ABSTRACT
Recent evidence shows that patients asymptomatically colonized with Clostridium difficile may contribute to the transmission of C. difficile in health care facilities. Additionally, these patients may have a higher risk of developing C. difficile infection. The aim of this study was to compare a commercially available PCR directed to both toxin A and B (artus C. difficile QS-RGQ kit CE; Qiagen), an enzyme-linked fluorescent assay to glutamate dehydrogenase (GDH ELFA) (Vidas, bioMérieux), and an in-house-developed PCR to tcdB, with (toxigenic) culture of C. difficile as the gold standard to detect asymptomatic colonization. Test performances were evaluated in a collection of 765 stool samples obtained from asymptomatic patients at admission to the hospital. The C. difficile prevalence in this collection was 5.1%, and 3.1% contained toxigenic C. difficile. Compared to C. difficile culture, the sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) of the C. difficile GDH ELFA were 87.2%, 91.2%, 34.7%, and 99.3%, respectively. Compared with results of toxigenic culture, the sensitivity, specificity, PPV, and NPV of the commercially available PCR and the in-house PCR were 95.8%, 93.4%, 31.9%, 99.9%, and 87.5%, 98.8%, 70%, and 99.6%, respectively. We conclude that in a low-prevalence setting of asymptomatically colonized patients, both GDH ELFA and a nucleic acid amplification test can be applied as a first screening test, as they both display a high NPV. However, the low PPV of the tests hinders the use of these assays as stand-alone tests.
KEYWORDS: Clostridium difficile, asymptomatic, carrier, diagnostics
INTRODUCTION
Clostridium difficile infection (CDI) is a leading cause of hospital-acquired diarrhea. The transmission of spores from symptomatic patients can spread C. difficile within health care facilities, with a subsequent development of more symptomatic patients and eventually clusters and outbreaks. However, recent data suggest that patients asymptomatically colonized with C. difficile also contribute to the spread of C. difficile spores to the environment and to other patients (1–3). Asymptomatic carriers shed spores into the environment to a lesser extent than CDI patients (3, 4), but by outnumbering the CDI patients, they can still play an important role in the transmission of the disease. This hypothesis has recently been supported in a Canadian study, where isolation of C. difficile-colonized patients significantly reduced the incidence of hospital-acquired CDI (5). A second new insight into the significance of asymptomatic colonization is that it may increase the risk of subsequent clinical disease in some colonized patients (6–10). Progression from colonization to CDI can be provoked by alterations of the microbiota and a subsequent decrease in secondary bile acids, which normally inhibit spore germination (11–13). But other factors, like preexisting antitoxin antibodies, may also play a role in protection from progression to CDI, although their exact roles need to be clarified.
Thus, recognition of asymptomatically colonized patients may be clinically relevant to reduction in nosocomial transmission and for protection from progression to symptomatic disease. Asymptomatic colonization of C. difficile varies widely between various patient populations studied. Approximately 5 to 15% of newly hospital admitted patients carry C. difficile in their feces (4, 5, 14–17). Carriage rates of residents in long-term-care facilities vary from 4 to 51% but in general tend to be higher than in hospitalized patients (3, 14, 18). Asymptomatic colonization of C. difficile in the pediatric populations is very high; approximately 37% of children are asymptomatic carriers in their first year of life, decreasing to 15% for children between 1 and 8 years of age (19).
A recently published European guidance document advises a two-stage algorithm to diagnose CDI using a toxin nucleic acid amplification test (NAAT) or glutamate dehydrogenase (GDH) enzyme immunoassay (EIA) as sensitive screening assay in combination with tests to detect the presence of free toxins in stools as a marker of disease activity (20). Samples without free toxins detected will largely represent asymptomatic carriers. However, this guideline addresses diagnostics of CDI in diarrheal patients and reviewed the literature of symptomatic patients with CDI. The optimal diagnostic test to detect C. difficile in asymptomatically colonized patients with normally formed stool is unknown. Therefore, the aim of this study was to compare the performances of a commercially available GDH EIA with that of a primary gold standard, a conventional culture of C. difficile in asymptomatically colonized patients at admission to three large hospitals in the Netherlands. Moreover, a commercially available PCR for tcdA and tcdB and in-house-developed PCR for detection of tcdB were compared with a secondary gold standard, toxigenic C. difficile culture (TC).
(Preliminary results from this study were presented at the European Congress of Clinical Microbiology and Infectious Diseases, 9 to 12 April 2016, Amsterdam, the Netherlands [21]).
RESULTS
In total, 765 feces samples from 581 unique patients were included in the evaluation, of which 39 samples (5.1%; 95% confidence interval [CI], 3.8 to 6.9%) were positive for the presence of C. difficile by culture; 24 samples (3.1%; 95% CI, 2.1 to 4.6%) contained toxigenic C. difficile. All 765 samples were tested by toxigenic culture, GDH enzyme-linked fluorescent assay (ELFA), and in-house PCR, but due to insufficient sample volume of one sample, 764 samples were tested with the artus PCR. The sensitivity, specificity, PPV, and NPV data of the various tests are depicted in Table 1. The artus PCR had the highest sensitivity, at 95.8%. The mean quantification cycle (Cq) values in true-positive samples were 27.5 for tcdA and 28.4 for tcdB. The in-house PCR showed a sensitivity of 87.5%, with a mean tcdB Cq value of 29.3 in true-positive samples. The difference in sensitivity between artus PCR and the in-house PCR was not significant (P = 0.5). The GDH ELFA had a sensitivity of 87.2%. The mean test value of the GDH ELFA in true positives was 11.7 relative fluorescent units (standard deviation [SD], 8.11). Specificities were 98.8%, 93.4%, and 91.2% for the in-house PCR, artus PCR, and GDH test, respectively. The specificity of the in-house PCR was significantly higher than that of the artus PCR (P < 0.000001). The NPV was in general very high and ranged from 99.3% to 99.9% for all assays. The PPV, on the other hand, was only 31.9% for the artus PCR and 34.7% for the GDH ELFA. In comparison to the artus PCR and GDH ELFA, the in-house PCR had a higher PPV of 70.0% (Table 1). Receiver operating characteristic (ROC) curves were made for the performances of the individual tests. For GDH ELFA, artus PCR, and in-house PCR, the diagnostic accuracy as given by the area under the curve was 0.8918, 0.9467, and 0.9314, respectively (Fig. S1).
TABLE 1.
Comparison of various C. difficile detection assays in comparison with culture of toxigenic and nontoxigenic C. difficile as gold standards
| Assay result | No. with toxigenic culture resulta: |
Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | PPV (%) | NPV (%) | |
|---|---|---|---|---|---|---|
| Pos | Neg | |||||
| GDH positive | 34 | 64b | 87.2 (72.6–95.7) | 91.2 (88.9–93.1) | 34.7 | 99.3 |
| GDH negative | 5 | 662 | ||||
| artus positive | 23 | 49b | 95.8 (78.9–99.9) | 93.4 (91.3–95.1) | 31.9 | 99.9 |
| artus negative | 1 | 691 | ||||
| In-house positive | 21 | 9b | 87.5 (67.6–97.3) | 98.8 (97.7–99.4) | 70 | 99.6 |
| In-house negative | 3 | 732 | ||||
GDH ELFA was compared with C. difficile culture, and artus PCR and in-house PCR were compared with toxigenic culture. Pos, positive; Neg, negative.
Four of the false-negative samples were positive in all tests (GDH, artus, and in-house PCR).
Of 623 samples tested with the in-house PCR without the addition of bovine serum albumin (BSA), 61 (9.8%) samples showed inhibition which disappeared at a 1:10 dilution of the sample. Of 142 samples tested by the in-house PCR with addition of BSA, no inhibition was observed. Of 764 samples tested by artus PCR, 40 (5.2%) samples showed inhibition which disappeared with repeated testing. Additionally, 26 (3.4%) invalid artus PCR results were obtained due to a tcdA Cq value above the accepted range (n = 4), a tcdB Cq value above the accepted range (n = 16), tcdA uncertain (n = 3), or tcdB uncertain (n = 3).
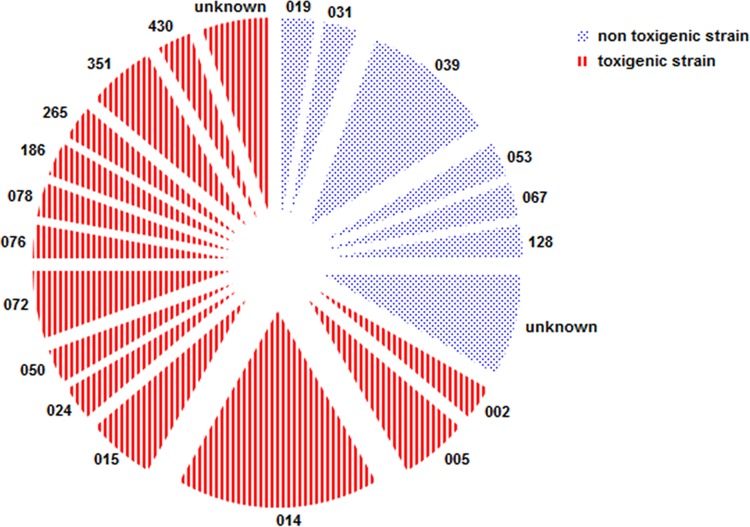
A discrepancy analysis was performed on discordant results and is displayed in Table 2. Four of 741 TC-negative samples tested positive with all three assays (GDH ELFA, in-house PCR, and artus PCR), suggesting a false-negative result of the TC. One of these 4 fecal samples was positive for culture of toxigenic C. difficile using the enriched TC method, suggesting that a very low number of C. difficile was present. Two other TC-negative results could be explained by vancomycin treatment at the time of fecal sampling, which inhibits the growth of C. difficile. No clear explanation was found for the remaining false-negative TC fecal sample. All 45 false-positive artus PCR samples tested negative with retesting. The artus PCR flags a result as real positive when one or both of the toxin genes are below a certain Cq value. Of these 45 TC-negative but positive-flagged artus PCR results, 21 results were positive only for tcdA and seven results only for tcdB, whereas 17 results were both tcdA and tcdB positive. The mean Cq values of the false positives were higher (tcdA, 33.1; tcdB, 33.4) than the Cq values of artus PCR true positives (tcdA, 27.8; tcdB, 28.4). No discrepancy analysis was performed on the remaining 60 GDH false-positive samples (Table 1; 60 − 4, as mentioned above) due to the expected low specificity. One feces sample tested negative by in-house PCR as well as the artus PCR and remained negative with retesting. However, the toxigenic cultured strain was positive tested by both PCRs, suggesting that a very low number of C. difficile was present in the feces. After the discrepancy analysis, the sensitivity, specificity, PPV, and NPV were 96.6%, 100%, 100%, and 99.9%, and 96.9%, 99.7%, 93.3%, and 99.7% for the artus PCR and in-house PCR, respectively. The distribution of PCR ribotypes isolated from asymptomatic patients in this cohort is displayed in Fig. 1. Five strains could not be ribotyped since the profiles of the corresponding strains were not present in the National Reference Laboratory of the Netherlands.
TABLE 2.
Results of first and second testsa
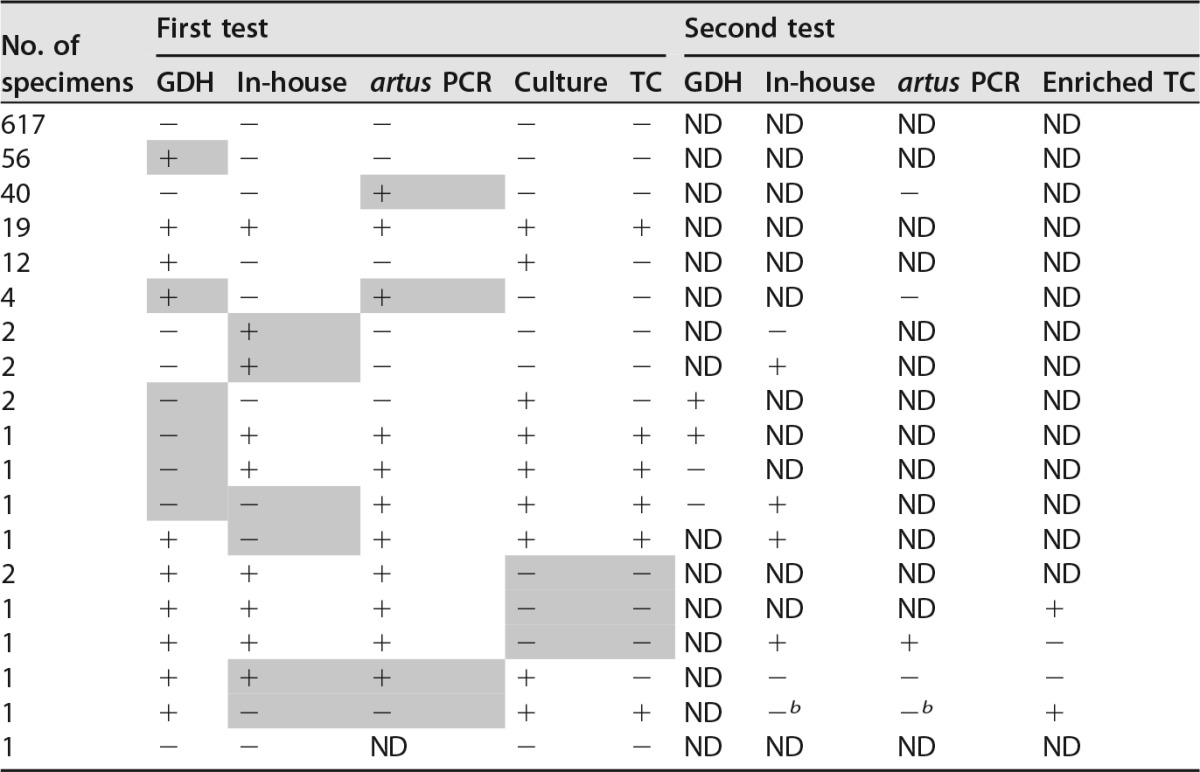
After resolving of the four TC-negative/other test-positive samples, results were as follows: two false-positive in-house PCR results retested positive (Cq values, 28 and 31.6, respectively), while three false-positive in-house PCR results could not be confirmed with retesting. All 45 false-positive artus samples retested negative. All remaining 60 GDH false-positive samples were not retested. Two out of three false-negative in-house PCR results retested positive. One in-house PCR and artus PCR false-negative sample remained negative upon retesting by both PCRs, while both in-house and artus PCR on the cultured strain were positive. Two GDH ELFA-negative samples retested positive, while two remained negative. Shading indicates a false (positive or negative) result. ND, not detected.
b In-house and artus PCR positive on strain, not on feces.
FIG 1.
Distribution of C. difficile ribotypes isolated from asymptomatic patients displayed in a pie chart. Indicated in red are the toxigenic strains, and indicated in blue are the nontoxigenic strains. The numbers indicate the corresponding ribotype number.
DISCUSSION
The aim of this study was to compare two molecular assays (artus C. difficile PCR and in-house tcdB PCR) and C. difficile GDH ELFA with (toxigenic) culture of C. difficile as gold standards to detect asymptomatic colonization.
In this study, 5.1% of the patients attending a tertiary-care hospital were positive with C. difficile, and 3.1% contained toxigenic C. difficile. Other studies testing fecal samples for the presence of asymptomatic colonization of C. difficile at admission (or collected feces within 72 h after attending the hospital) reported a higher prevalence of 7.5% to 15.7% toxigenic C. difficile (1, 2, 10, 22–25). The lower prevalence rate in our study is probably related to the overall low prevalence of C. difficile and CDI in the Netherlands. A recently completed cross-sectional study among 2,494 healthy adults in the Netherlands revealed a prevalence of toxigenic C. difficile of 1.2% in the community (unpublished data).
The sensitivity and specificity of the automated Vidas GDH ELFA in comparison to C. difficile culture were 87.2% and 91.2%, respectively. Davies et al. studied the performance of the same GDH ELFA in diarrheal samples submitted for C. difficile testing and reported a higher sensitivity of 95.8% and a similar specificity of 91% (40). The lower sensitivity found in our study could be due to presence of lower numbers of C. difficile in fecal samples of asymptomatically colonized patients than in patients with CDI (18). However, we do not exclude the possibility that the percentages change when larger number of fecal samples are tested. An alternative explanation for the lower sensitivity rates in our study are the formed fecal samples that we included instead of diarrheal samples.
The artus PCR and the in-house PCR were compared with TC and revealed sensitivities of 95.8% and 87.5%, respectively, although this difference in sensitivity rate was not significant. In contrast, the artus PCR was statistically less specific than the in-house PCR (93.8% versus 98.8%, respectively). Since the artus PCR-positive, TC-negative samples could not be confirmed by retesting, the results indicate false positivity. This was supported by the considerably higher Cq values of tcdA and tcdB for the false-positive test results than for true positives. A hypothetical algorithm to enhance the specificity of artus PCR is considering the artus PCR result positive only when tcdA as well as tcdB are positive. This resulted in a specificity of 97% and a subsequent PPV of 51%, while remaining the high sensitivity of 95.8%. However, in the rare event a patient is colonized with a toxin A-negative, toxin B-positive C. difficile strain (26, 27), the new strategy will not identify this strain. The test results of artus PCR showed lower inhibition rates than the in-house PCR, at 8% versus 5.3%, respectively; however, inhibition of the in-house PCR was overcome by adding the PCR enhancer BSA (28). An additional 3.5% of the artus PCR-tested samples gave invalid test results, largely because of Cq values above the accepted range. Because of invalid or inhibited results, 8.7% of the fecal samples needed retesting by the artus PCR. The performance of the artus PCR in this study resembled the results in loose-stool samples submitted for CDI testing, as reported by Jazmati et al. (29). In a collection of 201 stool specimens, all 28 TC-positive samples were detected by the artus PCR (sensitivity, 100%), but the specificity, like that in this study, was relatively low (89.5%) (29). They stated that the lower specificity could largely be explained by a higher sensitivity of the artus PCR than TC. However, we did not share this observation. Our hypothesis is that the false-positive results were based on DNA contamination, as none of the other diagnostic tests were positive in these samples. The contamination route can be explained by the manual handling of the tubes containing the isolated DNA and PCR mixture to the Rotor-Gene, which required placing caps on tiny tubes, arranged very close to each other. The sensitivity of both PCRs and specificity of the in-house PCR are in agreement with the NAAT performance in symptomatic C. difficile patients, as mentioned in the recently published European Society of Clinical Microbiology and Infectious Diseases (ESCMID) guidance document, with an overall sensitivity of 95% and specificity of 98% in comparison with TC (20). A discrepancy analysis was performed, mainly to clarify why conflicting test results were obtained. The test characteristics that were calculated after resolving discrepant results could thereby be biased in favor of the index tests and should be considered with caution.
For CDI diagnosis, the use of a two-step algorithm is recommended (27). After a first sensitive test, which reliably classifies nondiseased patients, a more specific test is applied as a second test to discern true positives from false positives. For the diagnosis of colonized patients, a similar approach could be used. All three assays that we analyzed in this study had high NPV and would therefore be useful as a first screening test. Thereafter, confirmation of positive samples by a specific test could be recommended. This specific test could be a NAAT or toxigenic culture when GDH was used as a first screening test. A second algorithm could be screening by NAAT and confirmation by TC.
The ribotypes of asymptomatic carriers found in this study do not differ from the ribotypes found among CDI patients (30). This supports the hypothesis that asymptomatic carriers and CDI patients share a source or transfer C. difficile to each other irrespective of the PCR ribotype. Furthermore, only one of 24 C. difficile strains belonged to the epidemic ribotype 078, and no 027 strains were detected. Other studies confirm this finding (31–33), supporting the hypothesis that epidemic strains seldom lead to asymptomatic colonization.
Many studies report on the performance of C. difficile diagnostic assays in patients with presumed CDI, but only a few report on the application of diagnostic tests in patients with asymptomatic C. difficile colonization (1, 2, 15, 22, 23). The studies in asymptomatically colonized patients vary greatly in patient inclusion criteria, tested material, and applied diagnostic and gold standard tests. For instance, a great number of the studies only test rectal swabs or use a combination of stool samples and rectal swabs (4, 8, 18, 22, 23, 31, 34). Guerrero et al. showed that asymptomatic carriers have lower numbers of C. difficile in their rectal swab than CDI patients, indicating that stool samples should be preferred (4). Furthermore, a mix of diagnostic screening tests have been applied to detect C. difficile, frequently subdivided into assays to recognize toxigenic or nontoxigenic strains (20). However, a comparison of various diagnostic tests with a reference method to detect asymptomatic colonization of C. difficile has not been studied before.
Our study has a few limitations. An important limitation is the low prevalence rate of asymptomatic C. difficile colonization, which resulted in a low PPV of 31.9 to 70% for the different tests. However, this prevalence rate provides the most precise information on the performance of the test in our patient population. All tests would have better PPVs in a population with higher prevalence rates of C. difficile colonization, or when a selection of samples is tested when a predictive model for C. difficile colonization becomes available. A second limitation may be the freeze-thaw step, which presumably can lower the sensitivity, although we performed all tests immediately after thawing, except for the discrepancy analysis. In addition, no published reports indicate that freeze-thawing affects the performances of diagnostic PCRs for bacterial pathogens.
In conclusion, this study is the first, to our knowledge, to evaluate the use of three different assays for the detection of asymptomatic C. difficile colonization in stool samples and compare it to their gold standards. In our setting of low endemicity of asymptomatically colonized patients, all three assays (i.e., GDH ELFA, artus PCR, and in-house PCR) can be applied as a first screening test, as they display a very high NPV. The positive predictive values of these tests were suboptimal, and therefore, these assays are not suitable as stand-alone tests in a low-prevalence setting.
MATERIALS AND METHODS
Study design.
A multicenter study was performed on fecal samples obtained between November 2014 and December 2015 in the Leiden University Medical Center, Leiden (623 samples); Amphia Hospital, Breda (72 samples); and Erasmus Medical Center, Rotterdam (70 samples) in the Netherlands. The study was designed to determine risk factors for asymptomatic C. difficile colonization at admission to the hospital (ZonMW 50-52200-98-035). The institutional review board judged that ethical approval was not required. Fecal samples were obtained from patients on admittance to internal medicine and surgical wards and from patients attending the kidney transplant outpatient clinic. If a patient was admitted twice in the study period, the patient remained eligible for this study.
Culture and characterization of C. difficile.
The samples were processed for C. difficile culture and TC within 72 h of arrival at the laboratory and were subsequently stored at −20°C without the addition of glycerol. Feces was inoculated on C. difficile selective agar (CLO medium; bioMérieux, Marcy l'Etoile, France) and CNA medium (colistin and nalidixic acid-containing agar; bioMérieux) with and without ethanol shock pretreatment (35). The media were incubated for 5 days in an anaerobic atmosphere at ±35°C. Gray-brown colonies with the characteristic horse manure odor were further tested by an in-house GDH PCR (36). C. difficile isolates were tested for the presence of toxin genes by PCR for toxin A (tcdA), toxin B (tcdB), and binary toxin (cdtA and cdtB) (36). Capillary gel-based electrophoresis PCR ribotyping was performed to characterize the isolates (37).
Diagnostic C. difficile tests.
After thawing the stored fecal samples, the GDH EIA and both NAATs were performed in bulk testing. The targets of the applied detection assays are depicted in Table 3. The GDH EIA was performed on an enzyme-linked fluorescent immunoassay (ELFA) platform (Vidas; bioMérieux, Marcy-l'Etoile, France), as previously described (38). A test value of ≥0.1 relative fluorescent units was regarded positive. Both GDH ELFA and artus PCR were performed according to the manufacturer's instructions. As both assays are not registered for use with formed stools from asymptomatic patients, instructions were modified for off-label use, in consultation with the manufacturer. For both tests, approximately the size of half a pea of feces (approximately 0.3 to 0.4 g), instead of 200 μl of liquid feces, was used, as this is routine practice in our laboratory for isolation of DNA of formed feces. For the artus PCR (artus C. difficile QS-RGQ kit CE; Qiagen, the Netherlands), feces was transferred into a test tube with 1,500 μl of tissue lysis buffer (ATL), vortexed, and centrifuged for a short amount of time. The tubes were then inserted into the QIAsymphony supplied with the artus C. difficile AS software, which regulates DNA isolation and preparation of PCR mixture. The PCR mixture was manually transferred to the Rotor-Gene Q MDx. Samples with invalid artus PCR results were retested until the result was valid, with a maximum of three testing rounds. For the in-house PCR, DNA extraction was performed using the MagNA Pure 96 system (Roche Diagnostics, Almere, the Netherlands). In short, approximately half a pea size of feces was resuspended in 1 ml of stool transport and recovery (S.T.A.R.) buffer (Roche Diagnostics), supplemented with Precellys beads (Bertin Technology, France), mixed thoroughly by shaking on a Vibrax shaker (5 min, 2,200 rpm), and centrifuged for 1 min at 14,000 rpm. Of the supernatant, 200 μl was used for nucleic acid (NA) extraction using the MP96 DNA and viral NA small-volume kit, yielding a final eluate of 100 μl. The in-house-developed real-time PCR for the specific detection of the tcdB gene was tested in a multiplex assay with phocine herpesvirus as an internal control, as described previously (39). Samples with a quantification cycle (Cq) value higher than 40 were considered negative. In addition, samples with an internal-control Cq value that deviated more than 3.3 Cq values from the internal-control Cq value of the negative control were considered inhibited. Due to a change in workflow of adding BSA to all our in-house PCRs with feces as sample material to decrease the inhibition rate, the last 142 samples were tested with addition of 5 mg/ml of the PCR enhancer bovine serum albumin (BSA) (28).
TABLE 3.
C. difficile detection assays included in this this studya
| Assay type | Assay | Target(s) | Supplier (reference) |
|---|---|---|---|
| Anaerobic culture | C. difficile culture | Identification by PCR with GDH as target | In-house (36) |
| Toxigenic culture | C. difficile culture and PCR for toxin genes | Multiplex PCR with tcdA, tcdB, and cdtA and cdtB (binary toxin) | In-house (36) |
| Automated immunoassay | Vidas GDH | GDH | bioMérieux, France |
| Nucleic acid amplification test | artus C. difficile QS-RGQ kit CE | tcdA and tcdB | Qiagen, Germany |
| Nucleic acid amplification test | In-house C. difficile PCR | tcdB | In-house (39) |
GDH, glutamate dehydrogenase; tcdA, toxin A; tcdB, toxin B; cdtA and cdtB, binary toxin.
Discrepancy analysis.
Samples with discordant results were retested, except for positive results of the GDH ELFA because of an expected low specificity. An enriched TC was performed when three diagnostic tests were positive and the TC was negative. For enriched TC, half a pea size of feces was suspended in a cycloserine-cefoxitin-mannitol broth with taurocholate, lysozyme, and cysteine (CCMB-TAL; Anaerobe Systems, Morgan Hill, CA). The enrichment broth was subcultured on CLO and CNA agar, as described above, on days 2 and 5.
Statistical analysis.
A GDH-positive result was considered true positive or true negative if the stool culture was positive or negative for C. difficile, respectively, irrespective of its toxin production. For both PCRs, a positive result was considered true positive or true negative if the stool culture was positive or negative for toxigenic C. difficile, respectively. False-positive and false-negative test results were defined as discrepant results compared to the gold standard. The sensitivity and specificity of the tests were determined by the proportion positive or or negative, respectively, correctly identified samples. The difference in both sensitivity and specificity between the toxin PCRs was determined using McNemar's test for paired proportions. The sensitivity and specificity data were used to calculate the positive predictive value (PPV) and negative predictive value (NPV). The Cq values of false-positive results were compared with Cq values of true-positive results using an independent Student t test. ROC curves were constructed for all index tests. Analyses were performed using SPSS 23.0 and STATA version 12.1 statistical software.
Supplementary Material
ACKNOWLEDGMENTS
Sample collection of this study was supported by the Netherlands Organization for Health Research and Development, ZonMW (grant 50-52200-98-035). An unrestricted grant was supplied by Qiagen. The funders had no role in the study design, data collection, interpretation of the data, or writing of the manuscript.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.01858-16.
REFERENCES
- 1.Curry SR, Muto CA, Schlackman JL, Pasculle AW, Shutt KA, Marsh JW, Harrison LH. 2013. Use of multilocus variable number of tandem repeats analysis genotyping to determine the role of asymptomatic carriers in Clostridium difficile transmission. Clin Infect Dis 57:1094–1102. doi: 10.1093/cid/cit475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eyre DW, Griffiths D, Vaughan A, Golubchik T, Acharya M, O'Connor L, Crook DW, Walker AS, Peto TE. 2013. Asymptomatic Clostridium difficile colonisation and onward transmission. PLoS One 8:e78445. doi: 10.1371/journal.pone.0078445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Riggs MM, Sethi AK, Zabarsky TF, Eckstein EC, Jump RL, Donskey CJ. 2007. Asymptomatic carriers are a potential source for transmission of epidemic and nonepidemic Clostridium difficile strains among long-term care facility residents. Clin Infect Dis 45:992–998. doi: 10.1086/521854. [DOI] [PubMed] [Google Scholar]
- 4.Guerrero DM, Becker JC, Eckstein EC, Kundrapu S, Deshpande A, Sethi AK, Donskey CJ. 2013. Asymptomatic carriage of toxigenic Clostridium difficile by hospitalized patients. J Hosp Infect 85:155–158. doi: 10.1016/j.jhin.2013.07.002. [DOI] [PubMed] [Google Scholar]
- 5.Longtin Y, Paquet-Bolduc B, Gilca R, Garenc C, Fortin E, Longtin J, Trottier S, Gervais P, Roussy JF, Levesque S, Ben-David D, Cloutier I, Loo VG. 2016. Effect of detecting and isolating Clostridium difficile carriers at hospital admission on the incidence of C. difficile infections: a quasi-experimental controlled study. JAMA Intern Med 176:796–904. doi: 10.1001/jamainternmed.2016.0177. [DOI] [PubMed] [Google Scholar]
- 6.Zacharioudakis IM, Zervou FN, Pliakos EE, Ziakas PD, Mylonakis E. 2015. Colonization with toxinogenic C. difficile upon hospital admission, and risk of infection: a systematic review and meta-analysis. Am J Gastroenterol 110:381–390, quiz 391. doi: 10.1038/ajg.2015.22. [DOI] [PubMed] [Google Scholar]
- 7.Knudsen JD. 2015. The significance of asymptomatic colonisation of Clostridium difficile, ECCMID oral presentation S116. Eur Cong Clin Microbiol Infect Dis, 25 to 28 April 2015, Copenhagen, Denmark. [Google Scholar]
- 8.Marciniak C, Chen D, Stein AC, Semik PE. 2006. Prevalence of Clostridium difficile colonization at admission to rehabilitation. Arch Phys Med Rehabil 87:1086–1090. doi: 10.1016/j.apmr.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 9.Lin HJ, Hung YP, Liu HC, Lee JC, Lee CI, Wu YH, Tsai PJ, Ko WC. 2015. Risk factors for Clostridium difficile-associated diarrhea among hospitalized adults with fecal toxigenic C. difficile colonization. J Microbiol Immunol Infect 48:183–189. doi: 10.1016/j.jmii.2013.08.003. [DOI] [PubMed] [Google Scholar]
- 10.Hung YP, Tsai PJ, Hung KH, Liu HC, Lee CI, Lin HJ, Wu YH, Wu JJ, Ko WC. 2012. Impact of toxigenic Clostridium difficile colonization and infection among hospitalized adults at a district hospital in southern Taiwan. PLoS One 7:e42415. doi: 10.1371/journal.pone.0042415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Buffie CG, Bucci V, Stein RR, McKenney PT, Ling L, Gobourne A, No D, Liu H, Kinnebrew M, Viale A, Littmann E, van den Brink MR, Jenq RR, Taur Y, Sander C, Cross JR, Toussaint NC, Xavier JB, Pamer EG. 2015. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature 517:205–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weingarden AR, Dosa PI, DeWinter E, Steer CJ, Shaughnessy MK, Johnson JR, Khoruts A, Sadowsky MJ. 2016. Changes in colonic bile acid composition following fecal microbiota transplantation are sufficient to control Clostridium difficile germination and growth. PLoS One 11:e0147210. doi: 10.1371/journal.pone.0147210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross CL, Spinler JK, Savidge TC. 2016. Structural and functional changes within the gut microbiota and susceptibility to Clostridium difficile infection. Anaerobe 41:36–43. doi: 10.1016/j.anaerobe.2016.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Arvand M, Moser V, Schwehn C, Bettge-Weller G, Hensgens MP, Kuijper EJ. 2012. High prevalence of Clostridium difficile colonization among nursing home residents in Hesse, Germany. PLoS One 7:e30183. doi: 10.1371/journal.pone.0030183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Loo VG, Bourgault AM, Poirier L, Lamothe F, Michaud S, Turgeon N, Toye B, Beaudoin A, Frost EH, Gilca R, Brassard P, Dendukuri N, Beliveau C, Oughton M, Brukner I, Dascal A. 2011. Host and pathogen factors for Clostridium difficile infection and colonization. N Engl J Med 365:1693–1703. doi: 10.1056/NEJMoa1012413. [DOI] [PubMed] [Google Scholar]
- 16.Leekha S, Aronhalt KC, Sloan LM, Patel R, Orenstein R. 2013. Asymptomatic Clostridium difficile colonization in a tertiary care hospital: admission prevalence and risk factors. Am J Infect Control 41:390–393. doi: 10.1016/j.ajic.2012.09.023. [DOI] [PubMed] [Google Scholar]
- 17.Agerso Y, Torpdahl M, Zachariasen C, Seyfarth A, Hammerum AM, Nielsen EM. 2012. Tentative colistin epidemiological cut-off value for Salmonella spp. Foodborne Pathog Dis 9:367–369. doi: 10.1089/fpd.2011.1015. [DOI] [PubMed] [Google Scholar]
- 18.Rogers DS, Kundrapu S, Sunkesula VC, Donskey CJ. 2013. Comparison of perirectal versus rectal swabs for detection of asymptomatic carriers of toxigenic Clostridium difficile. J Clin Microbiol 51:3421–3422. doi: 10.1128/JCM.01418-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Enoch DA, Butler MJ, Pai S, Aliyu SH, Karas JA. 2011. Clostridium difficile in children: colonisation and disease. J Infect 63:105–113. doi: 10.1016/j.jinf.2011.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Crobach MJ, Planche T, Eckert C, Barbut F, Terveer EM, Dekkers OM, Wilcox MH, Kuijper EJ, European Society of Clinical Microbiology and Infectious Diseases. 2016. European Society of Clinical Microbiology and Infectious Diseases: update of the diagnostic guidance document for Clostridium difficile infection. Clin Microbiol Infect 20(Suppl 2):1–26. doi: 10.1016/j.cmi.2016.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Terveer EM, Sanders IMJG, Crobach MJT, Vos MC, Verduin C, Kuijper EJ. 2016. Clostridium difficile diagnostics in asymptomatic carriers admitted to the hospital. abstr 5224. 26th European Clinical Congress of Clinical Microbiology and Infectious Diseases, 9 to 12 April 2016, Amsterdam, the Netherlands. [Google Scholar]
- 22.Alasmari F, Seiler SM, Hink T, Burnham CA, Dubberke ER. 2014. Prevalence and risk factors for asymptomatic Clostridium difficile carriage. Clin Infect Dis 59:216–222. doi: 10.1093/cid/ciu258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dubberke ER, Reske KA, Seiler S, Hink T, Kwon JH, Burnham CA. 2015. Risk factors for acquisition and loss of Clostridium difficile colonization in hospitalized patients. Antimicrob Agents Chemother 59:4533–4543. doi: 10.1128/AAC.00642-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruminhent J, Wang ZX, Hu C, Wagner J, Sunday R, Bobik B, Hegarty S, Keith S, Alpdogan S, Carabasi M, Filicko-O'Hara J, Flomenberg N, Kasner M, Outschoorn UM, Weiss M, Flomenberg P. 2014. Clostridium difficile colonization and disease in patients undergoing hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 20:1329–1334. doi: 10.1016/j.bbmt.2014.04.026. [DOI] [PubMed] [Google Scholar]
- 25.Koo HL, Van JN, Zhao M, Ye X, Revell PA, Jiang ZD, Grimes CZ, Koo DC, Lasco T, Kozinetz CA, Garey KW, DuPont HL. 2014. Real-time polymerase chain reaction detection of asymptomatic Clostridium difficile colonization and rising C. difficile-associated disease rates. Infect Control Hosp Epidemiol 35:667–673. doi: 10.1086/676433. [DOI] [PubMed] [Google Scholar]
- 26.Janezic S, Marin M, Martin A, Rupnik M. 2015. A new type of toxin A-negative, toxin B-positive Clostridium difficile strain lacking a complete tcdA gene. J Clin Microbiol 53:692–695. doi: 10.1128/JCM.02211-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Elliott B, Squire MM, Thean S, Chang BJ, Brazier JS, Rupnik M, Riley TV. 2011. New types of toxin A-negative, toxin B-positive strains among clinical isolates of Clostridium difficile in Australia. J Med Microbiol 60:1108–1111. doi: 10.1099/jmm.0.031062-0. [DOI] [PubMed] [Google Scholar]
- 28.Jensen MB, Olsen KE, Nielsen XC, Hoegh AM, Dessau RB, Atlung T, Engberg J. 2015. Diagnosis of Clostridium difficile: real-time PCR detection of toxin genes in faecal samples is more sensitive compared to toxigenic culture. Eur J Clin Microbiol Infect Dis 34:727–736. doi: 10.1007/s10096-014-2284-7. [DOI] [PubMed] [Google Scholar]
- 29.Jazmati N, Wiegel P, Licanin B, Plum G. 2015. Evaluation of the Qiagen artus C. difficile QS-RGQ kit for detection of Clostridium difficile toxins A and B in clinical stool specimens. J Clin Microbiol 53:1942–1944. doi: 10.1128/JCM.00613-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hensgens MP, Dekkers OM, Demeulemeester A, Buiting AG, Bloembergen P, van Benthem BH, Le Cessie S, Kuijper EJ. 2014. Diarrhoea in general practice: when should a Clostridium difficile infection be considered? Results of a nested case-control study. Clin Microbiol Infect 20:O1067–O1074. [DOI] [PubMed] [Google Scholar]
- 31.Sall O, Johansson K, Noren T. 2015. Low colonization rates of Clostridium difficile among patients and healthcare workers at Orebro University Hospital in Sweden. APMIS 123:240–244. doi: 10.1111/apm.12353. [DOI] [PubMed] [Google Scholar]
- 32.Miyajima F, Roberts P, Swale A, Price V, Jones M, Horan M, Beeching N, Brazier J, Parry C, Pendleton N, Pirmohamed M. 2011. Characterisation and carriage ratio of Clostridium difficile strains isolated from a community-dwelling elderly population in the United Kingdom. PLoS One 6:e22804. doi: 10.1371/journal.pone.0022804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galdys AL, Nelson JS, Shutt KA, Schlackman JL, Pakstis DL, Pasculle AW, Marsh JW, Harrison LH, Curry SR. 2014. Prevalence and duration of asymptomatic Clostridium difficile carriage among healthy subjects in Pittsburgh, Pennsylvania. J Clin Microbiol 52:2406–2409. doi: 10.1128/JCM.00222-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samore MH, DeGirolami PC, Tlucko A, Lichtenberg DA, Melvin ZA, Karchmer AW. 1994. Clostridium difficile colonization and diarrhea at a tertiary care hospital. Clin Infect Dis 18:181–187. doi: 10.1093/clinids/18.2.181. [DOI] [PubMed] [Google Scholar]
- 35.Borriello SP, Honour P. 1981. Simplified procedure for the routine isolation of Clostridium difficile from faeces. J Clin Pathol 34:1124–1127. doi: 10.1136/jcp.34.10.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Paltansing S, van den Berg RJ, Guseinova RA, Visser CE, van der Vorm ER, Kuijper EJ. 2007. Characteristics and incidence of Clostridium difficile-associated disease in The Netherlands, 2005. Clin Microbiol Infect 13:1058–1064. doi: 10.1111/j.1469-0691.2007.01793.x. [DOI] [PubMed] [Google Scholar]
- 37.Fawley WN, Knetsch CW, MacCannell DR, Harmanus C, Du T, Mulvey MR, Paulick A, Anderson L, Kuijper EJ, Wilcox MH. 2015. Development and validation of an internationally-standardized, high-resolution capillary gel-based electrophoresis PCR-ribotyping protocol for Clostridium difficile. PLoS One 10:e0118150. doi: 10.1371/journal.pone.0118150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shin BM, Lee EJ, Moon JW, Lee SY. 2016. Evaluation of the Vidas glutamate dehydrogenase assay for the detection of Clostridium difficile. Anaerobe 40:68–72. doi: 10.1016/j.anaerobe.2016.06.001. [DOI] [PubMed] [Google Scholar]
- 39.Knetsch CW, Bakker D, de Boer RF, Sanders I, Hofs S, Kooistra-Smid AM, Corver J, Eastwood K, Wilcox MH, Kuijper EJ. 2011. Comparison of real-time PCR techniques to cytotoxigenic culture methods for diagnosing Clostridium difficile infection. J Clin Microbiol 49:227–231. doi: 10.1128/JCM.01743-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Davies KA, Berry CE, Morris KA, Smith R, Young S, Davis TE, Fuller DD, Buckner RJ, Wilcox MH. 2015. Comparison of the Vidas C. difficile GDH automated enzyme-linked fluorescence immunoassay (ELFA) with another commercial enzyme immunoassay (EIA) (Quik Chek-60), two selective media, and a PCR assay for gluD for detection of Clostridium difficle in fecal samples. J Clin Microbiol 53:1931–1934. doi: 10.1128/JCM.00649-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.