Abstract
The molecular mechanism underlying non‐alcoholic fatty liver disease progression to hepatocellular carcinoma (HCC) remains unknown. In this study, immunohistochemistry staining results showed that NS5ABP37 protein, which is in a state of lower expression in tumor tissues, decreased with increasing degree of HCC malignancy. Two cell models, HepG2 and L02, were used to analyze the mechanism between NS5ABP37 and HCC. In agreement, NS5ABP37 protein overexpression significantly suppressed cell proliferation, caused G1/S cell cycle arrest, and induced apoptosis by increasing caspase‐3/7 activity and cleaved caspase‐3 levels. In addition, NS5ABP37 overexpression resulted in decreased intracellular triglyceride and total cholesterol contents, with level reduction in sterol regulatory element‐binding proteins (SREBPs) and downstream effectors. Furthermore, NS5ABP37 overexpression decreased SREBP1c and SREBP2 levels by reducing their respective promoters. Finally, reactive oxygen species levels and endoplasmic reticulum stress were both induced by NS5ABP37 overexpression. These findings together indicate that NS5ABP37 inhibits cancer cell proliferation and promotes apoptosis, by altering SREBP‐dependent lipogenesis and cholesterogenesis in HepG2 and L02 cells and inducing oxidative stress and endoplasmic reticulum stress.
Keywords: ER stress, lipid metabolism, liver cancer, NS5ABP37, oxidative stress
Hepatocellular carcinoma (HCC) is the sixth most common malignant cancer worldwide, and the third leading cause of cancer‐related death.1, 2 Hepatocellular carcinoma still has poor outcome, with no effective therapy available for advanced liver cancer. For a long time, research focus was placed on different genetic alterations caused by viral infections, alcohol consumption, and intake of aflatoxin‐contaminated food that ultimately leads to hepatocarcinogenesis.3 Currently, carcinogenesis is also thought to be closely related with lipid metabolism. Increasing epidemiologic evidence indicates a close relationship between non‐alcoholic fatty liver disease (NAFLD) and HCC.4, 5, 6, 7 However, the underlying mechanisms are largely unknown, due in part to disease complexity. Therefore, novel targets that could block lipogenesis and cholesterogenesis should be studied in HCC.
NS5ABP37 is a novel hepatitis C virus non‐structural protein (NS)5A‐associated binding protein that was isolated using the yeast two‐hybrid method by our group in 2003 (GenBank Accession No. AF543840.1). It is also known as FNDC3B, FAD104, PRO4979, and YVTM2421. Based on the NCBI database, the FNDC3B gene is located at 3q26.31 and comprises 3615 bp that encode a protein with 1204 amino acid residues; NS5ABP37 is the same protein as FNDC3B.
Recent studies reported that NS5ABP37 plays an important role in HCC. Indeed, NS5ABP37 was identified in HCC in an oncogenomic screen. It was shown that FNDC3B overexpression induces epithelial–mesenchymal transition and activates several cancer pathways, including phosphatidylinositol 3‐kinase/protein kinase B, retinoblastoma 1, and transforming growth factor‐β signaling.8 Study of factor for adipocyte differentiation‐104 (FAD104)‐deficient mice indicated that this protein is essential for newborn survival, and important for cell proliferation, adhesion, spreading, and migration.9 However, other studies indicated that FNDC3B may be a tumor inhibitor. It was shown that microRNA‐143 upregulation promotes cancer cell invasion/migration and tumor metastasis by repressing FNDC3B expression.10, 11 Melanoma cells stably expressing FAD104 showed reduced formation of lung colonization.12 Taken together, it is essential to comprehensively assess the role of NS5ABP37 in liver cancer development and the underlying mechanism.
It was also reported that the FAD104 gene is involved in adipogenesis. Indeed, it was demonstrated that FAD104 expression levels quickly increase in the early stage of adipogenesis, and differ between adipocytes and non‐adipogenic cells.13 Liver is an important organ for fatty synthesis. However, studies assessing the role of NS5ABP37 in liver lipid metabolism are scarce.
Our interest in the role of NS5ABP37 as a bridge between liver cancer and lipid metabolism initially stemmed from the yeast two‐hybrid system and suppression subtractive hybridization data, which showed that NS5ABP37 may be involved in cell growth regulation, cell apoptosis, glycometabolism, and lipid metabolism.14 In this context, we assessed a possible role of NS5ABP37 in both HCC development and liver lipid metabolism. Interestingly, NS5ABP37 inhibited cell proliferation and promoted apoptosis by altering sterol regulatory element‐binding protein (SREBP)‐dependent lipogenesis and cholesterogenesis in HepG2 cells, and inducing oxidative and endoplasmic reticulum (ER) stresses. Overall, these findings reveal a novel role for NS5ABP37 in regulating liver lipid metabolism and cancer progression.
Materials and Methods
Immunohistochemistry
A tissue microarray containing specimens from 110 HCC cases of different stages and 10 normal pancreatic tissue samples was purchased from US Biomax (BC03119a, Rockville, USA). All study protocols for clinical specimen collection were approved by the Ethics Committee of Beijing Ditan Hospital (Beijing, China) for Clinical Research. Immunohistochemistry staining for NS5ABP37 detection was carried out with routine procedures using anti‐NS5ABP37 antibody (PAB20193, 1:20; Abnova, Colorado, USA) overnight at 4°C, and HRP‐conjugated anti‐rabbit antibody (PV‐6001; ZSGB‐BIO, Beijing, China) for 30 min. Slides were counterstained with hematoxylin and analyzed under a microscope (BX51; Olympus, Tokyo, Japan).
Cell culture and transfection
HepG2 cells and L02 cells were separately cultured in DMEM supplemented with 10% FBS at 37°C with 5% CO2. A total of 200 000 cells (in 2 mL cell medium) were seeded per well in 6‐well plates, and allowed to grow to 60–80% confluency. Cells were then transiently transfected with plasmid or siRNA using jetPRIME (Polyplus‐transfection, Eastern France) according to the manufacturer's protocol. After 24 h of incubation at 37°C in 5% CO2, the transfection medium was replaced.
Plasmids and siRNA oligonucleotides
The pENTER (pNC) and Penter‐NS5ABP37 (pNS5ABP37) plasmids were purchased from Vigene (ShanDong, China); siRNA targeting NS5ABP37 and a negative control siRNA were purchased from GenePharma (Jiangsu, China) (Table 1). The pGL4.10 vector‐SREBP1c promoter and pGL4.10 vector‐SREBP2 were kind gifts from Dr. Lili Gao and Dr. Min Li.15, 16
Table 1.
Small interfering RNA oligonucleotides used in the study
| Gene | RNA duplex sequences (5′–3′) |
|---|---|
| siRNA‐NS5ABP37 | |
| Sense | GCAGCUGCACAACAGUAUATT |
| Anti‐sense | UAUACUGUUGUGCAGCUGCTT |
| Negative control | |
| Sense | UUCUCCGAACGUGUCACGUTT |
| Anti‐sense | ACGUGACACGUUCGGAGAATT |
RNA isolation and quantitative RT‐PCR
Total RNA from transfected HepG2 cells and L02 cells were separately prepared using a Total RNA Kit (R6834; Omega, Georgia, USA) according to the manufacturer's instructions, and reverse transcribed into first‐strand cDNA with the PrimeScript RT reagent kit (DRR027A; TaKaRa, Dalian, China). Then cDNA was subjected to quantitative PCR (4472908; ABI, Massachusetts, USA) amplification using specific primers. The relative mRNA amounts of SREBP1c, SREBP2, HMG‐CoA reductase (HMGCR), and fatty acid synthase (FASN) were determined by the comparative Ct method (ΔΔCt), and normalized to β‐actin levels. The primers used are listed in Table 2 along with their sequences.
Table 2.
Optimized primers used in the study
| Genes (all human) | Primer sequence (5′–3′) |
|---|---|
| β‐actin | |
| Sense | GAGGGATGGAAGGGTCTAAG |
| Anti‐sense | GCCTGGGCTGTGGTAAGT |
| NS5ABP37 | |
| Sense | GAGCATGCTGCATCAGTACC |
| Anti‐sense | GTGCGAACAGGGAGACTTTC |
| SREBP1c | |
| Sense | GGAGGGGTAGGGCCAACGGCCT |
| Anti‐sense | CATGTCTTCGAAAGTGCAATCC |
| SREBP2 | |
| Sense | CCCTTCAGTGCAACGGTCATTCAC |
| Anti‐sense | TGCCATTGGCCGTTTGTGTC |
| FASN | |
| Sense | AGCTGCCAGAGTCGGAGAAC |
| Anti‐sense | TGTAGCCCACGAGTGTCTCG |
| HMGCR | |
| Sense | CTTGTGTGTCCTTGGTATTAGAGC |
| Anti‐sense | ATCATCTTGACCCTCTGAGTTACAG |
| XBP‐1 | |
| Sense | AAACAGAGTAGCAGCTCAGACGC |
| Anti‐sense | TCCTTCTGGGTAGACCTCTGGGAG |
Western blot analysis
Cells were adequately lysed in lysis buffer (Thermo Fisher Scientific, Massachusetts, USA) containing a protease inhibitor cocktail (04693132001; Roche, Basel, Switzerland) for 30 min on ice. The resulting lysates were cleared at 12 000g for 15 min at 4°C. Protein concentrations were determined by the Pierce BCA assay (23225; Thermo Fisher Scientific). Equal amounts of protein from each sample were separated by 12% SDS‐PAGE (NP0341BOX; Invitrogen, California, USA) and transferred onto PVDF membranes (03010104000; Roche, Basel, Switzerland). After blocking with 5% non‐fat dry milk (2321000; Becton Dickinson, New Jersey, USA) for 2 h, the membranes were incubated overnight at 4°C with primary anti‐SREBP1c (SC‐8984; Santa Cruz Biotechnology, California, USA), anti‐SREBP2 (ab30682; Abcam, Cambridge, USA), anti‐HMGCR (ab74830; Abcam), anti‐FASN (ab128870; Abcam), anti‐cleaved caspase‐3 (9661s; Cell Signaling Technology, Boston, USA), anti‐glucose regulated protein (GRP)78 (3177; Cell Signaling Technology), anti‐inositol‐requiring enzyme (IRE)‐1a (ab37073; Abcam, USA), anti‐phospho (p)IRE‐1a (ab124945; Abcam), anti‐NS5ABP37 (NBP1‐90495; Novus, Colorado, USA), and anti‐GAPDH (5174; Cell Signaling Technology) antibodies, respectively. Membranes were then washed three times with TBS‐Tween, and incubated with anti‐rabbit or anti‐mouse secondary antibodies for 1 h at room temperature, followed by three washes with TBS‐Tween. Protein bands were detected using an enhanced chemiluminescence system (32209; Thermo Fisher Scientific) and analyzed with Bio1D software (Vilber, S: 11. 640150, Paris, France).
Apoptosis assessment
Cells were harvested 48 h after transfection, washed twice with cold BioLegend cell staining buffer (cat # 420201), and resuspended in annexin V binding buffer (cat. #422201) at a density of 106 cells/mL. Then 4 μL each of FITC, annexin V, and 7‐amino‐actinomycin D (7‐AAD; cat. #420401) were added successively. Before analysis by flow cytometry on FACSCalibur (BD Biosciences, New Jersey, USA), the cells were gently vortexed and incubated for 15 min at room temperature (25°C) in the dark.
Cell cycle analysis
Cells were harvested 48 h after transfection and washed with cold 1 × PBS. Cells were fixed with 70% ethanol for 24 h at 4°C. The next day, they were stained with 4 μL 7‐AAD reagent (cat. #420401). After incubation for 15 min at room temperature (25°C) in the dark, cell cycle distribution was analyzed by flow cytometry using CellQuest software (Becton Dickinson).
Caspase‐ 3/7 activity detection
Cells were seeded at a density of 10 000 cells/well (100 μL DMEM containing 10% FBS) in 96‐well plates 24 h before transfection. Then 100 μL caspase‐Glo 3/7 reagent (G8092; Promega, Madison, WI 53711, USA) were added to each well 48 h after transfection. The cells were lysed at room temperature for 1 h. Luminescence was measured using a Veritas plate‐reading luminometer (Turner Biosystems, Sunnyvale, CA, California, USA).
Cell proliferation assay
Cells were seeded into 96‐well plates at a density of 10 000 cells per well before transfection. For each well containing 100 μL culture medium, 10 μL CCK‐8 solution (EQ645; Dojindo, Kumamoto, Japan) was added 24, 48, and 72 h post‐transfection. After 40 min of incubation at 37°C, optical density values were obtained at 450 nm (Varioskan Flash; Thermo Fisher Scientific).
Oil red O staining
Cells were plated in 12‐well culture dishes in DMEM containing 10% FBS before transfection. Forty‐eight hours after transfection, HepG2 cells were washed three times with PBS and fixed with 4% formaldehyde for 30 min. Cells were then incubated with 60% filtered Oil red O solution (O0625; Sigma‐Aldrich, Missouri, Saint Louis, USA) for 30 min at room temperature after three washes with distilled water. After a brief washing step with 60% isopropanol, cells were stained with hematoxylin (Sigma‐Aldrich) and fixed with neutral balata. Oil red O stained cells were analyzed under a microscope (BX51; Olympus).
Determination of total intracellular triglyceride amounts
Cells were plated in 12‐well culture dishes in DMEM medium containing 10% FBS before transfection. Forty‐eight hours after transfection, intracellular triglyceride (TG) contents were measured using an Adipogenesis Assay Kit (MAK040; Sigma‐Aldrich) according to the manufacturer's instructions, and normalized to total protein concentrations. Intracellular TG levels were expressed in nmol/μg protein.
Determination of intracellular total cholesterol levels
Cells were plated in 12‐well culture dishes in DMEM medium containing 10% FBS before transfection. Cells were counted before the use of a Cholesterol/Cholesteryl Ester Quantitation Kit (K603‐100; Biovision, Milpitas, CA, USA) to extract and quantitate intracellular total cholesterol (TC) levels, according to the manufacturer's instructions, 48 h after transfection. Intracellular TC amounts were expressed in μg/106 cells.
Luciferase reporter assay
Transfection assays were carried out using jetPRIME (Polyplus‐transfection) in 48‐well culture dishes, with 125 ng SREBP1c or SREBP2 promoter reporter plasmids (pGL4.10‐SREBP1c‐P or pGL4.10‐SREBP1c‐P) and 8 ng Renilla luciferase vector (pRL‐TK) DNA 24 h after seeding. In some experiments, cells were cotransfected with 125 ng indicated expression vector (pENTER, pENTER‐NS5ABP37, siNC, or siNS5ABP37). After 24 h of transfection, cells were lysed in Passive Lysis Buffer (E1941; Promega). SREBP1c or SREBP2 promoter activities were measured on a microplate luminometer, according to the technical manual for the Dual‐Luciferase Reporter Assay System (E1910; Promega).
Quantitation of intracellular levels of reactive oxygen species
Intracellular reactive oxygen species (ROS) levels were assessed by using the oxidation‐sensitive fluorescent probe 2,7‐dichlorofluorescein diacetate (DCFH‐DA) (S0033; Beyotime, Shanghai, China). Forty‐eight hours after transfection, cells were washed twice in PBS and incubated with 10 μmol/L DCFH‐DA at 37°C for 20 min, according to the manufacturer's instructions. The DCFH‐DA is deacetylated intracellularly by a non‐specific esterase, and further oxidized by ROS to yield the fluorescent compound 2,7‐dichlorofluorescein (DCF). The DCF fluorescence was measured on a FACSCalibur flow cytometer (BD Biosciences). A total of 20 000 events were assessed per sample.
X‐box binding protein‐1 mRNA splicing assay
X‐box binding protein‐1 (XBP‐1) mRNA was amplified from 50 ng cDNA using specific primers (0.6 μM), MgCl2 (250 mM), and Simpler Red Taq DNA polymerase (0.25 U) from Applied Biosystems (Foster City, CA, USA), in a final volume of 25 μL at an annealing temperature of 66°C for 35 cycles. The primers used in this study are listed in Table 2 along with their sequences. The PCR products were digested with PstI and separated on a 3% agarose gel. A 448‐bp amplicon indicated spliced XBP‐1).
Statistical analysis
The experiments were carried out in triplicate, and repeated a total of three times. Data were compared by paired Student's t‐test using spss software version 16.0 (SPSS, Chicago, IL, USA). Data are mean ± SD, and P < 0.05 was considered statistically significant.
Results
NS5ABP37 levels decrease with increasing degree of HCC malignancy. To assess NS5ABP37 protein expression differences between HCC and normal liver tissues, a tissue microarray containing 110 specimens from HCC liver cases (2 stage I, 49 stage II, 57 stage III, and 2 stage IV cases) alongside 10 normal liver tissue samples was analyzed by immunohistochemistry. NS5ABP37 immunoreactivity was mainly distributed in the cytoplasm whereas limited signals were found in the nucleus. Interestingly, the NS5ABP37 protein was overexpressed in 9 (90%) normal liver tissue samples, whereas high NS5ABP37 expression was detected in only 1 (50%) stage I, 16 (32.7%) stage II, and 18 (31.6%) stage III HCC liver tissues. In the two stage IV HCC liver tissue samples, NS5ABP37 expression was not detected (Fig. 1).
Figure 1.
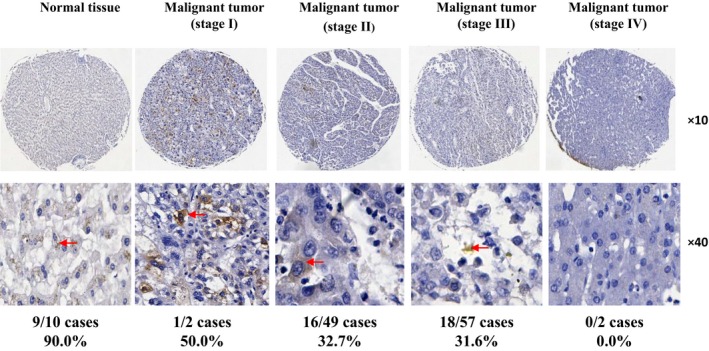
NS5ABP37 levels decrease with increasing degree of hepatocellular carcinoma (HCC) malignancy. The tissue microarray used in this study was purchased from US Biomax (BC03119a). Immunohistochemistry staining of NS5ABP37 in liver specimens from patients with different stages of HCC was carried out according to routine protocols. Representative images for HCC and normal liver tissues are shown.
Expression of NS5ABP37 mRNA and protein were downregulated in HCC cell lines. To determine the expression of NS5ABP37 in HCC cell lines, quantitative PCR and Western blot analysis were used to examine the mRNA and protein levels. The results showed that both the mRNA and protein of NS5ABP37 were detectable in HepG2, Huh7, and L02 cell lines. However, the expression of NS5ABP37 mRNA and protein were downregulated in HepG2 and Huh7 cells compared with that in L02 cells (Fig. 2).
Figure 2.
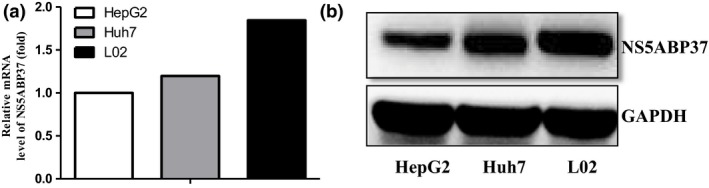
Expressions of NS5ABP37 mRNA and protein were downregulated in hepatocellular carcinoma cell lines. The mRNA and protein were extracted from HepG2, Huh7 and L02 cells. The mRNA level (A) and protein level (B) were detected by RT‐PCR and Western blot analysis, respectively.
Successful overexpression and silencing of NS5ABP37 in HepG2 cells and L02 cells. To assess the potential role of NS5ABP37 in cellular functions, the NS5ABP37 plasmid and siRNA were purchased. HepG2 and L02 cells were transiently transfected with the overexpression plasmid pENTER‐NS5ABP37 (control vector pENTER) or siRNA‐NS5ABP37 (siRNA‐NC). Compared with the control groups (pENTER), NS5ABP37 mRNA and protein levels were increased in cells transfected with the pENTER‐NS5ABP37 plasmid (Figure 3A and 3B), while NS5ABP37 mRNA and protein levels were decreased in siRNA‐NS5ABP37 transfected cells (Figure 3C and 3D ).
Figure 3.
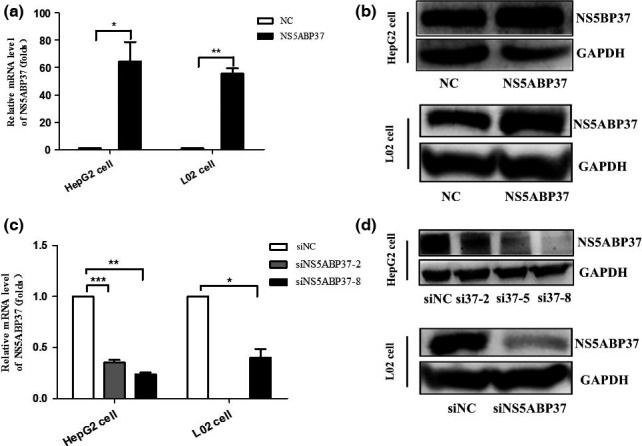
Successful overexpression and silencing of NS5ABP37 in HepG2 and L02 hepatocellular carcinoma cell lines. HepG2 cells and L02 cells were transiently transfected with pENTER (normal control [NC]), pENTER‐NS5ABP37, siRNA control (siNC), or siRNA‐NS5ABP37. After 48 h, total RNA (A,C) and protein (B,D) were extracted and analyzed for NS5ABP37 expression quantitation. Data are mean ± SD from triplicate experiments. *P < 0.05, **P < 0.005, ***P < 0.0001.
Effects of NS5ABP37 on cell proliferation, apoptosis, and cycle distribution. Previous studies reported that NS5ABP37 is associated with human cancers such as HCC,8 esophageal squamous cell cancer,17 glioblastoma,18 primary urethral clear‐cell adenocarcinoma,19 chronic lymphocytic leukemia,20 and prostate cancer.11 Therefore, we attempted to further study the effect of NS5ABP37 on the growth of cancer cells as well as normal hepatocytes. First, NS5ABP37 was overexpressed and silenced in HepG2 cells and L02 cells, as described above. As shown in Figure 4(A), NS5ABP37 overexpression significantly suppressed cell proliferation, which was promoted by its silencing, compared with the respective negative control groups.
Figure 4.
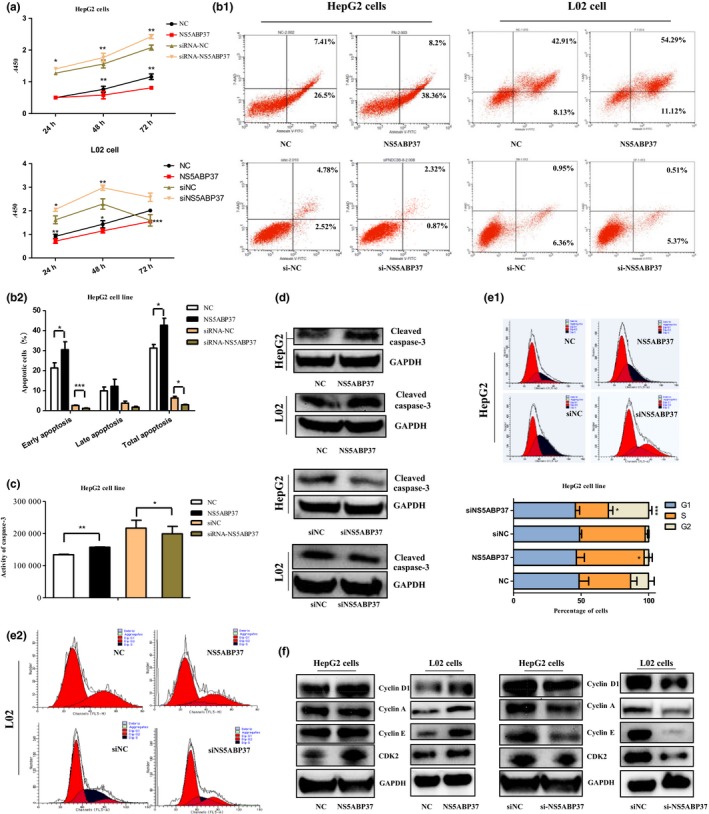
Effects of NS5ABP37 on cell proliferation, apoptosis, and cycle distribution in hepatocellular carcinoma cell lines. HepG2 and L02 cells were transiently transfected with pENTER (normal control [NC]), pENTER‐NS5ABP37, siRNA control (siNC), or siRNA‐NS5ABP37 for 48 h. (A) Cell proliferation was measured using a CCK‐8 kit. Cell apoptosis was assessed with an annexin V‐FITC/7‐AAD kit by flow cytometry (B1), and quantitated (B2). Caspase‐3 activity was detected by caspase‐3/7 activity assay (C) and Western blot using anti‐cleaved caspase‐3 antibody (D). (E1,E2) Cell cycle distribution was measured by flow cytometry and analyzed with ModFit software. (F) Cell cycle‐related proteins, including cyclin D1, cyclin A, cyclin E, and CDK2 were detected by Western blotting. Data are mean ± SD from triplicate experiments. *P < 0.05, **P < 0.005, ***P < 0.0001.
Next, annexin V/7‐AAD staining and caspase‐3/7 activity measurements were used to quantify cell apoptosis in the two cell lines. Interestingly, the result showed that NS5ABP37 overexpression increased the rates of apoptotic cells and caspase‐3/7 activity levels, which were decreased by NS5ABP37 silencing (Fig. 4B,C). As an effector of cell apoptosis, cleaved caspase‐3 is a sign of cell apoptosis. In this study, we showed that NS5ABP37 overexpression increased cleaved caspase‐3 amounts, which were decreased in the knockdown group (Fig. 4D).
In addition, we assessed whether NS5ABP37 also affects cell cycle distribution, by flow cytometry. As shown in Figure 4(E), cell cycle distribution analysis indicated that NS5ABP37 overexpression resulted in reduced cell population in the G2 phase, and increased S phase cells. NS5ABP37 silencing using siRNA caused increased numbers of cells in the G2 phase. Cyclin A, cyclin D1, cyclin E, and CDK2 are cell cycle regulators that promote cells’ entry into S stage from G1. Interestingly, Western blot analysis showed that cyclin D1, cyclin A, cyclin E and CDK2 protein amounts were increased by NS5ABP37 overexpression, and decreased in the siRNA‐NS5ABP37 group (Fig. 4F). The results showed that NS5ABP37 induced G1/S cell cycle arrest in HepG2 cells and L02 cells.
NS5ABP37 reduces triglyceride and cholesterol levels in HepG2 cells and L02 cells. As a novel gene named FAD104, NS5ABP37 is a critical factor during adipocyte differentiation. To assess NS5ABP37's function in liver lipid metabolism in liver cells, we first used staining and quantification assays to determine the changes of intracellular TG and TC levels caused by NS5ABP37. Oil red O staining results in HepG2 and L02 cells indicated that lipid droplet accumulation in cells overexpressing NS5ABP37 was decreased in comparison with negative controls, and increased in the knockdown group (Fig. 5A). In agreement, NS5ABP37 overexpression markedly decreased intracellular TG (Fig. 5B,C) and TC (Fig. 5D,E) levels compared with the control group, whereas intracellular TG and TC contents were both significantly increased following NS5ABP37 silencing. The downregulation of TG and TC levels by FNDC3B revealed that FNDC3B might be associated with lipid metabolism.
Figure 5.
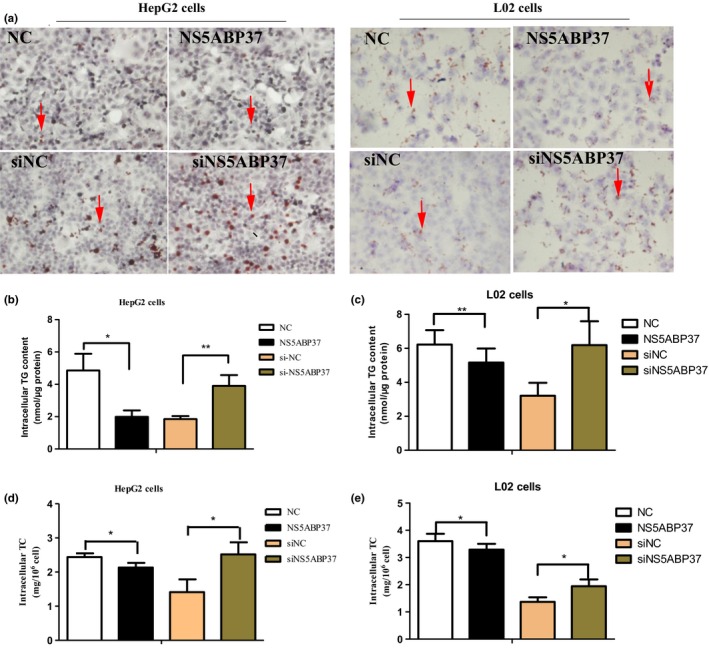
NS5ABP37 reduces triglyceride (TG) and cholesterol (TC) levels in HepG2 and L02 hepatocellular carcinoma cells. HepG2 and L02 cells were transiently transfected with pENTER (normal control [NC]), pENTER‐NS5ABP37, siRNA control (siNC), or siRNA‐NS5ABP37. (A) After 48 h, fatty acid levels were determined by Oil red O staining. Lipid droplets in HepG2 cells are shown by red arrows. (B,C) Cellular triglyceride levels were assessed using a triglyceride estimation kit. (D,E) Intracellular total cholesterol amounts were measured with a cholesterol estimation kit. Data are mean ± SD from triplicate experiments. *P < 0.05, **P < 0.005.
NS5ABP37 inhibits SREBP transcription activity in HepG2 and L02 cells. To further study the role of NS5ABP37 in lipid metabolism, the expression levels of genes involved in TG and TC anabolism were assessed. As shown in Figure 6, the mRNA (Fig. 6A,B) and protein (Fig. 6C,D) levels of both SREBP‐1c and SREBP‐2, as well as their downstream effectors FASN and HMGCR, were all decreased by NS5ABP37 overexpression in both HepG2 and L02 cells. Conversely, NS5ABP37 silencing resulted in increased mRNA (Fig. 6A,B) and protein (Fig. 6C,D) expression levels of these genes. As it was shown in Figure 1 that limited NS5ABP37 amounts were located in the nucleus, this protein may possess transcriptional activity. To assess whether NS5ABP37 directly regulates the transcriptional activity of SREBP1c and SREBP2 promoters, pGL4.10‐SREBP1c (nt −1005 to +24) or pGL4.10‐ SREBP2 (nt −933 to +263) promoter was cotransfected with pENTER‐NS5ABP37 (pENTER) and siRNA‐NS5ABP37 (siRNA‐NC), respectively. As shown in Figure 6(E,F), NS5ABP37 overexpression significantly reduced both SREBP1c and SREBP2 promoter activities compared with the control group; NS5ABP37 silencing resulted in the opposite effects.
Figure 6.
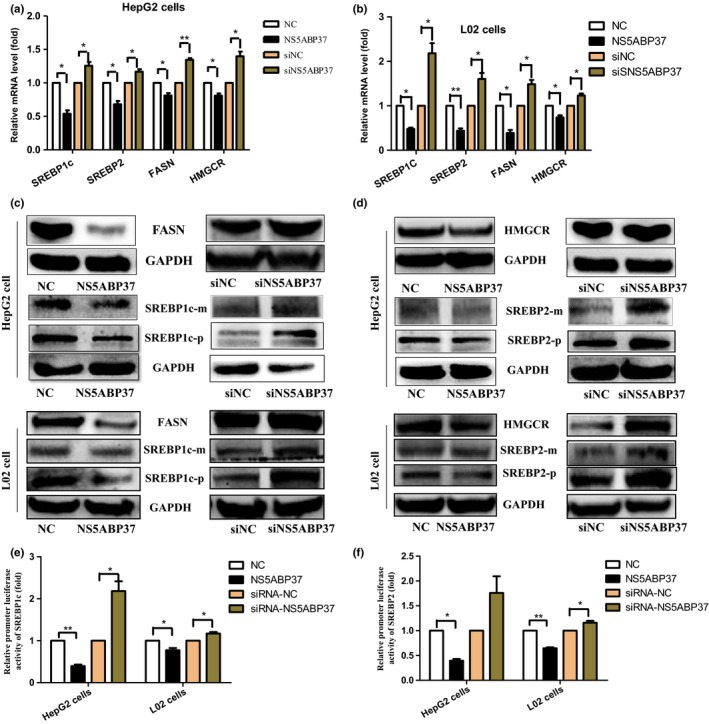
NS5ABP37 inhibits sterol regulatory element‐binding protein (SREBP) transcription activity in HepG2 and L02 hepatocellular carcinoma cells. HepG2 and L02 cells were transiently transfected with pENTER (normal control [NC]), pENTER‐NS5ABP37, siRNA control (siNC), or siRNA‐NS5ABP37. After 48 h, the mRNAand protein were extracted. The mRNA (A,B) and protein (C,D) levels of SREBP2, HMG‐CoA reductase (HMGCR), SREBP1c, and fatty acid synthase (FASN) were measured by RT‐PCR and Western blot analysis, respectively. Finally, HepG2 and L02 cells were transiently cotransfected with SREBP1c or SREBP2 promoter and plasmid pENTER, pENTER‐NS5ABP37, siRNA control, or siRNA‐NS5ABP37. After 24 h, SREBP1c and SREBP2 promoter activities were determined in a microplate luminometer (E,F). Data are mean ± SD from triplicate experiments. *P < 0.05, **P < 0.005.
Taken together, these data suggested that inhibition of SREBPs by NS5ABP37 caused decreased TG and TC levels in HepG2 and L02 cells.
NS5ABP37 induces oxidative stress and alters the expression of genes involved in ER stress response. Reactive oxygen species‐mediated oxidative stress is a common pathogenic feature in hepatic pathogenesis, including NAFLD and HCC.21, 22, 23 As the production of ROS is a significant contributor in cell proliferation and survival, we assessed whether NS5ABP37 could affect ROS production in different hepatocytes. As shown in Figure 7(A), NS5ABP37 overexpression increased ROS levels compared with the negative control group, whereas its silencing resulted in decreased ROS levels.
Figure 7.
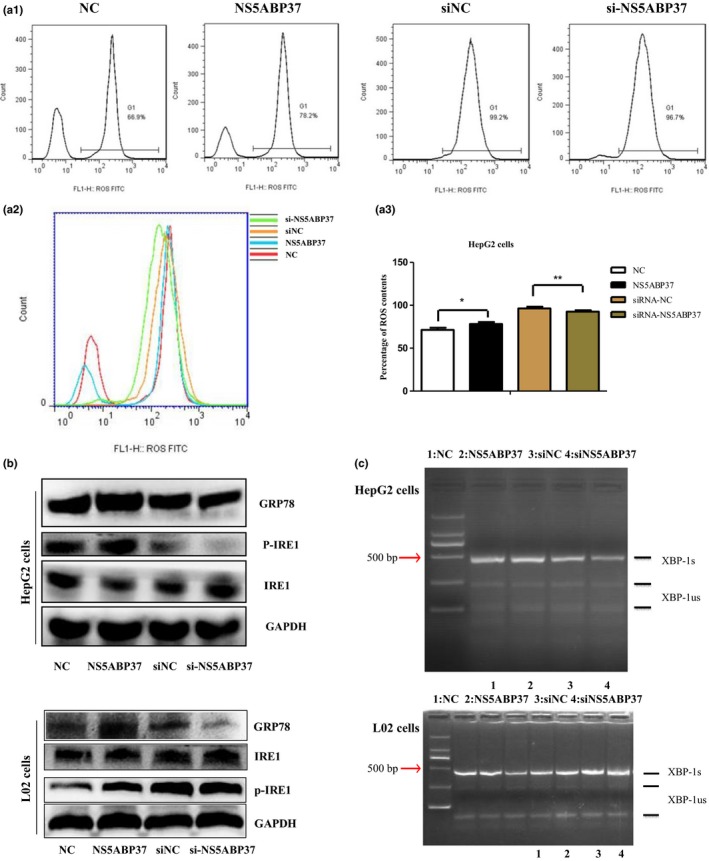
NS5ABP37 induces oxidative stress and alters the expression of genes involved in endoplasmic reticulum stress response. HepG2 cells were transiently transfected with pENTER (normal control [NC]), pENTER‐NS5ABP37, siRNA control (siNC), or siRNA‐NS5ABP37. After 48 h, intracellular reactive oxygen species (ROS) levels were detected with an oxidation‐sensitive fluorescent probe (2,7‐dichlorofluorescein diacetate) (A1,A2) and quantified (A3). (B) Protein levels of endoplasmic reticulum stress‐related genes in HepG2 and L02 cells were detected by Western blot analysis. (C) Splicing of X‐box binding protein‐1 (XBP‐1) was determined by RT‐PCR. Bands representing the unspliced (XBP‐1us) and spliced (XBP‐1s) transcripts are labeled. Data are mean ± SD from triplicate experiments. *P < 0.05, **P < 0.005.
Endoplasmic reticulum stress, also referred to as unfolded protein response, is a stress pathway activated in response to the accumulation of misfolded proteins in the ER. By Western blot analysis we found that ER stress‐related proteins, such as GRP78 and pIRE‐1a, were both increased by NS5ABP37 overexpression (Fig. 7B). Splicing of XBP‐1 was also assessed by RT‐PCR. As shown in Figure 7(C), XBP‐1 mRNA splicing was increased in different hepatocytes transfected with the pENTER‐NS5ABP37 plasmid. NS5ABP37 silencing resulted in decreased XBP‐1 mRNA splicing, indicating that NS5ABP37 played a key role in ER stress.
Discussion
NS5ABP37, also named FNDC3B or FAD104, is an important gene in both cancer and adipogenesis.8, 10, 11, 13 Using the yeast two‐hybrid method, NS5ABP37 was identified by our team. Further research using the yeast two‐hybrid system and suppression subtractive hybridization techniques found that MMP25, glutamate‐ammonia ligase, and protein kinase C are potential proteins interacting with NS5ABP37.14 These findings indicated that NS5ABP37 may be involved in cell growth, cell apoptosis, glycometabolism, and lipid metabolism. An oncogenomic screen indicated that NS5ABP37 overexpression induces epithelial–mesenchymal transition and activates several cancer pathways.8
Researchers have revealed that NS5ABP37 is important for cell proliferation, adhesion, spreading, and migration.9 It was also reported that NS5ABP37, downregulated by microRNA‐143, promotes cancer cell invasion/migration and tumor metastasis.10, 11 In this study, we found lower NS5ABP37 expression levels in HCC tissue samples compared with normal liver tissue specimens, and NS5ABP37 amounts decreased with increasing degree of HCC malignancy. In vitro, NS5ABP37 overexpression inhibited cell proliferation by inducing apoptosis and G1/S cell cycle arrest.
Recently, an increasing number of cancer studies have focused on metabolic processes. Indeed, lipid metabolism is a key player in metabolic reprogramming,24, 25, 26 including lipogenesis and cholesterol biosynthesis as two main processes. Sterol regulatory element‐binding proteins, especially SREBP‐1c and SREBP‐2, are key transcription factors controlling lipogenesis and cholesterogenesis. Interesting data in previous studies indicated that high levels of SREBPs are significantly associated with aggressive pathologic features in human cancer.27, 28, 29, 30
Based on NS5ABP37's role in adipogenesis,13 this study assessed its function in liver lipogenesis and cholesterol biosynthesis. As shown above, NS5ABP37 overexpression resulted in decreased intracellular TG and TC levels.
Fatty acids are indispensable elements of biological membrane lipids, and important substrates for energy metabolism. Studies in the past decades have acknowledged that SREBP‐1 may serve as a prognostic marker in HCC and promote tumor progression by inducing cell growth and metastasis.30 In addition, SREBP activity contributes to protein kinase B‐dependent cell growth, which requires mammalian target of rapamycin complex 1 activity.28, 29 As a key enzyme in the lipogenesis pathway, FASN is considered a metabolic oncogene. Its overexpression is a molecular marker for poor prognosis and malignant phenotype in many cancers, playing an active part in the development, maintenance, and metastatic progression of human cancer.25, 31, 32 This study showed that NS5ABP37 downregulated SREBP1c at the mRNA and protein levels, by inhibiting its promoter activity. In addition, the expression of FASN, the downstream effector of SREBP1c, was also decreased by NS5ABP37.
Cholesterol, an important ingredient of lipid rafts and the cell membrane, acts as the assembly center for signaling molecules.33 Hence, rapidly proliferating cancer cells, such as liver cancer cells, are inclined to have superior requirement for cholesterol.34 A clinical trial showed that low serum cholesterol level and body mass index could serve as prognostic factors to predict postoperative outcomes in HCC patients undergoing surgical hepatectomy.35 As a key factor in cholesterol metabolism, SREBP2 plays a role in sterol feedback regulation loss in prostate cancer cells.27 As shown above, NS5ABP37 downregulated SREBP2 at the mRNA and protein levels by inhibiting its promoter activity; this resulted in decreased expression of HMGCR, a downstream effector of SREBP2.
Reactive oxygen species play a very important role in apoptosis induction under both physiological and pathological conditions.36 Recent studies revealed relationships between oxidative stress and liver pathogenesis, including NAFLD and HCC.21, 23 It is well established that ER stress alters hepatic steatosis, and diverse ER stress signaling molecules play important roles in the regulation of lipid metabolism.37 To further assess the role of NS5ABP37 in lipid metabolism and cell apoptosis, oxidative and ER stresses were assessed in this study. Interestingly, NS5ABP37 overexpression resulted in increased intracellular ROS levels. NS5ABP37 also increased the protein levels of ER stress marker GRP78, and also affected the IRE‐1a/XBP‐1 pathway in ER stress.
In conclusion, this study unveiled the molecular mechanisms by which NS5ABP37 inhibits liver cancer growth and tumorigenicity. Indeed, NS5ABP37 inhibits cell proliferation by inducing cell apoptosis, effects likely mediated by SREBP‐dependent TG and TC metabolic impairment as well as oxidative and ER stresses (Fig. 8). Thus, these findings suggest that NS5ABP37 is a potential link between NAFLD and HCC.
Figure 8.
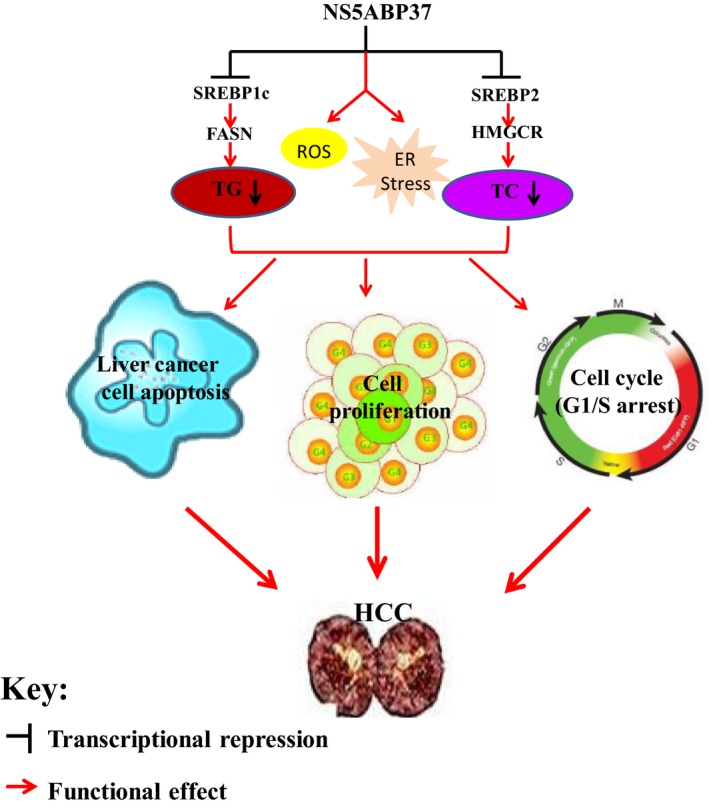
Schematic representation of the mechanisms underlying the effects of NS5ABP37 in hepatocellular carcinoma (HCC). NS5ABP37 inhibits cell proliferation by inducing cell apoptosis, which may be affected by sterol regulatory element‐binding protein (SREBP)‐dependent triglyceride (TG) and cholesterol (TC) metabolic impairment, as well as oxidative and endomplasmic reticulum (ER) stresses. FASN, fatty acid synthase; HMGCR, HMG‐CoA reductase; ROS, reactive oxygen species.
Disclosure Statement
The authors have no conflict of interest.
Acknowledgements
This research was supported by Special Funding Support of the Beijing Municipal Administration of Hospitals (ZYLX201402 and DFL20151701).
Cancer Sci 108 (2017) 12–22
Funding Information
Beijing Municipal Administration of Hospitals (ZYLX201402 and DFL20151701).
Contributor Information
Shunai Liu, Email: liusa1031@sina.com.
Jun Cheng, Email: jun.cheng.ditan@gmail.com.
References
- 1. Livraghi T, Makisalo H, Line PD. Treatment options in hepatocellular carcinoma today. Scand J Surg 2011; 100: 22–9. [DOI] [PubMed] [Google Scholar]
- 2. Nishikawa H, Arimoto A, Wakasa T, Kita R, Kimura T, Osaki Y. Surgical resection for hepatocellular carcinoma: clinical outcomes and safety in elderly patients. Eur J Gastroenterol Hepatol 2013; 25: 912–19. [DOI] [PubMed] [Google Scholar]
- 3. McGlynn KA, London WT. The global epidemiology of hepatocellular carcinoma: present and future. Clin Liver Dis 2011; 15: 223–43, vii‐x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Baffy G. Hepatocellular carcinoma in non‐alcoholic fatty liver disease: epidemiology, pathogenesis, and prevention. J Clin Transl Hepatol 2013; 1: 131–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mittal S, El‐Serag HB, Sada YH, et al Hepatocellular carcinoma in the absence of cirrhosis in United States veterans is associated with nonalcoholic fatty liver disease. Clin Gastroenterol Hepatol 2015; 14: 124–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Oda K, Uto H, Mawatari S, Ido A. Clinical features of hepatocellular carcinoma associated with nonalcoholic fatty liver disease: a review of human studies. Clin J Gastroenterol 2015; 8: 1–9. [DOI] [PubMed] [Google Scholar]
- 7. Perumpail RB, Liu A, Wong RJ, Ahmed A, Harrison SA. Pathogenesis of hepatocarcinogenesis in non‐cirrhotic nonalcoholic fatty liver disease: Potential mechanistic pathways. World J Hepatol 2015; 7: 2384–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cai C, Rajaram M, Zhou X, et al Powers, Activation of multiple cancer pathways and tumor maintenance function of the 3q amplified oncogene FNDC3B. Cell Cycle 2012; 11: 1773–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nishizuka M, Kishimoto K, Kato A, et al Disruption of the novel gene fad104 causes rapid postnatal death and attenuation of cell proliferation, adhesion, spreading and migration. Exp Cell Res 2009; 315: 809–19. [DOI] [PubMed] [Google Scholar]
- 10. Zhang X, Liu S, Hu T, Liu S, He Y, Sun S. Up‐regulated microRNA‐143 transcribed by nuclear factor kappa B enhances hepatocarcinoma metastasis by repressing fibronectin expression. Hepatology 2009; 50: 490–9. [DOI] [PubMed] [Google Scholar]
- 11. Fan X, Chen X, Deng W, Zhong G, Cai Q, Lin T. Up‐regulated microRNA‐143 in cancer stem cells differentiation promotes prostate cancer cells metastasis by modulating FNDC3B expression. BMC Cancer 2013; 13: 61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Katoh D, Nishizuka M, Osada S, Imagawa M. Fad104, a positive regulator of adipocyte differentiation, suppresses invasion and metastasis of melanoma cells by inhibition of STAT3 activity. PLoS ONE 2015; 10: e0117197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tominaga K, Kondo C, Johmura Y, Nishizuka M, Imagawa M. The novel gene fad104, containing a fibronectin type III domain, has a significant role in adipogenesis. FEBS Lett 2004; 577: 49–54. [DOI] [PubMed] [Google Scholar]
- 14. Lei Z, Ma QY, Meng XK, Li K, Cheng J. Screening of hepatocyte proteins binding to NS5ABP37 protein by yeast‐two hybrid system. Acad J Xi'an Jiaotong University 2009; 21: 234–7. [Google Scholar]
- 15. Gao LL, Li M, Wang Q, Liu SA, Zhang JQ, Cheng J. HCBP6 modulates triglyceride homeostasis in hepatocytes Via the SREBP1c/FASN Pathway. J Cell Biochem 2015; 116: 2375–84. [DOI] [PubMed] [Google Scholar]
- 16. Li M, Wang Q, Liu SA, et al MicroRNA‐185‐5p mediates regulation of SREBP2 expression by hepatitis C virus core protein. World J Gastroenterol 2015; 21: 4517–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Yang Y, Li D, Yang Y, Jiang G. An integrated analysis of the effects of microRNA and mRNA on esophageal squamous cell carcinoma. Mol Med Rep 2015; 12: 945–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Stangeland B, Mughal AA, Grieg Z, et al Combined expressional analysis, bioinformatics and targeted proteomics identify new potential therapeutic targets in glioblastoma stem cells. Oncotarget 2015; 6: 26192–215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mehra R, Vats P, Kalyana‐Sundaram S, et al Primary urethral clear‐cell adenocarcinoma: comprehensive analysis by surgical pathology, cytopathology, and next‐generation sequencing. Am J Pathol 2014; 184: 584–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Rajasagi M, Shukla SA, Fritsch EF, et al Systematic identification of personal tumor‐specific neoantigens in chronic lymphocytic leukemia. Blood 2014; 124: 453–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ha HL, Shin HJ, Feitelson MA, Yu DY. Oxidative stress and antioxidants in hepatic pathogenesis. World J Gastroenterol 2010; 16: 6035–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Choi J, Corder NL, Koduru B, Wang Y. Oxidative stress and hepatic Nox proteins in chronic hepatitis C and hepatocellular carcinoma. Free Radic Biol Med 2014; 72: 267–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gentric G, Maillet V, Paradis V, et al Oxidative stress promotes pathologic polyploidization in nonalcoholic fatty liver disease. J Clin Invest 2015; 125: 981–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kuhajda FP. Fatty acid synthase and cancer: new application of an old pathway. Cancer Res 2006; 66: 5977–80. [DOI] [PubMed] [Google Scholar]
- 25. Menendez JA, Lupu R. Fatty acid synthase and the lipogenic phenotype in cancer pathogenesis. Nat Rev Cancer 2007; 7: 763–77. [DOI] [PubMed] [Google Scholar]
- 26. Peck B, Schulze A. Lipid Desaturation: the next step in targeting lipogenesis in cancer. FEBS J 2016; 283: 2767–78. [DOI] [PubMed] [Google Scholar]
- 27. Chen Y, Hughes‐Fulford M. Human prostate cancer cells lack feedback regulation of low‐density lipoprotein receptor and its regulator, SREBP2. Int J Cancer 2001; 91: 41–5. [DOI] [PubMed] [Google Scholar]
- 28. Porstmann T, Santos CR, Griffiths B, et al SREBP activity is regulated by mTORC1 and contributes to Akt‐dependent cell growth. Cell Metab 2008; 8: 224–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Griffiths B, Lewis CA, Bensaad K, et al Sterol regulatory element binding protein‐dependent regulation of lipid synthesis supports cell survival and tumor growth. Cancer Metab 2013; 1: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li C, Yang W, Zhang J, et al SREBP‐1 has a prognostic role and contributes to invasion and metastasis in human hepatocellular carcinoma. Int J Mol Sci 2014; 15: 7124–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu H, Liu JY, Wu X, Zhang JT. Biochemistry, molecular biology, and pharmacology of fatty acid synthase, an emerging therapeutic target and diagnosis/prognosis marker. Int J Biochem Mol Biol 2010; 1: 69–89. [PMC free article] [PubMed] [Google Scholar]
- 32. Wu X, Qin L, Fako V, Zhang JT. Molecular mechanisms of fatty acid synthase (FASN)‐mediated resistance to anti‐cancer treatments. Adv Biol Regul 2014; 54: 214–21. [DOI] [PubMed] [Google Scholar]
- 33. Simons K, Toomre D. Lipid rafts and signal transduction. Nat Rev Mol Cell Biol 2000; 1: 31–9. [DOI] [PubMed] [Google Scholar]
- 34. Lu M, Hu XH, Li Q, et al A specific cholesterol metabolic pathway is established in a subset of HCCs for tumor growth. J Mol Cell Biol 2013; 5: 404–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Lee YL, Li WC, Tsai TH, Chiang HY, Ting CT. Body mass index and cholesterol level predict surgical outcome in patients with hepatocellular carcinoma in taiwan ‐ a cohort study. Oncotarget 2016; 7: 22948–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Li JJ, Tang Q, Li Y, Xiang BR, et al Role of oxidative stress in the apoptosis of hepatocellular carcinoma induced by combination of arsenic trioxide and ascorbic acid. Acta Pharmacol Sin 2006; 27: 1078–84. [DOI] [PubMed] [Google Scholar]
- 37. Zhang XQ, Xu CF, Yu CH, Chen WX, Li YM. Role of endoplasmic reticulum stress in the pathogenesis of nonalcoholic fatty liver disease. World J Gastroenterol 2014; 20: 1768–76. [DOI] [PMC free article] [PubMed] [Google Scholar]


