ABSTRACT
Bedaquiline (BDQ), a diarylquinoline antibiotic that targets ATP synthase, is effective for the treatment of Mycobacterium tuberculosis infections that no longer respond to conventional drugs. While investigating the off-label use of BDQ as salvage therapy, seven of 13 patients with Mycobacterium intracellulare lung disease had an initial microbiological response and then relapsed. Whole-genome comparison of pretreatment and relapse isolates of M. intracellulare uncovered mutations in a previously uncharacterized locus, mmpT5. Preliminary analysis suggested similarities between mmpT5 and the mmpR5 locus, which is associated with low-level BDQ resistance in M. tuberculosis. Both genes encode transcriptional regulators and are adjacent to orthologs of the mmpS5-mmpL5 drug efflux operon. However, MmpT5 belongs to the TetR superfamily, whereas MmpR5 is a MarR family protein. Targeted sequencing uncovered nonsynonymous mmpT5 mutations in isolates from all seven relapse cases, including two pretreatment isolates. In contrast, only two relapse patient isolates had nonsynonymous changes in ATP synthase subunit c (atpE), the primary target of BDQ. Susceptibility testing indicated that mmpT5 mutations are associated with modest 2- to 8-fold increases in MICs for BDQ and clofazimine, whereas one atpE mutant exhibited a 50-fold increase in MIC for BDQ. Bedaquiline shows potential for the treatment of M. intracellulare lung disease, but optimization of treatment regimens is required to prevent the emergence of mmpT5 variants and microbiological relapse.
KEYWORDS: bedaquiline, Mycobacterium avium, Mycobacterium intracellulare, antibiotic resistance
INTRODUCTION
The diarylquinoline bedaquiline (BDQ; Sirturo) represents a new class of antimycobacterial agents. Primarily developed for the treatment of Mycobacterium tuberculosis infections, BDQ has been successfully used to treat multidrug-resistant (MDR) and extensively drug-resistant (XDR) strains (1–4) and is approved by the U.S. Food and Drug Administration (FDA) for MDR tuberculosis (TB) treatment in combination with other active tuberculosis drugs (5). BDQ is an ATP synthase inhibitor that is bactericidal for M. tuberculosis (6). Functional and structural studies have confirmed that diarylquinolines abolish ATP synthesis by binding to the subunit c rotor ring complex, encoded by atpE (7). Consistent with this, BDQ resistance was first associated with mutations in the atpE gene (8, 9). However, atpE mutations are rare in vivo, and when strains with elevated MICs to BDQ were selected in vitro, only 28% (15/53 strains) exhibited changes in atpE (10). Subsequent studies demonstrated that modest (≤8-fold) increases in MIC to BDQ are associated with mutations in two nontarget loci, Rv0678 (mmpR5) (11, 12) and Rv2535c (pepQ) (13). MmpR5 is a transcriptional regulator that controls the expression of genes encoding the MmpS5-MmpL5 efflux pump. Upregulation of MmpS5-MmpL5 (e.g., due to inactivation of MmpR5) is associated with low-level resistance to various drugs, including the antimycobacterial agent clofazimine (CLF). PepQ, a putative Xaa-Pro aminopeptidase, also confers low-level resistance to BDQ, but the underlying mechanism has not been elucidated.
Although it is FDA approved only for the treatment of MDR-TB, BDQ shows activity against many nontuberculous mycobacteria (NTM), including species of the Mycobacterium avium complex (MAC) (8, 9). When used alone, the drug is not bactericidal against MAC in vitro or in vivo (minimum bactericidal concentration [MBC], >128 μg/ml). However, when BDQ is given in combination with amikacin or amikacin plus clarithromycin, bactericidal activity is observed after 3 to 4 months of treatment (14). In 2015, the first study describing the off-label use of BDQ for the treatment of patients with refractory MDR NTM was reported by Philley et al. (15). It included six patients with MAC lung disease. All had a measurable response to therapy, and two-thirds developed one or more negative sputum culture. Unfortunately, several of the patients who initially responded to BDQ experienced microbiological relapse while on therapy. Reduced susceptibility to BDQ was suspected, but the phenotypic and genotypic characteristics of the pretreatment and relapse isolates were not assessed.
Phenotypic susceptibility testing remains the gold standard for assessing drug resistance among NTM. However, in vitro results do not always correlate with in vivo performance, and breakpoints for many drugs, including BDQ, have not yet been established (16). Nonetheless, we have recently implemented a BDQ susceptibility testing method for MAC that can identify isolates with elevated MICs (defined as MICs higher than those of untreated or wild strains) (17). Here, we examine MAC isolates recovered before, during, and after BDQ therapy and describe a novel genetic locus associated with microbiological relapse.
RESULTS
Patient population.
A total of 16 BDQ-treated MAC patients from University of Texas Health Science Center at Tyler (UTHSCT) were identified, including six patients previously described in the study by Philley et al. (15). Species-level identification of MAC isolates by partial 16S rRNA gene sequencing revealed that 13 of 16 cases involved M. intracellulare. Of these, eight patients experienced microbiological relapse, despite an initial positive response to BDQ treatment. In five cases, the acid-fast bacilli (AFB) smear and culture results became negative before relapsing to 3+/4+ levels. Patient 6 became smear negative and culture status improved (i.e., counts as low as 8 CFU on Middlebrook 7H11 agar) but remained positive. Patient 5 exhibited only minor declines in smear and culture counts, but these were considered significant, since prior to BDQ treatment, the patient had produced 20 consecutive 4+ cultures. Closer examination of the eighth case, including variable-number tandem-repeat (VNTR) analysis (18), revealed that relapse was due to a new infection with a genotypically distinct strain of M. intracellulare and not a mutation of the pretreatment isolate. The clinical characteristics of the seven patients who experienced true relapses are summarized in Table 1. Four patients had nodular bronchiectasis disease, and three patients had fibronodular upper-lobe disease. Five patients also had features of cavitary disease. Prior to the addition of BDQ, the patients had been on MAC treatment for 20 to 96 months. All seven patients were heavily AFB smear and culture positive, and four patients exhibited macrolide resistance. Microbiological relapse occurred after the patients had been on BDQ treatment for ≥3 months.
TABLE 1.
Clinical characteristics of seven M. intracellulare patients who responded to BDQ and then relapsed
| Patient | Age (yr) | Disease typea | Duration of prior treatment (mo) | Companion drugs while on BDQb | Resistance |
AFB smear status |
7H11 agar culture status |
|||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Macrolide | Amikacin | Pretreatment | Lowest on BDQ | Relapse | Pretreatment | Lowest on BDQ | Relapse | |||||
| 1c | 54 | FN + C | 96 | EMB, RFB, STR | Yes | No | 4+ | Negative | 3+ | 4+ | Negative | 4+ |
| 2c | 60 | NB | 25 | AZM, EMB, RFB, STR | No | No | 4+ | Negative | 3+ | 4+ | Negative | 4+ |
| 3 | 68 | FN + C | 48 | AMK, EMB, RFB | Yes | No | 3+ | Negative | 2+ | 4+ | Negative | 2+ |
| 4 | 71 | NB + C | AMK, AZM, EMB, STR | No | No | 3+ | Negative | 2+ | 4+ | Negative | 3+ | |
| 5c | 58 | NB | 78 | EMB, RFB, STR | Yes | Yes | 4+ | 2+ | 4+ | 4+ | 3+ | 4+ |
| 6 | 66 | NB + C | 44 | AMK, EMB, RFB | Yes | Yes | 3+ | Negative | 2+ | 3+ | +d | 3+ |
| 7 | 64 | FN + C | 20 | CLR, EMB, RFB, STR | No | No | 3+ | Negative | 3+ | 4+ | Negative | 3+ |
FN, fibronodular; NB, nodular bronchiectasis; C, cavitary.
AMK, amikacin; AZM, azithromycin; CLR, clarithromycin; EMB, ethambutol; RFB, rifabutin; STR, streptomycin.
Previously described by Philley et al. (15).
Only 8 CFU were observed.
Susceptibility testing.
The antibiotic susceptibility testing results for BDQ, CLF, amikacin (AMK), and clarithromycin (CLR) are summarized in Table 2. In most cases, the MICs for pretreatment strains were 0.004 μg/ml for BDQ and ≤0.06 μg/ml for CLF, whereas the MICs for relapse isolates were typically ≥0.008 μg/ml for BDQ and ≥0.12 μg/ml for CLF. The BDQ MICs of pretreatment isolates from patients 3 and 6 were 0.008 μg/ml.
TABLE 2.
Antibiotic susceptibility profiles and genotypic characteristics of pretreatment and relapse isolates of M. intracellularea
| Isolate by patient | Treatment status (mo) | MIC (μg/ml)b |
Susceptibilityc |
mmpT5 allele | MmpT5 sequenced | AtpE sequenced | ||
|---|---|---|---|---|---|---|---|---|
| BDQ | CLF | AMK | CLR | |||||
| 1 | ||||||||
| 1A | Pretreatment | 0.004 | ≤0.06 | R | R | 3.1 | WT | WT |
| 1B | On BDQ (5) | 0.008 | 0.12 | R | R | 3.2 | Gly66→fs | WT |
| 1F | Post-BDQ (3) | 0.004 | ≤0.06 | R | R | 3.3 | Arg25→Pro | Ala65→Pro |
| 2 | ||||||||
| 2A | Pretreatment | 0.004 | ≤0.06 | R | S | 7.1 | WT | WT |
| 2C | On BDQ (9) | 0.03 | ≤0.06 | R | S | 7.2 | Val46→Gly | WT |
| 2D | Post-BDQ (6) | 0.03 | 0.12 | R | S | 7.2 | Val46→Gly | WT |
| 3 | ||||||||
| 3B | Pretreatment | 0.004 | 0.12 | S | S | 5.1 | Glu177→Lys | WT |
| 3F | On BDQ (10) | 0.008 | 0.25 | R | S | 5.2 | Ala23→Pro + Glu177→Lys | WT |
| 3G | On BDQ (11) | 0.008 | 0.5 | R | S | 5.2 | Ala23→Pro + Glu177→Lys | WT |
| 3H | On BDQ (15) | 0.008 | ≤0.06 | R | S | 5.1 | Glu177→Lys | WT |
| 4 | ||||||||
| 4A | Pretreatment | 0.004 | ≤0.06 | S | S | 4.1 | WT | WT |
| 4C | On BDQ (4.5) | 0.015 | 0.25 | S | S | 4.2 | Pro104→fs | WT |
| 5 | ||||||||
| 5A | Pretreatment | 0.004 | ≤0.06 | R | R | 1.1 | WT | WT |
| 5C | On BDQ (8) | 0.004 | ≤0.06 | R | R | 1.2 | Ala162→Pro | WT |
| 5D | On BDQ (15) | 0.008 | ≤0.06 | R | R | 1.2 | Ala162→Pro | WT |
| 5E | On BDQ (23) | 2 | ND | R | R | 1.1 | WT | Ala65→Pro |
| 5F | On BDQ (28) | 2 | ≤0.06 | R | R | 1.1 | WT | Ala65→Pro |
| 6 | ||||||||
| 6A | Pretreatment | 0.008 | 0.12 | R | R | 5.1 | Glu177→Lys | WT |
| 6B | On BDQ (7) | 0.008 | 0.25 | R | R | 5.3 | Ile19→Ser + Glu177→Lys | WT |
| 6C | Post-BDQ (4.5) | 0.008 | 0.25 | R | R | 5.4 | Val35→Gly + Glu177→Lys | WT |
| 6E | Post-BDQ (8.5) | 0.03 | 0.5 | R | R | 5.1 | Glu177→Lys | WT |
| 7 | ||||||||
| 7A | Pretreatment | 0.004 | ≤0.06 | S | S | 7.1 | WT | WT |
| 7C | Post-BDQ (5) | 0.015 | 0.5 | S | S | 7.3 | Pro104→fs | WT |
| 7D | Post-BDQ (5) | 0.03 | 0.5 | S | S | 7.3 | Pro104→fs | WT |
| 7E | Post-BDQ (10) | 0.03 | 0.5 | S | S | 7.3 | Pro104→fs | WT |
For the complete list of pretreatment and relapse isolates, see Table S1.
CLF, clofazimine; ND, not determined.
AMK, amikacin; CLR, clarithromycin; R, resistant; S, susceptible.
WT, wild-type sequence; fs, frameshift mutation.
Whole-genome sequencing.
Whole-genome sequencing of five M. intracellulare isolates (1A and 1B from patient 1, and isolates 2A, 2B, and 2C from patient 2) generated a median number of 2.25 million reads per isolate, at a median depth of 120-fold coverage. Reference-based mapping of sequencing data to available MAC genomes confirmed that the strains were M. intracellulare. However, the comparison also revealed extensive genetic diversity (e.g., insertions, deletions, and single nucleotide polymorphisms) among the sequenced and reference genomes, which precluded the use of a reference mapping approach for the identification of candidate BDQ resistance loci. Instead, de novo contig assemblies were generated for all 5 strains. Sequencing reads were mapped to these de novo draft genomes, and high-quality single nucleotide variants (SNVs) were identified. The pretreatment and relapse isolates from patient 1 differed by only five high-quality SNVs (variant frequency, >95%; quality = 35). A comparison of strains from patient 2 revealed six SNVs, with five SNVs shared by both postrelapse isolates (i.e., 2B and 2C) and one SNV unique to isolate 2C. No atpE mutations were found, and no SNVs were shared by all five isolates. However, all three relapse isolates (i.e., 1B, 2B, and 2C) had nonsynonymous mutations in a previously uncharacterized open reading frame adjacent to the mmpS5-mmpL5 operon. This locus is predicted to encode a transcriptional regulator and control expression of the mmpS5-mmpL5 operon. Notably, this regulator is not a homolog of MmpR5, which occupies the equivalent position in the M. tuberculosis genome. To emphasize this distinction, we have named the MAC locus mmpT5.
Targeted gene sequencing. (i) mmpT5.
A phylogenetic comparison of mmpT5 gene sequences from MAC type and reference strains indicates that M. intracellulare sequences are most similar to those from Mycobacterium chimaera, Mycobacterium yongonense, and the recently validated MAC species M. paraintracellulare (19), whereas sequences from M. avium and Mycobacterium colombiense are more distant (Fig. S1A). To assess the variability of mmpT5 among clinical isolates, targeted gene sequencing was performed on 50 M. intracellulare isolates from patients in our study group (Table S1). A comparison with M. intracellulare ATCC 13950T revealed 19 variable sites, including 10 sites associated with synonymous substitutions and 9 sites with nonsynonymous mutations (Table 3). Among pretreatment isolates, 8 different mmpT5 alleles were identified. These differed by 0 to 5 single nucleotide variants (SNVs) from the ATCC 13950T mmpT5 gene (Tables 3 and S1, and Fig. S1B). With the exception of G529→A (Glu177→Lys), which was present in all isolates from patients 3 and 6, the pretreatment alleles only contained synonymous substitutions relative to the ATCC 13950T mmpT5 sequence. In contrast, isolates associated with microbiological relapse contained nonsynonymous mutations. Most SNVs were restricted to individual patients, although the same frameshift (fs) mutation (G311_312→insC; Pro104→fs) occurred independently in patients 4 and 7 (Tables 2, 3, and S1).
TABLE 3.
Allelic diversity of mmpT5 among clinical isolates of M. intracellulare
| mmpT5 position |
mmpT5 allelea |
Nucleotide change | Statusb | Amino acid change | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1.1 | 1.2 | 2.1 | 3.1 | 3.2 | 3.3 | 4.1 | 4.2 | 5.1 | 5.2 | 5.3 | 5.4 | 6.1 | 7.1 | 7.2 | 7.3 | 8.1 | ||||
| 56 | T | • | • | • | • | • | • | • | • | • | G | • | • | • | • | • | • | T56→G | NS | Ile19→Ser |
| 67 | G | • | • | • | • | • | • | • | • | C | • | • | • | • | • | • | • | G67→C | NS | Ala23→Pro |
| 72 | A | • | • | • | • | • | • | • | • | • | • | • | G | G | G | G | G | A72→G | Syn | Val24→Val |
| 74 | G | • | • | • | • | C | • | • | • | • | • | • | • | • | • | • | • | G74→C | NS | Arg25→Pro |
| 104 | T | • | • | • | • | • | • | • | • | • | • | G | • | • | • | • | • | T104→G | NS | Val35→Gly |
| 129 | A | • | • | • | • | • | G | G | • | • | • | • | • | • | • | • | • | A129→G | Syn | Glu43→Glu |
| 137 | T | • | • | • | • | • | • | • | • | • | • | • | • | • | G | • | • | T137→G | NS | Val46→Gly |
| 196/197 | • | • | • | C | • | • | • | • | • | • | • | • | • | • | • | • | C196_197→insCc | FS | Gly66→fs | |
| 311/312 | • | • | • | • | • | • | C | • | • | • | • | • | • | • | C | • | C311_312→insC | FS | Pro104→fs | |
| 378 | G | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | A | G378→A | Syn | Ala126→Ala |
| 429 | C | • | • | • | • | • | • | • | • | • | • | • | T | T | T | T | • | C429→T | Syn | Val143→Val |
| 450 | C | • | • | • | • | • | • | • | • | • | • | • | T | • | • | • | • | C450→T | Syn | Pro150→Pro |
| 471 | G | • | • | A | A | A | A | A | A | A | A | A | • | • | • | • | • | G471→A | Syn | Leu157→Leu |
| 484 | G | C | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | G484→C | NS | Ala162→Pro |
| 504 | G | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | • | G504→Z | Syn | Ser168→Ser |
| 522 | G | • | • | • | • | • | • | • | • | • | • | • | • | A | A | A | • | G522→A | Syn | Ala174→Ala |
| 528 | C | • | T | T | T | T | T | T | T | T | T | T | T | T | T | T | • | C528→T | Syn | Gly176→Gly |
| 529 | G | • | • | • | • | • | • | • | A | A | A | A | • | • | • | • | • | G529→A | NS | Glu177→Lys |
| 582 | G | • | • | • | • | • | • | • | • | • | • | • | A | A | A | • | G582→A | Syn | Ala194→Ala | |
• identical to allele 1.1 (mmpT5 sequence from M. intracellulare ATCC 13950T).
Syn, synonymous change; NS, nonsynonymous change; FS, frameshift mutation.
insC, insertion of C.
(ii) atpE.
Pretreatment isolates from all 13 patients with M. intracellulare infections had wild-type atpE sequences. Among relapse cases, atpE variants were observed only twice. Three months after the end of BDQ treatment, isolates containing mutations in both mmpT5 (G74→C and Arg25→Pro) and atpE (G221→C and Ala65→Pro) were recovered from patient 1. After 2 years of BDQ treatment, multiple isolates with the same atpE mutation (atpE G221→C and Ala65→Pro) were recovered from patient 5 (Tables 2 and S1).
DISCUSSION
Bedaquiline (BDQ) is a welcome addition to the antimycobacterial arsenal. Despite initial concerns about side effects (especially cardiac toxicity), BDQ-containing regimens have proven to be well tolerated and efficacious for the treatment of drug-resistant M. tuberculosis infections (4). For patients with NTM infections, preliminary findings are promising, but more studies are required. In a recent uncontrolled study of patients with MAC or Mycobacterium abscessus lung disease refractory to prior drug therapy, the off-label use of BDQ was well tolerated. No serious adverse effects or abnormal electrocardiogram (ECG) changes occurred (15). Five of the six MAC patients showed symptomatic improvement (e.g., less cough, less sputum production, and/or improved energy level). None achieved sputum conversion (i.e., three consecutive AFB-negative cultures) during the 6-month study period, but four patients developed one or more negative cultures (i.e., no growth on Middlebrook 7H11 agar) despite having heavily positive (4+) pretreatment cultures. Four of the MAC patients eventually experienced microbiological relapse. Unfortunately, Philley et al. (15) were unable to perform therapeutic drug monitoring or antibiotic susceptibility testing or evaluate the possibility of emerging BDQ resistance among relapse isolates. In the current study, we revisited the issue of microbiological relapse. M. intracellulare isolates associated with microbiological relapse after ≥3 months of therapy exhibited elevated MICs for BDQ. Modest (i.e., 2- to 8-fold) elevation of MICs to BDQ and CLF is associated with the acquisition of nonsynonymous mutations in the previously uncharacterized gene mmpT5.
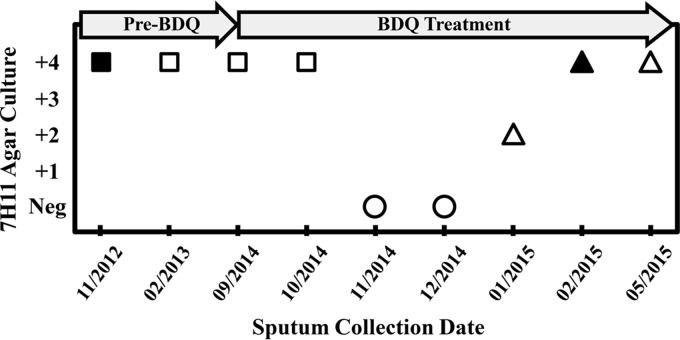
Of the 16 patients initially enrolled in this study, 13 patients had lung disease due to M. intracellulare, including seven who experienced true microbiological relapse. The experience of a typical patient is shown in Fig. 1. Despite combination therapy with multiple drugs, sputum cultures were heavily positive for M. intracellulare throughout the 20 months prior to BDQ initiation. Following the addition of BDQ, two consecutive specimens were culture negative on Middlebrook 7H11 agar, but this response was short-lived. Microbiological relapse was associated with a ≥2-fold increase in MICs for BDQ and CLF. Analysis of the relapse strain revealed a mutation in the mmpT5 gene.
FIG 1.
Representative microbiological response of a patient to BDQ treatment. Sputum samples for M. intracellulare culture were collected on the dates indicated (month/year). Targeted sequencing of mmpT5 was performed on positive cultures. Circles, negative, no AFB recovered from agar culture; squares, wild-type mmpT5 allele; triangles, mutant mmpT5 allele. MICs (in micrograms per milliliter) were determined for ■ (BDQ, 0.004; CLF, ≤0.06) and ▲ (BDQ, 0.008; CLF, 0.12).
Pretreatment MICs of M. intracellulare strains for BDQ were 0.004 μg/ml (six patients) to 0.008 μg/ml (one patient; see Table 2). This range is consistent with the results of a recent study conducted by members of our group (17). Briefly, of 103 MAC isolates tested by broth microdilution, 90 (87%) isolates showed BDQ MICs of ≤0.008 μg/ml, and only one isolate had an MIC of ≥0.015. These values are also similar to the 0.007 to 0.010 μg/ml range reported for 7 MAC strains using a Bactec system but lower than the ≥0.03 μg/ml reported for BDQ-susceptible M. tuberculosis strains (8, 20). After treatment, M. intracellulare isolates with elevated MICs were obtained from all relapse patients. We recognize that there is some variability associated with in vitro susceptibility testing and that 2-fold changes in MIC must be interpreted with caution. However, when assessing serial isolates from individuals on BDQ treatment, even small MIC differences may be relevant.
Loci associated with BDQ resistance have been reported for M. tuberculosis but not previously described for the MAC. The FoF1 ATP synthase subunit c targeted by BDQ is conserved in the MAC, but only a subset of BDQ-resistant M. tuberculosis strains contain atpE mutations (10). Comparative genomics indicate that the Rv0678 (mmpR5) locus, which is associated with low-level BDQ resistance in M. tuberculosis (11, 12), has no ortholog in the MAC. In order to identify M. intracellulare loci associated with elevated BDQ MICs, pretreatment and relapse isolates from two patients were subjected to whole-genome sequencing. Due to the extensive variability that is characteristic of MAC strains (21, 22), it was not feasible to identify candidate loci by comparing these sequences with publicly available M. intracellulare genomes. Instead, a de novo genome assembly approach was used. Sequences from isolates 1A and 1B (i.e., strain BDQ1) are most similar to the genome of Mycobacterium sp. strain MOTT36Y (NC_017904), whereas the 2A, 2B, and 2C isolates (i.e., strain BDQ2) appear to represent a different lineage of M. intracellulare. The BDQ1 strain is predicted to contain a large (∼100-kb) plasmid that is similar to a conjugative plasmid that is widely distributed among other NTM species, including the MAC (23). A comparison of the BDQ1 pretreatment and relapse isolates revealed five SNVs. Among the BDQ2 isolates, five SNVs were shared by both relapse isolates, and a sixth SNV was only found in the 2C genome. Although no SNVs were shared by all three relapse isolates, all had mutations in an uncharacterized locus upstream of the mmpS5-mmpL5 operon. Expression of the MmpS5-MmpL5 efflux system is associated with low-level resistance to antibiotics, including BDQ, CLF, and azole compounds (24). In M. tuberculosis, overexpression of MmpS5-MmpL5 can be caused by mutations in MmpR5, a MarR family transcriptional regulator that is encoded by the Rv0678 locus, which is adjacent to mmpS5-mmpL5. As previously indicated, M. intracellulare does not possess an ortholog of MmpR5. Instead, sequence analysis indicates that the uncharacterized M. intracellulare locus encodes a transcriptional regulator of the TetR family. The position of this gene and its association with BDQ resistance suggests that it controls the MAC mmpS5-mmpL5 operon. Because of its predicted role in transcription of the mmp5 operon, we have named this previously uncharacterized gene mmpT5. Experiments to formally assess the role of this regulator on mmpS5-mmpL5 transcription are under way.
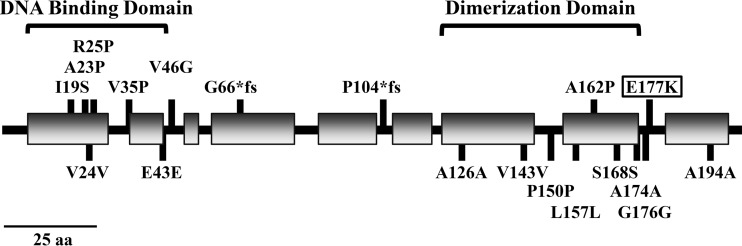
Targeted sequencing of clinical M. intracellulare strains revealed the mmpT5 gene to be polymorphic (Tables 3 and S1). Analysis of pretreatment isolates uncovered eight BDQ alleles composed of 10 synonymous and 1 nonsynonymous SNV. Eight additional polymorphisms, including six nonsynonymous SNVs and two frameshift mutations, were identified through sequencing of relapse isolates. TetR family regulators are homodimers. Each α-helical monomer features an N-terminal DNA-binding domain and a C-terminal dimerization/regulatory region (25). Protein modeling with Phyre2 (26) and JPred4 (27) predicts that five of the nonsynonymous SNVs are located in the DNA-binding domain of MmpT5, and two SNVs are located in the dimerization domain (Fig. 2). Mutations in these domains may impair MmpT5 homodimer formation and repression of (i.e., binding to) the mmpS5-mmpL5 operon, but functional studies are required to define roles for the individual substitutions. The frameshift mutations, which are predicted to abrogate MmpT5 activity, are consistently associated with increased MICs for both BDQ and CLF.
FIG 2.
Features of MmpT5. Protein modeling predicts that MmpT5 belongs to the TetR superfamily of transcriptional regulators. It features an N-terminal DNA-binding domain and a C-terminal dimerization domain. Shaded rectangles represent predicted α-helical regions. Mutations associated with M. intracellulare pretreatment, and relapse isolates are shown. Mutations were found throughout MmpT5, but the majority of nonsynonymous substitutions occur in the DNA-binding domain. The E177K substitution (boxed) at the edge of the predicted dimerization domain was present in pretreatment isolates from two patients.
Although mmpT5 mutations were common among relapse isolates, atpE mutations were only observed in two cases. Isolates containing mutations in both mmpT5 (G74→C; Arg25→Pro) and atpE (G221→C; Ala65→Pro) were obtained from patient 1, and isolates that were wild type for mmpT5 but exhibited the atpE G221→C (Ala65→Pro) mutation were recovered from patient 5 (Tables 2 and S1). The M. intracellulare mutation is equivalent to the Ala63→Pro change, which has been observed in BDQ-resistant M. tuberculosis strains (28). In contrast to mmpT5 mutations, which appeared in these patients within five (patient 1) to 8 months (patient 5) of BDQ initiation, the atpE mutations emerged much later, i.e., after patient 1 stopped therapy, and almost 2 years into patient 5's BDQ treatment.
BDQ is a promising agent for the treatment of MAC lung disease. In this study, even patients who eventually experienced microbiological relapse initially showed clinical improvement and reduced AFB burdens. Of concern, M. intracellulare isolates with new mmpT5 mutations and elevated MICs emerged within 3 to 8 months, much more rapidly than has been observed in MDR-TB trials. The relapse isolates showed only modest increases (i.e., ≤8-fold) in BDQ MICs, and in vitro values were ≤0.03 μg/ml, much lower than the BDQ concentrations expected to be maintained during therapy. For example, Diacon et al. (29) treated MDR-TB patients with a regimen that included thrice-weekly doses of 200 mg of BDQ. Throughout the dosing period, serum levels of BDQ remained above 0.6 μg/ml. Therapeutic drug monitoring was not available for our patients, but an equivalent dosing strategy was used. Additional studies are required to determine if this phenomenon of microbiological relapse is widespread during BDQ treatment of MAC disease, more common with M. intracellulare infections, or restricted to the challenging MDR strains found in our patient population.
A confounding factor may be the use of rifabutin (RFB) as a companion drug. Rifamycins induce the CYP450 system, which is the pathway for metabolism of BDQ. Thus, RFB cotreatment may compromise therapy by accelerating drug clearance and lowering BDQ exposure (30). Notably, of the seven cases with microbiological relapse, six were cotreated with RFB. Relapse isolates from those patients had elevated BDQ MICs and mmpT5 mutations, but the highest value (MIC, 2 μg/ml) was associated with an atpE mutant obtained from patient 5, who did not receive RFB. The impact of companion drugs and dosing schedules on the clinical efficacy of BDQ deserves further investigation, as the optimization of BDQ-containing regimens may slow or prevent the emergence of mmpT5 variants and improve treatment outcomes for all patients with MAC lung disease.
MATERIALS AND METHODS
Patients.
Patients for this study were identified from the NTM/Bronchiectasis Clinic of the University of Texas Health Science Center at Tyler (UTHSCT). All had treatment-refractory MDR-MAC lung disease and were approved for off-label therapy with BDQ. The treatment details for some patients have been presented previously (15). For the first 2 weeks of treatment, BDQ was dosed at 400 mg per day. The dose was then decreased to 200 mg three times per week. The BDQ treatment period and routine acid-fast bacilli (AFB) sputum smear and culture results were collected from chart reviews (Table 1). A decline in AFB smear and culture statuses was interpreted as a microbiological response to BDQ treatment. A patient was considered to have experienced a microbiological relapse if, after an initial response to treatment, their AFB smear and culture (i.e., CFU counts on Middlebrook 7H11 agar) statuses increased.
Informed consent.
Retrospective chart reviews and molecular analysis (i.e., DNA sequencing) of patient isolates were approved by the institutional review board at UTHSCT.
Bacterial strains.
Patients undergoing treatment for MAC infection were asked to submit sputum samples once per month. Samples were processed and cultured on liquid and solid media using standard methods (31) and then stored at −70°C or maintained at room temperature (20 to 22°C) in their original broth culture bottles. Cultures were identified as MAC using AccuProbe tests (Hologic, Inc., MA, USA). Partial 16S rRNA gene sequencing allowed differentiation of MAC species. Sequences were analyzed using the MicroSeq system, as described previously (32). Fifty clinical isolates of M. intracellulare, including BDQ pretreatment, untreated, and BDQ relapse strains, were chosen for further study. For cases involving BDQ therapy with microbiological relapse, multiple isolates were examined, including at least one pretreatment and one relapse isolate. These isolates are described in Tables 2 and S1.
Phenotypic susceptibility testing.
MICs to BDQ (0.004 μg/ml to 8 μg/ml) and CLF (0.06 μg/ml to 2 μg/ml) were determined using serial two-fold dilutions in cation-adjusted Mueller-Hinton broth plus 5% oleic acid-albumin-dextrose-catalase broth. Because the Clinical and Laboratory Standards Institute (CLSI) has not yet addressed susceptibility testing of M. intracellulare with BDQ or CLF, there are no CLSI-recommended breakpoints or established quality control procedures for these drugs. However, our testing procedure is consistent with published recommendations (33). Quality control for CLF was performed with M. avium ATCC 700898, as previously recommended (34). Quality control for BDQ was performed using M. avium ATCC 700898, and ranges of MICs were recorded in order to assess precision of testing and develop future standards for testing of the drug. Isolates were also tested against clarithromycin (CLR; 0.06 μg/ml to 64 μg/ml) and amikacin (AMK; 1 μg/ml to 64 μg/ml) using the CLSI guidelines for clarithromycin and the breakpoints for amikacin suggested by Brown-Elliott et al. (35).
Whole-genome sequencing and bioinformatic analyses.
Whole-genome sequencing of M. intracellulare isolates 1A, 1B, 2A, 2B, and 2C was performed on an Illumina MiSeq platform. Briefly, strains were cultured on Middlebrook 7H11 agar (BD-Canada, Ontario, Canada), and genomic DNA was extracted from a loopful of cells using the MasterPure DNA purification kit (Epicentre, WI, USA). To facilitate lysis, cells were first heated at 80°C for 20 min and then mixed with 600-μm glass beads and vortexed for 5 min. RNA-free genomic DNA was sheared by sonication with a Bioruptor Standard (Diagenode, NJ, USA), and size selection was performed using Agencourt AMPure beads (Beckman Coulter, Ontario, Canada). Paired-end libraries were generated with the NEBNext Ultra DNA library prep kit and NEBNext multiplex oligonucleotides for Illumina (New England BioLabs, Ontario, Canada). A final combined 4 nM library was prepared, denatured, diluted to 14 pM, and sequenced using a MiSeq reagent version 3 kit (600 cycle), according to the manufacturer's guidelines (Illumina, Inc., CA, USA). The SPAdes genome assembler (version 3.6.2) was used for de novo generation of contigs (36). Additional analyses, including reference-based mapping of sequencing reads to contigs and identification of high-quality single nucleotide variants (SNVs), were performed with the Geneious R7 package (Biomatters, Auckland, New Zealand) (37). MEGA5 was used for phylogenetic analyses (38).
Targeted gene sequencing.
DNA sequences of the mmpT5 and atpE genes were determined by targeted gene sequencing. M. intracellulare strains were cultured on agar medium, and genomic DNA was extracted using the PrepMan Ultra reagent (Life Technologies, CA, USA). Briefly, a loopful of bacteria was suspended in 100 μl of preparation reagent. Samples were held for 30 s, heated for 10 min at 100°C, allowed to cool at room temperature (20 to 22°C) for 2 min, and then centrifuged at maximum speed in a microcentrifuge for 2 min. The DNA-containing supernatant (50 μl) was retained for PCR and sequencing.
PCR amplification of the mmpT5 gene was performed in a 20-μl reaction using 1× FailSafe Premix I, 1.25 U of FailSafe enzyme mix (Epicentre, WI, USA), and 10 μM each primer (mmpT5_F, 5′-GATGGCACCTTTTGACTGC-3′; mmpT5_R, 5′-GCTGGTGTTTCAGGTCACTTC-3′). The PCR conditions included an initial denaturation of 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing of 54°C for 1 min, and elongation at 72°C for 1 min, with a final elongation at 72°C for 5 min.
PCR amplification of the atpE gene was performed in a 20-μl reaction using 1× FailSafe Premix I, 1.25 U of FailSafe enzyme mix (Epicentre, WI, USA), and a 10 μM concentration of each primer (atpE_F, 5′-CCCTACCAGATATCAAGGAGGATAAG-3′; atpE _R, 5′-CACCCATCACAGCGAACTAG-3′). The atpE-specific PCR conditions included an initial denaturation of 95°C for 1 min, followed by 30 cycles of denaturation at 95°C for 30 s, annealing of 54°C for 30 s, and elongation at 72°C for 30 s, plus a final elongation at 72°C for 7 min.
Agarose gel electrophoresis was used to check that amplicons had been generated and were of the correct size (615 bp for mmpT5 and 298 bp for atpE). In preparation for sequencing, 8 μl of PCR product was combined with 2 μl of ExoSAP-IT reagent (Affymetrix, CA, USA). Sequencing was performed using the BigDye Terminator version 3.1 cycle sequencing kit and the mmpT5_F/mmpT5_R or atpE_F/atpE_R primer pairs on an ABI 3500 genetic analyzer (Thermo Fisher Scientific, CA, USA). DNA sequence analysis was performed using RipSeq software (Pathogenomix, CA, USA). Phylogenetic analysis was performed using MEGA5 (38). The genome of M. intracellulare ATCC 13950T (GenBank accession no. GCA_000172115.1) was designated the wild type.
Accession number(s).
Data associated with this study have been registered as BioProject ID PRJNA339272. Raw sequencing data from M. intracellulare isolates 1A, 1B, 2A, 2B, and 2C have been deposited in the Sequence Read Archive (http://www.ncbi.nlm.nih.gov/sra) as BioSample SAMN05770403 through SAMN05770407. Nucleotide sequences of 17 representative mmpT5 alleles have been deposited in GenBank as accession numbers KX826800 through KX826816. Additional mmpT5 sequences were derived from the following publically available data sets: M. intracellulare ATCC 13950T (GenBank accession no. GCA_000172115.1), M. intracellulare GMI 1956 (GenBank accession no. GCA_000523815.1), M. intracellulare GMI 1280 (GenBank accession no. GCA_000524015.1), M. intracellulare M.i.198 (GenBank accession no. GCA_000309055.1), M. intracellulare MOTT-02 (GenBank accession no. GCA_000277145.1), M. yongonense 05-1390T (accession no. NC_021715.1), M. paraintracellulare MOTT-64T (accession no. NC_016948.1), M. chimaera MCIMRL2 (GenBank accession no. GCA_001307335.1), Mycobacterium sp. strain H4Y (GenBank accession no. GCA_000364405.1), Mycobacterium sp. MOTT36Y (GenBank accession no. GCA_000262165.1), M. colombiense CECT 3035T (GenBank accession no. GCA_000222105.4), M. avium subsp. avium ATCC 25291T (GenBank accession no. GCA_000174035.1), M. avium subsp. silvaticum ATCC 49884T (GenBank accession no. GCA_000504975.1), M. avium subsp. paratuberculosis ATCC 19698T (GenBank accession no. GCA_000240525.2), and “M. avium subsp. hominissuis” 104 (accession no. NC_008595.1).
Supplementary Material
ACKNOWLEDGMENTS
This study was funded in part by the Amon G. Carter Foundation and donations by individual MAC patients, and by funding (to A.D.S.C. and D.C.A.) from the Saskatchewan Health Research Foundation (grant 3378) and (to A.D.S.C.) from the Natural Sciences and Engineering Research Council of Canada (Discovery grant RGPIN-435784-2013).
We acknowledge the support of Janssen Pharmaceuticals for providing access to bedaquiline.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JCM.02087-16.
REFERENCES
- 1.Diacon AH, Donald PR, Pym A, Grobusch M, Patientia RF, Mahanyele R, Bantubani N, Narasimooloo R, De Marez T, van Heeswijk R, Lounis N, Meyvisch P, Andries K, McNeeley DF. 2012. Randomized pilot trial of eight weeks of bedaquiline (TMC207) treatment for multidrug-resistant tuberculosis: long-term outcome, tolerability, and effect on emergence of drug resistance. Antimicrob Agents Chemother 56:3271–3276. doi: 10.1128/AAC.06126-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Diacon AH, Pym A, Grobusch MP, de los Rios JM, Gotuzzo E, Vasilyeva I, Leimane V, Andries K, Bakare N, De Marez T, Haxaire-Theeuwes M, Lounis N, Meyvisch P, De Paepe E, van Heeswijk RP, Dannemann B, TMC207-C208 Study Group. 2014. Multidrug-resistant tuberculosis and culture conversion with bedaquiline. N Engl J Med 371:723–732. doi: 10.1056/NEJMoa1313865. [DOI] [PubMed] [Google Scholar]
- 3.Guglielmetti L, Le Du D, Jachym M, Henry B, Martin D, Caumes E, Veziris N, Métivier N, Robert J, MDR-TB Management Group of the French National Reference Center for Mycobacteria and the Physicians of the French MDR-TB Cohort. 2015. Compassionate use of bedaquiline for the treatment of multidrug-resistant and extensively drug-resistant tuberculosis: interim analysis of a French cohort. Clin Infect Dis 60:188–194. doi: 10.1093/cid/ciu786. [DOI] [PubMed] [Google Scholar]
- 4.Pontali E, Sotgiu G, D'Ambrosio L, Centis R, Migliori GB. 2016. Bedaquiline and multidrug-resistant tuberculosis: a systematic and critical analysis of the evidence. Eur Respir J 47:394–402. doi: 10.1183/13993003.01891-2015. [DOI] [PubMed] [Google Scholar]
- 5.Cohen J. 2013. Infectious disease. Approval of novel TB drug celebrated–with restraint. Science 339:130. [DOI] [PubMed] [Google Scholar]
- 6.Koul A, Dendouga N, Vergauwen K, Molenberghs B, Vranckx L, Willebrords R, Ristic Z, Lill H, Dorange I, Guillemont J, Bald D, Andries K. 2007. Diarylquinolines target subunit c of mycobacterial ATP synthase. Nature Chemical Biology 3:323–324. doi: 10.1038/nchembio884. [DOI] [PubMed] [Google Scholar]
- 7.Preiss L, Langer JD, Yildiz O, Eckhardt-Strelau L, Guillemont JE, Koul A, Meier T. 2015. Structure of the mycobacterial ATP synthase Fo rotor ring in complex with the anti-TB drug bedaquiline. Sci Adv 1:e1500106. doi: 10.1126/sciadv.1500106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Andries K, Verhasselt P, Guillemont J, Gohlmann HW, Neefs JM, Winkler H, Van Gestel J, Timmerman P, Zhu M, Lee E, Williams P, de Chaffoy D, Huitric E, Hoffner S, Cambau E, Truffot-Pernot C, Lounis N, Jarlier V. 2005. A diarylquinoline drug active on the ATP synthase of Mycobacterium tuberculosis. Science 307:223–227. doi: 10.1126/science.1106753. [DOI] [PubMed] [Google Scholar]
- 9.Huitric E, Verhasselt P, Andries K, Hoffner SE. 2007. In vitro antimycobacterial spectrum of a diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother 51:4202–4204. doi: 10.1128/AAC.00181-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huitric E, Verhasselt P, Koul A, Andries K, Hoffner S, Andersson DI. 2010. Rates and mechanisms of resistance development in Mycobacterium tuberculosis to a novel diarylquinoline ATP synthase inhibitor. Antimicrob Agents Chemother 54:1022–1028. doi: 10.1128/AAC.01611-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Andries K, Villellas C, Coeck N, Thys K, Gevers T, Vranckx L, Lounis N, de Jong BC, Koul A. 2014. Acquired resistance of Mycobacterium tuberculosis to bedaquiline. PLoS One 9:e102135. doi: 10.1371/journal.pone.0102135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hartkoorn RC, Uplekar S, Cole ST. 2014. Cross-resistance between clofazimine and bedaquiline through upregulation of MmpL5 in Mycobacterium tuberculosis. Antimicrob Agents Chemother 58:2979–2981. doi: 10.1128/AAC.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Almeida D, Ioerger T, Tyagi S, Li SY, Mdluli K, Andries K, Grosset J, Sacchettini J, Nuermberger E. 2016. Mutations in pepQ confer low-level resistance to bedaquiline and clofazimine in Mycobacterium tuberculosis. Antimicrob Agents Chemother 60:4590–4599. doi: 10.1128/AAC.00753-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lounis N, Gevers T, Van den Berg J, Vranckx L, Andries K. 2009. ATP synthase inhibition of Mycobacterium avium is not bactericidal. Antimicrob Agents Chemother 53:4927–4929. doi: 10.1128/AAC.00689-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Philley JV, Wallace RJ Jr, Benwill JL, Taskar V, Brown-Elliott BA, Thakkar F, Aksamit TR, Griffith DE. 2015. Preliminary results of bedaquiline as salvage therapy for patients with nontuberculous mycobacterial lung disease. Chest 148:499–506. doi: 10.1378/chest.14-2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Griffith DE, Aksamit T, Brown-Elliott BA, Catanzaro A, Daley C, Gordin F, Holland SM, Horsburgh R, Huitt G, Iademarco MF, Iseman M, Olivier K, Ruoss S, von Reyn CF, Wallace RJ Jr, Winthrop K, ATS Mycobacterial Diseases Subcommittee, American Thoracic Society, Infectious Disease Society of America. 2007. An official ATS/IDSA statement: diagnosis, treatment, and prevention of nontuberculous mycobacterial diseases. Am J Respir Crit Care Med 175:367–416. doi: 10.1164/rccm.200604-571ST. [DOI] [PubMed] [Google Scholar]
- 17.Brown-Elliott BA, Philley JV, Griffith DE, Thakkar F, Wallace RJ Jr. 21 November 2016. In vitro susceptibility testing of bedaquiline against Mycobacterium avium complex. Antimicrob Agents Chemother doi: 10.1128/AAC.01798-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Iakhiaeva E, McNulty S, Brown-Elliott BA, Falkinham JO III, Williams MD, Vasireddy R, Wilson RW, Turenne C, Wallace RJ Jr. 2013. Mycobacterial interspersed repetitive-unit-variable-number tandem-repeat (MIRU-VNTR) genotyping of Mycobacterium intracellulare for strain comparison with establishment of a PCR-based database. J Clin Microbiol 51:409–416. doi: 10.1128/JCM.02443-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee SY, Kim BJ, Kim H, Won YS, Jeon CO, Jeong J, Lee SH, Lim JH, Lee SH, Kim CK, Kook YH, Kim BJ. 2016. Mycobacterium paraintracellulare sp. nov., for the genotype INT-1 of Mycobacterium intracellulare. Int J Syst Evol Microbiol 66:3132–3141. doi: 10.1099/ijsem.0.001158. [DOI] [PubMed] [Google Scholar]
- 20.Torrea G, Coeck N, Desmaretz C, Van De Parre T, Van Poucke T, Lounis N, de Jong BC, Rigouts L. 2015. Bedaquiline susceptibility testing of Mycobacterium tuberculosis in an automated liquid culture system. J Antimicrob Chemother 70:2300–2305. doi: 10.1093/jac/dkv117. [DOI] [PubMed] [Google Scholar]
- 21.Semret M, Zhai G, Mostowy S, Cleto C, Alexander DC, Cangelosi G, Cousins D, Collins DM, van Soolingen D, Behr MA. 2004. Extensive genomic polymorphism within Mycobacterium avium. J Bacteriol 186:6332–6334. doi: 10.1128/JB.186.18.6332-6334.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cayrou C, Turenne C, Behr MA, Drancourt M. 2010. Genotyping of Mycobacterium avium complex organisms using multispacer sequence typing. Microbiology 156:687–694. doi: 10.1099/mic.0.033522-0. [DOI] [PubMed] [Google Scholar]
- 23.Ummels R, Abdallah AM, Kuiper V, Aajoud A, Sparrius M, Naeem R, Spaink HP, van Soolingen D, Pain A, Bitter W. 2014. Identification of a novel conjugative plasmid in mycobacteria that requires both type IV and type VII secretion. mBio 5(5):e01744-14. doi: 10.1128/mBio.01744-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milano A, Pasca MR, Provvedi R, Lucarelli AP, Manina G, Ribeiro AL, Manganelli R, Riccardi G. 2009. Azole resistance in Mycobacterium tuberculosis is mediated by the MmpS5-MmpL5 efflux system. Tuberculosis 89:84–90. doi: 10.1016/j.tube.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 25.Ramos JL, Martinez-Bueno M, Molina-Henares AJ, Teran W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. 2005. The TetR family of transcriptional repressors. Microbiol Mol Biol Rev 69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJ. 2015. The Phyre2 Web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Drozdetskiy A, Cole C, Procter J, Barton GJ. 2015. JPred4: a protein secondary structure prediction server. Nucleic Acids Res 43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrella S, Cambau E, Chauffour A, Andries K, Jarlier V, Sougakoff W. 2006. Genetic basis for natural and acquired resistance to the diarylquinoline R207910 in mycobacteria. Antimicrob Agents Chemother 50:2853–2856. doi: 10.1128/AAC.00244-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diacon AH, Pym A, Grobusch M, Patientia R, Rustomjee R, Page-Shipp L, Pistorius C, Krause R, Bogoshi M, Churchyard G, Venter A, Allen J, Palomino JC, De Marez T, van Heeswijk RP, Lounis N, Meyvisch P, Verbeeck J, Parys W, de Beule K, Andries K, Mc Neeley DF. 2009. The diarylquinoline TMC207 for multidrug-resistant tuberculosis. N Engl J Med 360:2397–2405. doi: 10.1056/NEJMoa0808427. [DOI] [PubMed] [Google Scholar]
- 30.Winter H, Egizi E, Murray S, Erondu N, Ginsberg A, Rouse DJ, Severynse-Stevens D, Pauli E. 2015. Evaluation of the pharmacokinetic interaction between repeated doses of rifapentine or rifampin and a single dose of bedaquiline in healthy adult subjects. Antimicrob Agents Chemother 59:1219–1224. doi: 10.1128/AAC.04171-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wallace RJ Jr, Brown-Elliott BA, McNulty S, Philley JV, Killingley J, Wilson RW, York DS, Shepherd S, Griffith DE. 2014. Macrolide/azalide therapy for nodular/bronchiectatic Mycobacterium avium complex lung disease. Chest 146:276–282. doi: 10.1378/chest.13-2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hall L, Doerr KA, Wohlfiel SL, Roberts GD. 2003. Evaluation of the MicroSeq system for identification of mycobacteria by 16S ribosomal DNA sequencing and its integration into a routine clinical mycobacteriology laboratory. J Clin Microbiol 41:1447–1453. doi: 10.1128/JCM.41.4.1447-1453.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2011. Susceptibility testing of mycobacteria, nocardiae, and other aerobic actinomycetes; approved standard, 2nd ed CLSI document M24-A2. Clinical and Laboratory Standards Institute, Wayne, PA. [PubMed] [Google Scholar]
- 34.van Ingen J, Egelund EF, Levin A, Totten SE, Boeree MJ, Mouton JW, Aarnoutse RE, Heifets LB, Peloquin CA, Daley CL. 2012. The pharmacokinetics and pharmacodynamics of pulmonary Mycobacterium avium complex disease treatment. Am J Respir Crit Care Med 186:559–565. doi: 10.1164/rccm.201204-0682OC. [DOI] [PubMed] [Google Scholar]
- 35.Brown-Elliott BA, Iakhiaeva E, Griffith DE, Woods GL, Stout JE, Wolfe CR, Turenne CY, Wallace RJ Jr. 2013. In vitro activity of amikacin against isolates of Mycobacterium avium complex with proposed MIC breakpoints and finding of a 16S rRNA gene mutation in treated isolates. J Clin Microbiol 51:3389–3394. doi: 10.1128/JCM.01612-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kearse M, Moir R, Wilson A, Stones-Havas S, Cheung M, Sturrock S, Buxton S, Cooper A, Markowitz S, Duran C, Thierer T, Ashton B, Meintjes P, Drummond A. 2012. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28:1647–1649. doi: 10.1093/bioinformatics/bts199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. 2011. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.