGN compartments and to induce tubulation during starvation, presumably as part of the Atg9 vesicle formation43. This role is consistent with the general paradigm of BAR-containing proteins regulating fission events at vesicle biogenesis sites. Fission also requires the interaction between Bif-1 and Dynamin-2 through sequences outside of the N-BAR44. Bif-1 may remain on these vesicles as time-lapse microscopy suggests colocalization of Bif-1 and Atg9 (and possibly fusion of independent vesicles)45. These early vesicles are very small, highly curved, and appear to adopt a crescent shape as fusion proceeds46. In addition to the Golgi, Bif-1 has been localized to mitochondria47, 48, and intriguingly also to phagophore initiation sites where it engages Beclin1 during starvation48. Collectively, these results suggest that Bif-1 has a role either direct or indirect in sensing and sculpting the emerging phagophore shape45.
Vps34 complex
Macroautophagic processes require the modification of local inositol lipids. Of the inositol ring’s three hydroxyl groups available for individual or combinatorial phosphorylation, autophagy occurs in the context of PI lipid singly phosphorylated at the third hydroxyl group. This species is termed phosphatidylinositol-3-phosphate, or “PI(3)P”, for short. In yeast, PI(3)P is generated only by the class III PI(3)P kinase, Vps34, whose co-factors determine target membrane specificity. That is, one Vps34 complex generates PI(3)P at the vacuole, while a second Vps34 complex-containing Atg14, Vps15, and Atg6, recruits Vps34 to the PAS. In mammalian cells, Vps34 knockout reduces but does not inhibit autophagy. In Vps34−/− cells, the class II PI(3)P kinase, PI2K–C2 also contributes autophagic PI(3)P; however whether this enzyme is directly recruited to the phagophore remains unclear49. Sufficient PI(3)P generated by PI2K–C2 at another compartment and routed to phagophores may instead be sufficient to compensate for Vps34 knockout.
The Vps34 complex is therefore the best candidate to generate local, starvation-induced PI(3)P, as its activity is stimulated by ULK1 kinase activity50 and its localization determined in large part by autophagy-specific co-factors including Atg14 and Beclin 1. VPS34 itself harbors an amphipathic helix and this motif contributes membrane curvature-dependency to the catalytic activity of the enzyme in vitro 51. Mammalian Atg14L/Barkor, homolog of yeast Atg14, is a member of the autophagy-specific Vps34 kinase complex that generates PI(3)P signaling lipid. Barkor targets the PI3K complex to early autophagic membranes, via a Barkor/Atg14(L) autophagosome targeting sequence (BATS) encoded by the protein’s C-terminal 80 amino acids52. This sequence includes an amphipathic alpha helix with a sequence distribution reminiscent of the curvature-sensing ALPs motif first described by Bruno Antonny (Figure 3). Zhong and colleagues demonstrated that the BATS motif preferentially binds 100nm liposomes over 800nm liposomes (indicating curvature-sensing) and that overexpression of this motif drives the formation of LC3-positive tubules in cells. Thus the Atg14L BATS domain displays hallmark characteristics of both curvature sensing and promoting autophagy The BATS motif also has affinity for the negatively charged phospholipids, PI(3)P and PI(4,5)P2 and in fact the curvature-sensitivity of BATS is lost on liposomes carrying a high molar fraction of PI(3)P or PI(4,5)P2. These two modes of binding may be in natural competition within the cell as mutation of the hydrophobic face of the amphipathic alpha helix within BATS re-distributes the domain from LC3-positive tubules to the cytoplasm and to the plasma membrane where PI(4,5)P2 is at a high surface density.
Interestingly, Barkor has also recently been identified as a component of the Stx17 complex53 involved in autophagosome-lysosome fusion54, 55. Barkor can homooligomerize into a dimer and in this conformation, the BATS domains support liposome-liposome tethering. This tethering depends upon the presence of either high curvature in the liposomes or phosphoinositides. The stimulation of fusion requires this tethering activity but also direct interactions with the t-SNARE complex, possibly stabilizing a fusion-active conformation of the SNAREs.
Atg14L’s binding partner, Beclin also carries a critical membrane-binding motif (the β-α-repeat autophagy domain or BARAD) 56, 57. Included within this domain is a short finger loop with several aromatic residues that is both necessary and sufficient to target Beclin or other proteins to acidic liposomes in vitro. At high concentrations, this motif also appears to deform liposomes, though no specific evidence of curvature induction is yet available. Thus the VPS34 complex includes multiple membrane interaction surfaces. How these surfaces are oriented with respect to one another could impart a second level of curvature dependence, possibly making the autophagosome-directed Beclin-containing complex significantly more dependent on local curvature 58, as seen when comparing activity on small vesicles or GUVs 59. In fact, the complex ordinarily exists as at least a heteropentamer (including the subunit NRBF2) 60, 61, and the heteropentamer organizes into a dimer around a motif in NRBF2 62, potentially allowing membrane tethering to be sensed by or driven by the VPS34 complex during autophagosome construction.
Atg5-Atg12/Atg16 complex
While the above complexes are thought to function predominantly during autophagy initiation, the Atg5-Atg12/Atg16 complex functions at later stages coincident with membrane expansion. Atg12 is a ubiquitin-like protein that becomes covalently attached to Atg53 (or in an unrelated complex with Atg34). The Atg5-Atg12 dimer binds Atg16L1 which itself can dimerize, such that the final membrane-associated complex is probably at least six unique polypeptides and possibly more (biochemistry suggests it forms a tetramer of heterotrimers 63, 64). This complex is proposed to support autophagosome growth by either direct activation of the lipidating enzyme Atg37 (via capture of Atg3 at autophagic membranes65, 66 or the induction of an active conformation on the enzyme 67), or through membrane scaffolding (where multimerization may shape autophagosomes 68). In either case, the complex must recognize a membrane and promote LC3 family lipidation(either directly or indirectly), driving the transition to autophagosome growth.
Yeast Atg5-Atg12/Atg16 binds membranes directly, but somewhat weakly, through motifs on Atg569, and is strongly recruited to Atg8-PE decorated membranes via an Atg8 interaction motif on Atg1268. Both sites appear to be crucial to support autophagy in vivo. In mammals, several other membrane-bound proteins also engage the complex and may influence its targeting. For example, WIPI2B is an Atg16L1-binding protein that also binds PI3P, is implicated in promoting LC3-lipidattion and is essential for growth of the autophagosome70. Atg16L1 also engages lipid-modified Rab proteins (especially Rab33)71, that may contribute membrane targeting.
The ability to form higher-order oligomers is suggestive of a coat-like behavior. Two excellent in vitro studies on Giant Unilamellar Vesicles (GUVs) have looked at how and whether membrane-associated Atg5-Atg12/Atg16 complex from yeast influences membrane structure68, 69. They have discovered that the Atg5-Atg12/Atg16 complex is sufficient to tether liposomes, suggesting an available interface even when bound to one membrane, 2) engages Atg8-PE and competes with cargo adaptors and Atg4 for Atg8-PE access, 3) organizes into very large higher-order oligomers in an Atg16 dimer-dependent manner, 4) rigidifies GUV membranes, creating deformations with very large radii of curvatures (on the micron scale) and thus stabilizing a molecularly-flat organization. Because mutants that disrupt the higher-order oligomers also impair autophagic flux in cells, Wollert and colleagues suggest that the ability to form an essentially flat architecture is a critical feature of phagophore development68. Importantly, this architecture retains sufficient plasticity to scaffold the wide-range of phagophores forming under different stress and cargo capture conditions.
Lipidation of Atg8 or LC3
Atg8 becomes covalently attached to the lipid phosphatidylethanolamine through ubiquitin like reactions involving the E1-like Atg7 and the E2-like Atg3. In mammals there are 6–8 homologs of Atg8, including LC3, GATE-16, GABARAP and GABARAP-L1 and each is also modified by PE through Atg7- and Atg3-dependent reactions. If lipidation is inhibited, autophagy largely aborts at the cup-shaped intermediate membrane. In fact, tomographic reconstructions of phagophores are often collected under these conditions because phagophores accumulate (e.g. 2). Thus, this structure, with its highly curved rim present, is likely the physiologic target of Atg3-dependent lipidation. In vitro, LC3 lipidation is highly curvature-sensitive thanks to an amphipathic alpha helix comprising most of the amino-terminus of Atg3 (Atg3helix)72. The helix is distinct from the ALPS motif and instead has two clear membrane delimiting lysines that likely engage lipid headgroups along with a poor hydrophobic face (Figure 4). Mutations introducing negative charge into the hydrophobic face of Atg3helix abrogated Atg3 liposome binding in vitro, and Atg3’s enzymatic activity in vitro and in vivo. Expanding the hydrophobic face of the helix makes Atg3 rely less on membrane defects and strongly supports a model where this helix is ordinarily tuned to a narrow range of lipid or membrane structural characteristics. A similar curvature dependency is found with the yeast Atg3 suggesting this sensitivity is conserved across organisms73. Although the amphipathic character of this helix is essential in vivo, where this amphipathic helix is used in cells is less clear. Attempts to localize Atg3 on membranes in mammalian cells have been largely unsuccessful, perhaps suggesting that Atg3 is only transiently associated with the developing autophagosome. In contrast, yeast Atg3 localizes uniformly along the phagophore surface74, 75. Furthermore Atg3 function in both species is dependent upon the Atg5-Atg12/Atg16 complex, and lipidation can be driven at heterologous membranes when this complex is artificially mislocalized 65. The Atg3helix may therefore primarily function as a sensor of the local membrane environment rather than as an essential targeting motif and limit lipidation to either curved portions of the phagophore or to unique lipid compositions consistent with autophagosome biogenesis.
Unlike all of the other proteins discussed above, Atg8-PE or LC3-PE represents a covalent linkage to the growing membrane and thus a significant body of work has centered on describing whether this unique protein-lipid adduct possesses inherent membrane remodeling capabilities. In a landmark paper on reconstitution, Atg8-PE was demonstrated to tether liposomes and even to drive hemi-fusion76. Subsequent papers established similar activities for LC3, GATE-16, and GABARAP-L177–80. Fusion however appears to be limited to membranes of very high and non-physiological cone-shaped lipid compositions as neither LC3-PE nor Atg8-PE are apparently fusion active on other mixtures69, 73, 78. Because we do not yet know the lipid composition of the phagophore it is impossible to predict whether fusion-promoting lipids are present, but intriguingly, highly curved membranes display the same physical characteristics that lower the fusion barrier (Figure 3B). Thus it is possible that Atg8-PE is simply only fusion-active at the rim of the phagophore or on highly curved donor vesicles and tubules. Within cells, LC3-PE can be found associated with membrane tubules formed by either BAR-domain proteins (e.g. 81) or phagophore-directed amphipathic helices (e.g. 82). In particular, the Snx-BAR protein Snx18 drives LC3-positive tubule formation at recycling endosomes and also coordinates the Atg5-Atg12/Atg16 complex at these sites, suggesting local membrane tubulation might be involved in autophagosome biogenesis at this site81, 83. Snx-BAR proteins have also been implicated to act directly upon the phagophore itself84. Enrichment of LC3-PE or Atg8-PE at the phagophore rim has not been observed in fluorescence or immuno-EM studies, but modest enrichment would be challenging to detect as we expect a high concentration of the protein to decorate the inner membrane where it engages cargo and on the outer membrane where cytoplasmic proteins are engaged. Indeed, Dimova and colleagues studied Atg8-PE partitioning on Giant Unilamellar Vesiclesj (GUVs), when both curved and flat membranes were available and suggest that enrichment will be only relatively modest 73. They observed that Atg8-PE partitions into curved membranes, but total enrichment is only about 2–3 fold on highly curved tubules compared to the molecularly flat surfaces of GUV membranes. Thus collectively, in vitro experiments are consistent with Atg8-PE (and LC3-PE) having the capacity to sense curvature and possibly to play a direct role in membrane remodeling but cellular relevance for these activities remain uncertain. What is lacking, fundamentally, is an example of an Atg8-PE mutant that accumulates at a tethered interface in cells, but does not support the fusion or fission reaction. This phenotype would establish which membranes are joined by Atg8-PE and at which point in autophagosome maturation such a tethering activity is required.
Autophagic Cargo
Why certain organelles or organelle subdomains are marked for macroautophagic clearance is not yet clear, but it is likely that this stage of quality control depends upon recognizing changes in both the protein and potentially the membrane components of these organelles. In this sense, we might expect that structural cues on the membrane, including those that sense whether the overall architecture is maintained, will play a vital role in initiating autophagic clearance. Perhaps the most structurally heterogeneous organelle in the cell is the endoplasmic reticulum, which consists of large areas of fully reticulated membrane, as well as relatively coherent and flattened sheets and elongated individual tubular protrusions. In a pair of exciting back-to-back papers last year, the Nakatagowa and Dikic labs described autophagy receptors for the clearance of ER from yeast and mammalian systems respectively12, 85. In yeast, two unrelated protein receptors were described that preferentially localized to either flattened peri-nuclear ER (Atg39) or to cortical tubular ER (Atg40)12. Atg40 includes a reticulon-like domain (Figure 3) and colocalizes with reticulon on tubular structures. Moreover, EM suggests that the autophagic bodies delivered by Atg40 are laden with complicated, often tubular, membrane material. All told, this suggests that Atg40-dependent ER-phagy is specific for the turnover of highly curved tubular ER. Intriguingly, although unrelated at the sequence level, the mammalian ER-phagy receptor discovered by the Dikic lab (FAM134B) also encodes a reticulon-like sequence85. Liposome experiments reveal that this sequence supports membrane remodeling in vitro, however in vivo the protein targets to both the flattened and tubular regions of the ER. The authors speculate that FAM134B plays a vital role in maintaining overall ER homeostasis, including the proper balance of architecturally distinct regions of the organelle.
Other phosphoinositide-binding proteins with in vitro curvature sensitivity
The Legionella pneumophila effector protein, RavZ, is an LC3-PE directed protease that inhibits autophagy by removing LC3 from the developing autophagosome86. A noteworthy feature of this activity is that RavZ does not cleave the soluble proform of LC3, and thus is exquisitely specific for the PE-modified form of LC3. This selectivity is encoded as multiple membrane-recognition motifs including a very short amphipathic helical motif, a unique PI3P binding protein fold, and general preference for membrane curvature87, thus RavZ encodes motifs that recognize each of the major membrane characteristics of the developing phagophore, aborting autophagy at the open cup-shaped membrane.
Likewise, the WIPI family of proteins also bind phosphoinositides to direct these proteins to early events in autophagic processes. In yeast, there are three members of this family, Atg18, Atg21, and Hsv2. Each has been shown to bind membranes via two phosphoinositide binding sites plus other sequences that insert in the membrane88. Hsv2 has also been examined on liposomes of different size and revealed to have a more than 10-fold increase in apparent affinity when the membranes become highly curved 89. The mechanism of curvature recognition for both RavZ and Hsv2 remains uncertain.
Future Perspectives
Phagophore biogenesis remains poorly understood. If this cup-like structure forms from the fusion of a few vesicles, then perhaps the crescent shape and the negligible luminal volume are simply a natural consequence of satisfying the changing volume to surface area as the organelle grows. In contrast, if the organelle is extruded from a pre-existing membrane, then it is likely that the lipid composition will impact membrane architecture, either by influencing the recruitment and activity of curvature-sensitive proteins (as in Figure 2) or by directly determining organelle structure (e.g. 90). Thus establishing the lipid composition of the developing membrane is a priority. Likewise, to the extent that proteins are involved, we remain relatively ignorant about the distribution of these proteins along the organelle membrane. The introduction of live fluorescence nanoscopy approaches should provide the first indications of protein distribution during organelle growth. These studies would benefit tremendously from fluorescent lipid analogs that collect at the phagophore and provide structural context to the protein distributions.
Research Highlights.
Autophagosome biogenesis involves significant structural remodeling of cellular membranes.
Proteins target these unique morphologies via curvature-sensing motifs.
Each of the major autophagy complexes utilizes one or more curvature-sensitive proteins.
Acknowledgments
This work was financially supported by the NIH (GM1000930, TJM; T32GM007223 and T32GM007223-S1, a training grant to NN and VS). Thank you also to Dr. Julia Dancourt who helped survey the literature leading to the production of Table I.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Citations
- 1.Deter RL, De Duve C. Influence of glucagon, an inducer of cellular autophagy, on some physical properties of rat liver lysosomes. The Journal of cell biology. 1967;33:437–449. doi: 10.1083/jcb.33.2.437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. A subdomain of the endoplasmic reticulum forms a cradle for autophagosome formation. Nat Cell Biol. 2009;11:1433–1437. doi: 10.1038/ncb1991. [DOI] [PubMed] [Google Scholar]
- 3.Israelachvili JN, Mitchell DJ. A model for the packing of lipids in bilayer membranes. Biochim Biophys Acta. 1975;389:13–19. doi: 10.1016/0005-2736(75)90381-8. [DOI] [PubMed] [Google Scholar]
- 4.Uemura T, Yamamoto M, Kametaka A, Sou YS, Yabashi A, Yamada A, Annoh H, Kametaka S, Komatsu M, Waguri S. A cluster of thin tubular structures mediates transformation of the endoplasmic reticulum to autophagic isolation membrane. Mol Cell Biol. 2014;34:1695–1706. doi: 10.1128/MCB.01327-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yla-Anttila P, Vihinen H, Jokitalo E, Eskelinen EL. 3D tomography reveals connections between the phagophore and endoplasmic reticulum. Autophagy. 2009;5:1180–1185. doi: 10.4161/auto.5.8.10274. [DOI] [PubMed] [Google Scholar]
- 6.Hayashi-Nishino M, Fujita N, Noda T, Yamaguchi A, Yoshimori T, Yamamoto A. Electron tomography reveals the endoplasmic reticulum as a membrane source for autophagosome formation. Autophagy. 2009;6:301–303. doi: 10.4161/auto.6.2.11134. [DOI] [PubMed] [Google Scholar]
- 7.Rez G, Meldolesi J. Freeze-fracture of drug-induced autophagocytosis in the mouse exocrine pancreas. Laboratory investigation; a journal of technical methods and pathology. 1980;43:269–277. [PubMed] [Google Scholar]
- 8.Hirsimaki Y, Hirsimaki P, Lounatmaa K. Vinblastine-induced autophagic vacuoles in mouse liver and Ehrlich ascites tumor cells as assessed by freeze-fracture electron microscopy. European journal of cell biology. 1982;27:298–301. [PubMed] [Google Scholar]
- 9.Cheng J, Fujita A, Yamamoto H, Tatematsu T, Kakuta S, Obara K, Ohsumi Y, Fujimoto T. Yeast and mammalian autophagosomes exhibit distinct phosphatidylinositol 3-phosphate asymmetries. Nat Commun. 2014;5:3207. doi: 10.1038/ncomms4207. [DOI] [PubMed] [Google Scholar]
- 10.Proikas-Cezanne T, Robenek H. Freeze-fracture replica immunolabelling reveals human WIPI-1 and WIPI-2 as membrane proteins of autophagosomes. J Cell Mol Med. 2011;15:2007–2010. doi: 10.1111/j.1582-4934.2011.01339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Webber JL, Tooze SA. New insights into the function of Atg9. FEBS letters. 2010;584:1319–1326. doi: 10.1016/j.febslet.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Mochida K, Oikawa Y, Kimura Y, Kirisako H, Hirano H, Ohsumi Y, Nakatogawa H. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature. 2015;522:359–362. doi: 10.1038/nature14506. [DOI] [PubMed] [Google Scholar]
- 13.Proikas-Cezanne T, Waddell S, Gaugel A, Frickey T, Lupas A, Nordheim A. WIPI-1alpha (WIPI49), a member of the novel 7-bladed WIPI protein family, is aberrantly expressed in human cancer and is linked to starvation-induced autophagy. Oncogene. 2004;23:9314–9325. doi: 10.1038/sj.onc.1208331. [DOI] [PubMed] [Google Scholar]
- 14.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Antonny B. Mechanisms of Membrane Curvature Sensing. Annu Rev Biochem. 2010 doi: 10.1146/annurev-biochem-052809-155121. [DOI] [PubMed] [Google Scholar]
- 16.Drin G, Antonny B. Amphipathic helices and membrane curvature. FEBS letters. 2010;584:1840–1847. doi: 10.1016/j.febslet.2009.10.022. [DOI] [PubMed] [Google Scholar]
- 17.Bhatia VK, Hatzakis NS, Stamou D. A unifying mechanism accounts for sensing of membrane curvature by BAR domains, amphipathic helices and membrane-anchored proteins. Semin Cell Dev Biol. 2010;21:381–390. doi: 10.1016/j.semcdb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 18.McMahon HT, Boucrot E. Membrane curvature at a glance. Journal of cell science. 2015;128:1065–1070. doi: 10.1242/jcs.114454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zimmerberg J, McLaughlin S. Membrane curvature: how BAR domains bend bilayers. Curr Biol. 2004;14:R250–R252. doi: 10.1016/j.cub.2004.02.060. [DOI] [PubMed] [Google Scholar]
- 20.Bigay J, Casella JF, Drin G, Mesmin B, Antonny B. ArfGAP1 responds to membrane curvature through the folding of a lipid packing sensor motif. The EMBO journal. 2005;24:2244–2253. doi: 10.1038/sj.emboj.7600714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goyal U, Blackstone C. Untangling the web: mechanisms underlying ER network formation. Biochim Biophys Acta. 2013;1833:2492–2498. doi: 10.1016/j.bbamcr.2013.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dancourt J, Melia TJ. Lipidation of the autophagy proteins LC3 and GABARAP is a membrane-curvature dependent process. Autophagy. 2014;10:1470–1471. doi: 10.4161/auto.29468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamamoto A, Masaki R, Tashiro Y. Characterization of the isolation membranes and the limiting membranes of autophagosomes in rat hepatocytes by lectin cytochemistry. J Histochem Cytochem. 1990;38:573–580. doi: 10.1177/38.4.2319125. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki K, Akioka M, Kondo-Kakuta C, Yamamoto H, Ohsumi Y. Fine mapping of autophagy-related proteins during autophagosome formation in Saccharomyces cerevisiae. Journal of cell science. 2013;126:2534–2544. doi: 10.1242/jcs.122960. [DOI] [PubMed] [Google Scholar]
- 25.Huang F, Sirinakis G, Allgeyer ES, Schroeder LK, Duim WC, Kromann EB, Phan T, Rivera-Molina FE, Myers JR, Irnov I, Lessard M, Zhang Y, Handel MA, Jacobs-Wagner C, Lusk CP, Rothman JE, Toomre D, Booth MJ, Bewersdorf J. Ultra-High Resolution 3D Imaging of Whole Cells. Cell. 2016;166:1028–1040. doi: 10.1016/j.cell.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seglen PO, Brinchmann MF. Purification of autophagosomes from rat hepatocytes. Autophagy. 2010;6:542–547. doi: 10.4161/auto.6.4.11272. [DOI] [PubMed] [Google Scholar]
- 27.Le Bars R, Marion J, Le Borgne R, Satiat-Jeunemaitre B, Bianchi MW. ATG5 defines a phagophore domain connected to the endoplasmic reticulum during autophagosome formation in plants. Nat Commun. 2014;5:4121. doi: 10.1038/ncomms5121. [DOI] [PubMed] [Google Scholar]
- 28.Mizushima N, Yamamoto A, Hatano M, Kobayashi Y, Kabeya Y, Suzuki K, Tokuhisa T, Ohsumi Y, Yoshimori T. Dissection of autophagosome formation using Apg5-deficient mouse embryonic stem cells. The Journal of cell biology. 2001;152:657–668. doi: 10.1083/jcb.152.4.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mizushima N, Kuma A, Kobayashi Y, Yamamoto A, Matsubae M, Takao T, Natsume T, Ohsumi Y, Yoshimori T. Mouse Apg16L, a novel WD-repeat protein, targets to the autophagic isolation membrane with the Apg12-Apg5 conjugate. Journal of cell science. 2003;116:1679–1688. doi: 10.1242/jcs.00381. [DOI] [PubMed] [Google Scholar]
- 30.Yamamoto H, Kakuta S, Watanabe TM, Kitamura A, Sekito T, Kondo-Kakuta C, Ichikawa R, Kinjo M, Ohsumi Y. Atg9 vesicles are an important membrane source during early steps of autophagosome formation. The Journal of cell biology. 2012;198:219–233. doi: 10.1083/jcb.201202061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orsi A, Razi M, Dooley HC, Robinson D, Weston AE, Collinson LM, Tooze SA. Dynamic and transient interactions of Atg9 with autophagosomes, but not membrane integration, are required for autophagy. Mol Biol Cell. 2012;23:1860–1873. doi: 10.1091/mbc.E11-09-0746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mari M, Griffith J, Rieter E, Krishnappa L, Klionsky DJ, Reggiori F. An Atg9-containing compartment that functions in the early steps of autophagosome biogenesis. The Journal of cell biology. 2010;190:1005–1022. doi: 10.1083/jcb.200912089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Young AR, Chan EY, Hu XW, Kochl R, Crawshaw SG, High S, Hailey DW, Lippincott-Schwartz J, Tooze SA. Starvation and ULK1-dependent cycling of mammalian Atg9 between the TGN and endosomes. Journal of cell science. 2006;119:3888–3900. doi: 10.1242/jcs.03172. [DOI] [PubMed] [Google Scholar]
- 34.Longatti A, Lamb CA, Razi M, Yoshimura S, Barr FA, Tooze SA. TBC1D14 regulates autophagosome formation via Rab11- and ULK1-positive recycling endosomes. The Journal of cell biology. 2012;197:659–675. doi: 10.1083/jcb.201111079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lamb CA, Nuhlen S, Judith D, Frith D, Snijders AP, Behrends C, Tooze SA. TBC1D14 regulates autophagy via the TRAPP complex and ATG9 traffic. The EMBO journal. 2016;35:281–301. doi: 10.15252/embj.201592695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reggiori F, Tucker KA, Stromhaug PE, Klionsky DJ. The Atg1-Atg13 complex regulates Atg9 and Atg23 retrieval transport from the pre-autophagosomal structure. Developmental cell. 2004;6:79–90. doi: 10.1016/s1534-5807(03)00402-7. [DOI] [PubMed] [Google Scholar]
- 37.Papinski D, Schuschnig M, Reiter W, Wilhelm L, Barnes CA, Maiolica A, Hansmann I, Pfaffenwimmer T, Kijanska M, Stoffel I, Lee SS, Brezovich A, Lou JH, Turk BE, Aebersold R, Ammerer G, Peter M, Kraft C. Early steps in autophagy depend on direct phosphorylation of Atg9 by the Atg1 kinase. Mol Cell. 2014;53:471–483. doi: 10.1016/j.molcel.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rao Y, Perna MG, Hofmann B, Beier V, Wollert T. The Atg1-kinase complex tethers Atg9-vesicles to initiate autophagy. Nat Commun. 2016;7:10338. doi: 10.1038/ncomms10338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ragusa MJ, Stanley RE, Hurley JH. Architecture of the Atg17 complex as a scaffold for autophagosome biogenesis. Cell. 2012;151:1501–1512. doi: 10.1016/j.cell.2012.11.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 41.He C, Baba M, Cao Y, Klionsky DJ. Self-interaction is critical for Atg9 transport and function at the phagophore assembly site during autophagy. Mol Biol Cell. 2008;19:5506–5516. doi: 10.1091/mbc.E08-05-0544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gao W, Kang JH, Liao Y, Ding WX, Gambotto AA, Watkins SC, Liu YJ, Stolz DB, Yin XM. Biochemical isolation and characterization of the tubulovesicular LC3-positive autophagosomal compartment. The Journal of biological chemistry. 2010;285:1371–1383. doi: 10.1074/jbc.M109.054197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Takahashi Y, Meyerkord CL, Hori T, Runkle K, Fox TE, Kester M, Loughran TP, Wang HG. Bif-1 regulates Atg9 trafficking by mediating the fission of Golgi membranes during autophagy. Autophagy. 2011;7:61–73. doi: 10.4161/auto.7.1.14015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Takahashi Y, Tsotakos N, Liu Y, Young MM, Serfass J, Tang Z, Abraham T, Wang HG. The Bif-1-Dynamin 2 membrane fission machinery regulates Atg9-containing vesicle generation at the Rab11-positive reservoirs. Oncotarget. 2016;7:20855–20868. doi: 10.18632/oncotarget.8028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Takahashi Y, Meyerkord CL, Wang HG. BARgaining membranes for autophagosome formation: Regulation of autophagy and tumorigenesis by Bif-1/Endophilin B1. Autophagy. 2008;4:121–124. doi: 10.4161/auto.5265. [DOI] [PubMed] [Google Scholar]
- 46.Takahashi Y, Meyerkord CL, Wang HG. Bif-1/endophilin B1: a candidate for crescent driving force in autophagy. Cell death and differentiation. 2009;16:947–955. doi: 10.1038/cdd.2009.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takahashi Y, Karbowski M, Yamaguchi H, Kazi A, Wu J, Sebti SM, Youle RJ, Wang HG. Loss of Bif-1 suppresses Bax/Bak conformational change and mitochondrial apoptosis. Mol Cell Biol. 2005;25:9369–9382. doi: 10.1128/MCB.25.21.9369-9382.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Farsad K, Ringstad N, Takei K, Floyd SR, Rose K, De Camilli P. Generation of high curvature membranes mediated by direct endophilin bilayer interactions. The Journal of cell biology. 2001;155:193–200. doi: 10.1083/jcb.200107075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Devereaux K, Dall’Armi C, Alcazar-Roman A, Ogasawara Y, Zhou X, Wang F, Yamamoto A, De Camilli P, Di Paolo G. Regulation of mammalian autophagy by class II and III PI 3-kinases through PI3P synthesis. PLoS One. 2013;8:e76405. doi: 10.1371/journal.pone.0076405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Russell RC, Tian Y, Yuan H, Park HW, Chang YY, Kim J, Kim H, Neufeld TP, Dillin A, Guan KL. ULK1 induces autophagy by phosphorylating Beclin-1 and activating VPS34 lipid kinase. Nat Cell Biol. 2013;15:741–750. doi: 10.1038/ncb2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Miller S, Tavshanjian B, Oleksy A, Perisic O, Houseman BT, Shokat KM, Williams RL. Shaping development of autophagy inhibitors with the structure of the lipid kinase Vps34. Science (New York, N.Y. 2010;327:1638–1642. doi: 10.1126/science.1184429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan W, Nassiri A, Zhong Q. Autophagosome targeting and membrane curvature sensing by Barkor/Atg14(L) Proceedings of the National Academy of Sciences of the United States of America. 2011;108:7769–7774. doi: 10.1073/pnas.1016472108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diao J, Liu R, Rong Y, Zhao M, Zhang J, Lai Y, Zhou Q, Wilz LM, Li J, Vivona S, Pfuetzner RA, Brunger AT, Zhong Q. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 2015;520:563–566. doi: 10.1038/nature14147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang P, Nishimura T, Sakamaki Y, Itakura E, Hatta T, Natsume T, Mizushima N. The HOPS complex mediates autophagosome-lysosome fusion through interaction with syntaxin 17. Mol Biol Cell. 2014;25:1327–1337. doi: 10.1091/mbc.E13-08-0447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Itakura E, Kishi-Itakura C, Mizushima N. The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 2012;151:1256–1269. doi: 10.1016/j.cell.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 56.Fogel AI, Dlouhy BJ, Wang C, Ryu SW, Neutzner A, Hasson SA, Sideris DP, Abeliovich H, Youle RJ. Role of membrane association and Atg14-dependent phosphorylation in beclin-1-mediated autophagy. Mol Cell Biol. 2013;33:3675–3688. doi: 10.1128/MCB.00079-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang W, Choi W, Hu W, Mi N, Guo Q, Ma M, Liu M, Tian Y, Lu P, Wang FL, Deng H, Liu L, Gao N, Yu L, Shi Y. Crystal structure and biochemical analyses reveal Beclin 1 as a novel membrane binding protein. Cell Res. 2012;22:473–489. doi: 10.1038/cr.2012.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mei Y, Su M, Sanishvili R, Chakravarthy S, Colbert CL, Sinha SC. Identification of BECN1 and ATG14 Coiled-Coil Interface Residues That Are Important for Starvation-Induced Autophagy. Biochemistry. 2016;55:4239–4253. doi: 10.1021/acs.biochem.6b00246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rostislavleva K, Soler N, Ohashi Y, Zhang L, Pardon E, Burke JE, Masson GR, Johnson C, Steyaert J, Ktistakis NT, Williams RL. Structure and flexibility of the endosomal Vps34 complex reveals the basis of its function on membranes. Science (New York, N.Y. 2015;350:aac7365. doi: 10.1126/science.aac7365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lu J, He L, Behrends C, Araki M, Araki K, Jun Wang Q, Catanzaro JM, Friedman SL, Zong WX, Fiel MI, Li M, Yue Z. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L–linked phosphatidylinositol-3 kinase III activity. Nat Commun. 2014;5:3920. doi: 10.1038/ncomms4920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao Y, Wang Y, Abi Saab WF, Yang F, Pessin JE, Backer JM. NRBF2 regulates macroautophagy as a component of Vps34 Complex I. The Biochemical journal. 2014;461:315–322. doi: 10.1042/BJ20140515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Young LN, Cho K, Lawrence R, Zoncu R, Hurley JH. Dynamics and architecture of the NRBF2-containing phosphatidylinositol 3-kinase complex I of autophagy. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:8224–8229. doi: 10.1073/pnas.1603650113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fujioka Y, Noda NN, Nakatogawa H, Ohsumi Y, Inagaki F. Dimeric coiled-coil structure of Saccharomyces cerevisiae Atg16 and its functional significance in autophagy. The Journal of biological chemistry. 2010;285:1508–1515. doi: 10.1074/jbc.M109.053520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuma A, Mizushima N, Ishihara N, Ohsumi Y. Formation of the approximately 350-kDa Apg12-Apg5.Apg16 multimeric complex, mediated by Apg16 oligomerization, is essential for autophagy in yeast. The Journal of biological chemistry. 2002;277:18619–18625. doi: 10.1074/jbc.M111889200. [DOI] [PubMed] [Google Scholar]
- 65.Fujita N, Itoh T, Omori H, Fukuda M, Noda T, Yoshimori T. The Atg16L complex specifies the site of LC3 lipidation for membrane biogenesis in autophagy. Mol Biol Cell. 2008;19:2092–2100. doi: 10.1091/mbc.E07-12-1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Walczak M, Martens S. Dissecting the role of the Atg12-Atg5-Atg16 complex during autophagosome formation. Autophagy. 2013;9:424–425. doi: 10.4161/auto.22931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Otomo C, Metlagel Z, Takaesu G, Otomo T. Structure of the human ATG12∼ATG5 conjugate required for LC3 lipidation in autophagy. Nat Struct Mol Biol. 2013;20:59–66. doi: 10.1038/nsmb.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Kaufmann A, Beier V, Franquelim HG, Wollert T. Molecular mechanism of autophagic membrane-scaffold assembly and disassembly. Cell. 2014;156:469–481. doi: 10.1016/j.cell.2013.12.022. [DOI] [PubMed] [Google Scholar]
- 69.Romanov J, Walczak M, Ibiricu I, Schuchner S, Ogris E, Kraft C, Martens S. Mechanism and functions of membrane binding by the Atg5-Atg12/Atg16 complex during autophagosome formation. The EMBO journal. 2012;31:4304–4317. doi: 10.1038/emboj.2012.278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dooley HC, Wilson MI, Tooze SA. WIPI2B links PtdIns3P to LC3 lipidation through binding ATG16L1. Autophagy. 2015;11:190–191. doi: 10.1080/15548627.2014.996029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Itoh T, Fujita N, Kanno E, Yamamoto A, Yoshimori T, Fukuda M. Golgi-resident small GTPase Rab33B interacts with Atg16L and modulates autophagosome formation. Mol Biol Cell. 2008;19:2916–2925. doi: 10.1091/mbc.E07-12-1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nath S, Dancourt J, Shteyn V, Puente G, Fong WM, Nag S, Bewersdorf J, Yamamoto A, Antonny B, Melia TJ. Lipidation of the LC3/GABARAP family of autophagy proteins relies on a membrane-curvature-sensing domain in Atg3. Nat Cell Biol. 2014;16:415–424. doi: 10.1038/ncb2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Knorr RL, Nakatogawa H, Ohsumi Y, Lipowsky R, Baumgart T, Dimova R. Membrane morphology is actively transformed by covalent binding of the protein Atg8 to PE-lipids. PLoS One. 2014;9:e115357. doi: 10.1371/journal.pone.0115357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sakoh-Nakatogawa M, Kirisako H, Nakatogawa H, Ohsumi Y. Localization of Atg3 to autophagy-related membranes and its enhancement by the Atg8-family interacting motif to promote expansion of the membranes. FEBS letters. 2015;589:744–749. doi: 10.1016/j.febslet.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 75.Ngu M, Hirata E, Suzuki K. Visualization of Atg3 during autophagosome formation in Saccharomyces cerevisiae. The Journal of biological chemistry. 2015;290:8146–8153. doi: 10.1074/jbc.M114.626952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nakatogawa H, Ichimura Y, Ohsumi Y. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 2007;130:165–178. doi: 10.1016/j.cell.2007.05.021. [DOI] [PubMed] [Google Scholar]
- 77.Jotwani A, Richerson DN, Motta I, Julca-Zevallos O, Melia TJ. Approaches to the study of Atg8-mediated membrane dynamics in vitro. Methods Cell Biol. 2012;108:93–116. doi: 10.1016/B978-0-12-386487-1.00005-5. [DOI] [PubMed] [Google Scholar]
- 78.Nair U, Jotwani A, Geng J, Gammoh N, Richerson D, Yen WL, Griffith J, Nag S, Wang K, Moss T, Baba M, McNew JA, Jiang X, Reggiori F, Melia TJ, Klionsky DJ. SNARE Proteins Are Required for Macroautophagy. Cell. 2011;146:290–302. doi: 10.1016/j.cell.2011.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Weidberg H, Shpilka T, Shvets E, Abada A, Shimron F, Elazar Z. LC3 and GATE-16 N Termini Mediate Membrane Fusion Processes Required for Autophagosome Biogenesis. Developmental cell. 2011;20:444–454. doi: 10.1016/j.devcel.2011.02.006. [DOI] [PubMed] [Google Scholar]
- 80.Landajuela A, Hervas JH, Anton Z, Montes LR, Gil D, Valle M, Rodriguez JF, Goni FM, Alonso A. Lipid Geometry and Bilayer Curvature Modulate LC3/GABARAP-Mediated Model Autophagosomal Elongation. Biophys J. 2016;110:411–422. doi: 10.1016/j.bpj.2015.11.3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Knaevelsrud H, Soreng K, Raiborg C, Haberg K, Rasmuson F, Brech A, Liestol K, Rusten TE, Stenmark H, Neufeld TP, Carlsson SR, Simonsen A. Membrane remodeling by the PX-BAR protein SNX18 promotes autophagosome formation. The Journal of cell biology. 2013;202:331–349. doi: 10.1083/jcb.201205129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martin DD, Heit RJ, Yap MC, Davidson MW, Hayden MR, Berthiaume LG. Identification of a post-translationally myristoylated autophagy-inducing domain released by caspase cleavage of huntingtin. Human molecular genetics. 2014;23:3166–3179. doi: 10.1093/hmg/ddu027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Knaevelsrud H, Carlsson SR, Simonsen A. SNX18 tubulates recycling endosomes for autophagosome biogenesis. Autophagy. 2013;9:1639–1641. doi: 10.4161/auto.26124. [DOI] [PubMed] [Google Scholar]
- 84.Zhao D, Liu XM, Yu ZQ, Sun LL, Xiong X, Dong MQ, Du LL. Atg20 and Atg24 family proteins promote organelle autophagy in fission yeast. Journal of cell science. 2016 doi: 10.1242/jcs.194373. [DOI] [PubMed] [Google Scholar]
- 85.Khaminets A, Heinrich T, Mari M, Grumati P, Huebner AK, Akutsu M, Liebmann L, Stolz A, Nietzsche S, Koch N, Mauthe M, Katona I, Qualmann B, Weis J, Reggiori F, Kurth I, Hubner CA, Dikic I. Regulation of endoplasmic reticulum turnover by selective autophagy. Nature. 2015;522:354–358. doi: 10.1038/nature14498. [DOI] [PubMed] [Google Scholar]
- 86.Choy A, Dancourt J, Mugo B, O’Connor TJ, Isberg RR, Melia TJ, Roy CR. The Legionella effector RavZ inhibits host autophagy through irreversible Atg8 deconjugation. Science (New York, N.Y. 2012;338:1072–1076. doi: 10.1126/science.1227026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Horenkamp FA, Kauffman KJ, Kohler LJ, Sherwood RK, Krueger KP, Shteyn V, Roy CR, Melia TJ, Reinisch KM. The Legionella Anti-autophagy Effector RavZ Targets the Autophagosome via PI3P- and Curvature-Sensing Motifs. Developmental cell. 2015;34:569–576. doi: 10.1016/j.devcel.2015.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Baskaran S, Ragusa MJ, Boura E, Hurley JH. Two-site recognition of phosphatidylinositol 3-phosphate by PROPPINs in autophagy. Mol Cell. 2012;47:339–348. doi: 10.1016/j.molcel.2012.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Busse RA, Scacioc A, Krick R, Perez-Lara A, Thumm M, Kuhnel K. Characterization of PROPPIN-Phosphoinositide Binding and Role of Loop 6CD in PROPPIN-Membrane Binding. Biophys J. 2015;108:2223–2234. doi: 10.1016/j.bpj.2015.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Knorr RL, Dimova R, Lipowsky R. Curvature of double-membrane organelles generated by changes in membrane size and composition. PLoS One. 2012;7:e32753. doi: 10.1371/journal.pone.0032753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Baba M, Takeshige K, Baba N, Ohsumi Y. Ultrastructural analysis of the autophagic process in yeast: detection of autophagosomes and their characterization. The Journal of cell biology. 1994;124:903–913. doi: 10.1083/jcb.124.6.903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. doi: 10.1247/csf.20.465. [DOI] [PubMed] [Google Scholar]
- 93.Rose TL, Bonneau L, Der C, Marty-Mazars D, Marty F. Starvation-induced expression of autophagy-related genes in Arabidopsis. Biol Cell. 2006;98:53–67. doi: 10.1042/BC20040516. [DOI] [PubMed] [Google Scholar]
- 94.Deter RL, Baudhuin P, De Duve C. Participation of lysosomes in cellular autophagy induced in rat liver by glucagon. The Journal of cell biology. 1967;35:C11–C16. doi: 10.1083/jcb.35.2.c11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mizushima N, Yamamoto A, Matsui M, Yoshimori T, Ohsumi Y. In vivo analysis of autophagy in response to nutrient starvation using transgenic mice expressing a fluorescent autophagosome marker. Mol Biol Cell. 2004;15:1101–1111. doi: 10.1091/mbc.E03-09-0704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Locke M, Collins JV. The Structure and Formation of Protein Granules in the Fat Body of an Insect. The Journal of cell biology. 1965;26:857–884. doi: 10.1083/jcb.26.3.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Yamano K, Fogel AI, Wang C, van der Bliek AM, Youle RJ. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. Elife. 2014;3:e01612. doi: 10.7554/eLife.01612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Dunn WA., Jr Studies on the mechanisms of autophagy: maturation of the autophagic vacuole. The Journal of cell biology. 1990;110:1935–1945. doi: 10.1083/jcb.110.6.1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kabeya Y, Mizushima N, Yamamoto A, Oshitani-Okamoto S, Ohsumi Y, Yoshimori T. LC3, GABARAP and GATE16 localize to autophagosomal membrane depending on form-II formation. Journal of cell science. 2004;117:2805–2812. doi: 10.1242/jcs.01131. [DOI] [PubMed] [Google Scholar]
- 100.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. The Journal of cell biology. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Duke EM, Razi M, Weston A, Guttmann P, Werner S, Henzler K, Schneider G, Tooze SA, Collinson LM. Imaging endosomes and autophagosomes in whole mammalian cells using correlative cryo-fluorescence and cryo-soft X-ray microscopy (cryo-CLXM) Ultramicroscopy. 2014;143:77–87. doi: 10.1016/j.ultramic.2013.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Baba M, Osumi M, Scott SV, Klionsky DJ, Ohsumi Y. Two distinct pathways for targeting proteins from the cytoplasm to the vacuole/lysosome. The Journal of cell biology. 1997;139:1687–1695. doi: 10.1083/jcb.139.7.1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Friedman JR, Lackner LL, West M, DiBenedetto JR, Nunnari J, Voeltz GK. ER tubules mark sites of mitochondrial division. Science (New York, N.Y. 2011;334:358–362. doi: 10.1126/science.1207385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hanson HH, Kang S, Fernandez-Monreal M, Oung T, Yildirim M, Lee R, Suyama K, Hazan RB, Phillips GR. LC3-dependent intracellular membrane tubules induced by gamma-protocadherins A3 and B2: a role for intraluminal interactions. The Journal of biological chemistry. 2010;285:20982–20992. doi: 10.1074/jbc.M109.092031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Fengsrud M, Erichsen ES, Berg TO, Raiborg C, Seglen PO. Ultrastructural characterization of the delimiting membranes of isolated autophagosomes and amphisomes by freeze-fracture electron microscopy. European journal of cell biology. 2000;79:871–882. doi: 10.1078/0171-9335-00125. [DOI] [PubMed] [Google Scholar]
- 106.Zeuschner D, Geerts WJ, van Donselaar E, Humbel BM, Slot JW, Koster AJ, Klumperman J. Immuno-electron tomography of ER exit sites reveals the existence of free COPII-coated transport carriers. Nat Cell Biol. 2006;8:377–383. doi: 10.1038/ncb1371. [DOI] [PubMed] [Google Scholar]
- 107.Polishchuk RS, San Pietro E, Di Pentima A, Tete S, Bonifacino JS. Ultrastructure of long-range transport carriers moving from the trans Golgi network to peripheral endosomes. Traffic. 2006;7:1092–1103. doi: 10.1111/j.1600-0854.2006.00453.x. [DOI] [PubMed] [Google Scholar]
- 108.Punnonen EL, Pihakaski K, Mattila K, Lounatmaa K, Hirsimaki P. Intramembrane particles and filipin labelling on the membranes of autophagic vacuoles and lysosomes in mouse liver. Cell Tissue Res. 1989;258:269–276. doi: 10.1007/BF00239447. [DOI] [PubMed] [Google Scholar]