ABSTRACT
A novel Bacteroides fragilis selective (BFS) medium, consisting of a brain heart infusion agar base supplemented with yeast extract, cysteine hydrochloride, bile salts, vitamin K, hemin, glucose, esculin, ferric ammonium citrate, bromothymol blue, gentamicin, kanamycin, and novobiocin, was evaluated. When BFS agar was tested with a collection of 303 bacteria of different genera, it allowed the growth of B. fragilis as large yellow colonies, with blackening of the medium after 48 h of anaerobic incubation, while the growth of most other anaerobes, facultative anaerobes, and aerobes was inhibited. In a prospective comparison of BFS agar with a routinely used medium (neomycin blood agar) in 1,209 clinical specimens, 60 B. fragilis bacteria were detected on BFS agar while 46 were detected on the routine agar (McNemar's test, P = 0.008). In conclusion, this novel medium may be added to improve the recovery of B. fragilis in clinical specimens and to facilitate surveillance of antimicrobial-resistant strains.
KEYWORDS: selective medium, Bacteroides, media
INTRODUCTION
Bacteroides species colonize the human gut as commensals, but they can also cause infections if translocated into the bloodstream or other tissues following disease, an operation, or trauma (1, 2). In the genus Bacteroides, Bacteroides fragilis is regarded as the most virulent species and has been implicated in various infections such as intra-abdominal sepsis, deep-seated abscesses, and necrotizing skin and soft tissue infections (2). In B. fragilis, emerging resistance to antibiotics is a big concern (3, 4). Among isolates from large hospitals in the United States and Europe, resistance rates for clindamycin and moxifloxacin are now >20% to 30% (3, 4). Currently, carbapenem and metronidazole resistance rates among B. fragilis isolates from the United States and Europe remain low (<1% to 2%) in general (3, 4). However, higher carbapenem resistance rates have been reported by investigators from Taiwan (7% to 12%) (5), Germany (4.9% to 5.3%) (1), and Canada (2.3% to 12.7%) (6), suggesting that this resistance phenotype may be emerging in certain hotspots in those countries. Most seriously, rare strains of multidrug-resistant B. fragilis with simultaneous resistance to carbapenems and metronidazole have also been identified (7, 8).
In B. fragilis, two divisions (I and II) have been identified by using matrix-assisted laser desorption ionization–time of flight mass spectrometry (MALDI-TOF MS) (9). Metronidazole is administered as an inactive prodrug, and reduction of the nitro group is required for activation to the active compounds. In B. fragilis, metronidazole resistance is usually attributed to acquisition of the nitroimidazole resistance gene (nim), encoding a nitroimidazole reductase which is presumed to modify metronidazole to an inactive compound (10). The acquisition of carbapenem resistance usually correlates with the activation of the carbapenemase gene, cfiA, in division II strains by an upstream insertion sequence (11). Most of the genetically characterized multidrug-resistant B. fragilis strains belong to division II strains, and some investigators have proposed to rename division II strains as a new Bacteroides genomospecies (7, 10, 12).
Although MALDI-TOF MS has facilitated the identification of B. fragilis and its subsequent differentiation into division I and II strains, without a selective medium, recognition of B. fragilis in mixed growth with other bacteria remains difficult (9, 13). Surveillance for drug-resistant B. fragilis strains in various patient populations also requires the availability of a selective and differential culture medium. While Bacteroides bile-esculin (BBE) agar supplemented with gentamicin has been developed for such a purpose, the selectivity of the medium is limited by increasing gentamicin resistance among enterococci and by the medium's inability to inhibit some lactobacilli (14–17). Furthermore, BBE agar may not produce large colonies for some B. fragilis strains, and culture media based on brain heart infusion have been found to be superior to BBE agar for recovery of B. fragilis (18, 19). Previous studies further reported that B. fragilis produced yellow colonies in the presence of glucose or glucuronic acid as a fermentable carbon source, and this may be a useful marker for enhancing recognition of the organism in mixed growth (18, 20). In an attempt to improve the detection of B. fragilis, the aforementioned principles were used to design a new medium. First, the selectivity of this Bacteroides fragilis selective (BFS) medium was validated by using a collection of ATCC strains and clinical isolates. Then, this medium was prospectively compared with an in-house method, based on neomycin blood agar for clinical specimens requiring culture for anaerobes.
RESULTS AND DISCUSSION
Recovery of B. fragilis and quality control.
Fourteen batches of BFS agar plates were tested, and all of them passed the quality control test. Large colonies of B. fragilis ATCC 25285 were consistently obtained in BFS plates. The log10 CFU counts (mean ± standard deviation) of B. fragilis ATCC 25285 bacteria in anaerobic blood agar and BFS plates exhibited only a small difference (3.44 ± 0.03 CFU and 3.23 ± 0.05 CFU, respectively; P = 0.001). No growth of Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 29212, and Lactobacillus rhamnosus ATCC 7469 was obtained when approximately 100,000 bacteria were inoculated onto BFS plates.
Validation of BFS agar with pure cultures.
In total, 104 B. fragilis clinical isolates (including 52 division I and 52 division II isolates), 63 other Bacteroides species, and 136 other bacteria were plated onto BFS medium and incubated anaerobically for 48 h (Table 1). All 104 isolates of B. fragilis grew well on BFS plates, with easy recognition of the typical yellow colonies and blackening of the surrounding medium (Fig. 1). Thirteen other Bacteroides species were tested. Five of them (B. caccae, B. nordii, B. ovatus, B. salyersiae, and B. thetaiotaomicron) grew as well as B. fragilis-like colonies while the remaining eight Bacteroides species (B. dorei, B. eggerthii, B. intestinalis, B. massiliensis, B. pyogenes, B. stercoris, B. uniformis, and B. vulgatus) yielded only pinpoint colonies on BFS plates (Table 1). Six isolates of Parabacteroides (two isolates each of Parabacteroides faecis, Parabacteroides distasonis, and Parabacteroides goldsteinii) were tested. The two P. faecis isolates yielded large colonies, but there was no blackening of the medium. Growth of the other two Parabacteroides species on BFS plates was poor. An isolate of Fusobacterium mortiferum also yielded large and yellow colonies in the medium, but there was no blackening of the medium. For the other anaerobes (Clostridium, Peptostreptococcus, and Prevotella species) as well as the aerobic and facultative anaerobic bacteria, growth was completely inhibited.
TABLE 1.
Growth characteristics of Bacteroides fragilis and other bacteria in BFS medium
| Isolate type and organism (no. of isolates)a | Colony profile on BFS agar (no. of isolates) |
|||||
|---|---|---|---|---|---|---|
| Esculin positivef | Growth after 48 h |
Yellow colonies | Blackening of the medium | |||
| Absent | Good | Poorg | ||||
| Anaerobic bacteria (215) | ||||||
| Bacteroides caccae | 4 (4) | 4 | 4 | 4 | ||
| Bacteroides dorei | 4 (4) | 4 | 4 | 4 | ||
| Bacteroides eggerthii | 1 (1) | 1 | 1 | 1 | ||
| Bacteroides fragilis (division I) | 52 (52) | 52 | 52 | 52 | ||
| Bacteroides fragilis (division II) | 52 (52) | 52 | 52 | 52 | ||
| Bacteroides intestinalis | 2 (2) | 2 | 2 | 2 | ||
| Bacteroides massiliensis | 1 (0) | 1 | 1 | |||
| Bacteroides nordii | 4 (4) | 4 | 4 | 4 | ||
| Bacteroides ovatus | 11 (11) | 11 | 11 | 11 | ||
| Bacteroides pyogenes | 2 (0) | 2 | 2 | |||
| Bacteroides salyersiae | 2 (2) | 2 | 2 | 2 | ||
| Bacteroides stercoris | 2 (2) | 2 | 2 | 2 | ||
| Bacteroides thetaiotaomicron | 10 (10) | 10 | 10 | 10 | ||
| Bacteroides uniformis | 10 (10) | 10 | 10 | 10 | ||
| Bacteroides vulgatus | 10 (4) | 10 | 10 | 4 | ||
| Clostridium species | 15 (3) | 15 | ||||
| Fusobacterium speciesb | 3 (0) | 2 | 1 | 1 | ||
| Parabacteroides distasonis | 2 (2) | 2 | 2 | 2 | ||
| Parabacteroides faecis | 2 (0) | 2 | 2 | |||
| Parabacteroides goldsteinii | 2 (0) | 2 | 2 | |||
| Peptostreptococcus species | 3 (0) | 3 | ||||
| Prevotella species | 16 (1) | 16 | ||||
| Other anaerobesc | 5 (0) | 5 | ||||
| Aerobic and facultative anaerobic bacteria (88) | ||||||
| Enterobacteriaceae | 16 (8) | 16 | ||||
| Enterococcus species | 6 (6) | 6 | ||||
| Lactobacillus species | 5 (5) | 5 | ||||
| Nonfermentersd | 7 (2) | 7 | ||||
| Staphylococcus species | 23 (0) | 23 | ||||
| Streptococcus species | 22 (4) | 22 | ||||
| Vibrio species | 6 (2) | 6 | ||||
| Other bacteriae | 3 (2) | 3 | ||||
| Total | 303 (193) | 129 | 138 | 36 | 174 | 160 |
The total includes 48 reference strains and 255 clinical isolates. The designations of the reference strains and the clinical isolates are listed in Tables S1 and S2 in the supplemental material.
Fusobacterium mortiferum (n = 1; good growth), F. varium (n = 1, absent), and F. nucleatum (n = 1, absent).
Bifidobacterium bifidum (n = 1), Finegoldia magna (n = 1), Parvimonas micra (n = 1), Propionibacterium acnes (n = 1), and Veillonella parvula (n = 1).
Acinetobacter baumannii (n = 2), Brevundimonas diminuta (n = 1), Flavobacterium meningosepticum (n = 1), Pseudomonas aeruginosa (n = 2), and Stenotrophomonas maltophilia (n = 1).
Aeromonas hydrophila (n = 1), Kocuria rosea (n = 1), and Listeria monocytogenes (n = 1).
Ability of the organisms to hydrolyze esculin was determined by separate testing.
Poor growth is defined by pinpoint-sized colonies after 48 h of incubation.
FIG 1.
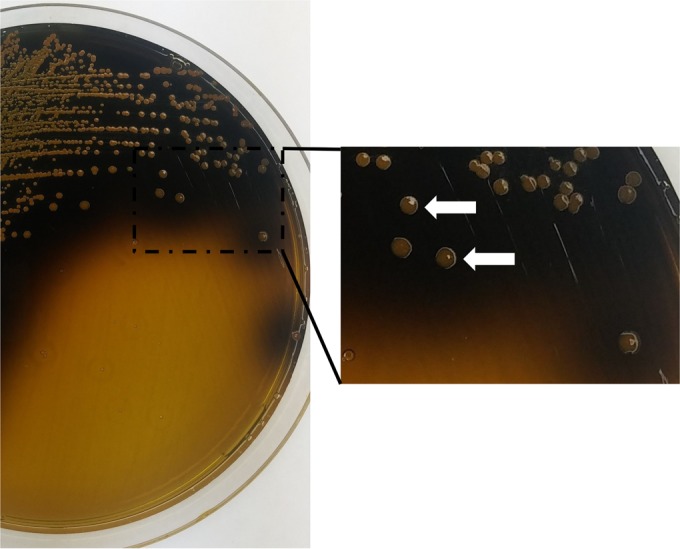
Culture on a plate of Bacteroides fragilis selective (BFS) medium after 48 h of anaerobic incubation at 35°C. B. fragilis (white arrows) appears as large yellow colonies (glucose fermentation) with blackening of the surrounding medium (esculin hydrolysis).
Validation of BFS agar with clinical specimens.
Of 1,209 clinical specimens, 540 yielded no growth on any plates, 513 showed growth only on plates using the routine agar (routine plates), and 156 had growth on both routine and BFS plates. Overall, B. fragilis was detected in 65 specimens (Table 2), of which 41 were detected by both routine and BFS plates, 5 were detected by routine plates only, and 19 were detected by BFS plates only (McNemar's test, P = 0.008). Colonies suggestive of B. fragilis on routine and BFS plates were tested with MALDI-TOF MS directly and after they were subcultured onto anaerobic blood agar plates. One to three colonies for each colony type were tested. All 106 cultures (46 from routine plates and 60 from BFS plates) were identified as B. fragilis with a Biotyper score of >2.0. Colony growth from BFS agar and anaerobic blood agar plates had Biotyper scores (mean ± standard deviation) of 2.227 ± 0.133 and 2.458 ± 0.111, respectively. A total of 106 colonies (one colony from each culture-positive plate) were further confirmed by species-specific PCR. The sensitivities of routine and BFS plates were 70.8% (46/65) and 92.3% (60/65), respectively. In the five specimens with B. fragilis that were missed by BFS plates, only a few colonies were found in the routine plates, and an inoculation problem may possibly explain the missed results. Of the 46 B. fragilis isolates from routine plates, 43 belonged to division I, and 3 belonged to division II. Of the 60 B. fragilis isolates from BFS plates, 54 belonged to division I, and 6 belonged to division II. In total, 59 specimens had B. fragilis division I isolates, and 6 specimens had B. fragilis division II isolates. No specimen had both B. fragilis division I and division II isolates. No difference in the identifications of the division of B. fragilis on routine and BFS plates was found. A total of 46 specimens were culture positive for Bacteroides species other than B. fragilis, including 53 isolates originating from BFS agar and 29 isolates from routine plates (Table 2). On BFS plates, 96 non-Bacteroides organisms were also recovered from 90 specimens. These include E. coli (n = 44), Enterobacter spp. (n = 5), Klebsiella spp. (n = 5), Citrobacter spp. (n = 4), Pseudomonas species (n = 13), Candida species (n = 15), Fusobacterium varium (n = 4), Aeromonas caviae (n = 1), Clostridium spp. (n = 1), and Enterococcus faecalis (n = 1). Besides Bacteroides species (Table 2), Fusobacterium varium was the only other species of anaerobe detected in mixed growth with B. fragilis (in 3 BFS agar plates).
TABLE 2.
Isolation of Bacteroides species from 1,209 clinical specimens plated onto Bacteroides fragilis selective (BFS) and routine anaerobic plates
| Bacteroides species | No. of isolates recovered on: |
|
|---|---|---|
| BFS plates | Routine plates | |
| B. fragilisa | 60 | 46 |
| B. caccae | 1 | 0 |
| B. nordii | 2 | 3 |
| B. ovatus | 16 | 6 |
| B. salyersiae | 3 | 1 |
| B. thetaiotaomicron | 29 | 12 |
| B. uniformis | 1 | 1 |
| B. vulgatus | 1 | 3 |
| B. stercoris | 0 | 2 |
| B. cellulosilyticus | 0 | 1 |
| Total | 113 | 75 |
In total, 65 B. fragilis isolates were detected: 41 by both methods, 5 by routine plates only, and 19 by BFS plates only. McNemar's test, P = 0.008 for comparison of results for B. fragilis detection by the two methods.
Our results showed that BFS medium supports the growth of B. fragilis, facilitates the differential recognition of the organism in specimens with mixed growth of other bacteria, and is significantly more sensitive than routine medium for the primary isolation of this organism. In addition, BFS agar is advantageous in that B. fragilis can be identified by MALDI-TOF MS directly using the growth in the agar plate. Among the Bacteroides species, some are less resistant to bile and can be susceptible to a high concentration of kanamycin (16, 21). This explains why BFS agar suppressed the growth of some Bacteroides species (Table 1). While BFS agar was designed to improve recovery of B. fragilis, the medium may also be used for isolation of several other Bacteroides species, such as B. thetaiotaomicron. In the latter species, metronidazole and carbapenem resistance have recently been described (22). Since the growth of B. vulgatus on BFS agar is inhibited (Table 1), the medium is not suitable for recovery of this antibiotic-resistant species (23). In the validation with clinical specimens, 59 of the 1,209 specimens had growth of Enterobacteriaceae, but their colony morphologies were different from the morphology of B. fragilis. Only a minority of the growth in BFS plates represented Candida species. According to Tierney et al., amphotericin B can be added without adversely affecting the recovery of B. fragilis (17). In our experience, the inclusion of amphotericin B is not essential.
A limitation of this study is that BBE agar was not included as a comparator. In addition, this study did not compare BFS agar against other nutritious agars such as brucella blood agar with vitamin K and hemin and chocolate agar containing hemin and supplemented with vitamin K and cysteine. For the test of the clinical specimens, the routine neomycin agar plates were processed by the on-duty laboratory technician in the usual manner while the BFS agar plates were followed by a research student. It is possible that the student exercised more diligence in following the BFS plates than would be expected with the routine handling of agar plates in a laboratory.
In conclusion, this study reports the evaluation of a new medium for recovery of B. fragilis from clinical specimens. BFS agar can be added to routine medium to improve recovery of B. fragilis and can potentially be supplemented with additional antibiotics (e.g., metronidazole and meropenem) for surveillance of isolates harboring resistance in different patient populations.
MATERIALS AND METHODS
Preparation of BFS agar.
Pilot studies were conducted to determine the optimal concentrations of glucose and bromothymol blue in the medium that would grow yellow colonies of B. fragilis. Several antibiotics (gentamicin, kanamycin, and novobiocin) were chosen based on their stability in prepared medium and antibacterial activities for common aerobes and facultative anaerobes that may hydrolyze esculin (19, 24). Concentrations of the antibiotics were chosen empirically after reviewing those that were previously used in various selective media (18, 19, 24). The final formulation of the new medium consisted of (per liter) 15 g of bacteriological agar (Oxoid agar no. 1), 37 g of brain heart infusion (CM1135; Oxoid), 5 g of yeast extract, 0.5 g of cysteine hydrochloride, 10 g of bile salts (LP0055; Oxoid), 0.05g of vitamin K, 0.5 g of hemin, 1 g of glucose, 1 g of esculin, 0.5 g of ferric ammonium citrate, 0.24 g of bromothymol blue (pH indicator), 4 mg of gentamicin, 100 mg of kanamycin, and 30 mg of novobiocin. All the ingredients, except the antibiotics and hemin-vitamin K components, were dissolved in deionized water, and the pH was titrated to 7.2 using hydrochloric acid. The medium was then autoclaved at 121°C for 15 min and cooled to 50°C. The hemin-vitamin K mixture and the antibiotics were then added aseptically. It was assumed that the ability of B. fragilis to ferment glucose would lower the pH of the medium and change the color of the indicator to yellow, thereby producing yellow colonies, while hydrolysis of esculin and subsequent reaction with iron in the medium would produce a diffusible black complex that would cause blackening of the medium surrounding the colonies (16, 18, 20). Antibiotics, including gentamicin, kanamycin, and novobiocin, were added to inhibit the growth of common bacteria (Enterobacteriaceae, enterococci, and lactobacilli) that are esculin positive, thereby facilitating the recognition of B. fragilis in mixed growth.
Recovery of B. fragilis and quality control.
The test strains were spread onto BFS plates at an inoculum of either 1,000 CFU (B. fragilis ATCC 25285) or 100,000 CFU (Escherichia coli ATCC 25922, Pseudomonas aeruginosa ATCC 27853, Enterococcus faecalis ATCC 29212, and Lactobacillus rhamnosus ATCC 7469). The inoculated BFS plates were incubated at 37°C in an anaerobic workstation (with 10% hydrogen, 10% carbon dioxide, and 80% nitrogen as the gas mixture) for 48 h, and the number of colonies in the plates was counted and compared against colony counts in aerobic or anaerobic blood agar plates (Becton Dickinson, Hong Kong) after aerobic (E. coli ATCC 25922, P. aeruginosa ATCC 27853, and E. faecalis ATCC 29212) or anaerobic (B. fragilis ATCC 25285 and Lactobacillus rhamnosus ATCC 7469) incubation. Differences between colony counts in the BFS and the blood agar plates were compared after conversion to a logarithmic scale, and counts are given as means and standard deviations. Batches of medium were considered suitable for use if they supported the growth of B. fragilis ATCC 25285 after 2 days of anaerobic incubation (colony counts on BFS plate ± 0.5 log CFU of that on the blood agar plate) while preventing the growth of the other four ATCC strains (complete absence of growth or >5 log CFU inhibition). The plates were stored at 4°C until use, and a shelf life of 10 days from quality control was assigned.
Validation of BFS agar with pure cultures.
Selectivity of the medium was evaluated by inoculation with pure cultures of different bacteria (see Tables S1 and S2 in the supplemental material). The bacterial collection included 48 ATCC strains and 255 clinical isolates that were chosen to represent aerobes, facultative anaerobes, and anaerobes that are commonly found in clinical specimens. The clinical isolates were recovered from specimens submitted to a clinical laboratory in 2013 to 2015 and had been identified to species level by MALDI-TOF MS (Bruker Daltonics, Bremen, Germany) (13, 25). All Bacteroides species were confirmed by conventional PCR assay using previously described species-specific primers or 16S rRNA gene sequencing (26). From a fresh subculture, the test organism was plated onto a BFS plate and a growth control plate (aerobic or anaerobic blood agar plates) by the standard four-quadrant method. The growth control plates were incubated under aerobic and anaerobic conditions as appropriate while all BFS plates were subjected to anaerobic incubation. Growth of colonies in the BFS plate was recorded as good, poor, or absent at 48 h. Poor growth was recorded when colonies remained pinpoint size.
Validation of BFS agar with clinical specimens.
The performance of BFS medium in the detection of B. fragilis in clinical specimens was prospectively examined. During a 3-month period from February to May 2016, 1,209 clinical specimens (793 pus and wound swabs and 214 bile and 202 peritoneal specimens) sent to a clinical laboratory for bacteriological investigation were cultured for the presence of B. fragilis. In the laboratory, these specimens were routinely inoculated onto three culture plates (blood agar and MacConkey agar plates for aerobic culture and anaerobic blood agar plates supplemented with 30 μg/ml neomycin for anaerobic culture). All of the routine media were obtained from Becton Dickinson, Hong Kong. Besides the routine screening, all specimens were inoculated onto one BFS plate and incubated anaerobically at 37°C for 48 h. Routine screening plates were processed by the on-duty laboratory technicians in the usual manner while the BFS plates were followed by a research student. The inoculated neomycin blood agar plates were incubated at 37°C in an anaerobic workstation (with 10% hydrogen, 10% carbon dioxide, and 80% nitrogen as the gas mixture) for 48 h before being read. Two independent personnel were engaged for the screening so that reading of the BFS plates would not be affected by results from readings of the routine plates and vice versa. After incubation of the plates, each colony type recovered in the agar plates was identified by a Bruker MALDI-TOF MS Biotyper system using the direct colony transfer and formic acid protocol (13). For bacterial identification, a reference library of 5,989 standard spectra (version 5.0.0.0) was used. Colony growth from the primary or subculture plates was transferred to a polished MSP 96 target (Bruker Daltonics) and overlaid with a saturated α-cyano-4-hydroxycinnamic acid (HCCA) matrix solution. Data were interpreted according to the manufacturer's criteria: species identification was made for a Biotyper score of ≥2.0, genus identification was made for a score range of ≥1.7 to <2.0, and no identification was made for a score of <1.7 (13). The ClinProTools software (Bruker Daltonics) was used for differentiation of B. fragilis into division I and II strains, and confirmation was obtained by PCR detection of the cfiA gene (27, 28). Results of the cultures from the routine processing and the BFS screening were kept blinded until the final analysis.
Statistical analysis.
Student's t test or McNemar's test was used for statistical analysis. A two-tailed P value of <0.05 was considered significant. All analyses were performed using GraphPad Prism, version 5 (GraphPad, La Jolla, CA).
Supplementary Material
ACKNOWLEDGMENTS
We thank the technical staff in Department of Microbiology, Queen Mary Hospital, University of Hong Kong, for assistance with the microbiological investigations.
We have no conflicts of interest to declare.
This work was supported by a grant from the Health and Medical Research Fund (HKM-15-M10) of the Food and Health Bureau of the Hong Kong Special Administrative Region.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01988-16.
REFERENCES
- 1.Seifert H, Dalhoff A, PRISMA Study Group. 2010. German multicentre survey of the antibiotic susceptibility of Bacteroides fragilis group and Prevotella species isolated from intra-abdominal infections: results from the PRISMA study. J Antimicrob Chemother 65:2405–2410. doi: 10.1093/jac/dkq321. [DOI] [PubMed] [Google Scholar]
- 2.Wexler HM. 2007. Bacteroides: the good, the bad, and the nitty-gritty. Clin Microbiol Rev 20:593–621. doi: 10.1128/CMR.00008-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nagy E, Urban E, Nord CE. 2011. Antimicrobial susceptibility of Bacteroides fragilis group isolates in Europe: 20 years of experience. Clin Microbiol Infect 17:371–379. doi: 10.1111/j.1469-0691.2010.03256.x. [DOI] [PubMed] [Google Scholar]
- 4.Snydman DR, Jacobus NV, McDermott LA, Golan Y, Goldstein EJ, Harrell L, Jenkins S, Newton D, Pierson C, Rosenblatt J, Venezia R, Gorbach SL, Queenan AM, Hecht DW. 2011. Update on resistance of Bacteroides fragilis group and related species with special attention to carbapenems 2006–2009. Anaerobe 17:147–151. doi: 10.1016/j.anaerobe.2011.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Liu CY, Huang YT, Liao CH, Yen LC, Lin HY, Hsueh PR. 2008. Increasing trends in antimicrobial resistance among clinically important anaerobes and Bacteroides fragilis isolates causing nosocomial infections: emerging resistance to carbapenems. Antimicrob Agents Chemother 52:3161–3168. doi: 10.1128/AAC.00355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Karlowsky JA, Walkty AJ, Adam HJ, Baxter MR, Hoban DJ, Zhanel GG. 2012. Prevalence of antimicrobial resistance among clinical isolates of Bacteroides fragilis group in Canada in 2010–2011: CANWARD surveillance study. Antimicrob Agents Chemother 56:1247–1252. doi: 10.1128/AAC.05823-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalapila A, Pergam S, Pottinger P, Butler-Wu S, Whimbey E. 2013. Multidrug-resistant Bacteroides fragilis, Seattle, Washington, 2013. MMWR Morb Mortal Wkly Rep 62:694–696. [PMC free article] [PubMed] [Google Scholar]
- 8.Soki J, Hedberg M, Patrick S, Balint B, Herczeg R, Nagy I, Hecht DW, Nagy E, Urban E. 2016. Emergence and evolution of an international cluster of MDR Bacteroides fragilis isolates. J Antimicrob Chemother 71:2441–2448. doi: 10.1093/jac/dkw175. [DOI] [PubMed] [Google Scholar]
- 9.Nagy E, Becker S, Soki J, Urban E, Kostrzewa M. 2011. Differentiation of division I (cfiA-negative) and division II (cfiA-positive) Bacteroides fragilis strains by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. J Med Microbiol 60:1584–1590. doi: 10.1099/jmm.0.031336-0. [DOI] [PubMed] [Google Scholar]
- 10.Husain F, Veeranagouda Y, Hsi J, Meggersee R, Abratt V, Wexler HM. 2013. Two multidrug-resistant clinical isolates of Bacteroides fragilis carry a novel metronidazole resistance nim gene (nimJ). Antimicrob Agents Chemother 57:3767–3774. doi: 10.1128/AAC.00386-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kato N, Yamazoe K, Han CG, Ohtsubo E. 2003. New insertion sequence elements in the upstream region of cfiA in imipenem-resistant Bacteroides fragilis strains. Antimicrob Agents Chemother 47:979–985. doi: 10.1128/AAC.47.3.979-985.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salipante SJ, Kalapila A, Pottinger PS, Hoogestraat DR, Cummings L, Duchin JS, Sengupta DJ, Pergam SA, Cookson BT, Butler-Wu SM. 2015. Characterization of a multidrug-resistant, novel Bacteroides genomospecies. Emerg Infect Dis 21:95–98. doi: 10.3201/eid2101.140662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen JH, Ho PL, Kwan GS, She KK, Siu GK, Cheng VC, Yuen KY, Yam WC. 2013. Direct bacterial identification in positive blood cultures by use of two commercial matrix-assisted laser desorption ionization-time of flight mass spectrometry systems. J Clin Microbiol 51:1733–1739. doi: 10.1128/JCM.03259-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cattoir V, Giard JC. 2014. Antibiotic resistance in Enterococcus faecium clinical isolates. Expert Rev Anti Infect Ther 12:239–248. doi: 10.1586/14787210.2014.870886. [DOI] [PubMed] [Google Scholar]
- 15.Klein G. 2011. Antibiotic resistance and molecular characterization of probiotic and clinical Lactobacillus strains in relation to safety aspects of probiotics. Foodborne Pathog Dis 8:267–281. doi: 10.1089/fpd.2010.0672. [DOI] [PubMed] [Google Scholar]
- 16.Livingston SJ, Kominos SD, Yee RB. 1978. New medium for selection and presumptive identification of the Bacteroides fragilis group. J Clin Microbiol 7:448–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tierney D, Copsey SD, Morris T, Perry JD. 2016. A new chromogenic medium for isolation of Bacteroides fragilis suitable for screening for strains with antimicrobial resistance. Anaerobe 39:168–172. doi: 10.1016/j.anaerobe.2016.04.003. [DOI] [PubMed] [Google Scholar]
- 18.Papastathopoulou A, Bezirtzoglou E, Legakis NJ. 1998. A new selective and differentiative medium for the isolation of Bacteroides fragilis. Microb Ecol Health Dis 10:110–113. doi: 10.1080/089106098435359. [DOI] [Google Scholar]
- 19.Pathela P, Hasan KZ, Roy E, Alam K, Huq F, Siddique AK, Sack RB. 2005. Enterotoxigenic Bacteroides fragilis-associated diarrhea in children 0–2 years of age in rural Bangladesh. J Infect Dis 191:1245–1252. doi: 10.1086/428947. [DOI] [PubMed] [Google Scholar]
- 20.Lyznicki JM, Busch EL, Blazevic DJ. 1982. Medium for selective isolation and presumptive identification of the Bacteroides fragilis group. J Clin Microbiol 15:123–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamazaki E, Sugimoto K, Niwano K, Okada J. 1982. A simple bile-disk method for the identification of Bacteroides fragilis. Microbiol Immunol 26:759–765. doi: 10.1111/j.1348-0421.1982.tb00222.x. [DOI] [PubMed] [Google Scholar]
- 22.Sadarangani SP, Cunningham SA, Jeraldo PR, Wilson JW, Khare R, Patel R. 2015. Metronidazole- and carbapenem-resistant Bacteroides thetaiotaomicron isolated in Rochester, Minnesota, in 2014. Antimicrob Agents Chemother 59:4157–4161. doi: 10.1128/AAC.00677-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Snydman DR, Jacobus NV, McDermott LA, Golan Y, Hecht DW, Goldstein EJ, Harrell L, Jenkins S, Newton D, Pierson C, Rihs JD, Yu VL, Venezia R, Finegold SM, Rosenblatt JE, Gorbach SL. 2010. Lessons learned from the anaerobe survey: historical perspective and review of the most recent data (2005–2007). Clin Infect Dis 50(Suppl 1):S26–S33. doi: 10.1086/647940. [DOI] [PubMed] [Google Scholar]
- 24.Jones RN. 1989. Should novobiocin be clinically re-evaluated? Diagn Microbiol Infect Dis 12:363–365. doi: 10.1016/0732-8893(89)90105-3. [DOI] [PubMed] [Google Scholar]
- 25.Luo Y, Siu GK, Yeung AS, Chen JH, Ho PL, Leung KW, Tsang JL, Cheng VC, Guo L, Yang J, Ye L, Yam WC. 2015. Performance of the VITEK MS matrix-assisted laser desorption ionization-time of flight mass spectrometry system for rapid bacterial identification in two diagnostic centres in China. J Med Microbiol 64:18–24. doi: 10.1099/jmm.0.080317-0. [DOI] [PubMed] [Google Scholar]
- 26.Tong J, Liu C, Summanen P, Xu H, Finegold SM. 2011. Application of quantitative real-time PCR for rapid identification of Bacteroides fragilis group and related organisms in human wound samples. Anaerobe 17:64–68. doi: 10.1016/j.anaerobe.2011.03.004. [DOI] [PubMed] [Google Scholar]
- 27.Eitel Z, Soki J, Urban E, Nagy E. 2013. The prevalence of antibiotic resistance genes in Bacteroides fragilis group strains isolated in different European countries. Anaerobe 21:43–49. doi: 10.1016/j.anaerobe.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Wybo I, De BA, Soetens O, Echahidi F, Vandoorslaer K, Van CM, Pierard D. 2011. Differentiation of cfiA-negative and cfiA-positive Bacteroides fragilis isolates by matrix-assisted laser desorption ionization-time of flight mass spectrometry. J Clin Microbiol 49:1961–1964. doi: 10.1128/JCM.02321-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


