ABSTRACT
The precise diagnosis of American tegumentary leishmaniasis (ATL) is an essential task due to the disease's associated morbidity. A noninvasive, extremely sensitive, and highly specific exam is critical, particularly for mucosal leishmaniasis (ML), in which a low parasite quantity is expected. We aimed to compare the diagnostic accuracy of swab and biopsy sample analysis using SYBR Green- and TaqMan-based real-time PCR (qPCR) assays with that of a composite reference standard consisting of the Montenegro skin test, serology, histopathology, smears, culture, and conventional PCR. In total, 55 patients with ATL (ML, 18 patients; cutaneous leishmaniasis [CL], 37 patients) and 36 patients without ATL were studied. qPCR analysis of swabs was more accurate when using SYBR Green (87.88%; 95% confidence interval [CI], 77.86 to 93.73 patients) than when using TaqMan (78.79%; 95% CI, 67.49 to 86.92%) (P = 0.031). SYBR Green (84.72%; 95% CI, 74.68 to 91.25%) was also more accurate than TaqMan (73.61%; 95% CI, 62.42 to 82.41%) for biopsy samples (P = 0.008). All qPCR methods were 100% specific. Swabs and biopsy specimens had similar sensitivity when using the same chemistry (P = 0.125 for SYBR Green and P = 0.625 for TaqMan). Moreover, qPCR achieved better performance than most existing techniques used for the diagnosis of ATL and also detected the Leishmania parasite in a greater proportion of patients than the associated histopathology, smear, culture, and conventional PCR techniques did. Swabs therefore represent a useful diagnostic tool because they not only are noninvasive but also can achieve an accuracy similar to that of biopsy samples. The high accuracy of SYBR Green-based qPCR may also reduce the requirement for associated parasitological tests for ATL diagnosis.
KEYWORDS: leishmaniasis, mucocutaneous, diagnosis, real-time PCR, kinetoplast DNA, real-time polymerase chain reaction
INTRODUCTION
Leishmaniasis is a neglected disease caused by a protozoan of the genus Leishmania. This disease is showing alarming growth, especially in periurban areas (1, 2). Leishmania (Viannia) braziliensis and Leishmania (Leishmania) amazonensis both have a tropism for the skin and mucosa and are endemic in Brazil (1). These parasites specifically stimulate an intense granulomatous reaction, resulting in disfiguring lesions in untreated patients (3).
The accurate diagnosis of American tegumentary leishmaniasis (ATL) is an essential task due to the disease's associated morbidity (1, 2) but is hindered by the lack of a gold-standard laboratory test (3–5). Currently available parasitological tests based on stained smears and culture offer low sensitivity, and other options, such as the Montenegro (leishmanin) skin test (MST), have high sensitivity but low specificity (3, 6, 7). Additionally, serological techniques may yield widely varied accuracy values (3). In contrast, although not completely sensitive, PCR can improve the accuracy of ATL diagnosis (5, 8, 9). Strategies that might improve the sensitivity of PCR include the use of reliable DNA isolation methods, primer pairs that amplify the kinetoplast DNA (kDNA) sequences of Leishmania, and real-time PCR (qPCR) techniques (5, 10, 11).
Swabs and cytology brush samples of localized cutaneous leishmaniasis (CL) are more sensitive than lesion aspirate and biopsy samples (12, 13). The utility of swab-based diagnosis in mucosal leishmaniasis (ML) has not been completely explored, as most studies have been conducted in highly controlled environments, usually using healthy controls for comparison (5). These shortcomings have precluded the use of this technique in clinical practice (5, 14).
In the present study, we examined the accuracy of qPCR using SYBR Green and TaqMan for the diagnosis of the clinical forms of CL and ML. In particular, we tested noninvasive (swabs) and invasive (biopsy) samples, aiming to simulate real situations experienced by clinicians (15).
RESULTS
In total, 91 patients were included in this study. After application of the composite reference standard (CRS), 55 patients were allocated to the ATL group (ML, 18 patients; CL, 37 patients). Additionally, 36 patients who were not diagnosed with active ATL were designated non-ATL patients. Among the non-ATL patients, 32 patients with skin lesions were diagnosed with vascular ulcers (n = 8), basal cell carcinoma (n = 1), sporotrichosis (n = 4), bacterial infection (n = 14), tuberculosis (n = 1), or nonspecific ulcers or infiltrations without an etiological diagnosis (n = 4). The other 4 non-ATL patients presented mucous lesions, and the final diagnosis confirmed leprosy (n = 1), epidermoid carcinoma (n = 1), or lesions without an etiological diagnosis (n = 2) in these patients.
Patients with ML were older than CL patients (P = 0.002) and non-ATL patients (P = 0.009). ML patients also presented a longer disease duration than CL patients (P < 0.001) and non-ATL patients (P < 0.001) (Table 1). The subgenus Leishmania (Viannia) was identified in 38 cases of ATL, and the subgenus Leishmania (Leishmania) was identified in 1 case of CL. The case of Leishmania (Leishmania) tested negative in both qPCR assays. No adverse events were reported during the sampling procedures.
TABLE 1.
Demographic characteristics and positive results from the exams used to define the composite reference standard (defined as the gold standard) for patients allocated to the mucosal and cutaneous groups of American tegumentary leishmaniasis and non-ATL patients
| Characteristic or resulta | Mucosal group | Cutaneous group | Non-ATL patients | P valueb |
|---|---|---|---|---|
| Male gender | 13/18 (72.22) | 22/37 (59.46) | 20/36 (55.55) | 0.492 |
| Age (mean [SD]) (yr) | 57.39 (10.47) | 42.56 (16.49) | 46.58 (14.56) | 0.005 |
| Disease duration (median [range]) (mo) | 48 (6–420) | 3 (0.5–63) | 4 (0.4–180) | <0.001 |
| MST positivity | 15/16 (93.75) | 24/30 (80.00) | 13/32 (40.63) | 0.001 |
| Serological positivity | 12/17 (70.58) | 13/32 (40.63) | 6/25 (24.00) | 0.004 |
| Smear positivity | 3/12 (25.00) | 8/25 (32.00) | 0/33 (0) | <0.001 |
| Culture positivity | 1/10 (10.00) | 10/23 (43.48) | 0/29 (0) | <0.001 |
| Histopathological positivityc | 2/16 (12.50) | 13/36 (36.11) | 0/35 (0) | <0.001 |
| PCR positivity using biopsy samples | 9/14 (64.29) | 26/32 (81.25) | 0/34 (0) | <0.001 |
| PCR positivity using swab samples | 8/14 (57.14) | 14/18 (77.77) | 0/34 (0) | <0.001 |
Data are shown as the no./total no. (%), unless otherwise indicated.
Comparison of the three groups simultaneously. The assessment of significant differences is presented in the Results.
For comparison purposes, positivity in histopathology was considered to be the presence of amastigote forms.
The swab-based qPCR was more accurate using SYBR Green (87.88%; 95% confidence interval [CI], 77.86 to 93.73%) than using TaqMan (78.79%; 95% CI, 67.49 to 86.92%) (P = 0.031). The accuracy of SYBR Green (84.72%; 95% CI, 74.68 to 91.25%) was also superior to that of TaqMan (73.61%; 95% CI, 62.42 to 82.41) for biopsy samples (P = 0.008). All qPCR methods were 100% specific. Swabs and biopsy specimens had similar sensitivity when using the same chemistry (P = 0.125 using SYBR Green and P = 0.625 using TaqMan) (Tables 2 and 3). The combination of index tests in parallel did not have a significant impact on diagnostic accuracy.
TABLE 2.
Test properties for ATL forms and controls
| Sample type/test | Mucosal leishmaniasis |
Cutaneous leishmaniasis |
American tegumentary leishmaniasisa |
||||
|---|---|---|---|---|---|---|---|
| Positivity (no./total no. tested) | Sensitivity (% [95% CI]) | Positivity (no./total no. tested) | Sensitivity (% [95% CI]) | Sensitivity (% [95% CI]) | Specificity (% [95% CI]) | Accuracy (% [95% CI]) | |
| Biopsy/SYBR Green | 9/13 | 69.23 (42.37–87.32) | 22/29 | 75.86 (57.89–87.78) | 73.81 (58.93–84.70) | 100 (86.65–100) | 84.72 (74.68–91.25) |
| Biopsy/ TaqMan | 6/13 | 46.15 (23.21–70.86) | 17/29 | 58.62 (40.74–74.49) | 54.76 (39.95–68.78) | 100 (88.65–100) | 73.61 (62.42–82.41) |
| Swab/SYBR Green | 7/14 | 50 (26.80–73.20) | 17/18 | 94.44 (74.24–99.01) | 75 (57.89–86.75) | 100 (89.85–100) | 87.88 (77.86–93.73) |
| Swab/TaqMan | 6/14 | 42.86 (21.38–67.41) | 12/18 | 66.67 (43.75–83.72) | 56.25 (39.33–71.83) | 100 (89.85–100) | 78.79 (67.49–86.92) |
CI, confidence interval; the American tegumentary leishmaniasis group represents test properties calculated for mucosal leishmaniasis patients and cutaneous leishmaniasis patients together.
TABLE 3.
Agreement between real-time PCR assays of biopsy samples and tests used to define the composite reference standarda
| Reference test | Biopsy |
Swab |
||||||
|---|---|---|---|---|---|---|---|---|
| SYBR Green |
TaqMan |
SYBR Green |
TaqMan |
|||||
| Overall agreement | Cohen's kappa | Overall agreement | Cohen's kappa | Overall agreement | Cohen's kappa | Overall agreement | Cohen's kappa | |
| MST | 61.29 (48.85 to 72.42) | 0.26 (0.04 to 0.49) | 54.84 (42.53 to 66.58) | 0.19 (0.00 to 0.39) | 53.47 (40.70 to 65.98) | 0.12 (−0.11 to 0.34) | 50 (37.33 to 62.67) | 0.07 (−0.14 to 0.28) |
| Serology | 53.33 (40.89 to 65.37) | 0.06 (−0.18 to 0.31) | 48.33 (36.17 to 60.69) | −0.06 (−0.31 to 0.18) | 50.98 (37.68 to 64.14) | −0.04 (−0.31 to 0.24) | 60 (46.18 to 72.39) | 0.12 (−0.16 to 0.39) |
| Smear | 81.36 (69.62 to 89.26) | 0.52 (0.30 to 0.74) | 85 (73.89 to 91.9) | 0.52 (0.28 to 0.77) | 85.42 (72.83 to 92.75) | 0.52 (0.27 to 0.77) | 89.58 (77.83 to 95.47) | 0.61 (0.35 to 0.87) |
| Culture | 84.62 (72.48 to 91.99) | 0.58 (0.33 to 0.83) | 88.46 (77.03 to 94.6) | 0.60 (0.33 to 0.87) | 87.5 (73.89 to 94.54) | 0.55 (0.28 to 0.83) | 92.50 (80.14 to 97.42) | 0.69 (0.39 to 0.98) |
| Histopathologyb | 73.91 (62.49 to 82.81) | 0.41 (0.22 to 0.60) | 73.91 (62.49 to 82.81) | 0.29 (0.07 to 0.50) | 75.81 (63.85 to 84.75) | 0.38 (0.16 to 0.60) | 80.33 (68.69 to 88.37) | 0.38 (0.14 to 0.63) |
| Biopsy sample/PCR | 95.83 (88.45 to 98.57) | 0.92 (0.68 to 1.14) | 87.5 (77.92 to 93.28) | 0.74 (0.52 to 0.96) | 91.38 (81.36 to 96.26) | 0.81 (0.56 to 1.06) | 86.21 (75.07 to 92.84) | 0.69 (0.44 to 0.94) |
| Swab sample/PCR | 90.38 (79.39 to 95.82) | 0.79 (0.53 to 1.05) | 86.54 (74.73 to 93.32) | 0.69 (0.42 to 0.96) | 90.91 (81.55 to 95.77) | 0.80 (0.56 to 1.04) | 90.91 (81.55 to 95.77) | 0.78 (0.55 to 1.02) |
| Biopsy sample/SYBR Green | 88.89 (79.58 to 94.26) | 0.77 (0.54 to 0.99) | 92.31 (81.83 to 96.97) | 0.84 (0.57 to 1.10) | 88.46 (77.03 to 94.6) | 0.75 (0.49 to 1.01) | ||
| Biopsy sample/TaqMan | 88.89 (79.58 to 94.26) | 0.77 (0.54 to 0.99) | 96.15 (87.02 to 98.94) | 0.91 (0.64 to 1.18) | 90.48 (77.93 to 96.23) | 0.61 (0.32 to 0.91) | ||
| Swab sample/SYBR Green | 92.31 (81.83 to 96.97) | 0.84 (0.57 to 1.11) | 96.15 (87.02 to 98.94) | 0.91 (0.64 to 1.18) | 90.91 (81.55 to 95.77) | 0.79 (0.56 to 1.02) | ||
| Swab sample/TaqMan | 88.46 (77.03 to 94.60) | 0.75 (0.49 to 1.01) | 92.31 (81.83 to 96.97) | 0.82 (0.55 to 1.09) | 90.91 (81.55 to 95.77) | 0.79 (0.56 to 1.02) | ||
All values are shown with the 95% confidence interval in parentheses.
For comparison purposes, positivity in histopathology was considered to be the presence of amastigote forms.
qPCR detected Leishmania in a greater proportion of ATL patients than histopathology, smears, or culture did when each of these tests was used either alone or in combination, resulting in poor diagnostic agreement (Tables 1 to 3). In contrast, SYBR Green qPCR and conventional PCR used alone had good agreement (Table 3). In 15 ATL patients (8 ML patients and 7 CL patients), there was no direct evidence for the presence of Leishmania using a combination of histopathology, smears, culture, and conventional PCR. However, these patients were allocated to the ATL group because they presented a positive MST or indirect immunofluorescence (IIF) value, along with histopathology showing granuloma formation, as well as an absence of any other identifiable causative agent. In this group, SYBR Green qPCR was positive for one case of ML when using a biopsy sample and for another case of ML when using a swab sample. However, TaqMan qPCR yielded negative results for this group of patients.
DISCUSSION
Reliable diagnostic examinations of ATL have been extensively studied (3, 16). A noninvasive, extremely sensitive, and highly specific exam is essential, particularly for ML, in which a low parasite quantity is expected (17, 18). In the current study, SYBR Green and TaqMan were 100% specific. High specificity is the most important characteristic of any ATL diagnostic test, as the unjustified prescription of pentavalent antimonials or amphotericin B for patients without ATL can be extremely harmful (19, 20).
After the high specificity of qPCR was established, its sensitivity was analyzed. SYBR Green qPCR exhibited greater sensitivity values (Table 2). Although TaqMan qPCR is generally more specific, proper standardization suppresses unspecific amplification in SYBR Green qPCR (21). Additionally, in the present study, swabs and biopsy samples had similar sensitivity values when the same chemistry was used for qPCR. These findings are in contrast to the higher accuracy of swab analysis in CL reported in previous studies (12, 13, 18).
The highest achieved sensitivity for ML in our study, or 69.23% (95% CI, 42.37 to 87.32%), was achieved using SYBR Green qPCR to analyze biopsy samples (Table 2) and is an important milestone (22). A recent meta-analysis stated that the sensitivity of conventional PCR in ML was approximately 70%; however, at least half of the analyzed references were case-control accuracy studies and did not use a blinded evaluator (5). As the methodology of the present study was based on a cross-sectional/cohort design and used blinded evaluators, we expected a different outcome from that of other methodologies.
Cultures, smears, and detection of amastigote forms via histopathology are clinically recognized to be highly specific (3, 23) and were defined as 100% specific in the current study once visualization of parasites ensured allocation to the ATL group. Although certain technique optimization methods, such as press imprint smears from tissue fragments (24) and microculture (16), can improve the positivity rate, these exams are frequently used in parallel to enhance sensitivity (3), which can also elevate costs. In the present study, in addition to its high specificity, qPCR yielded positive results in certain patients in whom none of the parasitological exams could detect Leishmania. qPCR was thus able to substitute for the use of parasitological tests alone or in parallel. Although SYBR Green qPCR detected 2 ATL cases among 15 cases in which Leishmania was not found using the CRS, conventional PCR and qPCR performed similarly (Table 3). The main advantage of qPCR is its greater automation, which results in time savings (25–27).
The MST is clinically recognized to be sensitive but not sufficiently specific to diagnose ATL alone (28). Here, the use of immunological exams in the CRS was crucial for the inclusion of patients in whom the parasite was not detected. This important group of patients is frequently neglected because most studies use only parasitological or molecular exams for CRS definition (5). In fact, these patients would most benefit from highly sensitive qPCR. The qPCR assessed here provided high sensitivity rates similar to those of the MST, but it was more specific. IIF detected 70% of ML patients but was positive for fewer than 50% of CL patients and for 24% of controls, confirming its controversial role in ATL diagnosis (Table 1) (3, 7).
A limitation of the present study is that the primer pairs used in both the SYBR Green and TaqMan assays were not capable of detecting Leishmania (L.) amazonensis (Fig. 1). Clinicians should be aware that in South America, approximately 3% of CL cases are caused by this species (29, 30). The standardization of multiplex qPCR capable of detecting both species might improve the sensitivity of qPCR. The primers used also target different, but adjacent, regions of the Leishmania (Viannia) kDNA. The present results do not indicate that SYBR Green is superior to TaqMan for ATL; the greater sensitivity observed for SYBR Green might be due to a better DNA target for the local Leishmania strains rather than the chemistry itself.
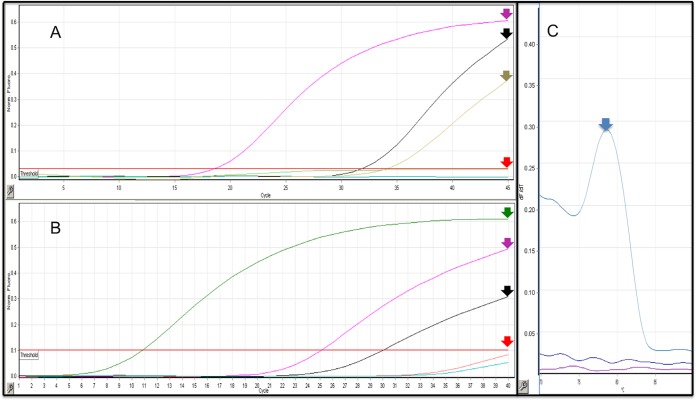
FIG 1.
(A) Linear amplification graph for the TaqMan-based assay. Leishmania (Viannia) braziliensis culture (pink arrow) (CT = 18.62), Leishmania (Viannia) guyanensis culture (black arrow) (CT = 31.86), Trypanosoma cruzi culture (brown arrow) (CT = 34.09). Leishmania amazonensis, DNA from healthy human skin, and a nontemplate control remained under the threshold line (red arrow) (CT = 0). y axis, normalized fluorescence; x axis, number of cycles. (B) Linear amplification graph for the SYBR Green-based assay. Leishmania (Viannia) braziliensis culture (green arrow) (CT = 10.84), Leishmania (Viannia) guyanensis culture (pink arrow) (CT = 25.13), and Trypanosoma cruzi culture (black arrow) (CT = 30.05). Leishmania amazonensis, DNA from healthy human skin and a nontemplate control remained under the threshold line (red arrow) (CT = 0). y axis, normalized fluorescence; x axis, number of cycles. (C) Melting curve for the SYBR Green-based assay. Expected melting curve analysis for Leishmania (Viannia) braziliensis culture (blue arrow). y axis, normalized fluorescence; x axis, temperature (°C). The images' scales were adjusted to fit the images in the panels.
Missing data is another limitation that can jeopardize results. In the present study, not all patients were subjected to all the tests partly because of difficulty in accessing certain laryngeal ML lesions and mainly because of an insufficient quantity of the extracted DNA for multiple PCR and qPCR tests. However, this limitation did not greatly affect the CRS because parasite detection by more than one technique was possible. With respect to the missing values for the index tests, this limitation was minimized by performing the agreement comparison for only those patients who were tested using all exams (Table 3).
We conclude that the performance of qPCR was superior to that of most of the techniques used for the detection of Leishmania. However, qPCR is not sufficiently accurate to be used as a single technique because its sensitivity is less than 100%. The inclusion of clinical data and the MST thus remained necessary in compatible cases in which the qPCR yielded negative results in our study. Nevertheless, the combination of swabs with qPCR is emerging as a powerful diagnostic tool because of its noninvasive yet simple collection method.
MATERIALS AND METHODS
Study location.
Patients were consecutively included at the Brasilia University Hospital, Brasilia, Federal District, Brazil, and at the University Hospital of Ribeirão Preto Medical School, University of São Paulo, Ribeirão Preto, São Paulo State, from 1 January 2012 to 30 August 2015.
Study population.
Patients who had clinical lesions consistent with ATL were included. All patients were tested using the MST, indirect immunofluorescence (IIF) analysis of Leishmania (L.) amazonensis culture (31), histopathologic examination of biopsy samples, culture of aspirate (Novy-MacNeal-Nicolle medium), smears from the lesion, conventional PCR of biopsy and swab samples, as previously described (8, 31), and qPCR of biopsy and swab samples.
Composite reference standard.
A composite reference standard (CRS) was defined. An ATL case was defined by detection of the parasite in at least one of the following examinations: histopathology, culture, smears, and conventional PCR of biopsy or swab samples, as described elsewhere (8). In the absence of parasite detection, ATL was diagnosed based on the MST or IIF positivity in combination with the presence of granulomas on histopathologic examination and a lack of identification of another etiological agent by special staining methods. The Leishmania subgenus was identified using conventional PCR coupled with restriction enzyme analysis, as described elsewhere (8).
After application of the CRS, the patients were prospectively allocated to two groups, consisting of ATL patients and patients without ATL. The ATL patients were grouped into ML and CL groups depending on the presence or absence, respectively, of mucous lesions, as previously described (8). In contrast, patients without ATL were not divided according to the presence of cutaneous or mucous lesions because of the small number of patients with mucous involvement.
Patients who were under 18 years old; who belonged to an indigenous community; or who presented any sign of immunosuppression, such as a positive HIV test, decompensated diabetes mellitus, or use of systemic corticosteroids or immunosuppressants, were not included.
Sampling (index tests).
Samples were collected in the following sequence: a sterile cotton swab (Absorve, São Paulo, Brazil) was rotated 360° 10 times against the internal border of the lesion, with slight pressure applied. The stem of the swab was then cut off using a sterile scalpel blade and stored in a 1.5-ml sterile microtube. A biopsy using a 4-mm punch was acquired from the border of a cutaneous or mucous lesion after proper cleansing and anesthesia and was then stored in a 1.5-ml sterile microtube.
All samples were stored at −80°C before and after DNA extraction.
DNA extraction and quantification (index tests).
DNA extraction was performed using the NucleoSpin tissue XS kit (Macherey-Nagel, GmbH & Co. KG, Düren, Germany), according to the manufacturer's instructions. After extraction, the samples were quantified in a NanoVue Plus spectrophotometer (GE Healthcare, Little Chalfont, United Kingdom) and diluted with ultrapure water to a concentration of 10 ng/μl for use as a DNA template.
Standardization tests (index tests).
Primer pair choices were based on previously described sequences that amplify fragments of the Leishmania (V.) braziliensis minicircle kDNA (5, 32–36). We evaluated primer and probe specificity for Leishmania (V.) braziliensis using NCBI BLAST. Two primer pairs were ultimately selected: MP1L/MP3H (for SYBR Green qPCR) (34) and L. braziliensis kDNA3 (for TaqMan qPCR) (33). For the L. braziliensis kDNA3 primer pair, we opted to use the described probe to enhance specificity. Standard curves were generated using Leishmania (V.) braziliensis (MHOM/BR/1994/M15176) DNA from 6 × 105 to 6 × 10−5 parasite DNA equivalents/reaction for MP1L/MP3H and from 2.4 × 105 to 2.4 × 10−5 parasite DNA equivalents/reaction for L. braziliensis kDNA3, as described elsewhere (33, 34).
Tests with Leishmania cultures and negative controls were performed prior to the processing of clinical samples, with the aim of improving the qualitative performance. This step was based on a comparison of Leishmania (V.) braziliensis (MHOM/BR/1994/M15176), Leishmania (Viannia) guyanensis (MHOM/BR/75/M4147), Leishmania (L.) amazonensis (IFLA/BR/67/PH8), and Trypanosoma cruzi (Y strain) cultures with negative controls, consisting of 10 healthy skin samples and controls in which the DNA template was omitted. The primer concentrations and annealing temperatures were optimized to reduce background signals, primer dimerization, nonspecific amplification, and false-positive results (Fig. 1).
Real-time qualitative PCR (index tests).
SYBR Green qPCR was performed with primers targeting the sequences of the kDNA of Leishmania of the subgenus Viannia, namely, MP1L-5′-TACTCCCCGACATGCCTCTG-3′ and MP3H-5′-GAACGGGGTTTCTGTATGC-3′, resulting in an amplified product of 70 bp (34). Reactions were performed in a final volume of 50 μl, consisting of 1× SYBR Green PCR master mix (Life Technologies, Carlsbad, CA, USA), 30 nM each primer, 19.7 μl of ultrapure water, and 50 ng of DNA template.
Amplification was performed at an initial holding temperature of 95°C for 10 min, followed by 40 cycles of 95°C for 20 s, 58°C for 20 s, and 72°C for 20 s and melting curve analysis in 1°C increments from 60°C to 99°C. All samples with a threshold cycle (CT) value greater than 35 and those with a melting curve not compatible with the expected pattern were considered negative. The SYBR Green qPCR resulted in a detection limit ranging from 6 × 105 to 6 × 10−2 parasite DNA equivalents/reaction (R2 = 0.999, efficiency = 90.238%, slope = −3.58).
TaqMan qPCR was performed with primers targeting sequences of the kDNA of Leishmania (Viannia) and a hydrolysis probe specific for Leishmania (V.) braziliensis, namely, primers 5′-TGCTATAAAATCGTACCACCCGACA-3′ and 5′-GAACGGGGTTTCTGTATGCCATTT-3′, and probe FAM-TTGCAGAACGCCCCTACCCAGAGGC-TAMRA (FAM, 6-carboxyfluorescein; TAMRA, 6-carboxytetramethylrhodamine), resulting in an amplified product of 83 bp (33). The reaction was performed in a final volume of 20 μl, consisting of 1× TaqMan Universal master mix (Life Technologies, Carlsbad, CA, USA), 400 nM each primer, 250 nM probe, 2 μl of ultrapure water, and 20 ng of DNA template.
Amplification was performed at two consecutive holding temperatures, or 50°C for 5 min and 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 60 s. The TaqMan qPCR resulted in a detection limit ranging from 2.4 × 105 to 2.4 × 102 parasite DNA equivalents/reaction (R2 = 0.993, efficiency = 90.782%, slope = −3.565).
Amplifications were performed in a Rotor-Gene Q (Qiagen, Hilden, Germany). The Dynamic Tube normalization option was selected to determine the average background of each sample. The threshold point of each reaction was defined manually in the region immediately above the nonspecific signal using a logarithmic amplification plot. All reactions were compared with a positive control, consisting of Leishmania (V.) braziliensis culture (MHOM/BR/1994/M15176), and negative controls, consisting of DNA extracted from a healthy skin sample and ultrapure water.
All reactions were performed twice. Divergent results were always considered negative in TaqMan assays and SYBR Green-based assays if the mean CT value of both reactions was greater than 35 cycles, without a compatible melting curve. All negative samples were analyzed for an endogenous gene by performing a qPCR assay using the C18X 343-bp 5′-GAAAGTGCCAGACCCGCCCCC-3′ and 5′-GCTGAAGCCACCGCCATAG-3′ primer pair specific for human keratin, as described elsewhere (8, 37). The reactions were performed without previous knowledge of the CRS, and the results were tabulated by a responsible party blinded to the final diagnosis of each patient.
Sample size.
An ATL prevalence of 60% was expected for the target population (patients attending the dermatology ambulatory clinic who had clinical lesions consistent with ATL). As this prevalence was >50%, we first estimated the number of controls based on an expected specificity of 95% (8, 38). Tables proposed by Flahault et al. were applied (39). Considering a minimal acceptable lower confidence limit of 0.75 (1 − α), the inclusion of 34 controls was recommended. Application of the following formula proposed by the same authors, or number of controls = number of cases [(1 − disease prevalence)/disease prevalence], indicated that the minimal number of ATL cases had to be 51 (39).
Statistical analysis.
Sensitivity was defined as the percentage of positive results in patients with ATL, and specificity was defined as the percentage of negative results in patients without ATL. Additionally, accuracy was defined as the sum of true-positive and true-negative results divided by the total number of patients tested. We assessed agreement between the qPCR and CRS exams based on the total agreement percentage, Cohen's kappa agreement, and McNemar's test. Missing variables were ignored, and only patients who underwent all exams were tested for agreement. We used the program SPSS 19.0 (IBM Corporation, Armonk, NY, USA). Statistical significance was defined as a P value of <0.05, and confidence intervals (CIs) were set at 95%.
Ethics.
Patients were included after signing an informed consent form. This study complied with the Declaration of Helsinki (40) and was approved by the Committee for Ethics in Research of the School of Medicine at the University of Brasilia and of the University Hospital of Ribeirão Preto Medical School, University of São Paulo (35611714.7.1001.5558,036/2011 and 35611714.7.2001.5440).
ACKNOWLEDGMENTS
We thank the clinical and laboratorial staff of the University of Brasília and the University Hospital of Ribeirão Preto Medical School for their aid in the performance of the exams used for patient diagnosis and clinical classification.
REFERENCES
- 1.Ministério da Saúde do Brasil–Secretaria de Vigilância em Saúde (SVS). 2007. Manual de vigilância da leishmaniose tegumentar americana, 2nd ed Ministério da Saúde, Brasília, Brazil. [Google Scholar]
- 2.World Health Organization. 2010. Control of the leishmaniases: report of a meeting of the WHO Expert Committee on the Control of Leishmaniases. World Health Organization, Geneva, Switzerland: http://apps.who.int/iris/bitstream/10665/44412/1/WHO_TRS_949_eng.pdf. [Google Scholar]
- 3.Gomes CM, Paula NA, Morais OO, Soares KA, Roselino AM, Sampaio RNR. 2014. Complementary exams in the diagnosis of American tegumentary leishmaniasis. An Bras Dermatol 89:701–709. doi: 10.1590/abd1806-4841.20142389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lima BSS, Pires SF, Fialho LC Jr, Oliveira EJ, Machado-de-Avila RA, Chávez-Olórtegui C, Chapeaurouge AD, Perales J, Andrade HM. 2017. A proteomic road to acquire an accurate serological diagnosis for human tegumentary leishmaniasis. J Proteomics 151:174–181. doi: 10.1016/j.jprot.2016.05.017. [DOI] [PubMed] [Google Scholar]
- 5.Gomes CM, Mazin SC, dos Santos ER, Cesetti MV, Bächtold GAB, de Freitas Cordeiro JH, Theodoro FCET, Damasco FDS, Carranza SAV, de Oliveira Santos A, Roselino AM, Sampaio RNR. 2015. Accuracy of mucocutaneous leishmaniasis diagnosis using polymerase chain reaction: systematic literature review and meta-analysis. Mem Inst Oswaldo Cruz 110:157–165. doi: 10.1590/0074-02760140280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reithinger R, Dujardin J-C, Louzir H, Pirmez C, Alexander B, Brooker S. 2007. Cutaneous leishmaniasis. Lancet Infect Dis 7:581–596. doi: 10.1016/S1473-3099(07)70209-8. [DOI] [PubMed] [Google Scholar]
- 7.Soares KA, Urdapilleta AAA, Santos GMD, Carneiro AL, Gomes CM, Roselino AM, Sampaio RNR. 2015. Field validation of a Leishmania (Leishmania) mexicana exo-antigens ELISA for diagnosing tegumentary leishmaniasis in regions of Leishmania (Viannia) predominance. Braz J Infect Dis 19:302–307. doi: 10.1016/j.bjid.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gomes CM, de Paula NA, Cesetti MV, Roselino AM, Sampaio RNR. 2014. Mucocutaneous leishmaniasis: accuracy and molecular validation of noninvasive procedures in a L. (V.) braziliensis-endemic area. Diagn Microbiol Infect Dis 79:413–418. doi: 10.1016/j.diagmicrobio.2014.05.002. [DOI] [PubMed] [Google Scholar]
- 9.Medeiros ACR, Rodrigues SS, Roselino AMF. 2002. Comparison of the specificity of PCR and the histopathological detection of leishmania for the diagnosis of American cutaneous leishmaniasis. Braz J Med Biol Res 35:421–424. doi: 10.1590/S0100-879X2002000400002. [DOI] [PubMed] [Google Scholar]
- 10.Eslami G, Hajimohammadi B, Jafari AA, Mirzaei F, Gholamrezai M, Anvari H, Khamesipour A. 2014. Molecular identification of Leishmania tropica infections in patients with cutaneous leishmaniasis from an endemic central of Iran. Trop Biomed 31:592–599. [PubMed] [Google Scholar]
- 11.Khosravi S, Hejazi SH, Hashemzadeh M, Eslami G, Darani HY. 2012. Molecular diagnosis of Old World leishmaniasis: real-time PCR based on tryparedoxin peroxidase gene for the detection and identification of Leishmania spp. J Vector Borne Dis 49:15–18. [PubMed] [Google Scholar]
- 12.Adams ER, Gomez MA, Scheske L, Rios R, Marquez R, Cossio A, Albertini A, Schallig H, Saravia NG. 2014. Sensitive diagnosis of cutaneous leishmaniasis by lesion swab sampling coupled to qPCR. Parasitology 141:1891–1897. doi: 10.1017/S0031182014001280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Suárez M, Valencia BM, Jara M, Alba M, Boggild AK, Dujardin J-C, Llanos-Cuentas A, Arevalo J, Adaui V. 2015. Quantification of Leishmania (Viannia) kinetoplast DNA in ulcers of cutaneous leishmaniasis reveals inter-site and inter-sampling variability in parasite load. PLoS Negl Trop Dis 9:e0003936. doi: 10.1371/journal.pntd.0003936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Korevaar DA, van Es N, Zwinderman AH, Cohen JF, Bossuyt PMM. 2016. Time to publication among completed diagnostic accuracy studies: associated with reported accuracy estimates. BMC Med Res Methodol 16:68. doi: 10.1186/s12874-016-0177-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Takwoingi Y, Leeflang MMG, Deeks JJ. 2013. Empirical evidence of the importance of comparative studies of diagnostic test accuracy. Ann Intern Med 158:544–554. doi: 10.7326/0003-4819-158-7-201304020-00006. [DOI] [PubMed] [Google Scholar]
- 16.Boggild AK, Miranda-Verastegui C, Espinosa D, Arevalo J, Martinez-Medina D, Llanos-Cuentas A, Low DE. 2008. Optimization of microculture and evaluation of miniculture for the isolation of Leishmania parasites from cutaneous lesions in Peru. Am J Trop Med Hyg 79:847–852. [PubMed] [Google Scholar]
- 17.Brelaz-de-Castro MCA, de Almeida AF, de Oliveira AP, de Assis-Souza M, da Rocha LF, Pereira VRA. 2012. Cellular immune response evaluation of cutaneous leishmaniasis patients cells stimulated with Leishmania (Viannia) braziliensis antigenic fractions before and after clinical cure. Cell Immunol 279:180–186. doi: 10.1016/j.cellimm.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 18.Boggild AK, Valencia BM, Veland N, Pilar Ramos A, Calderon F, Arevalo J, Low DE, Llanos-Cuentas A. 2011. Non-invasive cytology brush PCR diagnostic testing in mucosal leishmaniasis: superior performance to conventional biopsy with histopathology. PLoS One 6:e26395. doi: 10.1371/journal.pone.0026395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lima MIS, Arruda VO, Alves EVC, de Azevedo APS, Monteiro SG, Pereira SRF. 2010. Genotoxic effects of the antileishmanial drug glucantime. Arch Toxicol 84:227–232. doi: 10.1007/s00204-009-0485-0. [DOI] [PubMed] [Google Scholar]
- 20.Meiring S, Fortuin-de Smidt M, Kularatne R, Dawood H, Govender NP, GERMS SA. 2016. Prevalence and hospital management of amphotericin B deoxycholate-related toxicities during treatment of HIV-associated cryptococcal meningitis in South Africa. PLoS Negl Trop Dis 10:e0004865. doi: 10.1371/journal.pntd.0004865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tajadini M, Panjehpour M, Javanmard SH. 2014. Comparison of SYBR Green and TaqMan methods in quantitative real-time polymerase chain reaction analysis of four adenosine receptor subtypes. Adv Biomed Res 3:85. doi: 10.4103/2277-9175.127998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Al-Qahtani MS, Malik NW, Jamil S, Mekki TE. 2012. Diagnostic dilemma of primary mucosal leishmaniasis. Saudi Med J 33:1234–1238. [PubMed] [Google Scholar]
- 23.Daneshbod Y, Oryan A, Davarmanesh M, Shirian S, Negahban S, Aledavood A, Davarpanah MA, Soleimanpoor H, Daneshbod K. 2011. Clinical, histopathologic, and cytologic diagnosis of mucosal leishmaniasis and literature review. Arch Pathol Lab Med 135:478–482. [DOI] [PubMed] [Google Scholar]
- 24.Sousa AQ, Pompeu MML, Frutuoso MS, Lima JWO, Tinel JMBM, Pearson RD. 2014. Press imprint smear: a rapid, simple, and cheap method for the diagnosis of cutaneous leishmaniasis caused by Leishmania (Viannia) braziliensis. Am J Trop Med Hyg 91:905–907. doi: 10.4269/ajtmh.14-0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ben Abda I, de Monbrison F, Bousslimi N, Aoun K, Bouratbine A, Picot S. 2011. Advantages and limits of real-time PCR assay and PCR-restriction fragment length polymorphism for the identification of cutaneous Leishmania species in Tunisia. Trans R Soc Trop Med Hyg 105:17–22. doi: 10.1016/j.trstmh.2010.09.003. [DOI] [PubMed] [Google Scholar]
- 26.Francino O, Altet L, Sánchez-Robert E, Rodriguez A, Solano-Gallego L, Alberola J, Ferrer L, Sánchez A, Roura X. 2006. Advantages of real-time PCR assay for diagnosis and monitoring of canine leishmaniosis. Vet Parasitol 137:214–221. doi: 10.1016/j.vetpar.2006.01.011. [DOI] [PubMed] [Google Scholar]
- 27.Pereira MR, Rocha-Silva F, Graciele-Melo C, Lafuente CR, Magalhães T, Caligiorne RB. 2014. Comparison between conventional and real-time PCR assays for diagnosis of visceral leishmaniasis. Biomed Res Int 2014:639310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fagundes A, Antônio L, Schubach A, Marzochi KBF, Fagundes A. 2012. Comparison between in vivo measurement of the Montenegro skin test and paper recording. Int J Dermatol 51:618–619. doi: 10.1111/j.1365-4632.2010.04530.x. [DOI] [PubMed] [Google Scholar]
- 29.Teles CBG, Medeiros JF, Santos AP, Freitas LAR, Katsuragawa TH, Cantanhêde LM, Ferreira R de GM, Camarg LMA. 2015. Molecular characterization of American cutaneous leishmaniasis in the tri-border area of Assis Brasil, Acre State, Brazil. Rev Inst Med Trop Sao Paulo 57:343–347. doi: 10.1590/S0036-46652015000400012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medeiros AR, Silva WA Jr, Roselino AM. 2008. DNA sequencing confirms the involvement of Leishmania (L.) amazonensis in American tegumentary leishmaniasis in the state of São Paulo, Brazil. Clinics 63:451–456. doi: 10.1590/S1807-59322008000400007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Silveira TGV, Arraes SMAA, Bertolini DA, Teodoro U, Lonardoni MVC, Roberto ACBS, Ramos M, Nerilo Sobrinho A, Ishikawa E, Shaw J. 1999. Observations on laboratory diagnosis and cutaneous leishmaniasis epidemiology in the State of Paraná, South of Brazil. Rev Soc Bras Med Trop 32:413–423. doi: 10.1590/S0037-86821999000400013. [DOI] [PubMed] [Google Scholar]
- 32.Cruz I, Millet A, Carrillo E, Chenik M, Salotra P, Verma S, Veland N, Jara M, Adaui V, Castrillón C, Arevalo J, Moreno J, Cañavate C. 2013. An approach for interlaboratory comparison of conventional and real-time PCR assays for diagnosis of human leishmaniasis. Exp Parasitol 134:281–289. doi: 10.1016/j.exppara.2013.03.026. [DOI] [PubMed] [Google Scholar]
- 33.Weirather JL, Jeronimo SMB, Gautam S, Sundar S, Kang M, Kurtz MA, Haque R, Schriefer A, Talhari S, Carvalho EM, Donelson JE, Wilson ME. 2011. Serial quantitative PCR assay for detection, species discrimination, and quantification of Leishmania spp. in human samples. J Clin Microbiol 49:3892–3904. doi: 10.1128/JCM.r00764-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jara M, Adaui V, Valencia BM, Martinez D, Alba M, Castrillón C, Cruz M, Cruz I, Van der Auwera G, Llanos-Cuentas A, Dujardin J-C, Arevalo J. 2013. Real-time PCR assay for detection and quantification of Leishmania (Viannia) organisms in skin and mucosal lesions: exploratory study of parasite load and clinical parameters. J Clin Microbiol 51:1826–1833. doi: 10.1128/JCM.00208-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ceccarelli M, Galluzzi L, Migliazzo A, Magnani M. 2014. Detection and characterization of Leishmania (Leishmania) and Leishmania (Viannia) by SYBR Green-based real-time PCR and high resolution melt analysis targeting kinetoplast minicircle DNA. PLoS One 9:e88845. doi: 10.1371/journal.pone.0088845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pita-Pereira D, Lins R, Oliveira MP, Lima RB, Pereira BAS, Moreira OC, Brazil RP, Britto C. 2012. SYBR Green-based real-time PCR targeting kinetoplast DNA can be used to discriminate between the main etiologic agents of Brazilian cutaneous and visceral leishmaniases. Parasit Vectors 5:15. doi: 10.1186/1756-3305-5-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lugassy J, Itin P, Ishida-Yamamoto A, Holland K, Huson S, Geiger D, Hennies HC, Indelman M, Bercovich D, Uitto J, Bergman R, McGrath JA, Richard G, Sprecher E. 2006. Naegeli-Franceschetti-Jadassohn syndrome and dermatopathia pigmentosa reticularis: two allelic ectodermal dysplasias caused by dominant mutations in KRT14. Am J Hum Genet 79:724–730. doi: 10.1086/507792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Name RQ, Borges KT, Nogueira LSC, Sampaio JHD, Tauil PL, Sampaio RNR. 2005. Clinical, epidemiological and therapeuthic [sic] study of 402 patients with American cutaneous leishmaniasis attended at University Hospital of Brasilia, DF, Brazil. An Bras Dermatol 80:249–254. doi: 10.1590/S0365-05962005000300004. [DOI] [Google Scholar]
- 39.Flahault A, Cadilhac M, Thomas G. 2005. Sample size calculation should be performed for design accuracy in diagnostic test studies. J Clin Epidemiol 58:859–862. doi: 10.1016/j.jclinepi.2004.12.009. [DOI] [PubMed] [Google Scholar]
- 40.World Medical Association. 2013. World Medical Association Declaration of Helsinki: ethical principles for medical research involving human subjects. JAMA 310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]