ABSTRACT
Hepatitis D virus (HDV) is responsible for fulminant hepatitis and liver failure and accelerates evolution toward cirrhosis and hepatocellular carcinoma in hepatitis B virus (HBV)-infected patients. To date, treatment relies upon long-term administration of pegylated alpha-interferon with a sustained virological response in 30% of the patients. Very recently, new, promising anti-HDV therapies have been developed and are already being used in clinical trials. HDV RNA viral load (HDVL) monitoring must be an integral part of the management of the infected patients. However, HDV genus is characterized by a high genetic variability into eight genotypes (HDV-1 to -8), and most available in-house or commercial assays are useful for only a limited subset of genotypes. Results of a comparison of the performance of a new kit for HDVL quantification with the consensus in-house assay of the French National Reference Laboratory for HDV developed in 2005 are reported here. A total of 611 clinical samples of all HDV genotypes with various HDVL values, including several consecutive samples over several years from 36 patients, were studied. A specificity, sensitivity, and reproducibility evaluation was conducted using HDV-positive clinical samples, hepatitis A, B, C and E (HAV, HBV, HCV, and HEV, respectively) and HIV mono-infected samples, and the WHO HDV RNA international standard. Overall results were strictly comparable between the two assays (median difference, 0.07 log IU/ml), with high diagnosis precision and capacity. In summary, this new kit showed high performance in detection/quantification of HDVL, regardless of the genotype of the infecting strain used, and seems to be a suitable tool for patient management.
KEYWORDS: HDV, RNA, genotypes, viral load, kit, quantification, real-time RT-PCR
INTRODUCTION
The World Health Organization (WHO) estimates that 240 million people are chronically infected with hepatitis B virus (HBV), of which approximately 20 million are also infected with hepatitis D virus (HDV). HDV is responsible for much more severe liver disease, with more frequent occurrence of fulminant hepatitis and more rapid evolution to cirrhosis and hepatocellular carcinoma (HCC) in HBV/HDV-infected patients (1–5). The diagnosis of HDV infection relies upon the detection of total anti-HDV antibodies. IgM antibodies are positive in acute infection and persist in chronic infection (6, 7). However, they are lacking in some African patients (8). The quantification of HDV RNA viral load (HDVL) is the only reliable marker of HDV replication and must be an integral part in the management of HDV patients worldwide.
During the last decade, several commercial and in-house assays have been developed. However, very recently we showed that most of them dramatically underestimated or failed to detect/quantify positive HDV RNA samples, especially from patients infected with strains of African origin (HDV-1 and HDV-5 to -8) (9–11), highlighting the lack of efficient tools to routinely monitor HDVL for therapeutic management of infected patients.
In addition, several new specific anti-HDV drugs, such as entry and farnesylation inhibitors and nucleic acid polymers (12, 13), have been developed and are currently in phase 2 clinical trials, with or without the classical alpha-interferon (IFN-α) therapy (14–16). Therefore, providing standardized kits for quantification of HDVL is urgently needed in order to precisely evaluate the efficiency of these new HDV therapies in large multicenter trials. This is a crucial step toward the definition of consensus guidelines in the management of HDV-infected patients.
The aim of the present work was to evaluate the performance of the Eurobioplex HDV kit (Eurobio Company), a new commercially available assay based on real-time one-step reverse transcription-PCR (RT-PCR) technology.
RESULTS
A total of 611 clinical samples together with dilutions of the WHO HDV international standard (IS) were considered for the kit evaluation. This sampling included all of the eight known HDV genotypes with various HDVLs ranging from values for positive unquantifiable specimens (<3 log) to 9.6 log IU/ml according to the French National Reference Laboratory (FNRL) assay.
Global characteristics of the Eurobioplex HDV kit.
A 100% efficacy (mean slope, −3.25) of the reverse transcription-quantitative PCR (RT-qPCR) was systematically observed. The values obtained for the five different points of the standard of quantification over 20 different runs were highly reproducible, with a coefficient of variation (CV) of <4.4%. Similarly, good stability and reproducibility were observed with the internal control (IC), with a median threshold cycle (CT) of 29 ± 1.2 with either high or low HDVL values. No problem of contamination was witnessed during the entire process of evaluation. Nevertheless, we observed a slight background in 6-carboxyfluorescein (FAM) signal while evaluating HDV-negative samples. This background allowed the determination of the threshold of this assay at about 1,000 relative fluorescence units (RFU) for FAM. This threshold was 300 RFU for hexachlorofluorescein (HEX).
Serum versus plasma specimens.
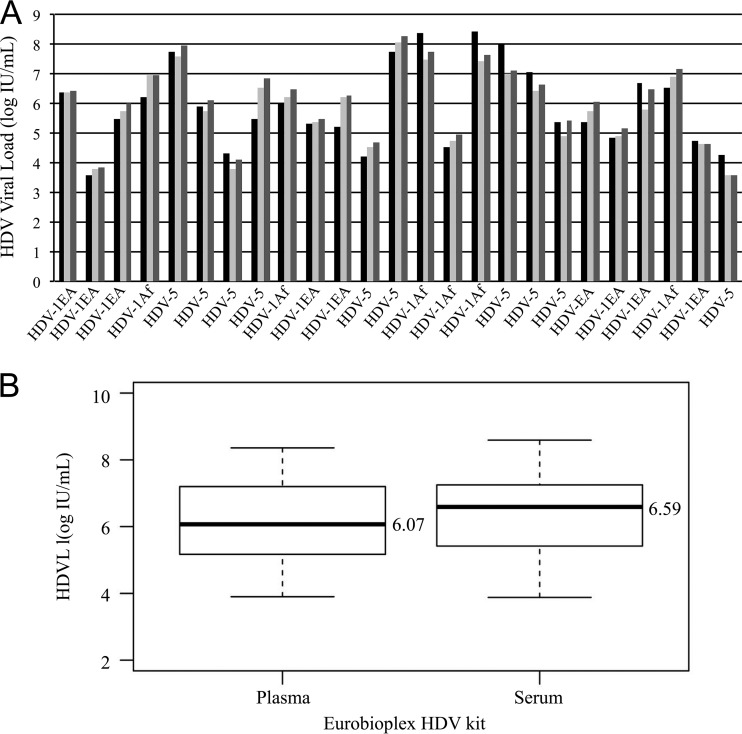
The first interesting point was that the Eurobioplex HDV kit found comparable results using either serum or plasma specimens, as assessed by the results obtained for paired serum/plasma samples from 25 patients infected with HDV-1 and HDV-5 strains (Fig. 1). The median HDVL values were, respectively, 6.07 and 6.59 log IU/ml for plasma and serum for the Eurobioplex kit, for an expected median value of 5.49 log IU/ml, according to HDV FNRL assay results on plasma samples (Fig. 1A and B).
FIG 1.
Comparison of HDVLs in serum versus plasma. (A) Histograms represent the HDVLs obtained in 25 pairs of serum and plasma samples of different HDV genotypes and HDVL values. Values obtained with the FNRL assay on plasma samples are shown in black; values obtained in plasma and serum samples with the Eurobioplex HDV kit are shown in light gray and dark gray, respectively. (B) Distribution of HDVL median values (in log IU/ml) obtained with the Eurobioplex HDV kit for the 25 pairs of specimens of plasma and serum samples.
Specificity.
The Eurobioplex HDV kit showed an excellent specificity (100%). Indeed, 105 negative anti-HDV antibody (Ab) samples positive for either for HAV IgM, HBV DNA, HCV RNA, HEV RNA, or HIV RNA were all found negative for HDV RNA.
Repeatability and reproducibility.
Repeatability was investigated using three samples, one each with a high, medium, and low HDVL value, together with one negative control as described in the Materials and Methods section. All replicates (3) of the negative control were found negative. For the three positive samples, the CV between the replicates was <3.4%, whatever the HDVL (Table 1). In addition, when dilutions of the WHO HDV IS were used, a CV of 2.8% was obtained (Table 1).
TABLE 1.
Repeatability using three clinical samples with high, intermediate, and low viral loads and two dilutions of the WHO HDV standard
| Parametera | Value for the parameterb |
||||
|---|---|---|---|---|---|
| High load | Intermediate load | Low load | WHO HDV IS at: |
||
| 1:30 | 1:500 | ||||
| No. of replicates | 3 | 3 | 30 | 100 | 80 |
| Q1 | NA | NA | 4.19 | 4.52 | 2.90 |
| Median | 7.55 | 6.36 | 4.26 | 4.57 | 2.98 |
| Q3 | NA | NA | 4.36 | 4.63 | 3.03 |
| CV (%) | 1.3 | 3.4 | 3.4 | 2.8 | 2.8 |
Q1, 25th percentile; Q3, 75th percentile.
NA, not applicable.
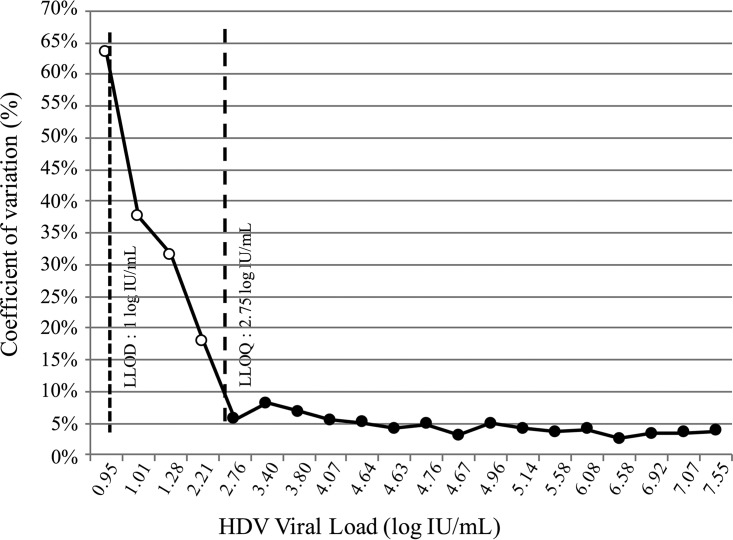
To evaluate reproducibility, four different technicians performed the experiments. The four negative samples were systematically found to be negative. When the results obtained with the set of positive samples (<3 to 7.55 log IU/ml) were analyzed, the CV values between the measures ranged from 2.5% to 8.1% for samples with an HDVL of >2.75 log IU/ml. This value defined the lower limit of quantification (LLOQ) of the kit (Fig. 2).
FIG 2.
Reproducibility of the Eurobioplex HDV kit. Mean HDVLs and coefficients of variation (CV) were calculated from HDVL values obtained with the kit by the four different operators. Dashed lines represent the LLOQ (2.75 log IU/ml) and LLOD (1 log IU/ml) of the assay, defined according to all our experiments. Filled circles, low CV values obtained for HDVL values ranging from 2.75 to 8.1%; open circles, high CV values obtained for very low HDVL values (below the LLOQ).
Linearity and sensitivity.
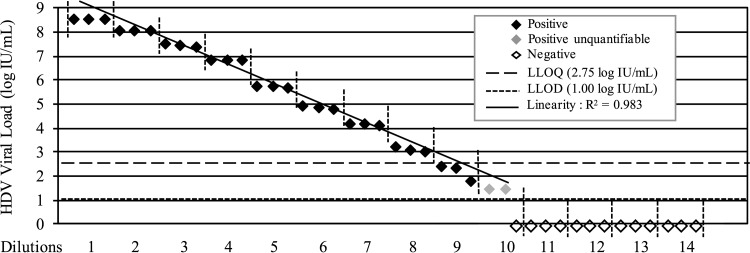
Serial dilutions (1:5) of a clinical sample with an HDVL of 8.5 log IU/ml were quantified each in triplicate. Results showed a linear quantification range (R2 = 0.983) between 2.75 log IU/ml and 8.5 log IU/ml and confirmed the LLOQ value of the kit at 2.75 log IU/ml (Fig. 3). Interestingly, the four samples below this LLOQ were found to be positive by the four technicians in the four different experiments with, however, high CV values between measures (15 to 65%). These results allowed us to define the lower limit of detection (LLOD) of the kit at around 1 log IU/ml (10 IU/ml) (Fig. 2 and 3). In addition, considering the samples overall (n = 611), an excellent detection sensitivity of 98.5% was found for the kit compared to results with the FNRL assay (Table 2).
FIG 3.
Linearity and sensitivity of the Eurobioplex HDV kit. The HDVL value for each of 14 serial dilutions (1:5) was quantified in triplicate, as indicated by the vertical dashed lines.
TABLE 2.
Sensitivity and specificity of the assays
Sample with a very low HDV viral load (<3 log IU/ml).
Influence of biochemical parameters.
The possible influence on the quantification results of the kit of triglycerides and bilirubin, two typically altered biochemical parameters in viral liver disease, was evaluated. No difference was observed over the 80 samples in which the same amount of HDV RNA (5 log IU/ml) was added, with a CV of around 3.1% (Fig. 4).
FIG 4.
Influence of triglyceride and bilirubin concentrations on HDVL quantification. Filled diamonds represent increasing concentrations of triglyceride (A) and bilirubin (B) in the samples, and open diamonds represent the HDV viral load values measured in these samples. Coefficients of variation (CVs) of the measures calculated from all HDVL values are indicated. Values shown on the x axis are the total number of samples considered.
Detection/quantification of the HDV genotypes.
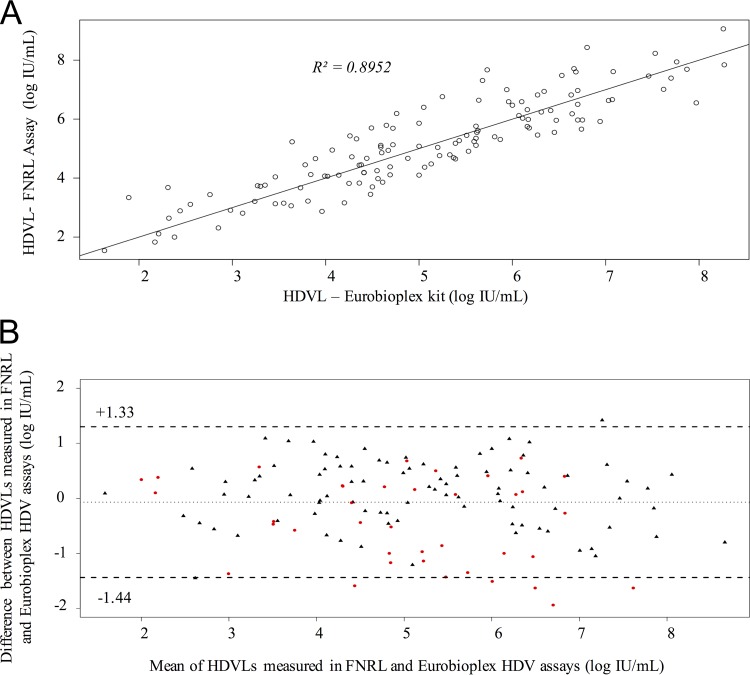
A total of 151 clinical samples of genotype HDV-1 to -8 were extracted and quantified with both assays. The 12 negative controls were found to be negative. For the 128/139 positive samples, results were comparable between the two assays, whatever the genotype or the viral load value (median difference, −0.11 log IU/ml; R2 = 0.8952) (Fig. 5A and Table 3). It was noteworthy that HDVL values obtained with the Eurobioplex HDV kit were quasi-systematically slightly higher for European/Asian or African HDV-1 strains (+0.25 log IU/ml; from 0.01 to 1.46) and lower for HDV-5 to -8 strains (−0.64 log IU/ml; from 0.07 to 1.95 log IU/ml) (Table 3). The Bland-Altman analysis confirmed the excellent agreement between the two assays with an intraclass coefficient correlation (ICC) of 0.894 (95% confidence interval [CI]), with HDVL values ranging between −1.44 and +1.33 log IU/ml (Fig. 5B). Of note, two HDV-1, four HDV-5, and one HDV-7 sample were outside this CI.
FIG 5.
Detection/quantification of the HDV genotypes. (A) Linear regression (R2) between quantified results for HDV with the Eurobioplex HDV kit and the FNRL assay. (B) Bland-Altman plots of data analyzed by the FNRL assay and Eurobioplex HDV kit. The x axis shows the means of any two measurements, and the y axis shows the difference between those two measurements. Black triangles represent measures of samples of HDV-1 genotype, and red circles represent measures of non-HDV-1 genotypes. Dashed lines represent the 95% limit of agreement (between −1.44 and +1.33).
TABLE 3.
Comparison of median HDV viral loads obtained by the HDV FNRL assay and Eurobioplex HDV kit according to genotype
| Test type | Median viral load by genotype(s) (log IU/ml) |
|||
|---|---|---|---|---|
| All | HDV-1 |
HDV-5 to HDV-8 | ||
| Europe/Asia | Africa | |||
| Eurobioplex kit | 5.05 | 5.87 | 4.36 | 4.74 |
| HDV FNRL | 5.16 | 5.61 | 4.11 | 5.38 |
| Difference between results | −0.11 | +0.26 | +0.25 | −0.64 |
On the other hand, the remaining 11 positive samples with low viral load (≤3log IU/ml) were detected either by the two assays for three samples or by one or the other assay for the remaining eight (five with the Eurobioplex HDV kit and three with the FNRL assay).
Longitudinal study.
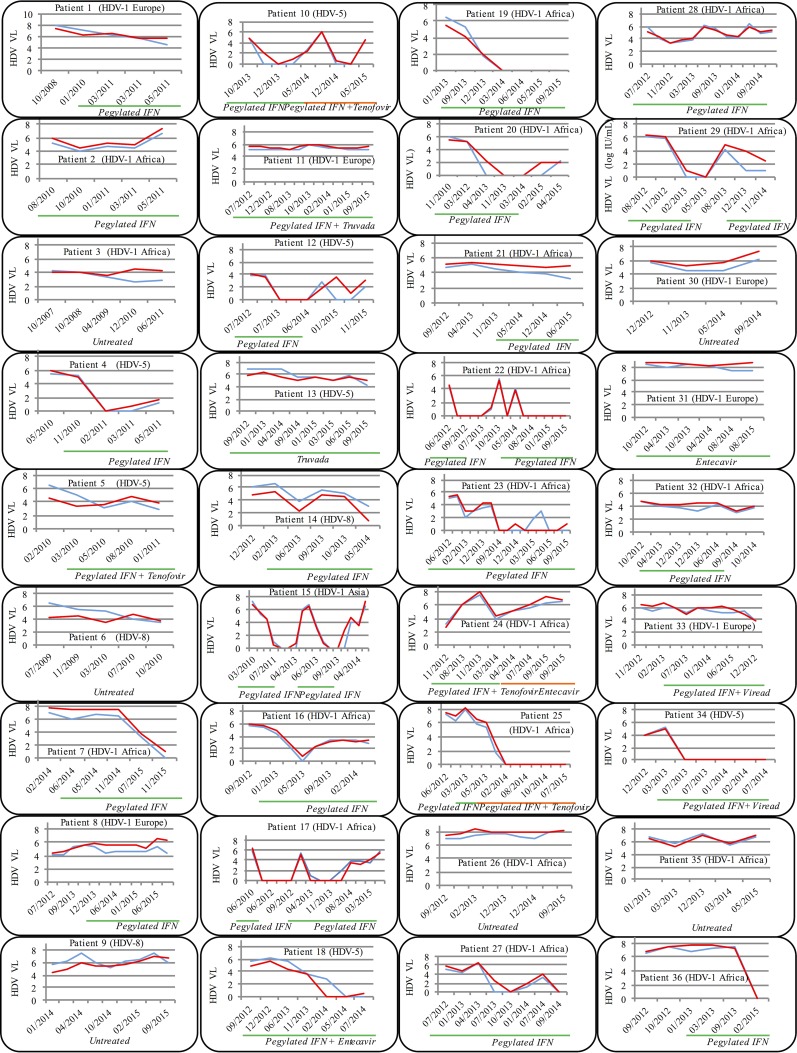
Several available consecutive blood samples (4 to 17) over several years of treated (30) and untreated (6) patients were analyzed. A total of 274 samples were quantified in parallel with the two assays. As shown in Fig. 6, we obtained strictly identical kinetic patterns of HDVL, whatever the HDV genotype or the treatment regimen. Among the 28 α-IFN-treated patients, 10 (35.8%) nonresponders (patients 1, 2, 5, 8, 11, 21, 24, 28, 32, and 33), 10 (35.8%) responders (patients 7, 14, 18, 19, 22, 25, 27, 29, 34, and 36), and 8 (28.5%) responders/relapsers (patients 4, 10, 12, 15, 16, 17, 20, and 23) could be individualized. Relapses occurred systematically after the α-IFN treatment was stopped for 1 year or more (patients 10, 12, 15, 17, 20, 22, and 29), and HDVL dropped back down at reintroduction of the treatment. HDVL rebounds could also be seen under α-IFN therapy (patients 4 and 16). Of note, after 2 years of IFN treatment, patients 18 (HDV-5-infected strain) and 23 (HDV-1 African) showed a very weak positivity (below the LLOQ of the assays) with the Eurobioplex HBV kit and not with the FNRL assay, confirming a high sensitivity of the kit, as mentioned above (Fig. 6). With respect to patients 13 and 31, nucleo(t)side analog ([NA] entecavir or tenofovir) monotherapy showed no efficiency, as described earlier, nor did nucleo(t)side analogs combined with IFN show any additional benefit in six out of eight patients (patients 5, 10, 11, 15, 24, and 33). Furthermore, no spontaneous resolution of the infection was observed in untreated patients (patients 3, 6, 9, 26, 30, and 35).
FIG 6.
Longitudinal study. Comparison of kinetic patterns of HDVL measured by both the FNRL assay (blue line) and Eurobioplex HDV kit (red line) in treated (30) or untreated (6) patients. Green and orange lines indicate the treatment regimen (IFN and/or nucleoside analogs) and duration. Genotypes of the HDV infecting strain and the dates of the different samples are indicated for each patient.
DISCUSSION
The main goal of this study was to evaluate a new commercial HDV kit for quantification of the HDVL in clinical blood samples. This is, to date, a key unsolved issue in HDV diagnosis and in the management of patients, as demonstrated by several recent studies (9–11).
One central challenge for an HDVL assay is to detect and quantify properly all different HDV genotypes. Particularly, African HDV genotypes (i.e., HDV-1 and HDV-5 to -8) are poorly quantified by almost all available assays and are either undetected or dramatically underestimated (11). These different genotypes originally had a specific geographic distribution; however, due to ancient or recent migrations of populations, this genetic variability is now encountered in different countries worldwide (9–11, 17–25). In this respect, France, where genotypes HDV-5 to -8 representing 20% of spreading strains were characterized, is an outstanding example (26, 27). Very interestingly, this new Eurobioplex HDV kit showed an excellent capacity to properly quantify HDVL in plasma and serum specimens, including strains with African genotypes (global median difference is −0.10 log IU/ml). Such capacity of the kit was also found in the longitudinal follow-up of infected patients (Fig. 6), where the HDVL kinetic patterns were strictly comparable. However, when we looked carefully, the median difference was −0.64 (from 0.07 to 1.95 log IU/ml) in samples of genotype HDV-5 to -8 (n = 36), and for 13 of them (36.1%) HDVL values were <1 log IU/ml with the Eurobio kit. This suggests that African patients, mainly those with low HDVL either before, during, or after the end of the treatment, should be carefully managed. Another clinical point is the sensitivity of the assay and its capacity to detect early rebounds of HDVL, as, unfortunately, relapses seem to be the rule at the end of the 12 or 18 months of pegylated α-IFN therapy. Considering these results, the Eurobioplex HDV kit seems to be highly suitable to monitor HDVL for patient management. Indeed, considering patients 18 and 23 (Fig. 6), the kit detected the rebound earlier than the FNRL assay, at a very low HDVL value (below the LLOQ of the two assays). These results confirmed the very good sensitivity of the kit. Of note, the manufacturer proposes a range of quantification between 3.53 and 8.53 log IU/ml regarding the concentration of the RNA standard of quantification. However, according to the experiments conducted here, we found an LLOQ of around 2.75 log IU/ml with very low CV, and more interestingly samples with very low HDVLs of around 1 log IU/ml were systematically detected by all operators. More experiments are needed with samples of African genotypes to confirm the low level of detection of the kit for all genotypes. Nevertheless, this kit can be used with assurance with an LLOQ and LLOD of 2.75 and 1 log IU/ml, respectively.
From technological and technical points of view, this multiplex real-time, one-step RT-PCR utilized primers and probe designed in the hepatitis D antigen (HDAg) coding region and not in the highest conserved ribozyme regions of the genome, as we proposed earlier (11). While this kit was in very good agreement with the FNRL assay, we noted that six samples of African genotypes (Fig. 4B) were under the 95% CI. This underestimation of >1.44 log IU/ml can be linked to the primer and probe sequence design, a critical point in the development of any assay (11). Unfortunately, we could not further address this point as primer/probe sequences are not available.
Another technical point of interest is the provision of an RNA internal control (IC) in this kit. In a recent study comparing almost all available HDVL quantification assays worldwide, 42% of them had no IC (11). More than 20 runs performed with this kit showed an excellent stability of this IC, without competition for high HDVL values, providing a robust tool to easily monitor all technical steps and to detect PCR inhibition events. Last, this assay runs on the CFX6 thermocycler (Bio-Rad) as recommended by the manufacturers. However, adaptation on fully automated devices might be conceivable and should be the next step for improvement of this kit in clinical practice.
In conclusion, the Eurobioplex HDV kit exhibited good specificity, sensitivity, precision, and clinical agreement with the FNRL assay and, most importantly, detected all HDV genotypes (although with a tendency to slightly underestimate HDV-5 to -8). This kit, which can be easily implemented in any clinical laboratory, will be a main and efficient tool for therapeutic management of patients. This constitutes a major step toward organization of large clinical studies, which will then issue guidelines, nonexistent to date, for monitoring and therapeutic management of HDV patients in the era of development of new potent anti-HDV therapies.
MATERIALS AND METHODS
Samples and study design.
All serum or plasma samples stored at −80°C, were selected from the French National Reference Laboratory for HDV (HDV FNRL) biobank. HDV genotype was determined as previously described by amplification and genotyping of the R0 region (26, 28). When necessary, sample dilutions were performed with hepatitis B surface antigen (HBsAg)-negative plasma kindly provided by the French National Institute for Blood Transfusion.
Serum and plasma samples.
First, we evaluated the performance of the kit on both serum and plasma samples. Quantification experiments were performed in pairs of serum and plasma specimens from 25 HDV-infected patients.
Specificity.
For specificity evaluation, 105 clinical samples negative for anti-HDV antibody (Ab) but positive for other hepatitis viruses and for the human immunodeficiency virus (HIV) were selected, including 5 positive IgM anti-HAV Ab, 75 positive HBV DNA, 10 positive HCV RNA, 5 positive HEV RNA, and 10 positive HIV RNA specimens.
Repeatability.
Repeatability evaluation was performed using three samples with high (>7 log IU/ml), medium (<6 log IU/ml), and low (4 log IU/ml) HDVL values. High, medium, and negative samples were extracted one time whereas the low sample was extracted 10 times. All samples were quantified in triplicate. In addition, two dilutions, 3 and 4.3 log IU/ml of the recently developed WHO HDV international standard (IS) titrated at 5.76 log IU/ml (29), were tested 80 and 100 times, respectively.
Reproducibility.
For reproducibility evaluation, four different technicians analyzed, in four different experiments, 4 negative samples and 20 positive samples, including 4 samples with detectable but unquantifiable HDVL (from 2 to 3 log IU/ml) and 16 samples with HDVLs ranging from 3 to 7.8 log IU/ml.
Linearity and sensitivity.
To determine the sensitivity of the new kit, we performed 14 serial dilutions (1:5) from a clinical sample of genotype HDV-1 quantified with the FNRL assay at 8.5 log IU/ml. Each diluted sample was extracted in the same series and quantified in triplicate in the same run.
Influence of biochemical parameters.
The influence of triglyceride and bilirubin in the blood samples was assessed to check for possible interference with quantification performance of the kit. The same amount, 5 log IU/ml (determined by the FNRL assay), of a plasma sample was added to 50 plasma samples with different triglyceride concentrations (1 to >5 millimoles/liter) and to 30 samples with different amounts of bilirubin (<20 to >50 micromoles/liter). All samples were then quantified with the Eurobioplex HDV kit.
Clinical samples of various genotypes and HDVLs.
The ability of the kit to detect and quantify strains with various HDVLs and genotypes was evaluated on 151 clinical samples including the following: 94 HDV-1 (33 African and 61 European/Asian patients); 1 HDV-2; 1 HDV-3; 1 HDV-4 (DNA plasmid containing one copy of the genome); 22 HDV-5, 7 HDV-6; 10 HDV-7; 3 HDV-8, and 12 negative controls.
Longitudinal study.
The kit was also evaluated in patient follow-up, using available consecutive serum/plasma samples (4 to 17) of treated (n = 30) or untreated (n = 6) patients. Patients were infected with strains of different genotypes: 26 with HDV-1 (from 19 African, 1 Asian, and 6 European patients), 7 with HDV-5, and 3 with HDV-8.
Ethics statements.
Evaluation of diagnosis tools, as well as prospective molecular characterization of all new HDV replicative strains, is part of the mission attributed to the HDV FNRL, associated to the French National Reference Centre for Hepatitis B, C and Delta, by the French Institut de Veille Sanitaire (InVS)/Santé Publique France.
Quantification experiments.
RNA extraction was performed on 500-μl patient samples, to which the RNA internal control (IC) of the Eurobioplex HDV kit had been previously added. The RNA extraction kit was used in strict accordance with the manufacturer's instructions on the m2000sp device (Abbott). RNA was eluted in 110 μl of sterile nuclease-free water and stored at −80°C until use.
The Eurobioplex HDV kit is a multiplex real-time one-step RT-qPCR, comprising an IC and a titrated RNA (8.53 log IU/ml) as a standard of quantification. Serial dilutions of this standard are made to create a 5-point standard curve of quantification ranging from 8.53 to 3.53 log IU/ml. The fluorophores used are FAM for the HDV target and HEX for the IC. Primers and probes are designed in the HDV antigen-coding region. Technical specifications indicated by the manufacturer were strictly followed. The amplification step was performed on a Bio-Rad CFX96 thermocycler.
The HDV FNRL consensus assay was developed in 2005 and since then has routinely been used as described previously (30), including with improvements that have been validated, such as addition of an RNA internal control, automated nucleic acid extraction using an Abbott m2000sp device, and the use of an ABI 7500 fast thermocycler. In addition, according to the WHO HDV IS, the LLOQ and the LLOD of the assay have been defined at 103 and 102 IU/ml, respectively.
Statistical analyses.
All results were expressed in international units per milliliter (IU/ml). Statistical analyses were performed on R software (version 3.2.1 [http://www-users.york.ac.uk/~mb55/meas/ba.htm]). The Pearson correlation coefficient (r) was calculated to determine the linear relationship between the two assays. The Bland-Altman method was used to assess the agreement between the assays. The means of all differences, the standard deviations (SD), and the intraclass correlation coefficient (ICC) were calculated.
ACKNOWLEDGMENTS
We express our thanks to the Eurobio Company for providing the kits and devices for implementation of this study and especially Marie-Claude Amoureux for technical and scientific support for result analyses. We thank Mei Chao and Chauting Yeh from Chang Gung University of Taiwan, China, who kindly provided us the HDV-4 plasmid. We thank Elhame Anouhal, Mirco Fabris, and Fernando Neri Pinto, technicians of the laboratory, who participated in the technical experiments.
F.L.G. and E.G. received grants from Eurobio Company to attend the 50th European association of the study of the liver (EASL) scientific meeting.
F.L.G. and E.G. were responsible for the study concept and design; F.L.G., A.G., and Z.B.A. were responsible for the technical process; F.L.G., A.G., D.R., S.D., and Z.B.A. were responsible for data acquisition of data; S.B., F.L.G., C.A., S.D., D.R., and E.G. were responsible for data analysis and interpretation; F.L.G., D.R., S.B., and E.G. drafted the manuscript; E.G. and F.L.G. supervised the study.
REFERENCES
- 1.Fattovich G, Boscaro S, Noventa F, Pornaro E, Stenico D, Alberti A, Ruol A, Realdi G. 1987. Influence of hepatitis delta virus infection on progression to cirrhosis in chronic hepatitis type B. J Infect Dis 155:931–935. doi: 10.1093/infdis/155.5.931. [DOI] [PubMed] [Google Scholar]
- 2.Fattovich G, Giustina G, Christensen E, Pantalena M, Zagni I, Realdi G, Schalm SW. 2000. Influence of hepatitis delta virus infection on morbidity and mortality in compensated cirrhosis type B. The European Concerted Action on Viral Hepatitis (Eurohep). Gut 46:420–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Manesis EK, Vourli G, Dalekos G, Vasiliadis T, Manolaki N, Hounta A, Koutsounas S, Vafiadis I, Nikolopoulou G, Giannoulis G, Germanidis G, Papatheodoridis G, Touloumi G. 2013. Prevalence and clinical course of hepatitis delta infection in Greece: a 13-year prospective study. J Hepatol 59:949–956. doi: 10.1016/j.jhep.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 4.Noureddin M, Gish R. 2014. Hepatitis delta: epidemiology, diagnosis and management 36 years after discovery. Curr Gastroenterol Rep 16:365. doi: 10.1007/s11894-013-0365-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Williams V, Brichler S, Khan E, Chami M, Deny P, Kremsdorf D, Gordien E. 2012. Large hepatitis delta antigen activates STAT-3 and NF-κB via oxidative stress. J Viral Hepat 19:744–753. doi: 10.1111/j.1365-2893.2012.01597.x. [DOI] [PubMed] [Google Scholar]
- 6.Mederacke I, Yurdaydin C, Dalekos GN, Bremer B, Erhardt A, Cakaloglu Y, Yalcin K, Gurel S, Zeuzem S, Zachou K, Bozkaya H, Dienes HP, Manns MP, Wedemeyer H, Hep-Net/International Delta Hepatitis Study Group. 2012. Anti-HDV immunoglobulin M testing in hepatitis delta revisited: correlations with disease activity and response to pegylated interferon-α2a treatment. Antivir Ther 17:305–312. doi: 10.3851/IMP1926. [DOI] [PubMed] [Google Scholar]
- 7.Hughes SA, Wedemeyer H, Harrison PM. 2011. Hepatitis delta virus. Lancet 378:73–85. doi: 10.1016/S0140-6736(10)61931-9. [DOI] [PubMed] [Google Scholar]
- 8.Mansour W, Malick FZ, Sidiya A, Ishagh E, Chekaraou MA, Veillon P, Ducancelle A, Brichler S, Le Gal F, Lo B, Gordien E, Lunel-Fabiani F. 2012. Prevalence, risk factors, and molecular epidemiology of hepatitis B and hepatitis delta virus in pregnant women and in patients in Mauritania. J Med Virol 84:1186–1198. doi: 10.1002/jmv.23336. [DOI] [PubMed] [Google Scholar]
- 9.Brichler S, Le Gal F, Butt A, Chevret S, Gordien E. 2013. Commercial real-time reverse transcriptase PCR assays can underestimate or fail to quantify hepatitis delta virus viremia. Clin Gastroenterol Hepatol 11:734–740. doi: 10.1016/j.cgh.2013.01.025. [DOI] [PubMed] [Google Scholar]
- 10.Brichler S, Le Gal F, Neri-Pinto F, Mansour W, Roulot D, Laperche S, Gordien E. 2014. Serological and molecular diagnosis of hepatitis delta virus infection: results of a French national quality control study. J Clin Microbiol 52:1694–1697. doi: 10.1128/JCM.03521-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Le Gal F, Brichler S, Sahli R, Chevret S, Gordien E. 2016. First international external quality assessment for hepatitis delta virus RNA quantification in plasma. Hepatology 64:1483–1494. doi: 10.1002/hep.28772. [DOI] [PubMed] [Google Scholar]
- 12.Lempp FA, Ni Y, Urban S. 2016. Hepatitis delta virus: insights into a peculiar pathogen and novel treatment options. Nat Rev Gastroenterol Hepatol 13:580–589. doi: 10.1038/nrgastro.2016.126. [DOI] [PubMed] [Google Scholar]
- 13.Vaillant A. 2016. Nucleic acid polymers: Broad spectrum antiviral activity, antiviral mechanisms and optimization for the treatment of hepatitis B and hepatitis D infection. Antiviral Res. 133:32–40. doi: 10.1016/j.antiviral.2016.07.004. [DOI] [PubMed] [Google Scholar]
- 14.Koh C, Canini L, Dahari H, Zhao X, Uprichard SL, Haynes-Williams V, et al. 2015. Oral prenylation inhibition with lonafarnib in chronic hepatitis D infection: a proof-of-concept randomised, double-blind, placebo-controlled phase 2A trial. Lancet Infect Dis 15:1167–1174. doi: 10.1016/S1473-3099(15)00074-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Noordeen F, Scougall CA, Grosse A, Qiao Q, Ajilian BB, Reaiche-Miller G, Finnie J, Werner M, Broering R, Schlaak JF, Vaillant A, Jilbert AR. 2015. Therapeutic antiviral effect of the nucleic acid polymer REP 2055 against persistent duck hepatitis B virus infection. PLoS One 10:e0140909. doi: 10.1371/journal.pone.0140909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bogomolov P, Alexandrov A, Voronkova N, Macievich M, Kokina K, Petrachenkova M, Lehr T, Lempp FA, Wedemeyer H, Haag M, Schwab M, Haefeli WE, Blank A, Urban S. 2016. Treatment of chronic hepatitis D with the entry inhibitor myrcludex B: first results of a phase Ib/IIa study. J Hepatol 65:490–498. doi: 10.1016/j.jhep.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 17.Manesis EK, Schina M, Le Gal F, Agelopoulou O, Papaioannou C, Kalligeros C, Arseniou V, Manolakopoulos S, Hadziyannis ES, Gault E, Koskinas J, Papatheodoridis G, Archimandritis AJ. 2007. Quantitative analysis of hepatitis D virus RNA and hepatitis B surface antigen serum levels in chronic delta hepatitis improves treatment monitoring. Antivir Ther 12:381–388. [PubMed] [Google Scholar]
- 18.Cross TJ, Rizzi P, Horner M, Jolly A, Hussain MJ, Smith HM, Vergani D, Harrison PM. 2008. The increasing prevalence of hepatitis delta virus (HDV) infection in South London. J Med Virol 80:277–282. doi: 10.1002/jmv.21078. [DOI] [PubMed] [Google Scholar]
- 19.Reinheimer C, Doerr HW, Berger A. 2012. Hepatitis delta: on soft paws across Germany. Infection. 40:621–625. doi: 10.1007/s15010-012-0287-9. [DOI] [PubMed] [Google Scholar]
- 20.De Paschale M, Ceriani C, Cerulli T, Cagnin D, Cavallari S, Ndayake J, Zaongo D, Priuli G, Vigano P, Clerici P. 2014. Prevalence of HBV, HDV, HCV, and HIV infection during pregnancy in northern Benin. J Med Virol 86:1281–1287. doi: 10.1002/jmv.23951. [DOI] [PubMed] [Google Scholar]
- 21.Genne D, Rossi I. 2011. Hepatitis delta in Switzerland: a silent epidemic. Swiss Med Wkly 141:w13176. doi: 10.4414/smw.2011.13176. [DOI] [PubMed] [Google Scholar]
- 22.Niro GA, Smedile A, Ippolito AM, Ciancio A, Fontana R, Olivero A, Valvano MR, Abate ML, Gioffreda D, Caviglia GP, Rizzetto M, Andriulli A. 2010. Outcome of chronic delta hepatitis in Italy: a long-term cohort study. J Hepatol 53:834–840. doi: 10.1016/j.jhep.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 23.Soriano V, Grint D, d'Arminio Monforte A, Horban A, Leen C, Poveda E, Antunes F, de Wit S, Lundgren J, Rockstroh J, Peters L. 2011. Hepatitis delta in HIV-infected individuals in Europe. AIDS 25:1987–1992. doi: 10.1097/QAD.0b013e32834babb3. [DOI] [PubMed] [Google Scholar]
- 24.Heidrich B, Deterding K, Tillmann HL, Raupach R, Manns MP, Wedemeyer H. 2009. Virological and clinical characteristics of delta hepatitis in Central Europe. J Viral Hepat. 16:883–894. doi: 10.1111/j.1365-2893.2009.01144.x. [DOI] [PubMed] [Google Scholar]
- 25.Santos MD, Gomes-Gouvea MS, Nunes JD, Barros LM, Carrilho FJ, Ferreira Ade S, Pinho JR. 2016. The hepatitis delta genotype 8 in Northeast Brazil: The North Atlantic slave trade as the potential route for infection. Virus Res 224:6–11. doi: 10.1016/j.virusres.2016.08.003. [DOI] [PubMed] [Google Scholar]
- 26.Radjef N, Gordien E, Ivaniushina V, Gault E, Anais P, Drugan T, Trinchet JC, Roulot D, Tamby M, Milinkovitch MC, Deny P. 2004. Molecular phylogenetic analyses indicate a wide and ancient radiation of African hepatitis delta virus, suggesting a deltavirus genus of at least seven major clades. J Virol 78:2537–2544. doi: 10.1128/JVI.78.5.2537-2544.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Le Gal F, Gault E, Ripault MP, Serpaggi J, Trinchet JC, Gordien E, Deny P. 2006. Eighth major clade for hepatitis delta virus. Emerg Infect Dis 12:1447–1450. doi: 10.3201/eid1209.060112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ivaniushina V, Radjef N, Alexeeva M, Gault E, Semenov S, Salhi M, Kiselev O, Deny P. 2001. Hepatitis delta virus genotypes I and II cocirculate in an endemic area of Yakutia, Russia. J Gen Virol 82:2709–2718. doi: 10.1099/0022-1317-82-11-2709. [DOI] [PubMed] [Google Scholar]
- 29.Chudy M, Hanschmann K-M, Bozdayi M, Kress J, Nubling CM, Collaborative Study Group. 2013. Collaborative study to establish a World Health Organization international standard for hepatitis D virus RNA for nucleic acid amplification technique (NAT)-based assays. Document WHO/BS/2013.2227. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 30.Le Gal F, Gordien E, Affolabi D, Hanslik T, Alloui C, Deny P, Gault E. 2005. Quantification of hepatitis delta virus RNA in serum by consensus real-time PCR indicates different patterns of virological response to interferon therapy in chronically infected patients. J Clin Microbiol 43:2363–2369. doi: 10.1128/JCM.43.5.2363-2369.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]