ABSTRACT
It has been hoped that the recent availability of WHO quantitative standards would improve interlaboratory agreement for viral load testing; however, insufficient data are available to evaluate whether this has been the case. Results from 554 laboratories participating in proficiency testing surveys for quantitative PCR assays of cytomegalovirus (CMV), Epstein-Barr virus (EBV), BK virus (BKV), adenovirus (ADV), and human herpesvirus 6 (HHV6) were evaluated to determine overall result variability and then were stratified by assay manufacturer. The impact of calibration to international units/ml (CMV and EBV) on variability was also determined. Viral loads showed a high degree of interlaboratory variability for all tested viruses, with interquartile ranges as high as 1.46 log10 copies/ml and the overall range for a given sample up to 5.66 log10 copies/ml. Some improvement in result variability was seen when international units were adopted. This was particularly the case for EBV viral load results. Variability in viral load results remains a challenge across all viruses tested here; introduction of international quantitative standards may help reduce variability and does so more or less markedly for certain viruses.
KEYWORDS: PCR, viral load, variability, quantitative, calibration, WHO
INTRODUCTION
The utility of viral load testing is well established in clinical practice and is generally applied either to patients with chronic viral disease (hepatitis B virus, hepatitis C virus, and human immunodeficiency virus [HBV, HCV, and HIV, respectively]) or to transplant-associated viruses (adenovirus [ADV], cytomegalovirus [CMV], Epstein-Barr virus [EBV], BK virus [BKV], human herpesvirus 6 [HHV6], and others). The dichotomy extends beyond patient population and virus of interest to typical assay characteristics. Tests for HBV, HCV, and HIV tend to be few in number, commercially produced, automated, and FDA cleared or approved, with long-available international quantitative standards (1). Testing for transplant-associated viruses is widely variable, comprised predominantly of laboratory-developed methods utilizing a wide variety of genetic targets, amplification chemistries, quantitative calibrators, and extraction methods. International standards for the latter group of viruses are either nonexistent or only recently available. Perhaps not surprisingly, result variability is greater for these methods than for hepatitis virus and HIV tests (2).
What is perhaps surprising is the degree of variability seen among laboratories testing identical samples. We and others have repeatedly demonstrated that the range of results produced for a given sample extends from 2 to 4 log10 copies/ml (3–5). That range of variability has likewise been shown to relate to several different aspects of nucleic acid amplification methodology (2). Such studies have spurred the recent development of World Health Organization (WHO) international quantitative standards, produced by the National Institute for Biological Standards and Control (Potters Bar, Hertfordshire, United Kingdom). As of this writing, WHO standards have been introduced for CMV, EBV, and BKV (6–8) (available in 2010, 2012, and 2016, respectively), with subsequent introductions for other viruses planned. These materials consist of lyophilized whole virus preparations, characterized by multicenter consensus studies, with designated concentration in international units/ml of reconstituted primary standard. Manufacturers are then expected to utilize this primary reference material to calibrate secondary standards, to be used in commercial assays or to be sold directly to users for direct use in viral load tests or for calibration of tertiary standards.
It has been hoped that the advent and broad introduction of WHO standards would markedly reduce the variability seen in corresponding viral load tests, despite the continued variation in sample preparation and testing methodologies. What remains unproven is whether such improvement has resulted. Earlier work by a College of American Pathologists (CAP) Microbiology Resource Committee (MRC) lead group was based on the evaluation of proficiency testing (PT) results from a CAP viral load survey (VLS). That group's analysis showed marked variability in results when common samples positive for EBV, CMV, or BKV were tested by large numbers of laboratories (2). Their work also showed that the quantitative calibrator in use accounted for some (though not all) of the quantitative variability seen, based on mathematical modeling. The current study seeks to expand on that previous work, including additional viruses (ADV and HHV6) and results from CAP surveys subsequent to the availability of WHO standards for CMV and EBV.
RESULTS
A total of 554 laboratories were included in the study. Among these, 504 (91%) reported results for CMV, 319 (58%) reported results for EBV, 339 (61%) reported results for BKV, 89 (16%) reported results for ADV, and 77 (14%) reported results for HHV6, including all challenges for each virus. In addition, 157 laboratories also reported CMV load in log10 international units (IU)/ml, while only 30 laboratories reported EBV load in log10 IU/ml.
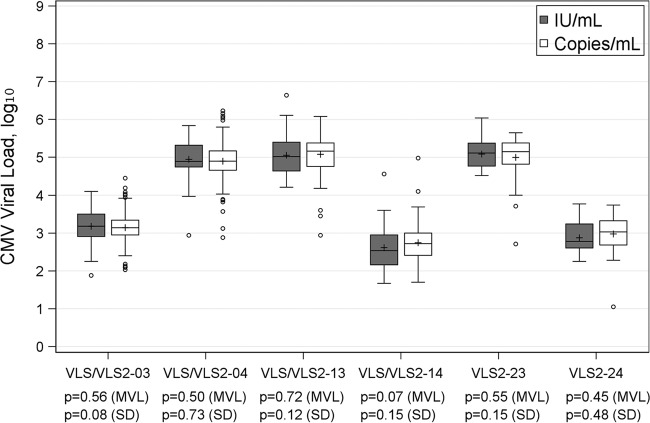
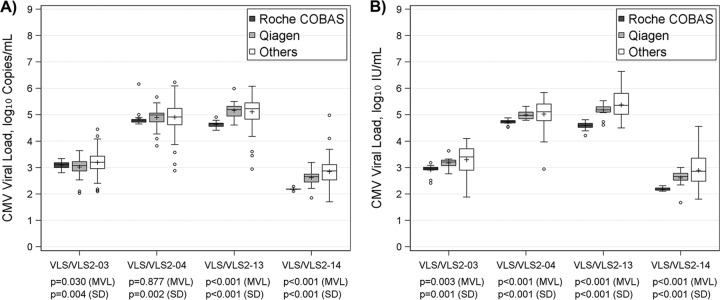
For CMV, 6 challenges had median reported viral loads ranging from 2.72 to 5.17 log10 copies/ml; the variability of results, characterized by interquartile range (IQR), ranged from 0.39 to 0.64 log10 copies/ml, with total ranges varying from 2.42 to 3.35 log10 copies/ml. When the viral load was reported in log10 IU/ml, 2 challenges (VLS/VLS2-14 and VLS2-24) had noticeably lower values than results from the same samples reported in copies/ml (Table 1). For the other 4 challenges, the values in IU/ml and those in copies/ml were close. Most challenges seemed to have a slightly larger variability in results in IU/ml than in copies/ml, suggested by a minor increase in IQR; however, the overall range was reduced when reported in IU/ml (1.52 to 2.90 log10 IU/ml), suggesting fewer outliers (Table 1). These trends are also revealed in Fig. 1. All challenges had a similar mean viral load between results reported in the two units, despite the above-noted differences in variability. As shown in Fig. 1, there were more extreme values reported by participating laboratories in copies/ml, although no significant heterogeneous variability was established by Levene's test. In comparison to laboratories using other systems, those using the Roche COBAS system tended to report a lower CMV value, followed by laboratories using Qiagen. Results from the Roche COBAS system had the least variability, and results using other systems had the greatest variability (Table 2). This is further demonstrated in Fig. 2, with the Roche COBAS system often reporting a significantly lower CMV load with less variability, irrespective of whether results were reported in copies/ml or IU/ml. Other systems had significantly more variable results, and the variability of Qiagen results fell in between.
TABLE 1.
Descriptive statistics of viral load for each virusa
| Virus | Challenge | n | log10 copies/ml |
n | log10 IU/mlb |
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Median (IQR) | Range | Mean (SD) | Median (IQR) | Range | ||||
| CMV | VLS/VLS2-03 | 174 | 3.14 (0.39) | 3.14 (0.39) | 2.42 | 55 | 3.18 (0.46) | 3.18 (0.59) | 2.22 |
| VLS/VLS2-04 | 182 | 4.90 (0.48) | 4.90 (0.51) | 3.35 | 56 | 4.95 (0.46) | 4.90 (0.58) | 2.90 | |
| VLS/VLS2-13 | 166 | 5.08 (0.46) | 5.17 (0.62) | 3.14 | 75 | 5.06 (0.50) | 5.02 (0.76) | 2.43 | |
| VLS/VLS2-14 | 134 | 2.75 (0.46) | 2.72 (0.59) | 3.28 | 66 | 2.61 (0.52) | 2.54 (0.79) | 2.89 | |
| VLS2-23 | 32 | 5.00 (0.64) | 5.15 (0.56) | 2.94 | 24 | 5.09 (0.38) | 5.12 (0.61) | 1.52 | |
| VLS2-24 | 28 | 2.98 (0.55) | 3.03 (0.64) | 2.69 | 24 | 2.87 (0.39) | 2.78 (0.64) | 1.52 | |
| EBV | VLS/VLS2-05 | 134 | 4.60 (0.58) | 4.57 (0.75) | 3.45 | 10 | 4.47 (0.51) | 4.52 (0.42) | 1.97 |
| VLS/VLS2-06 | 96 | 2.99 (0.56) | 3.00 (0.57) | 3.11 | 8 | 2.58 (0.39) | 2.55 (0.69) | 1.00 | |
| VLS/VLS2-15 | 103 | 2.89 (0.52) | 2.92 (0.65) | 2.70 | 15 | 2.53 (0.34) | 2.60 (0.63) | 1.04 | |
| VLS/VLS2-16 | 136 | 3.59 (0.56) | 3.65 (0.67) | 2.78 | 15 | 3.44 (0.30) | 3.45 (0.62) | 0.95 | |
| VLS2-25 | 23 | 2.63 (0.45) | 2.68 (0.40) | 2.33 | 5 | 2.04 (0.24) | 2.00 (0.08) | 0.63 | |
| BKV | VLS/VLS2-01 | 146 | 5.74 (0.54) | 5.72 (0.56) | 3.71 | ||||
| VLS/VLS2-11 | 149 | 4.50 (0.46) | 4.53 (0.57) | 2.69 | |||||
| VLS/VLS2-12 | 123 | 3.02 (0.45) | 3.00 (0.60) | 3.15 | |||||
| VLS2-21 | 41 | 4.34 (0.69) | 4.23 (1.05) | 2.99 | |||||
| VLS2-22 | 42 | 6.33 (0.67) | 6.25 (0.92) | 2.74 | |||||
| ADV | VLS2-07 | 26 | 4.57 (0.62) | 4.50 (0.79) | 2.91 | ||||
| VLS2-08 | 26 | 5.31 (1.07) | 5.04 (1.46) | 3.60 | |||||
| VLS2-17 | 31 | 5.33 (0.65) | 5.32 (0.91) | 2.91 | |||||
| VLS2-18 | 31 | 5.85 (1.03) | 5.94 (0.91) | 5.66 | |||||
| VLS2-27 | 32 | 4.78 (0.72) | 4.80 (0.59) | 4.50 | |||||
| VLS2-28 | 32 | 6.66 (1.09) | 6.62 (1.39) | 4.34 | |||||
| HHV6 | VLS2-09 | 25 | 5.15 (0.85) | 5.48 (0.98) | 3.51 | ||||
| VLS2-10 | 23 | 3.90 (0.66) | 4.00 (0.72) | 2.83 | |||||
| VLS2-20 | 27 | 4.49 (0.65) | 4.56 (0.78) | 2.73 | |||||
| VLS2-29 | 25 | 5.40 (0.94) | 5.53 (0.63) | 4.29 | |||||
| VLS2-30 | 25 | 3.91 (0.79) | 3.91 (0.63) | 4.08 | |||||
n, number of labs; SD, standard deviation; IQR, interquartile range.
In the version of this article published on 25 January 2017, this heading incorrectly read “log10 copies/ml.” This was changed in the version published on 22 November 2017.
FIG 1.
Box plot of CMV viral load (IU/ml versus copies/ml); Levene's test was used to test for equality of variances, and one-way ANOVA was applied to compare mean viral loads (MVL). SD, standard deviation.
TABLE 2.
CMV viral load by assaya
| Challenge | Assay | n | log10 copies/ml |
n | log10 IU/mlb |
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Median (IQR) | Range | Mean (SD) | Median (IQR) | Range | ||||
| VLS/VLS2-03 | Roche COBAS | 18 | 3.09 (0.14) | 3.11 (0.18) | 0.54 | 13 | 2.91 (0.22) | 2.97 (0.10) | 0.77 |
| Qiagen | 39 | 3.02 (0.35) | 3.07 (0.35) | 1.61 | 11 | 3.17 (0.23) | 3.21 (0.21) | 0.87 | |
| Others | 117 | 3.19 (0.42) | 3.20 (0.47) | 2.36 | 31 | 3.29 (0.54) | 3.40 (0.81) | 2.22 | |
| VLS/VLS2-04 | Roche COBAS | 18 | 4.86 (0.34) | 4.80 (0.10) | 1.51 | 13 | 4.72 (0.10) | 4.74 (0.09) | 0.35 |
| Qiagen | 40 | 4.89 (0.34) | 4.99 (0.33) | 1.85 | 11 | 4.99 (0.16) | 4.97 (0.25) | 0.53 | |
| Others | 124 | 4.91 (0.53) | 4.92 (0.62) | 3.35 | 32 | 5.02 (0.58) | 5.11 (0.63) | 2.90 | |
| VLS/VLS2-13 | Roche COBAS | 15 | 4.64 (0.12) | 4.66 (0.12) | 0.50 | 27 | 4.59 (0.12) | 4.60 (0.15) | 0.60 |
| Qiagen | 38 | 5.16 (0.26) | 5.19 (0.37) | 1.38 | 14 | 5.18 (0.26) | 5.19 (0.20) | 0.92 | |
| Others | 113 | 5.11 (0.51) | 5.23 (0.62) | 3.14 | 34 | 5.37 (0.49) | 5.36 (0.79) | 2.14 | |
| VLS/VLS2-14 | Roche COBAS | 8 | 2.18 (0.05) | 2.18 (0.01) | 0.18 | 20 | 2.18 (0.06) | 2.14 (0.08) | 0.21 |
| Qiagen | 35 | 2.62 (0.26) | 2.65 (0.30) | 1.34 | 14 | 2.61 (0.33) | 2.66 (0.26) | 1.33 | |
| Others | 91 | 2.84 (0.50) | 2.87 (0.58) | 3.28 | 32 | 2.89 (0.56) | 2.86 (0.88) | 2.76 | |
n, number of labs; SD, standard deviation; IQR, interquartile range.
In the version of this article published on 25 January 2017, this heading incorrectly read “log10 copies/ml.” This was changed in the version published on 22 November 2017.
FIG 2.
Box plot of CMV viral load by assay in IU/ml (A) and copies/ml (B); Levene's test was used to test for equality of variances, and one-way ANOVA was applied to compare mean viral loads (MVL).
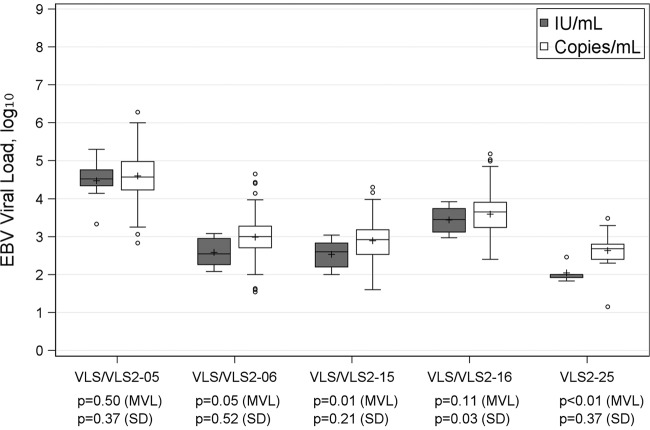
In tests for EBV, switching to the use of IU/ml led to lower EBV values (Table 1). The median EBV load ranged from 2.00 to 4.52 log10 IU/ml, while in comparison, the median in log10 copies/ml was from 2.68 to 4.57. The use of IU/ml in place of copies/ml also seemed to reduce result variability, with results in IU/ml usually having a smaller IQR and a markedly lower range. Figure 3 confirms this observation. Three challenges (VLS/VLS2-06, VLS/VLS2-15, and VLS2-25) had significantly lower reported EBV loads when results were reported in IU/ml. As seen with CMV, all challenges of EBV had more extreme reported values reported in copies/ml, suggesting a trend of lower variability when reported in IU/ml, although only results for VLS/VLS2-16 in IU/ml showed a significantly lower variability (P = 0.03). Unlike the case for CMV, no significant differences in either mean viral load or variability in copies/ml were found for EBV viral load when results were stratified by assay manufacturer (see Table S1 in the supplemental material). Because too few laboratories reported EBV load in IU/ml, no variability comparison across manufacturers was made.
FIG 3.
Box plot of EBV viral load (IU/ml versus copies/ml); Levene's test was used to test for equality of variances, and one-way ANOVA was applied to compare mean viral loads (MVL).
At the time of data collection for this publication, no international quantitative standards had yet been produced for BKV, ADV, or HHV6. Therefore, results in log10 IU/ml were available only for CMV and EBV (Table 1). The IQRs for BKV, ADV, and HHV6 were from 0.56 to 1.05, 0.59 to 1.46, and 0.63 to 0.98 log10 copies/ml, respectively. The range was dramatically higher in some cases, up to 3.71, 5.66, and 4.29 log10 copies/ml, respectively. Mean values and variabilities of BKV challenges were not significantly different among manufacturers (Table S2). For ADV and HHV6, due to the small number of laboratories stratified by manufacturer, no further statistical comparison was made (Tables S3 and S4).
DISCUSSION
Previous work has shown a wide variety of factors that contribute to the variability of viral load test results (2). The increasing availability of commercially produced assays (particularly for CMV) and the development of international quantitative standards (for CMV and EBV) have represented significant steps forward in addressing this challenge. Here we present some of the first data incorporating these advances, showing some improvement in result variability, particularly for EBV load testing, when assays are reported in IU/ml rather than in copies/ml.
Yet, the improvement is not as marked as one might have hoped. In the case of CMV, while the use of IU did reduce outlier spread, as demonstrated by reduced overall range, variability as assessed by IQR was unchanged or somewhat increased. More remarkable, in the case of CMV, was the improved agreement when results from those using a common, automated system are viewed. This is not completely unexpected, as the use of common reagents in an automated system has been shown to be associated with reduced variability of other viral load assays (2). Variability is likely the result of numerous factors, of which differing calibrators is only one. Use of an automated system encompasses common primers, probes, cycling, amplification conditions, chemistries, and extraction methods. At the same time, the use of robotics potentially reduces the variability attributable to manual pipetting of samples. So, while the production and utilization of international standards are a major step forward toward improved interlaboratory agreement, many other potential opportunities for further improvements remain. Increasing the availability of commercially available assays that have been cleared for diagnostic use is a primary step forward in the field, as has been demonstrated with HIV and the hepatitis viruses, where such progress, together with the availability of international standards, has been associated with a marked reduction in interlaboratory result variability. At the time of data collection for the present study, only a single system was available for clinical diagnostic testing of CMV viral load. The Qiagen viral load test has subsequently become commercially available. It is important to note that the Qiagen testing data contained herein represents an analyte-specific-reagent (ASR) version of that test. This may have resulted in more variability, due to differences in nucleic acid extraction, calibration, and other aspects of testing, than what might be seen with the now-available in vitro diagnostic (IVD) version of the test.
It can also not be assumed that in all cases conversion to IU alone is sufficient to remove calibration as a source of dissimilar results. Laboratories typically use secondary or tertiary standards for clinical testing rather than primary material from the WHO. It has been previously demonstrated that secondary materials do not always contain equal copy numbers of target for a given nominal value (9). Quantitative discrepancies among secondary standards may significantly undermine any attempt to foster agreement by adaptation of IU, as laboratories may be normalizing results to differing reference standards. Beyond this is the question of commutability, which should be demonstrated for each assay/standard system (10–13). That is, standards should behave in a like manner to patient samples, and that quantitative relationship should be the same among different assays. Noncommutable standards may actually worsen the divergence of results between assays rather than improving agreement (14). Recent work has indicated that not all assays show the same degree of commutability with WHO standards (15, 16). This may not reflect an intrinsic problem with the WHO material but rather greater or lesser compatibility with specific constellations of sample preparation, target sequences, primers, probes, amplification conditions, and other variables that might alter the consistent relationship between how clinical samples and standards behave in a given assay system.
Moreover, it is clear that in some cases, the introduction of IU has a more dramatic impact than it does in others. In EBV, the reduction in result variability was more marked than it was for CMV, as were the changes in mean and median results. This reinforces the notion that common international quantitative standards can be of great benefit in improving consensus among testing laboratories (1, 17, 18). That CMV did not have as dramatic an impact is more an illustration of the multiple factors responsible for result variability than a reason not to press on with the standardization process. In fact, the variability evident among viral load assays without a currently available international standard speaks to the continuing need for development of such material. With result ranges of up to >5 log units, it is easy to see why efforts must be made to improve viral load consistency. As previously mentioned, such improvement will likewise improve result portability, determination of thresholds for treatment or treatment endpoints, and comparative interpretation of the literature, which is currently quite difficult, given the lack of quantitative standardization among centers engaged in studying clinical correlates of viral load.
We hope that the improvement that we have seen here since our previous analysis of proficiency testing result agreement and variability is only the first in a multistep process of bringing consistency and improved clinical value to this field. We hope to monitor this progress in future publications, as more international standards are produced and as we learn to better characterize and utilize those standards in order to derive their maximum value.
MATERIALS AND METHODS
This study included results from 554 laboratories participating in the CAP VLS and VLS2 PT surveys during the 2013 calendar year. This included two VLS mailings with 2 challenges per mailing each for CMV, EBV, and BKV and three VLS2 mailings with 2 challenges per mailing each for CMV, EBV, BKV, ADV, and HHV6. The CMV, EBV, and BKV challenges were the same for both the VLS and VLS2 mailings. Participating laboratories performed testing only for those viruses that they routinely assayed for clinical purposes, using only their routine testing practices. Each laboratory provided information on manufacturers of reagents and quantitative calibrators and on the type of calibration material (whole organism quantified by various means or synthetic oligonucleotide) used. Each PT sample was comprised of purified, intact virus particles that were chemically modified to render them noninfectious and refrigerator stable (Zeptometrix, Buffalo, NY). These purified, inactivated virus particles were diluted in defribrinated K3 EDTA plasma treated with 0.9% sodium azide and stored at 2 to 8°C. Viral concentrations were specified for each challenge prior to preparation, such that the range of sample concentrations sent over the course of the year represented typically clinically useful viral loads that most laboratories might expect to encounter. Typically, this corresponded to approximately 3 to 6 log10 copies/ml. The PT samples were quantitated by real-time PCR and shipped overnight to participants on cool packs. Aggregate data were collected by the CAP and extracted for this analysis without attached laboratory identifiers. Result variability among all responding laboratories was determined for each challenge sample individually and across each virus. Variability was assessed overall and then stratified by manufacturer of both assay and calibrator. Where available (CMV and EBV), results and variability were further stratified by those calibrating to IU/ml and those continuing to report in copies/ml. Improvement in variability based on introduction of international standards was based on comparisons of measures of variability (the range, interquartile range, and standard deviation) when the present study results in log10 copies/ml were compared with those results in log10 IU/ml. Finally, in the case of CMV, results were stratified to those reporting by the single FDA-cleared assay at the time of data collection (Roche COBAS) and all others.
Statistical methods.
All analyses were performed using SAS 9.3 (SAS Institute, Cary, NC). Levene's test (19) and one-way analysis of variance (ANOVA) were, respectively, applied to compare variabilities and means across different groups. For comparisons in which Levene's test found significant evidence of nonequal variance across groups, Welch's ANOVA (20) was used compare means across groups. Two-sided P values of <0.05 were considered statistically significant. For consistency, we adopted previously used data inclusion criteria (2). Namely, only positive results from laboratories which answered all questions for any given virus were included in the analysis. In addition, positive results reported below or above detection limits were assigned as the limits.
Supplementary Material
ACKNOWLEDGMENTS
We thank Pamela Provax from the College of American Pathologists for her assistance with data abstraction and Stanley Pounds from the St. Jude Children's Research Hospital Department of Biostatistics for his assistance with statistical analysis and explanation.
Randall Hayden has served on a Roche Molecular advisory board. Angela Caliendo has served on advisory boards for Roche Molecular and Cepheid. Stephen A. Young has served on an advisory board for Roche Molecular. David Hillyard has served on an advisory board for Roche Diagnostics. Gary W. Procop has served on an advisory board for Roche Diagnostics and receives research funding from Qiagen and Hologic.
This work was supported in part by ALSAC.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.02044-16.
Note Added after Publication
In the version of this article published on 25 January 2017, the heading spanning the last three columns of Table 1 and the heading spanning the last three columns of Table 2 incorrectly read “log10 copies/ml.” These headings were changed to “log10 IU/ml” in the version published on 22 November 2017.
REFERENCES
- 1.Madej RM, Davis J, Holden MJ, Kwang S, Labourier E, Schneider GJ. 2010. International standards and reference materials for quantitative molecular infectious disease testing. J Mol Diagn 12:133–143. doi: 10.2353/jmoldx.2010.090067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hayden RT, Yan X, Wick MT, Rodriguez AB, Xiong X, Ginocchio CC, Mitchell MJ, Caliendo AM. 2012. Factors contributing to variability of quantitative viral PCR results in proficiency testing samples: a multivariate analysis. J Clin Microbiol 50:337–345. doi: 10.1128/JCM.01287-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hayden RT, Hokanson KM, Pounds SB, Bankowski MJ, Belzer SW, Carr J, Diorio D, Forman MS, Joshi Y, Hillyard D, Hodinka RL, Nikiforova MN, Romain CA, Stevenson J, Valsamakis A, Balfour HH Jr, U. S. EBV Working Group. 2008. Multicenter comparison of different real-time PCR assays for quantitative detection of Epstein-Barr virus. J Clin Microbiol 46:157–163. doi: 10.1128/JCM.01252-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoffman NG, Cook L, Atienza EE, Limaye AP, Jerome KR. 2008. Marked variability of BK virus load measurement using quantitative real-time PCR among commonly used assays. J Clin Microbiol 46:2671–2680. doi: 10.1128/JCM.00258-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pang XL, Fox JD, Fenton JM, Miller GG, Caliendo AM, Preiksaitis JK. 2009. Interlaboratory comparison of cytomegalovirus viral load assays. Am J Transplant 9:258–268. doi: 10.1111/j.1600-6143.2008.02513.x. [DOI] [PubMed] [Google Scholar]
- 6.WHO. 2013. 1st WHO international standard for Epstein-Barr virus for nucleic acid amplification techniques. NIBSC code 09/260. National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom. [Google Scholar]
- 7.WHO. 2016. 1st WHO international standard for BK virus DNA. NIBSC code 14/212. National Institute for Biological Standards and Control, Potters Bar, Hertfordshire, United Kingdom. [Google Scholar]
- 8.Fryer JF, Heath AB, Anderson R, Minor PD, Collaborative Study Group. 2010, posting date Collaborative study to evaluate the proposed 1st WHO international standard for human cytomegalovirus (HCMV) for nucleic acid amplification (NAT)-based assays. WHO Expert Committee on Biological Standardization, Geneva, Switzerland: http://apps.who.int/iris/handle/10665/70521. [Google Scholar]
- 9.Hayden RT, Gu Z, Sam SS, Sun Y, Tang L, Pounds S, Caliendo AM. 2015. Comparative evaluation of three commercial quantitative cytomegalovirus standards using digital and real-time PCR. J Clin Microbiol 53:1500–1505. doi: 10.1128/JCM.03375-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Caliendo AM, Shahbazian MD, Schaper C, Ingersoll J, Abdul-Ali D, Boonyaratanakornkit J, Pang XL, Fox J, Preiksaitis J, Schonbrunner ER. 2009. A commutable cytomegalovirus calibrator is required to improve the agreement of viral load values between laboratories. Clin Chem 55:1701–1710. doi: 10.1373/clinchem.2009.124743. [DOI] [PubMed] [Google Scholar]
- 11.Cattozzo G, Franzini C. 2013. Commutability: a peculiar property of calibration and control materials. Definition and evaluation. Clin Chem Lab Med 51:e167-8. doi: 10.1515/cclm-2013-0065. [DOI] [PubMed] [Google Scholar]
- 12.Gantzer ML, Miller WG. 2012. Harmonisation of measurement procedures: how do we get it done? Clin Biochem Rev 33:95–100. [PMC free article] [PubMed] [Google Scholar]
- 13.Vesper HW, Miller WG, Myers GL. 2007. Reference materials and commutability. Clin Biochem Rev 28:139–147. [PMC free article] [PubMed] [Google Scholar]
- 14.Hayden RT, Shahbazian MD, Valsamakis A, Boonyaratanakornkit J, Cook L, Pang XL, Preiksaitis JK, Schonbrunner ER, Caliendo AM. 2013. Multicenter evaluation of a commercial cytomegalovirus quantitative standard: effects of commutability on interlaboratory concordance. J Clin Microbiol 51:3811–3817. doi: 10.1128/JCM.02036-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Abeynayake J, Johnson R, Libiran P, Sahoo MK, Cao H, Bowen R, Chan KC, Le QT, Pinsky BA. 2014. Commutability of the Epstein-Barr virus WHO international standard across two quantitative PCR methods. J Clin Microbiol 52:3802–3804. doi: 10.1128/JCM.01676-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hayden RT, Preiksaitis J, Tong Y, Pang X, Sun Y, Tang L, Cook L, Pounds S, Fryer J, Caliendo AM. 2015. Commutability of the first World Health Organization international standard for human cytomegalovirus. J Clin Microbiol 53:3325–3333. doi: 10.1128/JCM.01495-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Holden MJ, Madej RM, Minor P, Kalman LV. 2011. Molecular diagnostics: harmonization through reference materials, documentary standards and proficiency testing. Expert Rev Mol Diagn 11:741–755. doi: 10.1586/erm.11.50. [DOI] [PubMed] [Google Scholar]
- 18.Kraft CS, Armstrong WS, Caliendo AM. 2012. Interpreting quantitative cytomegalovirus DNA testing: understanding the laboratory perspective. Clin Infect Dis 54:1793–1797. doi: 10.1093/cid/cis212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Levene H. 1960. Robust tests for equality of variances, p 278–292. In Olkin I. (ed), Contributions to probability and statistics: essays in honor of Harold Hotelling. Stanford University Press, Palo Alto, CA. [Google Scholar]
- 20.Welch BL. 1951. On the comparison of several mean values—an alternative approach. Biometrika 38:330–336. doi: 10.1093/biomet/38.3-4.330. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.