Abstract
Bacteriocins related to colicin M, acting via cleavage of the cell wall precursor lipid II, have been characterized in γ- and β-proteobacteria. Depending on the species, immunity is provided by either an inner membrane-anchored periplasmic protein or by an integral membrane protein. In Pseudomonas however, the immunity partner of colicin M-like bacteriocins remains unknown. Based on an in silico analysis in pseudomonad genomes, we here identify a gene encoding a putative immunity partner that represents a novel type of integral membrane protein (PmiA, Pseudomonas colicin M-like immunity type A). By heterologous expression of pmiA genes in susceptible strains, we show that immunity to colicin M-like bacteriocins is indeed provided by the cognate PmiA. Sequence homology among PmiA proteins is essentially absent, except for a short motif with a conserved periplasm-exposed aspartate residue. However, PmiA's protective function is not abolished by changing this acidic residue to the uncharged alanine. Immunity by PmiAs appears promiscuous to the extent that PmiA homologs from a clade sharing <40% pairwise amino acid identity, equally provide protection against the bacteriocin linked to the original PmiA. This study shows that multiple immunity factors have evolved independently to silence lipid II-targeting enzymatic bacteriocins. Their relaxed bacteriocin immunization capacity contrasts to the strict specificity of immunity proteins shielding the enzymatic domain of nuclease bacteriocins. The nature of associated immune functions needs consideration when using such natural protein antibiotics or designing novel variants.
Keywords: colicin M, PmiA, PmiB, pyocin, peptidoglycan, lipid II
Introduction
Antibacterial proteins are currently subject of renewed interest in the search for antimicrobials with novel modes of action. As yet poorly explored potential sources of such alternative protein-based antibiotics include lytic bacteriophages (Gerstmans et al., 2016) and bacteriocinogenic strains (Schulz et al., 2015). Bacteriocins are key mediators of intra-species competition between bacteria and combine high specificity with single-hit killing potency. Prominent bacteriocin producers that target human and plant pathogenic Gram-negative bacteria are found among enterobacteria (Cascales et al., 2007; Holt et al., 2013; Grinter et al., 2014; Schulz et al., 2015) and pseudomonads (Ghequire and De Mot, 2014).
Pseudomonas bacteriocins are very diverse with respect to structure and mode of action. R-type and F-type tailocins are multiprotein complexes that are evolutionary related to bacteriophage tails (Ghequire and De Mot, 2015a; Ghequire et al., 2015; Hockett et al., 2015). Lectin-like bacteriocins are constituted of a tandem of monocot mannose-binding lectin domains, and kill target cells via an unknown mechanism (Ghequire et al., 2013, 2014; McCaughey et al., 2014). S-type bacteriocins are multi-domain toxins with a modular structure similar to colicins, the bacteriocins from Escherichia coli (Cascales et al., 2007; Ghequire and De Mot, 2014; Dingemans et al., 2016; McCaughey et al., 2016). Self-inhibition of Pseudomonas aeruginosa strains producing a S-type bacteriocin (S pyocin) is circumvented by an immunity protein that transiently impedes the bacteriocin's toxic function. This immunity complement is co-expressed with the bacteriocin, and is typically encoded downstream of the pyocin gene, on the same or opposite strand. In the case of S-type bacteriocins with nuclease activity, bacteriocins are secreted as high-affinity bacteriocin-immunity protein complexes (Joshi et al., 2015). Pseudomonas colicin M-like bacteriocins (PseuMs) host a lipid II-cleaving domain, homologous to the catalytic domain of colicin M (ColM; Barreteau et al., 2009, 2012; Grinter et al., 2012b). However, in contrast to S pyocin-immunity complexes, it is not clear how PseuM producers protect themselves. Inspection of their pseuM genomic context did not reveal homologs of cmi, the cognate immunity partner for ColM in E. coli, nor were such homologs detected elsewhere in their genomes (Barreteau et al., 2009).
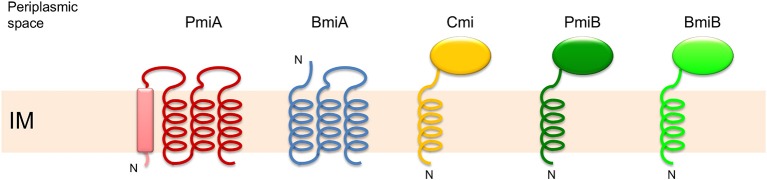
In E. coli, the cmi gene is located downstream of the bacteriocin gene on the opposite strand (Olschläger et al., 1984; Olschläger and Braun, 1987). Located in the periplasm, Cmi is anchored to the inner membrane by an amino-terminal hydrophobic α-helix, assisting in secretion but not being cleaved off. There, Cmi transiently impedes colicin M's activity during secretion and inactivates imported ColM (Olschläger and Braun, 1987; Olschläger et al., 1991; Gross and Braun, 1996). The crystal structure of Cmi revealed the presence of four α-helices and four β-strands, and is exemplary of a YebF fold (Gérard et al., 2011; Usón et al., 2012). In Pectobacterium a cmi-like gene is located downstream and opposite of the pectocin M gene, but its protective functionality remains to be demonstrated (Grinter et al., 2012a). In most strains of Burkholderia producing colicin M-like burkhocins, immunity is mediated by bmiB genes that give rise to proteins with a small size and predicted topology similar to Cmi (Ghequire and De Mot, 2015b). However, the amino acid sequence of the periplasmic moiety of BmiB is unrelated to Cmi and lacks the characteristic YebF domain. Fewer strains carry a second type of burkhocin immunity gene (bmiA) that encodes a small integral membrane protein comprising three transmembrane helices.
In this study we investigated the genomic context of pseuM genes, in search for candidate immunity partners. We demonstrate that a gene downstream of pseuM provides cognate immunity. Apart from sharing a common membrane topology reminiscent of BmiA proteins, the encoded proteins display very poor sequence conservation across PseuM-bacteriocinogenic pseudomonads. The possible role of a short semi-conserved motif in these proteins is explored, as well as the capability of PmiA proteins to immunize targeted cells in an (a)specific way.
Materials and methods
Genome searches and phylogenetic analysis
PseuM bacteriocin genes in Pseudomonas genomes were identified by homology searches using the National Center for Biotechnology (NCBI) non-redundant database. The Pfam Colicin_M module PF14859 was used as a query. Genes downstream of unique pseuM bacteriocin gene products were selected for further analysis. Multiple sequence alignments were generated with MUSCLE and phylogenetic reconstructions were carried out with PhyML (1000 bootstrap replicates), implemented in Geneious v7.1.7. The JTT substitution model (Jones et al., 1992) was used for studying the evolution of a large number of amino acid sequences. Amino-terminal signal sequences were predicted by PRED-TAT (http://www.compgen.org/tools/PRED-TAT) and TOPCONS (http://topcons.cbr.su.se/). Transmembrane regions and topology were predicted by TMHMM (http://www.cbs.dtu.dk/services/TMHMM/), and TOPCONS.
Bacterial strains and media
Strains used in this study are listed in Table S1. E. coli was grown in LB (MP Biomedicals) at 37°C, whereas Pseudomonas strains were grown with shaking at 200 rpm in Trypticase Soy Broth (TSB, BD Biosciences) and Casamino Acids Medium (CAA, BD Biosciences) at 30°C, except for P. aeruginosa that was grown at 37°C. Growth media were supplemented with agar (1.5%, Invitrogen), filter-sterilized isopropyl-β-D-thiogalactopyranoside (IPTG, 20 μg/ml, ForMedium), kanamycin (50 μg/ml, Sigma-Aldrich), or tetracycline (15–150 μg/ml, Sigma-Aldrich) when required. Strains were kept on plate at 4°C, or at −80°C in glycerol (25% v/v, Sigma-Aldrich).
DNA methods and construction of plasmids
Genomic DNA was collected with the Puregene Yeast/Bact. Kit B (Qiagen). Synthetic genes and primers were obtained from IDT DNA. Genes encoding bacteriocins (ERS445055_00256 from P. aeruginosa NCTC10332, PFLQ8_1129 from Pseudomonas fluorescens Q8r1-96 (Landa et al., 2003) and PSPTO_0572 from Pseudomonas syringae pv. tomato DC3000) and putative immunity proteins, encoded downstream, were amplified by polymerase chain reaction (PCR) with Q5 polymerase (New England Biolabs), with a T100™ Thermal cycler (Bio-Rad). Q5 polymerase was used according to the supplier's specifications. All primers are listed in Table S2. PCR fragments were purified with the Genelute PCR clean-up kit (Sigma-Aldrich), and double digested with NcoI/XhoI (1 h at 37°C, New England Biolabs) in the case of the bacteriocin genes, and with PstI/XbaI (1 h at 37°C, New England Biolabs) in the case of the putative immunity genes. The immunity gene amplicon of P. aeruginosa NCTC10332 was digested with PstI/EcoRI (1 h at 37°C, New England Biolabs). Purified bacteriocin genes were ligated with T4 DNA ligase (1 h at 37°C, Invitrogen) in pET28a(+) and putative immunity genes in shuttle vector pJB3Tc20. Ligation products were transformed to E. coli DH5α via heat shock. Standard methods were used for the preparation of competent E. coli cells and heat shock transformation of E. coli (Green and Sambrook, 2012). Transformants were initially verified for the presence of insert by PCR with Taq polymerase (New England Biolabs), using conditions as specified by the supplier. Plasmids were subsequently harvested with the Genelute HP plasmid miniprep kit (Sigma-Aldrich) and sequence confirmed for the presence of correct insert (GATC Biotech, Constance, Germany). Cloned immunity genes encompassed a 19-bp upstream region containing the predicted ribosome binding sites. Resulting plasmids are summarized in Table S1. The pET28a constructs were transformed to E. coli BL21/DE3 via heat shock, and pJB3Tc20 constructs electroporated to PseuM-susceptible Pseudomonas strains, and selected on the proper medium. Immunity genes encoding a mutation in a conserved aspartate residue were constructed via splicing by overlap extension, using pCMPG6251 (encoding the putative immunity complement of PseuMDC3000), pCMPG6252 (complement of PseuMNCTC10332), and pCMPG6269 (complement of PseuMQ8r1-96) as a template.
Expression and purification of recombinant bacteriocins
Recombinant carboxy-terminally His-tagged bacteriocins were generated in E. coli BL21(DE3) carrying pCMPG6248 (encoding PseuM from P. syringae DC3000), pCMPG6250 (encoding PseuM from P. aeruginosa NCTC10332) or pCMPG6271 (encoding PseuM from P. fluorescens Q8r1-96). Five milliliters of overnight cultures were transferred to 500 ml LB Erlenmeyer flasks and incubated at 37°C until OD600 reached 0.7. After, cultures were cooled and supplemented with IPTG (1 mM final concentration) and incubated at 20°C for 16 h. Next day, cells were harvested via centrifugation (20 min, 5000 g; Beckman X-15R) and frozen overnight (−20°C). Subsequently, the cell pellets were thawed, resuspended in lysis buffer (5 ml/g cell pellet; 300 mM NaCl, 50 mM NaH2PO4, 10 mM imidazole, pH 8.0) and sonicated (amplitude 20%, 10 cycles of 30 s on/off; Branson Digital Sonifier). Samples were treated with nuclease (0.01 U/μl, 37°C, 1 h; Invitrogen), remaining debris and insoluble proteins removed via centrifugation (30 min, 10000 g), and supernatants filtered (0.20 μm, Sarstedt). Soluble fractions containing the recombinant proteins were loaded on a 5-ml HisTrap HP column (GE Healthcare) and purified by nickel affinity chromatography with an Äkta Purifier (GE Healthcare). Matrix-bound proteins were eluted with a linear gradient of imidazole (10–500 mM) in lysis buffer. Fractions eluted at high imidazole concentrations were validated for the presence of recombinant protein, pooled and dialyzed to Tris buffer (50 mM, NaCl 200 mM, pH 7.5). Concentration of purified PseuMs was determined with a Genesys 10S UV-Vis Spectrophotometer (Thermo Scientific). Calculated extinction coefficients were 30035 mol/L−1 cm−1, 73340 mol/L−1 cm−1, and 32890 mol/L−1 cm−1 for the His-tagged PseuM bacteriocins from strains DC3000, NCTC10332, and Q8r1-96, respectively.
Bacteriocin assay
Bacteriocin activity was determined via spot assay. Filter-sterilized 20-μl volumes of purified recombinant proteins (1 mg/ml) were spotted onto bacterial cell lawns. After, samples were air-dried and incubated overnight. Dialysis buffer was used as a negative control. Following day, petri dishes were scored for zones lacking bacterial growth (halos).
Nucleotide accession number
GenBank accession number of the nucleotide sequence of the pseuM-pmiA pair from P. putida RW10S2 (Rokni-Zadeh et al., 2012) is KX086738.
Results
In silico search for candidate genes providing immunity to Pseudomonas colicin M-like bacteriocins
Based on the genetic architecture of modular bacteriocins in Gram-negatives, it is anticipated that bacteriocin immunity genes are likely to be found in proximity of their corresponding toxin genes. By scrutiny of their genomic contexts we observed that the characterized Pseudomonas colicin M-like bacteriocin genes, as well as several other predicted PseuM-encoding genes (Figure S1A), are consistently followed on the same DNA strand—at variable distances but within <200 nucleotides—by an open reading frame, but infrequently annotated as a coding region (Table S3). Typically, G+C content of these regions (~1.8 kb) is significantly lower (41–52%) than the Pseudomonas genome averages (P. aeruginosa: 66.2%, 1647 genomes; P. fluorescens: 60.2%, 92 genomes; P. syringae: 58.7%, 153 genomes). None of the deduced protein sequences contains the YebF domain signature (Pfam PF13995) as found in Cmi and the putative pectocin M immunity proteins.
In 14 Pseudomonas strains, mostly P. syringae pathovars, the encoded proteins (encompassing eight unique but very similar sequences of ~100 AAs) show only borderline homology with the burkhocin-associated BmiB immunity proteins (12–24% pairwise AA identity). However, these pseudomonad proteins, designated PmiBs, share with BmiBs two perfectly conserved cysteines (Figure S2). The PmiB and BmiB proteins are predicted to be translocated to the periplasm, albeit this appears to be achieved in different ways: instead of the BmiB amino-terminal membrane-anchoring segment or Sec-dependent cleavable signal sequence, a cleavable lipoprotein signal sequence typifies PmiB. This common feature is reflected in the phylogeny of ColM domains that reveals a well-separated clade harboring both the burkhocins and this small subset of PseuM proteins (Figure S1B).
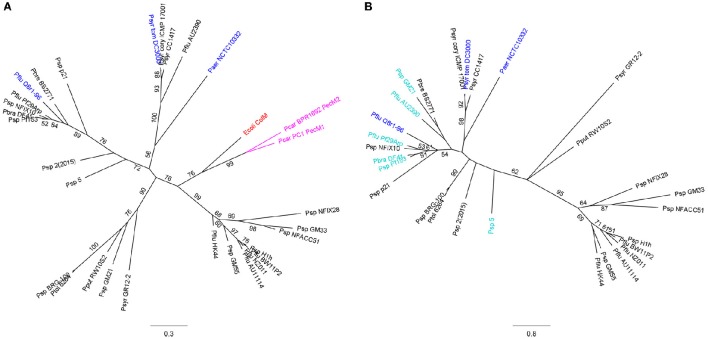
The majority of PseuM-derived ColM domains constitutes a separate clade of much more diversified sequences that are evolutionarily related to the corresponding domains in colicin M and colicin M domain-containing pectocins (Figure 1A, Figure S1B). In this second subset of PseuM-bacteriocinogenic strains the bacteriocin gene is linked with a gene encoding a protein unrelated to PmiB (~137 AAs; Figure S3). This protein family, designated PmiA, features four transmembrane helices, of which the first is predicted to represent a candidate Sec- or Tat-dependent cleavable signal sequence (Figure 2). The inferred topology, with two moieties facing the periplasm and two stretches (including the carboxy-terminus) residing in the cytoplasm, is reminiscent of the one adopted by BmiA proteins, albeit the latter lack an equivalent secretory signal sequence (Ghequire and De Mot, 2015b). No sequence homology can be detected between BmiAs and PmiAs however. Notably, four phytopathogenic strains of the P. syringae group each carry two unlinked PseuM-encoding paralogues, one from each PseuM clade and clustered with, respectively, a pmiA and pmiB gene. The low level of sequence identity between these bacteriocins (~24%) combined with the different nature of the respective candidate immunity proteins argues against a common ancestry and favors their independent acquisition.
Figure 1.
Maximum likelihood phylogenetic trees of ColM domains from pmiA-associated PseuMs and from related γ-proteobacterial bacteriocins (A), and of associated Pseudomonas PmiA immunity proteins (B). Characterized PseuM bacteriocins and cognate PmiAs are colored blue, colicin M (ColM) from E. coli is shown in red, and colicin M domain-containing pectocins (PecM1 and PecM2) in pink. PmiAs that were studied for promiscuity are shown in teal. The scale bars represent 0.3 (A) and 0.8 (B) substitutions per site. Bootstrap values (percentages of 1000 replicates) higher than 50 are shown at the branches. (A) For highly homologous colicin M domains (>95% pairwise AA identity), only one representative was included for phylogenetic tree construction. Species abbreviations: Ecoli, Escherichia coli; Paer, Pseudomonas aeruginosa; Pbra, Pseudomonas brassicacearum; Pbre, Pseudomonas brenneri; Pcar, Pectobacterium carotovorum; Pflu, Pseudomonas fluorescens; Pput, Pseudomonas putida; Psp, Pseudomonas sp.; Psyr, Pseudomonas syringae; Psyr cory, P. syringae pv. coryli; Psyr tom, P. syringae pv. tomato; Ptol, Pseudomonas tolaasii.
Figure 2.
Membrane topology model of integral membrane proteins PmiA and BmiA, and membrane-anchored proteins Cmi, PmiB, and BmiB, providing immunity to the activity of the ColM domain. PmiA from Pseudomonas has four predicted transmembrane helices, the first of which may represent a Sec- or Tat-dependent signal sequence (box). BmiA from Burkholderia carries three transmembrane helices. Cmi has a periplasmic YebF domain and is anchored in the inner membrane (IM) via an amino-terminal helix. PmiB and BmiB are equally anchored in the IM, and carry homologous periplasmic domains. “N” depicts the amino-terminus.
In different strains, the pseuM-pmiA gene pair has been recruited to diverse genomic locations. Several of these regions correspond to mobile elements. In P. aeruginosa, the gene couple generally occurs on ExoU island A, as has previously been noted for the pseuM of P. aeruginosa JJ692 (Barreteau et al., 2009). In some strains from the P. syringae group, such as isolates 642 and LMG 2367, pseuM-pmiA combinations appear as a cargo gene couple in a trpE/trpG-integrated Rp4 tailocin cluster (Hockett et al., 2015). In other P. syringae pathovar strains, such as DC3000 and ICMP 3923, the cargo is equally part of a Rp4-type sequence, although incomplete and apparently linked with diverged sequences. In some P. fluorescens group strains, such as Pseudomonas brassicacearum NFM421 and 51MFVCI2.1, and P. fluorescens Q8r1-96, the gene pair has been recruited by a Rp4 tailocin cluster as well, sandwiched between mutS and cinA (Ghequire et al., 2015; Hockett et al., 2015). For other strains, such as P. veronii R4, Pseudomonas sp. BRG-100 and Pseudomonas tolaasii 6264, regions flanking the pseuM-pmiA couples correspond to an unassigned prophage. The presence of bacteriocin cargo genes on mobile elements has previously been noted for lectin-like bacteriocins embedded in Pseudomonas Rp2 tailocin clusters (Ghequire et al., 2015).
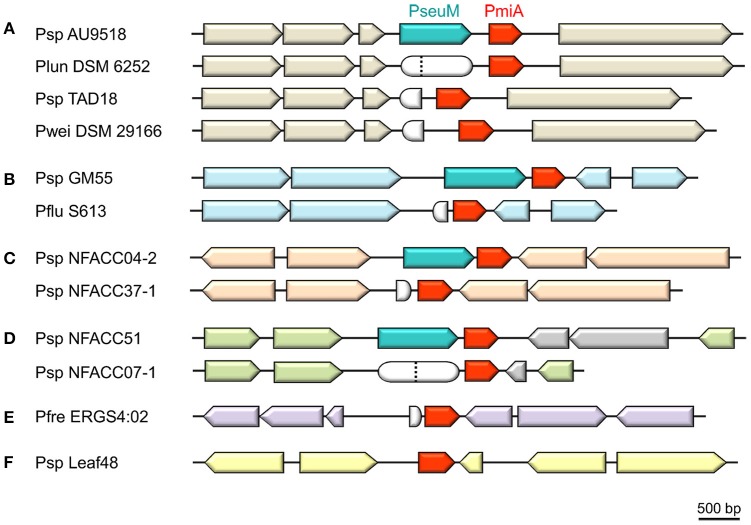
Some Pseudomonas strains carry a pmiA gene that is not linked to a cognate pseuM gene. The presence of pseuM remnant sequences upstream of such a pmiA orphan indicates that these strains may represent so-called cheaters that have lost the capacity of bacteriocin production but retain the associated immune function (Figure 3). For instance, in the type strain Pseudomonas lundensis (DSM 6252; De Jonghe et al., 2011) a frameshift impairs functionality of a pseuM gene that has remained intact in Pseudomonas sp. AU9518. From the equivalent genomic region of type strain Pseudomonas weihenstephanensis (DSM 291166; von Neubeck et al., 2016) and some related strains (TAD18; TAA207 and DSM 28140, not included in Figure 3), most of the 3′ region of a pseuM gene has been deleted, while their pmiA genes are well-conserved (>92% AA identity). No obvious pseuM sequences remain adjacent to the pmiA gene of Arabidopsis isolate Pseudomonas sp. Leaf48 (Bai et al., 2015), that is flanked by prophage genes.
Figure 3.
Genomic context of orphan pmiA genes in pseudomonads. Syntenic regions carrying a pmiA homolog associated with a full-length PseuM-encoding gene are shown for Pseudomonas sp. strains AU9518 (A), GM55 (B), NFACC04-2 (C), and NFACC51 (D). Synteny between these predicted bacteriocin producers and phylogenetically related cheaters lacking a (functional) bacteriocin gene is highlighted by upstream and downstream ORFs (arrows) shown in the same color. The gray-colored ORFs (D) reflect a local lack of synteny. Remnants of the respective pseuM genes are represented by white-colored rounded shapes corresponding to a frameshifted pseuM (position marked by a dotted vertical line) or a truncated residual N- or C-terminal fragment. For Pseudomonas strains ERGS4:02 (E) and Leaf48 (F), no strain with a closely related pmiA homolog and syntenic flanking region could be identified. Species abbreviations: Pfre, Pseudomonas frederikbergensis; Plun, Pseudomonas lundensis; Pwei, Pseudomonas weihenstephanensis. Other abbreviations are as in Figure 1.
PmiA proteins are cognate PseuM immunity partners
Previously described PseuM bacteriocins from P. aeruginosa NCTC10332 (PaeM), P. fluorescens Q8r1-96 (PflM), and P. syringae pv. tomato DC3000 (PsyM), each with a downstream pmiA gene (as identified by in silico analysis described in Section In silico Search for Candidate Genes Providing Immunity to Pseudomonas Colicin M-like Bacteriocins), were recombinantly expressed in E. coli and the His-tagged proteins purified by Ni-NTA affinity chromatography. Strain NCTC10332 encodes a PseuM bacteriocin that shares 90% AA identity with PaeM from P. aeruginosa JJ692 (Barreteau et al., 2009). Antibacterial activity of the recombinant proteins was challenged via spot-on-lawn assay against a panel of Pseudomonas strains, and strains with a clear halo susceptibility phenotype were selected for further experiments. Such indicator strains identified for PaeM, PflM, and PsyM were P. aeruginosa CPHL12447, P. fluorescens F113, and P. syringae pv. lachrymans LMG 5456 (Grinter et al., 2012b), respectively.
Next, the putative immunity genes from strains NCTC10332, Q8r1-96 and DC3000 were cloned in shuttle vector pJB3Tc20, and introduced in the corresponding PseuM-susceptible strains. Transformants were tested for altered PseuM sensitivity, using strains carrying empty vectors as negative controls (Table 1). When equipped with the cognate pmiA, transformants became fully insensitive to the respective bacteriocin (tested at 1 mg/ml by spot assay). When indicator strains were provided with either of the other two immunity genes, no diminished PseuM inhibition was observed. Together this indicates that PmiAs are cognate immunity partners of the bacteriocins they are associated with. Purified membrane fragments with the gene product of pmiADC3000 were analyzed via Maldi-MSMS (Poetsch et al., 2008), but no peptide fragments assignable to the PmiA of interest could be retrieved (data not shown).
Table 1.
Heterologous expression of pmiA genes in PseuM-susceptible strains.
| Indicator strain | PmiA immunity protein | PseuM bacteriocin | |||
|---|---|---|---|---|---|
| Species | Gene product | PaeM P. aeruginosa NCTC10332 | PflM P. fluorescens Q8r1-96 | PsyM P. syringae pv. tomato DC3000 | |
| P. aeruginosa CPHL12447 | P. aeruginosa | PmiANCTC10332 | − | ||
| PmiA*NCTC10332 | − | ||||
| P. fluorescens | PmiAQ8r1-96 | + | |||
| P. syringae pv. tomato | PmiADC3000 | + | |||
| NA | Control | + | |||
| P. fluorescens F113 | P. aeruginosa | PmiANCTC10332 | + | ||
| P. brassicacearum | PmiADF41 | − | |||
| P. fluorescens | PmiAQ8r1-96 | − | |||
| PmiA*Q8r1-96 | − | ||||
| PmiAAU2390 | + | ||||
| PmiAPf29Arp | − | ||||
| P. syringae pv. syringae | PmiADC3000 | + | |||
| Pseudomonas sp. | PmiA5 | T | |||
| PmiAGM21 | + | ||||
| PmiAPf153 | − | ||||
| NA | Control | + | |||
| P. syringae LMG 5456 | P. aeruginosa | PmiANCTC10332 | + | ||
| P. syringae pv. tomato | PmiADC3000 | − | |||
| PmiA*DC3000 | − | ||||
| NA | Control | + | |||
Indicator strains were transformed with shuttle vector pJB3Tc20 (control, without insert) or with pJB3Tc20-derived plasmids equipped with different pmiA genes, and tested for (altered) susceptibility to their respective PseuM via bacteriocin spot assay. +, sensitive; −, insensitive; T, turbid halo. The species used as source of immunity protein are specified with the respective strain names indicated as PmiA subscript (NA, not applicable). Asterisks define mutated immunity proteins (Asp to Ala). Strain LMG 5456 carrying a plasmid equipped with pmiAQ8r1−96 could not be obtained.
Since colicin M and colicin M-like bacteriocins domains exert their action in the periplasm, it is plausible that (at least part of) the periplasm-exposed moieties of PmiA proteins participate in the immunity function, directly or indirectly impeding the catalytic function of the ColM domain. Conserved residues in the immunity module may be critical in providing such property. In the mature PmiA proteins, a short well-conserved stretch with consensus motif D[T/S]XGXP precedes the second predicted transmembrane segment (Figure 4, Figure S3). No obvious sequence conservation among PmiA proteins can be detected for the shorter second periplasm-exposed loop. To further scrutinize the possible role of the first periplasmic stretch, the conserved Asp residue was mutated to Ala in the PmiAs of strains NCTC10332, Q8r1-96 and DC3000. Upon introduction of the PmiA variants in the respective indicator strains, the immunity phenotype to the respective PseuMs was evaluated. Halos were absent for all three indicator strains after bacteriocin spotting, showing the same phenotype as caused by the native immunity gene products (Table 1). This suggests that the conserved Asp residue is not instrumental to PmiA's immunity function.
Figure 4.
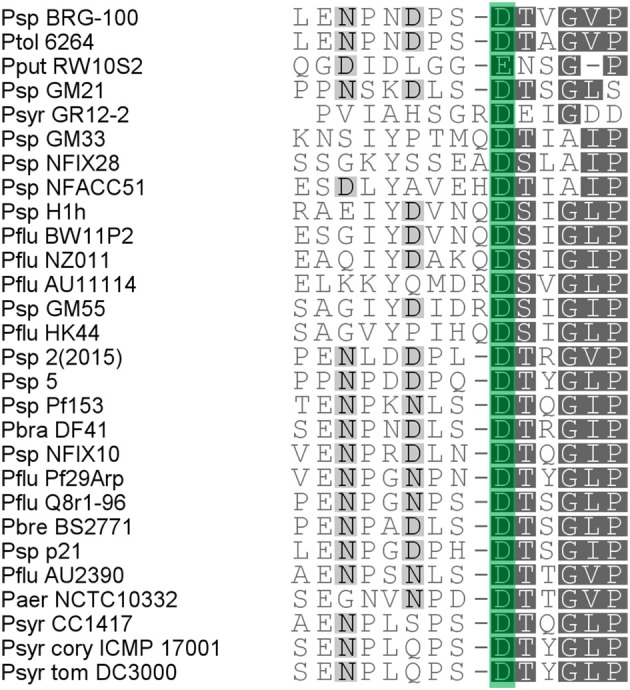
Multiple sequence alignment of the semi-conserved stretch in PmiA proteins, predicted to be oriented toward the periplasmic space. Pseudomonas species abbreviations are as in Figure 1. Gray shading reflects the degree of conservation. The conserved Asp residue that was mutated is boxed in green.
Immune promiscuity among sequence-diverged PmiAs
Noteworthy, sequence divergence of the PseuM-PmiA toxin-immunity pairs seems to be most pronounced among strains belonging to the P. fluorescens group. In contrast, for P. syringae pathovar isolates, high conservation of the ColM domain (at least 94% pairwise AA identity; Figures S1A,B) is reflected in well-conserved immunity proteins (at least 84% pairwise AA identity). Previously, it was demonstrated that immunity proteins sharing high homology may provide pyocin-silencing functionality of another partner (Dingemans et al., 2016).
To examine to what extent sequence divergence limits the immune function, PmiA homologs from the cluster with P. fluorescens Q8r1-96 were selected (P. brassicacearum DF41, P. fluorescens Pf29Arp, and Pseudomonas sp. Pf153), as well as some more distant PmiAs (Pseudomonas sp. strains 5 and GM21, P. fluorescens AU2390; Figure 1B). The PmiAs from the Q8r1-96 cluster share 36–40% AA identity with PmiAQ8r1-96. This level of sequence conservation stems mainly from the periplasm-exposed regions as well as the transmembrane helices, whereas the amino-terminal signal sequence/transmembrane helix and the cytoplasm-exposed stretches exhibit extensive sequence divergence. By comparison, the ColM domains of the PseuMs of strains DF41, Pf29Arp and Pf153, display considerably higher sequence conservation (~69% AA identity) with PflM. Pairwise identity with the more distant PmiAs ranges from 18 to 31%, corresponding with more diversified ColM domains (35–46% AA identity with PflM).
These six pmiA genes were cloned in pJB3Tc20 and introduced in P. fluorescens F113, and tested for their capacity to protect from PflM activity, using empty vector and the native pmiAQ8r196 immunity gene as controls for susceptibility and immunity, respectively (Table 1). Despite only moderate pairwise identities to PmiAQ8r1-96, PmiADF41, PmiAPf153, and PmiAPf29Arp provided full cross-immunity to PflM. The more distantly related PmiAs from strains AU2390 and GM21 (with 30.9 and 23.4% AA identity, respectively) could not immunize cells against the bacteriocin, resulting in a similar phenotype as the PmiAs from P. aeruginosa NCTC10332 and P. syringae DC3000, and the control. Despite barely showing homology with PmiAQ8r1-96 (18.4% AA identity), PmiA from Pseudomonas sp. 5 conferred an intermediate immunity phenotype (turbid halo).
Discussion
In this study we demonstrated that cognate immunity to a large subset of ColM domain-containing Pseudomonas bacteriocins is provoked by the poorly conserved transmembrane protein PmiA, unrelated in sequence and topology to the Cmi protein providing immunity to colicin M in E. coli. The pmiA gene, consistently located downstream of pseuM on the same strand, is rarely annotated (Table S3). Previously, immunity to ColM domains in Burkholderia burkhocins was equally associated with a transmembrane protein (BmiA; Ghequire and De Mot, 2015b). However, since ColM domains from bmiA- and pmiA-associated bacteriocins constitute phylogenetically distant clades, the immunity mechanism from these integral membrane proteins may be different. Further support for the existence of such evolutionarily distinct lipid II-targeting bacteriocin subsets is provided by the observation that a minor group of pseudomonad genes shares a similar immunity gene, as well as a similar ColM-domain-encoding gene, with Burkholderia. The gene organization of different ColM domain-hosting bacteriocins and their (putative) immunity genes is summarized in Figure 5.
Figure 5.
Schematic gene organizations of ColM domain-containing bacteriocins and (putative) immunity genes (if present). The arrows correspond with the gene orientations and the color legend describes the indicated domains and type of immunity gene. Predicted immunity genes, based on homology with characterized immunity factors, are delineated by a dashed line. Species abbreviations are as in Figure 1. Bamb, Burkholderia ambifaria; Bglu, Burkholderia glumae.
A conserved Asp residue located in a semi-conserved periplasm-exposed stretch proved not pivotal for the immunity phenotype of PmiA. Possibly, it is required for structural integrity or stability of the immunity protein, and structural analysis can give more insight in this issue. Since the periplasm-exposed moieties of PmiA likely contribute to immunity, it was investigated whether cross-immunity between proteins with different extent of sequence conservation in those regions, may occur. Since this was found to be the case, even between PmiAs sharing <40% pairwise amino acid identity, it will be of future interest to determine which residues or structural elements actually contribute to the immunity phenotype. This will render a first indication on how and where the immunity mechanism to this toxin module may be provoked. Previously, attempts to co-crystallize colicin M and Cmi to enlighten how Cmi neutralizes ColM-mediated toxicity proved unsuccessful (Usón et al., 2012).
From an evolutionary point of view, promiscuous protection by immunity genes sharing low homology may have far-reaching consequences toward competitive advantage for producer strains. It may explain why genome analysis in pseudomonads indicated that PseuMs are rather rare, as compared to DNase and RNase bacteriocins (Ghequire and De Mot, 2014). In contrast, immunity to the latter bacteriocins is provided by immunity proteins forming very specifically high-affinity bacteriocin-immunity complexes, stabilized by conserved hydrogen bonds and hydrophobic interactions (Li et al., 2004; Levin et al., 2009; Meenan et al., 2010; Joshi et al., 2015; Kleanthous et al., 2016). PseuM bacteriocin production may therefore provide limited benefit in most environments, and hence result in lower selection pressure to maintain these bacteriocins in genomes. In addition to this, genome mining also indicated that orphan pmiA-like genes occasionally occur in Pseudomonas genomes, although it remains to be investigated whether these enable cheating. A similar observation was made for bmiA genes in Burkholderia (Ghequire and De Mot, 2015b) and S-type pyocin immunity genes (Ghoul et al., 2015).
Author contributions
MG, RD conceived and designed the experiments; MG and LK performed the experiments; MG, LK, and RD analyzed the data; MG and RD wrote the paper; all authors approved the final version of the manuscript.
Funding
MG is the recipient of a postdoctoral fellowship from FWO-Vlaanderen (12M4615N).
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The author wish to thank Dr. Linda Thomashow (Washington State University) for kindly providing P. fluorescens Q8r1-96. The authors wish to thank Prof. Bart Devreese (Ghent University) for MS analysis and suggestions for improving this manuscript.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00093/full#supplementary-material
References
- Bai Y., Müller D. B., Srinivas G., Garrido-Oter R., Potthoff E., Rott M., et al. (2015). Functional overlap of the Arabidopsis leaf and root microbiota. Nature 528, 364–369. 10.1038/nature16192 [DOI] [PubMed] [Google Scholar]
- Barreteau H., Bouhss A., Fourgeaud M., Mainardi J. L., Touzé T., Gérard F., et al. (2009). Human- and plant-pathogenic Pseudomonas species produce bacteriocins exhibiting colicin M-like hydrolase activity towards peptidoglycan precursors. J. Bacteriol. 191, 3657–3664. 10.1128/JB.01824-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barreteau H., Tiouajni M., Graille M., Josseaume N., Bouhss A., Patin D., et al. (2012). Functional and structural characterization of PaeM, a colicin M-like bacteriocin produced by Pseudomonas aeruginosa. J. Biol. Chem. 287, 37395–37405. 10.1074/jbc.M112.406439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cascales E., Buchanan S. K., Duché D., Kleanthous C., Lloubès R., Postle K., et al. (2007). Colicin biology. Microbiol. Mol. Biol. Rev. 71, 158–229. 10.1128/MMBR.00036-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jonghe V., Coorevits A., Van Hoorde K., Messens W., Van Landschoot A., De Vos P., et al. (2011). Influence of storage conditions on the growth of Pseudomonas species in refrigerated raw milk. Appl. Environ. Microbiol. 77, 460–470. 10.1128/AEM.00521-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dingemans J., Ghequire M. G., Craggs M., De Mot R., Cornelis P. (2016). Identification and functional analysis of a bacteriocin, pyocin S6, with ribonuclease activity from a Pseudomonas aeruginosa cystic fibrosis clinical isolate. Microbiologyopen 5, 413–423. 10.1002/mbo3.339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gérard F., Brooks M. A., Barreteau H., Touzé T., Graille M., Bouhss A., et al. (2011). X-ray structure and site-directed mutagenesis analysis of the Escherichia coli colicin M immunity protein. J. Bacteriol. 193, 205–214. 10.1128/JB.01119-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstmans H., Rodríguez-Rubio L., Lavigne R., Briers Y. (2016). From endolysins to Artilysin®s: novel enzyme-based approaches to kill drug-resistant bacteria. Biochem. Soc. Trans. 44, 123–128. 10.1042/BST20150192 [DOI] [PubMed] [Google Scholar]
- Ghequire M. G., De Mot R. (2014). Ribosomally encoded antibacterial proteins and peptides from Pseudomonas. FEMS Microbiol. Rev. 38, 523–568. 10.1111/1574-6976.12079 [DOI] [PubMed] [Google Scholar]
- Ghequire M. G., De Mot R. (2015a). The tailocin tale: peeling off phage tails. Trends Microbiol. 23, 587–590. 10.1016/j.tim.2015.07.011 [DOI] [PubMed] [Google Scholar]
- Ghequire M. G., De Mot R. (2015b). Distinct colicin M-like bacteriocin-immunity pairs in Burkholderia. Sci. Rep. 5:17368. 10.1038/srep17368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G., Dillen Y., Lambrichts I., Proost P., Wattiez R., De Mot R. (2015). Different ancestries of R tailocins in rhizospheric Pseudomonas isolates. Genome Biol. Evol. 7, 2810–2828. 10.1093/gbe/evv184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G., Dingemans J., Pirnay J. P., De Vos D., Cornelis P., De Mot R. (2014). O serotype-independent susceptibility of Pseudomonas aeruginosa to lectin-like pyocins. Microbiologyopen 3, 875–884. 10.1002/mbo3.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghequire M. G., Garcia-Pino A., Lebbe E. K., Spaepen S., Loris R., De Mot R. (2013). Structural determinants for activity and specificity of the bacterial toxin LlpA. PLoS Pathog. 9:e1003199. 10.1371/journal.ppat.1003199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoul M., West S. A., Johansen H. K., Molin S., Harrison O. B., Maiden M. C., et al. (2015). Bacteriocin-mediated competition in cystic fibrosis lung infections. Proc. Biol. Sci. 282, 1814. 10.1098/rspb.2015.0972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green M. R., Sambrook J. (2012). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor. [Google Scholar]
- Grinter R., Josts I., Zeth K., Roszak A. W., McCaughey L. C., Cogdell R. J., et al. (2014). Structure of the atypical bacteriocin pectocin M2 implies a novel mechanism of protein uptake. Mol. Microbiol. 93, 234–246. 10.1111/mmi.12655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter R., Milner J., Walker D. (2012a). Ferredoxin containing bacteriocins suggest a novel mechanism of iron uptake in Pectobacterium spp. PLoS ONE 7:e33033. 10.1371/journal.pone.0033033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grinter R., Roszak A. W., Cogdell R. J., Milner J. J., Walker D. (2012b). The crystal structure of the lipid II-degrading bacteriocin syringacin M suggests unexpected evolutionary relationships between colicin M-like bacteriocins. J. Biol. Chem. 287, 38876–38888. 10.1074/jbc.M112.400150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gross P., Braun V. (1996). Colicin M is inactivated during import by its immunity protein. Mol. Gen. Genet. 251, 388–396. 10.1007/BF02172531 [DOI] [PubMed] [Google Scholar]
- Hockett K. L., Renner T., Baltrus D. A. (2015). Independent co-option of a tailed bacteriophage into a killing complex in Pseudomonas. MBio 6:e00452. 10.1128/mBio.00452-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt K. E., Thieu Nga T. V., Thanh D. P., Vinh H., Kim D. W., Vu Tra M. P., et al. (2013). Tracking the establishment of local endemic populations of an emergent enteric pathogen. Proc. Natl. Acad. Sci. U.S.A. 110, 17522–17527. 10.1073/pnas.1308632110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D. T., Taylor W. R., Thornton J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8, 275–282. 10.1093/bioinformatics/8.3.275 [DOI] [PubMed] [Google Scholar]
- Joshi A., Grinter R., Josts I., Chen S., Wojdyla J. A., Lowe E. D., et al. (2015). Structures of the ultra-high-affinity protein-protein complexes of pyocins S2 and AP41 and their cognate immunity proteins from Pseudomonas aeruginosa. J. Mol. Biol. 427, 2852–2866. 10.1016/j.jmb.2015.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleanthous C., Klein A., Wojdyla J., Joshi A., Josts I., McCaughey L. C., et al. (2016). Structural and biophysical analysis of nuclease proteins antibiotics. Biochem. J. 473, 2799–2812. 10.1042/BCJ20160544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landa B. B., Mavrodi D. M., Thomashow L. S., Weller D. M. (2003). Interactions between strains of 2,4-diacetylphloroglucinol-producing Pseudomonas fluorescens in the rhizosphere of wheat. Phytopathology 93, 982–994. 10.1094/PHYTO.2003.93.8.982 [DOI] [PubMed] [Google Scholar]
- Levin K. B., Dym O., Albeck S., Magdassi S., Keeble A. H., Kleanthous C., et al. (2009). Following evolutionary paths to protein-protein interactions with high affinity and selectivity. Nat. Struct. Mol. Biol. 16, 1049–1055. 10.1038/nsmb.1670 [DOI] [PubMed] [Google Scholar]
- Li W., Keeble A. H., Giffard C., James R., Moore G. R., Kleanthous C. (2004). Highly discriminating protein-protein interaction specificities in the context of a conserved binding energy hotspot. J. Mol. Biol. 337, 743–759. 10.1016/j.jmb.2004.02.005 [DOI] [PubMed] [Google Scholar]
- McCaughey L. C., Grinter R., Josts I., Roszak A. W., Waløen K. I., Cogdell R. J., et al. (2014). Lectin-like bacteriocins from Pseudomonas spp. utilise D-rhamnose containing lipopolysaccharide as a cellular receptor. PLoS Pathog. 10:e1003898. 10.1371/journal.ppat.1003898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCaughey L. C., Josts I., Grinter R., White P., Byron O., Tucker N. P., et al. (2016). Discovery, characterisation and in vivo activity of pyocin SD2, a protein antibiotic from Pseudomonas aeruginosa. Biochem. J. 473, 2345–2358. 10.1042/BCJ20160470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meenan N. A., Sharma A., Fleishman S. J., Macdonald C. J., Morel B., Boetzel R., et al. (2010). The structural and energetic basis for high selectivity in a high-affinity protein-protein interaction. Proc. Natl. Acad. Sci. U.S.A. 107, 10080–10085. 10.1073/pnas.0910756107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olschläger T., Braun V. (1987). Sequence, expression, and localization of the immunity protein for colicin M. J. Bacteriol. 169, 4765–4769. 10.1128/jb.169.10.4765-4769.1987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olschläger T., Schramm E., Braun V. (1984). Cloning and expression of the activity and immunity genes of colicins B and M on ColBM plasmids. Mol. Gen. Genet. 196, 482–487. 10.1007/BF00436196 [DOI] [PubMed] [Google Scholar]
- Olschläger T., Turba A., Braun V. (1991). Binding of the immunity protein inactivates colicin M. Mol. Microbiol. 5, 1105–1111. 10.1111/j.1365-2958.1991.tb01883.x [DOI] [PubMed] [Google Scholar]
- Poetsch A., Schlüsener D., Florizone C., Eltis L., Menzel C., Rögner M., et al. (2008). Improved identification of membrane proteins by MALDI-TOF MS/MS using vacuum sublimated matrix spots on an ultraphobic chip surface. J. Biomol. Tech. 19, 129–138. [PMC free article] [PubMed] [Google Scholar]
- Rokni-Zadeh H., Li W., Sanchez-Rodriguez A., Sinnaeve D., Rozenski J., Martins J. C., et al. (2012). Genetic and functional characterization of cyclic lipopeptide white-line-inducing principle (WLIP) production by rice rhizosphere isolate Pseudomonas putida RW10S2. Appl. Environ. Microbiol. 78, 4826–4834. 10.1128/AEM.00335-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulz S., Stephan A., Hahn S., Bortesi L., Jarczowski F., Bettmann U., et al. (2015). Broad and efficient control of major foodborne pathogenic strains of Escherichia coli by mixtures of plant-produced colicins. Proc. Natl. Acad. Sci. U.S.A. 112, E5454–E5460. 10.1073/pnas.1513311112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usón I., Patzer S. I., Rodríguez D. D., Braun V., Zeth K. (2012). The crystal structure of the dimeric colicin M immunity protein displays a 3D domain swap. J. Struct. Biol. 178, 45–53. 10.1016/j.jsb.2012.02.004 [DOI] [PubMed] [Google Scholar]
- von Neubeck M., Huptas C., Glück C., Krewinkel M., Stoeckel M., Stressler T., et al. (2016). Pseudomonas helleri sp. nov. and Pseudomonas weihenstephanensis sp. nov., isolated from raw cow's milk. Int. J. Syst. Evol. Microbiol. 66, 1163–1173. 10.1099/ijsem.0.000852 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.