Abstract
The pathogenesis of childhood-onset nephrotic syndrome (NS), disparity in incidence of NS among races, and variable responses to therapies in children with NS have defied explanation to date. In the last twenty years over 50 genetic causes of steroid resistant nephrotic syndrome (SRNS) have been identified and at least two disease loci for two pathologic variants of SRNS (FSGS and membranous nephropathy) have been defined. However, the genetic causes and risk loci for steroid sensitive nephrotic syndrome (SSNS) remain elusive partly because SSNS is relatively rare and also because cases of SSNS vary widely in phenotypic expression over time. A recent study of a well-defined modest cohort of children with SSNS identified variants in HLA-DQA1 as a risk factor for SSNS. This article reviews what is currently known about the genetics of SSNS and also discusses how recent careful phenotypic and genomic studies reinforce the role of adaptive immunity in the molecular mechanisms of SSNS.
Keywords: Nephrotic syndrome children, Genomics, Genetic risks, GWAS
Introduction
Nephrotic syndrome (NS) is the most common glomerular disorder of childhood with an estimated incidence of two to seven cases per 100,000 children and a prevalence rate of 16 cases per 100,000 children [1]. It is defined by the clinical and biochemical constellation of massive proteinuria, hypoalbuminemia, edema, and hypercholesterolemia. Based on the pattern of response to corticosteroids, nephrotic syndrome is subdivided into steroid-sensitive NS (SSNS) and steroid resistant NS (SRNS). In children over one year of age, SSNS is responsible for 80% of all cases of the disease [2]. Among patients with SSNS, the clinical course can be variable with differing relapse rates and overall dependence on steroid administration. It is estimated that approximately 50% of patients with SSNS will have a frequently-relapsing (FR) and/or steroid-dependent (SD) course [3, 4]. The biological basis for the pattern of response to corticosteroid remains unknown.
Etiology and pathogenesis of SSNS
The etiology of most cases of SSNS is unknown and are therefore given the label “idiopathic or primary SSNS”. The definitive reason why some patients respond to corticosteroids and others do not escapes explanation, but proposed mechanisms underlying the pathogenesis of SSNS have sought to clarify this variability in pattern of response. Shalhoub and colleagues [5] in the 1970s proposed that SSNS is the result of a primary T-cell dysfunction based on the evidence that (1) nephrotic syndrome responds to immunosuppressive agents like corticosteroids and calcineurin inhibitors which modulate T-cell function (2) there are reported cases of patients achieving remission following diseases, such as measles or malaria that are known to suppress cell-mediated immunity, and (3) there are patients who develop nephrotic syndrome as part of a paraneoplastic process in malignancies known to affect T-cell function such as Hodgkins lymphoma. Subsequent studies have explored changes in T-cell surface expression, function, and cytokine release in the setting of nephrotic syndrome, however experimental recapitulation of these findings in multiple studies have been lacking [6–10].
Another proposed mechanism of disease in SSNS includes the notion that soluble mediators play a role in altering capillary wall permeability, the so-called “circulating factor” theory. This model of nephrotic syndrome proposes that in patients with NS, normal kidneys exist in an abnormal environment [11]. Circulating stimuli provoke the glomeruli to leak protein. This theory has been particularly studied in SRNS and is supported by evidence that (1) serum from patients with SRNS when injected into normal rats has resulted in increased glomerular permeability and proteinuria, (2) some patients with recurrent SRNS post-transplant are responsive to plasmapheresis, and (3) urgent re-transplant of a kidney from a patient who had immediate recurrence of SRNS into a recipient without NS resulted in resolution of proteinuria [12–14]. While this theory has been particularly studied in SRNS, it has also been noted that a kidney allograft from a donor with biopsy-proven active SSNS into a recipient without the disease resulted in resolution of foot process effacement [15]. Several candidate circulating factors for both SRNS and SSNS have been reported in the literature including, but not limited to, hemopexin, vascular permeability factor (VPF), vascular endothelial growth factor (VEGF), reactive oxygen species, soluble urokinase-type plasminogen activator receptor (suPAR), interleukin-13 (IL-13), interleukin-18 (IL-18), tumor necrosis factor alpha (TNFα), and cardiotrophin-like cytokine factor 1 (CLC-1) [16–27]. While differential expression of these molecules have been reported during periods of relapse and remission, results have not been consistent across studies. The role of these factors in the pathogenesis of idiopathic NS was recently reviewed by Davin [19].
A third mechanism for SSNS proposes that structural defects of the podocyte and other components of the glomerular filtration barrier would explain the loss of filtration barrier integrity and resultant proteinuria. Altered expression of glomerular Podocyte B7-1 (CD80), Sphingomyelin phosphodiesterase acid –like 3b (SMPDL3b), and Angiopoietin-like 4 (Angptl4) have been reported in NS models and also addressed in the review by Darvin [19, 28–30]. This concept of structural changes is also supported by the fact that over 50 genes which localized to the podocyte and its slit diaphragm have been identified as causes of SRNS [27]. Identification of genetic defects associated with hereditary SSNS and identification of genetic variants associated with more common idiopathic SSNS may confirm or disprove some of the disease mechanisms described in this section.
Is SSNS a genetic disease?
There is incontrovertible evidence in the literature that some forms of SRNS have strong genetic underpinnings. Depending on the population being studied, it is estimated that 2–30% of all cases of SRNS are due to a single gene defect [31, 32]. In addition, mutations in over 50 genes have been associated with SRNS [33]. Furthermore, genetic association studies have established risk alleles for some pathologic variants of SRNS. For example, using admixture linkage analysis, G1 and G2 variants in APOL1 were identified as a risk allele for FSGS in African Americans, and variants in HLA-DQA1 and PLA2R1 were found to confer a robust risk for membranous nephropathy in Europeans [34–36].
While data on the genetics of SSNS are limited, there is, however, epidemiologic evidence to suggest that some SSNS may be linked to genetic defects in one or multiple genes. Firstly, there are ethnic trends seen among patients with nephrotic syndrome. In a limited epidemiologic study out of England, Feehally and others showed that Asian children develop SSNS seven times more commonly than their non-Asian counterparts [37]. In other studies, more protracted and challenging clinical courses characterized by frequent relapses and/or steroid dependence have been seen in patients of African American or Hispanic descent [38, 39]. Secondly, it has been estimated that 3% of children with SSNS may have an affected sibling or first degree relative, and there have been numerous published case reports of familial SSNS [40–45]. In addition, in our World-Wide cohort of over 600 patients with SSNS, there are at least 30 siblings or parent-child pairs with the disease (unpublished observation). Knowing these observed ethnic trends and familial predispositions, exploring genetic explanations for SSNS follows reason.
SSNS due to single gene defects
Over 50 highly penetrant monogenic mutations cause familial SRNS [46]. However, to date, no single gene has been identified as a cause of monogenic SSNS, despite reports of multiple families with multiple generations affected by SSNS, and inheritance patterns suggestive of a penetrant single gene mutation [42, 44, 45]. Wide variability in phenotypic expression of SSNS may explain why single genes have been elusive. SSNS is a disease that is characterized by relapse and remission and tends to get better with age. It is therefore possible to obtain falsely negative family histories during evaluation. The implication of this is that ascertainment of large pedigrees with enough power for genome-wide linkage studies may be difficult. Notwithstanding, even in situations where large pedigrees have been ascertained and a definite locus defined, causative genes have not been identified. Ruf et al. [47] investigated a consanguineous family in which three children were diagnosed with SSNS. They identified a genome-wide significant locus on chromosome 2p, however fine mapping of the locus and candidate gene analysis has not yielded insight into a causal mutation. In addition, the locus has not been reproduced in other studies.
Given the similarities between SRNS and SSNS in clinical manifestations and changes to the glomerular filtration barrier, it is tempting to speculate that the genetic architecture of SRNS and SSNS may be similar. In some forms of familial SRNS, individuals have been identified with a SSNS phenotype. In 2006, Hinkes et al. [48] identified two children with truncating mutations in Phospholipase C epsilon 1 (PLCE1) who responded to corticosteroid and cyclosporine therapy. The reason for this is unclear, however the authors suggest that there may be a critical time window in glomerular development during which treatment with immunosuppressive medications may overcome a putative defect imposed by PLCE1 loss of function.
More recently, Gee et al. [40] described an autosomal recessive form of SSNS linked to mutations in epithelial membrane protein type 2 (EMP2), however, a homozygous mutation in the same gene was also identified in a family with SRNS. EMP2 gene encodes for a protein that localizes to both the podocyte and glomerular endothelial cells and regulates caveolin-1 expression [40]. Increased caveolin-1 expression has been reported in EMP2 knock-out models and in patients with glomerular diseases [40, 49]. The mechanisms by which mutations in EMP2 may cause SSNS and SRNS is still under investigation, however overexpression of caveolin-1 in zebrafish embryo resulted in an edema phenotype that was rescued by glucocorticoids. These findings suggest that caveolin-1 may represent a novel therapeutic target for both SSNS and SRNS [50].
Additionally, mutations in nephrin (NPHS1) have been reported in rare cases of SSNS suggesting a possible genetic overlap with SRNS [51, 52]. Follow up studies by the same group that reported these cases suggest that the NPHS1 variants in patients with SSNS may represent a hypomorphic mutation that confers increased vulnerability to immunogenic stimuli reversible by immunomodulation [52].
In another study, using homozygosity mapping and whole-exome sequencing, recessive mutations in KANK 1 and 2, genes encoding for kidney ankyrin repeat-containing protein, have been identified in two families with both SSNS and SRNS [53]. Mutations in KANK resulted in defective podocyte cell signaling and function [53]. Subsequent evaluation of more than 1000 individuals with nephrotic syndrome using high-throughput next generation sequencing failed to identify additional patients with mutations in the gene suggesting that single gene causes of SSNS are exceedingly rare [53].
Steroid-sensitive nephrotic syndrome may also present as part of syndromes due to single gene defects. Study of such syndromes may provide insight into the genetics and pathogenesis of SSNS. For example, Exostosin-1 (EXT1) mutations, which result in autosomal dominant multiple exostoses (OMIM 133700), have been associated with SSNS [54]. EXT1 is involved in the synthesis and expression of heparan sulfate glycosaminoglycans, a key structural component of the glomerular basement membrane. Reduction in heparan sulfate under conditions of EXT1 mutation may render the glomerulus vulnerable in situations where there is risk for further loss of heparan sulfate. Thus, nephrotic syndrome could be theorized to result in situations where heparanse activity is increased. Heparanse is expressed by peripheral T lymphocytes, thus providing a link between immune abnormalities and glomerular basement membrane changes. Increased T-cell heparanase presence combined with structural changes to the glomerulus may explain the steroid-sensitive profile in patients with EXT1 associated nephrotic syndrome.
In another report, patients with Immunodysregulation, Polyendocrinophaty, Enteropathy, X-Linked (IPEX: OMIM 34790), a rare X-linked recessive life-threatening disorder caused by mutations in forkhead box p3 (FOXP3) gene, have been known to develop nephrotic syndrome [55]. The glomerular histopathology and steroid responsiveness of NS in the setting of IPEX can vary widely. Park et al [55] described two siblings with IPEX and nephrotic syndrome. One child had steroid- and calcineurin-resistant minimal change disease, while the other child responded to combination immunomodulatory therapy. Thus, in patients with IPEX, T-cell dysregulation may be implicated in the development of nephrotic syndrome, given that FOXP3 is required for the development of regulatory T-cells. However, the vastly different clinical courses of patients with IPEX, and the variable response to immunosuppression speaks to a more complex inheritance pattern involving environmental factors and multiple genes with highly variable penetrance in the development of SSNS.
Table 1 shows a list of genes that have been associated with mixed SSNS/SRNS phenotypes. It should be noted however that greater than 90% of individuals with monogenic NS will have a steroid-resistant disease course. Additionally, mutations known to lead to SRNS are extremely rare in patients with SSNS [56, 57]. Furthermore, genetic risk factors identified for some pathologic types of SRNS such as APOL1 have not been associated with SSNS (Unpublished observation). Thus, the genetics of SSNS and SRNS may be more disparate than overlapping.
Table 1.
Mendelian disease genes associated with SSNS/MCD
| GENE | LOCUS | Type of mutation |
Protein localization |
Associated with SRNS Y/N |
Extra renal manifestations |
References |
|---|---|---|---|---|---|---|
| EMP2 | 16p13 | Missense Truncating |
Podocyte Endothelial cells |
Y | No | Gee et al [40] |
| EXT1 | 8q23 | Missense | Glomerular basement membrane |
N | Multiple exostoses | Robert et al [54] |
| FOXP3 | Xp11 | Missense | Immune cells | Y | Immunodeficiency Polyendocrinopathy Enteropathy |
Park et al [55] |
|
KANK1 KANK2 |
9p24 19p13 |
Missense | Podocyte | Y | No | Gee et al [53] |
| NPHS1 | 19q12 | Missense | Podocyte and slit diaphragm |
Y | No | Kitamura et al and Lahdenkari et al [51, 52] |
| PLCE1 | 10q23 | Truncating | Podocyte and slit diaphragm |
Y | No | Hinkes et al [48] |
SSNS steroid sensitive nephrotic syndrome, MCD minimal change disease
Complex inheritance
Whereas Mendelian traits are characterized by a strong phenotypic-genotypic correlation, complex genetic traits typically exhibit weak correlations between genotype and phenotype. Diseases arising from complex or polygenic inheritance are believed to stem from variation in multiple genes and the interaction among these genes. They are also influenced by behavioral and environmental factors. The heterogeneity of SSNS would fit with this pattern of inheritance and may be an additional reason why genes causing monogenic SSNS have been elusive. Genetic heterogeneity, small effects of disease alleles on risk, and the confounding effects of multiple interactions among genes, and between genes and the environment, make identification of genes involved in polygenic disease more challenging. While single family studies are sufficient to study genes inherited in a Mendelian pattern, genetic determinants for complex traits require larger cohorts of patients. Given the rarity of SSNS, most studies investigating the genetic risk factor have relied on small cohort of patients, which limits the ability to identify a complex inheritance pattern explaining SSNS.
Because immune dysregulation has been implicated in the pathogenesis of SSNS, most studies exploring complex inheritance have focused on determining the role of variants in Human Leucocyte Antigen (HLA) genes and loci as genetic risk factors for SSNS. In a study of class II HLA antigens in French and German children with nephrotic syndrome, Konrad et al [58] reported an association between SSNS and variants in HLA-DQB and HLA-DQA. A study of UK children of white European descent also reported an association between SSNS and variants in HLA-DR7 and HLA-DQW2 [59]. Similarly, Lagueruela et al found an increased frequency of HLA-DQW2 and two extended haplotypes, [HLA-Al, B8, DR3, DRW52, SCO1] and [HLA-B44, DR7, DRW53, FC31] in American Caucasian children with steroid-sensitive nephrotic syndrome [60]. Also, two small Japanese studies (sample sizes 24 and 30 children, respectively) reported an association between alleles in HLA-DQA1 and HLA-DQB1 and risk of SSNS [61, 62]. Studies in children of Chinese descent have also reported similar association with alleles in HLA-DQB1 and different loci on HLA-DR [63, 64]. One of the studies found that risk alleles may differ in children with a frequently-relapsing course compared to those with infrequent relapses [64]. A summary of studies using a candidate approach to identify genetic risk loci for SSNS is summarized in Table 2. Sample sizes in most of these studies were small and genetic analysis were limited to a few markers in the HLA locus. Of note, most of the loci associations have not been replicated in independent cohorts of patients with SSNS. It is therefore difficult to determine the significance of these risk loci on a genome-wide scale or the relevance to cohorts that differ from the discovery group.
Table 2.
Candidate risk loci for SSNS
| Study population | Cohort size | Gene | References |
|---|---|---|---|
| French and German | 161 |
HLA-DQB HLA-DQA |
Konrad et al 1995 [58] |
| UK Caucasian | 40 |
HLA-DR7 HLA-DQW2 |
Clark et al 1990 [59] |
| US Caucasian | 32 | HLA-DQW2 | Lagueruela et al 1990 [60] |
| Japanese | 30 |
HLA-DQA1 HLA-DQB1 |
Kobayashi et al 1995 [62] |
| Chinese (Taiwan) | 59 |
HLA-DQB1 HLA-DR |
Huang et al 2009 [63] |
| South Asia | 76 |
HLA DRB1 HLA DQB1 |
Ramanathan et al 2015 [72] |
| South Asia USA white |
214 100 |
HLA-DQA1 PLCG2 |
Gbadegesin et al 2015 [65]* |
SSNS steroid sensitive nephrotic syndrome
Exome-wide study
On the other hand, genetic mapping through genome-wide association studies eliminates the need for a priori knowledge of candidate genes and provides an unbiased probe of the entire genome. In conditions where complex genetics drives inheritance, genome wide association studies (GWAS) provide hope for identifying genetic links, but require multiple cohorts to validate findings. This approach is relevant in diseases like SSNS where the phenotype is well defined and risk of misclassification is low. In GWAS on SSNS, risks for misclassification are lessened further by using a study population focused on young children who are unlikely to have been exposed to multiple environmental factors which can modify or mimic the phenotype of interest. In a recent paper, we used an exome array study to determine genetic risk factors for SSNS [65]. We hypothesized that SSNS is likely due to coding and non-coding, common and rare variants in the genome, and that these variants interact with the glomerular components, such as the podocyte, and predispose to SSNS. The discovery cohort included children from South Asia and gender-matched adult controls from the same population. We identified multiple variants in Major Histocompatibility Complex (MHC) genes, however only two HLA-DQA1 missense variants reached significance level and were replicated in an independent cohort of children of European descent with SSNS [65]. The HLA-DQA1 variant amino acid residues are located near the dimer interface and may disrupt the assembly of the antigen recognition domain. In silico modeling showed that the missense variants perturbed secondary protein structure and may therefore affect antigen presentation. Apart from being the first unbiased exome-wide study of genetic risk factors for SSNS, the study also confirmed previous candidate approach studies and reinforced the role of adaptive immunity in the pathogenesis of SSNS. The same locus has been replicated in a limited cohort of patients with SSNS or MCD in Europe [66]. The HLA-DQA1 region appeared to be pleotropic for different glomerular diseases because other variants in the gene have been associated with IgA and membranous nephropathy [58, 67]. However, this locus explained only about 4.6% of the risk for SSNS in the discovery cohort suggesting that there are most likely other risk variants yet to be discovered.
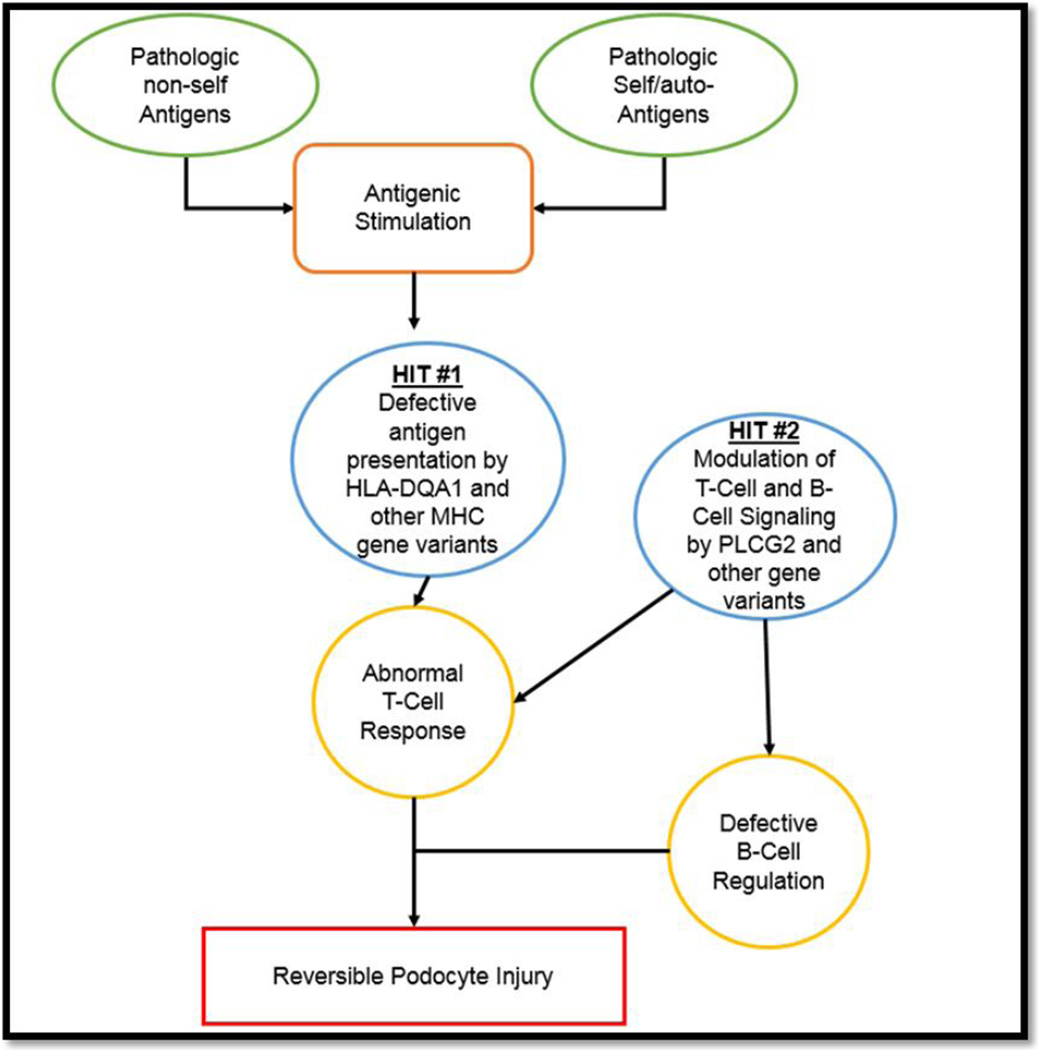
In the same study, rare variants (minor allele frequency <5%) in the gene PLCG2 encoding for the protein phospholipase c gamma 2 (PLCγ2) were found to be associated with SSNS in South Asian children. PLCG2 is involved in adaptive immunity and B cell signaling making it another plausible risk locus for SSNS. Based on these findings, it is tempting to speculate that the pathogenesis of SSNS may be explained by the combination of variations in HLA-DQA1 and other MHC genes, environmental factors, and rare variants in other genes such as PLCG2 that may serve as second-hit for the disease (Figure 1). This model has been shown to be relevant in other immune-mediated glomerular disease such as IgA nephropathy (HLA-DQA1 as the initiator and genes that are important for gut immunity as second hit) [68], membranous glomerulopathy (HLA-DQA1 as the initiator and PLA2R1 and thrombospondin type-1 domain-containing 7A (THSD7A) as second hit) [36, 69, 70], and ANCA-associated vasculitis (HLA-DP as initiator, and genes encoding α 1-antitrypsin (SERPINA1) and proteinase 3 (PRTN3) as second hit) [71]. However, the variants in PLCG2 have not been replicated in other ethnic groups and future studies will determine the significance of these findings and identification of rare variants in other genes.
Figure 1. Proposed mechanisms for complex inheritance in SSNS.
Variation in genetic make-up can predispose to a cascade towards reversible podocyte injury phenotype by aberrant adaptive immune cells, stimulation by non-self antigens such as infective agents, or self/auto-antigens.
Conclusion
Over the last twenty years, rapid advances in genomic science and technology has improved our understanding of the molecular pathogenesis of nephrotic syndrome. Over 50 genes are mutated in SRNS and multiple risk loci have been identified for different pathologic variants of SRNS. However, monogenic causes of SSNS have remained elusive due to wide variability in the clinical course of SSNS. More recently, an exome-wide study using a carefully phenotyped cohort of children with SSNS identified human MHC gene HLA-DQA1 as a risk allele for SSNS. These findings reinforce the role of adaptive immunity in the pathogenesis of SSNS. This locus explains only 4% of heritability of SSNS suggesting that there are other genes and loci associated with SSNS. Although we focused only on genomic studies in this review, based on findings from other studies [69], we believe utilization and integration of multiple platforms such as genomics, proteomics, metabolomics and epigenetic studies may help accelerate the discovery of further risk factors, broaden our understanding of the pathogenesis of SSNS, and identify specific and novel drug targets.
Acknowledgments
R.A.G. is supported by National Institutes of Health (NIH)/National Institutes of Diabetes and Digestive and Kidney Diseases (NIDDK) grant DK098135-01A1, and DK094987.
REFERENCES
- 1.Eddy AA, Symons JM. Nephrotic syndrome in childhood. Lancet. 2003;362:629–639. doi: 10.1016/S0140-6736(03)14184-0. [DOI] [PubMed] [Google Scholar]
- 2.Primary nephrotic syndrome in children: clinical significance of histopathologic variants of minimal change and of diffuse mesangial hypercellularity. A Report of the International Study of Kidney Disease in Children. Kidney Int. 1981;20:765–771. doi: 10.1038/ki.1981.209. [DOI] [PubMed] [Google Scholar]
- 3.Koskimies O, Vilska J, Rapola J, Hallman N. Long-term outcome of primary nephrotic syndrome. Arch Dis Child. 1982;57:544–548. doi: 10.1136/adc.57.7.544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lombel RM, Gipson DS, Hodson EM Kidney Disease: Improving Global O. Treatment of steroid-sensitive nephrotic syndrome: new guidelines from KDIGO. Pediatr Nephrol. 2013;28:415–426. doi: 10.1007/s00467-012-2310-x. [DOI] [PubMed] [Google Scholar]
- 5.Shalhoub RJ. Pathogenesis of lipoid nephrosis: a disorder of T-cell function. Lancet. 1974;2:556–560. doi: 10.1016/s0140-6736(74)91880-7. [DOI] [PubMed] [Google Scholar]
- 6.Garin EH. Circulating mediators of proteinuria in idiopathic minimal lesion nephrotic syndrome. Pediatr Nephrol. 2000;14:872–878. doi: 10.1007/s004679900269. [DOI] [PubMed] [Google Scholar]
- 7.Jamin A, Dehoux L, Dossier C, Fila M, Heming N, Monteiro RC, Deschenes G. Toll-like receptor 3 expression and function in childhood idiopathic nephrotic syndrome. Clin Exp Immunol. 2015;182:332–345. doi: 10.1111/cei.12659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Topaloglu R, Saatci U, Arikan M, Canpinar H, Bakkaloglu A, Kansu E. T-cell subsets, interleukin-2 receptor expression and production of interleukin-2 in minimal change nephrotic syndrome. Pediatr Nephrol. 1994;8:649–652. doi: 10.1007/BF00869075. [DOI] [PubMed] [Google Scholar]
- 9.van den Berg JG, Weening JJ. Role of the immune system in the pathogenesis of idiopathic nephrotic syndrome. Clin Sci. 2004;107:125–136. doi: 10.1042/CS20040095. [DOI] [PubMed] [Google Scholar]
- 10.Yan K, Nakahara K, Awa S, Nishibori Y, Nakajima N, Kataoka S, Maeda M, Watanabe T, Matsushima S, Watanabe N. The increase of memory T cell subsets in children with idiopathic nephrotic syndrome. Nephron. 1998;79:274–278. doi: 10.1159/000045049. [DOI] [PubMed] [Google Scholar]
- 11.Coward RJ, Foster RR, Patton D, Ni L, Lennon R, Bates DO, Harper SJ, Mathieson PW, Saleem MA. Nephrotic plasma alters slit diaphragm-dependent signaling and translocates nephrin, Podocin, and CD2 associated protein in cultured human podocytes. J Am Soc Nephrol. 2005;16:629–637. doi: 10.1681/ASN.2004030172. [DOI] [PubMed] [Google Scholar]
- 12.Dantal J, Bigot E, Bogers W, Testa A, Kriaa F, Jacques Y, Hurault de Ligny B, Niaudet P, Charpentier B, Soulillou JP. Effect of plasma protein adsorption on protein excretion in kidney-transplant recipients with recurrent nephrotic syndrome. N Engl J Med. 1994;330:7–14. doi: 10.1056/NEJM199401063300102. [DOI] [PubMed] [Google Scholar]
- 13.Gallon L, Leventhal J, Skaro A, Kanwar Y, Alvarado A. Resolution of recurrent focal segmental glomerulosclerosis after retransplantation. N Engl J Med. 2012;366:1648–1649. doi: 10.1056/NEJMc1202500. [DOI] [PubMed] [Google Scholar]
- 14.Zimmerman SW. Increased urinary protein excretion in the rat produced by serum from a patient with recurrent focal glomerular sclerosis after renal transplantation. Clin Nephrol. 1984;22:32–38. [PubMed] [Google Scholar]
- 15.Ali AA, Wilson E, Moorhead JF, Amlot P, Abdulla A, Fernando ON, Dorman A, Sweny P. Minimal-change glomerular nephritis Normal kidneys in an abnormal environment? Transplantation. 1994;58:849–852. [PubMed] [Google Scholar]
- 16.Bakker WW, van Dael CM, Pierik LJ, van Wijk JA, Nauta J, Borghuis T, Kapojos JJ. Altered activity of plasma hemopexin in patients with minimal change disease in relapse. Pediatr Nephrol. 2005;20:1410–1415. doi: 10.1007/s00467-005-1936-3. [DOI] [PubMed] [Google Scholar]
- 17.Bertelli R, Trivelli A, Magnasco A, Cioni M, Bodria M, Carrea A, Montobbio G, Barbano G, Ghiggeri GM. Failure of regulation results in an amplified oxidation burst by neutrophils in children with primary nephrotic syndrome. Clin Exp Immunol. 2010;161:151–158. doi: 10.1111/j.1365-2249.2010.04160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brenchley PEC. Vascular permeability factors in steroid-sensitive nephrotic syndrome and focal segmental glomerulosclerosis. Nephrol Dial Transplant. 2003;18:21–25. doi: 10.1093/ndt/gfg1057. [DOI] [PubMed] [Google Scholar]
- 19.Davin JC. The glomerular permeability factors in idiopathic nephrotic syndrome. Pediatr Nephrol. 2016;31:207–215. doi: 10.1007/s00467-015-3082-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garin EH, West L, Zheng W. Interleukin-8 alters glomerular heparan sulfate glycosaminoglycan chain size and charge in rats. Pediatr Nephrol. 2000;14:284–287. doi: 10.1007/s004670050760. [DOI] [PubMed] [Google Scholar]
- 21.Horita Y, Miyazaki M, Koji T, Kobayashi N, Shibuya M, Razzaque MS, Cheng M, Ozono Y, Kohno S, Taguchi T. Expression of vascular endothelial growth factor and its receptors in rats with protein-overload nephrosis. Nephrol Dial Transplant. 1998;13:2519–2528. doi: 10.1093/ndt/13.10.2519. [DOI] [PubMed] [Google Scholar]
- 22.Lennon R, Singh A, Welsh GI, Coward RJ, Satchell S, Ni L, Mathieson PW, Bakker WW, Saleem MA. Hemopexin induces nephrin-dependent reorganization of the actin cytoskeleton in podocytes. J Am Soc Nephrol. 2008;19:2140–2149. doi: 10.1681/ASN.2007080940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maas RJ, Deegens JK, Wetzels JF. Serum suPAR in patients with FSGS: trash or treasure? Pediatr Nephrol. 2013;28:1041–1048. doi: 10.1007/s00467-013-2452-5. [DOI] [PubMed] [Google Scholar]
- 24.Maas RJ, Deegens JK, Wetzels JF. Permeability factors in idiopathic nephrotic syndrome: historical perspectives and lessons for the future. Nephrol Dial Transplant. 2014;29:2207–2216. doi: 10.1093/ndt/gfu355. [DOI] [PubMed] [Google Scholar]
- 25.McCarthy ET, Sharma M, Savin VJ. Circulating permeability factors in idiopathic nephrotic syndrome and focal segmental glomerulosclerosis. Clin J Am Soc Nephrol. 2010;5:2115–2121. doi: 10.2215/CJN.03800609. [DOI] [PubMed] [Google Scholar]
- 26.Raveh D, Shemesh O, Ashkenazi YJ, Winkler R, Barak V. Tumor necrosis factor-alpha blocking agent as a treatment for nephrotic syndrome. Pediatr Nephrol. 2004;19:1281–1284. doi: 10.1007/s00467-004-1573-2. [DOI] [PubMed] [Google Scholar]
- 27.Saleem MA, Kobayashi Y. Cell biology and genetics of minimal change disease. F1000Res. 2016;5 doi: 10.12688/f1000research.7300.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Clement LC, Avila-Casado C, Mace C, Soria E, Bakker WW, Kersten S, Chugh SS. Podocyte-secreted angiopoietin-like-4 mediates proteinuria in glucocorticoid-sensitive nephrotic syndrome. Nat Med. 2011;17:117–122. doi: 10.1038/nm.2261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fornoni A, Sageshima J, Wei C, Merscher-Gomez S, Aguillon-Prada R, Jauregui AN, Li J, Mattiazzi A, Ciancio G, Chen L, Zilleruelo G, Abitbol C, Chandar J, Seeherunvong W, Ricordi C, Ikehata M, Rastaldi MP, Reiser J, Burke GW., 3rd Rituximab targets podocytes in recurrent focal segmental glomerulosclerosis. Sci Transl Med. 2011;3:85ra46. doi: 10.1126/scitranslmed.3002231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Garin EH, Mu W, Arthur JM, Rivard CJ, Araya CE, Shimada M, Johnson RJ. Urinary CD80 is elevated in minimal change disease but not in focal segmental glomerulosclerosis. Kidney Int. 2010;78:296–302. doi: 10.1038/ki.2010.143. [DOI] [PubMed] [Google Scholar]
- 31.Lovric S, Fang H, Vega-Warner V, Sadowski CE, Gee HY, Halbritter J, Ashraf S, Saisawat P, Soliman NA, Kari JA, Otto EA, Hildebrandt F Nephrotic Syndrome Study G. Rapid detection of monogenic causes of childhood-onset steroid-resistant nephrotic syndrome. Clin J Am Soc Nephrol. 2014;9:1109–1116. doi: 10.2215/CJN.09010813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sadowski CE, Lovric S, Ashraf S, Pabst WL, Gee HY, Kohl S, Engelmann S, Vega-Warner V, Fang H, Halbritter J, Somers MJ, Tan W, Shril S, Fessi I, Lifton RP, Bockenhauer D, El-Desoky S, Kari JA, Zenker M, Kemper MJ, Mueller D, Fathy HM, Soliman NA, Group SS, Hildebrandt F. A single-gene cause in 29.5% of cases of steroid-resistant nephrotic syndrome. J Am Soc Nephrol. 2015;26:1279–1289. doi: 10.1681/ASN.2014050489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rheault MN, Gbadegesin RA. The Genetics of Nephrotic Syndrome. J Pediatr Genet. 2016;5:15–24. doi: 10.1055/s-0035-1557109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bullich G, Ballarin J, Oliver A, Ayasreh N, Silva I, Santin S, Diaz-Encarnacion MM, Torra R, Ars E. HLA-DQA1 and PLA2R1 polymorphisms and risk of idiopathic membranous nephropathy. Clin J Am Soc Nephrol. 2014;9:335–343. doi: 10.2215/CJN.05310513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR. Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science. 2010;329:841–845. doi: 10.1126/science.1193032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, Voinescu C, Patel N, Pearce K, Hubank M, Stephens HA, Laundy V, Padmanabhan S, Zawadzka A, Hofstra JM, Coenen MJ, den Heijer M, Kiemeney LA, Bacq-Daian D, Stengel B, Powis SH, Brenchley P, Feehally J, Rees AJ, Debiec H, Wetzels JF, Ronco P, Mathieson PW, Kleta R. Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med. 2011;364:616–626. doi: 10.1056/NEJMoa1009742. [DOI] [PubMed] [Google Scholar]
- 37.Feehally J, Kendell NP, Swift PG, Walls J. High incidence of minimal change nephrotic syndrome in Asians. Arch Dis Child. 1985;60:1018–1020. doi: 10.1136/adc.60.11.1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bonilla-Felix M, Parra C, Dajani T, Ferris M, Swinford RD, Portman RJ, Verani R. Changing patterns in the histopathology of idiopathic nephrotic syndrome in children. Kidney Int. 1999;55:1885–1890. doi: 10.1046/j.1523-1755.1999.00408.x. [DOI] [PubMed] [Google Scholar]
- 39.Kim JS, Bellew CA, Silverstein DM, Aviles DH, Boineau FG, Vehaskari VM. High incidence of initial and late steroid resistance in childhood nephrotic syndrome. Kidney Int. 2005;68:1275–1281. doi: 10.1111/j.1523-1755.2005.00524.x. [DOI] [PubMed] [Google Scholar]
- 40.Gee HY, Ashraf S, Wan X, Vega-Warner V, Esteve-Rudd J, Lovric S, Fang H, Hurd TW, Sadowski CE, Allen SJ, Otto EA, Korkmaz E, Washburn J, Levy S, Williams DS, Bakkaloglu SA, Zolotnitskaya A, Ozaltin F, Zhou W, Hildebrandt F. Mutations in EMP2 cause childhood-onset nephrotic syndrome. Am J Hum Genet. 2014;94:884–890. doi: 10.1016/j.ajhg.2014.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kari JA, Sinnott P, Khan H, Trompeter RS, Snodgrass GJ. Familial steroid-responsive nephrotic syndrome and HLA antigens in Bengali children. Pediatr Nephrol. 2001;16:346–349. doi: 10.1007/s004670000549. [DOI] [PubMed] [Google Scholar]
- 42.Landau D, Oved T, Geiger D, Abizov L, Shalev H, Parvari R. Familial steroid-sensitive nephrotic syndrome in Southern Israel: clinical and genetic observations. Pediatr Nephrol. 2007;22:661–669. doi: 10.1007/s00467-006-0409-7. [DOI] [PubMed] [Google Scholar]
- 43.Moncrieff MW, White RH, Glasgow EF, Winterborn MH, Cameron JS, Ogg CS. The familial nephrotic syndrome II A clinicopathological study. Clin Nephrol. 1973;1:220–229. [PubMed] [Google Scholar]
- 44.Motoyama O, Sugawara H, Hatano M, Fujisawa T, Iitaka K. Steroid-sensitive nephrotic syndrome in two families. Clin Exp Nephrol. 2009;13:170–173. doi: 10.1007/s10157-008-0117-7. [DOI] [PubMed] [Google Scholar]
- 45.Xia Y, Mao J, Jin X, Wang W, Du L, Liu A. Familial steroid-sensitive idiopathic nephrotic syndrome: seven cases from three families in China. Clinics (Sao Paulo) 2013;68:628–631. doi: 10.6061/clinics/2013(05)08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saleem MA. New developments in steroid-resistant nephrotic syndrome. Pediatr Nephrol. 2013;28:699–709. doi: 10.1007/s00467-012-2239-0. [DOI] [PubMed] [Google Scholar]
- 47.Ruf RG, Fuchshuber A, Karle SM, Lemainque A, Huck K, Wienker T, Otto E, Hildebrandt F. Identification of the first gene locus (SSNS1) for steroid-sensitive nephrotic syndrome on chromosome 2p. J Am Soc Nephrol. 2003;14:1897–1900. doi: 10.1097/01.asn.0000070070.03811.02. [DOI] [PubMed] [Google Scholar]
- 48.Hinkes B, Wiggins RC, Gbadegesin R, Vlangos CN, Seelow D, Nurnberg G, Garg P, Verma R, Chaib H, Hoskins BE, Ashraf S, Becker C, Hennies HC, Goyal M, Wharram BL, Schachter AD, Mudumana S, Drummond I, Kerjaschki D, Waldherr R, Dietrich A, Ozaltin F, Bakkaloglu A, Cleper R, Basel-Vanagaite L, Pohl M, Griebel M, Tsygin AN, Soylu A, Muller D, Sorli CS, Bunney TD, Katan M, Liu J, Attanasio M, O'Toole J, F, Hasselbacher K, Mucha B, Otto EA, Airik R, Kispert A, Kelley GG, Smrcka AV, Gudermann T, Holzman LB, Nurnberg P, Hildebrandt F. Positional cloning uncovers mutations in PLCE1 responsible for a nephrotic syndrome variant that may be reversible. Nat Genet. 2006;38:1397–1405. doi: 10.1038/ng1918. [DOI] [PubMed] [Google Scholar]
- 49.Moriyama T, Tsuruta Y, Shimizu A, Itabashi M, Takei T, Horita S, Uchida K, Nitta K. The significance of caveolae in the glomeruli in glomerular disease. J Clin Pathol. 2011;64:504–509. doi: 10.1136/jcp.2010.087023. [DOI] [PubMed] [Google Scholar]
- 50.Wan X, Chen Z, Choi WI, Gee HY, Hildebrandt F, Zhou W. Loss of Epithelial Membrane Protein 2 Aggravates Podocyte Injury via Upregulation of Caveolin-1. J Am Soc Nephrol. 2015;27:1066–1075. doi: 10.1681/ASN.2014121197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kitamura A, Tsukaguchi H, Hiramoto R, Shono A, Doi T, Kagami S, Iijima K. A familial childhood-onset relapsing nephrotic syndrome. Kidney Int. 2007;71:946–951. doi: 10.1038/sj.ki.5002110. [DOI] [PubMed] [Google Scholar]
- 52.Lahdenkari AT, Kestila M, Holmberg C, Koskimies O, Jalanko H. Nephrin gene (NPHS1) in patients with minimal change nephrotic syndrome (MCNS) Kidney Int. 2004;65:1856–1863. doi: 10.1111/j.1523-1755.2004.00583.x. [DOI] [PubMed] [Google Scholar]
- 53.Gee HY, Zhang F, Ashraf S, Kohl S, Sadowski CE, Vega-Warner V, Zhou W, Lovric S, Fang H, Nettleton M, Zhu JY, Hoefele J, Weber LT, Podracka L, Boor A, Fehrenbach H, Innis JW, Washburn J, Levy S, Lifton RP, Otto EA, Han Z, Hildebrandt F. KANK deficiency leads to podocyte dysfunction and nephrotic syndrome. J Clin Invest. 2015;125:2375–2384. doi: 10.1172/JCI79504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roberts IS, Gleadle JM. Familial nephropathy and multiple exostoses with exostosin-1 (EXT1) gene mutation. J Am Soc Nephrol. 2008;19:450–453. doi: 10.1681/ASN.2007080842. [DOI] [PubMed] [Google Scholar]
- 55.Park E, Chang HJ, Shin JI, J JH, Lee KB, Moon KC, Kang HG, Ha I, Cheong HI. Familial IPEX syndrome: Different glomerulopathy in two siblings. Pediatr Int. 2015;57:e59–e61. doi: 10.1111/ped.12570. [DOI] [PubMed] [Google Scholar]
- 56.Fuchshuber A, Gribouval O, Ronner V, Kroiss S, Karle S, Brandis M, Hildebrandt F. Clinical and genetic evaluation of familial steroid-responsive nephrotic syndrome in childhood. J Am Soc Nephrol. 2001;12:374–378. doi: 10.1681/ASN.V122374. [DOI] [PubMed] [Google Scholar]
- 57.Gbadegesin R, Hinkes B, Vlangos C, Mucha B, Liu J, Hopcian J, Hildebrandt F. Mutational analysis of NPHS2 and WT1 in frequently relapsing and steroid-dependent nephrotic syndrome. Pediatr Nephrol. 2007;22:509–513. doi: 10.1007/s00467-006-0377-y. [DOI] [PubMed] [Google Scholar]
- 58.Konrad M, Mytilineos J, Bouissou F, Scherer S, Gulli MP, Meissner I, Cambon-Thomsen A, Opelz G, Scharer K. HLA class II associations with idiopathic nephrotic syndrome in children. Tissue Antigens. 1994;43:275–280. doi: 10.1111/j.1399-0039.1994.tb02340.x. [DOI] [PubMed] [Google Scholar]
- 59.Clark AG, Vaughan RW, Stephens HA, Chantler C, Williams DG, Welsh KI. Genes encoding the beta-chains of HLA-DR7 and HLA-DQw2 define major susceptibility determinants for idiopathic nephrotic syndrome. Clin Sci. 1990;78:391–397. doi: 10.1042/cs0780391. [DOI] [PubMed] [Google Scholar]
- 60.Lagueruela CC, Buettner TL, Cole BR, Kissane JM, Robson AM. HLA extended haplotypes in steroid-sensitive nephrotic syndrome of childhood. Kidney Int. 1990;38:145–150. doi: 10.1038/ki.1990.179. [DOI] [PubMed] [Google Scholar]
- 61.Abe KK, Michinaga I, Hiratsuka T, Ogahara S, Naito S, Arakawa K, Tsuru N, Tokieda K. Association of DQB1*0302 alloantigens in Japanese pediatric patients with steroid-sensitive nephrotic syndrome. Nephron. 1995;70:28–34. doi: 10.1159/000188540. [DOI] [PubMed] [Google Scholar]
- 62.Kobayashi T, Ogawa A, Takahashi K, Uchiyama M. HLA-DQB1 allele associates with idiopathic nephrotic syndrome in Japanese children. Acta Paediatr Jpn. 1995;37:293–296. doi: 10.1111/j.1442-200x.1995.tb03317.x. [DOI] [PubMed] [Google Scholar]
- 63.Huang YY, Lin FJ, Fu LS, Lan JL. HLA-DR, -DQB typing of steroid-sensitive idiopathic nephrotic syndrome children in Taiwan. Nephron Clin Pract. 2009;112:c57–c64. doi: 10.1159/000213082. [DOI] [PubMed] [Google Scholar]
- 64.Zhou GP, Guo YQ, Ji YH, Zhang GL. Major histocompatibility complex class II antigens in steroid-sensitive nephrotic syndrome in Chinese children. Pediatr Nephrol. 1994;8:140–141. doi: 10.1007/BF00865460. [DOI] [PubMed] [Google Scholar]
- 65.Gbadegesin RA, Adeyemo A, Webb NJ, Greenbaum LA, Abeyagunawardena A, Thalgahagoda S, Kale A, Gipson D, Srivastava T, Lin JJ, Chand D, Hunley TE, Brophy PD, Bagga A, Sinha A, Rheault MN, Ghali J, Nicholls K, Abraham E, Janjua HS, Omoloja A, Barletta GM, Cai Y, Milford DD, O'Brien C, Awan A, Belostotsky V, Smoyer WE, Homstad A, Hall G, Wu G, Nagaraj S, Wigfall D, Foreman J, Winn MP Mid-West Pediatric Nephrology C. HLA-DQA1 and PLCG2 Are Candidate Risk Loci for Childhood-Onset Steroid-Sensitive Nephrotic Syndrome. J Am Soc Nephrol. 2015;26:1701–1710. doi: 10.1681/ASN.2014030247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sekula P, Li Y, Stanescu HC, Wuttke M, Ekici AB, Bockenhauer D, Walz G, Powis SH, Kielstein JT, Brenchley P, Investigators G, Eckardt K-U, Kronenberg F, Kleta R, Köttgen A. Genetic risk variants for membranous nephropathy: extension of and association with other chronic kidney disease aetiologies. Nephrol Dial Transplant. 2016 doi: 10.1093/ndt/gfw001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fennessy M, Hitman GA, Moore RH, Metcalfe K, Medcraft J, Sinico RA, Mustonen JT, D'Amico G. HLA-DQ gene polymorphism in primary IgA nephropathy in three European populations. Kidney Int. 1995;49:477–480. doi: 10.1038/ki.1996.67. [DOI] [PubMed] [Google Scholar]
- 68.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Battaglia GG, Garozzo M, Maixnerova D, Tesar V, Eitner F, Rauen T, Floege J, Kovacs T, Nagy J, Mucha K, Paczek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Barratt J, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, Gharavi AG. Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet. 2014;46:1187–1196. doi: 10.1038/ng.3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Beck LH, Jr, Bonegio RG, Lambeau G, Beck DM, Powell DW, Cummins TD, Klein JB, Salant DJ. M-type phospholipase A2 receptor as target antigen in idiopathic membranous nephropathy. N Engl J Med. 2009;361:11–21. doi: 10.1056/NEJMoa0810457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tomas NM, Beck LH, Jr, Meyer-Schwesinger C, Seitz-Polski B, Ma H, Zahner G, Dolla G, Hoxha E, Helmchen U, Dabert-Gay AS, Debayle D, Merchant M, Klein J, Salant DJ, Stahl RA, Lambeau G. Thrombospondin type-1 domain-containing 7A in idiopathic membranous nephropathy. N Engl J Med. 2014;371:2277–2287. doi: 10.1056/NEJMoa1409354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lyons PA, Rayner TF, Trivedi S, Holle JU, Watts RA, Jayne DRW, Baslund B, Brenchley P, Bruchfeld A, Chaudhry AN, Tervaert JWC, Deloukas P, Feighery C, Gross WL, Guillevin L, Gunnarsson I, Harper L, Hruskova Z, Little MA, Martorana D, Neumann T, Ohlsson S, Padmanabhan S, Pusey CD, Salama AD, Sanders JSF, Savage CO, Segelmark M, Stegeman CA, Tesar V, Vaglio A, Wieczorek S, Wilde B, Zwerina J, Rees AJ, Clayton DG, Smith KGC. Genetically Distinct Subsets within ANCA-Associated Vasculitis. New Engl J Med. 2012;367:214–223. doi: 10.1056/NEJMoa1108735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramanathan AS, Senguttuvan P, Chinniah R, Vijayan M, Thirunavukkarasu M, Raju K, Mani D, Ravi PM, Rajendran P, Krishnan JI, Karuppiah B. Association of HLA-DR/DQ Alleles and Haplotypes with Nephrotic Syndrome. Nephrology. 2015 doi: 10.1111/nep.12669. [DOI] [PubMed] [Google Scholar]