Abstract
Rho family GTPases are signaling molecules that orchestrate cytoskeletal dynamics in a variety of cellular processes. Because they effect localized changes to the cytoskeleton only in their active (GTP-bound) conformation, the ability to monitor the active state of Rho GTPases in space and time is critical for understanding their function. Here, we summarize popular tools used for live imaging of active Rho GTPases, outlining advantages and drawbacks of these approaches. Additionally, we highlight key features of the Xenopus laevis embryo that make it well-suited for epithelial cell biology and discuss how application of Rho activity reporters in the Xenopus laevis embryo led to the discovery of a novel phenomenon, junctional Rho flares.
Keywords: Amphibian, Early development, Signaling
Why are Rho GTPases important in health and disease?
Rho family GTPases coordinate essential events in the life of a cell, from birth (cell division) to death (apoptosis) and many events in between (migration, adhesion, polarity), including a number of specialized functions (dendritic spine and immune synapse formation). As such, dysregulation of Rho GTPases is implicated in a number of disease processes, including tumor formation and metastasis, neurodegenerative diseases, and bacterial infection, among others (Boettner and Van Aelst 2002; Alan and Lundquist 2013; Cook et al. 2014). In cancer, transforming mutations in Rho GTPases are rare; however, mutations in other proteins that cause GTPases to be overexpressed or hyperactivated are frequently associated with disease, highlighting the need for strict regulation of these proteins (Haga and Ridley 2016).
Rho GTPase Basics
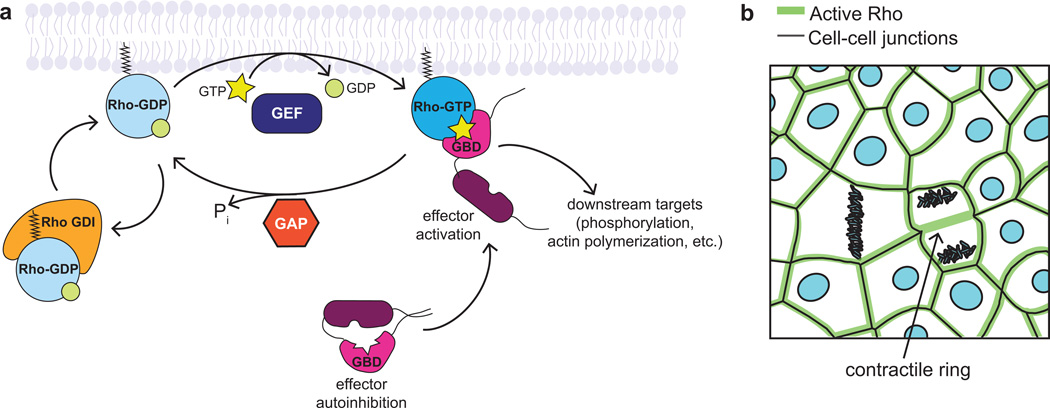
Rho GTPases are a family of 20 small GTPases divided into two classes: typical (12 members) and atypical (8 members) (Heasman and Ridley 2008). Typical GTPases, including RhoA, Rac1, and Cdc42, cycle between an active, GTP-bound state, and an inactive, GDP-bound state (Figure 1a). When in their active conformation, Rho GTPases are associated with the plasma membrane and can interact with and activate effector proteins, resulting in localized effects on the cytoskeleton (Figure 1a). For example, properly localized RhoA activity is required to direct formation of contractile actomyosin arrays during cytokinesis and at cell-cell junctions (Figure 1b) (Bement et al. 2005; Miller and Bement 2009; Reyes et al. 2014; Breznau et al. 2015). Thus, in order to understand the cellular consequences of Rho GTPase activity, one must understand localization of the active GTPase in space and time, making live imaging an ideal way to study the dynamics of these proteins (Pertz 2010). To distinguish between active and inactive populations of Rho GTPases, researchers have used GTPase binding domains (GBDs) of effector proteins, which are specific for the GTPase of interest and bind only the active conformation. Biochemical assays, such as affinity pull-downs using a GST-tagged GBD, can reveal the amount of active GTPase compared to the total (Figure 2a) (Ren et al. 1999; Boulter et al. 2010), but lack spatial information about where the GTPases are active. Imaging techniques that rely on specialized fixation to image active Rho GTPases (Yonemura et al. 2004; Ratheesh et al. 2012) only capture a snapshot in time. Live imaging offers a more nuanced approach to understanding these highly spatiotemporally regulated signaling molecules.
Figure 1. Rho GTPase activity is tightly regulated to create distinct zones of activation.
a. Typical Rho family GTPases cycle between an active, GTP-bound state and an inactive, GDP-bound state. GEFs promote the active state by exchanging GDP for GTP, while GAPs inactivate GTPases by stimulating GTP hydrolysis. Rho GDI sequesters Rho-GDP in the cytoplasm, protecting it from degradation and preventing its activation. In the active conformation, Rho GTPases activate effectors through direct binding, usually by relieving an autoinhibited conformation, allowing them to act on their downstream targets.
b. RhoA is active in distinct zones in epithelia: at cell-cell junctions and at the contractile ring of dividing cells.
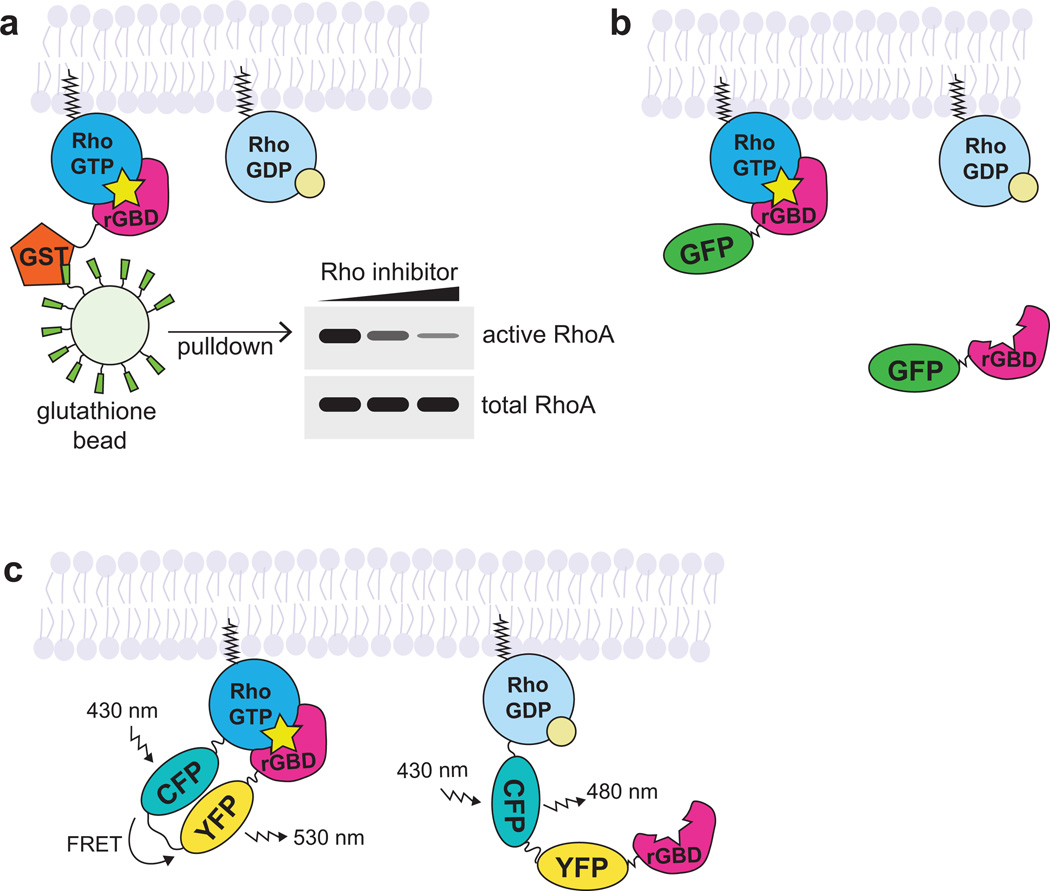
Figure 2. Approaches for studying active Rho GTPases.
a. In the GBD affinity pull-down approach, the active GTPase is pulled down with a GST-tagged effector GBD specific for the GTPase of interest (rGBD = Rhotekin GBD, which binds RhoA, B, and C). The amount of GTPase in the pull-down is compared with total GTPase in the sample to approximate the pool of active GTPase in the sample.
b. In the effector translocation (GBD probe) approach, the effector GBD is fluorescently-tagged (with GFP) and binds to the endogenous active GTPase. Local increase in fluorescence intensity over background is interpreted as increased active GTPase.
c. In the GTPase-effector FRET biosensor (FRET biosensor) approach, the GTPase is tagged with a donor fluorophore (CFP) and the effector GBD is tagged with an acceptor fluorophore (YFP). A unimolecular Rho biosensor is shown here. When the GTPase is inactive, the donor fluorophore emits light. When the GTPase is active, the donor fluorophore excites the acceptor fluorophore.
Advantages and drawbacks of current tools for live imaging of active Rho GTPases
The first live imaging studies of Rho GTPases simply tagged the GTPases with a fluorescent protein. However, these studies were hampered by high background due to the large pool of the inactive GTPase and by localization patterns that did not reflect the localization of the endogenous protein (Yonemura et al. 2004). Currently, there are two popular types of tools used to study active Rho GTPase dynamics with live imaging: 1) effector translocation probes (referred to as “GBD probes” hereafter) and 2) GTPase-effector biosensors (referred to as “FRET biosensors” hereafter) that employ Förster Resonance Energy Transfer (FRET). Both of these approaches take advantage of specific effector GBDs, as described above.
In GBD probe approach, the GBD is fluorescently tagged and observed by traditional confocal microscopy (Figure 2b) (Kraynov et al. 2000; Srinivasan et al. 2003; Benink and Bement 2005). Fluorescence intensity over background indicates the localization of the endogenous population of the active GTPase. This method offers several advantages, such as easy set-up, simple data interpretation, and the ability to co-image multiple proteins. Drawbacks of this method include potential dominant negative effects if the probe is expressed at too high a level, lack of specificity for a single GTPase (some effectors can bind multiple GTPases, though with varying affinities), and poor signal over background in some cell types.
In the FRET biosensor approach, a FRET donor fluorophore-tagged GTPase of interest and its corresponding FRET acceptor fluorophore-tagged GBD are introduced into the cell, either as individual molecules or as part of a unimolecular biosensor (Figure 2c) (Kraynov et al. 2000; Pertz et al. 2006; Pertz 2010). When the GTPase is GTP-bound, the GTPase will bind the GBD, generating FRET signal. FRET ratios (increase in the level of the FRET acceptor signal and decrease in the level of the FRET donor signal) across the cell can be compared and interpreted to determine areas of increased active GTPase. Benefits of this approach include improved GTPase specificity, as only the GTPase of interest will have FRET signal when activated, and lower potential for dominant negative effects as the GBD and GTPase are introduced in equal ratios. However, notable drawbacks of this approach stem from overexpressing tagged GTPases, which may not localize or function the same as endogenous GTPases (Yonemura et al. 2004, Bendezú et al. 2015). Additionally, overexpression of tagged GTPases could introduce artifacts, such as displacing other Rho GTPases from the limiting supply of Rho GDI, leading to their degradation (Boulter et al. 2010) or titrating out GEFs, which could keep them from activating other Rho GTPases. Furthermore, FRET requires a specialized microscope setup, careful controls to avoid artifacts, and complicated analysis (Spiering et al. 2013). Nonetheless, these probes have been successfully used to observe active Rho GTPase dynamics in many types of cultured cells.
Case study for live imaging of active Rho in Xenopus laevis embryos: junctional Rho flares
GBD probes have been used to study active Rho GTPase dynamics in several cellular processes at a range of X. laevis developmental stages including: 1) wound healing in oocytes and embryos (Benink and Bement 2005; Clark et al. 2009), 2) cortical excitability in oocytes and embryos (Bement et al. 2015), 3) cytokinesis in blastula-staged (Bement et al. 2005; Miller and Bement 2009) and gastrula-staged (Breznau et al. 2015) embryos as well as in a reconstituted system with egg extracts (Nguyen et al. 2014), and 4) cell-cell junctions in gastrula-staged embryos (Reyes et al. 2014; Breznau et al. 2015). Below, we highlight the features of the X. laevis gastrula-staged embryo that make it well-suited for studying epithelial cell-cell junctions, and how the use of GBD probes described above led to the discovery of Rho flares, dynamic accumulations of active Rho at cell-cell junctions.
The gastrula-staged embryo (Nieuwkoop and Faber stage 10–12) is covered with a fully polarized epithelium, with the apical cell-cell junctions facing the outer surface of the embryo, making it accessible for live imaging without having to dissect the embryo. This feature means that the epithelium can be studied in its native mechanical and biological context. Other intrinsic features of the epithelium that make it well-suited for live imaging include its relatively large cells (~20–40 µm in diameter) and, when slightly compressed between two coverslips, a flat imaging plane, meaning that apical junctions can be captured in relatively few z-slices. mRNA encoding fluorescent proteins can be injected into the early embryo, either globally or to create a mosaic pattern (e.g., to distinguish between the contributions of two neighboring cells to the junctional population of a protein (Shindo and Wallingford 2014) or to have groups of control and treated cells within the same embryo (Breznau et al. 2015; Higashi et al. 2016). Table 1 lists many useful fluorescent probes that can be used in Xenopus to study Rho GTPases and the cytoskeleton.
Table 1. Probes for live imaging of Rho GTPases and the cytoskeleton in Xenopus.
A non-exhaustive list of validated probes available for live imaging in Xenopus. References for pCS2+-based plasmids are listed. “*” denotes that the plasmid is available through Addgene.
| Probe based on: | pCS2+ vectors, citations, availability through Addgene (*) |
|
|---|---|---|
| Active Rho GTPase Probes | ||
| Active Rho | rGBD (GBD of Rhotekin) |
GFP†-rGBD (Benink and Bement 2005)* 3xGFP-rGBD (Bement et al. 2015) mCherry-2xrGBD (Davenport et al. 2016) |
| Active Cdc42 | wGBD (GBD of N-WASP) |
GFP-wGBD (Sokac et al. 2003)* mRFP-wGBD (Benink and Bement 2005)* |
| Active Rac‡ | pGBD (GBD of PAK3) |
GFP-pGBD (Benink and Bement 2005; Miller and Bement 2009)* |
| Actin Probes | ||
| Filamentous (F-) actin | UtrCH (Calponin Homology domain of Utrophin) |
mRFP-UtrCH (Burkel et al. 2007)* GFP-UtrCH (Burkel et al. 2007)* mCherry-UtrCH (Miller and Bement 2009) |
| Globular (G-) and F-actin§ | Lifeact (First 17 amino acids of abp140 from S. cerevisiae) |
Lifeact-GFP (Riedl et al. 2008; Bement et al. 2015; Higashi et al. 2016) Lifeact-mRFP (Riedl et al. 2008; Bement et al. 2015; Higashi et al. 2016) |
| Microtubule Probes | ||
| Polymerized microtubules | EMTB (Ensconsin microtubule binding domain) |
EMTB-3xGFP (Miller and Bement 2009)* EMTB-mCherry (Miller and Bement 2009) EMTB-2xmCherry (vonDassow et al. 2009) |
| Total tubulin** | α-tubulin | GFP-tubulin (Woolner et al. 2008) mCherry-tubulin (Woolner et al. 2008) |
| Microtubule plus ends | EB3 (End binding 3) |
EB3-GFP (Shindo et al. 2008) |
| Membrane Probes | ||
| Membrane | Farnesylation (Farnesylation sequence from human Ras) |
GFP-farnesyl (Reyes et al. 2014) mCherry-farnesyl (Reyes et al. 2014) |
| Membrane | Myristolation (2xmyristolation sequence from Lyn kinase) |
Mem-TagBFP (Higashi et al. 2016) |
| DNA Probe | ||
| Chromatin | H2B (Histone 2B) |
mCherry-H2B (Reyes et al. 2014) |
GFP refers to Enhanced GFP (EGFP).
pGBD can bind to Rac and Cdc42, so controls to determine specificity of response should be performed (Breznau et al. 2015).
Lifeact binds to F-actin with ~30-fold higher affinity than G-actin (Riedl et al. 2008).
Using fluorescently-tagged tubulin may result in poor signal over background; however, EMTB binds microtubules in a cell cycle-dependent manner, meaning that signal becomes significantly weaker during mitosis.
The importance of Rho GTPases in regulating epithelial cell-cell junctions has been evident from the mid-1990s, when constitutively active and dominant negative Rho GTPases were used to test their effects on cell-cell junction architecture and perijunctional actin assembly (reviewed in Citi et al. 2014; Quiros and Nusrat 2014). In the decades since, biochemical and immunostaining methods have been applied to learn more about regulation of junctional Rho GTPases and their downstream consequences. However, it is only within the past five years that active RhoA has been observed at cell-cell junctions of cultured epithelial cells with the FRET biosensors described above (Terry et al. 2011; Ratheesh et al. 2012). Interestingly, the authors of these studies reported only snapshots in time, depicting a stable population of active RhoA. In contrast, when the GBD probe was observed in the X. laevis epithelium with high resolution live imaging, Reyes et al. observed a baseline level of Rho activity around cell-cell junctions as well as transient, localized accumulations, or “flares” of active Rho (Reyes et al. 2014). Furthermore, forthcoming work from our lab demonstrates that Rho flares occur in response to local discontinuities in the tight junction proteins ZO-1 and Occludin, which are locally reinforced following the flare (Stephenson and Miller, unpublished). As Rho flares occur primarily on junctions that are experiencing changes in tension (e.g., near dividing cells), we propose that Rho flares represent a mechanism by which cells are able to rapidly repair and reinforce their junctions in response to cell shape changes, creating a robust and flexible barrier.
Summary and Future Directions
Fluorescent reporters for active Rho family GTPases have become key tools for uncovering the spatiotemporal dynamics of active Rho GTPases in a variety of cellular contexts. To extend this work, it will be important to create new probes to study the less-characterized members of the Rho GTPase family. Additionally, expanding the range of Rho GTPase-mediated cellular processes examined with these Rho probes is an essential goal. To this end, Xenopus is a versatile model system, where intact embryos can be used to investigate basic cell and developmental biology questions as well as to model human developmental defects and disease. Furthermore, Xenopus tissue explants can be employed to study specific organs or tissue mechanics, and Xenopus egg extracts can recapitulate many cellular activities, and are easily manipulated biochemically. To further advance this field, it will be useful to apply advanced imaging approaches such as Total Internal Reflection Fluorescence (TIRF) microscopy, which reduces background fluorescence outside the focal plane, making it possible to use GBD probes in cell types where high background fluorescence has been a challenge, or optogenetic tools, which can allow experimenter-controlled activation of Rho GTPases. These advances, coupled with new knowledge of biological process-specific GEFs, GAPs, and effector proteins will enhance our mechanistic understanding of how Rho family GTPases function in health and disease.
Acknowledgments
We thank Elaina Breznau for acquiring the microscopy image in Figure 3a and Andrew Goryachev, Tomohito Higashi, and Torey Arnold for critically reading the manuscript. Research in the Miller lab is supported by grants from the NIH (R01 GM112794) and the NSF (Award number: 1615338). R.E.S. is supported by the NSF Graduate Research Fellowship (DGE #1256260).
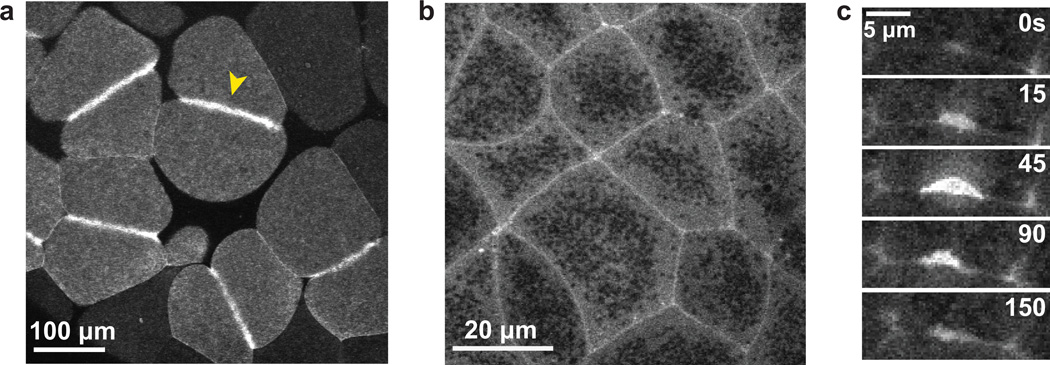
Figure 3. Rho zones and Rho flares in X. laevis embryos.
a. GFP-rGBD (GBD probe for active Rho) in the large blastomeres of the early X. laevis embryo. A distinct zone of active Rho specifies the position of the contractile ring (yellow arrowhead).
b. GFP-rGBD in epithelial cells of the gastrula-stage X. laevis embryo. Zones of active Rho encircle the perimeter of each epithelial cell.
c. A montage depicting a Rho flare over time. These transient accumulations of active Rho at cell-cell junctions were first observed in the X. laevis embryo (Reyes et al. 2014).
References
- Alan JK, Lundquist EA. Mutationally activated Rho GTPases in cancer. Small GTPases. 2013;4:159–163. doi: 10.4161/sgtp.26530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Benink HA, vonDassow G. A microtubule-dependent zone of active RhoA during cleavage plane specification. The Journal of cell biology. 2005;170:91–101. doi: 10.1083/jcb.200501131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Leda M, Moe AM, Kita AM, Larson ME, Golding AE, Pfeuti C, Su K-C, Miller AL, Goryachev AB, vonDassow G. Activator-inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nature Cell Biology. 2015;17:1471–1483. doi: 10.1038/ncb3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú FO, Vincenzetti V, Vavylonis D, Wyss R, Vogel H, Martin SG. Spontaneous Cdc42 Polarization Independent of GDI-Mediated Extraction and Actin-Based Trafficking. PLoS Biology. 2015;13(4):1–30. doi: 10.1371/journal.pbio.1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benink HA, Bement WM. Concentric zones of active RhoA and Cdc42 around single cell wounds. The Journal of cell biology. 2005;168:429–439. doi: 10.1083/jcb.200411109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettner B, Van Aelst L. The role of Rho GTPases in disease development. Gene. 2002;286:155–174. doi: 10.1016/s0378-1119(02)00426-2. [DOI] [PubMed] [Google Scholar]
- Boulter E, Garcia-Mata R, Guilluy C, Dubash A, Rossi G, Brennwald PJ, Burridge K. Regulation of Rho GTPase crosstalk, degradation and activity by RhoGDI1. Nature Cell Biology. 2010;12:477–483. doi: 10.1038/ncb2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breznau EB, Semack AC, Higashi T, Miller AL. MgcRacGAP restricts active RhoA at the cytokinetic furrow and both RhoA and Rac1 at cell-cell junctions in epithelial cells. Molecular Biology of the Cell. 2015;26:2439–2455. doi: 10.1091/mbc.E14-11-1553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burkel BM, vonDassow G, Bement WM. Versatile fluorescent probes for actin filaments based on the actin-binding domain of utrophin. Cell motility and the cytoskeleton. 2007;64:822–832. doi: 10.1002/cm.20226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citi S, Guerrera D, Spadaro D, Shah J. Epithelial junctions and Rho family GTPases: the zonular signalosome. Small GTPases. 2014;5:1–15. doi: 10.4161/21541248.2014.973760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark AG, Miller AL, Vaughan E, Yu H-YE, Penkert R, Bement WM. Integration of single and multicellular wound responses. Current biology. 2009;19:1389–1395. doi: 10.1016/j.cub.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook DR, Rossman KL, Der CJ. Rho guanine nucleotide exchange factors: regulators of Rho GTPase activity in development and disease. Oncogene. 2014;33:4021–4035. doi: 10.1038/onc.2013.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davenport NR, Sonnemann KJ, Eliceiri KW, Bement WM. Membrane dynamics during cellular wound repair. Molecular Biology of the Cell. 2016;27:2272–2285. doi: 10.1091/mbc.E16-04-0223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haga RB, Ridley AJ. Rho GTPases: Regulation and roles in cancer cell biology. Small GTPases. 2016:1–15. doi: 10.1080/21541248.2016.1232583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heasman SJ, Ridley AJ. Mammalian Rho GTPases: new insights into their functions from in vivo studies. Nature reviews. Molecular cell biology. 2008;9:690–701. doi: 10.1038/nrm2476. [DOI] [PubMed] [Google Scholar]
- Higashi T, Arnold TR, Stephenson RE, Dinshaw KM, Miller AL. Maintenance of the Epithelial Barrier and Remodeling of Cell-Cell Junctions during Cytokinesis. Current biology. 2016;26:1829–1842. doi: 10.1016/j.cub.2016.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraynov VS, Chamberlain C, Bokoch GM, Schwartz MA, Slabaugh S, Hahn KM. Localized Rac activation dynamics visualized in living cells. Science. 2000;290:333–337. doi: 10.1126/science.290.5490.333. [DOI] [PubMed] [Google Scholar]
- Miller AL, Bement WM. Regulation of cytokinesis by Rho GTPase flux. Nature Cell Biology. 2009;11:71–77. doi: 10.1038/ncb1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen PA, Groen AC, Loose M, Ishihara K, Wühr M, Field CM, Mitchison TJ. Spatial organization of cytokinesis signaling reconstituted in a cell-free system. Science. 2014;346:244–247. doi: 10.1126/science.1256773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertz O. Spatio-temporal Rho GTPase signaling - where are we now? Journal of cell science. 2010;123:1841–1850. doi: 10.1242/jcs.064345. [DOI] [PubMed] [Google Scholar]
- Pertz O, Hodgson L, Klemke RL, Hahn KM. Spatiotemporal dynamics of RhoA activity in migrating cells. Nature. 2006;440:1069–1072. doi: 10.1038/nature04665. [DOI] [PubMed] [Google Scholar]
- Quiros M, Nusrat A. RhoGTPases, actomyosin signaling and regulation of the epithelial Apical Junctional Complex. Seminars in cell & developmental biology. 2014;36:194–203. doi: 10.1016/j.semcdb.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratheesh A, Gomez GA, Priya R, Verma S, Kovacs EM, Jiang K, Brown NH, Akhmanova A, Stehbens SJ, Yap AS. Centralspindlin and α-catenin regulate Rho signalling at the epithelial zonula adherens. Nature Cell Biology. 2012;14:818–828. doi: 10.1038/ncb2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren XD, Kiosses WB, Schwartz MA. Regulation of the small GTP-binding protein Rho by cell adhesion and the cytoskeleton. The EMBO journal. 1999;18:578–585. doi: 10.1093/emboj/18.3.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes CC, Jin M, Breznau EB, Espino R, Delgado-Gonzalo R, Goryachev AB, Miller AL. Anillin Regulates Cell-Cell Junction Integrity by Organizing Junctional Accumulation of Rho-GTP and Actomyosin. Current Biology. 2014;24:1263–1270. doi: 10.1016/j.cub.2014.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riedl J, Crevenna AH, Kessenbrock K, Yu JH, Neukirchen D, Bista M, Bradke F, Jenne D, Holak TA, Werb Z, Sixt M, Wedlich-Soldner R. Lifeact: a versatile marker to visualize F-actin. Nature Methods. 2008;5:605–607. doi: 10.1038/nmeth.1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, Wallingford JB. PCP and septins compartmentalize cortical actomyosin to direct collective cell movement. Science. 2014;343:649–652. doi: 10.1126/science.1243126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shindo A, Yamamoto TS, Ueno N. Coordination of cell polarity during Xenopus gastrulation. PloS one. 2008;3:e1600. doi: 10.1371/journal.pone.0001600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokac AM, Co C, Taunton J, Bement W. Cdc42-dependent actin polymerization during compensatory endocytosis in Xenopus eggs. Nature Cell Biology. 2003;5:727–732. doi: 10.1038/ncb1025. [DOI] [PubMed] [Google Scholar]
- Spiering D, Bravo-Cordero JJ, Moshfegh Y, Miskolci V, Hodgson L. Quantitative ratiometric imaging of FRET-biosensors in living cells. Methods in cell biology. 2013;114:593–609. doi: 10.1016/B978-0-12-407761-4.00025-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S, Wang F, Glavas S, Ott A, Hofmann F, Aktories K, Kalman D, Bourne HR. Rac and Cdc42 play distinct roles in regulating PI(3,4,5)P3 and polarity during neutrophil chemotaxis. The Journal of cell biology. 2003;160:375–385. doi: 10.1083/jcb.200208179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terry SJ, Zihni C, Elbediwy A, Vitiello E, Leefa Chong San IV, Balda MS, Matter K. Spatially restricted activation of RhoA signalling at epithelial junctions by p114RhoGEF drives junction formation and morphogenesis. Nature Cell Biology. 2011;13:159–166. doi: 10.1038/ncb2156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- vonDassow G, Verbrugghe KJC, Miller AL, Sider JR, Bement WM. Action at a distance during cytokinesis. The Journal of cell biology. 2009;187:831–845. doi: 10.1083/jcb.200907090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolner S, O'Brien LL, Wiese C, Bement WM. Myosin-10 and actin filaments are essential for mitotic spindle function. The Journal of cell biology. 2008;182:77–88. doi: 10.1083/jcb.200804062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonemura S, Hirao-Minakuchi K, Nishimura Y. Rho localization in cells and tissues. Experimental cell research. 2004;295:300–314. doi: 10.1016/j.yexcr.2004.01.005. [DOI] [PubMed] [Google Scholar]