LETTER
Vibrio parahaemolyticus is the leading seafood-transmitted bacterial pathogen worldwide. It causes gastroenteritis and, rarely, lethal septicemia. The estimated 45,000 annual cases of foodborne V. parahaemolyticus infections in the United States are concerning because their incidences are rising despite control measures, in part due to the impact of changing climate on pathogen abundance and distribution (1; https://www.cdc.gov/vibrio/). Although the pandemic complex of strains of sequence type 3 (ST3) (serotype O3:K6) has dominated infections worldwide (2), in the United States and Canada, the most prevalent clinical strains are of ST36 (O4:K12), which recently spread from the Pacific into the Atlantic (3–8).
Here we report that a new lineage of V. parahaemolyticus, identified as ST631, is rapidly emerging as the predominant pathogenic clade endemic to the Atlantic coast of North America (3, 4, 8). The first reported ST631 genome came from a clinical case that occurred in Louisiana in 2007 and was traced to oysters from Florida (8). In 2009, a second ST631 clinical isolate was reported in Prince Edward Island, Canada (O11:KUT) (4). From 2010 to 2015, the incidence of infections by strains of ST631 has increased, with 35 confirmed cases reported in four Atlantic coastal U.S. states (Table 1), where they are second only to ST36 strains in prevalence. Due to the self-limiting nature of infections and underreporting (9), ST631 infections may be more widespread.
TABLE 1.
ST631 isolates with relevant information
| Isolate | SRA or GenBank accession no.a | State of isolationb | Trace-back locationb | Yr of isolation | Reporting countryc | Sourced | Geographic locatione |
|---|---|---|---|---|---|---|---|
| VP2007-095 | SRR869104 | LA | FL | 2007 | USA | C | FL |
| 09-4436 | LRAJ01000000 | PEI | PEI | 2009 | Canada | C | PEI |
| S487-4 | LFZE01000000 | NA | Canada | 2013 | Canada | E | PEI |
| MAVP-A | SRR4032168 | MA | NA | 2010 | USA | C | |
| MAVP-E | SRR1952988 | MA | MA | 2010 | USA | C | GOM |
| MAVP-P | SRR4032175 | MA | NA | 2010 | USA | C | |
| MAVP-T | SRR4032176 | MA | NA | 2010 | USA | C | |
| MAVP-L | SRR4032169 | MA | MA | 2011 | USA | C | GOM |
| MAVP-Q | SRR4035056 | MA | MA | 2011 | USA | C | GOM |
| MAVP-4 | SRR4032177 | MA | NA | 2013 | USA | C | |
| MAVP-30 | SRR4032178 | MA | NA | 2013 | USA | C | |
| MAVP-39 | SRR4032179 | MA | NA | 2013 | USA | C | |
| MAVP-56 | SRR4032180 | MA | PEI | 2013 | USA | C | PEI |
| MAVP-74 | SRR4032181 | MA | CT or PEI | 2014 | USA | C | LIS or PEI |
| MAVP-75 | SRR4032182 | MA | CT or MA | 2014 | USA | C | GOM or LIS |
| MAVP-78 | SRR4032170 | MA | MA | 2014 | USA | C | GOM |
| MAVP-90 | SRR4032171 | MA | CT | 2015 | USA | C | LIS |
| MAVP-94 | SRR4032172 | MA | MA | 2015 | USA | C | GOM |
| MAVP-109 | SRR4032173 | MA | MA | 2015 | USA | C | GOM |
| MAVP-112 | SRR4032174 | MA | MA | 2015 | USA | C | GOM |
| VP1 | SRR4032354 | MD | VA | 2012 | USA | C | MAC |
| VP8 | SRR4032362 | MD | NA | 2012 | USA | C | |
| VP9 | SRR4032363 | MD | NJ | 2012 | USA | C | MAC |
| VP31 | SRR4032355 | MD | NJ | 2013 | USA | C | MAC |
| VP35 | SRR4032356 | MD | NA | 2013 | USA | C | |
| VP41 | SRR4032357 | MD | NA | 2013 | USA | C | |
| VP44 | SRR4032358 | MD | NA | 2013 | USA | C | |
| VP45 | SRR4032359 | MD | CT or VA | 2013 | USA | C | LIS or MAC |
| VP47 | SRR4032360 | MD | NA | 2013 | USA | C | |
| VP55 | SRR4032361 | MD | NA | 2014 | USA | C | |
| PNUSAV000012 | SRR4016797 | MD | CT, MA, or ME | 2015 | USA | C | GOM or LIS |
| PNUSAV000015 | SRR4016801 | MD | CT, MA, NY, PEI, or VA | 2015 | USA | C | GOM, LIS, MAC, or PEI |
| PNUSAV00021 | SRR4018053 | MD | NA | 2015 | USA | C | |
| CTVP27C | SRR4090622 | CT | CT or VA | 2013 | USA | C | LIS or MAC |
| CTVP31C | SRR4090623 | CT | NA | 2013 | USA | C | |
| CTVP34C | SRR4090624 | CT | NA | 2013 | USA | C | |
| MEVP-12 | SRR4090625 | ME | NA | 2015 | USA | C | |
| MEVP-14 | SRR4090626 | ME | NA | 2015 | USA | C |
Massachusetts, Connecticut, and Maine isolates were sequenced using the Illumina HiSeq2500 sequencer at the Hubbard Center for Genomic Studies at the University of New Hampshire, whereas Maryland isolates were sequenced using the Illumina MiSeq sequencer at the Center for Food Safety and Applied Nutrition, Food and Drug Administration, Maryland, or at the Department of Health and Hygiene, Maryland.
Where available, the U.S. state or Canadian location of isolation and infection is identified. For multisource traces, all possible sources are listed. CT, Connecticut; FL, Florida; LA, Louisiana; MA, Massachusetts; ME, Maine; NA, information was not available or was not determined; NJ, New Jersey; NY, New York; PEI, Prince Edward Island; VA, Virginia.
The country which reported the isolate.
C, clinical isolate; E, environmental isolate (specifically, from an oyster).
The geographic locations of the sources corresponding to those identified in Fig. 1A. These include Florida (FL), the Gulf of Maine (GOM), Long Island Sound (LIS), the Mid-Atlantic Coast (MAC), and Prince Edward Island (PEI).
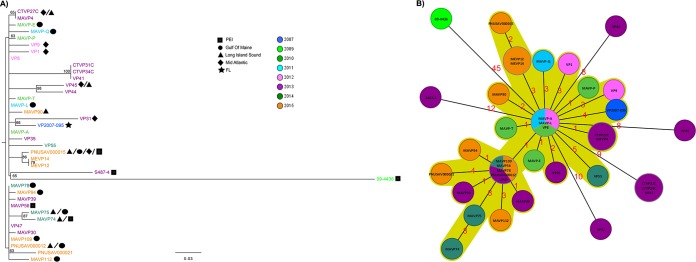
Genome comparisons were used to understand the potential relationships of ST631 strains, which share no recent ancestry with and differ substantially from ST36 and ST3 strains (>3,600 out of 3,909 shared genes contained variation). ST631 has a virulence gene profile similar to that of ST36 in that it harbors tdh, trh, and a type 3 secretion system (T3SS2) and is urease positive. We applied a core genome multilocus typing (cgMLST) scheme to draft genomes of 37 clinical isolates and 1 environmental isolate (Table 1) representing the geographic and time spans of infections. This analysis identified 132 single nucleotide polymorphisms (SNPs) in the population and confirmed that clinical ST631 isolates are clonal, with limited diversification (Fig. 1). Within the ST631 population, 97% of the core genes are identical, whereas less than 8% of the core genes are identical between ST631, ST36, and ST3 strains. Both maximum-likelihood phylogeny and minimum spanning tree analysis indicated a mixed population (Fig. 1A and B). Most isolates grouped within one clonal complex, with only a few divergent isolates (Fig. 1B). This population structure suggests that this pathogenic lineage recently evolved and that its distribution may have expanded along the North American Atlantic Coast (10).
FIG 1.
Phylogenetic relationships among ST631 isolates traced to the northwestern Atlantic (2007 to 2015). (A) A maximum-likelihood tree constructed with the core genome SNPs identified from cluster analysis (described below) of 35 newly sequenced clinical isolates reported in Massachusetts, Maryland, Maine, and Connecticut and 3 isolates whose draft genomes were publicly available in the NCBI database (strains 09-4436, S487-4, and VP2007-95) (Table 1) demonstrates the highly clonal nature of pathogenic ST631 isolates, which are colored by year and marked by geographic distribution. The scale bar represents the average number of nucleotide substitutions per site, and branches with greater than 60% bootstrap support are labeled. (B) A minimum spanning tree analysis reflecting the relationships among ST631 isolates based on core gene SNPs differences further demonstrates the clonal population structure. The numbers above the connected lines (not to scale) represent SNP differences. The isolates are colored by year of isolation using the same color scheme as in panel A. Cluster analysis of ST631 was performed using a custom cgMLST analysis using Ridom SeqSphere+ software v3.2.1 (Ridom GmbH, Münster, Germany). Briefly, the cgMLST software first defines a cgMLST scheme using the cgMLST target definer tool with default settings. MAVP-Q was used as the reference genome (4,568 genes). Then, five other V. parahaemolyticus genomes (strains BB22OP, CDC_K4557, FDA_R31, RIMD 2210633, and UCM-V493) were used for comparison with the reference genome to establish the core and accessory genome genes. Genes that are repeated in more than one copy in any of the six genomes were removed from the analysis. Subsequently, a task template that contains both core and accessory genes was created. Each individual gene locus from MAVP-Q was assigned allele number 1. Then each individual ST631 V. parahaemolyticus genome assembly was queried against the task template, during which any locus that differed from the reference genome or any other queried genome was assigned a new allele number. For the cgMLST, a gene-by-gene analysis of all core genes (excluding accessory genes) was performed and SNPs were identified within different alleles to establish genetic distance calculations. PEI, Prince Edward Island; FL, Florida.
The fact that an increasing number of cases tracing to sources in the northwestern Atlantic suggests that ST631 poses a mounting public health threat and calls for surveillance of this lineage to reduce illnesses. That its emergence coincided with warming ocean trends in some areas of the northwestern Atlantic (2) and invasion by a nonresident pathogen indicates that a changing climate may be driving pathogen dynamics (1, 2, 3, 7). However, this does not eliminate the potential of anthropogenic influences on the dissemination of ST631 strains, whose continued population expansion may increase human health risk beyond North America.
Accession number(s).
Sequences were deposited in the Sequence Read Archive under accession numbers SRR1952988, SRR4016797, SRR4016801, SRR4018053, SRR4032168 to SRR4032182, SRR4032354 to SRR4032363, SRR4035056, and SRR4090622 to SRR4090626.
ACKNOWLEDGMENTS
Feng Xu and Cheryl A. Whistler declare a potential conflict of interest in the form of a pending patent application (U.S. patent application 62/128,764).
Partial funding for this work was provided by the USDA National Institute of Food and Agriculture (Hatch projects NH00574, NH00609 [accession number 233555], and NH00625 [accession number 1004199]). Additional funding was provided by the National Oceanic and Atmospheric Administration College Sea Grant program and grants R/CE-137, R/SSS-2, and R/HCE-3. Support was also provided through the National Institutes of Health (1R03AI081102-01), the National Science Foundation (EPSCoR IIA-1330641), and the National Science Foundation (DBI 1229361 NSF MRI). N.G.-E. was funded through the FDA Foods Science and Research Intramural Program.
REFERENCES
- 1.Baker-Austin C, Trinanes JA, Taylor NG, Hartnell R, Siitonen A, Martinez-Urtaza J. 2013. Emerging Vibrio risk at high latitudes in response to ocean warming. Nat Clim Change 3:73–77. [Google Scholar]
- 2.Vezzulli L, Grande C, Reid P, Helaouet P, Edwards M, Hofle MG, Brettard I, Colwell RR, Pruzzo C. 2016. Climate influence on Vibrio and associated human diseases during the past half-century in the coastal North Atlantic. Proc Natl Acad Sci U S A 13:E5062–E5071. doi: 10.1073/pnas.1609157113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu F, Ilyas S, Hall JA, Jones SH, Cooper VS, Whistler CA. 2015. Genetic characterization of clinical and environmental Vibrio parahaemolyticus from the Northeast U.S.A. reveals emerging resident and non-indigenous pathogen lineages. Front Microbiol 6:272. doi: 10.3389/fmicb.2015.00272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banerjee SK, Kearney AK, Nadon CA, Peterson C-L, Tyler K, Bakouche L, Clark CG, Hoang L, Gilmour MW, Farber JM. 2014. Phenotypic and genotypic characterization of Canadian clinical isolates of Vibrio parahaemolyticus collected from 2000 to 2009. J Clin Microbiol 52:1081–1088. doi: 10.1128/JCM.03047-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Turner JW, Paranjpye RN, Landis ED, Biryukov SV, González-Escalona N, Nilsson WB, Strom MS. 2013. Population structure of clinical and environmental Vibrio parahaemolyticus from the Pacific Northwest coast of the United States. PLoS One 8:e55726. doi: 10.1371/journal.pone.0055726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.García K, Bastías R, Higuera G, Torres R, Mellado A, Uribe P, Espejo RT. 2013. Rise and fall of pandemic Vibrio parahaemolyticus serotype O3:K6 in southern Chile. Environ Microbiol 15:527–534. doi: 10.1111/j.1462-2920.2012.02883.x. [DOI] [PubMed] [Google Scholar]
- 7.Martinez-Urtaza J, Baker-Austin C, Jones JL, Newton AE, Gonzalez-Aviles GD, DePaola A. 2013. Spread of Pacific Northwest Vibrio parahaemolyticus strain. N Engl J Med 369:1573–1574. doi: 10.1056/NEJMc1305535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haendiges J, Timme R, Allard MW, Myers RA, Brown EW, Gonzalez-Escalona N. 2015. Characterization of Vibrio parahaemolyticus clinical strains from Maryland (2012–2013) and comparisons to locally and globally diverse V. parahaemolyticus strains by whole-genome sequence analysis. Front Microbiol 6:125. doi: 10.3389/fmicb.2015.00125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Scallan E, Hoekstra RM, Angulo FJ, Tauxe RV, Widdowson M-A, Roy SL, Jones JL, Griffin PM. 2011. Foodborne illnesses acquired in the United States—major pathogens. Emerg Infect Dis 17:7–15. doi: 10.3201/eid1701.P11101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jonsson BF, Watson JR. 2016. The timescale of global surface ocean connectivity. Nat Commun 7:11239. doi: 10.1038/ncomms11239. [DOI] [PMC free article] [PubMed] [Google Scholar]